Abstract
The prion agent is unique in biology and is comprised of prion protein scrapie (PrPSc), a self-templating conformational variant of the host encoded prion protein cellular (PrPC). The deposition patterns of PrPSc in the CNS can vary considerably from a diffuse synaptic pattern to large plaque-like aggregates. Alterations of PrPC posttranslational processing can change PrPSc deposition patterns; however, the mechanism underlying these observations is unclear. In this issue of the JCI, Sevillano and authors determined that parenchymal PrPSc plaques of the mouse brain preferentially incorporated underglycosylated PrPC that had been liberated from the cell surface by the metalloproteinase, ADAM-10, in combination with heparan sulfate. These results provide mechanistic insight into the formation of PrPSc plaques and suggest that PrP posttranslational modifications direct pathogenicity as well as the rate of disease progression.
Protein-only infectious agents
Prions are protein-only infectious agents that subvert the normal form of the host cellular prion protein, PrPC, to the self-templating scrapie prion form, PrPSc (1, 2). Due to the unique nature of prions, the properties of both the host PrPC and agent PrPSc contribute to the pathogenesis of disease. Specifically, strains of prions result in distinct patterns of PrPSc deposition and neurodegeneration in the CNS that are encoded by strain-specific conformations of PrPSc (3, 4). Interestingly, changes in the host PrPC can result in alteration of the phenotype of disease, including neuropathology, in the absence of altering the prion strain. For example, murine PrPC is polymorphic with two alleles, and inoculation of ME7 from one PrPC genetic background to the other resulted in a change in both the incubation period of disease and the distribution of neurodegeneration (5). Importantly, back passage of ME7 to the original PrPC genetic background resulted in an incubation period and neuropathology indistinguishable from the original PrPC allele (i.e., class 1 strain) (5). This seminal work indicated that differences in the amino acid sequence of PrPC contribute to the pathogenesis of disease independently of the prion strain.
Altering prion pathogenesis
Posttranslational modifications of PrPC can alter prion pathogenesis. PrPC is tethered to the cell membrane by a glycophosphatidylinositol (GPI) anchor (6). In transgenic mice, deleting the PrPC GPI anchor results in PrPC retrafficking into the intracellular space. Subsequent prion inoculation resulted in PrPSc formation, indicating that prion conversion is independent of the GPI anchor (7). Importantly, these mice showed substantial PrPSc distribution differences in the whole body as well as differences in the CNS. In the CNS, the pattern of PrPSc deposition was characterized by dense plaque-like formations, compared with the more diffuse synaptic pattern of PrPSc deposition in nontransgenic WT animals. While prion infection of PrPC GPI anchorless mice resulted in prion formation, these animals did not develop clinical signs of prion disease. It is unclear if their healthy phenotype is due to a failure of PrPSc to gain access to the areas of the CNS responsible for the onset of clinical signs, if the PrPSc generated was less neurotoxic, or if the absence of symptoms was a property of the strain tested.
In addition to a GPI anchor, PrPC contains two N-linked glycosylation sites that have variable occupancy (8, 9). Transgenic mice in which the PrPC lacked both N-linked glycosylation sites still supported prion formation. PrPSc from these mice transmits prion disease to WT mice, indicating that glycosylation is not required for prion formation or transmission (10). In the strains tested, however, the absence of PrPC glycosylation resulted in an increase in incubation period and PrPSc amyloid plaque deposits compared with WT animals that have a full repertoire of PrPC glycosylation. Shifting of PrPSc to a more amyloid plaque structure can reduce the pathogenicity of PrPSc by extraneuronal routes of infection, consistent with the hypothesis that small, soluble PrPSc oligomers are the preferred species of PrPSc for retrograde transport in the peripheral nervous system that facilitates neuroinvasion (11, 12). Both GPI anchorless PrPC and underglycosylated PrPC share a common neuropathological feature of parenchymal PrPSc plaques. These studies indicate that alteration of PrPC posttranslational modifications can result in profound changes in the neuropathology of prion disease. The mechanism responsible for these observations is, however, unclear.
How PrPC posttranslational modifications may alter neuropathology
The work of Sevillano et al. in this issue of the JCI examines the mechanism of how PrPC posttranslational modifications alter neuropathology (13). Mice expressing unglycosylated PrPC were generated (Prnp180Q/196Q) and were shown to have normal PrPC expression patterns that, importantly, failed to spontaneously develop neurodegeneration. The Prnp180Q/196Q mice were challenged with four murine-adapted prion strains, three of which (RML, 22L, and ME7) are categorized as subfibrillar strains with a predominately diffuse synaptic pattern of PrPSc deposition in the CNS, and one of which (mCWD) is a fibrillar strain that is characterized by large plaques of PrPSc in the CNS. All of the strains tested resulted in the Prnp180Q/196Q animals developing prion disease. Notably, the mice inoculated with the subfibrillar strains developed plaque-like deposits. This altered neuropathology is consistent with previous studies indicating that reduced PrPC glycosylation results in an increase in PrPSc plaque formation (10). To examine the composition of these plaque-like deposits in the Prnp180Q/196Q mice, the authors used an anti-PrP antibody that specifically recognizes PrP that has been cleaved from the cell surface by the metalloproteinase ADAM-10 (14). It was determined that the PrPSc plaques were enriched for ADAM-10–cleaved PrP. Next, the authors identified that heparan sulfate (HS) colocalized to the plaques but was restricted from areas with diffuse aggregates of PrPSc. Consistently, HS specifically bound to PrP that had been cleaved by ADAM-10. Further, the HS/ADAM-10–cleaved PrP interaction was influenced by the glycosylation state of PrP where a progressive reduction in the glycosylation of PrP resulted in a corresponding increase in affinity of PrP to bind HS. These data suggest that underglycosylated PrPC that is cleaved from the cell surface in combination with HS produces large plaque-like structures.
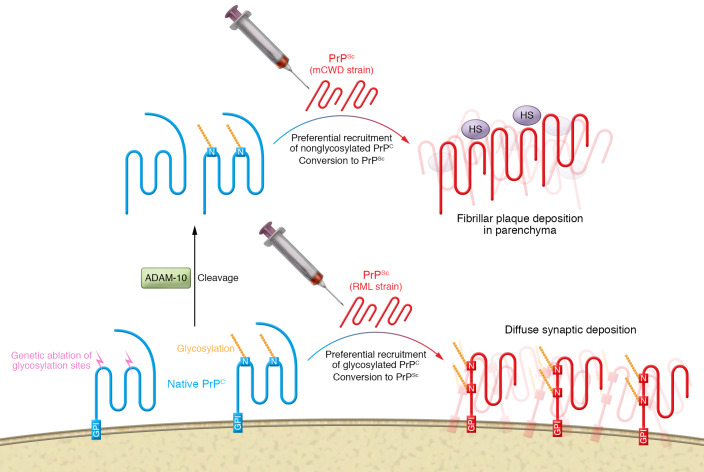
To test the hypothesis that glycosylation influences plaque-like pathology, Sevillano and colleagues generated novel transgenic mice that expressed PrPC with three N-linked glycosylation sites (Prnp187N) (13). Inoculation of the Prnp187N mice with the plaque-forming strain mCWD resulted in a neuropathology that lacked plaques and instead was comprised of diffuse PrPSc deposits compared with WT mice that contain 2 N-linked PrPC glycosylation sites. This observation is consistent with the premise that PrP glycosylation inhibits PrPSc fibril formation by interfering with binding to HS. Overall, a model for prion formation suggests that underglycosylated PrP released from the cell surface associates with HS to form large parenchymal deposits, whereas cell-bound fully glycosylated PrPSc preferentially forms small diffuse aggregates (Figure 1 and ref. 13).
Figure 1. Role of posttranslational processing of PrPC in the formation of prion aggregates.
Fibrillar prion strains such as mCWD preferentially incorporate underglycosylated PrPC (blue) that is released from the cell surface by ADAM-10 into plaques that form in association with HS in the parenchyma. Subfibrillar prion stains, such as RML, preferentially incorporate glycosylated PrPC into diffuse synaptic deposits of PrPSc (red) without a HS scaffold. Genetic ablation of N-linked glycosylation sites (N) can switch in nonfibrillar strain pathology from synaptic diffuse deposition to plaque-like deposition of PrPSc in the parenchyma.
The role of the prion strain in the preponderance to form plaques versus diffuse deposits of PrPSc is only partially understood. Transmission of all four prion strains to Prnp180Q/196Q mice resulted in an increase in plaque-like PrPSc deposits in the CNS. However, the degree to which plaque-like deposits increased was not uniform between the strains tested. The mechanism behind this observation remains unclear (13). It is possible that the strain-specific differences in the preferred PrPC substrate for conversion, as has been identified using in vitro studies, may play a role in this observation (15). In addition, strain-specific rates of formation and clearance may favor specific patterns of prion accumulation (16).
Changing the incubation period of prion disease
Strain-specific responses in the alteration of the incubation period of disease in the Prnp180Q/196Q mice were observed. Specifically, the incubation period of RML-infected WT or Prnp180Q/196Q mice was similar. This was in contrast to that of 22L-infected animals, where the incubation period of Prnp180Q/196Q-infected mice was longer compared with WT mice. Importantly, the extended incubation period of RML-infected Prnp180Q/196Q mice was preserved upon second passage. One potential explanation for this observation is that differences in the PrP amino acid sequence between the WT and Prnp180Q/196Q mice may cause a species barrier–like phenomenon that can alter strain properties. Alternatively, while evidence suggests that glycosylation is not required on the inoculum PrPSc to maintain strain properties (17), it is possible the host PrPC glycosylation directly or indirectly via interactions with cellular cofactors plays a role in maintenance of strain properties during prion conversion (18). The small differences in incubation period between WT and Prnp180Q/196Q mice infected with RML suggest that glycosylation may not substantially affect prion formation in contrast to what has been observed in vitro where glycosylation inhibits prion formation (19, 20). Several scenarios could explain this discrepancy. It is possible that the strains tested in this study respond differently to changes in glycosylation compared with strains that were used in the in vitro studies (13, 19, 20). Many factors in addition to the kinetics of PrPSc accumulation can influence the incubation period of disease. The relative rate of PrPSc formation, clearance, and transport to populations of neurons that are susceptible to PrPSc neurotoxicity are only a few of the factors that contribute to the incubation period of disease.
When Sevillano and colleagues introduced a third glycosylation site in PrPC, the incubation period shortened. Further, there was a corresponding increase in diffuse PrPSc aggregates and an absence of PrPSc plaques (13). These findings suggest that a more rapid disease course corresponds with diffuse PrPSc deposits. While more work is needed to further characterize the contributions of posttranslational modifications of PrPC and the prion strain to prion pathogenesis, the Sevillano et al. study illustrates that shifting diffuse synaptic PrPSc deposits to plaques is a viable therapeutic modality to slow the progression of prion disease.
Acknowledgments
JCB is supported by the National Institute of Allergy and Infectious Diseases and the National Institute of Neurological Disorders and Stroke of the NIH via grants P01 0011877A, R01 NS103763, and R01 NS107246, and by award CWD20-018 from the State of Michigan.
Version 1. 01/27/2020
Electronic publication
Version 2. 01/28/2020
Corrected heparan typographical error
Version 3. 03/02/2020
Print issue publication
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(3):1087–1089. https://doi.org/10.1172/JCI134842.
See the related article at Prion protein glycans reduce intracerebral fibril formation and spongiosis in prion disease.
References
- 1.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USA. 2007;104(23):9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216(4542):136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 3.Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68(12):7859–7868. doi: 10.1128/JVI.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser H, Dickinson AG. Distribution of experimentally induced scrapie lesions in the brain. Nature. 1967;216(5122):1310–1311. doi: 10.1038/2161310a0. [DOI] [PubMed] [Google Scholar]
- 5. Bruce ME, Dickinson AG. Biological stability of different classes of scrapie agent. In: Prusiner SB, Hadlow WJ, eds. Slow transmissible diseases of the nervous system. Vol 2. New York, New York, USA: Academic Press; 1979:71–86. [Google Scholar]
- 6.Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51(2):229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 7.Chesebro B, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308(5727):1435–1439. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 8.Endo T, Groth D, Prusiner SB, Kobata A. Diversity of oligosaccharide structures linked to asparagines of the scrapie prion protein. Biochemistry. 1989;28(21):8380–8388. doi: 10.1021/bi00447a017. [DOI] [PubMed] [Google Scholar]
- 9.Oesch B, et al. A cellular gene encodes scrapie PrP 27-30 protein. Cell. 1985;40(4):735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 10.Tuzi NL, et al. Host PrP glycosylation: a major factor determining the outcome of prion infection. PLoS Biol. 2008;6(4):e100. doi: 10.1371/journal.pbio.0060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bett C, et al. Enhanced neuroinvasion by smaller, soluble prions. Acta Neuropathol Commun. 2017;5(1):32. doi: 10.1186/s40478-017-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancellotti E, et al. Glycosylation of PrPC determines timing of neuroinvasion and targeting in the brain following transmissible spongiform encephalopathy infection by a peripheral route. J Virol. 2010;84(7):3464–3475. doi: 10.1128/JVI.02374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sevillano, et al. Prion protein glycans reduce intracerebral fibril formation and spongiosis in prion disease. J Clin Invest. 2020;130(3):1350–1362. doi: 10.1172/JCI131564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linsenmeier L, et al. Structural and mechanistic aspects influencing the ADAM10-mediated shedding of the prion protein. Mol Neurodegener. 2018;13(1):18. doi: 10.1186/s13024-018-0248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katorcha E, Makarava N, Savtchenko R, D’Azzo A, Baskakov Sialylation of prion protein controls the rate of prion amplification, the cross-species barrier, the ratio of PrPSc glycoform and prion infectivity. PLoS Pathog. 2014;10(9):e1004366. doi: 10.1371/journal.ppat.1004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shikiya RA, et al. PrPSc formation and clearance as determinants of prion tropism. PLoS Pathog. 2017;13(3):e1006298. doi: 10.1371/journal.ppat.1006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piro JR, et al. Prion protein glycosylation is not required for strain-specific neurotropism. J Virol. 2009;83(11):5321–5328. doi: 10.1128/JVI.02502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deleault NR, et al. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci USA. 2012;109(28):E1938–E1946. doi: 10.1073/pnas.1206999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camacho MV, Telling G, Kong Q, Gambetti P, Notari S. Role of prion protein glycosylation in replication of human prions by protein misfolding cyclic amplification. Lab Invest. 2019;99(11):1741–1748. doi: 10.1038/s41374-019-0282-1. [DOI] [PubMed] [Google Scholar]
- 20.Makarava N, Savtchenko R, Baskakov Selective amplification of classical and atypical prions using modified protein misfolding cyclic amplification. J Biol Chem. 2013;288(1):33–41. doi: 10.1074/jbc.M112.419531. [DOI] [PMC free article] [PubMed] [Google Scholar]