Bunyaviruses are significant human pathogens, causing diseases ranging from hemorrhagic fevers to encephalitis. Among these viruses, La Crosse virus (LACV), a member of the California serogroup, circulates in the eastern and midwestern United States. While LACV infection is often asymptomatic, dozens of cases of encephalitis are reported yearly. Unfortunately, no antivirals have been approved to treat LACV infection. Here, we developed a method to rapidly test potential antivirals against LACV infection.
KEYWORDS: bunyaviruses, La Crosse virus, antivirals, ionophores
ABSTRACT
Bunyaviruses are significant human pathogens, causing diseases ranging from hemorrhagic fevers to encephalitis. Among these viruses, La Crosse virus (LACV), a member of the California serogroup, circulates in the eastern and midwestern United States. While LACV infection is often asymptomatic, dozens of cases of encephalitis are reported yearly. Unfortunately, no antivirals have been approved to treat LACV infection. Here, we developed a method to rapidly test potential antivirals against LACV infection. From this screen, we identified several potential antiviral molecules, including known antivirals. Additionally, we identified many novel antivirals that exhibited antiviral activity without affecting cellular viability. Valinomycin, a potassium ionophore, was among our top targets. We found that valinomycin exhibited potent anti-LACV activity in multiple cell types in a dose-dependent manner. Valinomycin did not affect particle stability or infectivity, suggesting that it may preclude virus replication by altering cellular potassium ions, a known determinant of LACV entry. We extended these results to other ionophores and found that the antiviral activity of valinomycin extended to other viral families, including bunyaviruses (Rift Valley fever virus, Keystone virus), enteroviruses (coxsackievirus, rhinovirus), flavirivuses (Zika virus), and coronaviruses (human coronavirus 229E [HCoV-229E] and Middle East respiratory syndrome CoV [MERS-CoV]). In all viral infections, we observed significant reductions in virus titer in valinomycin-treated cells. In sum, we demonstrate the importance of potassium ions to virus infection, suggesting a potential therapeutic target to disrupt virus replication.
INTRODUCTION
Bunyaviruses are the largest family of viruses, composed of hundreds of members. These segmented, negative-sense RNA viruses are transmitted primarily by an arthropod vector, and several family members pose significant threats to human health. Rift Valley fever virus (RVFV) frequently infects humans and ruminants, resulting in severe morbidity and mortality. While RVFV is primarily transmitted in Africa and the Middle East, the threat of global spread looms, and recent examples of Zika (1) and chikungunya (2) viruses illustrate this tangible hazard. In addition to RVFV, several other bunyaviruses infect humans and are associated with severe pathologies as well. La Crosse virus (LACV), distantly related to RVFV, is a bunyavirus present primarily in the midwestern and eastern United States (3). LACV is transmitted by Aedes triseriatus, though Aedes albopictus (4–6) efficiently carries the virus as well. While relatively unknown and frequently undiagnosed, LACV infects and causes neuroinvasive disease in dozens of people every year. In fact, from 2009 to 2018, 679 such cases were reported according to the Centers for Disease Control and Prevention (7). Further, LACV cases recently have emerged in the southeastern United States, suggesting spread of the virus (8). The ability of LACV to infect Aedes albopictus mosquitos will likely lead to its continuing spread. Despite this spread and the severe disease associated with LACV infection, no antivirals or vaccines are available to prevent or treat infection. Thus, LACV presents a significant threat to human health.
Despite the prevalence of bunyaviruses, no antivirals are available to treat bunyavirus infection, and palliative care is given to patients presenting with LACV encephalitis. While several vaccine candidates have been developed to target RVFV (9–11), no similar effort has been invested in anti-LACV therapeutics. With the increasing availability of drug panels to screen molecules for antivirals, rapid investigation and deployment of antivirals for emerging viruses is possible. Several prior screens for antivirals have successfully identified lead molecules and highlighted cellular pathways critical to virus infection. For example, recent screens with chikungunya virus highlighted berberine, abamectin, and ivermectin as promising antivirals (12). Additionally, Zika virus screens have uncovered known and novel antivirals (13–15), including mycophenolic acid and daptomycin, among others. More closely related to LACV, an RVFV screen highlighted azauridine and mitoxantrone (16). The ability to rapidly screen molecules highlights an opportunity to identify both unique virus-specific and broad-spectrum antivirals, as these reports have highlighted.
Antivirals may target diverse viral and cellular processes (17). Host-directed antivirals have gained appreciation recently, as interfering with host processes crucial to viral processes has several benefits, including potential for broad-spectrum activity and a heightened requirement for antiviral resistance beyond minor mutations in the virus. Additionally, numerous approved and available drugs are already available that can be repurposed as antivirals (18, 19). While significant work remains to be done to identify and verify such antivirals, rapidly screening compounds can provide insight.
Using the NIH’s Developmental Therapeutics Program (DTP), we obtained and screened >500 compounds for activity against LACV. We identified several known antivirals, including deoxyuridine and quinonone. Importantly, we also identified a variety of novel classes of antivirals, including metal ion chelators. Valinomycin, a top hit in our screen, functions by transporting potassium ions against the electrochemical gradient. Potassium plays important roles in bunyavirus entry (20–22), highlighting the potential of targeting viral entry. We investigated the antiviral activity of valinomycin, observing that valinomycin exhibits antiviral activity in several cellular systems in a dose-dependent manner and independent of treatment time. We also found that valinomycin does not directly inactivate viral particles, highlighting a cellular role for potassium ions in virus infection. We expanded our results to additional ionophores, observing that some but not all effectively blocked LACV replication. Finally, we determined that valinomycin is broadly antiviral, as it reduced the replication of several viruses from diverse families, including flaviviruses and enteroviruses. Together, these data highlight the utility of rapid screening of antiviral molecules as well as a crucial role for potassium ions in LACV infection.
RESULTS
Development of rapid screening of NIH DTP compounds active against LACV.
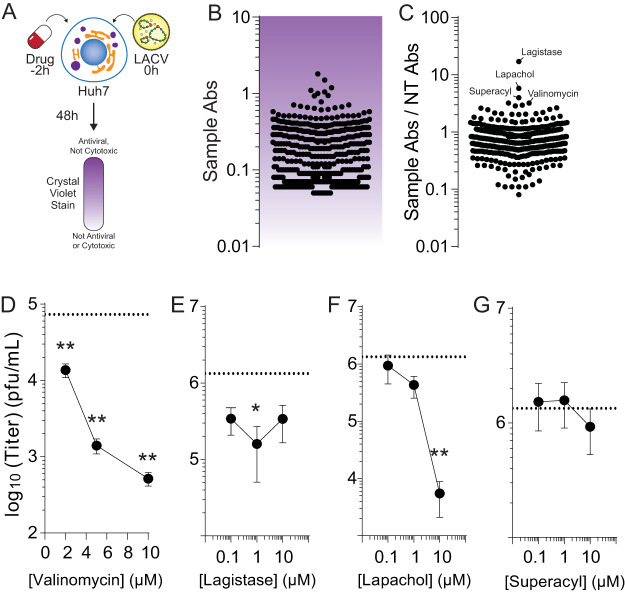
We developed a simple, rapid assay to measure antiviral activity in Huh7 cells (Fig. 1A). We plated Huh7 cells to confluence in 96-well plates, to which we added 2 μM drug from the NIH NCI Development Therapeutics Program (DTP). Two hours later, cells were infected at a multiplicity of infection (MOI) of 0.1 plaque-forming units (PFU) per cell. At 48 h postinfection (hpi), cells were fixed with formalin and stained with crystal violet stain. Because viable cells robustly stain with crystal violet, while dead cells do not, we could discriminate between live and dead cells. Importantly, any cytotoxic molecule would not stain with crystal violet; thus, stained cells indicate antiviral molecules that are not cytotoxic at 2 μM. Further, this assay cannot distinguish proviral molecules from cytotoxic molecules. Stain was subsequently resuspended in 10% acetic acid and absorbance read at 595 nm. To control for interplate variability, each plate contained an untreated and infected (low survival), untreated and uninfected (high survival), and ribavirin-treated (400 μM) control (high survival). The absorbances of drug-treated and infected wells (Fig. 1B) were compared to untreated and uninfected controls by dividing their absorbance values (Fig. 1C). This ratio highlighted several candidate antivirals, including lagistase, lapachol, superacyl, and valinomycin. Interestingly, we identified several known antivirals in our screen, including deoxyuridine and nelarabine (summarized in Table 1). Thus, our assay identified molecules with novel activity against LACV, including some recognized antivirals.
FIG 1.
Rapid screening for LACV antivirals. Schematic of the screen performed in these studies. (A) Potential antivirals were added to cells 2 h prior to infection with La Crosse virus at an MOI of 0.01. At 48 hpi, cells were fixed and stained with crystal violet stain, which was then quantified, with darker-staining cells surviving drug treatment and virus infection. (B) Quantification of antiviral activity. Each dot represents a single compound analyzed in our assay. (C) The quantification of antiviral activity as represented in panel B was compared to control cells that were not treated (sample Abs/NT Abs) to obtain the relative antiviral activity. The top hits in this screen are indicated. (D to G) Huh7 cells were pretreated for 2 h with increasing doses of (D) valinomycin, (E) lagistase, (F) lapachol, and (G) superacyl and subsequently infected. Viral titers were determined at 24 hpi. The dotted line indicates titers from untreated samples.
TABLE 1.
Top hits in primary antiviral screen
| DTP NSC no.a | Primary screen hit value | Antiviral name | Description | Prior antiviral activity against:b |
|---|---|---|---|---|
| 407286 | 16.97 | Lagistase | Naturally occurring plant phenol; antioxidant and metabolism-modulating properties | HIV (34), HRV (35), HBV (36) |
| 11905 | 5.76 | Lapachol | Naphthoquinone from the lapacho tree; anti-inflammatory and antiproliferation agent | EBV (37), EV (38) |
| 41833 | 3.97 | Superacyl | Base analog | |
| 122023 | 3.19 | Valinomycin | Potassium ionophore | HRV (39), SARS-CoV (28), RSV (40), PV (41) |
| 51787 | 3.19 | 5-Isoquinolinol | Metal chelating agent | |
| 24819 | 2.93 | β-peltatin | Glycoside from Podophyllum roots | MV, HSV (42) |
| 401005 | 2.84 | Pleurotine | Antifungal agent; thioredoxin inhibitor | |
| 44175 | 2.59 | Polyporin | Benzoquinone isolated from Hapalopilus nidulans; dihydroorotate dehydrogenase inhibitor | |
| 712807 | 2.48 | Capecitabine | Antimetabolite antineoplastic agent | |
| 15307 | 2.45 | Quininone | Quinine metabolite | |
| 36398 | 2.20 | Taxifolin | Polyphenol from milk thistle seeds | HAV (43), HIV (44) |
| 38010 | 2.15 | Chaulmoogric acid ethyl ester | Ethyl ester of long-chain fatty acid from chaulmoogra seeds | |
| 747972 | 2.10 | Lenalidomide | Antiangiogenic factor, used to treat multiple myeloma | |
| 718781 | 2.04 | Tarceva | Epidermal growth factor receptor inhibitor antineoplastic agent, used to treat pancreatic and non-small cell lung cancer | |
| 301683 | 2.02 | Daphnetin | Antioxidant agent; coumarin; used in traditional Chinese medicine to treat cardiovascular disease | |
| 755985 | 1.88 | Nelarabine | Antimetabolite antineoplastic agent, used to treat T-cell acute lymphoblastic leukemia | |
| 5366 | 1.81 | Noscapine | Antitussive agent | |
| 279836 | 1.76 | Mitoxantrone | Topoisomerase inhibitor; antineoplastic agent | HIV (45), VACV (46), HCV (47), HSV (48), RVFV (16) |
| 11440 | 1.64 | Protopine | Calcium channel blocker and anti-inflammatory agent | |
| 11926 | 1.60 | Tardolyt | Carcinogenic molecule from birthwort plants | HSV (49) |
NSC, National Service Center.
HIV, human immunodeficiency virus; HRV, human rhinovirus; HBV, hepatitis B virus; EBV, Epstein-Barr virus; EV, enterovirus; SARS-CoV, severe acute respiratory syndrome coronavirus; RSV, respiratory syncytial virus; PV, poliovirus; MV, measles virus; HSV, herpes simplex virus; HAV, hepatitis A virus; VACV, vaccinia virus.
Valinomycin restricts LACV replication.
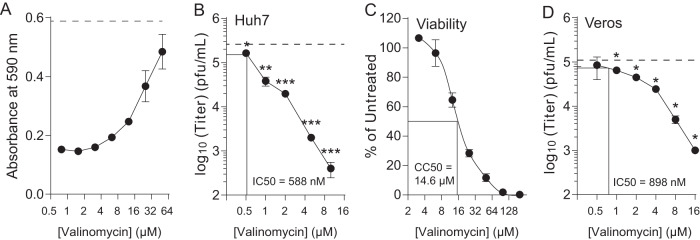
We chose four compounds from our screen for secondary screening. We performed a dose response with valinomycin, lagistase, lapachol and superacyl infected for 24 h and measured infectious virus with a plaque assay (Fig. 1D to G). We observed that valinomycin, lapachol, and lagistase reduced titers. In contrast, superacyl showed no antiviral activity. We chose valinomycin to investigate further because of its strong phenotype, because of its known effect on potassium ions in the cell relevant to bunyavirus infection, and because it was not previously described to have antiviral activity against LACV. To better characterize valinomycin’s antiviral activity, we initially measured the ability of a range of doses to inhibit virus replication. Cells were seeded to confluence, treated with increasing doses of valinomycin, from 1 to 64 μM, and infected at an MOI of 0.1. At 48 h, cells were fixed and stained with crystal violet, and stain was quantified by absorbance reading. We observed that valinomycin exhibited antiviral activity at doses above 10 μM, as crystal violet staining was stronger, suggesting more surviving cells (Fig. 2A; the dashed line is the absorbance from untreated, uninfected cells). Doses as high as 64 μM did not affect crystal violet stain, suggesting that cellular viability was not compromised. To confirm this phenotype with titers, we treated cells with increasing doses of valinomycin 2 h prior to infection at an MOI of 0.1 and measured titers by plaque assay at 48 hpi. We observed that viral titers were significantly decreased compared to untreated controls (Fig. 2B, dashed line) at concentrations above 1 μM. In fact, viral titers were reduced over 100-fold at 10 μM. We calculated a 50% inhibitory concentration (IC50) value of 588 nM (Table 2). To confirm that cellular viability was not compromised, we used a fluorescence assay to measure cellular ATP content after treatment with increasing doses of valinomycin. We observed cellular toxicity at doses at and above 16 μM (Fig. 2C; 50% cytotoxic concentration [CC50] value of 14.7 μM), though no toxicity was observed using either cellular morphology or fluorescence ATP assay below 10 μM. The selectivity index for valinomycin is thus 25.
FIG 2.
Valinomycin is antiviral. Huh7 cells were treated with increasing doses of valinomycin for 2 h prior to LACV infection. (A) At 48 hpi, cells were stained with crystal violet and quantified. (B) Viral titers were measured with a plaque assay. The dashed line indicates the titer of untreated controls. Lines indicate the IC50 value. (C) Viability was measured using a fluorescence intracellular ATP assay. Lines indicate the CC50 value. (D) Vero cells were treated and infected as described for the Huh7 cells. Viral titers were determined at 48 hpi. Lines indicate the IC50 value. *, P < 0.05; **, P < 0.01; ***, P < 0.0001 using two-tailed Student’s t test; n = 3. Error bars represent one standard error of the mean.
TABLE 2.
Antiviral activity of valinomycin against diverse viruses
| Virus | Cell type | IC50 value (nM) |
|---|---|---|
| LACV | Huh7 | 588 |
| LACV | Vero-E6 | 898 |
| MP12 | Huh7 | 41 |
| HRV2 | Huh7 | 610 |
| CVB3 | Huh7 | 971 |
| ZIKV | Huh7 | 78 |
| KEYV | Huh7 | 156 |
| HCoV-229E | Huh7 | 67 |
| MERS-CoV | Vero-E6 | 5 |
As further confirmation of valinomycin’s antiviral activity, we measured cell-associated viral genomes. Huh7 cells were treated and infected as described above, and cell-associated RNA was collected at 48 hpi. RNA was purified, reverse transcribed, and analyzed via quantitative PCR (qPCR) for small, medium, and large genome segments, normalizing to cellular β-actin. Paralleling our titer data, valinomycin treatment reduced viral genome content in a dose-dependent manner (Fig. 2D). Viral genome content was reduced upward of 100-fold with 10 μM valinomycin treatment. To extend these results to other cell types, we treated and infected Vero-E6 cells as described above and determined viral titers with a plaque assay at 48 hpi. Again, we observed a significant reduction in viral titer, with an IC50 value of 898 nM. In sum, our data suggest that valinomycin is antiviral at noncytotoxic doses in these distinct cell types, reducing both viral titers and cell-associated viral genomes in a dose-dependent manner.
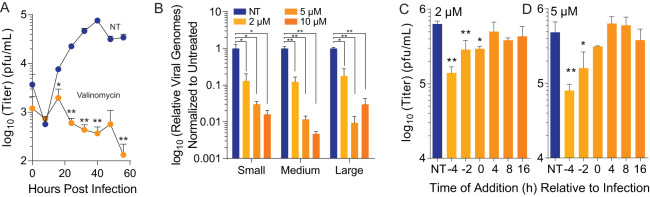
Valinomycin is antiviral over multiple rounds of infection.
Our initial assays were performed at low MOI, and viral titer was measured at 48 hpi. To determine if valinomycin was antiviral over several rounds of replication, we treated Huh7 cells with 2 μM valinomycin 2 h prior to infection at an MOI of 0.1 and subsequently collected samples to titer every 8 h for 56 h total. We found that LACV titers were significantly reduced at all times after 8 hpi (Fig. 3A); in fact, virus failed to replicate above the input virus titers (0 h). To confirm that valinomycin was reducing infectious virus production, we measured viral RNA genomes. To this end, we treated cells with increasing doses of valinomycin, infected them with LACV, and collected cell-associated RNA in TRIzol at 24 hpi. After purifying and reverse-transcribing, we performed qPCR using primers specific to the small, medium, and large genome segments. We observed that treatment with valinomycin significantly reduced the number of viral genomes by >90% with treatment and that no individual genome segment was affected more than another (Fig. 3B). Together, these data suggest that valinomycin blocks virus replication and reduces viral RNA accumulation. To determine at what times during LACV infection valinomycin was able to reduce viral titers, valinomycin was added before, during, or after infection at a high MOI of 5 with a concentration of 2 or 5 μM valinomycin. Cellular supernatant was then collected at 24 hpi, and virus was quantified via plaque assay. Viral titers were significantly decreased by 4.5-fold when cells were treated 4 h before infection compared to no treatment with 2 μM valinomycin (Fig. 3C). Viral titers were also reduced when cells were pretreated 2 h before infection as well as during infection when treated with 2 μM valinomycin, but when it was added any time after infection, it failed to reduce viral titers. A similar pattern was seen when cells were treated with 5 μM valinomycin (Fig. 3D). These data suggest that valinomycin prevents the early stages of LACV infection within cells, such as entry.
FIG 3.
Valinomycin is antiviral over multiple rounds of infection. Huh7 cells were treated with 2 μM valinomycin for 2 h prior to infection at an MOI of 0.01. (A) Cellular supernatant was collected at the times indicated, and viral titers were determined with a plaque assay. (B) Viral RNA was purified from infected cell supernatant at 48 hpi and quantified by qPCR using primers specific to each genome segment. (C and D) Huh7 cells were treated with (C) 2 μM and (D) 5 μM valinomycin at various times prior to and after infection. Virus titers were determined with a plaque assay at 24 hpi. *, P < 0.05; **, P < 0.01 using two-tailed Student’s t test; n = 2 or 3. Error bars represent one standard error of the mean.
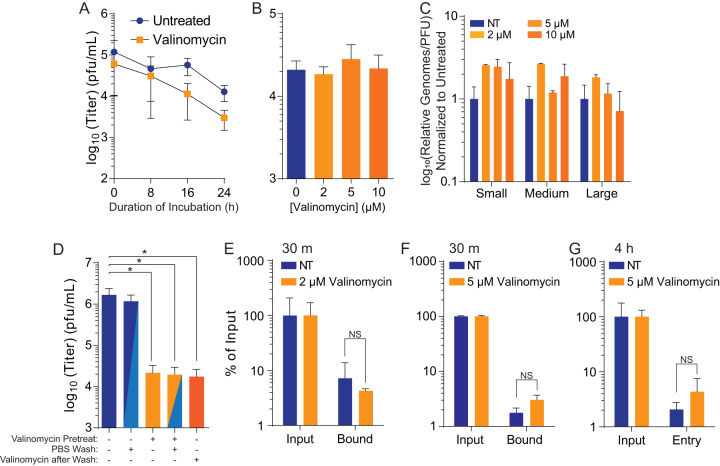
Valinomycin does not reduce viral particle infectivity.
Because we observed significant reductions in LACV titers with valinomycin treatment, we hypothesized that valinomycin might directly affect cellular processes to reduce virus infection. Nonetheless, valinomycin is a cyclic peptide and could potentially directly inactivate viral particles, as seen previously (23). To test whether valinomycin directly reduced virus infectivity, we directly incubated LACV with 2 μM valinomycin for 24 h and directly determined the titer of the surviving virus at regular intervals. We found that valinomycin did not significantly alter viral titer over the time examined (Fig. 4A), suggesting that valinomycin does not directly inactivate viral particles. We further examined the capacity of valinomycin to inactivate particles by incubating the virus with increasing doses, up to 10 μM valinomycin, for 24 h prior to directly determining the titer. As in our time course, we observed no significant change in viral titers at any dose (Fig. 4B), again suggesting that valinomycin does not directly inactivate LACV particles. As a final confirmation of this phenotype, we measured viral RNA in viruses exposed to increasing doses of valinomycin. We then compared the relative number of genomes to the titer to calculate the genome-to-PFU ratio. We observed that this number did not change with valinomycin treatment, suggesting no change in specific infectivity (Fig. 4C). In sum, these data suggest that valinomycin does not affect virus infectivity by directly acting on the virion.
FIG 4.
Valinomycin does not directly reduce particle infectivity. (A) Stock LACV was incubated with 2 μM valinomycin at 37°C for the times indicated before virus titer was determined with a plaque assay. (B) Stock virus was incubated with increasing doses of valinomycin at 37°C for 24 h prior to determination of the titer. (C) Viral genomes from panel B were quantified from purified RNA and compared to viral titers to calculate the relative viral genomes compared to infectious virus (PFU). No significant differences were observed. (D) Huh7 cells were treated with 2 μM valinomycin and subsequently washed with PBS and replenished with fresh medium as indicated. Fresh valinomycin was added as indicated following a PBS wash and medium replenishment. (E and F) Cells were similarly treated with (E) 2 μM or (F) 5 μM valinomycin and infected for 30 min before analysis of attached viral genomes, normalizing to input. (G) A similar analysis was also performed at 4 hpi. *, P < 0.05. NS, not significant using two-tailed Student’s t test; n ≥ 2. Error bars represent one standard error of the mean.
We thus hypothesized that valinomycin’s antiviral activity was due to its effect on the cell. The role of potassium in bunyavirus infection has been well documented, and bunyavirus entry is potassium dependent (21, 22). To test if valinomycin was affecting the cell rather than the virus, we treated Huh7 cells with 2 μM valinomycin, and immediately before infection, we washed away the drug. As a control, we maintained valinomycin on cells or replaced the valinomycin after washing away the initial treatment. We observed that even after removing and washing valinomycin from the cells, the antiviral activity persisted, as viral titers remained reduced to the same level as when valinomycin treatment is concurrent with infection (Fig. 4D). Together, these data suggest that valinomycin does not directly inactivate viral particles but that treatment of cells reduces LACV infection.
Because valinomycin affects potassium ion gradients, which are important for bunyavirus entry, we measured the effect of valinomycin on the ability of LACV to bind and enter cells. Cells treated with 2 μM valinomycin were exposed to LACV at an MOI of 1 and were either left unwashed for input samples or washed with phosphate-buffered saline (PBS) after a 30-minute incubation to measure bound virus. Cells and viruses were then collected for RNA purification and reverse transcription. When quantifying bound virus, normalizing to cellular β-actin and input virus, we found that valinomycin had no effect on LACV binding (Fig. 4E). We repeated this assay with 5 μM valinomycin and again observed no change in virus binding (Fig. 4F). We also collected samples at 4 hpi, after virus entry, and measured cell-associated viral RNA and again observed no change in viral RNA levels. Together, these data suggest that valinomycin does not affect virus binding to susceptible cells.
Ionophores are selectively antiviral.
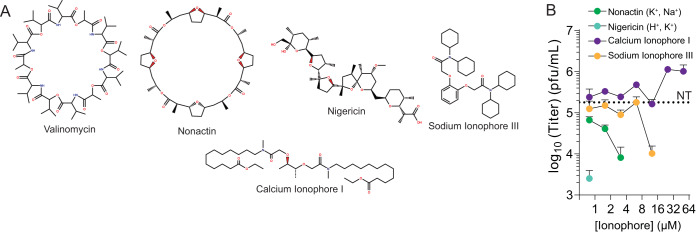
Given that valinomycin is a potassium ionophore, we wished to investigate whether other ionophores, for potassium or otherwise, were antiviral. Potassium ions play a crucial role in cellular entry; however, a role for sodium or calcium ions is not as well described. Marituba virus (MTBV) infection results in a sodium ion influx (24), but the origin or function of these ionic changes are not known. To determine if other ionophores might exhibit antiviral activity, we treated cells with increasing doses of nonactin (potassium and sodium ionophore), nigericin (hydrogen and potassium ionophore), calcium ionophore I, and sodium ionophore III (Fig. 5A). Two hours later, we infected the cells with LACV at an MOI of 0.01 and measured viral titers at 48 hpi. Both nonactin and nigericin exhibited significant antiviral activity, and viral titers were not measurable above 4 and 1 μM, respectively (Fig. 5B). Treatment with sodium ionophore III resulted in a dose-dependent decrease in viral titers, and virus was not recovered above 10 μM. Interestingly, treatment with calcium ionophore I showed no changes in viral titer, even at the highest dose. Thus, we observe that LACV replication is disrupted by several ionophores, especially potassium ionophores, highlighting the role for potassium in virus infection.
FIG 5.
Ionophores are selectively antiviral. (A) Structures of valinomycin, nonactin, calcium ionophore I, nigericin, and sodium ionophore III. (B) Huh7 cells were treated with increasing doses of the iophores for 2 h prior to LACV infection. Virus titers were determined at 48 hpi with a plaque assay. The dotted line indicates titers from untreated (NT) control cells. n = 3. Error bars represent one standard error of the mean.
Valinomycin is broadly antiviral.
LACV is related to several other medically relevant bunyaviruses, including Keystone virus and Rift Valley fever virus. To determine whether these viruses respond to valinomycin treatment, we treated and infected Huh7 cells with these viruses and measured viral titers at 48 hpi. As with LACV, we observed a dose-dependent decrease in viral titers, with titers decreasing greater than 100-fold at concentrations above 2 μM (Fig. 6A). To expand beyond bunyaviruses, we performed the same analysis with Zika virus (flavivirus) infection (Fig. 6B), coxsackievirus B3 and human rhinovirus 2 (picornaviruses) (Fig. 6C), and human coronavirus 229E (HCoV-229E) and Middle East respiratory syndrome (MERS) coronaviruses (Fig. 6D). Different concentrations of valinomycin were used, as the viruses exhibited different sensitivities. In all cases, valinomycin significantly reduced viral titers. Zika virus titers were the most sensitive to valinomycin treatment, and virus was not recovered above 500 nM valinomycin treatment. Similarly, valinomycin treatment significantly disrupted both HCoV-229E and MERS-CoV infection. Thus, valinomycin exhibits broad antiviral activity, highlighting cellular potassium as a conserved and crucial host factor in virus replication.
FIG 6.
Valinomycin is broadly antiviral. (A to D) Huh7 cells were treated with increasing doses of valinomycin for 2 h prior to infection at an MOI of 0.1 with (A) bunyaviruses KEYV and RVFV (strain MP12) for 48 h, (B) flavivirus ZIKV for 48 h, (C) enteroviruses HRV2 and CVB3 for 24 h, and (D) coronaviruses HCoV-229E and MERS-CoV for 24 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using two-tailed Student’s t test; n ≥ 2. Error bars represent one standard error of the mean.
DISCUSSION
Treatment for virus-infected patients, including encephalitic patients, is limited in the scope of available therapeutics. La Crosse virus lacks any approved treatment, and antiviral development could benefit not only LACV-infected patients but perhaps patients infected with related bunyaviruses. Unfortunately, antiviral development is a nonlinear process that has provided no promising targets for this specific virus to date. Our screen is the first to investigate a large number (>500) of compounds in their ability to reduce LACV infection in vitro, and our hits demonstrate that several known antivirals may exhibit anti-LACV activity. Additionally, the identification of novel antivirals, such as valinomycin, provides novel avenues of antiviral development.
In our screens, valinomycin consistently exhibited antiviral activity and was selected for follow-up experiments. Based on our results, valinomycin is a host-targeted antiviral with broad-spectrum activity. In fact, our tests against eight distinct viruses and four diverse families of virus suggest (i) that valinomycin has activity against evolutionarily distant viruses (IC50 values summarized in Table 2) and (ii) that these viruses rely on this conserved host process for efficient replication. Precisely why each virus is sensitive to valinomycin is likely virus specific, though mechanisms for bunyaviruses have been described (20, 22). As a potassium ionophore, valinomycin disrupts potassium ion gradients, resulting in aberrant cellular events, including endocytosis. This potassium-dependent endocytosis is required for efficient bunyavirus entry (20). Potassium affects endosomal maturation (25) and chloride concentrations (26), which may impact viral entry. Additionally, a study using Hazara virus, a distantly related bunyavirus, demonstrated that the viral fusion spike conformation was potassium sensitive (21). Thus, valinomycin’s antiviral activity fits with the prescribed role of potassium ions during virus infection, highlighting both the importance of this pathway for virus infection and its potential as an antiviral target.
Valinomycin is a naturally occurring cycling peptide, synthesized by Streptomyces species as an antibiotic (27). In our assays, we did not observe significant cellular toxicity, as measured either by gross cellular morphology or by cellular ATP levels, until doses above 10 μM (CC50 value of 14), and our IC50 of 588 nM suggests there is a window of therapeutic potential. However, given the drug’s toxicity in vivo, modification of the valinomycin structure may tune toxicity while maintaining antiviral activity. Thus, valinomycin is not FDA-approved for human use; however, other ionophores are less toxic and also show antiviral activity. Thus, using other, less toxic potassium ionophores may similarly function to inhibit LACV infection, as we observed with nonactin and nigericin. For outbreak viruses such as severe acute respiratory syndrome CoV (SARS-CoV) or Zika virus (ZIKV), however, limited valinomycin usage might be beneficial. Interestingly, valinomycin was identified in an earlier screen for anti-SARS-CoV molecules (28), underscoring the antiviral potential of valinomycin despite its negative characteristics. Importantly, the emergence of a new group 2B CoV, SARS-CoV-2, signals the ongoing threat and need to rapidly respond to novel emergent viruses. Additional in vivo testing, combined with medicinal chemistry approaches to structural modification, would be necessary prior to clinical use.
In addition to valinomycin, our screen identified several tantalizing antiviral candidates. In fact, several known antivirals were identified (summarized in Table 1). Dasatinib has previously been described to inhibit dengue virus and HIV infection (29, 30). Quininone has activity against enterovirus proteases as well as reverse transcriptases (31, 32). As mentioned previously, mitoxantrone was identified in a screen for anti-RVFV molecules (16). Additionally, we identified a number of cancer therapeutics, targeting processes such as angiogenesis and topoisomerase, that show significant promise. Nonetheless, further verification of antiviral activity and investigation of the mechanisms of action is necessary. Regardless, the development of novel antivirals is crucial to combat virus infection and to respond to the possibility of rapid virus dissemination and evolution. LACV is a significant threat to human health, and continued development of novel antivirals may prove fruitful if the virus were to spread, as arboviruses are wont to do.
MATERIALS AND METHODS
Cell culture.
Cells were maintained at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) with bovine serum and penicillin-streptomycin. Vero cells (BEI Resources) were supplemented with 10% newborn calf serum (NBCS; Thermo Fisher), and Huh7 cells, kindly provided by Susan Uprichard, were supplemented with 10% fetal bovine serum (FBS; Thermo Fisher).
Drug treatment.
For standard treatment experiments, Huh7 cells were infected at a multiplicity of infection (MOI) of 0.01 PFU/cell, unless otherwise indicated, with LACV, Keystone virus (KEYV), RVFV MP-12, and ZIKV and were concurrently treated with ionophores (valinomycin, nigericin, nonactin, calcium ionophore I, and sodium ionophore III; Cayman Chemical) dissolved in dimethyl sulfoxide (DMSO). Cells were then incubated at 37°C in 5% CO2 for 48 h. For direct incubation experiments, 100 μl of LACV stock virus was incubated with increasing concentrations of valinomycin over various time periods at 37°C. For wash-away experiments, cells were seeded as stated above for standard treatment experiments. Cells were treated 4 h prior to removing medium and washing with phosphate-buffered saline (PBS). Medium containing LACV was then replaced, and cells were incubated for 48 h. Chemical structures were recreated using MarvinSketch 19.26 (ChemAxon Ltd.).
Rapid screening of antiviral compounds.
Huh7 cells were seeded in 96-well plates, infected with LACV at an MOI of 0.01, and concurrently treated with 100 μM of each compound from the NIH DTP compound plates. Cells were incubated at 37°C in 5% CO2 for 48 h. Medium was aspirated, cells were fixed with 4% formalin, and live cells were stained with crystal violet solution (Sigma-Aldrich). Excess stain was removed in a mild bleach solution, and the cells were allowed to dry for 24 h. The remaining crystal violet stain was resuspended in 10% acetic acid. The absorbance at 590 nm was detected using a BioTek Synergy H1 plate reader.
Infection and enumeration of viral titers.
RVFV (strain MP-12 [33]), LACV (product no. NR-540; BEI Resources), and KEYV (strain B64-5587.05; product no. NR537; BEI Resources) were derived from the first passage of virus in Huh7 cells. ZIKV (strain MR766) was derived from the first passage of virus in Vero cells. CVB3 (Nancy strain) and HRV2 were derived from the first passage of virus in HeLa cells. LACV and KEYV were obtained from Biodefense and Emerging Infections (BEI) Research Resources. HCoV-229E and MERS-CoV were propagated and quantitated via standard methods as previously described and under biosafety level 3 (BSL3) conditions (23). Drug was maintained throughout infection as designated. Viral stocks were maintained at –80°C. For infection, virus was diluted in serum-free DMEM for a multiplicity of infection (MOI) of 0.1 on Huh7 cells unless otherwise indicated. Viral inoculum was overlaid on cells for 10 to 30 min, and the cells were washed with PBS before replenishment of medium. Supernatants were collected at the times specified. Dilutions of cell supernatant were prepared in serum-free DMEM and used to inoculate a confluent monolayer of Vero cells for 10 to 15 min at 37°C. Cells were overlaid with 0.8% agarose in DMEM containing 2% NBCS. CVB3 and HRV2 samples were incubated for 2 days, RVFV MP-12, ZIKV, and LACV samples were incubated for 4 days, and KEYV samples were incubated for 5 days at 37°C. Following appropriate incubation, cells were fixed with 4% formalin and revealed with crystal violet solution (10% crystal violet; Sigma-Aldrich). Plaques were enumerated and used to back-calculate the number of plaque-forming units (PFU) per milliliter of collected volume.
RNA purification and cDNA synthesis.
Medium was cleared from cells, and TRIzol reagent (Zymo Research) was directly added. Lysate was then collected, and RNA was purified according to the manufacturer’s protocol utilizing the Direct-zol RNA miniprep plus kit (Zymo Research). Purified RNA was subsequently used for cDNA synthesis using high-capacity cDNA reverse transcription kits (Thermo-Fischer) according to the manufacturer’s protocol with 10 to 100 ng of RNA and random hexamer primers.
Viral genome quantification.
Following cDNA synthesis, reverse transcription-quantitative PCR (qRT-PCR) was performed using QuantStudio3 (Applied Biosystems by Thermo-Fischer) and SYBR green mastermix (DOT Scientific). Samples were held at 95°C for 2 min prior to 40 cycles of 95°C for 1 s and 60°C for 30 s. Primers were verified for linearity using 8-fold serial-diluted cDNA and checked for specificity via melt curve analysis followed by agarose gel electrophoresis. All samples were used to normalize to total RNA using the ΔCT method.
Genome-to-PFU ratio calculations.
The number of viral genomes quantified as described above were divided by the viral titer, as determined by plaque assay, to measure the genome-to-PFU ratio. The values obtained were normalized to untreated conditions to obtain the relative genome-to-PFU ratio. The primers used were small 5′-GGC-AGG-TGG-AGG-TTA-TCA-AT-3′ (forward), 5′-AAG-GAC-CCA-TCT-GGC-TAA-ATA-C-3′ (reverse); medium 5′-CCT-GCC-TAG-AGA-CTG-AGA-GTA-T-3′ (forward), 5′-GAG-TTG-CAA-TGT-TGG-TGT-AAG-G-3′ (reverse); large 5′-ACT-GGA-AGG-TCG-AGG-ATC-TAA-3′ (forward), 5′-GTC-GCT-TGT-CTC-ACC-CAT-AAT-A-3′ (reverse); and GAPDH 5′-GAT-TCC-ACC-CAT-GGC-AAA-TTC-3′ (forward), 5′-CTG-GAA-GAT-GGT-GAT-GGG-ATT-3′ (reverse).
Statistical analysis.
Prism 6 (GraphPad) was used to generate graphs and perform statistical analysis. For all analyses, one-tailed Student’s t test was used to compare groups, unless otherwise noted, with α = 0.05. For tests of sample proportions, P values were derived from calculated Z scores with two tails and α = 0.05.
ACKNOWLEDGMENTS
We are grateful to Susan Uprichard, Justin Harbison, and Gail Reid for critical discussion of the data and manuscript. We also thank Susan Uprichard for the Huh7 cells, Shinji Makino and Kaori Terasaki for generously providing the MP-12 strain of RVFV, and Bill Jackson for HRV2.
This research was supported by a grant from the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH (U19AI100625 to V.D.M.). The research was also supported by a STARs award provided by the University of Texas System to V.D.M. and trainee funding provided by the Graduate School of Biomedical Sciences at UTMB.
REFERENCES
- 1.Wikan N, Smith DR. 2016. Zika virus: history of a newly emerging arbovirus. Lancet Infect Dis 16:e119–e126. doi: 10.1016/S1473-3099(16)30010-X. [DOI] [PubMed] [Google Scholar]
- 2.Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney M-C, Lavenir R, Pardigon N, Reynes J-M, Pettinelli F, Biscornet L, Diancourt L, Michel S, Duquerroy S, Guigon G, Frenkiel M-P, Bréhin A-C, Cubito N, Desprès P, Kunst F, Rey FA, Zeller H, Brisse S. 2006. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 3:e263. doi: 10.1371/journal.pmed.0030263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans AB, Peterson KE. 2019. Throw out the map: neuropathogenesis of the globally expanding California serogroup of orthobunyaviruses. Viruses 11:794. doi: 10.3390/v11090794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westby KM, Fritzen C, Paulsen D, Poindexter S, Moncayo AC. 2015. La Crosse encephalitis virus infection in field-collected Aedes albopictus, Aedes japonicus, and Aedes triseriatus in Tennessee. J Am Mosq Control Assoc 31:233–241. doi: 10.2987/moco-31-03-233-241.1. [DOI] [PubMed] [Google Scholar]
- 5.Jackson BT, Brewster CC, Paulson SL. 2012. La Crosse virus infection alters blood feeding behavior in Aedes triseriatus and Aedes albopictus (Diptera: Culicidae). J Med Entomol 49:1424–1429. doi: 10.1603/ME12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bewick S, Agusto F, Calabrese JM, Muturi EJ, Fagan WF. 2016. Epidemiology of La Crosse virus emergence, Appalachia region, United States. Emerg Infect Dis 22:1921–1929. doi: 10.3201/eid2211.160308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC. 2019. La Crosse encephalitis: epidemiology & geographic distribution. https://www.cdc.gov/lac/tech/epi.html.
- 8.Curren EJ, Lehman J, Kolsin J, Walker WL, Martin SW, Staples JE, Hills SL, Gould CV, Rabe IB, Fischer M, Lindsey NP. 2018. West Nile virus and other nationally notifiable arboviral diseases: United States, 2017. MMWR Morb Mortal Wkly Rep 67:1137–1142. doi: 10.15585/mmwr.mm6741a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caplen H, Peters CJ, Bishop DH. 1985. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J Gen Virol 66:2271–2277. doi: 10.1099/0022-1317-66-10-2271. [DOI] [PubMed] [Google Scholar]
- 10.Faburay B, LaBeaud AD, McVey DS, Wilson WC, Richt JA. 2017. Current status of Rift Valley fever vaccine development. Vaccines 5:29. doi: 10.3390/vaccines5030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikegami T. 2017. Rift Valley fever vaccines: an overview of the safety and efficacy of the live-attenuated MP-12 vaccine candidate. Expert Rev Vaccines 16:601–611. doi: 10.1080/14760584.2017.1321482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varghese FS, Kaukinen P, Gläsker S, Bespalov M, Hanski L, Wennerberg K, Kümmerer BM, Ahola T. 2016. Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antiviral Res 126:117–124. doi: 10.1016/j.antiviral.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 13.Micewicz ED, Khachatoorian R, French SW, Ruchala P. 2018. Identification of novel small-molecule inhibitors of Zika virus infection. Bioorg Med Chem Lett 28:452–458. doi: 10.1016/j.bmcl.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 14.Dong S, Kang S, Dimopoulos G. 2019. Identification of anti-flaviviral drugs with mosquitocidal and anti-Zika virus activity in Aedes aegypti. PLoS Negl Trop Dis 13:e0007681. doi: 10.1371/journal.pntd.0007681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrows NJ, Campos RK, Powell ST, Prasanth KR, Schott-Lerner G, Soto-Acosta R, Galarza-Muñoz G, McGrath EL, Urrabaz-Garza R, Gao J, Wu P, Menon R, Saade G, Fernandez-Salas I, Rossi SL, Vasilakis N, Routh A, Bradrick SS, Garcia-Blanco MA. 2016. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe 20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang Y, Li Y, Jasperson D, Henningson J, Lee J, Ma J, Li Y, Duff M, Liu H, Bai D, McVey S, Richt JA, Ikegami T, Wilson WC, Ma W. 2019. Identification and evaluation of antivirals for Rift Valley fever virus. Vet Microbiol 230:110–116. doi: 10.1016/j.vetmic.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 17.Lou Z, Sun Y, Rao Z. 2014. Current progress in antiviral strategies. Trends Pharmacol Sci 35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Serradilla M, Risco C, Pacheco B. 2019. Drug repurposing for new, efficient, broad spectrum antivirals. Virus Res 264:22–31. doi: 10.1016/j.virusres.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercorelli B, Palù G, Loregian A. 2018. Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol 26:865–876. doi: 10.1016/j.tim.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hover S, Foster B, Fontana J, Kohl A, Goldstein SAN, Barr JN, Mankouri J. 2018. Bunyavirus requirement for endosomal K+ reveals new roles of cellular ion channels during infection. PLoS Pathog 14:e1006845. doi: 10.1371/journal.ppat.1006845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Punch EK, Hover S, Blest HTW, Fuller J, Hewson R, Fontana J, Mankouri J, Barr JN. 2018. Potassium is a trigger for conformational change in the fusion spike of an enveloped RNA virus. J Biol Chem 293:9937–9944. doi: 10.1074/jbc.RA118.002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlton FW, Hover S, Fuller J, Hewson R, Fontana J, Barr JN, Mankouri J. 2019. Cellular cholesterol abundance regulates potassium accumulation within endosomes and is an important determinant in Bunyavirus entry. J Biol Chem 294:7335–7347. doi: 10.1074/jbc.RA119.007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson BA, Hage A, Kalveram B, Mears M, Plante JA, Rodriguez SE, Ding Z, Luo X, Bente D, Bradrick SS, Freiberg AN, Popov V, Rajsbaum R, Rossi S, Russell WK, Menachery VD. 2019. Peptidoglycan-associated cyclic lipopeptide disrupts viral infectivity. J Virol 93:e01282-19. doi: 10.1128/JVI.01282-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frugulhetti ICI, Rebello MA. 1989. Na+ and K+ Concentration and Regulation of Protein Synthesis in L-A9 and Aedes albopictus Cells Infected with Marituba Virus (Bunyaviridae). J Gen Virol 70:3493–3499. doi: 10.1099/0022-1317-70-12-3493. [DOI] [PubMed] [Google Scholar]
- 25.Lukacs GL, Chang XB, Kartner N, Rotstein OD, Riordan JR, Grinstein S. 1992. The cystic fibrosis transmembrane regulator is present and functional in endosomes. Role as a determinant of endosomal pH. J Biol Chem 267:14568–14572. [PubMed] [Google Scholar]
- 26.Hammond TG, Goda FO, Navar GL, Campbell WC, Majewski RR, Galvan DL, Pontillon F, Kaysen JH, Goodwin TJ, Paddock SW, Verroust PJ. 1998. Membrane potential mediates H(+)-ATPase dependence of “degradative pathway” endosomal fusion. J Membr Biol 162:157–167. doi: 10.1007/s002329900353. [DOI] [PubMed] [Google Scholar]
- 27.Matter AM, Hoot SB, Anderson PD, Neves SS, Cheng Y-Q. 2009. Valinomycin biosynthetic gene cluster in Streptomyces: conservation, ecology and evolution. PLoS One 4:e7194. doi: 10.1371/journal.pone.0007194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu C-Y, Jan J-T, Ma S-H, Kuo C-J, Juan H-F, Cheng Y-S, Hsu H-H, Huang H-C, Wu D, Brik A, Liang F-S, Liu R-S, Fang J-M, Chen S-T, Liang P-H, Wong C-H. 2004. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc Natl Acad Sci U S A 101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bermejo M, López-Huertas MR, García-Pérez J, Climent N, Descours B, Ambrosioni J, Mateos E, Rodríguez-Mora S, Rus-Bercial L, Benkirane M, Miró JM, Plana M, Alcamí J, Coiras M. 2016. Dasatinib inhibits HIV-1 replication through the interference of SAMHD1 phosphorylation in CD4+ T cells. Biochem Pharmacol 106:30–45. doi: 10.1016/j.bcp.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 30.de Wispelaere M, LaCroix AJ, Yang PL. 2013. The small molecules AZD0530 and dasatinib inhibit Dengue virus RNA replication via Fyn kinase. J Virol 87:7367–7381. doi: 10.1128/JVI.00632-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hafuri Y, Takemori E, Oogose K, Inouye Y, Nakamura S, Kitahara Y, Nakahara S, Kubo A. 1988. Mechanism of inhibition of reverse transcriptase by quinone antibiotics. II. Dependence on putative quinone pocket on the enzyme molecule. J Antibiot (Tokyo) 41:1471–1478. doi: 10.7164/antibiotics.41.1471. [DOI] [PubMed] [Google Scholar]
- 32.Jung E, Lee J-Y, Kim HJ, Ryu C-K, Lee K-I, Kim M, Lee C-K, Go YY. 2018. Identification of quinone analogues as potential inhibitors of picornavirus 3C protease in vitro. Bioorg Med Chem Lett 28:2533–2538. doi: 10.1016/j.bmcl.2018.05.046. [DOI] [PubMed] [Google Scholar]
- 33.Ikegami T, Won S, Peters CJ, Makino S. 2006. Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J Virol 80:2933–2940. doi: 10.1128/JVI.80.6.2933-2940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Promsong A, Chuenchitra T, Saipin K, Tewtrakul S, Panichayupakaranant P, Satthakarn S, Nittayananta W. 2018. Ellagic acid inhibits HIV-1 infection in vitro: potential role as a novel microbicide. Oral Dis 24:249–252. doi: 10.1111/odi.12835. [DOI] [PubMed] [Google Scholar]
- 35.Park SW, Kwon MJ, Yoo JY, Choi H-J, Ahn Y-J. 2014. Antiviral activity and possible mode of action of ellagic acid identified in Lagerstroemia speciosa leaves toward human rhinoviruses. BMC Complement Altern Med 14:171. doi: 10.1186/1472-6882-14-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin MS, Kang EH, Lee YI. 2005. A flavonoid from medicinal plants blocks hepatitis B virus-e antigen secretion in HBV-infected hepatocytes. Antiviral Res 67:163–168. doi: 10.1016/j.antiviral.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Sacau EP, Estévez-Braun A, Ravelo AG, Ferro EA, Tokuda H, Mukainaka T, Nishino H. 2003. Inhibitory effects of lapachol derivatives on Epstein-Barr virus activation. Bioorg Med Chem 11:483–488. doi: 10.1016/s0968-0896(02)00542-4. [DOI] [PubMed] [Google Scholar]
- 38.do Carmo Lagrota MH, Wigg MD, Aguiar AN, Pinto AV, Pinto Mdo C. 1986. Antiviral activity of naphthoquinones. I. Lapachol derivatives against enteroviruses. Rev Latinoam Microbiol 28:221–225. [PubMed] [Google Scholar]
- 39.Berka U, Khan A, Blaas D, Fuchs R. 2009. Human rhinovirus type 2 uncoating at the plasma membrane is not affected by a pH gradient but is affected by the membrane potential. J Virol 83:3778–3787. doi: 10.1128/JVI.01739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norris MJ, Malhi M, Duan W, Ouyang H, Granados A, Cen Y, Tseng Y-C, Gubbay J, Maynes J, Moraes TJ. 2018. Targeting intracellular ion homeostasis for the control of respiratory syncytial virus. Am J Respir Cell Mol Biol 59:733–744. doi: 10.1165/rcmb.2017-0345OC. [DOI] [PubMed] [Google Scholar]
- 41.Irurzun A, Carrasco L. 2001. Entry of poliovirus into cells is blocked by valinomycin and concanamycin A. Biochemistry 40:3589–3600. doi: 10.1021/bi002069p. [DOI] [PubMed] [Google Scholar]
- 42.Bedows E, Hatfield GM. 1982. An investigation of the antiviral activity of Podophyllum peltatum. J Nat Prod 45:725–729. doi: 10.1021/np50024a015. [DOI] [PubMed] [Google Scholar]
- 43.Biziagos E, Crance JM, Passagot J, Deloince R. 1987. Effect of antiviral substances on hepatitis A virus replication in vitro. J Med Virol 22:57–66. doi: 10.1002/jmv.1890220108. [DOI] [PubMed] [Google Scholar]
- 44.Min BS, Lee HK, Lee SM, Kim YH, Bae KH, Otake T, Nakamura N, Hattori M. 2002. Anti-human immunodeficiency virus-type 1 activity of constituents from Juglans mandshurica. Arch Pharm Res 25:441–445. doi: 10.1007/bf02976598. [DOI] [PubMed] [Google Scholar]
- 45.Fesen MR, Kohn KW, Leteurtre F, Pommier Y. 1993. Inhibitors of human immunodeficiency virus integrase. Proc Natl Acad Sci U S A 90:2399–2403. doi: 10.1073/pnas.90.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng L, Dai P, Ciro A, Smee DF, Djaballah H, Shuman S. 2007. Identification of novel antipoxviral agents: mitoxantrone inhibits vaccinia virus replication by blocking virion assembly. J Virol 81:13392–13402. doi: 10.1128/JVI.00770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furuta A, Tsubuki M, Endoh M, Miyamoto T, Tanaka J, Salam KA, Akimitsu N, Tani H, Yamashita A, Moriishi K, Nakakoshi M, Sekiguchi Y, Tsuneda S, Noda N. 2015. Identification of hydroxyanthraquinones as novel inhibitors of hepatitis C virus NS3 helicase. Int J Mol Sci 16:18439–18453. doi: 10.3390/ijms160818439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Q, Hou J, Yang P, Yan J, Yu X, Zhuo Y, He S, Xu F. 2019. Antiviral activity of mitoxantrone dihydrochloride against human herpes simplex virus mediated by suppression of the viral immediate early genes. BMC Microbiol 19:274. doi: 10.1186/s12866-019-1639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Möse JR, Stünzner D, Zirm M, Egger-Büssing C, Schmalzl F. 1980. Effect of aristolochic acid on herpes simplex infection of the rabbit eye. Arzneimittelforschung 30:1571–1573. (In German.) [PubMed] [Google Scholar]