Abstract
Hematopoietic stem cell (HSC) attrition is considered the key event underlying progressive BM failure (BMF) in Fanconi anemia (FA), the most frequent inherited BMF disorder in humans. However, despite major advances, how the cellular, biochemical, and molecular alterations reported in FA lead to HSC exhaustion remains poorly understood. Here, we demonstrated in human and mouse cells that loss-of-function of FANCA or FANCC, products of 2 genes affecting more than 80% of FA patients worldwide, is associated with constitutive expression of the transcription factor microphthalmia (MiTF) through the cooperative, unscheduled activation of several stress-signaling pathways, including the SMAD2/3, p38 MAPK, NF-κB, and AKT cascades. We validated the unrestrained Mitf expression downstream of p38 in Fanca–/– mice, which display hallmarks of hematopoietic stress, including loss of HSC quiescence, DNA damage accumulation in HSCs, and reduced HSC repopulation capacity. Importantly, we demonstrated that shRNA-mediated downregulation of Mitf expression or inhibition of p38 signaling rescued HSC quiescence and prevented DNA damage accumulation. Our data support the hypothesis that HSC attrition in FA is the consequence of defects in the DNA-damage response combined with chronic activation of otherwise transiently activated signaling pathways, which jointly prevent the recovery of HSC quiescence.
Keywords: Cell Biology, Hematology
Keywords: Bone marrow, DNA repair, Genetic diseases
Introduction
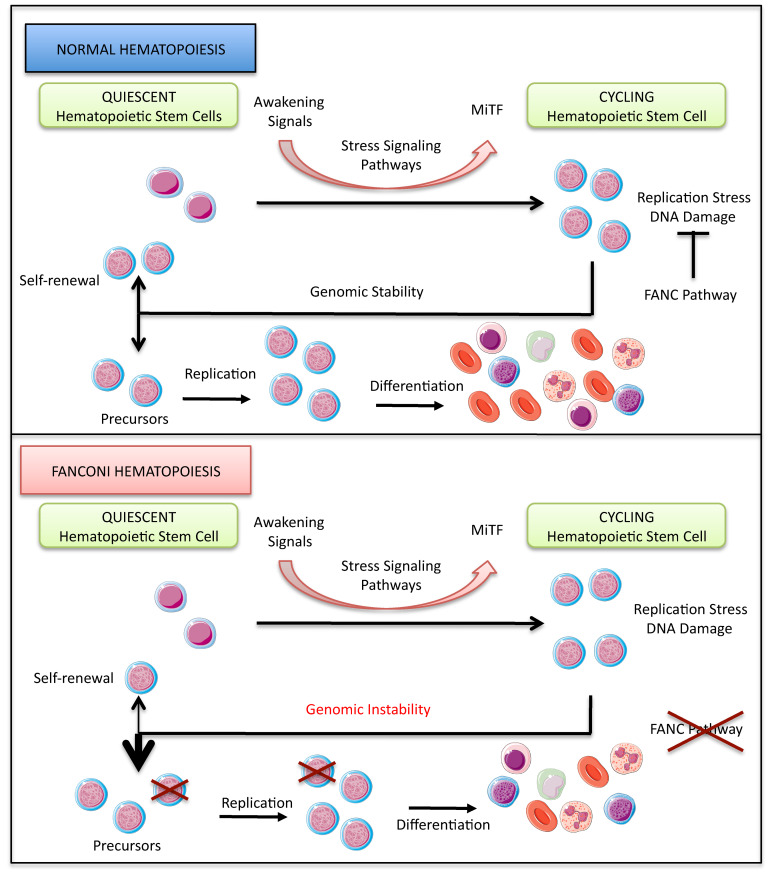
Hematopoiesis is a fine-tuned process in which a small pool of specialized cells, hematopoietic stem cells (HSCs), provide the BM and the bloodstream with undifferentiated and differentiated cells throughout a lifetime, both during homeostasis and under health-threatening stress conditions, which include DNA damage, bleeding, infection, and BM transplantation; this latter process is also defined as stress hematopoiesis (1–3). Indeed, HSCs sustain hematopoiesis by ensuring both the generation of more differentiated cells and the maintenance of their stemness via the reacquisition of dormancy (self-renewal) (4, 5). Signals transmitted by several factors, including IFNs, stem cell factor (SCF), thrombopoietin (TPO), TGF family members, TNF-α, and several ILs, transiently upregulate the activity of various intrinsic pathways to control the quiescence-awakening cycle of HSCs. The SMAD (6–8), p38 MAPK (9), and NF-κB (10) signaling cascades are among the major intrinsic pathways that govern several common and alternative downstream effectors and determine BM behavior during both homeostatic and stress hematopoiesis. However, to be beneficial, the action of such pathways must be finely tuned in time and intensity; otherwise, it can constitute a major threat to cell and BM homeostasis (4, 5). Indeed, unrestrained activation of the SMAD (7, 8), p38 (9, 11), or NF-κB (10, 12) cascade has been associated with reduced HSC quiescence, BM failure (BMF), and/or leukemia.
Known for its involvement in melanocyte, melanoma, mast cell, and osteoclast biology, and targeted by the SMAD, p38 MAPK, and NF-κB pathways (13–15), the transcription factor microphthalmia (MiTF) has recently been recognized as a key factor in sustaining DNA replication downstream of p38 signaling in awakened HSCs (16). MiTF is also a direct transcriptional regulator of several FANC genes (17, 18) whose inactivation leads to Fanconi anemia (FA), an inherited BMF disease that presents predisposition to acute myeloid leukemia (AML) and chromosome fragility (19, 20). The FANC genes encode proteins that have been functionally and biochemically subdivided into 3 major groups: the FANC core complex, the ID2 complex (FANCD2 and FANCI), and a third group that includes factors involved in homologous recombination (HR). Stalled replication forks trigger FANC core complex–mediated monoubiquitination of FANCD2 and FANCI and their recruitment to subnuclear foci, allowing the ID2 heterodimer to coordinate DNA repair and replication restart via HR and protecting cells against replication-associated genetic instability (21–23). Consequently, FANC pathway inactivation leads to cellular and chromosomal hypersensitivity to DNA lesions and stresses that impinge on replication fork progression (20, 22). Moreover, FA is also associated with altered expression and/or responses to growth factors and cytokines as well as the aberrant expression/activity of several signaling pathways, including the SMAD, p38 MAPK, and NF-κB cascades (24–33). However, the impact of the overactivation of such signaling pathways in FA cells as well as in BMF remains poorly understood.
The p38 and SMAD signaling pathways (a) are chronically active in FA (24, 31, 32), (b) cooperate in some settings (34, 35), and(c) are involved in the expression and activity of MiTF (16, 36), which is able to transcriptionally induce several FANC genes (17, 18). In addition, consistent with these reports, (d) the inhibition of these pathways rescued some FA-associated hematopoietic abnormalities (31, 37, 38). Thus, we were motivated to add insight into the relationship between stress signaling pathways and MiTF in FA to identify new events that, along with defects in DNA repair and genetic stability maintenance, contribute to BMF in this disease. Since approximately 70% of FA patients harbor FANCA biallelic mutations, supporting the importance of the encoded protein in the BM, and approximately 90% of FA patients have inactivating mutations in components of FANC core complex physiology (20, 22), we mainly focused on the loss of function of FANCA and FANCC (other components of the FANC core complex) to connect alterations in the p38/MiTF axis to the FA phenotype.
Results
FANC core complex loss of function leads to MiTF overexpression in human and mouse cells.
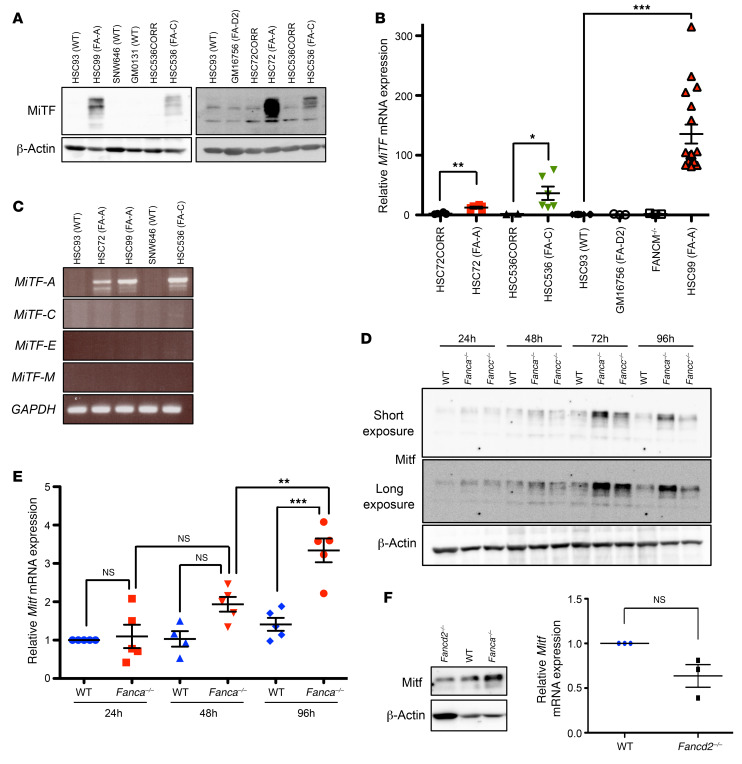
To determine whether MiTF expression is altered downstream of FANC pathway loss of function, we first analyzed exponentially growing EBV-immortalized lymphoblasts derived from FA patients and healthy donors. Immunoblot analysis of extracts from FANCA–/– (HSC99 and HSC72) or FANCC–/– (HSC536) cells revealed strong expression of MiTF, which was almost undetectable in FANC pathway–proficient cells (HSC93, SNW646, and GM0131) (Figure 1A). Moreover, the ectopic expression of the corresponding WT FANC gene in HSC72 or HSC536 cells normalized MiTF expression, establishing a robust link between the loss-of-function of FANCA or FANCC and the overexpression of MiTF (Figure 1A). Subsequently, we performed qRT-PCR analysis on RNA isolated from FANC pathway–deficient or FANC pathway–proficient cells and established that high expression of MiTF in FA is associated with an increased level of MiTF RNA (Figure 1B). To extend our observations beyond the FANC core complex, to which FANCA and FANCC belong, we analyzed 2 FA cell lines bearing mutations in FANCM (FANCM–/–) or FANCD2 (GM16756), which are FANC core complex upstream and downstream proteins, respectively. In both cases, MiTF expression was similar to that observed in FANC pathway–proficient cells, suggesting that its overexpression is mainly associated with FANC core complex abnormalities (Figure 1, A and B).
Figure 1. FANC core complex loss of function is associated with MiTF overexpression.
(A) Representative Western blots illustrating MiTF expression in lymphoblasts from WT healthy (HSC93, SNW646, GM0131) or FA donors HSC99 (FANCA–/–, FA-A), HSC536 (FANCC–/–, FA-C), HSC72 (FANCA–/–, FA-A), GM16756 (FANCD2–/–, FA-D2), or FA cells complemented with the corresponding WT FANC gene (HSC536CORR and HSC72CORR). We performed at least 5 individual experiments for each cell line, with similar results. (B) qRT-PCR of MiTF mRNA expression in the indicated cell lines. In each experiment, MiTF expression was first normalized to that of Actin (internal control) and then normalized to the MiTF/Actin ratio in control HSC93 cells, which was set as 1 in each experiment. Data are shown as mean ± SEM of n = 3 (FANCM–/–, GM16756), n = 6 (HSC72, HSC72CORR, HSC536, HSC536CORR), and n = 18 (HSC93, HSC99) experiments. (C) RT-PCR analysis of MiTF-A, MiTF-C, MiTF-E, and MiTF-M isoforms in cells from WT (HSC93, SNW646) or FA (HSC72, HSC99, HSC536) donors. (D) Representative Western blots (n = 3) showing Mitf expression in WT, Fanca–/–, or Fancc–/– MEFs collected at the indicated time points after seeding. (E) qRT-PCR of Mitf expression in the same cells and conditions described in D. qRT-PCR data were normalized against Oaz1 RNA and normalized to the Mitf/Oaz1 ratio in WT cells at 24 hours, which was set as 1. Data are shown as mean ± SEM of n = 5 experiments. (F) Representative Western blot (n = 2, left) and qRT-PCR data (right) showing Mitf expression in WT, Fanca–/–, or Fancd2–/– MEFs 72 hours after seeding. RNA samples were analyzed as described in E. Data are shown as mean ± SEM of n = 3 experiments. β-Actin was used as a loading control for A, D, and F. Statistical significance was assessed using an unpaired 2-tailed t test with Welch’s correction (B and F) or 1-way ANOVA with Bonferroni’s correction (E). *P < 0.05; **P < 0.01; ***P < 0.001.
The MiTF locus can generate several isoforms that share exons 2 to 9 (39–41), which are all targeted by the previously used antibodies and primers. By reverse transcriptase PCR (RT-PCR) analysis, we demonstrated that, among the principal MiTF isoforms, MiTF-A is the only one that is robustly expressed in FA-deficient lymphoblasts (Figure 1C). The several bands observed in the immunoblots of Figure 1A are the consequence of either the expression of other minor not analyzed isoforms of MiTF or due to posttranslational modifications of the encoded proteins (42).
To extend our observations, we subsequently analyzed Mitf expression in immortalized mouse embryo fibroblasts (MEFs) from Fanca-KO (Fanca–/–) or Fancc-KO (Fancc–/–) mice. Confluent MEFs were subcultured at a dilution of 1/10, and proteins and RNAs were extracted and analyzed daily for 4 successive days. One day after cell seeding, we observed no major differences in Mitf expression at the protein or mRNA level between WT and Fanca–/– or Fancc–/– MEFs (Figure 1, D and E). However, later, Mitf expression increased progressively in both Fanca–/– and Fancc–/– MEFs, whereas in WT cells, it remained barely detectable (Figure 1, D and E, and data not shown). Finally, as observed in human lymphoblasts, Mitf expression was indistinguishable between Fancd2–/– and WT MEFs, but clearly lower in these cells than in Fanca–/– or Fancc–/– MEFs (Figure 1F and data not shown).
Together, our previous data robustly demonstrate that loss of function of key components of the FANC core complex is associated with high expression of MiTF at both the RNA and protein levels.
SMAD and p38 signaling pathways are necessary but not sufficient to activate MiTF expression in FA MEFs.
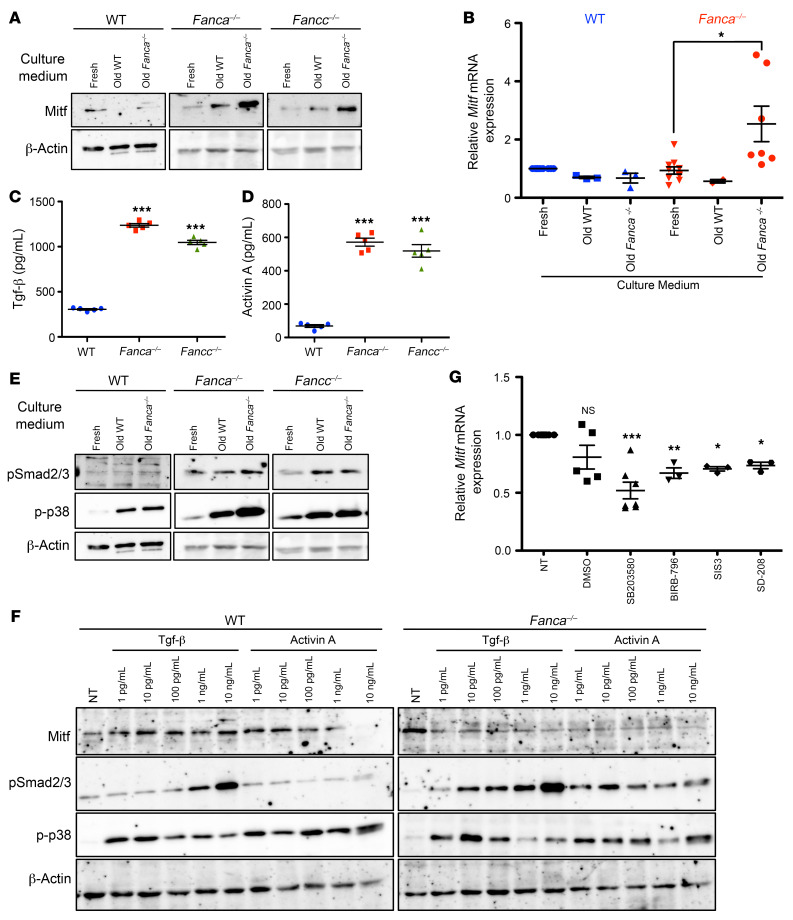
The increase in Mitf expression in FANC pathway–deficient MEFs with increasing time in the same culture medium (Figure 1, D and E) could be induced by cell-to-cell contacts or in an autocrine manner by cell-secreted factor or factors. Consistent with the latter hypothesis, we observed that Mitf expression is robustly induced at both the protein and RNA levels by 24 hours after replating FANC pathway–deficient MEFs in a “conditioned” medium in which the same cells previously grew for 72 hours (Figure 2, A and B, and data not shown). Notably, WT-derived 72-hour-old conditioned medium was unable to significantly induce Mitf in Fanca–/– or Fancc–/– MEFs. Similarly, Fanca–/–– or Fancc–/––derived conditioned medium was unable to induce Mitf in WT MEFs (Figure 2, A and B, and data not shown).
Figure 2. Mitf expression in FA is dependent on cell-secreted factor(s).
(A) Representative Western blots illustrating Mitf expression in extracts from WT, Fanca–/–, or Fancc–/– MEFs collected 24 hours after seeding in fresh or 72-hour conditioned (old) medium from WT or Fanca–/– MEF cultures. β-Actin was used as a loading control (n = 3). (B) qRT-PCR of Mitf expression in WT or Fanca–/– MEFs. MiTF expression in each cell line was first normalized to that of Oaz1 (internal control) and then to the Mitf/Oaz1 ratio in WT MEFs, which was set as 1 in each experiment. Data are shown as mean ± SEM of n experiments (for WT cells, n = 10 [fresh] and n = 3 [old WT and old Fanca–/–]; for Fanca–/– cells, n = 10 [Fresh], n = 2 [Old WT] and n = 7 [Old Fanca–/–]). (C and D) TGF-β (C) or activin A (D) concentrations (pg/mL) measured by ELISA in the supernatants of WT, Fanca–/–, or Fancc–/– MEFs maintained for 48 hours in medium without serum. Data are shown as mean ± SEM (n = 5). (E) Representative Western blots illustrating pSmad2/3 or p-p38 expression in the same extracts and conditions described in A. Since these are the same cell extracts analyzed in A, the β-actin blot is the same. (F) Representative Western blots of WT and Fanca–/– MEFs treated with the indicated concentrations of TGF-β or activin A and collected 24 hours later. β-Actin was used as a loading control (n = 3). (G) Mitf expression evaluated by qRT-PCR in Fanca–/– MEFs 2 hours following treatment with solvent (DMSO), p38 inhibitors (SB203580 [10 μM] or BIRB-796 [10 μM]), or pSmad2/3 inhibitors (SIS3 [2.5 μM] or SD208 [10 μM]). Data are shown as mean ± SEM of n = 5 (NT, DMSO, SB203580) or n = 3 (BIRB-796, SIS3, and SD208) experiments. Statistical significance was assessed using an unpaired 2-tailed t test with Welch’s correction (B) or 1-way ANOVA with Dunnett’s correction (C, D, and G). *P < 0.05; **P < 0.01; ***P < 0.001.
Thus, it is conceivable that Mitf is induced in response to the accumulation in the culture medium of factor or factors secreted by the Fanc-deficient cells themselves. FA cells appear able to secrete and respond to such factor(s), whereas WT cells are poor secretors and nonresponders.
Next, we performed ELISA to determine the presence, if any, of known extracellular factors in the culture medium of MEFs. To better perform our analysis, 2 days after dilution, the complete culture medium was washed out, and the cells were fed 2 more days with medium without serum (a longer incubation affects cell morphology, indicating the presence of stress and survival). The serum-free medium was collected and filtered to eliminate detached cells and cellular debris to perform ELISA. We failed to detect TNF-α or IL-1β in FA-derived conditioned medium, but revealed 4 to 6 times higher concentrations of 2 TGF family members (43, 44), TGF-β and activin A (Figure 2, C and D). Finally, the conditioned medium was analyzed by mass spectrometry. Again, we identified high levels of both TGF-β and activin A in the culture medium isolated from FANC pathway–deficient MEFs, but did not distinguish other growth factors differentially expressed between WT and FA MEFs (data not shown). Thus, 2 alternative approaches validated that cultured FANC pathway–deficient MEFs secrete TGF-β and activin A.
TGF-β and activin A determine cell fate and hematopoiesis by signaling mainly through the Smad and p38 pathways (8, 43–46), which were previously described as chronically activated in FA cells (24, 32). In Fanca–/– or Fancc–/– MEFs maintained for 24 hours in 72-hour-old conditioned medium, we observed a modest but reproducible increase in Smad2/3 phosphorylation and extremely strong phosphorylation of p38 (Figure 2E). The culture of WT MEFs in conditioned medium (both from FANC pathway–proficient or FANC pathway–deficient cells) induced p38 but not Smad2/3 phosphorylation (Figure 2E). Given our observations, we decided to treat MEFs with increasing doses of TGF-β or activin A and determine Mitf expression as well as p38 and Smad2/3 phosphorylation 24 hours later. Since ELISA estimated the concentration of TGF-β and activin A in the 72-hour-old medium at approximately 1 ng/mL, we tested doses ranging from 1 pg/mL to 10 ng/mL for each growth factor (Figure 2F). Phosphorylation of p38 was similarly observed in response to TGF- or activin A in both WT and Fanca–/– MEFs. Following TGF-β exposure, Smad2/3 phosphorylation was induced by 1 pg/mL in Fanca–/– MEFs and by 1 ng/mL in WT MEFs, whereas in response to activin A, it was observed only in Fanca–/– MEFs. Finally, whereas in TGF-β or activin A–treated WT cells, Mitf slightly increased, we did not observe induction of Mitf, but rather its downregulation in Fanca–/– MEFs exposed to TGF-β or activin A (Figure 2F).
Therefore, exposure to TGF-β, activin A, or both (data not shown) in FA cells is not sufficient for Mitf induction, despite the induction of pSmad2/3 and phosphorylated p38 (p-p38), suggesting that Mitf expression in FA MEFs requires the presence of still undetermined factors secreted into the culture medium by FANC pathway–deficient cells or that exposure to TGF-β and/or activin A also induces Mitf inhibitor(s). However, to more directly ascertain the involvement of pSmad2/3 and/or p-p38 in MiTF expression in FA, we evaluated the expression of the transcription factor by quantitative RT-PCR (qRT-PCR) in MEFs exposed to either p38 MAPK inhibitors SB203580 and BIRB-796 or pSmad2/3 inhibitors SIS3 and SD208. All inhibitors reduced Mitf expression (Figure 2G), supporting the involvement of both signaling pathways in Mitf expression downstream of FANC pathway loss of function.
Since the observed strong p38 and Smad2/3 activation induced by exogenous exposure to TGF-β or activin A failed to induce Mitf (Figure 2F), whereas the inhibition of their activity significantly reduced Mitf expression (Figure 2G), it appears that p38 and Smad2/3 signaling are necessary but not sufficient to induce Mitf expression. Alternatively, exposure to exogenous TGF-β or activin A could also activate repressor(s) of Mitf expression.
Multiple routes lead to MiTF induction in FA cells.
As in MEFs, high MiTF expression in human FA lymphoblasts (Figure 1, A and B, and Figure 3A) is also associated with increased phosphorylation of SMAD2/3 and p38 (Figure 3, B–D), and MiTF is downregulated by their inhibition (Figure 3, E and H, and data not shown). Subsequently, we examined the presence of TNF-α, TGF-β, and/or activin A in the culture medium of human lymphoblasts. As expected (24, 29), we identified increased levels of TNF-α in FA-derived conditioned medium, but we did not detect differences in either TGF-β or activin A levels in the culture medium from FANC pathway–proficient or FANC pathway–deficient cells (Figure 3F and data not shown). Moreover, exposure to TGF-β, activin A, or TNF-α was unable to induce or modify MiTF expression in WT, FANCA–/–, or FANCC–/– cells (Figure 3, C, D, and G, and data not shown). Thus, as previously observed in MEFs, other still-undetermined factors contribute together with TGF-β, activin A, or TNF-α to MiTF expression, or the exogeneous treatments also induce unidentified MiTF inhibitor(s).
Figure 3. Multiple routes lead to MiTF expression.
(A and B) Relative MiTF (A) or pSMAD2/3 (B) levels evaluated by FACS in indicated cell lines. Data are shown as mean ± SEM. (A) n = 14 (HSC93, HSC99), n = 4 (HSC72, HSC536). (B) n = 6 (HSC93, HSC99), n = 4 (HSC72), and n = 3 (HSC536). (C and D) Western blots illustrating MiTF, pSMAD2/3, and p-p38 expression in extracts from WT, FANCA–/–, or FANCC–/– cells, treated (6 hours) or not treated with 10 ng/mL of TGF-β, activin A, or TNF-α. Vertical lines (C) separate noncontiguous samples migrated on the same gel. Vertical lines in D were added to improve readability. (E) Western blots showing MiTF and p-p38 expression in FANCA–/– or FANCC–/– cells treated with the p-p38 inhibitor SB203580. β-Actin was used as a loading control. (F) TNF-α concentration (pg/mL) measured by ELISA in supernatants from indicated WT or FANC pathway–deficient cells 48 hours after seeding. Data are shown as mean ± SEM (n = 5). (G) Relative MiTF expression evaluated by qRT-PCR in FANCA–/– cells (HSC99) 6 hours following exposure to 10 ng/mL TGF-β, activin A, or TNF-α. Data are shown as mean ± SEM (n = 3). (H) Relative MiTF expression evaluated by qRT-PCR in FANCA–/– lymphoblasts treated for 2 hours with DMSO, SB203580 (10 μM), BIRB-796 (10 μM), SIS3 (2.5 μM), SD208 (10 μM), curcumin (50 μM), LY294002 (10 μM), and NBD (1 μM). The results were normalized against Actin expression and calculated relative to untreated cells (n = 5 [NT, DMSO, SB203580, BIRB-796, SIS3, SD208, SB203580 + SIS3], n = 4 [curcumin], and n = 3 [LY294002 and NBD]). (I) Western blots showing MiTF and pAKT expression in WT and FANC pathway–deficient cells. β-Actin was used as a loading control. (J) Summary of our previous observations combined with data from the literature. Statistical significance was assessed using an unpaired 2-tailed t test with Welch’s correction (F) or 1-way ANOVA with Dunnett’s correction (A, B, G, and H). *P < 0.05; **P < 0.01; ***P < 0.001.
TNF-α oversecretion in FA was associated with activation of NF-κB (24, 47), which is known to target the MiTF gene downstream of TNF-α (48). Thus, we examined MiTF expression by qRT-PCR in cells treated not only with p38 and/or pSMAD2/3 inhibitors, but also with curcumin or NEMO-binding domain binding peptide (NBD), NF-κB inhibitors. In response to p38 inhibition by SB203580 or SMAD2/3 inhibition by SIS3, NF-κB inhibition by curcumin or NBD decreased MiTF expression (Figure 3H). Notably, the combined inhibition of SMAD, p38, and/or NF-κB signaling had minimal additional effects (Figure 3H and data not shown). Finally, we also treated FA cells with LY294002, which is an inhibitor of AKT, a kinase that is overactivated in FA (Figure 3I and ref. 27). AKT inhibition also downregulated MiTF expression (Figure 3H).
Thus, it seems that several intrinsic pathways converge and collaborate to allow the unrestrained maintenance of MiTF expression (Figure 3J), which, in light of its involvement in melanocytes, mast cells, osteoclasts, and HSC physiology, could markedly contribute to the FA phenotype.
p38 Is constitutively activated in HSCs from Fanca–/– mice.
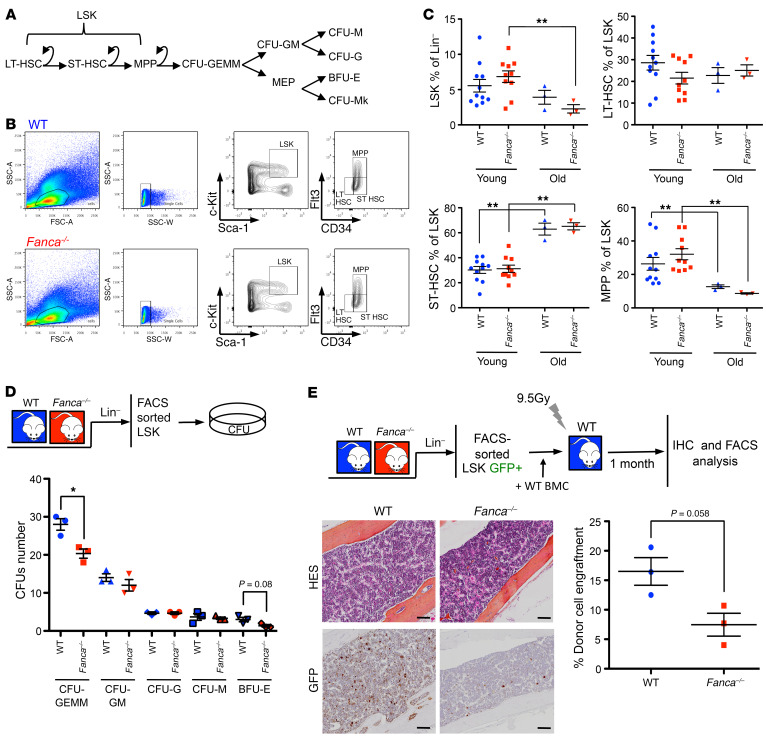
Human immortalized lymphoblasts and the previously used MEFs represent useful experimental models, but they remain a surrogate for real life. Therefore, to extend our observations to in vivo settings and to identify their pathological consequences, we chose to monitor Mitf expression in BM-derived cells from Fanca–/– mice and its eventual dependency on p38 signaling. It is generally accepted that FANC pathway–deficient mice present mild hematopoietic phenotypes (49). The Fanca–/– model we used (50) is known to present reduced platelet counts, which mimics patient thrombocytopenia, and altered immunoglobulin formation in association with alterations in the DNA damage response and B cell differentiation (51–53), abnormalities already reported in FA patients (54). Nevertheless, Fanca–/– and WT mice, independently of their ages (3–6 months old, i.e., young, or more than 1 year old, i.e., old) present similar levels of Lin–Sca+Kit+ (LSK) cells and their 3 subpopulations, long-term HSCs (LT-HSCs, Flt3–CD34–), short-term HSCs (ST-HSCs, Flt3–CD34+), and multipotent progenitors (MPPs, Flt3+CD34+), on which the maintenance of hematopoiesis depends (Figure 4, A–C). Nevertheless, compared with their WT counterparts, LSK cells from Fanca–/– mice presented a decreased ability to form colony-forming unit–granulocyte, erythroid, macrophage, megakaryocyte (CFU-GEMM), and burst-forming unit–erythroid (BFU-E) colonies (Figure 4D). We subsequently performed a competitive transplantation assay using GFP transgenic Fanca–/– and Fanca+/+ mice as donors and non-GFP mice on the same genetic background as recipients. Four weeks later, the presence of GFP-positive cells in the BM was visualized by IHC staining, and the frequency of circulating GFP-positive cells was determined by FACS analysis, demonstrating that, compared with WT cells, Fanca–/– hematopoietic stem and progenitor cell (HSPCs) have a significantly reduced ability to reconstitute the peripheral blood (Figure 4E).
Figure 4. Fanca–/– mice display altered BM homeostasis.
(A) Schematic representation of the HSC hierarchy and colony stages. (B) Gating strategy for the analysis of the LSK fraction and its 3 subpopulations, long-term HSCs (LT-HSCs, Flt3–CD34–), short-term HSCs (ST-HSCs, Flt3–CD34+), and multipotent progenitors (MPPs, Flt3+CD34+). (C) Percentages of LSK cells, LT-HSCs, ST-HSCs, and MPPs calculated among the total numbers of Lin– cells in young (3 to 6 months old) and old (more than 1 year old) WT and Fanca–/– mice. Each point represents a single mouse. (D) WT and Fanca–/– CFUs of different colony types after LSK plating and 7 days of culture (n = 3). (E) Bottom left: representative images after immunohistochemical staining of GFP-positive cells in the BM sections of recipients. Bottom right: graph of donor chimerism in the circulating blood 4 weeks after LSK+ competitor transplants (n = 3). Scale bars: 50 μm. Statistical significance was assessed using an unpaired 2-tailed t test with Welch’s correction (C, D, and E). *P < 0.05; **P < 0.01.
Given our previously published data (51–53) and observations from other FA mouse models (55, 56), it appears that Fanca deficiency restrains the clonal growth of the branch that leads from long-term HSCs (LT-HSCs) to BFU-E and CFU-megakaryocyte (CFU-Mk) colonies via CFU-GEMM formation (Figure 4A), even if the observed abnormalities have no major consequences on BM homeostasis during the mouse lifetime.
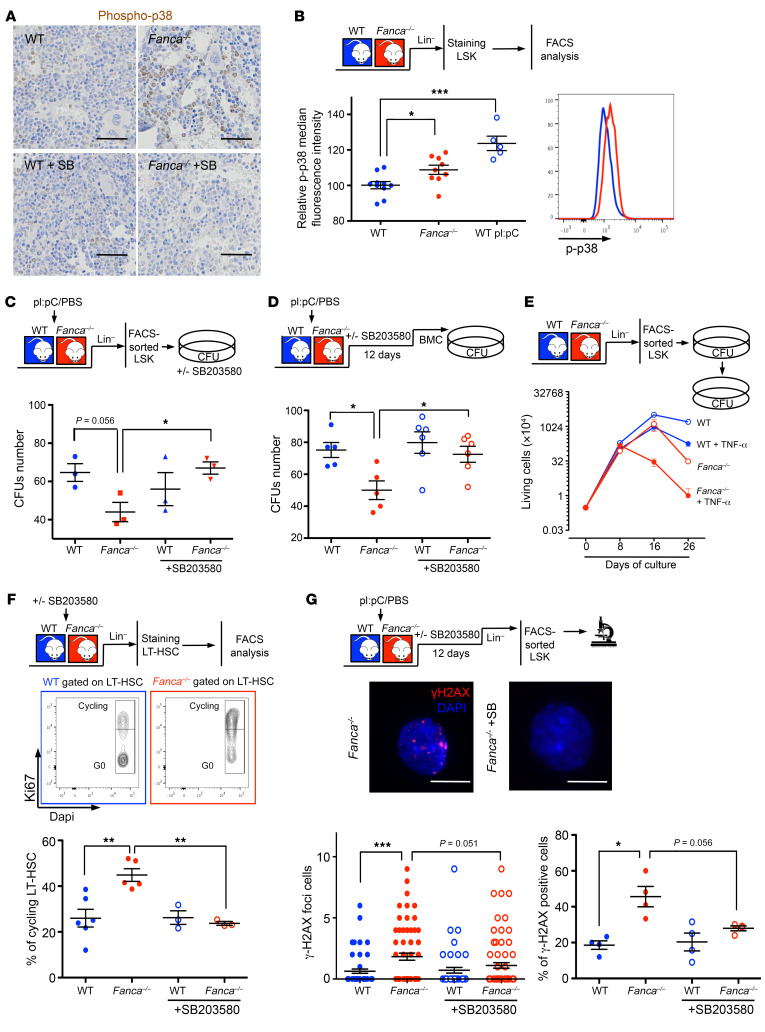
Next, we evaluated p-p38 levels in BM and LSK cells from WT and Fanca–/– mice. IHC analysis of BM sections demonstrated a stronger p-p38 signal in Fanca–/– mice than in their WT counterparts, and this signal decreased following SB203580 treatment of animals (Figure 5A). FACS analysis of the Lin–-enriched population detected a p-p38 signal in Fanca–/– LSK cells, whose intensity was intermediate between that observed in WT LSK cells before and after exposure to poly(I:C) (pI:pC), a widely used inducer of hematopoietic stress that awakens and mobilizes LT-HSCs (Figure 5B and ref. 57). Compared with those from their WT counterparts, Fanca–/– LSK cells isolated from pI:pC-treated mice demonstrated reduced colony formation proficiency, a defect that was rescued by in vitro culture in the presence of the p38 inhibitor SB203580 (Figure 5C). Subsequently, we administered the p38 inhibitor to pI:pC-treated mice for 12 days before isolating BM cells to monitor their capacity to form CFUs in vitro, which appeared to be largely rescued (Figure 5D).
Figure 5. p38 Abnormalities in HSCs from Fanca–/– mice.
(A) Immunohistochemical staining of p-p38 in BM sections of WT and Fanca–/– mice treated or not with 50 mg/kg SB203580 i.p. daily for 12 days. Scale bars: 100 μm. (B) p-p38 Protein expression level measured by FACS (right) in LSK cells from untreated WT and Fanca–/– mice or from WT mice treated i.p. with pI:pC (5 mg/kg) 24 hours before. Each point represents a single mouse. (C) Colony-forming hematopoietic progenitor frequency counted 7 days after plating of LSK cells from WT and Fanca–/– mice (n = 3) treated 24 hours before with pI:pC (5 mg/kg). LSK cells were cultured with or without SB203580 (20 μM). (D) WT and Fanca–/– colony-forming hematopoietic progenitor frequency after a dose of pI:pC and in vivo treatment with SB203580 (50 mg/kg). Each point represents a single mouse. (E) Successive seeding of WT and Fanca–/– progenitors with or without TNF-α in MethoCult 3434 medium after replating at 3 × 103 cells per dish in triplicate. Each point represents the mean value of 3 independent experiments started with 3 different mice. (F) Middle: Gating strategy to assess the percentage of cycling cells through the use of the Ki67+ intracellular marker. Bottom: percentages of cycling LT-HSCs from WT and Fanca–/– mice treated or not with a dose of SB203580 (50 mg/kg) for 24 hours. Each point represents a single mouse. (G) Bottom: Quantification of γ-H2ax foci per cell (left) or γ-H2ax-positive LSK cells (right) sorted from mice treated with a dose of pI:pC (5 mg/kg), followed or not by successive treatment with SB203580. Middle: representative image of Fanca–/– or Fanca–/– LSK cells treated with SB203580. Scale bars: 10 μm. Each point represents a single mouse. Statistical significance was assessed using an unpaired 2-tailed t test with Welch’s correction (C, D, F, and G) or 1-way ANOVA with Dunnett’s correction (B). *P < 0.05; **P < 0.01; ***P < 0.001.
Finally, we performed IHC analysis on BM sections from Fanca–/– and WT mice, looking at the presence of TNF-α, TGF-β, and/or IL-1β as possible inducers of p38, and we failed to observe differences between the 2 genotypes (data not shown). However, in vitro–cultured LSKs from Fanca–/– mice demonstrated higher sensitivity to chronic exposure to TNF-α than their WT counterparts, mimicking human patients and demonstrating that the cells are sensitive to this cytokine (Figure 5E).
In search for a signal or an event responsible for the observed constitutive p38 activation in FA, we reasoned that it could be a “pathological” response to an otherwise physiological input. Resident BM LT-HSCs exist mostly in a quiescent state (58) that they leave to maintain both steady-state and stress-induced hematopoiesis, ensuring BM and/or bloodstream cell replenishment. Although dispensable for steady-state hematopoiesis, p38 activation appears necessary for stress hematopoiesis and exhibits a transient increase in cycling LT-HSCs (16). In accordance with the idea that FANC pathway loss of function leads to a “hematopoietic stress situation,” and as previously observed by others (32, 57), the frequency of cycling LT-HSCs was significantly higher in Fanca–/– mice than in their WT counterparts (Figure 5F). LSKs from Fanca–/– mice also presented an increased level of spontaneous DNA damage (Figure 5G), which was recognized as a key factor underlying defects in regaining quiescence (57, 59, 60). Similar to observations in Fancd2-deficient mice in response to Smad signaling inhibition (32), p38 inhibition rescued both dormancy and DNA damage in Fanca–/– mice (Figure 5, F and G).
Thus, we validated that the p38 signaling pathway is chronically activated downstream of FANC pathway deficiency and is associated with alterations in colony-forming capability, quiescence, and proliferation as well as with the accumulation of DNA damage, all of which are rescued by p38 inhibition.
p38-Dependent Mitf overexpression participates in BMF in Fanca–/– mice.
Therefore, we established that p38 inhibition normalizes HSC proliferation and colony-forming capability and limits DNA damage accumulation. Knowing that Mitf transient activation is necessary for HSC proliferation during stress hematopoiesis (16) and having identified it as a downstream target of p38 in FA cells, we investigated its upregulation in Fanca–/– BM cells and its potential involvement in the hematopoietic impairment of Fanca–/– mice.
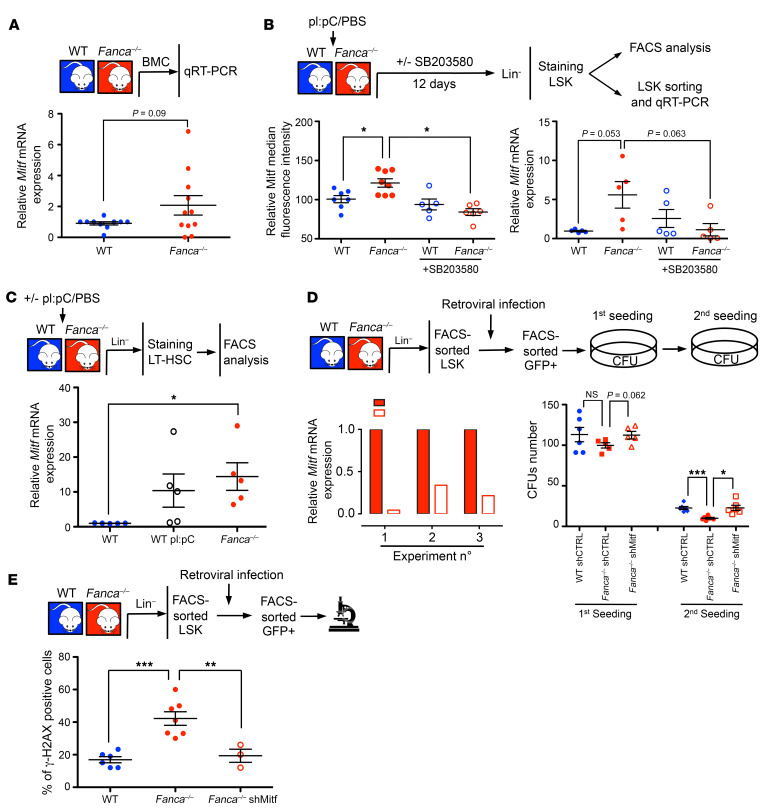
First, by qRT-PCR analysis, we demonstrated slightly increased expression of Mitf RNA in BM cells from Fanca–/– mice compared with BM cells from their WT counterparts (Figure 6A). Moreover, Mitf overexpression was also observed at both the protein and RNA levels in Fanca–/– LSK cells, and it was normalized by in vivo exposure to SB203580 (Figure 6B). Finally, in accordance with previously published data (16) and in association with p-p38 induction (Figure 5B), pI:pC treatment also increased Mitf expression in LSK cells isolated from WT mice to a level similar to that observed in Fanca–/– cells (Figure 6C).
Figure 6. Mitf overexpression participates in altered BM homeostasis in Fanca–/– mice.
(A) qRT-PCR analysis of Mitf expression in BM from WT and Fanca–/– mice. The results were calculated relative to WT BM and normalized against Oaz1. Each point represents a single mouse. (B) Left: Mitf protein expression measured by FACS in LSK cells from mice treated with a single dose of pI:pC (5 mg/kg) followed or not by SB203580 (50 mg/kg) injections once a day for 12 days. Right: qRT-PCR analysis of Mitf gene expression in sorted LSK cells from mice treated with a single dose of pI:pC (5 mg/kg) followed or not by SB203580 (50 mg/kg) injections once a day for 12 days. The results were calculated relative to WT LSK cells and normalized against Oaz1. Each point represents a single mouse. (C) Mitf expression in WT, WT pI:pC-treated, and Fanca–/– mice. (D) Left: validation of Mitf silencing after retroviral infection with a GFP-tagged shMitf-containing vector and GFP-positive LSK cells sorted 48 hours after infection. Right: colony-forming hematopoietic progenitor frequency from LSK cells isolated from WT and Fanca–/– animals infected with retroviruses expressing shScramble-GFP or shMitf-GFP (n = 6). (E) Quantification of γ-H2AX–positive LSK cells isolated from WT and Fanca–/– animals infected with retroviruses expressing shMitf-GFP (n = 6). Statistical significance was assessed using an unpaired 2-tailed t test with Welch’s correction (A, B, D, and E) or 1-way ANOVA with Dunnett’s correction (C). *P < 0.05; **P < 0.01; ***P < 0.001.
Next, we isolated LSK cells from Fanca–/– mice, infected them with a retroviral construct bearing a shRNA sequence targeting Mitf, and analyzed the effect of Mitf downregulation (Figure 6D) on colony formation by a replica plating approach (Figure 6D) and on DNA damage by examining the presence and level of γ-H2ax foci (Figure 6E). As previously observed following p38 (Figure 5G) or Smad signaling (32) inhibition, shRNA-infected, Mitf-downregulated Fanca–/– LSK cells exhibited recovered colony-formation capabilities (Figure 6D) and rescued DNA damage (Figure 6E).
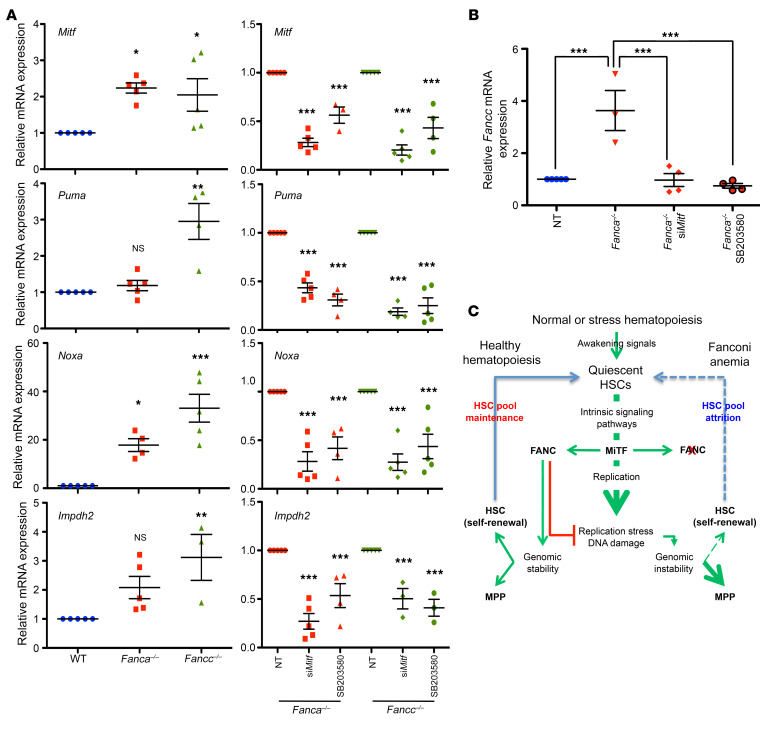
To add insight into the role of Mitf in FA, we looked at several downstream end points. The siRNA-mediated downregulation of Mitf or the inhibition of p38 signaling significantly reduced the expression of both Noxa and Puma (Figure 7A), 2 p53 targets potentially involved in the proapoptotic phenotype that characterizes FA cells in vitro (61, 62) and in vivo (32, 57, 63). Mitf downregulation or the inhibition of p38 signaling also reduced the expression of Impdh2, which is involved in purine metabolism (16), reported as deficient in FA (64) and possibly associated with the replication abnormalities of the FANC pathway–deficient cells. Notably, in accordance with the direct involvement of Mitf in Fanc gene expression (17, 18), the level of Fancc mRNA in Fanca–/– cells (Figure 7B) or of Fanca mRNA in Fancc–/– cells (data not shown) was higher than in WT cells; again, this change is normalized by Mitf downregulation or p38 inhibition.
Figure 7. Mitf downregulation or p38 inhibition normalizes gene expression in Fanca–/– or Fancc–/– cells.
(A) qRT-PCR analysis of Mitf, Puma, Noxa, and Impdh2 expression in WT, Fanca–/–, or Fancc–/– MEFs under basal conditions after siRNA-mediated MiTF downregulation or a 2-hour treatment with the p38 inhibitor SB203580. In each individual experiment, Mitf, Puma, Noxa, and Impdh2 expression were first normalized to that of Oaz1 (internal control) and then normalized to the Mitf, Puma, Noxa, or Impdh2/Oaz1 ratio of the WT MEFs, which was set as 1 in each experiment. Data are shown as mean ± SEM of n = 5 experiments. (B) qRT-PCR analysis of Fancc expression in WT or in Fanca–/– MEFs under basal conditions after siRNA-mediated Mitf downregulation or a 2-hour treatment with the p38 inhibitor SB203580. (C) Schematic model combining our observations with state-of-the-art knowledge concerning the effects of FANC pathway inactivation on BM homeostasis. Statistical significance was assessed using 1-way ANOVA with Dunnett’s (A) or Bonferroni’s (B) correction. *P < 0.05; **P < 0.01; ***P < 0.001.
Thus, Mitf overexpression may appear as a strategy for the FANC-deficient cell to override its own replication problems, sustaining the production of proteins involved in synthesizing DNA (Impdh2) and safeguarding replication (the FANC proteins, obviously futilely), which exacerbates, in a pernicious cycle, the replication stress of the cells. Indeed, as described following Smad2/3 inhibition (32), p38 inhibition and/or Mitf downregulation reduce this pernicious cycle and, consequently, the level of DNA damage and the “toxic” DNA damage response associated with loss of function of the FANC pathway (Figure 7C).
Discussion
In the present work, we investigated the impact of the constitutive activation of stress signaling pathways on HSC maintenance and function in FA by mainly focusing on p38 and one of its known targets, MiTF. Our observations indicate that unrestrained expression of MiTF characterizes FANC core complex–deficient FA cells in vitro and affects the repopulation capacities of the HSCs in Fanca–/– mice. In accordance with previous observations, we demonstrated that MiTF overexpression is associated with the aberrant, constitutive activation of several intrinsic cellular pathways, including the p38 MAPK, SMAD, NF-κB, and AKT cascades, and with the excessive secretion of some cytokines and growth factors, including TGF-β, activin A, and/or TNF-α. Although exogenous exposure to TGF-β, activin A, or TNF-α was unable to induce MiTF in FA cells, the pharmacological inhibition of p38, SMAD2/3, NF-κB, and/or AKT appeared sufficient for its downregulation in vitro, linking MiTF overexpression in FA to the unscheduled activation of stress pathways. We demonstrated that p38 inhibition or Mitf downregulation rescues the dormancy and repopulation capacities of HSCs and reduces DNA damage accumulation. Having demonstrated that SMAD2/3 or p38 inhibition restrains Mitf expression in vitro, it is likely that the recovery of HSC homeostasis in FA mice treated with SMAD (32) or p38 (this work) inhibitors can be ascribed to Mitf downregulation.
Previous works suggested that BMF in FA is dependent upon the attrition of the HSC pool due to excessive accumulation of unrepaired DNA damage in association with slight but robust activation of the growth-inhibitory p53-to-p21 axis and increased cell death (23, 57). Notably, shRNA-mediated downregulation of the TGF-β pathway (32), p38 inhibition, or MiTF silencing reduced p53 activity/activation. This supports otherwise compelling data suggesting that pathways that exert both positive and negative effects on HSC homeostasis, including TGF-β– and TNF-α–associated signaling pathways, have important consequences on FA, not only on BM physiology, but also on the chromosomal fragility of the cells. Indeed, TGF-β or TNF-α signaling inhibition reverses BMF in Fancd2–/– mice, rescues erythroid failure in cultured BM from FA patients, and reduces DNA damage accumulation and chromosomal fragility in FANC pathway–deficient cells (29, 32, 33).
Thus, several events cooperate to create chronic hematopoietic stress conditions that corrupt BM homeostasis in FA by switching on and maintaining constitutive activation of intracellular signaling pathways that are otherwise transiently activated and tightly regulated. Moreover, in FA patients, natural infections or bleeding lead to proinflammatory and cytostatic/cytotoxic signals, from which mouse models largely escape, and these signals could accelerate/exacerbate BM decline, fostering stress hematopoiesis. Notably, the FA mouse model or cells treated with PS or TNF-α, reproducing “natural” human situations, rapidly develop BMF and eventually preleukemia (65, 66).
When dormant HSCs are mobilized, p38 and other signaling pathways are transiently activated to support proliferation and MiTF expression, which sustains purine metabolism (16), to fuel DNA synthesis and induces FANC gene expression (17, 18) to help replication and cell division, preventing genetic instability, senescence, and cell death (21, 67, 68). The activated FANC pathway, which participates in the DNA damage response and in genetic stability maintenance, allows the awakened HSCs to undergo asymmetric division, generating a daughter cell that progresses to hematopoietic differentiation, whereas a sister cell would reacquire dormancy. This self-renewal process allows maintenance of the stem cell reservoir (Figure 7C). Even in healthy individuals, when the level of DNA damage surpasses a determined threshold, both sister cells eventually differentiate or succumb (59), limiting self-renewal. Thus, we speculated that the DNA damage spontaneously generated during the physiological process of DNA replication that accompanies the mobilization of HSCs cannot be correctly repaired as a consequence of FANC pathway loss of function, leading to an activated DNA damage response; i.e., the observed activation of the ATM/ATR-p53-p21 cascade and the accumulation of genetic instability pushed the cells toward differentiation at the expense of quiescence (Figure 7C). To equilibrate the loss of quiescent cells in FA, the BM continues to mobilize dormant HSCs to improve their pool by self-renewal, promoting persistent activity of the p38-MiTF axis, which maintains the cells in a pernicious circle characterized by (a) the accumulation of DNA damage, (b) impairment of dormancy reestablishment, (c) progressive HSPC attrition, and eventually, (d) AML caused by increased genetic instability in cycling HSCs. Accordingly, pharmacological or genetic inhibition of growth and proinflammatory signals as well as intracellular signaling pathways ameliorates DNA damage and genetic instability by limiting the proliferative potential of HSCs in FA. Thus, FANC pathway activation might be part of a feedback loop that restrains MiTF expression and proliferation and allows the recovery of homeostasis (Figure 7C).
Finally, our findings have key implications for the care of FA patients. Despite enormous progress in the exploration of FA pathogenesis, BM transplantation, which is associated with a high risk of complications, remains the only therapy. Therefore, new strategies are urgently needed to enhance the functions of the BM in FA patients. The potential to selectively reverse BMF by inhibiting p38 has a marked advantage over the use of proinflammatory cytokine inhibitors by themselves. Notably, targeting p38 has already been validated in different clinical trials for the treatment of rheumatoid arthritis and advanced cancers (38). Here, we demonstrated that p38 inhibition increased the overall fitness and function of FA HSCs and enhanced their quiescence. The beneficial action of SB203580 appears to be specific to mutant FA cells exhibiting high levels of MiTF, indicating that MiTF is a sensitive biomarker for predicting the amelioration of hematopoiesis by p38 inhibitors in FA. Clearly, further studies are required to demonstrate whether targeting stress signaling pathways or MiTF activity truly represents a new approach to treat some clinical features of FA patients.
Methods
Cell lines and culture conditions.
Human lymphoblasts from healthy donors (HSC93 [RRID:CVCL_G049], SNW646 [gift of V.A. Bohr, NIH, Baltimore, Maryland, USA], and GM0131 [Coriell GM17202, RRID:CVCL_7275]) and FA patients (HSC72 [RRID:CVCL_AK37] and HSC99 [RRID:CVCL_G050]), both the cells lines bearing inactivating mutations in FANCA (HSC536 [RRID:CVCL_G045] [FANCC mutated], GM16756 [FANCD2 mutated, RRID:CVCL_G041], and FANCM–/–; ref. 69) and the corrected cell lines [HSC72CORR and HSC536CORR], were routinely maintained in RPMI 1640/GlutaMAX (Gibco; Life Technologies) supplemented with 13% FCS and 100 U/mL penicillin/50 μg/mL streptomycin at 37°C in a humidified atmosphere containing 5% CO2. All HSC cell lines, a gift from M. Buchwald (Hospital for Sick Children, Toronto, Ontario, Canada) (70), have been routinely used in our laboratory for at least 20 years. The FANCM–/– cell line was derived in our laboratory (69). MEFs from WT, Fanca–/–, Fancc–/–, and Fancd2–/– mice (gift from the Fanconi Anemia Research Fund [FARF], Eugene, Oregon, USA) were grown in DMEM (Gibco; Life Technologies) supplemented with 10% FCS, 100 units/mL penicillin, and 50 μg/mL streptomycin at 37°C in a humidified atmosphere containing 5% CO2. Cultures were routinely tested for mycoplasma contamination.
Chemicals, peptides, and recombinant proteins.
We used SB203580, a p38 MAPK inhibitor (Calbiochem, 559389); Doramapimod (BIRB-796), a p38 MAPK inhibitor (Selleckchem, S1574); SIS3-HCl, a SMAD3 inhibitor (Selleckchem, S7959); SD-208, a TGF-βRI (ALK5) inhibitor (Selleckchem, S7624); curcumin, an NF-κB inhibitor (MilliporeSigma, 239802); NBD (MilliporeSigma, 480025); LY294002, a pAKT inhibitor (Calbiochem, 440202); pI:pC (InvivoGen, 31852-29-6); recombinant murine TNF-α (Peprotech, 315-01A); recombinant murine IL-3 (Peprotech, 213-13); recombinant murine IL-6 (Peprotech, 216-16); recombinant murine SCF (Peprotech, 250-03); recombinant murine Flt3L (Peprotech, 250-31L); recombinant human TGF-β1 (HEK-293 derived, Peprotech, 100-21); recombinant human/murine/rat activin A (E. coli derived, Peprotech, 120-14E); and recombinant human TNF-α (E. coli derived, Peprotech, 300-01A).
Protein extraction and analysis.
The collected cells were disrupted in lysis buffer (50 mM Tris-HCl pH 7.9, 40 mM NaCl, 1 mM MgCl2, 0.1% SDS, and 1% Benzonase [Merck, 70746-3] supplemented with protease and phosphatase inhibitors [Roche, 05892791001 and 04906837001]). After incubation for 20 minutes at room temperature, protein concentrations were determined using the Bradford assay (Bio-Rad, 500-0006), and the samples were combined with 4× Laemmli buffer containing β-mercaptoethanol and denatured via boiling at 98°C. The proteins (20 μg) were separated using SDS-PAGE, transferred onto a nitrocellulose membrane (GE Healthcare Life Sciences, Amersham Protran 0.2 μM NC, 10600001), and then exposed to the appropriate antibodies. The proteins were visualized using an enhanced chemiluminescence system (WesternBright ECL by Advansta, K-12045-050) and an ImageQuant LAS 4000 system (GE Healthcare Life Sciences). We used rabbit anti-FANCA (Bethyl, A301-980A), mouse anti-MiTF (C5, Abcam, ab12039), mouse anti-Vinculin (Abcam, ab18058), rabbit anti–phospho-p38 MAPK (pT180/pY182, Cell Signaling, 9211S), rabbit anti–phospho-SMAD2(S465/467)/SMAD3(S423/425) (D27F4, Cell Signaling, 8828S), rabbit anti–phospho-AKT(Ser473) (Cell Signaling, 9271), goat anti–β-actin (Abcam, ab8229), HRP-conjugated goat anti-mouse IgG (Bethyl, A90-516P), HRP-conjugated donkey anti-rabbit IgG (Bethyl, A120-108P), and HRP-conjugated donkey anti-goat IgG (Bethyl, A50-101P).
siRNA transfection.
MEFs at 50% confluence were transfected using INTERFERin (Polyplus) according to the manufacturer’s instructions. MiTF siRNA was purchased from Invitrogen: 5′-GGUGAAUCGGAUCAUCAAG-3′.
RT-PCR and qRT-PCR analysis.
Total RNA was isolated using the Maxwell RSC simplyRNA Cell Kit (Promega, AS1390) and reverse-transcribed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, K1622). qRT-PCR was performed using Maxima SYBR Green/ROX qRT-PCR Master Mix (Thermo Scientific, K0222) and a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). PCR was performed on cDNA using DreamTaq Green DNA Polymerase (Thermo Scientific, EPO713). The following primers (Eurogentec) were used for qRT-PCR: Pan-MiTF (forward, 5′-CGAGCTCATGGACTTTCCCTTA-3′ and reverse, 5′-CTTGATGATCCGATTCACCAAA-3′), actin (forward, 5′-GACGGCCAGGTCATCACTATTG-3′ and reverse, 5′-AGGAAGGCTGGAAAAGAGCC-3′), Oaz1 (forward, 5′-TTTCAGCTAGCATCCTGTACTCC-3′ and reverse, 5′-GACCCTGGTCTTGTCGTTAGA-3′), IMPDH2 (forward, 5′-CTTGCTGGTGTGGATGTAGTGG-3′ and reverse, 5′-GCCTCCAATGACCTGTAGACTG-3′), PUMA (forward, 5′-GCGGCGGAGACAAGAAGA-3′ and reverse, 5′-AGTCCCATGAAGAGATTGTACATGAC-3′), NOXA (forward, 5′-TCGCAAAAGAGCAGGATGAG-3′ and reverse, 5′-CACTTTGTCTCCAATCCTCCG-3′), FANCA (forward, 5′-CCCTGACTTCTTAAACAGCCTC-3′ and reverse, 5′-CTGGAATGTGAAACACGTACCT-3′), and FANCC (forward, 5′-TGCCTGAGAAGGATGGAGTGCT-3′ and reverse, 5′-CCGTCAGTTTCCAGCAGCACAA-3′). The following primers (Eurogentec) were used for PCR: GAPDH (forward, 5′-GAAGGTGAAGGTCGGAGT-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′), MiTF-A (forward, 5′-TGAAGAGCCCAAAACCTATTACGA-3′), MiTF-C (forward, 5′-CTTCAGTGGTTTTCCCACGAGCT-3′), MiTF-E (forward, 5′-AGTAGCAGGGGTTAGTAGGTGGAT-3′), and MiTF-M (forward, 5′-CCTTCTCTTTGCCAGTCCATCTTC-3′). A common MiTF reverse primer (5′-GATCAATCAAGTTTCCCGAGACAG-3′) was used for all MiTF isoforms.
ELISA.
TGF-β (MB100B and DB100B), activin A (DAC00B), TNF-α (MTA00B and DTA00D), and IL-1β (MLBOOC and DLB50) were measured by sandwich ELISA according to the manufacturer’s instructions (R&D Systems), and the analyses were performed using a VICTOR Multilabel Plate Reader (PerkinElmer).
Mice and genotyping.
Fanca–/– mice on an FVB background were described previously (50, 52). The mice were a gift of F. Arwert (Free University Medical Center, Amsterdam, Netherlands) and have been routinely maintained in our laboratory for more than 10 years. Fanca+/– mice were backcrossed with WT FVB/N mice (>10 generations). As Fanca–/– mice show severely reduced fertility, the WT and Fanca–/– mice used for analysis correspond to siblings derived from crossbreeding of heterozygous mice. Mouse genotyping was performed using DreamTaq Green DNA Polymerase (Thermo Fisher Scientific). The following primers (Eurogentec) were used: FANCA (forward, 5′-GGATCAGGCCTCGAGGCTGG-3′, reverse, 5′-TGCAGTAGCTCCTGTAGGCT-3′) and NEO (forward, 5′-GACTGGGCACAACAGACAATCGGCT-3′, reverse, 5′-TGATATTCGGCAAGCAGGCATCGCC-3′).
Flow cytometry and HSPC culture.
BM was harvested from mice by flushing tibias and femurs. RBCs were eliminated using EasySep Red Blood Cell Lysis Buffer (Stem Cell Technologies, 20110). Lin– progenitors were isolated using biotinylated antibodies from a lineage cell depletion kit (Miltenyi Biotec, 130-090-858). LSK cells and their subpopulations were analyzed by immunophenotyping using APC-Cy7- or PE–anti-c-Kit (BD Biosciences — Pharmingen, 560185 and 553869), PE-Cy7- or FITC-anti-Sca-1 (eBioscience, 25-5981-81 and 11-5981-82), eFluor450-anti-CD34 (eBioscience, 48-0341-80), or PerCP/Cy5.5-anti-CD34 (BioLegend, 128607), and APC–anti-Flt3 (BioLegend, 135309) antibodies. Intracellular protein levels were assessed after cell permeabilization (eBioscience Staining Buffer Set, 00-5523) using PE–anti-phospho-p38 (pT180/pY182) (BD Biosciences, 562065), Alexa Fluor 488–anti-MiTF (Abcam, ab201675), or PE-CF594 mouse anti-Smad2/Smad3 (BD Biosciences, 562697). Cell cycle analysis was performed after cell permeabilization using PE-Cy7-anti-Ki67 (eBioscience, 25-5698-80) and DAPI. Analysis was performed using an LSRII flow cytometer or BDAccuri C6 together with FACSDiva (BD Biosciences) or FlowJo (TreeStar Inc) software. Cells were sorted using an ARIA3 cell sorter (BD Biosciences). LSK cells were grown in StemSpan SFEM Medium (Stem Cell Technologies, 09600) supplemented with IL-3 (10 ng/mL), IL-6 (20 ng/mL), SCF (50 ng/mL), and Flt3L (35 ng/mL), with or without the p38 inhibitor SB203580 (20 μM). qRT-PCR was performed on mouse cells as previously described in RT-PCR and qRT-PCR analysis.
Immunofluorescence.
Immunofluorescence was performed using cytospun cells on poly-l-lysine–coated slides. The antibodies used were mouse anti-γH2AX (Abcam, ab26350). Incubations with donkey anti-mouse Alexa Fluor 488 secondary antibodies (Thermo Fisher Scientific, R37114) were performed at room temperature for 1 hour in the dark. Slides were air dried, mounted in Vectashield (Vector Laboratories) with DAPI to stain nuclei, and visualized using an Axio Observer Z1 (Carl Zeiss) microscope equipped with a ×63 oil-immersion objective and a charge-coupled device camera, AxioCam MRm (Carl Zeiss). Pictures were analyzed using ImageJ software (NIH).
CFU assay.
LSK cells (1 × 103) or BMCs (5 × 104) isolated from WT or Fanca–/– mice were cultured in methylcellulose-based medium (Stem Cell Technologies, MethoCult M3434) and scored for CFUs after 7 days. For replating, all colonies were mixed, the cells were counted, and 3 × 103 cells were cultured as described previously on 35 mm dishes.
Mitf expression inhibition by shRNA.
The 2 different shMitf purchased from Origene (TG514672A/GI584594: ATGCTAGAATACAGTCACTACCAGGTGCA and TG514672C/GI584596: CCAACTTGGAAGACATCCTGATGGACGAT) were mixed equally to prepare retroviral particles. The production and titration of retroviral particles were performed as described (71). The infection of isolated LSK cells was performed on retronectin-coated plates (TaKaRa Bio). Forty-eight hours later, 800 GFP+LSK cells per point were sorted, plated on MethoCult (Stem Cell Technologies, MethoCult M3434), and scored for CFUs after 10 days.
In vivo p38 MAPK inhibition.
One day following i.p. injection of a single dose of pI:pC (5 mg/kg), WT and Fanca–/– mice were administered SB203580 (50 mg/kg) or vehicle (SB203580 diluent) i.p. once a day for 12 days. The SB203580 diluent was composed of 4% DMSO, 30% PEG400, 5% Tween 80, and H2O.
Statistics.
Results are expressed as mean values ± SEM. Data were analyzed using Prism (GraphPad software, version 5). A 2-tailed, unpaired t test with Welch’s correction was used to assess statistical significance between 2 groups. ANOVA with Dunnett’s or Bonferroni’s correction for multiple comparisons was used to examine differences between more than 2 groups. A P value of less than 0.05 was considered statistically significant. A P value greater than 0.05 was considered not significant; however, P values between 0.05 and 0.1 are indicated.
Study approval.
This project was officially approved by the Animal Experimentation Ethics Committee (no. 26) of Gustave Roussy and conducted in accordance with French laws and regulations.
Author contributions
PP and FR conceptualized the study. AO, JB, ER, and PP investigated. FR acquired funding. PP and FR supervised. PP and FR wrote the paper.
Acknowledgments
This work was funded by grants from La Ligue Contre Le Cancer (to FR), INCa-DGOS-INSERM 6043 (to FR) and GEFLUC Ile-de-France (to PP). The authors thank individuals from the UMR8200-CNRS research unit for helpful discussions and Yann Lecluse and Philippe Rameau from the Imaging and Cytometry Platform of Gustave Roussy Cancer Center, UMS3655/US23, as well as Olivia Bawa from the Experimental and Translational Platform of Pathology AMMICa INSERM US23/UMS CNRS3655.
Version 1. 12/26/2019
In-Press Preview
Version 2. 02/17/2020
Electronic publication
Version 3. 03/02/2020
Print issue publication
Footnotes
JB’s present address is: Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom.
ER’s present address is: Cell Biology Unit, University Medical Center of the Johannes Gutenberg University Mainz, Mainz, Germany.
PP’s present address is: INSERM U981 “Identification of Molecular Predictors and new Targets for Cancer Treatment,” and Circulating Tumor Cells Translational Platform, CNRS UMS3655 INSERM US23AMMICA, Gustave Roussy, Villejuif, France.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(3):1377–1391.https://doi.org/10.1172/JCI131540.
Contributor Information
Alessia Oppezzo, Email: Alessia.OPPEZZO@gustaveroussy.fr.
Julie Bourseguin, Email: julie.bourseguin@gmail.com.
Emilie Renaud, Email: renaud.emilie@gmail.com.
Patrycja Pawlikowska, Email: Patrycjamarta.pawlikowska@gustaveroussy.fr.
Filippo Rosselli, Email: Filippo.ROSSELLI@gustaveroussy.fr.
References
- 1.Blanpain C, Mohrin M, Sotiropoulou PA, Passegué E. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8(1):16–29. doi: 10.1016/j.stem.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453(7193):306–313. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
- 4.Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol. 2006;6(2):93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 5.Trumpp A, Essers M, Wilson A. Awakening dormant haematopoietic stem cells. Nat Rev Immunol. 2010;10(3):201–209. doi: 10.1038/nri2726. [DOI] [PubMed] [Google Scholar]
- 6.Larsson J, Karlsson S. The role of Smad signaling in hematopoiesis. Oncogene. 2005;24(37):5676–5692. doi: 10.1038/sj.onc.1208920. [DOI] [PubMed] [Google Scholar]
- 7.Blank U, Karlsson S. The role of Smad signaling in hematopoiesis and translational hematology. Leukemia. 2011;25(9):1379–1388. doi: 10.1038/leu.2011.95. [DOI] [PubMed] [Google Scholar]
- 8.Blank U, Karlsson S. TGF-β signaling in the control of hematopoietic stem cells. Blood. 2015;125(23):3542–3550. doi: 10.1182/blood-2014-12-618090. [DOI] [PubMed] [Google Scholar]
- 9.Geest CR, Coffer PJ. MAPK signaling pathways in the regulation of hematopoiesis. J Leukoc Biol. 2009;86(2):237–250. doi: 10.1189/jlb.0209097. [DOI] [PubMed] [Google Scholar]
- 10.Bottero V, Withoff S, Verma IM. NF-kappaB and the regulation of hematopoiesis. Cell Death Differ. 2006;13(5):785–797. doi: 10.1038/sj.cdd.4401888. [DOI] [PubMed] [Google Scholar]
- 11.Navas TA, et al. Inhibition of overactivated p38 MAPK can restore hematopoiesis in myelodysplastic syndrome progenitors. Blood. 2006;108(13):4170–4177. doi: 10.1182/blood-2006-05-023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa MM, Chen H, Rathinam CV. Constitutive activation of NF-κB pathway in hematopoietic stem cells causes loss of quiescence and deregulated transcription factor networks. Front Cell Dev Biol. 2018;6:143. doi: 10.3389/fcell.2018.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakami A, Fisher DE. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab Invest. 2017;97(6):649–656. doi: 10.1038/labinvest.2017.9. [DOI] [PubMed] [Google Scholar]
- 14.Nechushtan H, Razin E. The function of MITF and associated proteins in mast cells. Mol Immunol. 2002;38(16-18):1177–1180. doi: 10.1016/S0161-5890(02)00059-7. [DOI] [PubMed] [Google Scholar]
- 15.Hershey CL, Fisher DE. MiTF and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone. 2004;34(4):689–696. doi: 10.1016/j.bone.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Karigane D, et al. p38α Activates purine metabolism to initiate hematopoietic stem/progenitor cell cycling in response to stress. Cell Stem Cell. 2016;19(2):192–204. doi: 10.1016/j.stem.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Bourseguin J, et al. FANCD2 functions as a critical factor downstream of MiTF to maintain the proliferation and survival of melanoma cells. Sci Rep. 2016;6:36539. doi: 10.1038/srep36539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strub T, et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene. 2011;30(20):2319–2332. doi: 10.1038/onc.2010.612. [DOI] [PubMed] [Google Scholar]
- 19.Savage SA, Dufour C. Classical inherited bone marrow failure syndromes with high risk for myelodysplastic syndrome and acute myelogenous leukemia. Semin Hematol. 2017;54(2):105–114. doi: 10.1053/j.seminhematol.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Bogliolo M, Surrallés J. Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev. 2015;33:32–40. doi: 10.1016/j.gde.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Naim V, Rosselli F. The FANC pathway and mitosis: a replication legacy. Cell Cycle. 2009;8(18):2907–2911. doi: 10.4161/cc.8.18.9538. [DOI] [PubMed] [Google Scholar]
- 22.Gueiderikh A, Rosselli F, Neto JBC. A never-ending story: the steadily growing family of the FA and FA-like genes. Genet Mol Biol. 2017;40(2):398–407. doi: 10.1590/1678-4685-gmb-2016-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17(6):337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 24.Briot D, Macé-Aimé G, Subra F, Rosselli F. Aberrant activation of stress-response pathways leads to TNF-alpha oversecretion in Fanconi anemia. Blood. 2008;111(4):1913–1923. doi: 10.1182/blood-2007-07-099218. [DOI] [PubMed] [Google Scholar]
- 25.Du W, Erden O, Pang Q. TNF-α signaling in Fanconi anemia. Blood Cells Mol Dis. 2014;52(1):2–11. doi: 10.1016/j.bcmd.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garbati MR, et al. Cytokine overproduction and crosslinker hypersensitivity are unlinked in Fanconi anemia macrophages. J Leukoc Biol. 2016;99(3):455–465. doi: 10.1189/jlb.3A0515-201R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibáñez A, Río P, Casado JA, Bueren JA, Fernández-Luna JL, Pipaón C. Elevated levels of IL-1beta in Fanconi anaemia group A patients due to a constitutively active phosphoinositide 3-kinase-Akt pathway are capable of promoting tumour cell proliferation. Biochem J. 2009;422(1):161–170. doi: 10.1042/BJ20082118. [DOI] [PubMed] [Google Scholar]
- 28.Matsushita N, et al. Direct inhibition of TNF-α promoter activity by Fanconi anemia protein FANCD2. PLoS ONE. 2011;6(8):e23324. doi: 10.1371/journal.pone.0023324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosselli F, Sanceau J, Gluckman E, Wietzerbin J, Moustacchi E. Abnormal lymphokine production: a novel feature of the genetic disease Fanconi anemia. II. In vitro and in vivo spontaneous overproduction of tumor necrosis factor alpha. Blood. 1994;83(5):1216–1225. [PubMed] [Google Scholar]
- 30.Schultz JC, Shahidi NT. Tumor necrosis factor-alpha overproduction in Fanconi’s anemia. Am J Hematol. 1993;42(2):196–201. doi: 10.1002/ajh.2830420211. [DOI] [PubMed] [Google Scholar]
- 31.Svahn J, et al. p38 Mitogen-activated protein kinase inhibition enhances in vitro erythropoiesis of Fanconi anemia, complementation group A-deficient bone marrow cells. Exp Hematol. 2015;43(4):295–299. doi: 10.1016/j.exphem.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, et al. TGF-β inhibition rescues hematopoietic stem cell defects and bone marrow failure in fanconi anemia. Cell Stem Cell. 2016;18(5):668–681. doi: 10.1016/j.stem.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dufour C, et al. TNF-alpha and IFN-gamma are overexpressed in the bone marrow of Fanconi anemia patients and TNF-alpha suppresses erythropoiesis in vitro. Blood. 2003;102(6):2053–2059. doi: 10.1182/blood-2003-01-0114. [DOI] [PubMed] [Google Scholar]
- 34.Hanafusa H, et al. Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor-beta-induced gene expression. J Biol Chem. 1999;274(38):27161–27167. doi: 10.1074/jbc.274.38.27161. [DOI] [PubMed] [Google Scholar]
- 35.Yu L, Hébert MC, Zhang YE. TGF-beta receptor-activated p38 MAP kinase mediates Smad-independent TGF-beta responses. EMBO J. 2002;21(14):3749–3759. doi: 10.1093/emboj/cdf366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J Biol Chem. 2002;277(13):11077–11083. doi: 10.1074/jbc.M111696200. [DOI] [PubMed] [Google Scholar]
- 37.Anur P, et al. p38 MAPK inhibition suppresses the TLR-hypersensitive phenotype in FANCC- and FANCA-deficient mononuclear phagocytes. Blood. 2012;119(9):1992–2002. doi: 10.1182/blood-2011-06-354647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patnaik A, et al. A first-in-human phase I study of the oral p38 MAPK inhibitor, ralimetinib (ly2228820 dimesylate), in patients with advanced cancer. Clin Cancer Res. 2016;22(5):1095–1102. doi: 10.1158/1078-0432.CCR-15-1718. [DOI] [PubMed] [Google Scholar]
- 39.Li XH, Kishore AH, Dao D, Zheng W, Roman CA, Word RA. A novel isoform of microphthalmia-associated transcription factor inhibits IL-8 gene expression in human cervical stromal cells. Mol Endocrinol. 2010;24(8):1512–1528. doi: 10.1210/me.2009-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bharti K, Liu W, Csermely T, Bertuzzi S, Arnheiter H. Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development. 2008;135(6):1169–1178. doi: 10.1242/dev.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibahara S, et al. Microphthalmia-associated transcription factor (MITF): multiplicity in structure, function, and regulation. J Investig Dermatol Symp Proc. 2001;6(1):99–104. doi: 10.1046/j.0022-202x.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- 42.Hartman ML, Czyz M. MITF in melanoma: mechanisms behind its expression and activity. Cell Mol Life Sci. 2015;72(7):1249–1260. doi: 10.1007/s00018-014-1791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol. 2013;2(1):47–63. doi: 10.1002/wdev.86. [DOI] [PubMed] [Google Scholar]
- 44.Massagué J. The transforming growth factor-beta family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 45.Vaidya A, Kale VP. TGF-β signaling and its role in the regulation of hematopoietic stem cells. Syst Synth Biol. 2015;9(1–2):1–10. doi: 10.1007/s11693-015-9161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pauklin S, Vallier L. Activin/Nodal signalling in stem cells. Development. 2015;142(4):607–619. doi: 10.1242/dev.091769. [DOI] [PubMed] [Google Scholar]
- 47.Zanier R, Briot D, Dugas du Villard JA, Sarasin A, Rosselli F. Fanconi anemia C gene product regulates expression of genes involved in differentiation and inflammation. Oncogene. 2004;23(29):5004–5013. doi: 10.1038/sj.onc.1207677. [DOI] [PubMed] [Google Scholar]
- 48.Zhou F, Xu X, Wang D, Wu J, Wang J. Identification of novel NF-κB transcriptional targets in TNFα-treated HeLa and HepG2 cells. Cell Biol Int. 2017;41(5):555–569. doi: 10.1002/cbin.10762. [DOI] [PubMed] [Google Scholar]
- 49.Bakker ST, de Winter JP, te Riele H. Learning from a paradox: recent insights into Fanconi anaemia through studying mouse models. Dis Model Mech. 2013;6(1):40–47. doi: 10.1242/dmm.009795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng NC, et al. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum Mol Genet. 2000;9(12):1805–1811. doi: 10.1093/hmg/9.12.1805. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen TV, Riou L, Aoufouchi S, Rosselli F. Fanca deficiency reduces A/T transitions in somatic hypermutation and alters class switch recombination junctions in mouse B cells. J Exp Med. 2014;211(6):1011–1018. doi: 10.1084/jem.20131637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pawlikowska P, Fouchet P, Vainchenker W, Rosselli F, Naim V. Defective endomitosis during megakaryopoiesis leads to thrombocytopenia in Fanca-/- mice. Blood. 2014;124(24):3613–3623. doi: 10.1182/blood-2014-01-551457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen TV, Pawlikowska P, Firlej V, Rosselli F, Aoufouchi S. V(D)J recombination process and the Pre-B to immature B-cells transition are altered in Fanca–/– mice. Sci Rep. 2016;6:36906. doi: 10.1038/srep36906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myers KC, et al. Impaired immune function in children and adults with Fanconi anemia. Pediatr Blood Cancer. 2017;64(11):e26599. doi: 10.1002/pbc.26599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aubé M, Lafrance M, Charbonneau C, Goulet I, Carreau M. Hematopoietic stem cells from fancc(-/-) mice have lower growth and differentiation potential in response to growth factors. Stem Cells. 2002;20(5):438–447. doi: 10.1634/stemcells.20-5-438. [DOI] [PubMed] [Google Scholar]
- 56.Barroca V, et al. Impaired functionality and homing of Fancg-deficient hematopoietic stem cells. Hum Mol Genet. 2012;21(1):121–135. doi: 10.1093/hmg/ddr447. [DOI] [PubMed] [Google Scholar]
- 57.Walter D, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 58.Flach J, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512(7513):198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santos MA, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514(7520):107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bakker ST, Passegué E. Resilient and resourceful: genome maintenance strategies in hematopoietic stem cells. Exp Hematol. 2013;41(11):915–923. doi: 10.1016/j.exphem.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ridet A, et al. Deregulated apoptosis is a hallmark of the Fanconi anemia syndrome. Cancer Res. 1997;57(9):1722–1730. [PubMed] [Google Scholar]
- 62.Rosselli F, Ridet A, Soussi T, Duchaud E, Alapetite C, Moustacchi E. p53-dependent pathway of radio-induced apoptosis is altered in Fanconi anemia. Oncogene. 1995;10(1):9–17. [PubMed] [Google Scholar]
- 63.Ceccaldi R, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11(1):36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Usai C, et al. Dysregulated Ca2+ homeostasis in Fanconi anemia cells. Sci Rep. 2015;5:8088. doi: 10.1038/srep08088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otsuki T, Nagakura S, Wang J, Bloom M, Grompe M, Liu JM. Tumor necrosis factor-alpha and CD95 ligation suppress erythropoiesis in Fanconi anemia C gene knockout mice. J Cell Physiol. 1999;179(1):79–86. doi: 10.1002/(SICI)1097-4652(199904)179:1<79::AID-JCP10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 66.Li J, et al. TNF-alpha induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J Clin Invest. 2007;117(11):3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giuliano S, et al. Microphthalmia-associated transcription factor controls the DNA damage response and a lineage-specific senescence program in melanomas. Cancer Res. 2010;70(9):3813–3822. doi: 10.1158/0008-5472.CAN-09-2913. [DOI] [PubMed] [Google Scholar]
- 68.Pichierri P, Rosselli F. Fanconi anemia proteins and the s phase checkpoint. Cell Cycle. 2004;3(6):698–700. [PubMed] [Google Scholar]
- 69.Fouquet B, et al. A homozygous FANCM mutation underlies a familial case of non-syndromic primary ovarian insufficiency. Elife. 2017;6:e30490. doi: 10.7554/eLife.30490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weksberg R, Buchwald M, Sargent P, Thompson MW, Siminovitch L. Specific cellular defects in patients with Fanconi anemia. J Cell Physiol. 1979;101(2):311–323. doi: 10.1002/jcp.1041010211. [DOI] [PubMed] [Google Scholar]
- 71.Hamelin V, Letourneux C, Romeo PH, Porteu F, Gaudry M. Thrombopoietin regulates IEX-1 gene expression through ERK-induced AML1 phosphorylation. Blood. 2006;107(8):3106–3113. doi: 10.1182/blood-2005-07-2953. [DOI] [PubMed] [Google Scholar]