Abstract
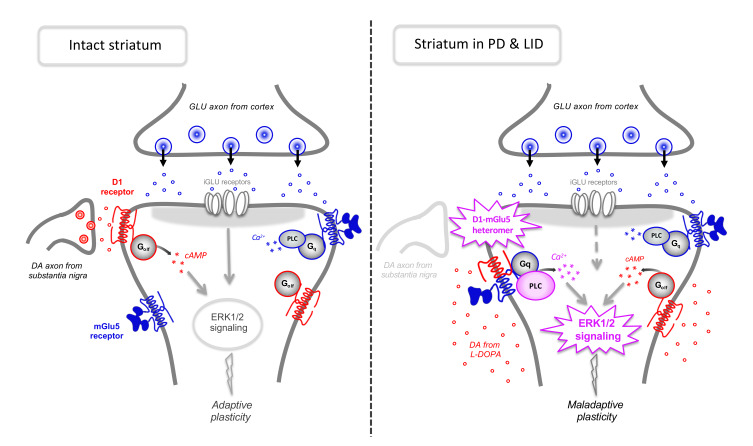
Dopamine receptor D1 modulates glutamatergic transmission in cortico-basal ganglia circuits and represents a major target of L-DOPA therapy in Parkinson’s disease. Here we show that D1 and metabotropic glutamate type 5 (mGlu5) receptors can form previously unknown heteromeric entities with distinctive functional properties. Interacting with Gq proteins, cell-surface D1-mGlu5 heteromers exacerbated PLC signaling and intracellular calcium release in response to either glutamate or dopamine. In rodent models of Parkinson’s disease, D1-mGlu5 nanocomplexes were strongly upregulated in the dopamine-denervated striatum, resulting in a synergistic activation of PLC signaling by D1 and mGlu5 receptor agonists. In turn, D1-mGlu5–dependent PLC signaling was causally linked with excessive activation of extracellular signal–regulated kinases in striatal neurons, leading to dyskinesia in animals treated with L-DOPA or D1 receptor agonists. The discovery of D1-mGlu5 functional heteromers mediating maladaptive molecular and motor responses in the dopamine-denervated striatum may prompt the development of new therapeutic principles for Parkinson’s disease.
Keywords: Neuroscience
Keywords: G-protein coupled receptors, Movement disorders, Parkinson’s disease
Introduction
Functional interactions between dopamine (DA) and glutamate (Glu) receptors are crucial to the striatum, a brain structure integrating dopaminergic and glutamatergic inputs to control action selection and behavioral plasticity (1, 2). In Parkinson’s disease (PD), a severe degeneration of dopaminergic nigrostriatal projections leads to impairments in striatal synaptic plasticity and the associated motor and cognitive functions (1). Pharmacological DA replacement with L-DOPA may initially correct some of these impairments (3, 4), but with the progression of PD, this treatment causes dyskinesia (abnormal involuntary movements [AIMs]) in a majority of patients. L-DOPA–induced dyskinesia (LID) depends on a dysfunctional interplay between DA and Glu signaling whose molecular basis is poorly understood. What is clear is that DA D1 receptors play a pivotal role in the abnormalities of striatal signaling and synaptic plasticity associated with LID (4–6).
Metabotropic Glu type 5 (mGlu5) receptors are coexpressed with D1 in about 50% of striatal neurons (7), and they are regarded as a potential therapeutic target for the treatment of LID (8). Indeed, pharmacological antagonism of mGlu5 improves peak-dose LID (9, 10) and counters an abnormal striatal activation of ERK1 and ERK2 downstream of D1 receptors (11).
We set out to address the hypothesis that the formation of signaling platforms involving both D1 and mGlu5 proteins may lead to cross-talk between these receptors in DA-denervated striatal neurons. Such a process could be favored by the relocalization of a postsynaptic pool of D1 receptors following DA denervation (3). Using bioluminescence resonance energy transfer (BRET) and bimolecular fluorescence complementation (BiFC), we here demonstrate for what we believe is the first time that mGlu5 and D1 receptors do form heteromers at the plasma membrane both in a heterologous expression system and in neurons. Interestingly, D1-mGlu5 receptor heteromers are Gq coupled and mediate a synergistic activation of PLC signaling and intracellular Ca2+ release by their respective receptor agonists. By applying a proximity ligation assay (PLA) to striatal tissue from adult rodents, we show that D1-mGlu5 complexes increase in abundance after DA denervation and persist in the face of DA replacement therapy. Accordingly, DA-denervated striata exhibit a synergistic potentiation of PLC signaling upon treatment with D1 and mGlu5 agonists. Finally, we provide evidence that D1- and mGlu-dependent PLC activation is causally linked with both striatal ERK1/2 signaling and dyskinetic behaviors in rodent models of PD. Taken together, these data indicate that an increased abundance of D1-mGlu5 heteromers is a molecular trait of DA-denervated striatal neurons favoring a development of dyskinesia when L-DOPA is used as a replacement therapy.
Results
D1 and mGlu5 receptors form heteromers in vitro.
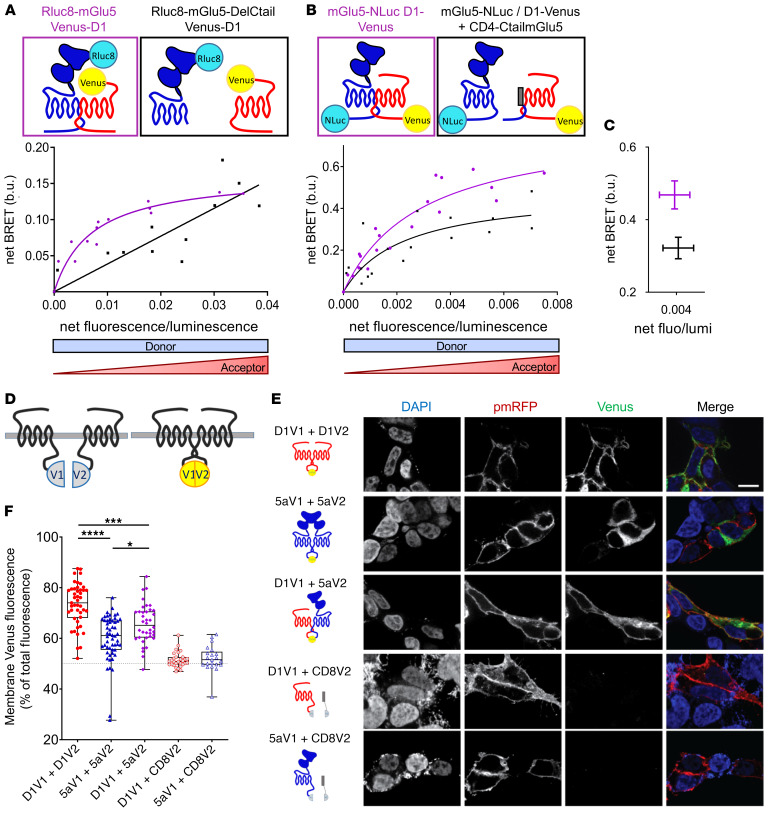
When coexpressed in HEK cells, D1 and mGlu5 receptors colocalized at the cell surface (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI126361DS1). To assess a possible direct physical interaction between D1 and mGlu5 receptors, we performed BRET assays in this cell population. The energy donor Renilla luciferase 8 (Rluc8) was fused to the N-terminus of mGlu5 and the acceptor yellow fluorescent protein Venus to the N-terminus of D1 receptor (Rluc8-mGlu5 and Venus-D1, respectively). At a constant level of Rluc8-mGlu5 expression, the BRET signal increased hyperbolically as a function of the Venus-D1 acceptor expression level (fluorescence/luminescence ratio) (Figure 1A). Saturation of the BRET signal when all Rluc8-mGlu5 was bound to Venus-D1 revealed a specific interaction between the mGlu5 and D1 receptors in living cells. As nonradiative energy transfer requires close proximity (<75 Å) between the acceptor and the donor (12), these BRET experiments demonstrate that mGlu5 and D1 receptors are close enough to form heteromers. This saturable interaction was further confirmed by measuring the BRET signal between full-length mGlu5 and D1 receptors tagged in a C-terminal position with Nanoluciferase (Nluc) and Venus (mGlu5-Nluc and D1-Venus, respectively; Figure 1B). The C-terminal tails of mGlu5 and D1 receptors have been reported to mediate interactions with other GPCRs, such as the μ opioid one (13), or with NMDA receptors (14, 15). Deletion of mGlu5 C-tail did not alter correct expression of the receptor at the cell surface (Rluc8-mGlu5DelCtail; Supplemental Figure 1B), but prevented its interaction with D1 (Figure 1A). Now the BRET signal between Rluc8-mGlu5DelCtail and Venus-D1 receptors increased linearly with the latter’s expression levels (Figure 1A), most likely reflecting random collision between mGlu5DelCtail and D1 receptors. Consistently, coexpression of a plasma membrane–targeted mGlu5 C-tail (CD4-CtailmGlu5) as a dominant negative peptide decreased the BRET signal (Figure 1C), confirming the involvement of mGlu5 C-tail in the receptor interaction. Using the same methodology, we found that this heteromerization process is not random and only occurs between specific partners (Supplemental Figure 2).
Figure 1. Heteromerization between mGlu5 and D1 receptors in living cells.
(A–C) BRET titration curves measured on HEK293 cells cotransfected with N- (A) or C-tagged (B) receptors. BRET signals were measured for increasing expression levels of acceptor (Venus-D1, A; or D1-Venus, B) at constant levels of donor expression (Rluc8-mGlu5, A; or mGlu5-Nluc, B). Results were analyzed by nonlinear regression on a pooled data set from 3 independent experiments, assuming a model with 1-site binding (GraphPad Prism 7). (A) BRET signals measured between Venus-D1 receptor and Rluc8-mGlu5 (purple curve) or Rluc8-mGlu5 deleted from its C-tail (Rluc8-mGlu5-DelCtail, black curve); b.u., BRET unit. (B) BRET signals measured between D1-Venus and mGlu5-Nluc, with (black curve) or without (purple curve) coexpression of mGlu5-Ctail fused to the CD4 membrane domain (CD4-CtailmGlu5). (C) Decrease of net BRET signal between D1-Venus and mGlu5-Nluc in cells coexpressing CD4-CtailmGlu5 (black) for identical D1-Venus/mGlu5-Nluc expression ratios. P = 0.0571, Mann-Whitney U test. (D–F) BiFC measured on transiently transfected HEK293 cells. (D) Schematic representation of BiFC principle. Nonfluorescent fragments from the Venus fluorescent protein (V1 and V2) are fused to putative interaction partners. Physical association triggers bimolecular fluorescent Venus complex. (E) BiFC images of receptors fused to nonfluorescent monomeric Venus split V1 or V2. Specificity was controlled in cells expressing D1-V1 or 5a-V1 together with CD8-V2 (last 2 rows). The green channel illustrates the expression of Venus complementation (V1 + V2), whereas DAPI and pmRFP fluorescences stain the nucleus and plasma membrane, respectively. Scale bar: 10 μm. (F) Quantification of complemented Venus fluorescence intensity at the membrane (colocalized with pmRFP) expressed as a percentage of whole-cell Venus fluorescence. Box and whiskers plots of 21 to 54 measurements. *P < 0.05; ***P < 0.001; ****P < 0.0001, Kruskal-Wallis test. Box and whiskers plots: in this and the following figures: line, median; bounds: 25th to 75th percentiles; whiskers, minimum to maximum.
We further assessed the cellular localization of the D1-mGlu5 heteromer by BiFC assays, a protein fragment complementation method appropriate to visualizing GPCR oligomerization (16, 17) (Figure 1D). We fused complementary Venus1 (V1) and Venus2 (V2) fragments to the C-termini of D1 and mGlu5 receptors (D1-V1, D1-V2, mGlu5-V1, and mGlu5-V2, respectively; Figure 1E). As expected from BRET results, coexpression of D1-V1 and mGlu5-V2 in HEK cells resulted in the reconstitution of a fluorescent Venus protein, confirming the ability of these receptors to form heteromers (Figure 1E). Colocalization of Venus-fluorescence intensity with a red fluorescent plasma membrane protein (pmRFP) further highlighted a preferential expression of D1-V1/mGlu5-V2 heteromers at the cell surface, as 65.66% ± 12.57 % of total Venus fluorescence colocalized with pmRFP (Figure 1F). This cell-surface preferential expression of the heteromer was significantly lower than that of D1-V1/D1-V2 (73.35% ± 12.46 %) and was higher than that of mGlu5-V1/mGlu5-V2 homodimers (60.04% ± 12.62%, Figure 1F). Together, these data disclose the existence of D1-mGlu5 heteromers, preferentially localized at the cell surface of HEK cells.
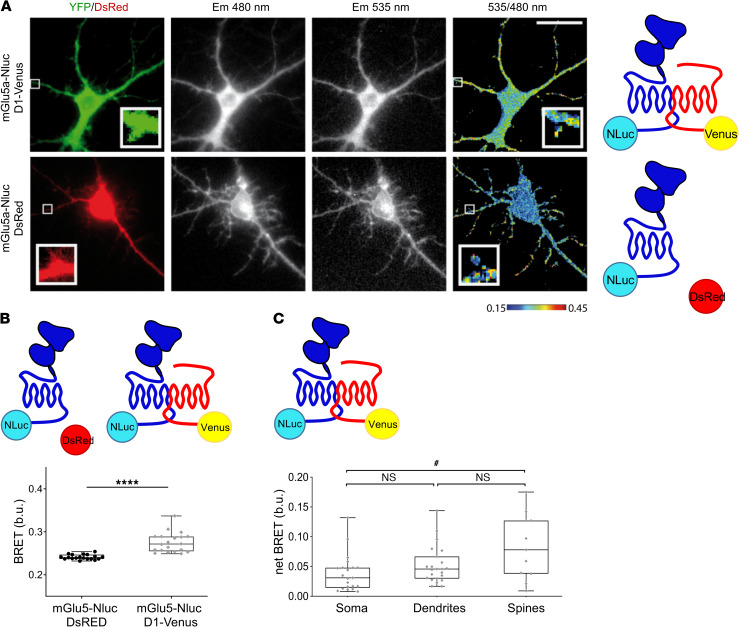
D1 and mGlu5 receptors also formed heteromers in primary, cultured hippocampal neurons (Figure 2), which endogenously express these receptors (Supplemental Figure 1C) (18, 19). The BRET signal between mGlu5-Nluc and D1-Venus in hippocampal neurons was indeed significantly higher than the basal BRET measured in cells expressing the BRET donor alone (Figure 2, A and B). Because cultured hippocampal neurons form dendrites and spines, we were able to assess the relative localization of D1-mGlu5 heteromers in different cellular compartments, finding a higher BRET signal in dendritic shafts and spines compared with that in the cell soma (Figure 2A; see 535/480 ratio image in upper row, and quantification in Figure 2C; P < 0.05 for spines vs. soma). This subcellular pattern is reminiscent of the preferential distribution of native D1 and mGlu5 in spines and dendrites relative to perikaryal regions (20–22). We did not carry out this experiment in primary striatal cultures because the striatal neurons expressing both D1 and mGlu5 receptors (so-called medium spiny neurons) are aspiny in culture.
Figure 2. D1 and mGlu5 receptors form heteromers in neurons.
(A–C) BRET imaging between mGlu5-Nluc and D1-Venus was measured in soma, dendrites, and spines of hippocampal neurons. (A) Single-cell BRET imaging in neurons expressing either mGlu5-Nluc and D1-Venus (top) or mGlu5-Nluc with DsRed as transfection reporter (bottom). Cells were identified by green or red fluorescence (left). Em480 and Em535 images were recorded, and the 535 nm/480 nm pseudo-colored ratio images were processed. Square areas are shown at a higher magnification in the insets, which are 3 μm × 3 μm. Cells are representative of 19 to 21 cells. Scale bar: 10 μm. (B) Quantification of the BRET signal intensity in soma from mGlu5-Nluc and D1-Venus transfected neurons compared with the basal BRET measured in neurons expressing mGlu5-Nluc alone (left). Box and whiskers plots of 19 to 20 measurements in the soma of neurons. ****P < 0.0001, Mann-Whitney U test. (C) netBRET between mGlu5-Nluc and D1-Venus in soma, dendrites, and spines. The average basal BRET in respective compartment has been subtracted from BRET measurements. Box and whiskers plots of n = 23 measurements in soma, n = 21 in dendrites, n = 11 in spines from neurons expressing mGlu5-Nluc and D1-Venus. #P < 0.05, Kruskal-Wallis test.
Functional properties of D1-mGlu5 heteromers.
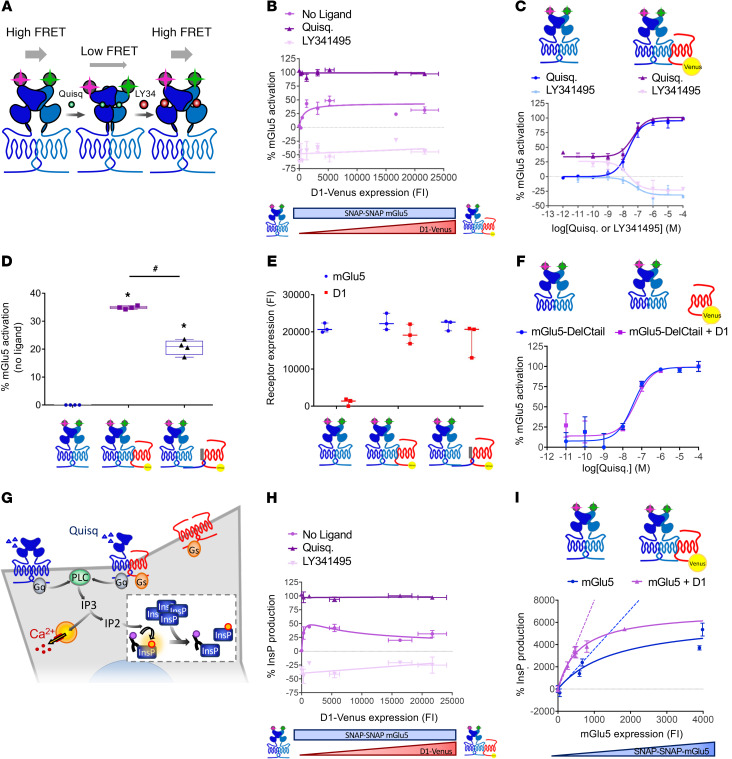
We next interrogated the functional consequences of D1-mGlu5 receptor heteromerization. Using a time-resolved fluorescence resonance energy transfer–based (TR-FRET–based) sensor, we measured extracellular conformational changes within the mGlu5 receptor homodimer (Figure 3A). Upon receptor and G-protein activation, the reorientation of the extracellular ligand–binding domains (ECDs) of the mGlu5 homodimer subunits resulted in a decrease of the FRET signal (23). Interestingly, in the absence of ligand, the proportion of mGlu5 receptors in active-like conformation increased hyperbolically as a function of the D1-Venus expression level (Figure 3B). This result revealed a D1 receptor–induced basal activation of the mGlu5 receptor that saturates (at around 30% of maximal mGlu5 activation rate) for high D1 receptor expression levels. D1 receptor coexpression did not affect the maximum effect (Emax) or the EC50 of mGlu5 agonist (quisqualic acid) or antagonist (LY341495) (Figure 3, B and C). This conformational change of mGlu5 was triggered by its physical interaction with D1. Impairment of D1-mGlu5 interaction by coexpression of mGlu5-Ctail fused to the plasma membrane (CD4-CtailmGlu5, Figure 1B) significantly reduced the amount of mGlu5 receptors displaying an active conformation (Figure 3D), while receptors expression was not changed (Figure 3E). Consistently, mGlu5 sensor deleted from its C-tail (which cannot interact with D1, cf. Figure 1A) was not affected by D1 coexpression (Figure 3F). Hence, under basal conditions, D1-mGlu5 heteromerization favors an active conformation of mGlu5.
Figure 3. D1-mGlu5 heteromer favors Ca2+ signaling in vitro by enhancing basal activation and basal signaling of mGlu5 receptor.
A TR-FRET–based sensor monitors mGlu5 receptor ECD conformation. (A) In the absence of ligand or in the presence of antagonist (LY341495), the proximity of TR-FRET donor and acceptor results in a high FRET signal. Agonist-induced (quisqualic acid [Quisq]) relative movement of the ECDs decreases the FRET signal (23). (B and C) Percentage of mGlu5 active conformation (B) with increasing amount of D1-Venus receptors in basal conditions (no ligand), with Quisq (10 μM) or with LY341495 (100 μM) (C) in the absence (blue) or presence (purple) of D1 receptor (corresponding to 279 ± 68 D1 fluorescence intensity [FI] on B) with increasing concentrations of Quisq (dark color) or LY341495 (light color). (D) mGlu5 basal activation when expressed alone (left), with D1 receptor (middle), or with D1 receptor and mGlu5Ctail as dominant negative peptide (right). *P < 0.05 vs. mGlu5 basal activation; #P < 0.05 vs. mGlu5 plus D1 receptor activation, unpaired t test (Mann-Whitney U test). Box and whiskers plots. (E) SNAP-mGlu5 and D1-Venus receptor expression levels were controlled by measurement of SNAP-Lumi4-Tb and Venus, respectively (FI). (F) Percentage of mGlu5-DelCtail active-conformation measured in absence (blue) or presence (purple) of D1 receptor with increasing concentrations of Quisq. (G) PLC activation is reported by new InsP production-induced decrease of HTRF signal between fluorescent-InsP and InsP-antibody (insert). (H) mGlu5-induced InsP production with increasing amount of D1 receptors in basal conditions (no ligand), with Quisq (10 μM), or with LY341495 (100 μM). InsP production was normalized to the maximal activity of mGlu5 in the presence of agonist. (I) Constitutive InsP production with increasing amount of mGlu5 receptor in absence (blue) or presence (purple) of a constant amount of D1 receptor (corresponding to 319 ± 4 D1 FI on H). Values are shown as mean ± SEM of 3 independent experiments.
We next studied the functional consequences of D1-induced enhancement of mGlu5 receptors in active conformation. Since mGlu5 canonical coupling to Gq induces PLC pathway activation, we assessed phosphoinositide (PI) hydrolysis by measuring the production of inositolmonophosphate (InsP), which originates from the sequential dephosphorylation of inositol-1,4,5-trisphosphate (Figure 3G). For a constant expression of mGlu5 receptors, basal InsP levels increased hyperbolically as a function of the D1-Venus expression levels to reach 30% of the maximal agonist-induced InsP production (Figure 3H). Moreover, the slope of constitutive InsP production induced by increasing expression levels of mGlu5 was higher in the presence than in the absence of D1 receptor (Figure 3I). Hence, D1-mGlu5 heteromerization not only enhanced the rate of mGlu5 receptors in active-like conformation, but also the basal InsP production.
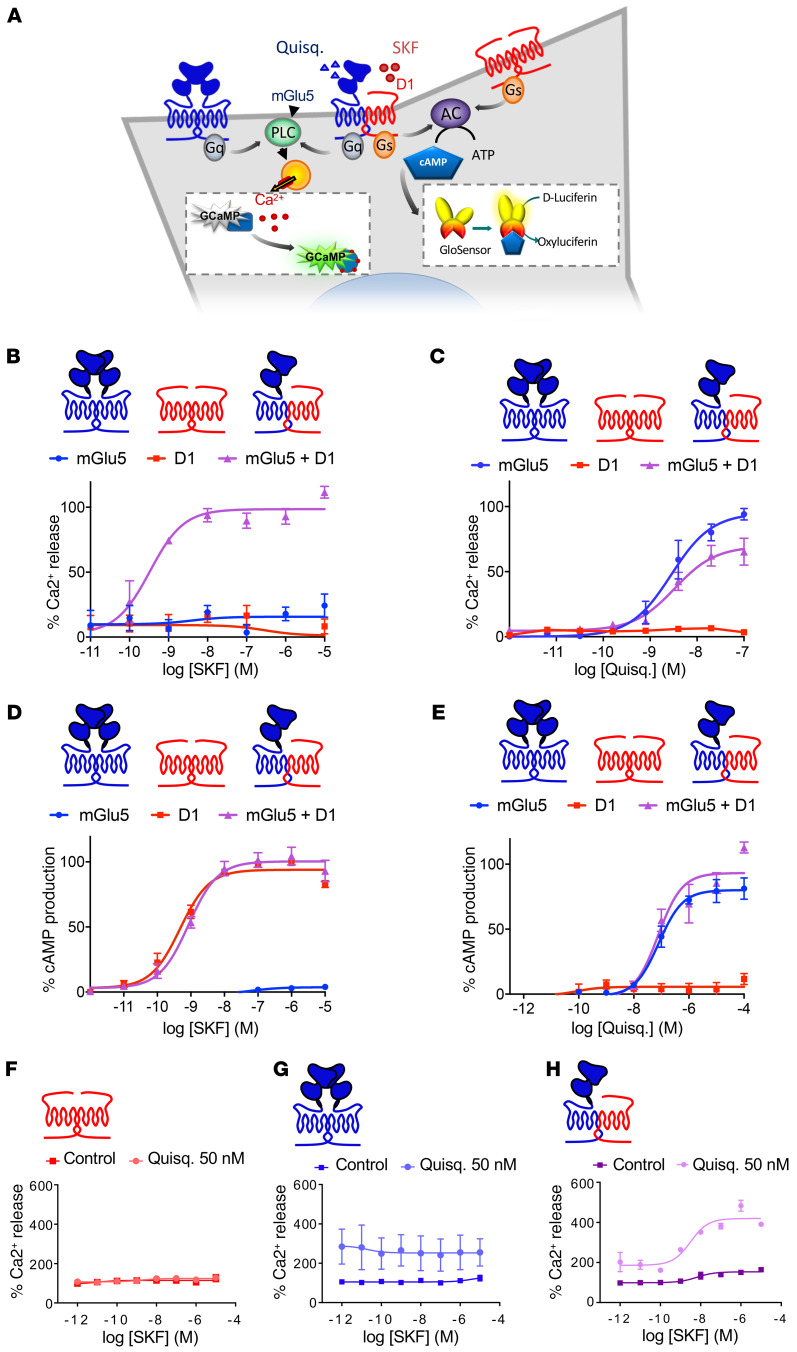
To further analyze the functional consequences of mGlu5 and D1 coexpression on their respective canonical signaling pathways (Gq- and Gs-preferential coupling, respectively), we generated a HEK cell line stably expressing 2 sensors, the GCaMP6 fluorescent calcium sensor and the Glo luminescent cAMP sensor (Figure 4A). While D1-like agonist (SKF81297) induced cAMP production but not Ca2+ release on cells expressing D1 alone (Figure 4, D and B, respectively), the coexpression of mGlu5 enabled a D1-like agonist-induced Ca2+ release (Figure 4B) without affecting the potency of SKF81297 to induce cAMP production (Figure 4D). This result suggested that an allosteric interaction between mGlu5 and D1 receptors allows the D1 receptor to signal through a Ca2+-release pathway. On the other side, the mGlu5 agonist quisqualate induced a similar Ca2+ response in the presence or absence of coexpressed D1 receptors (Figure 4C). In agreement with recent reports (24), we noticed that the mGlu5 agonist induced cAMP production when mGlu5 was expressed alone, and this property was not affected by D1 coexpression (Figure 4E). The application of SKF81297 did not induce Ca2+ release in cells expressing only D1 receptors (Figure 4F) or mGlu5 receptors (Figure 4G). In contrast, when both receptors were coexpressed, the coapplication of SKF81297 and a mGlu5 agonist (quisqualate 50 nM) produced an over 3-fold increase in Ca2+ release above baseline levels (Figure 4H). Taken together, these results suggest that the D1-mGlu5 heteromer has a signaling bias toward Ca2+-dependent pathways and mediates a synergistic activation of such pathways upon stimulation.
Figure 4. D1-mGlu5 heteromer creates an atypical D1 receptor–mediated intracellular Ca2+ release.
(A) HEK cell line stably expressing GcAMP6 calcium indicator and Glo cAMP-sensor, cotransfected with mGlu5 and D1 receptors. (B–E) Ca2+ release and cAMP production were measured in cells transfected with mGlu5, D1, or mGlu5 plus D1 receptors. Values represent mean ± SEM of 3 or 4 independent experiments. Data are expressed as a percentage of the maximal Quisq- or SKF-induced effect. (B and C) Ca2+ release dose-response curve induced by (B) D1 agonist (SKF81297) and (C) mGlu5 agonist Quisq. (D and E) cAMP production dose response curve induced by (D) D1 agonist SKF81297 and (E) mGlu5 agonist Quisq. (F–H) Synergistic activation of Ca2+ release induced by D1 and mGlu5 receptors. Cells were transfected with D1 (F), mGlu5 (G), or both receptors (H). Ca2+ release is here expressed as a percentage of basal levels in each transfection condition. Values represent mean ± SEM of 2 to 4 independent experiments.
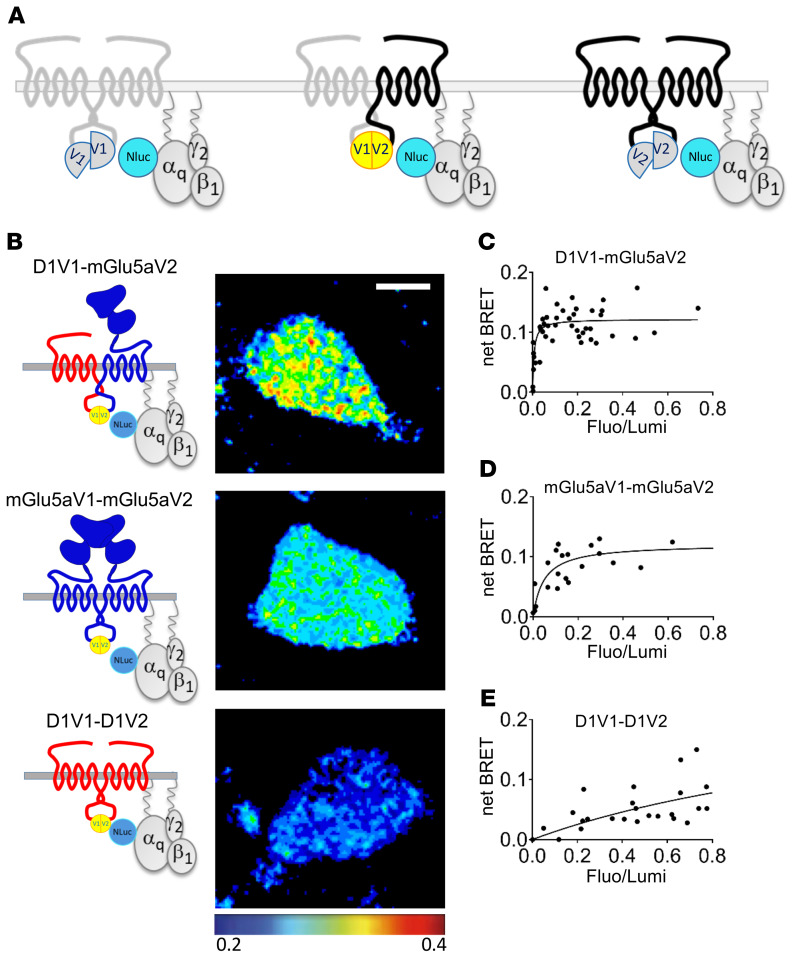
The biased Ca2+ signaling associated with D1 and mGlu5 coexpression could be driven by activation of Gq-type proteins by the heteromer. To specifically study Gq protein binding to the heterodimer, we used complemented donor-acceptor resonance energy transfer (CODA-RET) (25) combining BiFC and BRET (Figure 5A). We took advantage of the BiFC combinations described in Figure 1D to follow exclusively one signaling at a time, coming from D1-V1/D1-V2, mGlu5-V1/mGlu5-V2, or D1-V1/mGlu5-V2 (Figure 5B). BRET imaging revealed an interaction of the D1-mGlu5 heteromer with the Nluc-Gαq subunit of the Gαqβ12 heterotrimer (Figure 5C). Consistent with the canonical G-protein coupling described for those receptors, BRET imaging also showed a Gq interaction with mGlu5 homomers (Figure 5D), but not with D1 homomers (Figure 5E).
Figure 5. D1-mGlu5 heteromer is coupled to Gq protein.
(A) CODA-RET enables exclusive monitoring of 1 dimer species of interest. (B–E) CODA-RET imaging monitored on HEK cells expressing the indicated complemented dimers. (B) The pseudocolor images quantify the BRET between the complemented Venus-dimer and Nluc-Gαq protein. (C–E) BRET titration curves were measured for increasing acceptor/donor ratio (Fluo/Lumi). Results were analyzed by nonlinear regression on a pooled data set from 3 independent experiments, assuming a model with 1-site binding (GraphPad Prism 7).
D1 and mGlu5 form abundant receptor complexes in the DA-denervated striatum.
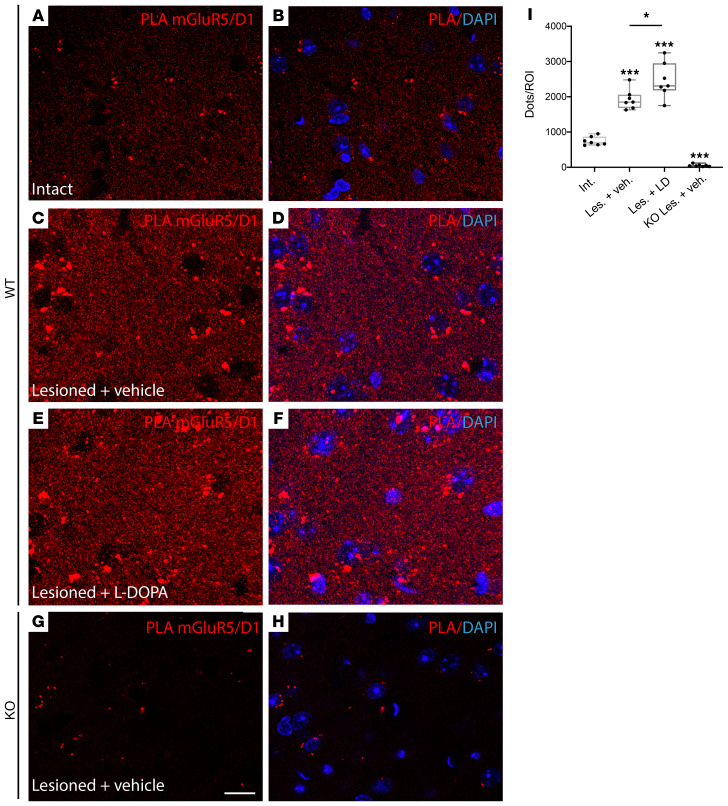
The biochemical properties of D1-mGlu5 heteromers are reminiscent of signaling features typical of DA-denervated striatal neurons, where D1 and mGlu5 functionally interact and activate ERK1 and ERK2 in a manner that requires PLC activity and Ca2+ release from intracellular stores (11). To search for D1-mGlu5 complexes in DA-denervated striatal neurons, we prepared mice with unilateral 6-hydroxydopamine (6-OHDA) lesions of the medial forebrain bundle (MFB) and then processed both intact and DA-denervated striatal tissue for PLA with antibodies against D1 and mGlu5. With in situ PLA, fluorescent-labeled complementary oligonucleotide probes bind to the amplicons (amplified DNA) at the sites of protein-protein interaction, producing distinct dots of high fluorescence on the tissue. Fluorescence dots were clearly detectable, although sparse in the intact striatum (Figure 6, A and B), and their abundance increased by approximately 2.5-fold in DA-denervated samples (Figure 6, C, D, and I; P < 0.05 for lesioned vs. intact striatum), indicating a higher association of mGlu5 with D1 receptors after nigrostriatal dopaminergic degeneration. The number of D1-mGlu5 puncta was further increased after a chronic dyskinesiogenic course of L-DOPA treatment (Figure 6, E, F, and I). Transgenic mice with a selective genetic ablation of mGlu5 in D1-expressing neurons (mGluR5KO-D1) did not exhibit a distinct signal (Figure 6, G–I), attesting the specificity of our PLA methodology.
Figure 6. Endogenous D1-mGlu5 clusters are increased in the DA-denervated striatum.
Protein ligation assay (PLA) experiments were performed on intact (A and B) and DA-denervated (C–H, lesioned) striata of WT mice (A–F, WT, n = 7 per treatment condition) and mice with a selective KO of mGluR5 in D1 receptor–expressing neurons (mGluR5KO-D1; n = 7) (G and H). DA-denervated mice received daily s.c. injection of either L-DOPA (6 mg/kg) (E and F) or vehicle (saline, C and D, G and H) for 3 weeks. All L-DOPA–treated mice developed dyskinesia. (A–H) PLA were obtained with primary antibodies directed against mGlu5 and D1. Plus and minus probes that correspond to secondary antibodies allow rolling-circle amplification and detection of the amplicons by a fluorescence labeled probe (red dots). Nuclei were counterstained with DAPI. Scale bar: 10 μm. (I) Box and whiskers plots of the number of PLA signals (reds dots) per presized zone from WT and KO striatal sections in indicated conditions. Six ROI were analyzed per striatum to measure a mean value per animal. *P < 0.05; ***P < 0.001, Mann-Whitney U test.
D1-mGlu5 synergistically activates PLC signaling in the DA-denervated striatum.
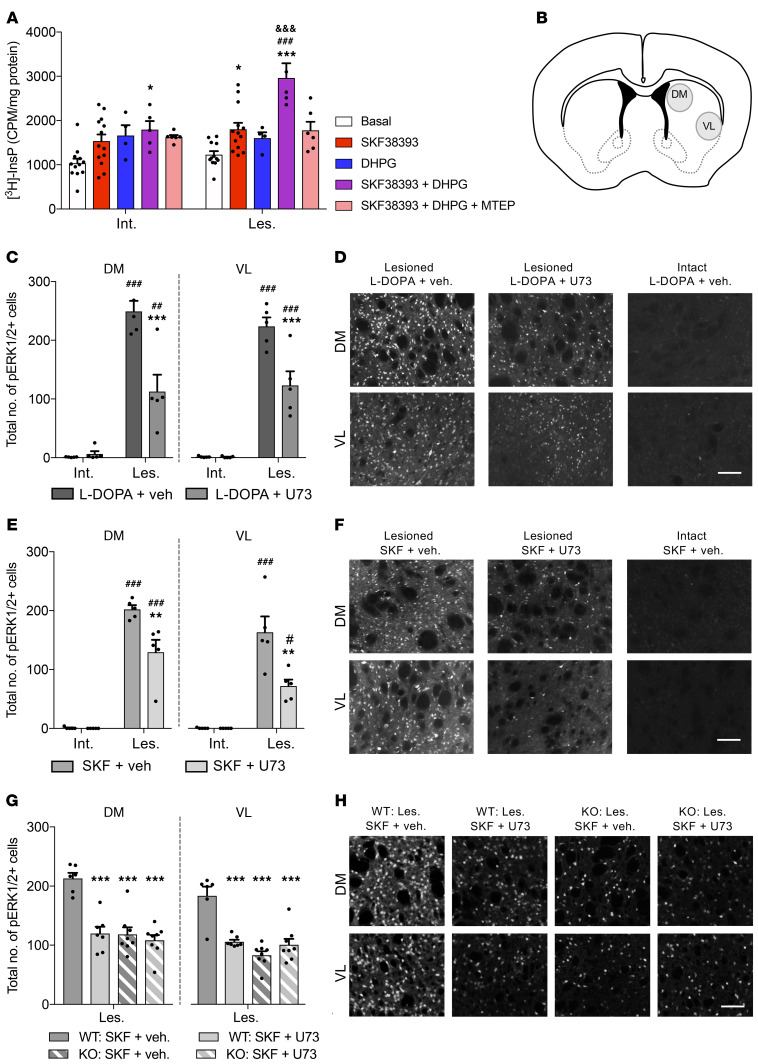
mGlu5 is a Gq-coupled receptor, and its stimulation leads to PI hydrolysis via PLCβ (26). While stimulation of bona fide D1 receptors does not activate PLC, our data so far indicate that such a response would occur in the presence of D1-mGlu5 heteromers. To test this prediction, we used an ex vivo assay of PLC activity measuring the production of 3H-InsP (27) and compared the effects of D1/mGlu5 stimulation in DA-denervated or intact striata. For this assay, we used 6-OHDA–lesioned rats rather than mice because we needed a large amount of striatal tissue (see Methods). Striatal samples were incubated with the D1-like agonist SKF38393, the mGlu1/5 receptor agonist DHPG, or a combination in the presence of mGlu1 blockade (achieved using saturating concentrations of JNJ16259685). When applied alone, both SKF38393 and DHPG tended to enhance 3H-InsP levels in both intact and DA-denervated striata, but only the effect of SKF38393 in the lesioned striatum reached statistical significance (Figure 7A; SKF38393 vs. basal: 46% increase, P < 0.05). The coapplication of SKF38393 and DHPG produced a significant increase in 3H-InsP production in both intact and denervated samples, but the effect of agonist coapplication was markedly larger in the presence of DA denervation (Figure 7A, 141% vs. 70% increase above basal levels in lesioned vs. intact striatum, respectively; P < 0.001). This effect was blocked by coincubation with the mGlu5 antagonist MTEP (Figure 7A). In DA-denervated samples, the effect of SKF38393-DHPG coapplication was over 2-fold larger than the sum of changes produced by each agonist alone, indicating a synergistic activation of PLC signaling (Figure 7A). These data suggest that a concomitant increase in DA and Glu levels in the parkinsonian striatum would lead to a strong activation of PLC in cells expressing both D1 and mGlu5 receptors.
Figure 7. mGlu5 and D1 receptors interact to induce PLC signaling and ERK1/2 activation in the DA-denervated striatum.
(A) InsP hydrolysis in intact (Int.) and lesioned (Les.) rat striatal slices. SKF38393 and DHPG synergistically activated PLC in the DA-denervated striatum. n = 4–13 tubes per treatment. Treatment: F(4,70) = 14.33, P < 0.001; side: F(1,70) = 9.40, P < 0.01; interaction: F(4,70) = 2.81, P < 0.05. Bonferroni’s test: *P < 0.05 and ***P < 0.001 vs. baseline of same group; ###P < 0.001 vs. all other treatments within same group (Les.); &&&P < 0.001 vs. same treatment of opposite group (Int.). (B) Sample areas for counting pERK1/2-positive cells in DM and VL striata (rat and mouse). (C–F) U73122 (U73) attenuates L-DOPA– (C and D) and SKF38393-induced pERK1/2 (E and F) in DA-denervated rat striatum (n = 5 per condition). (D and F) pERK1/2-immunostained rat striatal sections. Scale bar: 200 μm. C-DM: treatment: F(1,16) = 14.64, P < 0.01; side: F(1,16) = 106.6, P < 0.001; interaction: F(1,16) = 17.13, P < 0.001. C-VL: treatment: F(1,16) = 12.57, P < 0.01; side: F(1.16) = 146.1, P < 0.001; interaction: F(1,16) = 12.36, P < 0.01. E-DM: treatment: F(1,16) = 10.67, P < 0.01; side: F(1,16) = 212.7, P < 0.001; interaction: F(1,16) = 10.21, P < 0.01. E-VL: treatment: F(1,16) = 9.99, P < 0.01; side: F(1,16) = 65.29, P < 0.001; interaction: F(1,16) = 9.82, P < 0.01). Bonferroni’s test: **P < 0.01 and ***P < 0.001 vs. L-DOPA/SKF38393 + vehicle of Les. side; #P < 0.05, ##P < 0.01, and ###P < 0.001 vs. L-DOPA/SKF38393 + vehicle/U73122 of Int. side. (G and H) U73122 attenuates D1-dependent pERK1/2 in DA-denervated striata from WT but not mGlu5KO-D1 mice (KO) (n = 6–8 mice per condition). (H) pERK1/2 immunostainings. Scale bar: 200 μm. G-DM: treatment: F(3,25) = 18.63, P < 0.001. G-VL: treatment: F(3,25) = 20.36, P < 0.001. Bonferroni’s test: ***P < 0.001 vs. WT: SKF38393 + vehicle.
Because striatal levels of both DA and Glu increase in PD shortly after the administration of L-DOPA (28, 29), we set out to examine the effects of PLC inhibition in unilaterally 6-OHDA–lesioned rats treated with L-DOPA using ERK1/2 activation as a molecular marker of aberrant D1-mGlu5–dependent signaling (11). Animals were perfusion fixed 30 minutes following the combined administration of L-DOPA and the systemically active, selective PLC inhibitor U73122, and cells immunoreactive for the active, phosphorylated form of ERK1/2 were counted in different regions of the motor striatum (Figure 7B, and see Methods). As expected (5), L-DOPA (6 mg/kg s.c.) produced a large induction of phosphorylated ERK1/2 in DA-denervated, but not intact, striata (Figure 7, C and D). Throughout the DA-denervated striatum, the effect of L-DOPA was substantially reduced in animals cotreated with U73122 (30 mg/kg i.p.; Figure 7, C and D). The magnitude of such reduction was comparable to that produced by MTEP in a previous study (30). In other groups of 6-OHDA–lesioned rats, we examined the effects of the selective D1- and D2-like agonists SKF38393 and quinpirole, respectively. While quinpirole did not have an inducing effect on ERK1/2 (data not shown), the D1 agonist SKF38393 (2 mg/kg s.c.) produced robust ERK1/2 activation throughout the DA-denervated striatum, and this response was substantially attenuated by U73122 (Figure 7, E and F). Next, we verified the effect of PLC inhibition on D1-dependent ERK1/2 activation using both WT and mGluR5KO-D1 mice. As expected (11), SKF38393 induced a large number of phosphorylated ERK1/2–positive (pERK1/2-positive) cells in the DA-denervated striatum of WT mice, and a significantly lower number in mGluR5KO-D1 mice (Figure 7, G and H). In WT mice, the effect of SKF38393 was significantly blunted by U73122 cotreatment (Figure 7, G and H). In contrast, there was no difference between vehicle and U73122 coadministration in lesioned mGluR5KO-D1 mice treated with SKF38393 (Figure 7G). These data indicate that the absence of mGlu5 receptor in D1-positive neurons occludes the molecular pathway through which U73122 attenuates the aberrant striatal activation of ERK1/2.
In summary, these data show that PLC signaling accounts for a considerable proportion of striatal ERK1/2 activation in parkinsonian animals treated with drugs that stimulate the D1 receptor. The residual, PLC-independent ERK1/2 activation most likely reflects the supersensitivity of D1-mediated cAMP/PKA signaling after DA denervation because this signaling response is not affected by the inhibition of either mGlu5 or PLC (11).
Inhibition of PLC or mGlu5 receptors reduces dyskinesias that depend on D1 receptor stimulation.
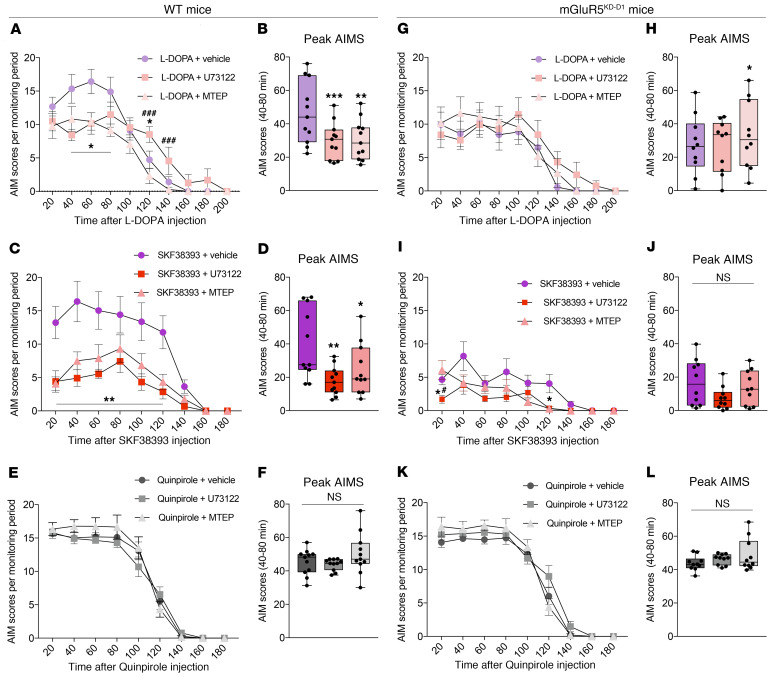
Marked striatal activation of ERK1/2 by L-DOPA is a maladaptive molecular response causally linked with dyskinesia (31), and cotreatment with mGlu5 antagonists blunts both L-DOPA–induced ERK1/2 activation and L-DOPA–induced peak AIMs, that is, the most disabling dyskinesias appearing when brain levels of L-DOPA are high (9, 10). We therefore set out to examine potential antidyskinetic effects of PLC inhibition in parkinsonian rodents chronically treated with dopaminergic agents. Mice with unilateral 6-OHDA lesions sustained 3 consecutive drug-treatment periods using escalating doses of L-DOPA (3 and 6 mg/kg), SKF38393 (3 and 6 mg/kg), or quinpirole (0.1 and 0.5 mg/kg). With this protocol, the 3 different dopaminergic drugs induced similarly severe AIMs by the end of each treatment period, at which point we compared the effects of PLC inhibition with U73122 or selective mGlu5 antagonism with MTEP.
The peak severity of L-DOPA–induced AIMs was reduced by both U73122 and MTEP (Figure 8A; P < 0.05 for each compound versus vehicle at 20-80 minutes after L-DOPA administration) to a comparable extent (Figure 8B). Treatment with the D1-like agonist SKF38393 induced severe and sustained AIMs, which were significantly attenuated by U73122 and MTEP with a comparable temporal course (Figure 8, C and D). The D2 receptor agonist quinpirole can induce dyskinesia in rodents primed with L-DOPA or D1 agonists (32). We exploited this phenomenon to investigate the efficacy of U73122 or MTEP on dyskinesias that are mediated via the D2 receptor. When mice were challenged with quinpirole, alone or combined with U73122 or MTEP, neither the PLC inhibitor nor the mGlu5 receptor antagonist had any antidyskinetic action (Figure 8, E and F). The pattern of antidyskinetic effects by PLC inhibition and mGlu5 antagonism was identical in the rat model of LID, in which we also verified that pharmacological inhibition of PLC did not interfere with the antiakinetic (therapeutic-like) effect of the dopaminergic agents (see rat AIMs and motor test in Supplemental Figure 3). Moreover, using this model, we showed that pharmacological inhibition of muscarinic receptors with dicyclomine (33) did not have any antidyskinetic effect (see Supplemental Figure 3, J and K). This experiment rules out that the PLC activity contributing to LID is downstream of the Gq-coupled muscarinic receptor M1, which is abundantly expressed in striatal projection neurons (i.e., cells expressing active ERK1/2 upon treatment with L-DOPA or SKF38393 (5, 11).
Figure 8. MTEP and U73122 improve D1 receptor–dependent dyskinesias in WT mice, but have no effect in mGluR5KD-D1 mice.
(A and B) L-DOPA–induced AIMs in WT mice. (A) Time course of AIMs (n = 11; repeated-measures [RM] 2-way ANOVA, treatment: F(2,20) = 13.6, P < 0.001; time: F(9,90) = 45.87, P < 0.001; interaction: F(18,180) = 4.49, P < 0.001). (B) Peak AIMs (n = 11; Friedman test (Fr) = 16.55, P < 0.001). (C and D) SKF38393-induced AIMs in WT. (C) Time course of AIMs (n = 11; RM 2-way ANOVA, treatment: F(2,20) = 17.28, P < 0.001; time: F(8,80) = 23, P < 0.001; interaction: F(16,160) = 5.05, P < 0.001). (D) Peak AIMs (n = 11; Fr = 11.45, P < 0.01). (E and F) Quinpirole-induced AIMs in WT. (E) Time course of AIMs (n = 11; RM 2-way ANOVA, treatment: F(2,20) = 1.48, P = 0.25; time: F(8,80) = 148.6, P < 0.001; interaction: F(16,160) = 2.16, P < 0.01). (F) Peak AIMs (n = 11; Fr = 4.97, P > 0.05). (G and H) L-DOPA–induced AIMs in mGluR5KD-D1 mice. (G) Time course of AIMs (n = 10; RM 2-way ANOVA, treatment: F(2,18) = 3.92, P = 0.31; time: F(9,81) = 20.56, P < 0.001; interaction: F(18,162) = 2.04, P < 0.05). (H) Peak AIMs (n = 10; Fr = 7.4, P < 0.05). (I and J) SKF38393-induced AIMs in mGluR5KD-D1 mice. (I) Time course of AIMs (n = 10; RM 2-way ANOVA, treatment: F(2,18) = 5.42, P < 0.05; time: F(8,72) = 17.33, P < 0.001; interaction: F(16,144) = 3.23, P < 0.001). (J) Peak AIMs (n = 10; Fr = 5, P > 0.05). (K and L) Quinpirole-induced AIMs in mGluR5KD-D1 mice. (K) Time course of AIMs (n = 10; RM 2-way ANOVA, treatment: F(2,18) = 2.51, P = 0.12; time: F(8,72) = 178.8, P < 0.001; interaction: F(16,144) = 2.98, P < 0.001). (L) Peak AIMs (n = 10; Fr = 1.4, P > 0.05). Bonferroni’s test or Dunn’s test (for peak AIMs): *P < 0.05, **P < 0.01 and ***P < 0.001 vs. DA receptor agonist + vehicle; #P < 0.05 and ###P < 0.001 vs. DA receptor agonist + MTEP.
We next set out to verify whether the antidyskinetic effects of U73122 or MTEP are mediated by neurons that coexpress both D1 and mGlu5 receptors. To this end, we applied the same protocol of 6-OHDA lesions and dopaminergic treatments to mGluR5KD-D1 transgenic mice, which display an almost complete loss of mGlu5 in striatal neurons expressing the D1 receptor (7) and a blunted striatal ERK1/2 response to D1 stimulation in a parkinsonian setting (11). Transgenic mGluR5KD-D1 mice responded to the 6-OHDA lesion in the same way as did WT mice (Supplemental Figure 4, A and B), and they also showed a similar antiakinetic response to L-DOPA in a test of forelimb use (Supplemental Figure 4C). However, compared with their WT littermates, transgenic mGluR5KD-D1 mice developed significantly milder dyskinesias when treated with L-DOPA or SKF38393 (see Supplemental Figure 4, D and E; and F and G for a direct genotype comparison). When challenged with either U73122 or MTEP, mGluR5KD-D1 mice showed no further reductions in peak AIM scores, as elicited by either L-DOPA (Figure 8, G and H) or SKF38393 (Figure 8, I and J), although the duration of SKF38393-induced dyskinesia appeared shorter after cotreatment with either compound (Figure 8I; P < 0.05 at 120 minutes for both U73122 and MTEP vs. vehicle cotreatment in the SKF38393 experiment). In contrast, quinpirole-induced AIMs were as severe in mGluR5KD-D1 mice as in WT controls (cf. Figure 8, E and K, and Supplemental Figure 4, H and I) and remained completely unaffected by U73122 or MTEP cotreatment (Figure 8, K and L).
Taken together, these results demonstrate that, in parkinsonian animals treated with dopaminergic agents, the same pattern of antidyskinetic effects is obtained by antagonizing either mGlu5 or PLC signaling. These antidyskinetic effects occur downstream of D1 receptor stimulation and are mediated by neurons that coexpress D1 and mGlu5 receptors.
Discussion
In cognitive, limbic, and motor brain networks, glutamatergic neurotransmission is under the pivotal control of dopaminergic afferents. The present work contributes to an improved understanding of the interplay between dopaminergic and glutamatergic signaling by revealing unheralded physical and functional interactions between D1 and mGlu5 receptors, 2 important therapeutic targets in several neuropsychiatric diseases (8, 34). Using recent improvements in single-cell BRET imaging (35), we demonstrate for what we believe is the first time the occurrence of D1-mGlu5 heteromers displaying atypical properties compared with the homomers. D1-mGlu5 heteromers interact with Gq proteins and enable a synergistic activation of PLC signaling and intracellular calcium release by their respective agonists. The native receptors in the striatum can form nanocomplexes revealed as fluorescent dots by the PLA assay, a method used to report a theoretical maximum distance of 30 nm between 2 target proteins (36). These receptor complexes become abundant and functionally crucial after a lesion of nigrostriatal dopaminergic projections mimicking PD. Accordingly, in the DA-denervated striatum, the costimulation of D1 and mGlu5 leads to a large activation of PLC. Inhibiting PLC signaling counters maladaptive molecular and behavioral responses to L-DOPA, the most important form of DA replacement therapy in PD.
In addition to reporting a physical interaction between D1 and mGlu5 receptors, the present study reveals the peculiar functional properties of this new signaling complex. Thus, in HEK cells expressing D1-mGlu5 heteromers, D1-like agonists trigger both the expected increase in cAMP production and an atypical release of intracellular Ca2+. Taken together, our cellular assays indicate that D1-mGlu5 heteromers can lead to a potentiation of PLC/Ca2+-dependent signaling both by increasing mGlu5 constitutive activity and by biasing a pool of D1 receptors toward activating this pathway. Moreover, our results predict that D1-mGlu5 heteromers would mediate a synergistic activation of PLC/Ca2+-dependent signaling in the presence of both receptor agonists.
The administration of L-DOPA results in a surge of striatal DA and Glu levels in both PD patients and animal models of PD (28, 29). These neurochemical changes concur to produce a strong striatal activation of ERK1/2 signaling, a master switch of synaptic, transcriptional, and epigenetic changes that mediate dyskinetic behaviors (31, 37–40). Despite the fact that NMDA and D1 receptors cooperate to activate ERK1/2 in the intact striatum (41, 42), NMDA receptor antagonists have no effect on the ERK1/2 response to L-DOPA in DA-denervated striatal neurons, which instead results from a functional interaction between D1 and mGlu5 receptors (11, 30). Using DA-denervated striatal slices incubated with SKF38393, we have previously shown that D1 agonist–induced ERK1/2 activation is both PLC and Ca2+ dependent (11). Taken together, these previous findings have suggested that the D1 receptor engages in a preferential crosstalk with mGlu5- and Gq-related signaling components in DA-denervated striatal neurons. The present study offers a plausible molecular explanation of such a phenomenon. Indeed, the results of our cellular assays reveal that D1-mGlu5 heteromers couple to Gq-type proteins and mediate a synergistic activation of PLC/Ca2+ signaling by the respective agonists. Consistent with these results, an assay of InsP production in rodent models reveals synergistic activation of PLC signaling by D1 and mGlu5 agonists specifically in the DA-denervated striatum. Accordingly, the PLA data (Figure 6) show that D1-mGlu5 receptor nanocomplexes become significantly more abundant in the striatum after DA denervation. Plausibly, the larger abundance of D1-mGlu5 receptor clusters in the DA-denervated striatum is linked to changes in D1 receptor localization occurring after a loss of dopaminergic inputs. In particular, a relocalization of D1 receptors away from synaptic membranes, as reported in DA-denervated rodents (3), would facilitate an association of D1 proteins with the mGlu5 receptor, which normally resides in perisynaptic membranes (20).
The heteromerization process increases the diversity of a receptor’s signaling through functional platforms (43). However, the possibilities of interaction are not random and their specificity seems tightly controlled. For example, we have previously shown 2 distinct groups of association compatibility between mGlu receptor subtypes (44): group I mGlu receptors (subtypes 1 and 5) interact together, but do not associate with others, while group II (subtypes 2 and 3) and III mGlu receptors (subtypes 4, 7, and 8) preferentially associate with each other, but not with those from group I. Similarly, we showed that group I mGlu1 receptors do not interact with GABAb receptors, which are, however, coexpressed in Purkinje neurons and functionally coupled (45). In the scope of the present work, we further tested other D1 and mGlu5 potential partners. We report no specific BRET signal between D1 and β2 adrenergic receptors, nor between mGlu5 and GABAb receptors, suggesting that those pairs of GPCRs cannot form heteromers, despite their demonstrated capacity for crossinteraction in neural modulation (46–49). In these cases, converging signaling pathways downstream of the receptors might be involved rather than a direct physical association. By opposition, we found specific BRET signals between D1 and mGlu1 receptors, which is a previously unknown interaction. Importantly, mGlu1 and mGlu5 receptors share a high degree of sequence homology and signal-transduction properties (50), but they differ substantially in terms of striatal expression levels (51). We furthermore found an interaction between mGlu5 and D2 receptors, which is in agreement with previous publications (52, 53). Beyond receptor structure, cellular context and physio/pathological conditions might or might not be permissive of specific interactions. Related to the striatum, a pathogenic role of heteromers involving D1 receptors has already been reported. In particular, it has been proposed that physical interactions between D1 and NMDA GluN1 (54) or D1 and D3 receptors (55) play a causal role in LID. However, these previously described heteromers were unequivocally induced by the chronic treatment with L-DOPA, often leading to an upregulation of the involved receptor proteins. In contrast, we here report that the D1-mGlu5 assembly is prompted by DA denervation and further maintained upon chronic L-DOPA treatment. Therefore, our results uncover a molecular determinant of denervation-induced signaling abnormalities that potently predispose to LID (56).
The present results have major implications for both understanding the striatal pathophysiology of PD and developing novel therapeutic options. On the one hand, our data point to D1-mGlu5 signaling complexes as mediators of maladaptive synaptic and molecular changes during the pharmacotherapy of PD (reviewed in refs. 6, 57). On the other hand, we here identify a mechanistic target for the treatment and prevention of LID, that is, a D1-mGlu5–dependent activation of PLC signaling. Indeed, we show that a pharmacological inhibition of PLC dampens the striatal activation of ERK1/2 and markedly improves dyskinesias induced by either L-DOPA or D1-like agonists without affecting the beneficial antiakinetic effect of these treatments. We furthermore show that a genetic knockdown of mGlu5 in D1-positive neurons occludes the effect of both PLC and mGlu5 inhibition on dyskinesias evoked by either L-DOPA or a D1 receptor agonist.
In line with the recent identification of GPCR heteromers as functional units and potential drug targets (58, 59), the present dissection of the D1-mGlu5 heteromer biochemical fingerprint could prompt a development of screening tools and ligands for therapeutic research. Compared with antagonizing ubiquitous D1 or mGlu5 receptors, disrupting the D1-mGlu5 heteromer is predicted to offer a better risk-benefit profile for the treatment and prevention of LID.
Methods
cDNA expression vectors.
For BRET experiments, receptors were tagged at the extracellular N-terminus or intracellular C-terminus part using standard molecular biology techniques employing PCR and fragment replacement strategies resulting in plasmids coding for Rluc8-mGlu5a, Venus-D1, mGlu5a-Nluc, and D1-Venus. The pRK5-HA-SNAP-mGlu5 plasmid was previously described (23). pRK5-HA-SNAP-mGlu5-DelCtail encodes for the mGlu5a receptor without C-terminal tail (until the GKSVT sequence at position 804). pRK5-cherryThr-CD4-mGlu5aCtail was obtained by substitution of mGlu1 C-tail coding sequence by mGlu5 C-tail coding sequence in the previously described pRK5-cherryThr-CD4-mGlu1Ctail plasmid (60). For BiFC, plasmids coding for D1-V1, D1-V2, CD8-V1, and CD8-V2 were provided by J. Javitch (Columbia University, New York, New York, USA). The splits of monomeric Venus were fused to the C-terminus of the D1, mGlu5a, or CD8. D1–V1 and CD8-V1 encoded the N-terminal split of mVenus (amino acids 1–155) fused to the D1 or CD8, respectively. D1–V2 and CD8-V2 express D1 receptor and CD8 respectively fused to the C-terminal split of mVenus (amino acids 156–240). The cDNA sequences coding for the splits of mVenus were amplified by PCR and subcloned in frame to the 3′ end coding sequence of mGlu5a without STOP codon in the pcDNA3.1-Myc-mGlu5a-Rluc8 plasmid to obtain plasmids coding for 5a-V1 and 5a-V2. The pmRFP plasmid coding for a MyrPalm tag corresponding to the N-terminal amino acid sequence (MGCIKSKRKDNLNDDE) from Lyn kinase fused to RFP protein was a gift from L. Hunday (Semmelweis University, Budapest, Hungary). pGloSensor 22F-cAMP plasmid was from Promega, and GcAMP6 coding plasmid was from Addgene.
Cell culture and transfection.
HEK293 cell culture and the calcium phosphate precipitation method were previously described (15). The double-stable HEK cell line coexpressing the Glo cAMP-sensor and the GcAMP6 Ca2+-sensor was created in HEK293 cells. Double antibiotic-resistant clones were selected with 250 μg/mL hygromycin + 500 μg/mL geneticin. The double-sensor stable cell line was plated in 96-well plates and transfected using Lipofectamine 2000 according to the manufacturer’s instructions (Sigma-Aldrich). To maintain a low concentration of ambient Glu, cells were cotransfected with the Glu transporter EAAC1 and incubated in serum-free DMEM Glutamax Medium (Life Technologies) at least 2 hours before the different assays were performed.
Primary hippocampal neurons culture.
Hippocampal neuronal primary cultures were prepared from embryonic day 17.5 rats as previously described (61). Hippocampal neurons were transfected according to the calcium phosphate transfection procedure previously described (62).
BiFC.
HEK cells were cotransfected with a 1:1 ratio of Venus splits V1 and V2 fusion proteins together with the pmRFP plasmid coding for a red fluorescent tag targeted to plasma membrane. Twenty-four hours after transfection, cells were washed with PBS and fixed with 4% paraformaldehyde for 15 minutes. Cell nuclei were stained with DAPI, and coverslips were mounted in Mowiol. Images were acquired using an Axio Observer Z.1 microscope and analyzed using ImageJ software (NIH). Red fluorescence images were thresholded to keep only the plasma membrane fluorescence area. The selection corresponding to this threshold was transferred on the Venus fluorescence images, and membranous fluorescence of complemented Venus was quantified in this area. Intracellular fluorescence was measured in a square intracellular area excluding the nucleus. The membrane/(membrane + intracellular) Venus fluorescence ratio was then calculated and expressed as a percentage of total fluorescence.
Cell population BRET measurements.
BRET measurements in cell populations were performed as previously described (63) using the Mithras LB 940 (Berthold Technologies). Briefly, HEK cells were transfected with a constant amount of the plasmid coding for the donor entity (1 μg of Rluc8-tagged donors or 100 ng of Nluc-tagged donors) combined with increasing quantities of the plasmid coding for the acceptor entity (0–4 μg). A suitable amount of the noncoding plasmid pcDNA3 was added to reach a total amount 5 μg of DNA per 100 mm dish. Forty-eight hours after transfection, cells were suspended in PBS with 0.1% (w/v) glucose at room temperature (RT). Cells were then distributed in 96-well microplates (Greiner) at a density of 100,000 cells/well with Rluc8 donor or 50,000 cells/well with Nluc donor. Before the BRET experiment, we measured the light emitted at 525 nm upon light excitation at 485 nm, indicative of the amount of Venus-tagged proteins. BRET (535 nm/485 nm ratio) was assessed by calculating the ratio of the light emitted by the acceptor entity (510–550 nm band-pass filter, Em535) to the light emitted by the donor (460–500 nm band-pass filter, Em480) after the addition of 5 μM of the Rluc8 substrate Coelenterazine H or 20 μM of the Nluc substrate Furimazine. Net BRET values were plotted as a function of total fluorescence/luminescence ratio, and saturation curves were drawn using the one-site specific binding nonlinear fit of GraphPad Prism 7 software (GraphPad Software Inc.)
Single-cell BRET imaging.
Single-cell BRET imaging experiments were performed and analyzed as previously described (35, 64, 65). Briefly, hippocampal neurons in culture were cotransfected at DIV10 to 12 either with plasmids coding for the Nluc-tagged donor together with the Venus-tagged acceptor or with plasmids coding for the donor only and DsRed as a transfection reporter. From 24 to 48 hours after transfection, neurons were recorded in the following external medium: 140 mM NaCl, 2 mM CaCl2, 3 mM KCl, 10 mM HEPES, 10 mM d-glucose, 0.01 mM glycine, pH 7.4, with an osmolarity of 330 mOsm.
TR-FRET measurements.
TR-FRET experiments with the SNAP-tagged mGlu5 receptor were performed as previously described for other mGluRs (23). HEK293 cells were cotransfected with plasmids coding for HA-SNAP-mGlu5 with or without D1-Venus.
InsP assay in cells.
Quantification of InsP levels were performed using the homogeneous time-resolved FRET (HTRF) assay IPone kit according to the manufacturer’s recommendations (Cisbio). HEK cells were plated in 96-well plates and transiently transfected with the indicated receptors together with the Glu transporter EAAC1. The day of the experiments, cells were incubated for 2 hours in serum-free DMEM Glutamax before stimulation with mGlu5 ligands for 30 minutes at 37°C (l-quisqualic acid [10 μM] or LY341495 [100 μM]). Cells were then lysed using the supplied lysis buffer containing d2-labeled InsP and Lumi4-Tb cryptate-labeled anti-InsP antibodies. The HTRF signal was measured after 1 hour of RT incubation in the dark using PHERAstar FS microplate reader. FRET signal was determined by measuring d2 acceptor emission (665 nm) and Tb donor emission (620 nm) using a 50 μs delay and a 450 μs integration upon excitation at 337 nm on a PHERAstar FS (BMG LabTech). TR-FRET (or HTRF) ratio (665 nm/620 nm × 104, Cisbio Bioassays patent US5,527,684) was calculated.
Intracellular calcium and cAMP measurements.
After 2 hours in serum-free DMEM Glutamax, stable double-sensor HEK cells were washed with Tag-Lite (Cisbio). Drug-triggered cytosolic calcium release was monitored by measurements of GcAMP6 fluorescence (excitation 485 nm and emission 525 nm) using a FlexStation (Molecular Devices). Increasing drug concentrations were injected after 20 seconds of recording. For basal controls, buffer alone was added after 20 seconds. For cAMP measurement, the GloSensor (Promega) assay is based on cAMP-induced conformational changes of a genetically modified firefly luciferase into which a cAMP-binding protein moiety has been inserted. Cells were incubated in Tag-Lite buffer with 450μg/mL of d-Luciferin during 1 hour at RT in the dark. After 5 minutes of temperature stabilization at 28°C in the plate reader, increasing doses of ligand were added to the cells with 500 μM of IBMX (cAMP/cGMP phosphodiesterases inhibitor). Drug-triggered cAMP production was assessed by luminescence measurements using the Mithras LB 940 plate-reader (no emission filter, 1-second integration time). For each drug concentration, the maximal amplitude of fluorescence or luminescence intensity increase was plotted with respect to the concentrations in a logarithmic scale. Dose-response curves were drawn using the “log (agonist) vs. response (3 parameters)” nonlinear fit of GraphPad Prism 7 software (GraphPad Software Inc.).
PLA.
Experiments were performed on intact and lesioned striatal sections of both WT and mGluR5KO-D1 mice following a period of daily s.c. treatment with either vehicle or L-DOPA (6 mg/kg) for 3 weeks. Coronal striatal sections, 50 μm thick, were blocked with 10% horse serum and permeabilized with 0.2% Triton X-100 (v/v). Experiments were then performed according to the manufacturer’s instructions (Duolink & PLA Technology, Olink-Bioscience ref. 36). Briefly, sections were incubated with D1 guinea pig primary antibodies (Frontier Institut Co., AB_2571595) directly conjugated to minus Duolink II PLA probes and with mGlu5 rabbit primary antibody (Millipore; AB5675) at 4°C for 36 to 48 hours. PLA probes anti-rabbit plus, which are secondary antibodies conjugated with oligonucleotides, were added and incubated for 1 hour at 37°C. Amplification template oligonucleotides were hybridized to pairs of PLA and circularized by ligation. The formed DNA circle was then amplified using rolling circle amplification, and detection of the amplicons was carried out using the 624 Duolink In Situ Detection Kits, resulting in red fluorescence signals. Sections were mounted and were analyzed under a ×40 oil immersion objective using a confocal microscope (Zeiss LSM 780 or Leica TCS SP8 X). All acquisitions were made the same day under the same microscope setting and laser power to minimize variability among samples. Distinct bright spots contained within an area of the section designated by the experimenter were counted using an ImageJ macro. Briefly, we determined a presized region of interest (ROI) and then performed segmentation by thresholding in order to generate binary images. Six ROIs were analyzed per striatum to measure a mean value per animal. To minimize experimental bias during the analysis, all images were processed blindly using the intermodes autothreshold function (66). With this approach, we observed minimum variability in the analysis of PLA quantification. Using the Analyze Particle function of ImageJ, the numbers of particles larger than 0.198 μm2 in the binary image were counted as clusters. Data were exported to Prism software for further analysis.
Animals.
Behavioral experiments were performed using female Sprague Dawley rats (200–225 g; Charles River), or WT mice and BAC transgenic mice having a selective knockdown of mGlu5 in D1 receptor–expressing neurons (the previously characterized mGluR5KD-D1 line; refs. 7, 11). For studying receptor complexes (Figure 6) and SKF38393-induced pERK1/2 activation (Figure 7, G and H), we used mice with a genetic ablation of the mGlu5 receptor in D1-expressing cells, obtained by crossbreeding homozygous mGluR5loxP/loxP mice (67) with heterozygous BAC-Drd1a-Cre mice (GENSAT project founder line EY262; ref. 68). All mice were on a C57BL/6 genetic background and approximately 10 weeks old at the beginning of the experiments; both sexes were used. Rats and mice were housed under a 12-hour light/12-hour dark cycle with access to water and food ad libitum.
DA-denervating lesions.
Unilateral DA-denervating lesions were produced by injecting 6-OHDA in the right MFB, according to methods that are well established in both in rats and mice (69, 70). Additional information is provided in Supplemental Methods.
Evaluation of 6-OHDA lesion in mice and rats.
Three weeks after 6-OHDA lesion, forelimb use asymmetry was evaluated in rats and mice using the cylinder test, revealing a significant reduction in the spontaneous use of the forelimb contralateral to the lesion (69–72). Briefly, mice and rats were placed individually in a glass cylinder (10 cm diameter and 14 cm height for mice; 21 cm diameter and 34 cm height for rats) and were videotaped for 5 minutes. The total number of supporting wall contacts performed independently with left and right forepaw were counted offline. Results were expressed as the percentage use of the forelimb contralateral to the lesion: (left forepaw contact/total number of wall contacts) × 100. The 6-OHDA–lesioned animals used in this study were selected using a stringent cutoff value of 25% or less contralateral paw usage. The cylinder test was also used to evaluate beneficial “therapeutic-like” effects of L-DOPA and rule out possible adverse effects by U73122 (rat data in Supplemental Figure 2; mouse data in Supplemental Figure 3). To assess the antiakinetic action of L-DOPA, animals were recorded in the cylinder test at 15 to 20 minutes after drug injection. In addition to the behavioral screening, tyrosine hydroxylase (TH) histochemistry was used to verify the extent of nigrostriatal DA degeneration at the end of the experiments (see below).
In vivo drug treatments.
L-DOPA methyl ester (3 and 6 mg/kg, Sigma-Aldrich) was always coadministered with a fixed dose of Benserazide-HCl (12 mg/kg in both rats and mice; Sigma-Aldrich). The drugs were freshly dissolved in physiological saline and injected s.c. All other drugs used and their doses and administration routes are reported in Supplemental Table 1.
AIM ratings.
AIMs were quantified by an experimentally blinded investigator using previously validated scales (71–73). Additional information is provided in Supplemental Methods.
Immunohistochemistry.
Rats and mice were anesthetized with pentobarbital (240 mg/kg, i.p., Apoteksbolaget), 30 minutes after the final injection of either L-DOPA or SKF38393, and were transcardially perfused with 4% ice-cold PFA (pH 7.4). Extracted brains were postfixed in 4% ice-cold PFA for 2 hours and subsequently transferred to a 25% sucrose solution for 24 to 36 hours. Brains were sectioned coronally on a freezing microtome at 30 μm thickness. Free-floating sections from the striatum were used for immunohistochemical detection of pERK1/2 and TH as previously (11). Additional information is provided in Supplemental Methods.
Counts of pERK1/2-positive cells.
Striatal sections immunostained for pERK1/2 were visualized under a ×20 objective using a Nikon eclipse 80i microscope merged with a Nikon DMX 1200F video camera (11). Sample areas (0.913 mm2) were acquired from the dorsomedial (DM) and ventrolateral (VL) quadrants of the motor striatum at 3 different coronal levels (rostral, mid, and caudal striatum) based on predefined anatomical coordinates. In each data set to be compared, all acquisitions were made on the same day under the same microscope setting. The digitized images were then converted to an 8-bit grayscale using the open source image processing program ImageJ (NIH). Background threshold was set to the same value in all samples, and immunostained cells were counted as single particles in an automated fashion (analyze particles plugin). Data were expressed as total number of immunoreactive cells per sample area (averaged for all sample areas per striatal quadrant per animal).
InsP assay in striatal slices.
InsP formation was measured in rat striatal slices prelabeled with a tritiated precursor as described previously (27). A total of 20 rats with unilateral 6-OHDA lesions were sacrificed by decapitation; striatal tissue was manually dissected out on ice and transferred on ice-cold Krebs-Henseleit buffer (118 mM NaCl, 4.7 mM KCl, 1.18 mM MgSO4, 1.18 mM KH2PO4, 24.8 mM NaHCO3, 1.2 mM CaCl2, 10 mM d-glucose) that had been pregassed with 95% O2 and 5% CO2 to pH 7.4. Slices (350 × 350 μm) were prepared using a McIlwain tissue chopper and randomly distributed into different tubes. A total of 40 μL of gravity packed slices/tube were incubated for 60 minutes in 350 μL buffer containing 1 μCi of myo-[3H]inositol. Slices were then incubated with LiCl (10 mM to block InsP degradation) and with the mGlu1 receptor antagonist JNJ16259685 (10 μM to block mGlu1 receptors). Thereafter, slices were stimulated with ligands of D1 and mGlu5 receptors at standard concentrations for this assay (74–75). The following treatments were compared: (i) vehicle (basal levels), (ii) the D1 receptor agonist SKF38393 (200 μM), (iii) the group I mGlu receptor agonist DHPG (200 μM), (iv) the combination of SKF38393 and DHPG, and (v) the same combination as in (iv) in the presence of the mGlu5 antagonist MTEP (10 μM). Incubations were stopped after 60 minutes by adding 900 μL methanol/chloroform (2:1). After further addition of 300 μL chloroform and 600 μL water, samples were centrifuged at low speed to facilitate phase separation, and the [3H]InsP present in the supernatant was separated by anion exchange chromatography. Samples were removed from their water phase, incubated with 0.5 N NaOH, and allowed to dry at 50°C for 2 hours. Proteins were measured as described by Lowry et al. (76). In the statistical analysis, n represents the number of tubes incubated with different treatments (each tube containing tissue from more than 1 rat, as in ref. 27).
Statistics.
Statistical analysis was carried out using GraphPad Prism 7 software (GraphPad Software Inc.). Statistical tests and post hoc comparisons are indicated in the figure legends. P < 0.05 was considered statistically significant. Data compared using parametric statistical tests are presented as group mean ± SEM, whereas data compared with nonparametric statistics are presented as group median and range.
Study approval.
All experiments were approved by the Malmö-Lund Ethical Committee on Animal Research, Lund District Court, Lund, Sweden.
Author contributions
IS, EG, MAC, and JP designed the experiments and analyzed and interpreted the data. IS performed all experiments in rats with the assistance of NM (behavioral pharmacology) and LDM (InsP hydrolysis assay). IS and LA performed behavioral-pharmacological and histopathological experiments in mice; EG performed most of the experiments on the physical and functional molecular interaction in vitro. DMD and JFI designed, performed, and analyzed the TR-FRET experiments and helped with the pharmacological data analysis. NB helped with the conception and production of Venus and Luciferase fusion BRET probes. EM performed BRET experiments in HEK cells. FA designed, performed, and analyzed PLA experiments. CJT performed PLA experiments. LA, JPP, LF, and FN provided help with experiment design, data analysis, and interpretation for different parts of the study. MAC and JP wrote the manuscript with input from IS and EG. IS and EG contributed equitably to this work. IS appears first in the author list because she handled the preparation and revision of the manuscript until publication.
Supplementary Material
Acknowledgments
This work was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation program (grant agreement no. 646788 to JP), the Agence Nationale de la Recherche (ANR-13-JSV4-0005-01 and ANR-17-CE11-0046 to JP), and the Région Languedoc-Roussillon (Chercheur d’Avenir) to JP; by grants from the Swedish Research Council, the Basal Ganglia Disorders Linnaeus Consortium (BAGADILICO), Multidisciplinary Research on Parkinson’s Disease (Multipark), the Swedish Brain Foundation, and Swedish Governmental Funds for Clinical Research (to MAC). IS is supported by a postdoctoral scholarship from the Swedish Brain Foundation. We thank J. Javitch for providing the Venus-split plasmids, Dr. C. Gales (Toulouse University, Toulouse, France) for providing the Nluc-Gαq plasmid, and L. Hunday for giving us the pmRFP plasmid. We thank the ARPEGE Pharmacology-Screening-Interactome platform facility (UMS Biocampus, Montpellier, France) for the use of fluorimeters for cell population BRET and functional assays, and Ann-Christin Lindh, Elena Espa, and Michael Sparrenius for excellent technical assistance.
Version 1. 02/10/2020
Electronic publication
Version 2. 03/02/2020
Print issue publication
Funding Statement
To JP
To JP
To JP
To MAC
To IS
To MAC
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(3):1168–1184.https://doi.org/10.1172/JCI126361.
Contributor Information
Irene Sebastianutto, Email: irene.sebastianutto@med.lu.se.
Elise Goyet, Email: elise.goyet@gmail.com.
Laura Andreoli, Email: laura.andreoli@med.lu.se.
Joan Font-Ingles, Email: Joan.Font-Ingles@igf.cnrs.fr.
David Moreno-Delgado, Email: david.Moreno@ucb.com.
Nathalie Bouquier, Email: Nathalie.Bouquier@igf.cnrs.fr.
Céline Jahannault-Talignani, Email: celine.Jahannault-Talignani@igf.cnrs.fr.
Enora Moutin, Email: Enora.Moutin@igf.cnrs.fr.
Luisa Di Menna, Email: luisa.dimenna@neuromed.it.
Natallia Maslava, Email: natallia.maslava@med.lu.se.
Jean-Philippe Pin, Email: jean-philippe.pin@igf.cnrs.fr.
Laurent Fagni, Email: laurent.fagni@igf.cnrs.fr.
Ferdinando Nicoletti, Email: nicoletti@neuromed.it.
Fabrice Ango, Email: fabrice.ango@igf.cnrs.fr.
M. Angela Cenci, Email: Angela.Cenci_Nilsson@med.lu.se.
Julie Perroy, Email: julie.perroy@igf.cnrs.fr.
References
- 1.Bagetta V, Ghiglieri V, Sgobio C, Calabresi P, Picconi B. Synaptic dysfunction in Parkinson’s disease. Biochem Soc Trans. 2010;38(2):493–497. doi: 10.1042/BST0380493. [DOI] [PubMed] [Google Scholar]
- 2.Surmeier DJ, Graves SM, Shen W. Dopaminergic modulation of striatal networks in health and Parkinson’s disease. Curr Opin Neurobiol. 2014;29:109–117. doi: 10.1016/j.conb.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiorentini C, et al. Loss of synaptic D1 dopamine/N-methyl-D-aspartate glutamate receptor complexes in L-DOPA-induced dyskinesia in the rat. Mol Pharmacol. 2006;69(3):805–812. doi: 10.1124/mol.105.016667. [DOI] [PubMed] [Google Scholar]
- 4.Picconi B, et al. Loss of bidirectional striatal synaptic plasticity in L-DOPA-induced dyskinesia. Nat Neurosci. 2003;6(5):501–506. doi: 10.1038/nn1040. [DOI] [PubMed] [Google Scholar]
- 5.Westin JE, Vercammen L, Strome EM, Konradi C, Cenci MA. Spatiotemporal pattern of striatal ERK1/2 phosphorylation in a rat model of L-DOPA-induced dyskinesia and the role of dopamine D1 receptors. Biol Psychiatry. 2007;62(7):800–810. doi: 10.1016/j.biopsych.2006.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murer MG, Moratalla R. Striatal signaling in L-DOPA-induced dyskinesia: common mechanisms with drug abuse and long term memory involving D1 dopamine receptor stimulation. Front Neuroanat. 2011;5:51. doi: 10.3389/fnana.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novak M, et al. Incentive learning underlying cocaine-seeking requires mGluR5 receptors located on dopamine D1 receptor-expressing neurons. J Neurosci. 2010;30(36):11973–11982. doi: 10.1523/JNEUROSCI.2550-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebastianutto I, Cenci MA. mGlu receptors in the treatment of Parkinson’s disease and L-DOPA-induced dyskinesia. Curr Opin Pharmacol. 2018;38:81–89. doi: 10.1016/j.coph.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Rylander D, et al. A mGluR5 antagonist under clinical development improves L-DOPA-induced dyskinesia in parkinsonian rats and monkeys. Neurobiol Dis. 2010;39(3):352–361. doi: 10.1016/j.nbd.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Iderberg H, Rylander D, Bimpisidis Z, Cenci MA. Modulating mGluR5 and 5-HT1A/1B receptors to treat l-DOPA-induced dyskinesia: effects of combined treatment and possible mechanisms of action. Exp Neurol. 2013;250:116–124. doi: 10.1016/j.expneurol.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Fieblinger T, et al. Mechanisms of dopamine D1 receptor-mediated ERK1/2 activation in the parkinsonian striatum and their modulation by metabotropic glutamate receptor type 5. J Neurosci. 2014;34(13):4728–4740. doi: 10.1523/JNEUROSCI.2702-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dacres H, Wang J, Dumancic MM, Trowell SC. Experimental determination of the Förster distance for two commonly used bioluminescent resonance energy transfer pairs. Anal Chem. 2010;82(1):432–435. doi: 10.1021/ac9022956. [DOI] [PubMed] [Google Scholar]
- 13.Juhasz JR, Hasbi A, Rashid AJ, So CH, George SR, O’Dowd BF. Mu-opioid receptor heterooligomer formation with the dopamine D1 receptor as directly visualized in living cells. Eur J Pharmacol. 2008;581(3):235–243. doi: 10.1016/j.ejphar.2007.11.060. [DOI] [PubMed] [Google Scholar]
- 14.Lee FJ, et al. Dual regulation of NMDA receptor functions by direct protein-protein interactions with the dopamine D1 receptor. Cell. 2002;111(2):219–230. doi: 10.1016/S0092-8674(02)00962-5. [DOI] [PubMed] [Google Scholar]
- 15.Perroy J, et al. Direct interaction enables cross-talk between ionotropic and group I metabotropic glutamate receptors. J Biol Chem. 2008;283(11):6799–6805. doi: 10.1074/jbc.M705661200. [DOI] [PubMed] [Google Scholar]
- 16.Vidi PA, Przybyla JA, Hu CD, Watts VJ. Visualization of G protein-coupled receptor (GPCR) interactions in living cells using bimolecular fluorescence complementation (BiFC) Curr Protoc Neurosci. 2010;Chapter 5:Unit 5.29. doi: 10.1002/0471142301.ns0529s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaeger WC, Armstrong SP, Hill SJ, Pfleger KD. Biophysical detection of diversity and bias in GPCR function. Front Endocrinol (Lausanne) 2014;5:26. doi: 10.3389/fendo.2014.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladepeche L, et al. Single-molecule imaging of the functional crosstalk between surface NMDA and dopamine D1 receptors. Proc Natl Acad Sci U S A. 2013;110(44):18005–18010. doi: 10.1073/pnas.1310145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kern A, Mavrikaki M, Ullrich C, Albarran-Zeckler R, Brantley AF, Smith RG. Hippocampal dopamine/DRD1 signaling dependent on the ghrelin receptor. Cell. 2015;163(5):1176–1190. doi: 10.1016/j.cell.2015.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lujan R, Nusser Z, Roberts JD, Shigemoto R, Somogyi P. Perisynaptic location of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur J Neurosci. 1996;8(7):1488–1500. doi: 10.1111/j.1460-9568.1996.tb01611.x. [DOI] [PubMed] [Google Scholar]
- 21.Sesack SR, Aoki C, Pickel VM. Ultrastructural localization of D2 receptor-like immunoreactivity in midbrain dopamine neurons and their striatal targets. J Neurosci. 1994;14(1):88–106. doi: 10.1523/JNEUROSCI.14-01-00088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu XY, et al. Modulation of D2R-NR2B interactions in response to cocaine. Neuron. 2006;52(5):897–909. doi: 10.1016/j.neuron.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Doumazane E, et al. Illuminating the activation mechanisms and allosteric properties of metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2013;110(15):E1416–E1425. doi: 10.1073/pnas.1215615110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasrallah C, et al. Direct coupling of detergent purified human mGlu5 receptor to the heterotrimeric G proteins Gq and Gs. Sci Rep. 2018;8(1):4407. doi: 10.1038/s41598-018-22729-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urizar E, Yano H, Kolster R, Galés C, Lambert N, Javitch JA. CODA-RET reveals functional selectivity as a result of GPCR heteromerization. Nat Chem Biol. 2011;7(9):624–630. doi: 10.1038/nchembio.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pin JP, Duvoisin R. The metabotropic glutamate receptors: structure and functions. Neuropharmacology. 1995;34(1):1–26. doi: 10.1016/0028-3908(94)00129-G. [DOI] [PubMed] [Google Scholar]
- 27.Nicoletti F, Meek JL, Iadarola MJ, Chuang DM, Roth BL, Costa E. Coupling of inositol phospholipid metabolism with excitatory amino acid recognition sites in rat hippocampus. J Neurochem. 1986;46(1):40–46. doi: 10.1111/j.1471-4159.1986.tb12922.x. [DOI] [PubMed] [Google Scholar]
- 28.de la Fuente-Fernández R, et al. Levodopa-induced changes in synaptic dopamine levels increase with progression of Parkinson’s disease: implications for dyskinesias. Brain. 2004;127(Pt 12):2747–2754. doi: 10.1093/brain/awh290. [DOI] [PubMed] [Google Scholar]
- 29.Ahmed I, et al. Glutamate NMDA receptor dysregulation in Parkinson’s disease with dyskinesias. Brain. 2011;134(Pt 4):979–986. doi: 10.1093/brain/awr028. [DOI] [PubMed] [Google Scholar]
- 30.Rylander D, Recchia A, Mela F, Dekundy A, Danysz W, Cenci MA. Pharmacological modulation of glutamate transmission in a rat model of L-DOPA-induced dyskinesia: effects on motor behavior and striatal nuclear signaling. J Pharmacol Exp Ther. 2009;330(1):227–235. doi: 10.1124/jpet.108.150425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fasano S, et al. Inhibition of Ras-guanine nucleotide-releasing factor 1 (Ras-GRF1) signaling in the striatum reverts motor symptoms associated with L-dopa-induced dyskinesia. Proc Natl Acad Sci U S A. 2010;107(50):21824–21829. doi: 10.1073/pnas.1012071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delfino MA, Stefano AV, Ferrario JE, Taravini IR, Murer MG, Gershanik OS. Behavioral sensitization to different dopamine agonists in a parkinsonian rodent model of drug-induced dyskinesias. Behav Brain Res. 2004;152(2):297–306. doi: 10.1016/j.bbr.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Ding Y, Won L, Britt JP, Lim SA, McGehee DS, Kang UJ. Enhanced striatal cholinergic neuronal activity mediates L-DOPA-induced dyskinesia in parkinsonian mice. Proc Natl Acad Sci U S A. 2011;108(2):840–845. doi: 10.1073/pnas.1006511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foster DJ, Conn PJ. Allosteric modulation of GPCRs: new insights and potential utility for treatment of schizophrenia and other CNS disorders. Neuron. 2017;94(3):431–446. doi: 10.1016/j.neuron.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goyet E, Bouquier N, Ollendorff V, Perroy J. Fast and high resolution single-cell BRET imaging. Sci Rep. 2016;6:28231. doi: 10.1038/srep28231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fredriksson S, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20(5):473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 37.Pavón N, Martín AB, Mendialdua A, Moratalla R. ERK phosphorylation and FosB expression are associated with L-DOPA-induced dyskinesia in hemiparkinsonian mice. Biol Psychiatry. 2006;59(1):64–74. doi: 10.1016/j.biopsych.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 38.Cerovic M, et al. Derangement of Ras-guanine nucleotide-releasing factor 1 (Ras-GRF1) and extracellular signal-regulated kinase (ERK) dependent striatal plasticity in L-DOPA-induced dyskinesia. Biol Psychiatry. 2015;77(2):106–115. doi: 10.1016/j.biopsych.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Heiman M, et al. Molecular adaptations of striatal spiny projection neurons during levodopa-induced dyskinesia. Proc Natl Acad Sci U S A. 2014;111(12):4578–4583. doi: 10.1073/pnas.1401819111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santini E, Heiman M, Greengard P, Valjent E, Fisone G. Inhibition of mTOR signaling in Parkinson’s disease prevents L-DOPA-induced dyskinesia. Sci Signal. 2009;2(80):ra36. doi: 10.1126/scisignal.2000308. [DOI] [PubMed] [Google Scholar]
- 41.Girault JA, Valjent E, Caboche J, Hervé D. ERK2: a logical AND gate critical for drug-induced plasticity? Curr Opin Pharmacol. 2007;7(1):77–85. doi: 10.1016/j.coph.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 42.Valjent E, Corvol JC, Pages C, Besson MJ, Maldonado R, Caboche J. Involvement of the extracellular signal-regulated kinase cascade for cocaine-rewarding properties. J Neurosci. 2000;20(23):8701–8709. doi: 10.1523/JNEUROSCI.20-23-08701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat Rev Neurosci. 2001;2(4):274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 44.Doumazane E, Scholler P, Zwier JM, Trinquet E, Rondard P, Pin JP. A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J. 2011;25(1):66–77. doi: 10.1096/fj.10-163147. [DOI] [PubMed] [Google Scholar]
- 45.Rives ML, et al. Crosstalk between GABAB and mGlu1a receptors reveals new insight into GPCR signal integration. EMBO J. 2009;28(15):2195–2208. doi: 10.1038/emboj.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei S. Cross interaction of dopaminergic and adrenergic systems in neural modulation. Int J Physiol Pathophysiol Pharmacol. 2014;6(3):137–142. [PMC free article] [PubMed] [Google Scholar]
- 47.Smith Y, Charara A, Hanson JE, Paquet M, Levey AI. GABA(B) and group I metabotropic glutamate receptors in the striatopallidal complex in primates. J Anat. 2000;196(Pt 4):555–576. doi: 10.1046/j.1469-7580.2000.19640555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gubellini P, Pisani A, Centonze D, Bernardi G, Calabresi P. Metabotropic glutamate receptors and striatal synaptic plasticity: implications for neurological diseases. Prog Neurobiol. 2004;74(5):271–300. doi: 10.1016/j.pneurobio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Dickerson JW, Conn PJ. Therapeutic potential of targeting metabotropic glutamate receptors for Parkinson’s disease. Neurodegener Dis Manag. 2012;2(2):221–232. doi: 10.2217/nmt.12.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abe T, Sugihara H, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. Molecular characterization of a novel metabotropic glutamate receptor mGluR5 coupled to inositol phosphate/Ca2+ signal transduction. J Biol Chem. 1992;267(19):13361–13368. [PubMed] [Google Scholar]
- 51.Testa CM, Standaert DG, Young AB, Penney JB. Metabotropic glutamate receptor mRNA expression in the basal ganglia of the rat. J Neurosci. 1994;14(5 Pt 2):3005–3018. doi: 10.1523/JNEUROSCI.14-05-03005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beggiato S, et al. Functional role of striatal A2A, D2, and mGlu5 receptor interactions in regulating striatopallidal GABA neuronal transmission. J Neurochem. 2016;138(2):254–264. doi: 10.1111/jnc.13652. [DOI] [PubMed] [Google Scholar]
- 53.Cabello N, et al. Metabotropic glutamate type 5, dopamine D2 and adenosine A2a receptors form higher-order oligomers in living cells. J Neurochem. 2009;109(5):1497–1507. doi: 10.1111/j.1471-4159.2009.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song L, et al. Targeting the D1-N-methyl-D-aspartate receptor complex reduces L-dopa-induced dyskinesia in 6-hydroxydopamine-lesioned Parkinson’s rats. Drug Des Devel Ther. 2016;10:547–555. doi: 10.2147/DDDT.S93487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farré D, et al. Stronger dopamine D1 receptor-mediated neurotransmission in dyskinesia. Mol Neurobiol. 2015;52(3):1408–1420. doi: 10.1007/s12035-014-8936-x. [DOI] [PubMed] [Google Scholar]
- 56.Nadjar A, Gerfen CR, Bezard E. Priming for l-dopa-induced dyskinesia in Parkinson’s disease: a feature inherent to the treatment or the disease? Prog Neurobiol. 2009;87(1):1–9. doi: 10.1016/j.pneurobio.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cenci MA, Konradi C. Maladaptive striatal plasticity in L-DOPA-induced dyskinesia. Prog Brain Res. 2010;183:209–233. doi: 10.1016/S0079-6123(10)83011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borroto-Escuela DO, et al. Understanding the role of GPCR heteroreceptor complexes in modulating the brain networks in health and disease. Front Cell Neurosci. 2017;11:37. doi: 10.3389/fncel.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuxe K, et al. Moonlighting proteins and protein-protein interactions as neurotherapeutic targets in the G protein-coupled receptor field. Neuropsychopharmacology. 2014;39(1):131–155. doi: 10.1038/npp.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raynaud F, et al. SNAP23-Kif5 complex controls mGlu1 receptor trafficking. J Mol Cell Biol. 2018;10(5):423–436. doi: 10.1093/jmcb/mjy031. [DOI] [PubMed] [Google Scholar]
- 61.Xu C, et al. REV, A BRET-based sensor of ERK activity. Front Endocrinol (Lausanne) 2013;4:95. doi: 10.3389/fendo.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goetze B, Grunewald B, Baldassa S, Kiebler M. Chemically controlled formation of a DNA/calcium phosphate coprecipitate: application for transfection of mature hippocampal neurons. J Neurobiol. 2004;60(4):517–525. doi: 10.1002/neu.20073. [DOI] [PubMed] [Google Scholar]
- 63.Perroy J, Pontier S, Charest PG, Aubry M, Bouvier M. Real-time monitoring of ubiquitination in living cells by BRET. Nat Methods. 2004;1(3):203–208. doi: 10.1038/nmeth722. [DOI] [PubMed] [Google Scholar]
- 64.Coulon V, et al. Subcellular imaging of dynamic protein interactions by bioluminescence resonance energy transfer. Biophys J. 2008;94(3):1001–1009. doi: 10.1529/biophysj.107.117275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chastagnier Y, Moutin E, Hemonnot AL, Perroy J. Image processing for bioluminescence resonance energy transfer measurement-BRET-analyzer. Front Comput Neurosci. 2017;11:118. doi: 10.3389/fncom.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prewitt JM, Mendelsohn ML. The analysis of cell images. Ann N Y Acad Sci. 1966;128(3):1035–1053. doi: 10.1111/j.1749-6632.1965.tb11715.x. [DOI] [PubMed] [Google Scholar]
- 67.Xu J, Zhu Y, Contractor A, Heinemann SF. mGluR5 has a critical role in inhibitory learning. J Neurosci. 2009;29(12):3676–3684. doi: 10.1523/JNEUROSCI.5716-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gong S, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27(37):9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cenci MA, Lundblad M. Ratings of L-DOPA-induced dyskinesia in the unilateral 6-OHDA lesion model of Parkinson’s disease in rats and mice. Curr Protoc Neurosci. 2007;Chapter 9:Unit 9.25. doi: 10.1002/0471142301.ns0925s41. [DOI] [PubMed] [Google Scholar]
- 70.Francardo V, Recchia A, Popovic N, Andersson D, Nissbrandt H, Cenci MA. Impact of the lesion procedure on the profiles of motor impairment and molecular responsiveness to L-DOPA in the 6-hydroxydopamine mouse model of Parkinson’s disease. Neurobiol Dis. 2011;42(3):327–340. doi: 10.1016/j.nbd.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 71.Sebastianutto I, Maslava N, Hopkins CR, Cenci MA. Validation of an improved scale for rating l-DOPA-induced dyskinesia in the mouse and effects of specific dopamine receptor antagonists. Neurobiol Dis. 2016;96:156–170. doi: 10.1016/j.nbd.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 72.Lundblad M, Andersson M, Winkler C, Kirik D, Wierup N, Cenci MA. Pharmacological validation of behavioural measures of akinesia and dyskinesia in a rat model of Parkinson’s disease. Eur J Neurosci. 2002;15(1):120–132. doi: 10.1046/j.0953-816x.2001.01843.x. [DOI] [PubMed] [Google Scholar]
- 73.Breger LS, Dunnett SB, Lane EL. Comparison of rating scales used to evaluate L-DOPA-induced dyskinesia in the 6-OHDA lesioned rat. Neurobiol Dis. 2013;50:142–150. doi: 10.1016/j.nbd.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 74.Romano MR, et al. Type-1, but not type-5, metabotropic glutamate receptors are coupled to polyphosphoinositide hydrolysis in the retina. Neurochem Res. 2016;41(4):924–932. doi: 10.1007/s11064-015-1775-y. [DOI] [PubMed] [Google Scholar]
- 75.Di Menna L, et al. Functional partnership between mGlu3 and mGlu5 metabotropic glutamate receptors in the central nervous system. Neuropharmacology. 2018;128:301–313. doi: 10.1016/j.neuropharm.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.