Spectinomycin is a ribosome-binding antibiotic that blocks the translocation step of translation. A prevalent resistance mechanism is modification of the drug by aminoglycoside nucleotidyl transferase (ANT) enzymes of the spectinomycin-specific ANT(9) family or by enzymes of the dual-specificity ANT(3ʺ)(9) family, which also acts on streptomycin. We previously reported the structural mechanism of streptomycin modification by the ANT(3ʺ)(9) AadA from Salmonella enterica.
KEYWORDS: antibiotic resistance, spectinomycin, enzyme structure, antimicrobial agents, X-ray crystallography
ABSTRACT
Spectinomycin is a ribosome-binding antibiotic that blocks the translocation step of translation. A prevalent resistance mechanism is modification of the drug by aminoglycoside nucleotidyl transferase (ANT) enzymes of the spectinomycin-specific ANT(9) family or by enzymes of the dual-specificity ANT(3ʺ)(9) family, which also acts on streptomycin. We previously reported the structural mechanism of streptomycin modification by the ANT(3ʺ)(9) AadA from Salmonella enterica. ANT(9) from Enterococcus faecalis adenylates the 9-hydroxyl of spectinomycin. Here, we present the first structures of spectinomycin bound to an ANT enzyme. Structures were solved for ANT(9) in apo form, in complex with ATP, spectinomycin, and magnesium, or in complex with only spectinomycin. ANT(9) shows an overall structure similar to that of AadA, with an N-terminal nucleotidyltransferase domain and a C-terminal α-helical domain. Spectinomycin binds close to the entrance of the interdomain cleft, while ATP is buried at the bottom. Upon drug binding, the C-terminal domain rotates 14 degrees to close the cleft, allowing contacts of both domains with the drug. Comparison with AadA shows that spectinomycin specificity is explained by a straight α5 helix and a shorter α5-α6 loop, which would clash with the larger streptomycin substrate. In the active site, we observed two magnesium ions, one of them in a previously unobserved position that may activate the 9-hydroxyl for deprotonation by the catalytic base Glu-86. The observed binding mode for spectinomycin suggests that spectinamides and aminomethyl spectinomycins, recent spectinomycin analogues with expansions in position 4 of the C ring, are also subjected to modification by ANT(9) and ANT(3ʺ)(9) enzymes.
INTRODUCTION
Spectinomycin is a broad-spectrum antibiotic that belongs to the aminocyclitol class and was first isolated from Streptomyces spectabilis in 1961 (1). It is included in the WHO model list of essential medicines because of its clinical use to treat gonorrheal infections (2) and is considered to have limited side effects. Spectinomycin is an inhibitor of bacterial protein synthesis. Structural and biochemical studies show that the drug binds to 16S rRNA helix 34 of the 30S subunit of the bacterial ribosome, where it blocks the translocation step of protein synthesis (3–5). Spectinomycin is usually bacteriostatic but can be bactericidal at high concentrations (6).
Bacteria have evolved three mechanisms of resistance to spectinomycin. The first is lowering the drug concentration with efflux transporters. This makes some bacteria, for example, Mycobacterium tuberculosis, intrinsically resistant to spectinomycin (7). The second is alteration of the drug-binding site through chromosomal mutations in the ribosomal 16S rRNA genes (8) or in the gene for ribosomal protein S5 (9). The third is modification of the drug by aminoglycoside-modifying enzymes (10–12). Two classes of enzymes modify spectinomycin using ATP: aminoglycoside O-phosphotransferases of the APH(9) class, which phosphorylate the antibiotic, and aminoglycoside O-nucleotidyl transferases of the ANT(9) and ANT(3ʺ)(9) classes, which instead adenylate the antibiotic.
Although spectinomycin is chemically dissimilar to the aminoglycoside streptomycin, both drugs can be adenylated by enzymes of the ANT(3ʺ)(9) [also called ANT(3ʺ)-Ia] class. Aiming to show how one of these, AadA from Salmonella enterica (13), recognizes the two chemically distinct molecules in the same active site, we previously solved crystal structures of AadA in its apo form and in complex with ATP, magnesium, and streptomycin (14, 15). We were able to explain how the enzyme coordinates ATP and streptomycin but failed to obtain a structure of AadA with spectinomycin. Furthermore, site-directed mutagenesis followed by in vivo resistance measurements and in vitro binding assays were used to demonstrate that AadA makes use of different parts of the active site to recognize its two drug substrates. A spectinomycin-binding site was proposed based on manual docking and molecular dynamics simulations. Based on structure-guided sequence analysis, we were able to confirm that ANT(9) enzymes, which are active only on spectinomycin, are likely to display a similar overall structure. We also identified sequence determinants that distinguish them from the ANT(3ʺ)(9) enzymes, which are active on both antibiotics (15).
Spectinomycin-specific ANT(9) enzymes have been experimentally verified in Enterococcus faecalis, Staphylococcus aureus, and Campylobacter jejuni and classified into the ANT(9)-Ia and ANT(9)-Ib groups (12, 16–18). Introduction of a plasmid encoding ANT(9) from E. faecalis into Escherichia coli leads to a 10,000-fold rise in the MIC of spectinomycin, from 10 μg/ml to 100 mg/ml. The ANT(9) enzymes show 29 to 37% sequence identity to the dual-specificity AadA enzyme from S. enterica. In this study, we set out to elucidate how ANT(9) enzymes recognize spectinomycin and how they structurally differ from the ANT(3ʺ)(9) enzymes. To this end, we have solved crystal structures of ANT(9)-Ib from the clinical strain E. faecalis LDR55 (16) in its apo form and in complex with spectinomycin [ANT(9)-spc] or with ATP and spectinomycin [ANT(9)-ATP-spc]. This structural information allowed detailed analysis of (i) how spectinomycin is recognized by the enzyme and (ii) the structural difference between the single- and dual-specificity enzymes of the same family. In the light of recent developments of novel spectinomycin analogues (19–21), it is most important to understand the molecular basis of the present resistance mechanisms.
RESULTS AND DISCUSSION
Protein production and crystallization.
ANT(9) from E. faecalis was expressed in E. coli with a C-terminal His tag. After size exclusion chromatography, the protein gradually precipitated. Hence, the protein was directly flash frozen in liquid nitrogen for storage or subjected to crystallization. Crystals appeared in a few hours under several crystallization conditions.
The apo- crystals of ANT(9) diffracted to 1.9 Å and belonged to space group I4, with one molecule in the asymmetric unit. The structure was solved by molecular replacement.
Overall structure of ANT(9).
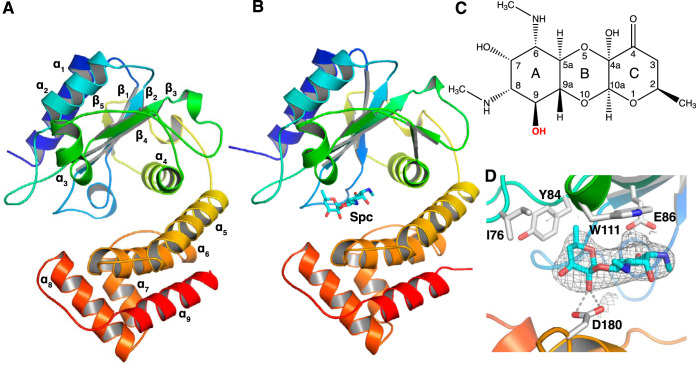
ANT(9) has a bilobed two-domain structure with approximate dimensions of 52 by 40 by 30 Å (Fig. 1A). The N-terminal domain is a nucleotidyltransferase domain composed of a central five-stranded β-sheet (β1 to β5) and four α-helices (α1 to α4). The C-terminal domain is an α-helical domain consisting of five α-helices (α5 to α9). A search for similar structures in the Protein Data Bank (PDB) using the DALI server (22) confirmed that AadA from S. enterica, which was used as a molecular replacement model, was the most similar available structure.
FIG 1.
Structure of ANT(9). (A) Apo structure of ANT(9) represented in rainbow colors going from the N terminus in blue to the C terminus in red. (B) Structure of ANT(9) in complex with spectinomycin (cyan) bound at the entry of the interdomain cleft. (C) Chemical structure of spectinomycin. The 9-OH modification site is in red bold. (D) Detailed interactions of spectinomycin (cyan) in the ANT(9)-spc structure. The Fo-Fc omit map of spectinomycin, contoured at 2.5σ, is shown as gray mesh.
Structures of ANT(9) with spectinomycin and ATP.
Attempts to cocrystallize ANT(9) with the substrates ATP and spectinomycin in the presence of magnesium failed. Therefore, pregrown ANT(9) crystals were soaked with ligands, which allowed two structures to be solved, one with only spectinomycin at pH 8.5 and the other with spectinomycin, ATP, and Mg at pH 7.5. A summary of crystallographic data and refinement statistics is shown in Table 1.
TABLE 1.
Summary of crystallographic data and refinement statistics
| Parameter | ANT(9) | ANT(9)-spc | ANT(9)-ATP-spc |
|---|---|---|---|
| Data collection | |||
| Beamline | DLS I04-1 | DLS I04 | DLS I04 |
| Wavelength (Å) | 0.915 | 0.979 | 0.979 |
| Space group | I4 | I4 | C121 |
| Unit cell parameters | |||
| a, b, c (Å) | 103.97, 103.97, 61 | 99.6, 99.6, 61.3 | 133.5, 69.3, 94.5 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 135, 90 |
| Resolution (Å)a | 36.8–2.1 (2.17–2.1) | 49.8–2.8 (2.9–2.8) | 55.9–3.0 (3.1–3.0) |
| Rmeas (%)a | 5.8 (161.8) | 14.7 (160.3) | 32.4 (145.7) |
| I/σIa | 25.3 (1.5) | 19.8 (2.2) | 5.4 (1.2) |
| CC ½ (%)a | 100 (79.5) | 99.9 (90.4) | 98 (47.2) |
| Completeness (%)a | 99.1 (97.3) | 99.57 (99.5) | 99.85 (99.9) |
| Redundancya | 13.7 (13.8) | 22.9 (23.6) | 6.3 (6.3) |
| Refinement | |||
| Resolution (Å) | 36.98–2.10 | 49.8–2.8 | 55.9–3.0 |
| Reflections/test set | 18,972/1,847 | 7,482/747 | 12,341/1,186 |
| Rwork/Rfree (%) | 23.0/27.2 | 24.3/29.6 | 24/27.1 |
| No. of atoms | |||
| Protein | 2,047 | 2,047 | 4,062 |
| Ligand/ion | 6 | 23 | 113 |
| Water | 16 | 12 | |
| B factors | |||
| Protein | 68.0 | 75.9 | 44.2 |
| Ligands | 78.4 | 71.0 | 38.4 |
| Solvent | 56.0 | 32.6 | |
| RMSD from ideal | |||
| Bond lengths (Å) | 0.005 | 0.003 | 0.004 |
| Bond angles (°) | 0.7 | 0.6 | 0.8 |
| Ramachandran plot | |||
| Preferred (%) | 95.6 | 96 | 94.3 |
| Allowed (%) | 4.4 | 4.0 | 5.7 |
| Outliers (%) | 0 | 0 | 0 |
| PDB accession number | 6SXJ | 6XZ0 | 6XXQ |
The highest-resolution shell is shown in parentheses.
The structure of ANT(9)-spc (Fig. 1B) was solved in the same space group (I4) as apo-ANT(9), but with slightly different cell dimensions. The structure was refined to a 2.8-Å resolution. We observed a strong difference density for spectinomycin (Fig. 1C and D) but not for ATP or magnesium, even though the soaking solution contained all ligands.
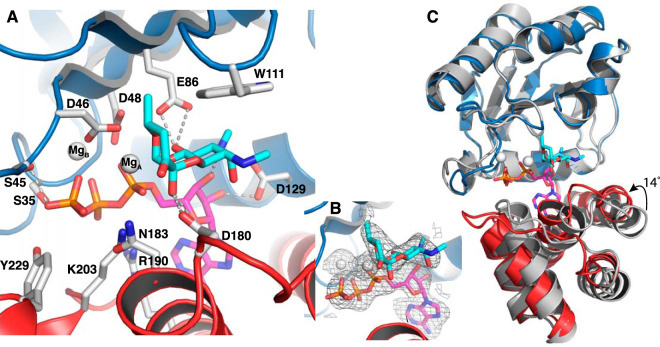
We suspected that the high pH (8.5) of the crystallization condition might not be optimal for ATP binding. Therefore, further soaking tests were done with crystals grown at pH 7.5. This resulted in the ANT(9)-ATP-spc structure, for which the space group shifted from I4 to C121 with two molecules in the asymmetric unit. The structure was refined to a 3-Å resolution and shows clear electron density for ATP, spectinomycin, and one or two magnesium ions in the different chains (Fig. 2A and B). The two molecules are nearly identical to each other with a root mean square deviation (RMSD) of 0.43 Å over 248 Cα atoms.
FIG 2.
Structure of ANT(9) in complex with spectinomycin (cyan), ATP (magenta), and magnesium (white). The N-terminal domain is in blue, and the C-terminal domain is in red. (A) Interactions of spectinomycin in the active site of ANT(9)-ATP-spc. Direct hydrogen bonds to spectinomycin are shown as dashed lines. (B) Fo-Fc omit maps of spectinomycin, ATP, and magnesium contoured at 2.5σ. (C) Superposition of the N-terminal domain of ANT(9) apo- (gray) with ANT9-ATP-spc. Upon ligand binding, the C-terminal domain rotates 14° toward the N-terminal domain.
The structures of the individual domains in the ligand complexes are very similar to the apo- structure, with an RMSD below 0.8 Å for 150 aligned Cα atoms in the N-terminal domain and an RMSD below 0.7 Å over 96 aligned Cα atoms in the C-terminal domain. Ligand binding is accommodated by a rotational shift of the second domain toward the first domain (Fig. 2C), a shift that also induces changes in cell dimensions and space group. Analysis with DynDom (23) shows that binding of spectinomycin induces a 5-degree rotation of the C-terminal domain. Binding of ATP leads to an additional 9-degree rotation, resulting in up to 4 Å movement at the outer edge of the domain.
In the ANT(9)-spc structure, spectinomycin binds between β4 and α3 from the N-terminal domain and α6 from the C-terminal domain, at the entry of the interdomain cleft. In the L-shaped spectinomycin molecule, the two first rings are coplanar. The A ring stacks against Trp-111 and is coordinated by a hydrogen bond between O9 and Glu-86, the assumed catalytic base (14). The C ring is positioned by a hydrogen bond between the hydroxyl in position 4a and Asp-180 and the insertion of the methyl group in position 2 into a hydrophobic pocket between Ile-76, Tyr-84, and Trp-111 (Fig. 1D).
In the ANT(9)-ATP-spc structure, ATP binds at the bottom of the interdomain cleft. In the N-terminal domain, Ser-35, Ser-45, Asp-46, and Asp-48 form hydrogen bonds with the phosphates of ATP, and Asp-129 hydrogen bonds to the ribose. In the C-terminal domain, Arg-190, Lys-203, and Tyr-229 form interactions with the ATP phosphates and Arg-190 forms a cation-pi interaction with the base (Fig. 2A). In both chains, one magnesium ion is coordinated by the three phosphates, Asp-46, and Asp-48. In chain A, an additional magnesium ion is coordinated by the alpha phosphate, Asp-46, and O-10 of spectinomycin.
In the presence of ATP, spectinomycin together with the C-terminal domain swings 2 Å further into the active site, maintaining the position of the C-ring methyl group. Asp-180 is within hydrogen bond distance of both 4a-OH and 4-O, while Asn-183 also comes within hydrogen bond distance of the 4a-OH. Because of the limited resolution, the exact hydrogen bond network is not clear. In the N-terminal domain, the side chain of Glu-86 moves along with the ligand, forms a short or very short (2.7 or 2.2 Å in the two molecules) hydrogen bond with 9-O, and is within hydrogen bond distance of the secondary amine in position 8. The same amine also forms a hydrogen bond to the 3′ hydroxyl of the ribose of ATP. There are only minor differences between the ligand interactions in the two molecules.
Isothermal titration calorimetry was performed to test the binding affinity of ANT(9) to its substrates and to confirm the specificity to spectinomycin. Due to precipitation problems, these experiments did not generate reproducible Kd and stoichiometry values but showed micromolar affinity binding to both ATP and spectinomycin. Despite our observation that spectinomycin could bind in the absence of ATP in the crystal structure, we did not observe spectinomycin binding in solution in the absence of ATP (data not shown). This suggests that in solution, binding of ATP may position the two domains for interaction with the antibiotic substrate, as previously observed for AadA (15).
Comparison of ANT(9) with the dual-specificity AadA.
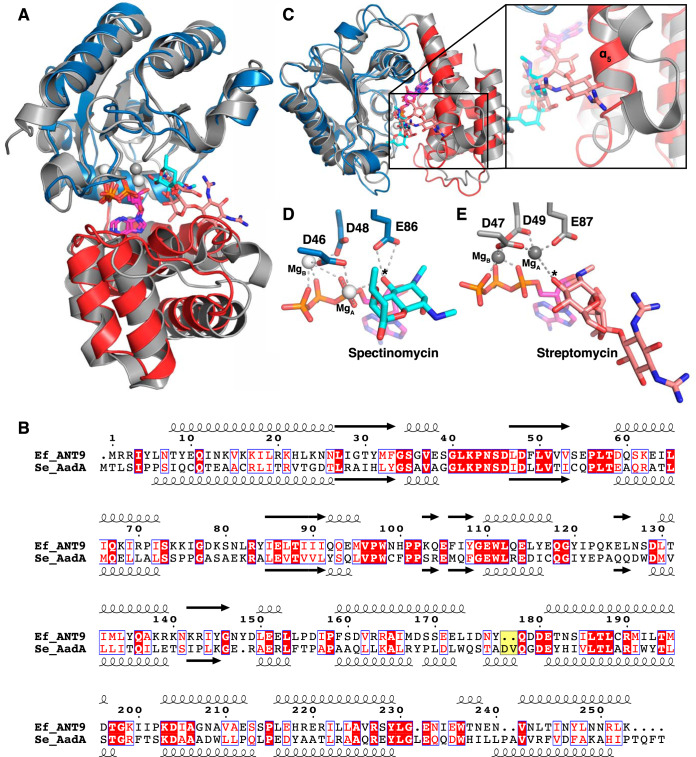
Comparison of ANT(9)-ATP-spc with the structure of the dual-specificity ANT(3ʺ)(9) AadA in complex with ATP and streptomycin (AadA-ATP-sry) shows that their overall structures are very similar (Fig. 3A). The N-terminal domains superpose with an RMSD of 0.95 Å over 110 Cα atoms and the C-terminal domains with an RMSD of 0.83 Å over 62 Cα atoms. The interdomain orientation is slightly different, leading to an RMSD of 1.5 Å over 230 Cα atoms when the complete structures are superposed.
FIG 3.
Comparison of ANT(9) and AadA. (A) Superposition of the N-terminal domain of ANT(9)-ATP-spc (colors as in Fig. 2) with AadA in complex with ATP and streptomycin (15) (gray; streptomycin in salmon). (B) Structure-guided sequence alignment of E. faecalis ANT(9) with S. enterica AadA (15, 42). The characteristic Asp-Val insertion in AadA is highlighted in yellow. (C) The straight α5 in ANT(9) would clash with streptomycin. AadA shows a kink in helix α5, and the insertion in the α5-α6 loop forms a short helix to accommodate streptomycin (salmon). The view is perpendicular to that in panel A. (D and E) Comparison of active-site and magnesium coordination in ANT(9)-ATP-spc (D) and AadA-ATP-sry (E). The asterisk indicates the substrate hydroxyl to be modified.
Previous structure-guided sequence alignments showed that the active-site residues that bind to ATP and spectinomycin are conserved between ANT(3ʺ)(9) and ANT(9) enzymes. Enzymes with activity on streptomycin contain a characteristic Asp-Val or Asp-Trp insertion in the loop after α5 (Fig. 3B). Site-directed mutagenesis showed that in this region, Trp-173 and Asp-178 are critical for binding and resistance to streptomycin but not spectinomycin (15).
When this region of the superposed structures is examined closely (Fig. 3C), a striking difference can be observed for amino acids 160 to 185 in the α5-α6 region of the C-terminal domain. In AadA-ATP-sry, Pro169 in helix α5 kinks the helix approximately 10 degrees away from the cleft. This, combined with the extension of the loop to form a short helix, allows the enzyme to accommodate the large streptomycin substrate. In contrast, the corresponding region of the C-terminal domain in ANT(9)-ATP-spc forms a straight helix α5 and a short loop followed by helix α6. In the loop, Asp-180 and Asn-183 hydrogen bond to spectinomycin in a position where they would clash with a larger substrate, such as streptomycin.
Previous site-directed mutagenesis of Asp-182 in AadA, the equivalent of Asp-180 in ANT(9), showed that the Asp182Asn mutation abolishes binding of spectinomycin to AadA while not affecting binding of ATP or streptomycin (15). In vivo, the Asp182Asn, Asp182Ala, Trp112Phe, and Trp112Ala mutations are more detrimental to resistance to spectinomycin than streptomycin (15). In the present ANT(9) complex structures, only three amino acids, Glu-86, Asp-180, and Asn-183 (Fig. 2A), form direct hydrogen bonds to spectinomycin, thus explaining why Asp-180 is critical for spectinomycin resistance. The small and rigid spectinomycin molecule requires closing of the interdomain cleft to allow snug packing against the N-terminal domain with only two hydrogen bonds to the C-terminal domain. Our analysis predicts that a similar domain closure would need to occur during binding of spectinomycin to AadA and that the binding sites are identical. Previous MD simulations of AadA with spectinomycin and ATP (15) were not long enough to capture any larger conformational changes of the protein as observed here.
The larger streptomycin molecule is instead positioned by stacking with the equivalent of Trp-111 and hydrogen bonds to three residues of the C-terminal domain up to 15 Å away from the catalytic base.
The only other structurally characterized resistance-mediating enzyme in complex with spectinomycin is APH(9)-Ia, which phosphorylates the hydroxyl group that ANT(9) adenylates. Compared to ANT(9), APH(9)-Ia forms more extensive hydrogen bond interactions with spectinomycin, at five different positions of the drug (24).
Catalytic site.
Previous in vivo MIC experiments and in vitro adenylation experiments on AadA demonstrated that the equivalent of Glu-86 in ANT(9) is the catalytic base. Adenyltransferase enzymes, similar to polymerases, make use of a two-metal-ion mechanism (15, 25). In ANT(9)-ATP-spc, one magnesium (MgB) (Fig. 2A) is positioned as previously observed in AadA and polymerases. Interestingly, the second magnesium (MgA), which displayed clear density in chain A but weak density in chain B, is shifted by 3 Å toward the substrate 9-hydroxyl compared to its position in AadA (Fig. 3D and E). The close proximity between MgA and the 9-hydroxyl (3.2 Å) likely lowers the pKa of the 9-hydroxyl to allow the catalytic base Glu-86 to accept its proton. The hydrogen bond between Glu86 and the 9-hydroxyl is very short, 2.2 Å. Computational prediction using H++ (26) suggests that the pKa of Glu-86 is substantially upshifted in the structural context of the active site, allowing its activity in acid-base catalysis at physiological pH. The distance between the 9 oxygen of spectinomycin and the alpha phosphorus of ATP that would be subjected to a nucleophilic attack is 3.9 to 4.3 Å in our structures (Fig. 3D), thus making this structural snapshot closer to the reactive state than previous structures of AadA-ATP-sry (Fig. 3E). The active site is organized in agreement with the single in-line displacement mechanism proposed for other adenylyltransferases, KNTase (27) and LinB (28), but the shifted metal ion site may represent a previously unobserved reactive state.
Conclusions and future outlook.
Here, we have shown for the first time how spectinomycin is recognized by ANT resistance enzymes and observed an active-site arrangement that is closer to the reactive state than what has previously been observed for ANT enzymes.
During the late 1980s, there were many attempts to make spectinomycin variants for treatment of bacterial infections (29, 30), but none of these drugs are in clinical use. Lately, however, semisynthetic modifications of spectinomycin, i.e., spectinamides and aminomethyl spectinomycins, have been proposed for treatment of multidrug-resistant tuberculosis and other infections (19, 21). In these molecules, added functional groups on the 4-O position of the C ring of spectinomycin mediate additional contacts in the spectinomycin-binding site of the ribosome and, importantly, prevent the drug from being exported by the intrinsic efflux pump of M. tuberculosis.
In APH(9)-Ia, the 4-O position resides within the active-site pocket and is involved in water-mediated contacts with the enzyme (24). However, in ANT(9), the 4-O is pointing out of the active site. Although promising, these modified drugs will probably still be subject to modification at position 9 by the ANT(9) and ANT(3ʺ)(9) classes of enzymes. To make sure that a new drug does not directly suffer from any of the present resistance mechanisms, it is important to map the contacts of all resistance enzymes on the drug, as was previously done for other aminoglycosides (31). In addition, the combination of few critical interactions between ANT(9) and its antibiotic substrate and a substrate-binding site that can be modulated in size based on interdomain flexibility indicates that this class of resistance enzymes may be prone to evolve in response to other new related antibiotic molecules.
MATERIALS AND METHODS
Protein expression and purification.
The codon-optimized genes encoding ANT(9) from E. faecalis (UniProtKB Q07448) and with a C-terminal His6 tag were synthesized by GenScript and cloned into the pET11a vector. Plasmids were transformed into Escherichia coli strain BL21 Star (DE3). A single colony was used to inoculate a 5-ml overnight culture of Luria-Bertani (LB) medium supplemented with 100 μg/ml ampicillin. The overnight culture was then added to a 1-liter culture and grown at 37°C until an optical density at 600 nm (OD600) of 0.5 was reached. Protein expression was induced with 1 mM isopropyl-β-thiogalactopyranoside (IPTG), and the culture was incubated at 18°C for 20 h. The cells were harvested by centrifugation, washed with 25 mM Tris-HCl (pH 8.0)–150 mM NaCl, and stored at –20°C.
All the protein purification steps were performed at 8°C. The cells were resuspended in buffer A (50 mM Tris-HCl [pH 8.5], 500 mM NaCl, 50 mM [each] glutamic acid and arginine, and 5 mM β-mercaptoethanol) with 10 mM imidazole and one mini-Complete protease inhibitor tablet and then lysed using a cell disruptor (Constant Systems Ltd., Daventry, United Kingdom). The lysate was cleared by centrifugation (23,500 × g) for 45 min at 4°C. The supernatant was loaded into a gravity flow column containing Ni Sepharose High Performance (GE Healthcare, Uppsala, Sweden) pre-equilibrated with buffer A. After incubation for 45 min, the column was washed with 50 ml of buffer A followed by 100 ml of buffer A with 40 mM imidazole. Finally, the protein was eluted with buffer A containing 500 mM imidazole. This fraction was loaded onto a HiLoad 16/60 Superdex 75 prep grade column (GE Healthcare, Uppsala, Sweden) equilibrated with buffer B (25 mM Tris-HCl [pH 8.5], 200 mM NaCl, 50 mM [each] glutamic acid and arginine) with 5 mM β-mercaptoethanol. The peak fractions containing ANT(9) were pooled and concentrated to 6 mg/ml using a Vivaspin concentrator with a 10-kDa cutoff (Sartorius AG, Göttingen, Germany).
Crystallization and data collection.
Crystallization trials were performed at 20°C and 8°C by sitting-drop vapor diffusion set up with a Mosquito crystallization robot (TTP Labtech, Melbourn, United Kingdom) in 200-nl drops. The sitting drops consisted of a 1:1 ratio of reservoir solution to protein, in which the reservoir solutions were the Morpheus HT-96 screen (Molecular Dimensions, Sheffield, United Kingdom). Hexagonal prism-shaped crystals of ANT(9) appeared at 8°C in 10% (wt/vol) polyethylene glycol (PEG) 4000, 20% (vol/vol) glycerol, amino acids (0.02 M [each] sodium l-glutamate, dl-alanine, glycine, dl-lysine, dl-serine), 0.1 M bicine-Trizma base, pH 8.5. These crystals were harvested directly from the drop and vitrified in liquid nitrogen. All X-ray data collection was done at 100 K at Diamond Light Source, Didcot, United Kingdom.
Soaking was performed to obtain structures of ANT(9) in complex with ligands. For ANT(9)-spc, crystals were grown in 10% (wt/vol) PEG 4000, 20% (vol/vol) glycerol, alcohols [0.02 M (each) 1,6-hexanediol and 1-butanol, 0.2 M (RS)-1,2-propanediol, 2-propanol, 1,4-butanediol, and 1,3-propanediol], and 0.1 M bicine-Trizma base, pH 8.5.
For the ANT(9)-ATP-spc complex, crystals were grown in 10% (wt/vol) PEG 4000, 20% (vol/vol) glycerol, monosaccharides (0.02 M [each] d-glucose, d-mannose, d-galactose, l-fucose, d-xylose, N-acetyl-d-glucosamine), and 0.1 M MOPS (morpholinepropanesulfonic acid)-HEPES-Na, pH 7.5. Soaking was done in a 10-μl drop of mother liquor, including 10 mM ATP, 10 mM magnesium chloride, and spectinomycin powder to saturation. The crystals were soaked for 30 s for ANT(9)-spc and 180 s for ANT(9)-ATP-spc before vitrification.
Structure determination and refinement.
All diffraction data were scaled and merged using XDS (32) and AIMLESS (33). Xtriage (34) was used to check the data quality and determine the Matthews coefficient. The CCP4 online pipeline MoRDa (35) was used to find a suitable search model and succeeded with PDB ID 5G4A (AadA in complex with ATP and magnesium [15]). The structure was solved by molecular replacement with Phaser (36). The output model from Phaser showed amino acids 8 to 154 and 166 to 255, and PHENIX AutoBuild (37) was used for completion of the missing regions between amino acids 154 to 166. Composite omit maps were calculated for the entire model to check model bias. Manual building was done in COOT (38) and refinement with phenix.refine (39). Protein geometry was validated in MolProbity (40).
The ligand-bound structures were solved by molecular replacement with Phaser (36) using the apo-ANT(9) structure as the template. The polder omit map (41) was calculated to confirm the densities for ATP and spectinomycin.
Isothermal titration calorimetry.
Binding studies were performed at 25°C using a MicroCal iTC200 instrument (GE Healthcare). ANT(9) was used right after the elution from size exclusion chromatography in buffer B with 5% glycerol and 1 mM tris(2-carboxyethyl)phosphine. Titration was done with 450 to 650 μM ATP and 200 to 1,000 μM spectinomycin. All ligands were freshly dissolved in the buffer prior to each experiment. For titration of spectinomycin in the presence of ATP, a first titration of ANT(9) with ATP to saturation was followed by a second titration with spectinomycin. The data were analyzed using the MicroCal Analysis plugin in Origin.
Data availability.
Coordinates and structure factors have been deposited in the PDB with accession codes 6SXJ [apo ANT(9)], 6XZ0 [ANT(9)-spc], and 6XXQ [ANT(9)-ATP-spc].
ACKNOWLEDGMENTS
This work was supported by grants 2017-03827 and 2016-06264 from the Swedish Research Council to M.S.
We are grateful for access to beamlines IO4 and IO4-1 at the Diamond Light Source, Didcot, United Kingdom (proposals MX15868 and MX22906). We thank Terese Bergfors for constructive comments on the manuscript.
REFERENCES
- 1.Bergy M, Eble TE, Herr RR. 1961. Actinospectacin, a new antibiotic. IV. Isolation, purification, and chemical properties. Antibiot Chemother Fortschritte Adv Progrès 11:661–664. [PubMed] [Google Scholar]
- 2.WHO. 2019. World Health Organization model list of essential medicines, 21st list, 2019. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 4.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 5.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate J. 2007. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol 2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovering AM, Reeves DS. 2011. Aminoglycosides and aminocyclitols, p 145–169. In Finch RG, Greenwood D, Norrby SR, Whitley RJ (ed), Antibiotic and chemotherapy, 9th ed Elsevier, New York, NY. [Google Scholar]
- 7.Ramón-García S, Martín C, De Rossi E, Aínsa JA. 2007. Contribution of the Rv2333c efflux pump (the Stp protein) from Mycobacterium tuberculosis to intrinsic antibiotic resistance in Mycobacterium bovis BCG. J Antimicrob Chemother 59:544–547. doi: 10.1093/jac/dkl510. [DOI] [PubMed] [Google Scholar]
- 8.Anderson P. 1969. Sensitivity and resistance to spectinomycin in Escherichia coli. J Bacteriol 100:939–947. doi: 10.1128/JB.100.2.939-947.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funatsu G, Nierhaus K, Wittmann-Liebold B. 1972. Ribosomal proteins. XXII. Studies on the altered protein S5 from a spectinomycin-resistant mutant of Escherichia coli. J Mol Biol 64:201–206. doi: 10.1016/0022-2836(72)90329-4. [DOI] [PubMed] [Google Scholar]
- 10.Davies J, Wright GD. 1997. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol 5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. doi: 10.1128/MMBR.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollingshead S, Vapnek D. 1985. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid 13:17–30. doi: 10.1016/0147-619X(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Näsvall J, Wu S, Andersson DI, Selmer M. 2015. Structure of AadA from Salmonella enterica: a monomeric aminoglycoside (3ʺ)(9) adenyltransferase. Acta Crystallogr D Biol Crystallogr 71:2267–2277. doi: 10.1107/S1399004715016429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stern AL, Van der Verren SE, Kanchugal SP, Näsvall J, Gutiérrez-de-Terán H, Selmer M. 2018. Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase from Salmonella enterica. J Biol Chem 293:11481–11490. doi: 10.1074/jbc.RA118.003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeBlanc DJ, Lee LN, Inamine JM. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother 35:1804–1810. doi: 10.1128/AAC.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3″)(9). Mol Gen Genet 200:33–39. doi: 10.1007/BF00383309. [DOI] [PubMed] [Google Scholar]
- 18.Nirdnoy W, Mason CJ, Guerry P. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob Agents Chemother 49:2454–2459. doi: 10.1128/AAC.49.6.2454-2459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee RE, Hurdle JG, Liu J, Bruhn DF, Matt T, Scherman MS, Vaddady PK, Zheng Z, Qi J, Akbergenov R, Das S, Madhura DB, Rathi C, Trivedi A, Villellas C, Lee RB, Rakesh Waidyarachchi SL, Sun D, McNeil MR, Ainsa JA, Boshoff HI, Gonzalez-Juarrero M, Meibohm B, Böttger EC, Lenaerts AJ. 2014. Spectinamides: a new class of semisynthetic antituberculosis agents that overcome native drug efflux. Nat Med 20:152–158. doi: 10.1038/nm.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scarff JM, Waidyarachchi SL, Meyer CJ, Lane DJ, Chai W, Lemmon MM, Liu J, Butler MM, Bowlin TL, Lee RE, Panchal RG. 2019. Aminomethyl spectinomycins: a novel antibacterial chemotype for biothreat pathogens. J Antibiot 72:693–701. doi: 10.1038/s41429-019-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruhn DF, Waidyarachchi SL, Madhura DB, Shcherbakov D, Zheng Z, Liu J, Abdelrahman YM, Singh AP, Duscha S, Rathi C, Lee RB, Belland RJ, Meibohm B, Rosch JW, Böttger EC, Lee RE. 2015. Aminomethyl spectinomycins as therapeutics for drug-resistant respiratory tract and sexually transmitted bacterial infections. Sci Transl Med 7:1–12. doi: 10.1126/scitranslmed.3010572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Laakso LM. 2016. Dali server update. Nucleic Acids Res 44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayward S, Lee RA. 2002. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model 21:181–183. doi: 10.1016/S1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 24.Fong DH, Lemke CT, Hwang J, Xiong B, Berghuis AM. 2010. Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila. J Biol Chem 285:9545–9555. doi: 10.1074/jbc.M109.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steitz TA. 1993. DNA- and RNA-dependent DNA polymerases. Curr Opin Struct Biol 3:31–38. doi: 10.1016/0959-440X(93)90198-T. [DOI] [Google Scholar]
- 26.Anandakrishnan R, Aguilar B, Onufriev AV. 2012. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res 40:W537–W541. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen LC, Benning MM, Holden HM. 1995. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry 34:13305–13311. doi: 10.1021/bi00041a005. [DOI] [PubMed] [Google Scholar]
- 28.Morar M, Bhullar K, Hughes DW, Junop M, Wright GD. 2009. Structure and mechanism of the lincosamide antibiotic adenylyltransferase LinB. Structure 17:1649–1659. doi: 10.1016/j.str.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Zurenko GE, Yagi BH, Vavra JJ, Wentworth BB. 1988. In vitro antibacterial activity of trospectomycin (U-63366F), a novel spectinomycin analog. Antimicrob Agents Chemother 32:216–223. doi: 10.1128/AAC.32.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barry AL, Jones RN, Thornsberry C. 1989. Antibacterial activity of trospectomycin (U-63366F) and initial evaluations of disk diffusion susceptibility tests. Antimicrob Agents Chemother 33:569–572. doi: 10.1128/AAC.33.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bassenden AV, Rodionov D, Shi K, Berghuis AM. 2016. Structural analysis of the tobramycin and gentamicin clinical resistome reveals limitations for next-generation aminoglycoside design. ACS Chem Biol 11:1339–1346. doi: 10.1021/acschembio.5b01070. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwart PH, Grosse-Kunstleve RW, Adams PD. 2005. Xtriage and Fest: automatic assessment of X-ray data and substructure structure factor estimation. CCP4 Newsl 43. [Google Scholar]
- 35.Vagin A, Lebedev A. 2015. MoRDa, an automatic molecular replacement pipeline. Acta Crystallogr A Found Adv 71:s19. doi: 10.1107/S2053273315099672. [DOI] [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liebschner D, Afonine PV, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD. 2017. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr D Struct Biol 73:148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Coordinates and structure factors have been deposited in the PDB with accession codes 6SXJ [apo ANT(9)], 6XZ0 [ANT(9)-spc], and 6XXQ [ANT(9)-ATP-spc].