Abstract
Background
In this study, we discovered and validated candidate microRNA (miRNA) biomarkers for coronary artery disease (CAD).
Method
Candidate tissue-derived miRNAs from atherosclerotic plaque material in patients with stable coronary artery disease (SCAD) (n=14) and unstable coronary artery disease (UCAD) (n=25) were discovered by qPCR-based arrays. We validated differentially expressed miRNAs, along with seven promising CAD-associated miRNAs from the literature, in the serum of two large cohorts (n=395 and n=1000) of patients with SCAD and UCAD and subclinical atherosclerosis (SubA) and controls, respectively.
Result
From plaque materials (discovery phase), miR-125b-5p and miR-193b-3p were most upregulated in SCAD, whereas miR-223-3p and miR-142-3p were most upregulated in patients with UCAD. Subsequent validation in serum from patients with UCAD, SCAD, SubA and controls demonstrated significant upregulation of miR-223-3p, miR-133a-3p, miR-146-3p and miR-155-5p. The ischaemia-related miR-499-5p was also highly upregulated in patients with UCAD compared with the other groups (SCAD OR 20.63 (95% CI 11.16 to 38.15), SubA OR 96.10 (95% CI 40.13 to 230.14) and controls OR 15.73 (95% CI 7.80 to 31.72)). However, no significant difference in miR-499-5p expression was observed across SCAD, SubA and controls. MiR-122-5p was the only miRNA to be significantly upregulated in the serum of both patients with UCAD and SCAD.
Conclusion
In conclusion, miR-122-5p and miR-223-3p might be markers of plaque instability.
Keywords: atherosclerosis, risk factors, genetics, metabolic medicine, myocardial ischaemia and infarction (IHD)
Key questions.
What is already known about this subject?
Several circulating microRNAs (miRNAs) have been reported to differentially express between healthy individuals and patients with coronary artery disease (CAD).
Unfortunately, none of the identified circulating miRNA biomarkers was strong enough to make it to clinical practice.
What does this study add?
In this large cross-sectional study, we were able to show that tissue-derived miR-223-3p and serum miR-122-5p are robust and promising biomarkers for unstable CAD disease, possibly reflecting plaque instability.
Besides, miR-122-5p could also reflect an adverse metabolic profile increases in the risk for atherosclerosis.
How might this impact on clinical practice?
Existing CAD risk algorithms do not take into the account that CAD can either remain stable (SCAD) or can become unstable (UCAD).
Therefore, these miRNAs could be helpful in risk prediction for (early) CAD as well as markers that can discriminate SCAD from UCAD.
Introduction
Cardiovascular disease is one of the leading causes of death worldwide.1 However, early detection of coronary artery disease (CAD) remains a challenge. Therefore, biomarkers are needed to improve prognostication and to individualise treatment. Nowadays, the risk of CAD is assessed by algorithms based on patients’ risk factors such as the American College of Cardiology/American Heart Association or Systematic COronary Risk Evaluation (SCORE) calculators.2 3 Unfortunately, these risk score algorithms provide a poor estimate of an individual’s risk, especially in young individuals. Besides, these algorithms do not take into account that CAD can either remain stable (SCAD) or can become unstable (UCAD). Although atherosclerosis is their common factor, SCAD and UCAD have completely different pathophysiology and outcome.4 SCAD can remain stable for many years and may cause cardiac complaints during exercise, yet it does not immediately jeopardise the myocardium. Whereas, in patients with UCAD, the atherosclerotic plaque may erode or even rupture and as such cause an acute coronary syndrome, which can lead to loss of myocardium by ischaemia and eventually heart failure and/or death.5 Therefore, for accurate cardiovascular risk prediction and early treatment, it is crucial to have markers that can discriminate SCAD from UCAD in an early phase.
miRNAs are short, non-coding RNAs that post-transcriptionally regulate gene expression6 by binding the 3′ untranslated region of target gene mRNAs.7 miRNAs are noted as key players in modulating the function of endothelial cells, smooth muscle cells and macrophages, regulating atherosclerosis pathogenesis.8–11 They are ideal biomarkers since they are quite tissue-specific and remain stable in the blood,12 13 but an optimal performing biomarker should only present in the circulation in the diseased state and absent in healthy controls. This poses a problem for CAD since one would ideally need biopsy material to be able to investigate the most abundant miRNAs for CAD. Nowadays, there are a few studies on miRNA expression profiles in atherosclerotic plaques,8 14 15 but because of the often small sample sizes, these studies are prone to bias, because of small sample errors and issues with reproducibility (ref: small sample size paper).
In this study, we aimed to overcome the issues of small sample error and reproducibility, by showing robust outcomes of miRNAs for either SCAD or UCAD from discovery to multiple validations. Explicitly this study was not intended to explore the mechanisms of the proposed miRNAs, but to explore the reproducibility of earlier discovered atherosclerotic plaque miRNAs, as a robust marker of SCAD or UCAD. We used an epidemiological approach using larger sample sizes and multiple validation methods, to overcome small sample errors. Therefore, this study, on miRNA biomarkers for SCAD and UCAD, comprises a discovery phase, a first validation of the discovery phase and a second validation to be able to further substantiate the first validation.
Methods
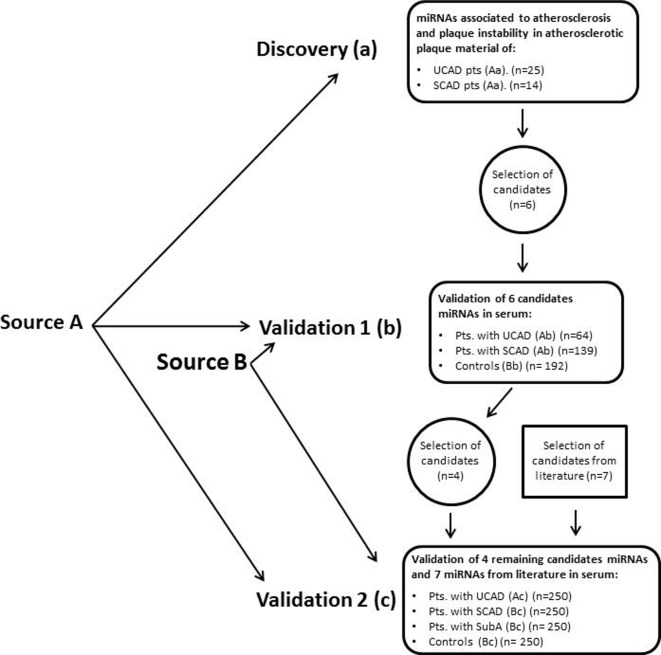
A flow chart of the distinct cohorts and their properties used in the discovery and validation phase of the study is presented in figure 1.
Figure 1.
Flow chart of the study outline. miRNA, microRNA; SCAD, stable coronary artery disease; SubA, subclinical atherosclerosis; UCAD, unstable coronary artery disease.
Source populations
For our study, we made use of two different source populations A and B. In short, source population A consisted of patients who underwent a percutaneous coronary intervention, electively or because of acute coronary syndrome. Source population B consisted of serum samples from a biobank of patients with premature atherosclerosis and their ‘healthy’ family members. From these two source populations, we selected three study populations (a, b and c). For details on source and study populations, see online supplementary methods.
openhrt-2019-001223supp001.pdf (306KB, pdf)
Study populations
Discovery cohort
The discovery cohort a (n=39) consisted of patients with an acute coronary syndrome (UCAD group; n=25; Aa) or stable angina (SCAD group; n=14; Aa) from whom tissue of either thrombectomy or atherectomy material from coronary angiography was available. Further detailed information on tissue collection is described in online supplementary methods.
First validation cohort
The first validation cohort b (n=395) consisted of patients with an acute coronary syndrome (UCAD group; n=64; Ab), or stable angina (SCAD group; n=139; Ab) and controls (control; n=192; Bb). Controls did not have any detectable atherosclerosis as indicated by an Agatston coronary artery calcium (CAC) score of 0 determined by coronary CT scan.
Second validation cohort
The second validation cohort c (n=1000) consisted of patients with an acute coronary syndrome (UCAD group; n=250; Ac), or patients with a history of CAD, without cardiac complains (SCAD group; n=250; Bc), or patients with subclinical atherosclerosis, as indicated by a CAC score of ≥1, without cardiac complains (SubA group; n=250; Bc) and controls with a CAC score of 0 (control; n=250; Bc).
Blood withdrawal
Serum samples were drawn before the administration of heparin and were stored in cryovials at –80°C as described in online supplementary methods.
Selection of candidates for the first validation phase
Tissue-derived miRNAs from the discovery cohort were ranked, as described in online supplementary methods, for the first validation candidate selection.
Selection of miRNAs for the second validation phase
To be able to show the robustness of our results, we decided to revalidate these results in a second validation phase. The first validation phase showed that miR-125b-5p, miR-223-3 p, miR-142-3 p and miR-193b-3p could be promising biomarkers for either stable or unstable cardiovascular disease. These miRNA together with the most promising miRNA biomarkers for cardiovascular disease published, namely miR-133a-3p, miR-146-3 p, miR-126-3 p, miR-145-5 p, miR-155-5 p and miR-122-5 p, were measured in the second validation phase. For the literature miRNAs, we conducted a literature search and selected all miRNAs that were consistently upregulated or downregulated in more than two studies.9 16–19 To be able to differentiate whether the candidate miRNAs were due to ischaemia, we also added an ischaemia-related miR-499–5 p to our panel.20
RNA isolation and quantification of miRNAs by RT-qPCR
Procedure on RNA isolation, complementary DNA synthesis, quantification, RT-qPCR data handling, and normalisation are described in online supplementary methods.
Biochemical parameters
Liver enzymes and lipids were measured as described in the online supplementary methods.
Coronary CT scan
Coronary artery CT scan was performed as described in the online supplementary methods.
Patient and public involvement
It was not appropriate or possible to involve patients or the public in the design, conduct, reporting, or dissemination plans of our research.
Statistical analysis
Baseline characteristics are expressed as mean±SD for continuous variables and number (%) for dichotomous variables, except when indicated otherwise. Analysis of variance with post hoc Student’s t-test, Mann-Whitney U test and Fisher’s exact test was used to calculate differences in baseline characteristics as appropriate. In the discovery phase, T-tests were used to calculate differences in array expression which were expressed in a mean difference of normalised Cq values. P values of the qPCR-based array were Benjamini-Hochberg corrected for multiple testing. In the validation phase, logistic regression was used to analyse the differences in miRNA expression between the SCAD, UCAD and the control group expressed as normalised log-transformed starting concentrations (N0) as calculated by LinRegPCR.21 In the validation cohort, multivariate analyses were performed on two different models to adjust for age and gender. All statistical analyses were performed using SPSS for Windows V.23. A p value <0.05 was considered to be statistically significant.
Results
Discovery phase
qPCR-based miRNA array on tissue samples
The clinical characteristics of the discovery cohort are shown in table 1. Of the 742 measurable tissue-derived miRNAs, 125 qualified for reliable analysis as described in the ‘Methods’ section and were included in the analysis. In total, 54 tissue-derived miRNAs were significantly upregulated in SCAD compared with UCAD and 50 tissue-derived miRNAs were significantly upregulated in UCAD compared with SCAD. For expression levels of all tissue-derived miRNAs, see online supplementary table 1.
Table 1.
Patient characteristics
| Discovery cohort (n=39) | Validation cohort 1 (n=395) | Validation cohort 2 (n=1000) | |||||||
| SCAD | UCAD | Control | SCAD | UCAD | Control | SCAD | UCAD | SubA | |
| N | 14 | 25 | 192 | 139 | 64 | 250 | 250 | 250 | 250 |
| Age, years±SD | 61.5±10.4 | 62.8±10.1 | 46.5±7.3 | 65.2±11.6* | 66.4±11.8* | 46.0±7.7 | 51.2±9.8* | 62.1±13.3*† | 53.7±9.8*†‡ |
| Gender, male, n (%) | 10 (71) | 18 (72) | 61 (32) | 89 (64)* | 41 (64)* | 83 (33) | 188 (75)* | 177 (71)* | 125 (50)*†‡ |
| BMI, kg/m2, n (%) | 24.9±2.2 | 26.1±3.4 | 27.1±4 | 27.1±4 | 26.9±3.9 | 26.5±4.5 | 27.7±4.5 | 27.0±4.3 | 26.9±6.9 |
| Diabetes, n (%) | 2 (14) | 2 (8) | 6 (3) | 31 (22)* | 12 (19)* | 4 (2) | 34 (14)* | 32 (13)* | 32 (13)* |
| Hypertension, n (%) | 3 (21) | 12 (48) | 38 (20) | 86 (62)* | 32 (50) * | 54 (22) | 98 (39)* | 86 (34)* | 110 (44)*‡ |
| Current smoker, n (%) | 3 (21) | 9 (36) | 42 (22) | 37 (27) | 19 (30) | 49 (20) | 56 (22) | 101 (40)*† | 54 (22)‡ |
| Hypercholesterolaemia, n (%) | 5 (36) | 8 (32) | 49 (26) | 54 (39)* | 21 (33) | 57 (23) | 90 (36)* | 54 (22)† | 86 (35)*‡ |
| Glucose, mmol/L | – | – | – | – | – | 5.2±0.5 | 6.3±4.9* | 8.5±3.3* † | 5.9±4.0 ‡ |
| History of | |||||||||
| MI, n (%) | 5 (36) | 5 (20) | 0 | 51 (37)* | 26 (41)* | – | 169 (68) | 35 (14)† | – |
| PCI, n (%) | 1 (7) | 4 (16) | 0 | 52 (37)* | 20 (31)* | – | 61 (24) | 24(10)† | – |
| CABG, n (%) | 2 (14) | 0 | 0 | 9 (6)* | 6 (9)* | – | 20 (8) | 5 (2)† | – |
| Type of disease (acute phase) | |||||||||
| SAP, n (%) | 14 (100) | 0† | – | 139 (100) | 0† | – | – | 0 (0) | – |
| UAP, n (%) | 0 | 0 | – | 0 | 59 (92)† | – | – | 0 (0) | – |
| NSTEMI, n (%) | 0 | 0 | – | 0 | 5 (8)† | – | – | 0 (0) | – |
| STEMI, n (%) | 0 | 25 (100)† | – | 0 | 0 | – | – | 250 (100) | – |
| Lipid profile | |||||||||
| ALAT (U/L) | – | – | – | – | – | 22.8±1.5 | 30.54±1.6* | 22.3±1.9† | 25.1±1.5†‡ |
| GGT (U/L) | – | – | – | – | – | 23.0±1.8 | 35.5±1.9* | 34.9±1.6* | 27.7±1.7*†‡ |
| Total cholesterol (mmol/L) | – | – | – | – | – | 5.24 (1.95–9.7) | 4.30 (2.32–8.20)* | 5.30 (1.90–9.30) † | 5.60 (2.79–8.80)† ‡ |
| Triglyceride (mmol/L) | – | – | – | – | – | 1.10 (0.20–8.90) | 1.29 (0.43–8.00) | 0.90 (0.30–4.70)† | 1.20 (0.44–5.80) ‡ |
| HDL cholesterol (mmol/L) | – | – | – | – | – | 1.40 (0.78–2.90) | 1.21 (0.60–2.40)* | 1.03 (0.53–2.09)*† | 1.39 (0.50–3.52)†‡ |
| LDL cholesterol (mmol/L) | – | – | – | – | – | 3.22 (1.29–7.53) | 2.39 (0.41–5.60)* | 3.70 (0.72–7.49)*† | 3.59 (1.08–6.57)†‡ |
All values are given as mean±SD for continuous variables and median for lipid profile or as absolute number (percentage) for dichotomous variable. P values were obtained using ANOVA with post hoc T-tests for numerical values, Kruskal-Wallis test for median and χ2 tests were performed for dichotomous and categorical variables.
*P<0.05 compared with control.
†P<0.05 compared with SCAD.
‡P<0.05 compared with UCAD.
ALAT, alanine aminotransferase; ANOVA, analysis of variance; BMI, body mass index; CABG, coronary artery bypass graft; CV medication, cardiovascular medication (ie, aspirin, beta-blocker, calcium antagonist, nitrates, ACE-inhibitors, angiotensin receptor blocker or statins); GGT, gamma-glutamyltransferase; HDL, high-density lipoprotein; LDL, low-density lipoprotein; MI, myocardial infarction; NSTEMI, non- ST-elevation myocardial infarction; PCI, percutaneous coronary intervention; SAP, stable angina pectoris; SCAD, stable coronary artery disease; STEMI, ST-elevation myocardial infarction; UAP, unstable angina pectoris; UCAD, unstable coronary artery disease.
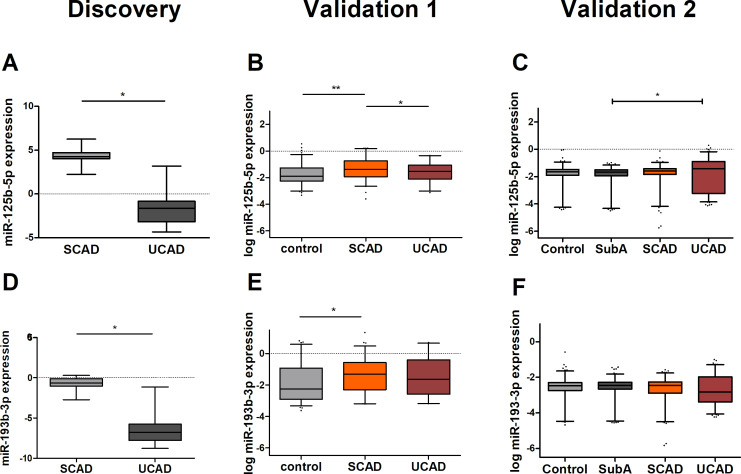
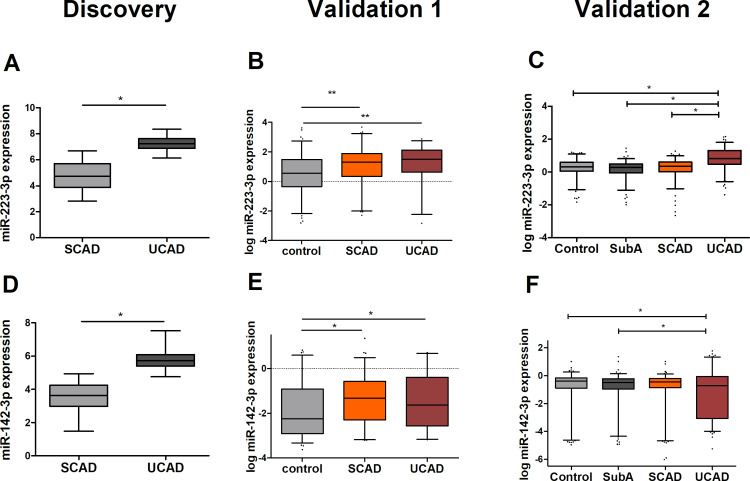
The top three most upregulated tissue-derived miRNAs in SCAD were miR-125b-5p (mean diff: 5.94), miR-455-3 p (mean diff: 5.66) and miR-193b-3p (mean diff: 5.66) and in UCAD were miR-223-3 p (mean diff: 2.55), miR-142-3 p (mean diff: 2.24) and miR-126-5 p (mean diff: 1.77).
Of these tissue-derived miRNAs, miR-455-3 p and miR-126-5 p could not be measured in serum. For miR-455-3 p, the expression level was undetectable and for miR-126-5 p the cDNA synthesis failed and there was not enough material to repeat the analyses. Therefore, four out of the six candidate tissue-derived miRNAs were validated: miR-125b-5p, miR-193b-3p, miR-223-3 p and miR-142-3 p.
First validation
Multivariate analysis of the four candidate tissue-derived miRNAs in serum
Clinical characteristics of validation cohort 1 are shown in table 1. We performed a multivariate analysis correcting for age and gender, in the first validation. We were able to validate, in serum, all four candidate tissue-derived miRNAs observed in our discovery experiment against controls. We were able to show that miR-125b-5p (OR 2.09, 95% CI 1.30 to 3.36), miR-193b-3p (OR 1.41, 95% CI 1.00 to 2.00), miR-223-3 p (OR 1.61, 95% CI 1.20 to 2.15) and miR-142-3 p (OR 1.38, 95% CI 1.05 to 1.82) (figures 2 and 3 and online supplementary table 3) were significantly upregulated in patients with SCAD as compared with controls. On the other hand, we were only able to validate the difference in SCAD versus UCAD for miR-125b-5p (OR 1.64, 95% CI 1.09 to 2.48). On the other hand, there was a slight non-significant higher expression of miR-193b-3p in SCAD as compared with UCAD, similar as observed in the discovery experiment. In addition, for patients with UCAD miR-223-3 p (OR 2.38, 95% CI 1.41 to 4.01) and miR-142–3 p (OR 1.62, 95% CI 1.12 to 2.35) also showed a significant difference with controls. Combined, these results suggest that all four tissue-derived miRNAs could be validated in serum and might serve as markers for the presence of coronary atherosclerosis and to a lesser extent, miR-223-3 p might serve as a marker for UCAD.
Figure 2.
Expression levels of microRNAs (miRNAs) upregulated in SCAD (miR-125b-5p and miR-193b-3p). Normalised expression levels are shown for both the discovery and the validation experiments. Expression levels of miR-125b-5p (A) and miR-193b-3p (D) in both atherectomy (SCAD, n=14) and thrombectomy material (UCAD, n=25) were determined by qPCR-based array in the discovery experiments. In the first validation phase, log-transformed expression levels of miR-125b-5p (B) and miR-193b-3p (E) were determined by serum qPCR measurement in patients with UCAD (n=64), SCAD (n=139) and controls (n=192). In the second validation phase, log-transformed expression levels of miR-125b-5p (C) and miR-193b-3p (F) were determined by serum qPCR in patients with UCAD (n=250), SCAD (n=250), SubA (n=250) and controls (n=250). Statistical analyses in the validation phases were corrected for age and gender. More detailed information on expression level differences is found in online supplementary table 3 (first validation) and online supplementary table 5 (second validation). SCAD, stable coronary artery disease, SubA, subclinical atherosclerosis (ie, coronary calcium score ≥1 on coronary CT scan); UCAD, unstable coronary artery disease. *P<0.05, **p<0.01.
Figure 3.
Expression levels of microRNAs (miRNAs) upregulated in UCAD (miR-223-3 p, miR-142-3 p). Normalised expression levels are shown for both the discovery and the validation experiments. Expression levels of miR-223-3 p (A) and miR-142-3 p (D) in both atherectomy (SCAD, n=14) and thrombectomy material (UCAD, n=25) were determined by qPCR-based array in the discovery experiments. In the first validation phase, log-transformed expression levels of miR-223-3 p (B) and miR-142-3 p (E) were determined by serum qPCR measurement in patients with UCAD (n=64), SCAD (n=139) and controls (n=192). In the second validation phase, log-transformed expression levels of miR-223-3 p (C) and miR-142-3 p (F) were determined by serum qPCR in patients with UCAD (n=250), SCAD (n=250), SubA (n=250) and controls (n=250). Statistical analyses in the validation phases were corrected for age and gender. More detailed information on expression level differences is found in online supplementary table 3 (first validation) and online supplementary table 5 (second validation). SCAD, stable coronary artery disease; SubA, subclinical atherosclerosis (ie, coronary calcium score ≥1 on coronary CT scan); UCAD, unstable coronary artery disease. *P<0.05, **p<0.01.
Second validation
The previous data imply that the discovered tissue-derived miRNA markers can also detect the presence of atherosclerosis in the serum of these patients. Since circulating miRNA biomarkers for the disease are often not being replicated in other studies, we deemed it necessary to try and replicate this finding ourselves in a second, larger cohort. Besides, for replication purposes, we added miRNAs from the literature which were suggested to be associated with CAD.
Multivariate analysis of the candidate miRNAs from the second validation study
The clinical characteristics of the validation cohort are shown in table 1. In our second validation study, in which we did a multivariate analysis correcting for age and gender, we were able to validate, in serum, one out of the four candidate tissue-derived miRNAs identified from our discovery experiment. MiR-223-3 p was significantly upregulated in the UCAD group compared with controls (OR 13.92 (95% CI 7.29 to 26.57) and patients with SCAD (10.75 (95% CI 6.46 to 17.89); figure 3 and online supplementary table 5). This miRNA was also significantly upregulated when compared with patients with SubA (OR 12.75 (95% CI 7.42 to 21.91). Concerning the other candidate miRNAs from the discovery experiment, miR-142-3 p showed an opposite effect, namely a significant downregulation as compared with controls (OR 0.90 (95% CI 0.75 to 1.07)) and patients with SubA (OR 0.83 (95% CI 0.72 to 0.96)) when compared with patients with UCAD.
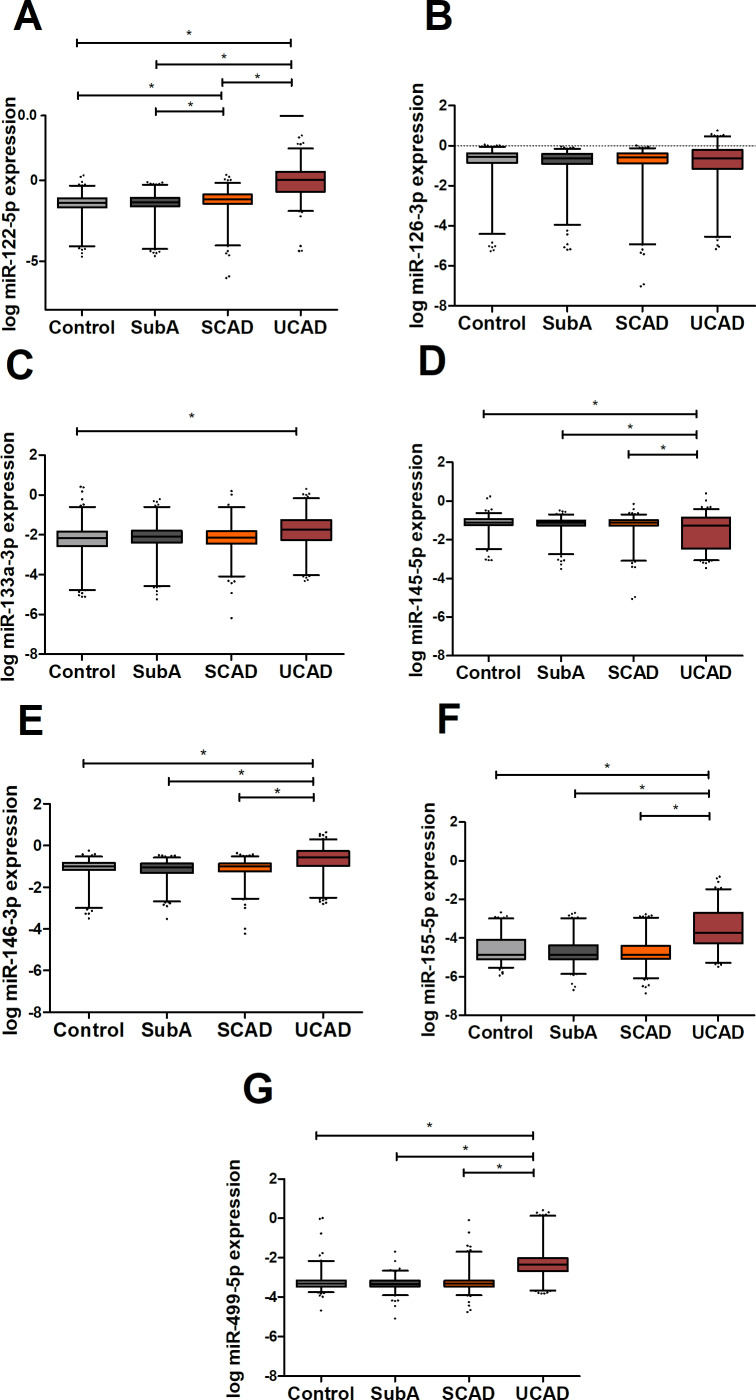
Multivariate analysis of the candidate miRNAs from literature
Age-corrected and gender-corrected multivariate analysis of the CAD-related miRNAs from the literature (figure 4, online supplementary table 5) showed an elevated expression, in serum, in patients with UCAD of miR-122-5 p, miR-146-3 p and miR-155-5 p with all other subgroups (OR and 95% CI, online supplementary table 5). On the other hand, miR-145-5 p was significantly downregulated, in serum, in patients with UCAD as compared with the other subgroups. Furthermore, miR-122-5 p was significantly upregulated in patients with UCAD versus the other groups, and patients with SCAD as compared with both SubA OR 1.38 (95% CI 1.07 to 1.77) and controls OR 1.45 (95% CI 1.09 to 1.93). The ischaemia-related miR-499-5 p was also highly upregulated in patients with UCAD compared with the other patient groups (SCAD OR 20.63 (95% CI 11.16 to 38.15), SubA OR 96.10 (95% CI 40.13 to 230.14)) and controls OR 15.73 (95% CI 7.80 to 31.72) and might indicate that most of the miRNAs that are upregulated in the patients with UCAD reflect acute ischaemia. Indeed, this could be the case for all miRNAs upregulated in patients with UCAD, except for miR-122-5 p, which was also upregulated in patients with SCAD. If the upregulation of miR-122-5 p in patients with UCAD would be an indication for acute ischaemia, this would mean that patients with SCAD with an upregulated miR-122-5 p would have acute ischaemia, although they did not have any complaints. Since miR-499-5 p was only upregulated in UCAD and not in SCAD, this seems unlikely. Furthermore, in a linear regression model, no significant association was noticed between hsTnI and miR-122-5 p (data not shown).
Figure 4.
MicroRNA (miRNA) expression levels of atherosclerosis-related miRNAs from literature. Graphs show log transformed and normalised concentrations of miR-122-5 p (A), miR126-3p (B), miR133a-3p (C), miR145-5p (D), miR-146-3 p (E), miR-155-5 p (F) and miR-499-5 p (G). SCAD, stable coronary artery disease; SubA, subclinical atherosclerosis (ie, coronary calcium score >0 on coronary CT scan); UCAD, unstable coronary artery disease. *P<0.05.
Finally, we also performed a receiver operating characteristic analysis in which we were able to show that miR-122–5 p and miR-223–3 p when considered together in a model that identified SCAD and UCAD with an area under the curve of 0.80 and 0.96, respectively (table 2).
Table 2.
AUC for ROC curve for SCAD compared with controls
| Model | AUC |
| Model I | |
| Age and gender | 0.79 |
| Model II | |
| miR-122-5 p | 0.63 |
| Model III | |
| miR-223-3 p | 0.50 |
| Model IV | |
| Age and gender+miR-122-5p+miR-223-3 p | 0.80 |
| AUC for ROC curve for UCAD compared with controls | |
| Model I | |
| Age and gender | 0.88 |
| Model II | |
| miR-122-5 p | 0.90 |
| Model III | |
| miR-223-3 p | 0.76 |
| Model IV | |
| Age and gender+miR-122-5p+miR-223-3 p | 0.96 |
AUC, area under the curve; ROC, receiver operating characteristic.
To conclude, the second validation, miR-122-5 p, miR-146-3 p, miR-155-5 p and miR-145-5 p could be promising markers for unstable CAD. Besides, since miR-122-5 p was also significantly upregulated in SCAD as compared with controls, this miRNA could also be a promising marker for atherosclerotic disease.
To substantiate that our findings are not merely due to chance, we wanted to show that at least one of these miRNAs showed similar results as expected from the literature. Mir-122-5 p is known to be a liver-specific miRNA. Therefore, we expect that elevated miR-122-5 p is associated with increased liver enzymes. We observed a significant positive correlation with increasing levels of miR-122-5 p with gamma-glutamyltransferase (gamma-GT) β 0.08 (95% CI 0.06 to 0.09; p<0.001) and ALAT β 0.05 (95% CI 0.03 to 0.06; p<0.001). Besides, we also observed that gamma-GT and ALAT were significantly elevated in SCAD (mean±SD; for gamma-GT 35.5±1.9 U/L; ALAT 30.5±1.6 U/L) as compared with SubA (gamma-GT 27.7±1.7 U/L; ALAT 25.1±1.5 U/L, p<0.05) and controls (gamma-GT 23.0±1.8 U/L; ALAT 22.8±1.5 U/L, p<0.05). Concerning the patients with UCAD, only gamma-GT (34.9±1.6 U/L) was significantly elevated as compared with SubA (27.7±1.7 U/L, p<0.05) and controls (23.0±1.8 U/L, p<0.05).
Discussion
In this study, we were able to show that tissue-derived miR-223-3 p and serum miR-122-5 p are robust and promising biomarkers for unstable CAD disease, possibly reflecting plaque instability. Besides, miR-122-5 p could also reflect an adverse metabolic profile increases the risk for atherosclerosis. The miRNAs miR-146-3 p, miR-155-5 p and miR-145-5 p might also be able to identify unstable CAD, however, the data were less robust, especially miR-233-3 p seems a promising biomarker for individuals at risk for an acute coronary syndrome. MiR-122-5 p could be used to identify individuals with an adverse metabolic profile, such as insulin resistance at risk for unstable CAD.
This study does not aim to unravel the biochemical process underlying these candidate miRNAs but merely tries to robustly show the epidemiological evidence that they are promising biomarkers. We show that miR-223-3 p both in tissue and in serum as well as in multiple independent cohorts show similar results. Many investigators have shown the relation between plaque-derived miRNAs or serum miRNAs in relation to CAD, but most of this research fails when it comes to validation, because of small sample errors and change finding. We managed to improve our miRNA measurement and analytical techniques as such22 that we were able to robustly validate the findings of miR-223-3 p. We only show that miR-223-3 p is related to the acute phase of CAD, while others have shown that miR-223-3 p could be released from vascular endothelial cells23 24 and that it might play a role in endothelial cell dysfunction, atherosclerotic lesion initiation, progression and possibly plaque rupture.25 26 Therefore, miR-223-3 p and maybe also miR-122-5 p, might originate from plaque tissue and directly reflect damaged endothelial cells of the plaque, on the edge of rupture. On the other hand, miR-122-5 p was the only miRNA that was also upregulated in SCAD. Therefore, miR-122-5 p might rather be a marker of the atherosclerotic process or even the unfavourable metabolic profile underlying atherosclerosis. In line with these results, Niculescu et al also found a significant upregulation of miR-122-5 p in patients with stable and unstable angina compared with controls.27 Besides, we were able to confirm the relationship of miR-122-5 p as a highly conserved liver miRNA regulating lipid metabolism28 29 and has been associated with liver diseases such as metabolic syndrome and type 2 diabetes, major risk factors for atherosclerosis. Its elevation among both the patients with SCAD and UCAD might reflect underlying liver pathology such as hepatic steatosis, which is related to atherosclerosis,30 31 but we did not investigate this. Whether miR-122-5 p has a direct or indirect effect on atherosclerosis is still unknown, since it has also been suggested that miR-122-5 p is released from both the liver and endothelial cells.32 33
Strengths and limitations
One might argue that our findings are contradictory to the claim the studies make, we selected our candidate literature miRNAs from. Namely, these studies claim to have investigated miRNAs related to SCAD instead of UCAD. On the other hand, the cohorts we selected our candidate literature miRNAs from, did not truly represent SCAD. In some of these cohorts, blood was drawn at the time that these patients had complaints of angina27 34–36 and interestingly, in one of these studies, CK-MB and troponin levels were even higher in the group of SCAD as compared with the group with UCAD.34 On the other hand, one might argue that also in our study, these markers simply reflect acute ischaemia, since blood was taken in the acute phase of an acute coronary syndrome. For most of our candidate miRNAs, this might very well be true, since miR-223,37 38 miR-133a-3p,20 miR-146-3p27 39 and miR-155-5p39 are known to be related to myocardial damage as could be shown by an elevation in the ischaemia markers miR-499-5 p and hsTnI levels. On the other hand, miR-122-5 p was apart from its upregulation in patients with UCAD also upregulated in patients with SCAD, and could not simply reflect ischaemia, since the miRNA-499-5 p and hsTnI levels were low in these patients.
Another limitation of our study is that the discovery phase was a comparison between miRNAs of plaque tissue of patients with SCAD and UCAD and not compared with the healthy vessel wall of controls. This is not possible, but on the other hand, this unique set-up is able to identify markers of plaque instability in a background of atherosclerosis. Regarding miR-223-3 p, this was the only miRNA that could be validated from the discovery phase, and might, therefore, reflect a true marker of plaque instability.
Most miRNAs form the discovery phase and could not be validated in the validation cohorts. This could be because miRNAs concentration is much higher in tissue as compared with blood and that the level in the circulation, although this might be higher as compared with controls, could still be too low to measure it reliably. If the lack of validation was due to technical differences in the measuring method, none of the miRNAs could have been measurable in the validation cohorts, which was not the case.
Another limitation comes from the fact that we used different source populations. Therefore, samples might differ, since they were handled differently and since the age of the source populations were different. We do not think this has affected the outcomes substantially since the expression levels of miR-125-5 p, miR-193-3 p and miR-126-3 p were quite similar among all the four groups. Besides, miRNAs are known for their stability. Therefore, longer storage times or slightly different handling of the samples are unlikely to affect miRNA expression levels. Finally, our data handling protocol takes into account technical differences.
One of our major strengths is that miRNA data handling is extremely thorough and handles missing values and technical difficulties in a systematic way, leading to more robust results.40 For instance, all miRNAs were measured in triplicates and interplate variance was taken into account as an extra normalisation procedure by the second validation study. Besides, normalisation was performed taking into account three endogenous miRNAs as well as two technical normalisers. Furthermore, extra quality checks were carried out by a prespecified data handling pipeline, which has been shown to increase both accuracy and precision when analysing circulating miRNAs.40 We think that the robust methods and large sample sizes used in this study make this one of the most extensive circulating miRNA studies in the cardiovascular research field so far. Moreover, the differences in miRNA expression between the multiple validation phases underscore the small sample error issues that circulating miRNA research often encounters and underscores the robustness of the miRNAs that could indeed be validated. Also in our study, validation of previous findings was difficult. In contrast, we were able to confirm most of the literature picked candidate miRNAs and we showed a consistent upregulation of miR-223-3 p from discovery to second validation, again underscoring the robustness of the results in this study.
Finally, we would like to emphasise that we did not include speculations on the mechanism since we believe many other studies are prone to small sample error and therefore, the quality and robustness of many experimental small miRNA studies is questionable. For true mechanistic investigations, animal experiments are the preferred method thereafter to be validated in sufficiently large enough human studies. Therefore, this study was planned as a clinical study investigating biomarkers for UCAD.
Conclusions
In conclusion, miR-223-3 p was the only miRNA that could be validated in both the validation cohorts and seems a promising marker for UCAD. Furthermore, miR-122-5 p and miR-223-3 p might even be markers of plaque instability and miR-122-5 p could also reflect an adverse metabolic profile that increases the risk for atherosclerosis.
Acknowledgments
The authors would like to thank Joost Leenders, Roelien Meijering, Zhen Liu from ACS Biomarker BV, the Netherlands (http://www.acsbiomarker.com) and Suzanne Battjes, AMC medical student, for the execution of the qPCR miRNA measurements in the validation experiments.
Footnotes
SS and MWJdR contributed equally.
Contributors: SS, MWJdR, S-JP-S, JCMM, EEC: conception and design of research. MWJdR, MGMK, BMS: material collection. MWJdR, MGMK, BMS: performed experiments. SS, MWJdR, MGMK, S-JP-S: analysis and interpretation. SS, MWJdR, MGMK: prepared figures. SS, MWJdR, MGMK: drafted manuscript. MAMB, RDW, ACvdW, BMS, JCMM, EEC, S-JP-S: edited and revised manuscript.
Funding: This study was funded by BDDA PoC grant (2012); validation of miRNA biomarkers for unstable plaque and The European Union: FP7 (BestAgeing, GA 306031).
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: This study complies with the Declaration of Helsinki. The study protocol for both biobank used in the first (MASS study biobank: 2013_220) and the second source population (biobank premature atherosclerosis: 2014_095) was approved by the biobank ethical committee of the Academic Medical Center in Amsterdam and all individuals gave written informed consent.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available on reasonable request.
References
- 1.Roth GA, Johnson C, Abajobir A, et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol 2017;70:1–25. 10.1016/j.jacc.2017.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goff DC, Lloyd-Jones DM, Bennett G, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American heart association Task force on practice guidelines. Circulation 2014;129:S49–73. 10.1161/01.cir.0000437741.48606.98 [DOI] [PubMed] [Google Scholar]
- 3.Piepoli MF, Hoes AW, Agewall S, et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts)Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR). Eur Heart J 2016;37:2315–81. 10.1093/eurheartj/ehw106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crea F, Liuzzo G. Pathogenesis of acute coronary syndromes. J Am Coll Cardiol 2013;61:1–11. 10.1016/j.jacc.2012.07.064 [DOI] [PubMed] [Google Scholar]
- 5.Libby P. Mechanisms of acute coronary syndromes and their implications for therapy. N Engl J Med 2013;368:2004–13. 10.1056/NEJMra1216063 [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 7.Chistiakov DA, Bobryshev YV, Orekhov AN. Changes in transcriptome of macrophages in atherosclerosis. J Cell Mol Med 2015;19:1163–73. 10.1111/jcmm.12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bildirici AE, Arslan S, Özbilüm Şahin N, et al. MicroRNA-221/222 expression in atherosclerotic coronary artery plaque versus internal mammarian artery and in peripheral blood samples. Biomarkers 2018;23:670–5. 10.1080/1354750X.2018.1474260 [DOI] [PubMed] [Google Scholar]
- 9.Romaine SPR, Tomaszewski M, Condorelli G, et al. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart 2015;101:921–8. 10.1136/heartjnl-2013-305402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522–31. 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 11.Kim JS, Pak K, Goh TS, et al. Prognostic value of microRNAs in coronary artery diseases: a meta-analysis. Yonsei Med J 2018;59:495–500. 10.3349/ymj.2018.59.4.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet 2010;3:484–8. 10.1161/CIRCGENETICS.110.958363 [DOI] [PubMed] [Google Scholar]
- 13.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–8. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parahuleva MS, Lipps C, Parviz B, et al. MicroRNA expression profile of human advanced coronary atherosclerotic plaques. Sci Rep 2018;8:7823. 10.1038/s41598-018-25690-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y, Thavarajah T, Gu W, et al. Impact of miRNA in atherosclerosis. Arterioscler Thromb Vasc Biol 2018;38:e159–70. 10.1161/ATVBAHA.118.310227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busch A, Eken SM, Maegdefessel L. Prospective and therapeutic screening value of non-coding RNA as biomarkers in cardiovascular disease. Ann Transl Med 2016;4:236. 10.21037/atm.2016.06.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahlin F, Arfvidsson J, Vargas KG, et al. MicroRNAs as circulating biomarkers in acute coronary syndromes: a review. Vascul Pharmacol 2016;81:15–21. 10.1016/j.vph.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 18.Schulte C, Zeller T. microRNA-based diagnostics and therapy in cardiovascular disease-Summing up the facts. Cardiovasc Diagn Ther 2015;5:17–36. 10.3978/j.issn.2223-3652.2014.12.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navickas R, Gal D, Laucevičius A, et al. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res 2016;111:322–37. 10.1093/cvr/cvw174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corsten MF, Dennert R, Jochems S, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet 2010;3:499–506. 10.1161/CIRCGENETICS.110.957415 [DOI] [PubMed] [Google Scholar]
- 21.Ruijter JM, Ramakers C, Hoogaars WMH, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 2009;37:e45. 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kok MGM, de Ronde MWJ, Moerland PD, et al. Small sample sizes in high-throughput miRNA screens: a common pitfall for the identification of miRNA biomarkers. Biomol Detect Quantif 2018;15:1–5. 10.1016/j.bdq.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabet F, Vickers KC, Cuesta Torres LF, et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun 2014;5:3292. 10.1038/ncomms4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan Y, Liang H, Liu H, et al. Platelet-secreted microRNA-223 promotes endothelial cell apoptosis induced by advanced glycation end products via targeting the insulin-like growth factor 1 receptor. J Immunol 2014;192:437–46. 10.4049/jimmunol.1301790 [DOI] [PubMed] [Google Scholar]
- 25.Gimbrone MA, García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res 2016;118:620–36. 10.1161/CIRCRESAHA.115.306301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damani S, Bacconi A, Libiger O, et al. Characterization of circulating endothelial cells in acute myocardial infarction. Sci Transl Med 2012;4:126ra33 10.1126/scitranslmed.3003451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niculescu LS, Simionescu N, Sanda GM, et al. MiR-486 and miR-92a identified in circulating HDL discriminate between stable and vulnerable coronary artery disease patients. PLoS One 2015;10:e0140958. 10.1371/journal.pone.0140958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norata GD, Sala F, Catapano AL, et al. MicroRNAs and lipoproteins: a connection beyond atherosclerosis? Atherosclerosis 2013;227:209–15. 10.1016/j.atherosclerosis.2012.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elmén J, Lindow M, Silahtaroglu A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res 2008;36:1153–62. 10.1093/nar/gkm1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willeit P, Skroblin P, Moschen AR, et al. Circulating microRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes 2017;66:347–57. 10.2337/db16-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai W-C, Hsu S-D, Hsu C-S, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 2012;122:2884–97. 10.1172/JCI63455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Yang Y, Wang L, et al. Plasma miR-122 and miR-3149 potentially novel biomarkers for acute coronary syndrome. PLoS One 2015;10:e0125430. 10.1371/journal.pone.0125430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X-D, Yang Y-J, Wang L-Y, et al. Elevated plasma miRNA-122, -140-3p, -720, -2861, and -3149 during early period of acute coronary syndrome are derived from peripheral blood mononuclear cells. PLoS One 2017;12:e0184256. 10.1371/journal.pone.0184256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Alessandra Y, Carena MC, Spazzafumo L, et al. Diagnostic potential of plasmatic microRNA signatures in stable and unstable angina. PLoS One 2013;8:e80345. 10.1371/journal.pone.0080345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong J, Liang Y-Z, Zhang J, et al. Potential role of Lipometabolism-Related microRNAs in peripheral blood mononuclear cells as biomarkers for coronary artery disease. J Atheroscler Thromb 2017;24:430–41. 10.5551/jat.35923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao W, He H-W, Wang Z-M, et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis 2012;11:55. 10.1186/1476-511X-11-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C, Fang Z, Jiang T, et al. Serum microRNAs profile from genome-wide serves as a fingerprint for diagnosis of acute myocardial infarction and angina pectoris. BMC Med Genomics 2013;6:16. 10.1186/1755-8794-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulte C, Molz S, Appelbaum S, et al. miRNA-197 and miRNA-223 predict cardiovascular death in a cohort of patients with symptomatic coronary artery disease. PLoS One 2015;10:e0145930. 10.1371/journal.pone.0145930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zidar N, Boštjančič E, Glavač D, et al. MicroRNAs, innate immunity and ventricular rupture in human myocardial infarction. Dis Markers 2011;31:259–65. 10.1155/2011/247654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Ronde MWJ, Ruijter JM, Lanfear D, et al. Practical data handling pipeline improves performance of qPCR-based circulating miRNA measurements. RNA 2017;23:811–21. 10.1261/rna.059063.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
openhrt-2019-001223supp001.pdf (306KB, pdf)