Polymyxins are increasingly used as the critical last-resort therapeutic options for multidrug-resistant Gram-negative bacteria. Unfortunately, polymyxin resistance has increased gradually over the past few years. Although studies on polymyxin mechanisms are expanding, systemwide analyses of the underlying mechanism for polymyxin resistance and stress response are still lacking. To understand how Klebsiella pneumoniae adapts to colistin (polymyxin E) pressure, we carried out proteomic analysis of a K. pneumoniae strain cultured with different concentrations of colistin.
KEYWORDS: polymyxin resistance, crrB mutation, Klebsiella pneumoniae, lipid A modification, proteomics, β-lactamase, multidrug efflux pump KexD
ABSTRACT
Polymyxins are increasingly used as the critical last-resort therapeutic options for multidrug-resistant Gram-negative bacteria. Unfortunately, polymyxin resistance has increased gradually over the past few years. Although studies on polymyxin mechanisms are expanding, systemwide analyses of the underlying mechanism for polymyxin resistance and stress response are still lacking. To understand how Klebsiella pneumoniae adapts to colistin (polymyxin E) pressure, we carried out proteomic analysis of a K. pneumoniae strain cultured with different concentrations of colistin. Our results showed that the proteomic responses to colistin treatment in K. pneumoniae involve several pathways, including (i) gluconeogenesis and the tricarboxylic acid (TCA) cycle, (ii) arginine biosynthesis, (iii) porphyrin and chlorophyll metabolism, and (iv) enterobactin biosynthesis. Interestingly, decreased abundances of class A β-lactamases, including TEM, SHV-11, and SHV-4, were observed in cells treated with colistin. Moreover, we present comprehensive proteome atlases of paired polymyxin-susceptible and -resistant K. pneumoniae strains. The polymyxin-resistant strain Ci, a mutant of K. pneumoniae ATCC BAA 2146, showed a missense mutation in crrB. This crrB mutant, which displayed lipid A modification with 4-amino-4-deoxy-l-arabinose (l-Ara4N) and palmitoylation, showed striking increases in the expression of CrrAB, PmrAB, PhoPQ, ArnBCADT, and PagP. We hypothesize that crrB mutations induce elevated expression of the arnBCADTEF operon and pagP via PmrAB and PhoPQ. Moreover, the multidrug efflux pump KexD, which was induced by crrB mutation, also contributed to colistin resistance. Overall, our results demonstrated proteomic responses to colistin treatment and the mechanism of CrrB-mediated colistin resistance, which may offer valuable information on the management of polymyxin resistance.
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) pose urgent threats to human health because they are resistant to nearly all antibiotics (1, 2). As a last resort for infections caused by multidrug-resistant bacteria, polymyxins (i.e., polymyxin B and colistin) are increasingly used as the therapeutic options in the clinic (3, 4). Unfortunately, polymyxin resistance has increased gradually with the growing and suboptimal use of polymyxins (5–7). In particular, the emergence of mcr-1, the first plasmid-mediated polymyxin resistance gene, indicates that polymyxin resistance can readily spread from one strain to another via horizontal gene transfer (8). Thus, there is an urgent need for increased knowledge to aid in the development of effective therapeutic methodologies for infections caused by these “superbugs.” Proteomics is a valuable technique for revealing unknown functional relationships. Comparative proteomic analysis of Klebsiella pneumoniae strains responding to colistin treatment may provide insight into the impact of polymyxin on bacteria, thus leading to the discovery of enhanced therapeutic strategies.
Previous studies have revealed that polymyxin resistance is mediated mainly by mechanisms such as lipid A modification and multidrug efflux pumps (9, 10). As a lipid component of lipopolysaccharide (LPS), lipid A is an important polymyxin-binding target. When lipid A is modified with 4-amino-4-deoxy-l-arabinose (l-Ara4N) by arnBCADTEF operon gene products, the interaction between polymyxin and lipid A is affected, resulting in polymyxin resistance. Mutations in two-component regulatory systems (TCRs), mainly PhoPQ and PmrAB, can trigger constitutive expression of the arnBCADTEF operon, leading to polymyxin resistance (11). Moreover, lipid A can also be modified by ept gene products, which are responsible for phosphoethanolamine (PEtN) addition (12). Recently, amino acid substitutions of crrB have been reported to be responsible for colistin resistance in K. pneumoniae (13, 14). Multidrug efflux pumps are also important mechanisms of polymyxin resistance. It has been reported that pumps such as Sap (sensitive antimicrobial peptides), the AcrAB-TolC complex, and KpnEF can confer tolerance to polymyxins (15–17). While all the mechanisms of resistance mentioned above focus on transcriptional regulatory networks, some studies have demonstrated that metabolic changes might contribute to decreased antibiotic susceptibility (18–20). For example, the ppGpp biochemical network has been found to support bacterial survival of antibiotic exposure (21). Variations in ATP levels have also been demonstrated to cause persister formation by decreasing the activity of antibiotic targets (22). For such reasons, metabolic changes have been considered to induce a protective state in bacteria by reducing the production of cytotoxic metabolic by-products (23).
However, the changes in protein expression associated with polymyxin treatment are poorly understood. To elucidate this process in a K. pneumoniae strain treated with polymyxin, we profiled proteome alterations in K. pneumoniae in the presence of different concentrations of colistin (polymyxin E). Meanwhile, we also investigated proteome changes between paired polymyxin-susceptible and -resistant strain in order to unravel the potential mechanisms associated with polymyxin resistance.
RESULTS
Genomic analysis.
The genomes of the wild-type (wt) K. pneumoniae strain ATCC BAA 2146 (colistin MIC, 2 μg/ml) and its paired mutant Ci (colistin MIC, >1,024 μg/ml) were sequenced and assembled. By comparison to the wild type, a ccrB missense mutation (locus, kpn2146_RS15700, encoding the HAMP domain-containing histidine kinase CrrB) and a silent satP mutation (locus, kpn2146_RS03945, encoding a succinate-acetate/proton symporter) were identified in Ci (see Table S1 in the supplemental material). The results showed that the mutant strain Ci encoded CrrB harboring a substitution of serine for proline at codon 151 (P151S). Previous studies have indicated that crrB substitutions contribute to colistin resistance (14, 24). Since the satP mutation does not alter the amino acid sequence, this mutation is considered nonfunctional in polymyxin resistance.
Deep proteomes of Klebsiella pneumoniae.
In order to explore the bacterial response to colistin, the proteomes of K. pneumoniae ATCC BAA 2146 cultured with different concentrations of colistin (low concentration, 0.5 μg/ml; high concentration, 4 μg/ml) were profiled. Additionally, K. pneumoniae ATCC BAA 2146 and its paired crrB mutant Ci were subjected to quantitative proteomic analysis for exploration of the underlying molecular mechanisms of polymyxin resistance (the colistin MICs and minimal bactericidal concentrations [MBCs] are shown in Table S3). In total, there were four groups in our experimental design (Fig. 1A), and 8-plex iTRAQ labeling was employed for protein quantification. To evaluate data reproducibility, we performed duplicate technical experiments from cell disruption to liquid chromatography–tandem mass spectrometry (LC–MS-MS) measurements for three biological replicates. Deep proteome measurements identified 3,174 proteins in all, with a false-discovery rate (FDR) of <1% for protein and peptide identification. Eventually, 2,851 proteins identified from at least two of the three replicates were collected for further bioinformatics analysis (Fig. 1B; Data Set S1). Principal-component analysis (PCA) indicated that different treatments showed clear separation with distinct features, while samples from the same group tend to be clustered together, suggesting that the differences among the three biological replicates were acceptable (Fig. 1C).
FIG 1.
Proteomic landscape of K. pneumoniae. (A) Experimental design and work flow of iTRAQ-based proteomics analysis in the study. (B) Venn diagram of proteins identified in three replicates (rep). (C) PCA of total proteome data from biological and technical replicates of the four different antibiotic treatments.
Proteomic response to colistin.
We profiled the proteome alterations of K. pneumoniae ATCC BAA 2146 in the presence of different concentrations (0.5 μg/ml, 4 μg/ml) of colistin, which may offer an opportunity to find the key biological pathways affected by colistin. Differentially expressed proteins were divided into four clusters according to dynamic changes of proteins at increased concentrations of colistin (Data Set S2). Proteins whose expression increased or declined with increasing colistin concentrations were assigned to cluster 1 or cluster 2, respectively. Proteins whose expression remained constant at the low concentration of colistin, but increased or decreased at the high concentration of colistin, were assigned to cluster 3 or cluster 4, respectively (Fig. 2A). In order to explore the significant pathways in cells treated with the low and high concentrations of colistin, the proteins in clusters 1 and 2 were combined into phase 1, which represented proteins that continually changed with increasing concentrations. Similarly, clusters 3 and 4 were incorporated into phase 2, which represented specific proteins differentially expressed at the high concentration.
FIG 2.
Functional analysis of cells treated with different concentrations of colistin. (A) In order to find out the pathways that changed dynamically with treatment with different concentrations of colistin, proteins that were differentially expressed in the colistin treatment groups relative to the wild type were divided into four clusters: proteins whose expression increased with increasing colistin concentrations (cluster 1) (red), proteins whose expression decreased with increasing colistin concentrations (cluster 2) (blue), proteins that were upregulated only with a high concentration of colistin (cluster 3) (orange), and proteins that were downregulated only with a high concentration of colistin (cluster 4) (purple). (B) Effects of colistin on the β-lactamases TEM, SHV-11, SHV-4, and NDM-1. (C and D) GO and KEGG analyses. In order to explore the significant pathways in cells treated with the low and high concentrations of colistin, proteins in clusters 1 and 2 were combined as phase 1, which represented proteins that continually changed with increasing colistin concentrations. Similarly, clusters 3 and 4 were incorporated into phase 2, which represented specific proteins differentially expressed at the high concentration.
Gene Ontology (GO) enrichment analysis and KEGG pathway analysis were performed to identify the representative biological significance for each phase. For phase 1, GO enrichment analysis showed that differentially expressed proteins were enriched in the process of translation (shown in red in Fig. 2C). KEGG pathway analysis revealed that proteins in phase 1 were associated with the metabolism and biosynthesis of amino acids, ABC transporters, and ribosomes (shown in red in Fig. 2D). For phase 2, GO enrichment analysis revealed that biosynthetic and metabolic processes of multiple metabolites, including heterocycles, nitrogen compounds, aromatic compounds, biogenic amines, and cofactors, were changed. Aerobic respiration and the tricarboxylic acid (TCA) cycle were also involved (shown in green in Fig. 2C). Proteins in phase 2 were characterized by pathways including the TCA cycle, porphyrin and chlorophyll metabolism, and propanoate metabolism (shown in green in Fig. 2D).
The principal pathways that responded to colistin treatment are shown in detail in Fig. 3 and Data Set S4. For cells treated with 0.5 μg/ml colistin, relative to untreated cells, the expression of maltose transporters was sharply downregulated (LamB, 0.33-fold; MalK, 0.37-fold), while arginine biosynthesis was moderately upregulated (ArgA, 1.36-fold; ArgB, 1.39-fold). When the concentration of colistin rose to 4 μg/ml, more changes appeared in maltose transporters (LamB, 0.21-fold; MalK, 0.34-fold) and arginine biosynthesis (ArgA, 1.96-fold; ArgB, 2.01-fold). Within the central metabolism, most of the proteins in the TCA cycle were highly expressed with colistin treatment. However, the fluxes flowing to gluconeogenesis seemed to be upregulated due to increased phosphate acetyltransferase (MaeB, 1.51-fold) and phosphoenolpyruvate carboxykinase (PckA, 1.33-fold) expression. The key gluconeogenesis enzymes, such as phosphofructokinase (PfkB, 3.36-fold) and fructose-bisphosphate aldolase (FbaB, 3.78-fold) were observed to be highly expressed. Moreover, porphyrin and chlorophyll metabolism, the process from precorrin-2 to cobyrinic acid a,c-diamide in particular, was likely repressed, since the expression of all proteins in this process was found to be downregulated. Increased biosynthesis of enterobactin, which has been reported to protect cells against oxidative stress (25–27), was also observed. Besides, we observed increased expression of thioredoxin/glutathione peroxidase (BtuE, 1.64-fold) and catalase (KatE, 3.6-fold), which can remove reactive oxygen species (ROS) to protect cells (28).
FIG 3.
Perturbations of biochemical pathways for K. pneumoniae ATCC BAA 2146 exposed to colistin, including gluconeogenesis (blue), the TCA cycle (purple), arginine biosynthesis (pink), enterobactin biosynthesis (yellow), porphyrin and chlorophyll metabolism (green), and ROS removal (orange). Notably, porphyrin and chlorophyll metabolism represents the biosynthesis of vitamin B12 but not chlorophyll. The colors of proteins correspond to their clusters (cluster 1, red; cluster 2, blue; cluster 3, orange; cluster 4, purple; unidentified, gray). The regulation of pathway reactions is presented as upregulation (green vector), downregulation (red segment), or no change (black vector). Data are shown in Data Set S4 in the supplemental material.
Interestingly, the expression of class A β-lactamases, including TEM, SHV-11, and SHV-4, was significantly downregulated in the presence of colistin (Fig. 2B). However, NDM-1 stayed the same in cells exposed to colistin. Meropenem-ceftazidime MICs were evaluated with and without colistin in order to explore the effect of colistin on β-lactamase. Unfortunately, the addition of colistin did not enhance the susceptibility of K. pneumoniae ATCC BAA 2146 to meropenem-ceftazidime (data not shown).
Comparative proteomics for polymyxin-resistant and -susceptible strains.
Comparative proteomic analysis of paired polymyxin-susceptible (K. pneumoniae ATCC BAA 2146) and -resistant (Ci, a crrB mutant of K. pneumoniae ATCC BAA 2146) strains was carried out to find the potential mechanism of crrB-mediated resistance to colistin in K. pneumoniae. Volcano plot analysis revealed that 301 proteins were differentially expressed in the mutant relative to the wild type (Fig. 4A; Data Set S3). KEGG pathway analysis found that the differentially expressed proteins were enriched in two-component systems (TCSs) and the porphyrin and chlorophyll metabolism pathways (Fig. 4B).
FIG 4.
Perturbations of the crrB mutant Ci relative to the wild type. (A) Volcano plot for screening of differentially expressed proteins. (B) KEGG enrichment analysis of the proteins differentially expressed in the crrB mutant Ci relative to the wild type. The gene ratio is the ratio of the differentially expressed proteins identified in a pathway to all proteins in the same pathway. (C) Expression of the two-component systems CrrAB, PmrAB, and PhoPQ was elevated in the crrB mutant Ci. (D) Proteins associated with lipid A modification were significantly upregulated in the crrB mutant strain Ci. (E) Expression of KexD, an RND efflux pump, and KPN_02063, a type of acetyltransferase, was markedly increased in the colistin-resistant mutant. Dashed lines mark the fold change threshold of 1.3. Data are shown in Table S2 in the supplemental material.
TCSs, which typically consist of a sensor kinase and a response regulator, are involved in the regulation of gene expression in response to environmental signals, such as antibiotic exposure (29). In the current study, TCSs, including CrrAB, PhoPQ, and PmrAB, were more highly expressed in the crrB mutant Ci than in the wild type (Fig. 4C). Increased abundances of ArnBCADT (>2.8-fold) and PagP (1.92-fold), which are responsible for LPS modification, were observed in the crrB mutant Ci (Fig. 4D). CrrB mutations have been reported to induce CrrC expression, thereby inducing elevated expression of the arnBCADTEF operon via PmrAB (14, 30). However, our results showed that PhoPQ and PagP expression was also elevated in the crrB mutant. Therefore, we hypothesize that the crrB mutant (P151S) may regulate the expression of the arnBCADTEF operon and pagP through PmrAB and PhoPQ.
Notably, the multidrug efflux pump KexD (KPN_02064, 6.76-fold) and a putative acetyltransferase (KPN_02063, 5.40-fold) were found to be the maximally increased proteins in the mutant, while neither was differentially expressed in cells treated with colistin (Fig. 4E). In agreement with our proteomic data, real-time PCR results also showed that the KPN_02063 and KexD genes were upregulated at the transcriptional level (Fig. S1). A previous study reported that a putative resistance-nodulation-division (RND)-type efflux pump, H239_3064, contributes to colistin resistance through CrrB (31). We found that the protein encoded by H239_3064 shared 100% amino acid identity with KexD. Accordingly, crrB mutation may induce KexD transcription, resulting in colistin resistance.
In addition, the expression of 14 proteins involved in porphyrin and chlorophyll metabolism, mainly enriched in vitamin B12 biosynthetic pathways from precorrin-2 to cobyrinic acid a,c-diamide, was found to be lower in the polymyxin-resistant mutant than in the wild type (32). Besides, the expression of maltose transporter proteins, including MalE (0.27-fold), MalF (0.36-fold), MalM (0.32-fold), MalK (0.25-fold), and LamB (0.27-fold), was significantly decreased in the polymyxin-resistant mutant, and a similar phenomenon was observed in K. pneumoniae exposed to doxycycline (20).
Validation of lipid A modification in the crrB mutant Ci.
The proteomic results reported above indicate that the lipid A of the mutant strain Ci is likely to be modified by ArnBCADT and PagP. To confirm this, the lipid A species of the wild type and its paired mutant Ci were characterized by mass spectrometry. Two ions, at m/z 919.6201 and m/z 1,840.2497, corresponding to the doubly deprotonated and singly deprotonated biphosphorylated lipid A molecule with hydroxylation on the C′-2 fatty acyl chain, were found to be dominant in the wild type, in agreement with the findings of previous studies (Fig. 5A) (33, 34). Also of note is m/z 1,760.2826, which represents lipid A containing a monophosphate group (annotated in orange in Fig. 5B). The loss of the phosphate moiety could be due to the nature of the extraction method (34). The mutant strain Ci predominantly produced lipid A species with one (m/z 919.6160, 985.1449, 1,840.2398, 1,971.2985) or two (m/z 919.6160, 1,050.6742, 1,840.2398, 2,102.3574) l-Ara4N modifications (Δm/z, 131.05 or 262.10, respectively) (annotated in red in Fig. 5D). In addition, lipid A modified with two l-Ara4N molecules and palmitoylation (m/z 1,169.7984) (shown in blue in Fig. 5D) was also observed in the polymyxin-resistant mutant. All the lipid A structures presented in the wt and the mutant strain Ci are shown in Fig. S2.
FIG 5.
MS characterization of lipid A profiles in the wild type and the crrB mutant Ci. (A and C) MS spectra of lipid A in the wild type (A) and in the crrB mutant Ci (C). (B and D) Proposed structures of lipid A in the wild type (B) and in the crrB mutant Ci (D). Orange indicates the loss of a phosphate moiety. Palmitoylation addition and l-Ara4N are indicated in blue and red, respectively.
The roles of the multidrug efflux pump KexD and the putative acetyltransferase KPN_02063 in polymyxin resistance.
As mentioned above in the presentation of the proteomic data, the expression of the multidrug efflux pump KexD (KPN_02064) and the putative acetyltransferase KPN_02063 was strongly upregulated in the crrB mutant Ci relative to the wild type. To investigate the role that the multidrug efflux pump played in polymyxin resistance, we measured the MIC of polymyxin against the mutant with the addition of the efflux pump inhibitor phenylalanine arginine β-naphthylamide (PAβN) (35, 36). The MIC of polymyxin against the mutant decreased from >1,024 μg/ml to 256 μg/ml in the presence of PAβN, suggesting that KexD might play an important role in polymyxin resistance (Table S3). To investigate whether KPN_02063 contributes to colistin resistance, KPN_02063 was knocked out in the mutant strain Ci. However, the Ci mutant without KPN_02063 did not show altered susceptibility to colistin, indicating that KPN_02063 has no link to colistin resistance (Table S3).
DISCUSSION
The current study shows the changes of pathways for strains treated with different concentrations of colistin and presents a comprehensive investigation of the proteomes of polymyxin-resistant and polymyxin-susceptible strains of K. pneumoniae, which should be useful for dissecting the mechanisms of antimicrobial resistance.
We present the main pathways that changed in cells treated with colistin. It has been found that bacterial metabolism plays a vital role in mediating the cellular response to antibiotic treatment (20). With regard to amino acid biosynthesis, arginine biosynthesis flux seemed to be upregulated after colistin treatment, since increased levels of the arginine-biosynthetic enzymes ArgABCDE, ArgI, ArgG, and ArgH were observed in cells treated with colistin (Fig. 3). This result is consistent with previous observations that Acinetobacter baumannii treated with colistin displayed increased arginine biosynthesis flux (37). A previous study reported that the ArgJ-mediated arginine biosynthesis pathway was critical for persister formation in Staphylococcus aureus after gentamicin treatment (38). The arginine deiminase (ADI) pathway catabolizes arginine to form ornithine, with carbon dioxide, ammonia, and ATP as by-products. Ammonia could further produce NH4+ and thus neutralize the cytoplasmic pH, thereby protecting the bacteria from the potentially lethal effects of acidic conditions (39, 40). Overall, K. pneumoniae may enhance arginine biosynthesis to mitigate damage from hydroxyl radicals via ammonia production (37, 41).
Expression of the maltose transporter LamB, which also serves as a porin involved in the influx of antibiotics, was decreased in cells treated with colistin. Negative regulation of LamB is a general response in resistance to different classes of antibiotics. Attenuation of LamB hampers the uptake of maltose, glucose, and antibiotics, resulting in decreased intracellular concentrations of sugar and antibiotics (42, 43). Gluconeogenesis seems to be upregulated due to increased levels of phosphofructokinase and fructose-bisphosphate aldolase, which are responsible for the rate-limiting step of gluconeogenesis. Increased gluconeogenesis may result from a lack of sugar substrates in the cytoplasm.
Within the central metabolism, all the proteins of the TCA cycle were highly expressed in cells treated with a high concentration of colistin. These results accord with earlier observations, which showed that most of the genes in the TCA cycle were upregulated at the transcriptional level, while the fluxes and metabolites through the TCA cycle declined (37). A previous study discovered that increased energy production, such as that from the TCA cycle, could be necessary for cell survival in the presence of ampicillin (44). Our observation indicates that K. pneumoniae may also regulate the TCA cycle in response to colistin treatment.
Moreover, porphyrin and chlorophyll metabolism, which represents the biosynthesis of vitamin B12 but not chlorophyll, was likely downregulated in cells treated with the high concentration of colistin. Similar results were observed for the polymyxin-resistant mutant relative to the wild type. Since the genes encoding the downregulated proteins CbiL, CbiH, CbiF, CbiG, CbiD, CbiJ, CbiC, and CbiA are in the same operon, it seems that the operon was repressed (45). However, it is difficult to explain the mechanism behind the phenomenon with limited information about the pathway.
Surprisingly, we discovered that the expression of class A β-lactamases, including TEM, SHV-11, and SHV-4, decreased in the presence of colistin (Fig. 2B). However, the meropenem-ceftazidime MIC for K. pneumoniae ATCC BAA 2146 showed no difference with or without colistin, perhaps due to the unaltered abundance of NDM-1. A previous study reported decreased expression levels of blaOXA-23 and blaADC-25 in A. baumannii MDR-ZJ06 with a subinhibitory concentration of tigecycline. Furthermore, the strain showed a lower ceftazidime MIC in the presence of tigecycline (30).
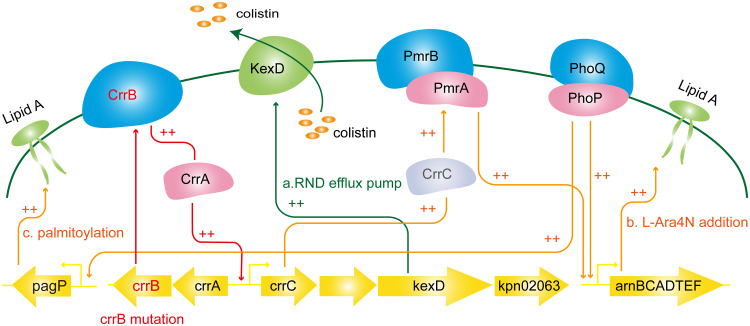
Comparative proteomic analysis of polymyxin-susceptible K. pneumoniae ATCC BAA 2146 and its paired crrB mutant Ci should provide useful information for the understanding of crrB-mediated colistin resistance (Fig. 6). The lipid A profiles of the crrB mutant Ci presented the addition of one or two l-Ara4N molecules and palmitoylation to the predominant lipid A species. Our proteomic results showed that CrrAB, PmrAB, and ArnBCADT levels were elevated in the crrB mutant. A previous study reported that a crrB missense mutation (N141I) induced CrrC expression, thereby inducing elevated expression of arnBCADTEF via PmrAB but not PhoPQ (14). However, our results showed that PhoPQ and PagP levels were also elevated in the crrB mutant (Fig. 4C and D). These results implied that the crrB mutant (P151S) not only regulated PmrAB but might also regulate PhoPQ, thereby inducing elevated expression of the arnBCADTEF operon and pagP (14, 31).
FIG 6.
Schematic diagram of the crrB-mediated colistin resistance mechanism. A crrB missense mutation induces the expression of CrrC and KexD through CrrA. Increased CrrC expression then induces PmrAB expression. The arnBCADTEF operon and pagP are activated by PmrAB and PhoPQ, resulting in increased resistance to colistin. Moreover, KexD, an RND efflux pump, also contributes to colistin resistance.
Moreover, the multidrug efflux pump KexD and the putative GNAT family N-acetyltransferase encoded by KPN_02063 were highly expressed in the crrB mutant. KexD is predicted to be an RND-type efflux pump. The colistin MIC for the crrB mutant decreased 4-fold in the presence of the efflux pump inhibitor PAβN, indicating that KexD contributed to colistin resistance. In contrast, KPN_02063 is irrelevant to colistin resistance, since the deletion of KPN_02063 in the crrB mutant Ci did not attenuate the colistin MIC. A crrB mutant has been reported to induce the expression of a putative RND-type efflux pump, H239_3064, through CrrA, contributing to colistin resistance (31). We found that H239_3064 shares 100% amino acid identity with the KexD pump. RND-type pumps commonly function as a complex with an inner membrane protein (RND), an outer membrane protein, and a periplasmic protein, but neither a periplasmic protein gene nor an outer membrane gene exists in the same operon with KexD. Previous functional studies of the multidrug efflux pump KexD reported that KexD could utilize TolC and AcrA from K. pneumoniae (31, 46).
In conclusion, our data highlight the significant pathways in K. pneumoniae associated with the response to colistin, including an activated TCA cycle, increased arginine biosynthesis, and decreased porphyrin and chlorophyll metabolism. crrB missense mutants display higher colistin resistance than strains harboring mutations in mgrB, pmrB, or phoQ (14). Our proteomic results suggest that the crrB mutant contributes to colistin resistance through LPS modification and the RND-type efflux pump KexD.
MATERIALS AND METHODS
Chemicals and reagents.
Colistin sulfate salt (C4461-1G) was purchased from Sigma-Aldrich. Lysyl endopeptidase (Lys-C; catalog no. 129-02541) was purchased from Wako. Trypsin (V511A) was purchased from Promega. All other reagents were from Sigma-Aldrich unless specified otherwise.
Bacterial strains and culture.
The K. pneumoniae ATCC BAA 2146 and Ci strains were provided by Xuefu You’s group (Institute of Medical Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China). The resistant strain Ci was a mutant of K. pneumoniae ATCC BAA 2146 induced by colistin. Strains were grown at 37°C in LB broth overnight. Cells were subcultured 1/100 into LB broth and were grown at 37°C with shaking until the optical density at 600 nm (OD600) reached 0.4 to 0.6.
Susceptibility testing process.
MICs were determined by the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (47). Briefly, the tests were performed in 96-well plates with concentrations from 0 to 1,024 mg/liter in cation-adjusted Mueller-Hinton broth (CaMHB). The MIC was defined as the lowest concentration that completely inhibited bacterial growth after a 16-h incubation at 37°C. To evaluate the effects of the efflux pump inhibitor on colistin resistance, the MIC of colistin in the presence of 25 μg/ml PAβN was determined.
DNA sequencing and genomic analysis.
Genomic DNA was isolated with TRIzol reagent (Thermo Fisher) according to the manufacturer’s protocol. Paired-end 150-bp DNA sequencing of the wt and Ci strains was performed on an Illumina HiSeq X Ten platform. The reads of Ci were aligned to the reference genome (GenBank assembly accession no. GCA_000364385.3) using the Burrows-Wheeler aligner. Gene variation was determined with the Genome Analysis Toolkit and was annotated with ANNOVAR.
Protein extraction and in-solution digestion.
Cell pellets were suspended in 800 μl lysis buffer (7 M urea, 2 M thiourea, 0.5% n-octyl-β-d-glucopyranoside, 1% 1,4-dithiothreitol [DTT], proteinase inhibitor cocktail) and were disrupted by a FastPrep (MP Biomedicals) instrument at a speed of 6.0 m/s twice for 40 s each time. The supernatants were collected by centrifugation at 1,000 rpm for 10 min, and the protein concentration was determined using the Bradford assay. Two hundred micrograms of each sample was alkylated with 45 mM iodoacetamide (IAM) for 30 min at room temperature in the dark. DTT (5 mM) was then added to react with the extra IAM. After that, samples were proteolyzed for 4 h at 37°C using sequencing-grade Lys-C at 1/200 (wt/wt). After dilution into 1.5 M urea, samples were further digested with trypsin (Promega) at a 1:50 enzyme/protein ratio at 37°C overnight. All peptide samples were desalted with self-packed reversed-phase columns.
Eight-plex iTRAQ labeling.
Peptides were labeled with the 8-plex iTRAQ reagent as shown in Fig. 1A. Labeling was performed according to the manufacturer’s instructions. After labeling, samples were combined and acidified, and the peptides were desalted with self-packed reversed-phase columns.
High-pH prefractionation.
iTRAQ-labeled peptides were fractionated on a Dionex UltiMate 3000 system equipped with an Acquity ultraperformance liquid chromatography (UPLC) M-class, CSH C18 reversed-phase analytical column (pore size, 130 Å; particle size, 1.7 μm; inside diameter [i.d.], 300 μm; length, 100 mm [Waters]). Mobile phase A contained 20 mM ammonium formate in water (pH 9.3), and mobile phase B was 80% acetonitrile (ACN)–20% solvent A (pH 9.3). Peptides were eluted with a multistep gradient system from 2% B to 40% B for 71 min, then to 50% B at 122 min, followed by a wash with 95% B for 10 min at a flow rate of 5 μl/min. In total, 20 fractions were collected.
LC-MS-MS analysis.
An Easy-nLC system coupled to a Q Exactive HF mass spectrometer was employed in the study. Each fraction was separated with a nano self-packed C18 trap column (i.d., 100 μm; length, 3 cm; particle size, 5 μm; ReproSil-Pur 120 C18-AQ; Dr. Maisch GmbH) and a nano self-packed C18 analytical column (i.d., 75 μm; length, 18 cm; particle size, 3 μm; Reprosil-Pur 120 C18-AQ; Dr. Maisch GmbH) using a multistep gradient as follows: 5% buffer B (95% ACN, 0.1% formic acid [FA]) increasing to 10% buffer B within 5 min, increasing to 34% B from min 5 to min 125, increasing to 50% buffer B from min 125 to min 135, and then to 100% buffer B at 140 min, followed by a wash with 100% buffer B for 8 min at a flow rate of 250 nl/min. For MS1, 3E6 ions were accumulated in the Orbitrap cell over a maximum time of 100 ms and were scanned at a resolution of 120,000 full width at half maximum (FWHM) from 350 to 1,600 m/z. Each MS1 was followed by 15 MS-MS scans of the most intense precursor ions with a 15-s dynamic exclusion time. The MS-MS survey scan was acquired in the Orbitrap analyzer at 30,000 FWHM, and 1E5 ions were accumulated over a maximum time of 120 ms. The normalized collision energy was set to 34 eV.
Protein identification and quantification.
The individual raw MS data files were processed with the commercially available software Proteome Discoverer, v. 2.2.0.388 (Thermo Fisher Scientific). Database searches were carried out against Klebsiella pneumoniae in the Swiss-Prot database, containing 1,706 entries, and against KLEP7 in UniProt, containing 5,127 entries (download on 24 September 2018), using the Sequest search engine. The search parameters were as follows. (i) Precursor mass tolerance was set to 10 ppm, and fragment mass tolerance was set to 0.02 Da. (ii) Carbamidomethylation of cysteine and iTRAQ modification of the N terminus and lysine residues were added to fixed modifications, and oxidation of methionine (M) and acetylation were set to variable modifications. (iii) Two missed cleavage sites were allowed. For reporter ion quantification, the coisolation threshold was set to 50, and the average reporter ion signal-to-noise (S/N) threshold was set to 10.
Bioinformatics and statistical analysis.
Two-way analysis of variance (ANOVA) and Student’s t test were performed for the treatment groups (no colistin or colistin treatment at 0.5 mg/liter or 4 mg/liter) and resistance groups (wt, Ci), respectively. Proteins with an absolute log2 fold change above 0.379 and a P value below 0.05 were considered differentially expressed proteins. Gene Ontology (GO) enrichment was based on DAVID (the Database for Annotation, Visualization, and Integrated Discovery) bioinformatics resources (48). KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis was based on the KEGG database (49). All data analyses were conducted with R (v. 3.6.1).
RT-qPCR.
Total RNA was isolated with TRIzol reagent (Thermo Fisher) according to the manufacturer’s protocol. Two micrograms of RNA was used for reverse transcription (RT) to obtain cDNA with the Reverse Transcription System (Promega). Quantitative PCR (qPCR) was performed with SYBR green on an Applied Biosystems StepOnePlus real-time PCR system, and products were analyzed by the 2–ΔΔCT method. 16S rRNA was applied as an internal control. All the primers used are shown in Table S4 in the supplemental material.
Isolation and structural characterization of lipid A.
Lipid A extraction was performed as described previously with some modifications (50). Cell pellets were suspended in phosphate-buffered saline (PBS), and then chloroform and methanol were added to the tube for a single-phase Bligh-Dyer mixture (chloroform-methanol-water, 1:2:0.8 [vol/vol/vol]). After incubation at room temperature for 20 min, the pellets were collected, resuspended in hydrolysis buffer (50 mM sodium acetate [pH 4.5], 1% SDS), assisted by sonication at a constant duty cycle for 5 s at 25% output (5-s/burst, ∼5 s between bursts, 5 min in total), and then kept in a water bath at 95°C for 1 h. The SDS solution was converted into a two-phase Bligh-Dyer mixture by adding chloroform and methanol to form a chloroform-methanol-water (2:2:1.8, vol/vol/vol) mixture. The lower phases were collected and were washed twice with the upper phase of a preequilibrated two-phase Bligh-Dyer mixture (2:2:1.8, vol/vol/vol), followed by drying under nitrogen. The dried lipids were redissolved in chloroform-methanol (1:1) and were characterized with a TripleTOF 5600 system (AB Sciex) in negative-ion mode.
Data availability.
The raw genomic data of the K. pneumoniae ATCC BAA 2146 and Ci strains have been deposited to NCBI with BioProject accession number PRJNA612645.
Supplementary Material
ACKNOWLEDGMENTS
We thank Arek Nawrocki and Eva Christina Østerlund of the University of Southern Denmark for assistance with technique. We are also grateful to Tingting Zhang and Na Li of the Institute of Biophysics, Chinese Academy of Sciences, for sharing their wisdom with us during this research.
This work was supported by grants from the National Key R&D Program of China (grant 2018YFA0507801) and the National Natural Science Foundation of China (grant 91640112).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Centers for Disease Control and Prevention (CDC). 2013. Antibiotic resistance threats in the United States, 2013. https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf.
- 2.Perez F, Bonomo RA. 2019. Carbapenem-resistant Enterobacteriaceae: global action required. Lancet Infect Dis 19:561–562. doi: 10.1016/S1473-3099(19)30210-5. [DOI] [PubMed] [Google Scholar]
- 3.Falagas ME, Kasiakou SK, Saravolatz LD. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 4.Tsuji BT, Pogue JM, Zavascki AP, Paul M, Daikos GL, Forrest A, Giacobbe DR, Viscoli C, Giamarellou H, Karaiskos I, Kaye D, Mouton JW, Tam VH, Thamlikitkul V, Wunderink RG, Li J, Nation RL, Kaye KS. 2019. International Consensus Guidelines for the Optimal Use of the Polymyxins: endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacotherapy 39:10–39. doi: 10.1002/phar.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Chaisiri K, Komalamisra C, Adelowo OO, Fagade OE, Banjo OA, Oke AJ, Adler A, Assous MV, Morand S, Raoult D, Rolain JM. 2014. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents 44:500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 6.Parisi SG, Bartolini A, Santacatterina E, Castellani E, Ghirardo R, Berto A, Franchin E, Menegotto N, De Canale E, Tommasini T, Rinaldi R, Basso M, Stefani S, Palù G. 2015. Prevalence of Klebsiella pneumoniae strains producing carbapenemases and increase of resistance to colistin in an Italian teaching hospital from January 2012 to December 2014. BMC Infect Dis 15:244. doi: 10.1186/s12879-015-0996-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tansarli GS, Papaparaskevas J, Balaska M, Samarkos M, Pantazatou A, Markogiannakis A, Mantzourani M, Polonyfi K, Daikos GL. 2018. Colistin resistance in carbapenemase-producing Klebsiella pneumoniae bloodstream isolates: evolution over 15 years and temporal association with colistin use by time series analysis. Int J Antimicrob Agents 52:397–403. doi: 10.1016/j.ijantimicag.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 9.Trimble MJ, Mlynárčik P, Kolář M, Hancock REW. 2016. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb Perspect Med 6:a025288. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baron S, Hadjadj L, Rolain JM, Olaitan AO. 2016. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 11.Cheng HY, Chen YF, Peng HL. 2010. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci 17:60. doi: 10.1186/1423-0127-17-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raetz CR, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem 76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayol A, Nordmann P, Brink A, Villegas MV, Dubois V, Poirel L. 2017. High-level resistance to colistin mediated by various mutations in the crrB gene among carbapenemase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 61:e01423-17. doi: 10.1128/AAC.01423-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng YH, Lin TL, Lin YT, Wang JT. 2016. Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob Agents Chemother 60:3709–3716. doi: 10.1128/AAC.00009-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srinivasan VB, Rajamohan G. 2013. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob Agents Chemother 57:4449–4462. doi: 10.1128/AAC.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telke AA, Olaitan AO, Morand S, Rolain JM. 2017. soxRS induces colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrAB-tolC efflux pump. J Antimicrob Chemother 72:2715–2721. doi: 10.1093/jac/dkx215. [DOI] [PubMed] [Google Scholar]
- 17.Parra-Lopez C, Baer MT, Groisman EA. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J 12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Acker H, Van Dijck P, Coenye T. 2014. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol 22:326–333. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Keasey SL, Suh MJ, Das S, Blancett CD, Zeng X, Andresson T, Sun MG, Ulrich RG. 2019. Decreased antibiotic susceptibility driven by global remodeling of the Klebsiella pneumoniae proteome. Mol Cell Proteomics 18:657–668. doi: 10.1074/mcp.RA118.000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amato SM, Orman MA, Brynildsen MP. 2013. Metabolic control of persister formation in Escherichia coli. Mol Cell 50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Shan Y, Brown Gandt A, Rowe SE, Deisinger JP, Conlon BP, Lewis K. 2017. ATP-dependent persister formation in Escherichia coli. mBio 8:e02267-16. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright MS, Suzuki Y, Jones MB, Marshall SH, Rudin SD, van Duin D, Kaye K, Jacobs MR, Bonomo RA, Adams MD. 2015. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 59:536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adler C, Corbalan NS, Seyedsayamdost MR, Pomares MF, de Cristobal RE, Clardy J, Kolter R, Vincent PA. 2012. Catecholate siderophores protect bacteria from pyochelin toxicity. PLoS One 7:e46754. doi: 10.1371/journal.pone.0046754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Zhu L, Pan D, Li S, Xiao H, Zhang Z, Shen X, Wang Y, Long M. 2018. Siderophore-mediated iron acquisition enhances resistance to oxidative and aromatic compound stress in Cupriavidus necator JMP134. Appl Environ Microbiol 85:e01938-18. doi: 10.1128/AEM.01938-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peralta DR, Adler C, Corbalan NS, Paz Garcia EC, Pomares MF, Vincent PA. 2016. Enterobactin as part of the oxidative stress response repertoire. PLoS One 11:e0157799. doi: 10.1371/journal.pone.0157799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X, Drlica K. 2014. Reactive oxygen species and the bacterial response to lethal stress. Curr Opin Microbiol 21:1–6. doi: 10.1016/j.mib.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhagirath AY, Li Y, Patidar R, Yerex K, Ma X, Kumar A, Duan K. 2019. Two component regulatory systems and antibiotic resistance in Gram-negative pathogens. Int J Mol Sci 20:E1781. doi: 10.3390/ijms20071781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hua X, Chen Q, Li X, Yu Y. 2014. Global transcriptional response of Acinetobacter baumannii to a subinhibitory concentration of tigecycline. Int J Antimicrob Agents 44:337–344. doi: 10.1016/j.ijantimicag.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 31.Cheng YH, Lin TL, Lin YT, Wang JT. 2018. A putative RND-type efflux pump, H239_3064, contributes to colistin resistance through CrrB in Klebsiella pneumoniae. J Antimicrob Chemother 73:1509–1516. doi: 10.1093/jac/dky054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore SJ, Lawrence AD, Biedendieck R, Deery E, Frank S, Howard MJ, Rigby SE, Warren MJ. 2013. Elucidation of the anaerobic pathway for the corrin component of cobalamin (vitamin B12). Proc Natl Acad Sci U S A 110:14906–14911. doi: 10.1073/pnas.1308098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung LM, Cooper VS, Rasko DA, Guo Q, Pacey MP, McElheny CL, Mettus RT, Yoon SH, Goodlett DR, Ernst RK, Doi Y. 2017. Structural modification of LPS in colistin-resistant, KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother 72:3035–3042. doi: 10.1093/jac/dkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu YY, Chandler CE, Leung LM, McElheny CL, Mettus RT, Shanks RMQ, Liu JH, Goodlett DR, Ernst RK, Doi Y. 2017. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrob Agents Chemother 61:e00580-17. doi: 10.1128/AAC.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasdemir UO, Chevalier J, Nordmann P, Pages JM. 2004. Detection and prevalence of active drug efflux mechanism in various multidrug-resistant Klebsiella pneumoniae strains from Turkey. J Clin Microbiol 42:2701–2706. doi: 10.1128/JCM.42.6.2701-2706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pages JM, Masi M, Barbe J. 2005. Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol Med 11:382–389. doi: 10.1016/j.molmed.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Y, Zhao J, Maifiah MHM, Velkov T, Schreiber F, Li J. 2019. Metabolic responses to polymyxin treatment in Acinetobacter baumannii ATCC 19606: integrating transcriptomics and metabolomics with genome-scale metabolic modeling. mSystems 4:e00157-18. doi: 10.1128/mSystems.00157-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yee R, Cui P, Xu T, Shi W, Feng J, Zhang W, Zhang Y. 2017. Identification of a novel gene argJ involved in arginine biosynthesis critical for persister formation in Staphylococcus aureus. bioRxiv 10.1101/114827. [DOI] [PubMed]
- 39.Ryan S, Begley M, Gahan CG, Hill C. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ Microbiol 11:432–445. doi: 10.1111/j.1462-2920.2008.01782.x. [DOI] [PubMed] [Google Scholar]
- 40.Xiong L, Teng JL, Botelho MG, Lo RC, Lau SK, Woo PC. 2016. Arginine metabolism in bacterial pathogenesis and cancer therapy. Int J Mol Sci 17:363. doi: 10.3390/ijms17030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindgren JK, Thomas VC, Olson ME, Chaudhari SS, Nuxoll AS, Schaeffer CR, Lindgren KE, Jones J, Zimmerman MC, Dunman PM, Bayles KW, Fey PD. 2014. Arginine deiminase in Staphylococcus epidermidis functions to augment biofilm maturation through pH homeostasis. J Bacteriol 196:2277–2289. doi: 10.1128/JB.00051-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin XM, Yang MJ, Li H, Wang C, Peng XX. 2014. Decreased expression of LamB and Odp1 complex is crucial for antibiotic resistance in Escherichia coli. J Proteomics 98:244–253. doi: 10.1016/j.jprot.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 43.Lin XM, Yang JN, Peng XX, Li H. 2010. A novel negative regulation mechanism of bacterial outer membrane proteins in response to antibiotic resistance. J Proteome Res 9:5952–5959. doi: 10.1021/pr100740w. [DOI] [PubMed] [Google Scholar]
- 44.Mathieu A, Fleurier S, Frenoy A, Dairou J, Bredeche MF, Sanchez-Vizuete P, Song X, Matic I. 2016. Discovery and function of a general core hormetic stress response in E. coli induced by sublethal concentrations of antibiotics. Cell Rep 17:46–57. doi: 10.1016/j.celrep.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. 2012. ProOpDB: Prokaryotic Operon DataBase. Nucleic Acids Res 40:D627–D631. doi: 10.1093/nar/gkr1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogawa W, Onishi M, Ni R, Tsuchiya T, Kuroda T. 2012. Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene 498:177–182. doi: 10.1016/j.gene.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Clinical and Laboratory Standards Institute (CLSI). 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard; 10th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 48.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 49.Kanehisa M, Sato Y. 2020. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci 29:28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henderson JC, O’Brien JP, Brodbelt JS, Trent MS. 2013. Isolation and chemical characterization of lipid A from gram-negative bacteria. J Vis Exp e50623. doi: 10.3791/50623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw genomic data of the K. pneumoniae ATCC BAA 2146 and Ci strains have been deposited to NCBI with BioProject accession number PRJNA612645.