Abstract
Despite advancements in targeting the immune checkpoints program cell death protein 1 (PD-1), programmed death ligand 1 (PD-L1), and cytotoxic T lymphocyte–associated protein 4 (CTLA-4) for cancer immunotherapy, a large number of patients and cancer types remain unresponsive. Current immunotherapies focus on modulating an antitumor immune response by directly or indirectly expanding antitumor CD8 T cells. A complementary strategy might involve inhibition of Tregs that otherwise suppress antitumor immune responses. Here, we sought to identify functional immune molecules preferentially expressed on tumor-infiltrating Tregs. Using genome-wide RNA-Seq analysis of purified Tregs sorted from multiple human cancer types, we identified a conserved Treg immune checkpoint signature. Using immunocompetent murine tumor models, we found that antibody-mediated depletion of 4-1BB–expressing cells (4-1BB is also known as TNFRSF9 or CD137) decreased tumor growth without negatively affecting CD8 T cell function. Furthermore, we found that the immune checkpoint 4-1BB had a high selectivity for human tumor Tregs and was associated with worse survival outcomes in patients with multiple tumor types. Thus, antibody-mediated depletion of 4-1BB–expressing Tregs represents a strategy with potential activity across cancer types.
Keywords: Immunology, Oncology
Keywords: Cancer immunotherapy, Immunotherapy, T cells
Introduction
Immune checkpoint molecules are a group of surface proteins expressed on various immune cell subsets that can function as either stimulatory or inhibitory mediators, depending upon cell-specific and contextual expression (1). Immunotherapy targeting immune checkpoints has revolutionized the treatment of cancer — yet a large majority of patients and cancer types remain refractory to current therapies (2). Approved therapies targeting the program cell death protein 1 (PD-1) and cytotoxic T lymphocyte–associated protein 4 (CTLA-4) axes likely function by reversing effector T cell exhaustion in the tumor microenvironment (TME) (3). The complex TME involves multiple suppressive mechanisms (4); among suppressive cell populations, targeting Tregs has potential as an evolving approach to enhancing antitumor immunity (5).
Immune checkpoint molecules have generally been studied based on their expression on effector T cells in the context of T cell exhaustion. Given these characteristics and the recent clinical successes targeting PD-1, programmed death ligand 1 (PD-L1), and CTLA-4, other immune checkpoints may represent more appropriate Treg targets. CTLA-4 is known to be important for Treg suppressive functions, and the CTLA-4 targeting antibody ipilimumab may have effects on Tregs (6). Preclinical models further demonstrated potent antitumor activity, with CTLA-4 monoclonal antibodies capable of efficiently depleting tumor Tregs. These observations prompted phase I clinical trials in patients using a variant of ipilimumab with enhanced ADCC (7, 8). CTLA-4 is expressed on Tregs in a variety of settings other than cancer; thus, therapies that are targeted to deplete CTLA-4–expressing Tregs may have unanticipated adverse effects (9). A number of other immune checkpoints and agonist molecules, including TIM-3, PD-1, TIGIT, and tumor necrosis factor receptor (TNFR) family members, have been reported to be important in context-specific Treg suppressive function (10–13). While much is known about each of these individual immune molecules expressed on Tregs, it remains unclear how the collective checkpoint landscape contributes to Treg function in the TME across different types of cancer in humans.
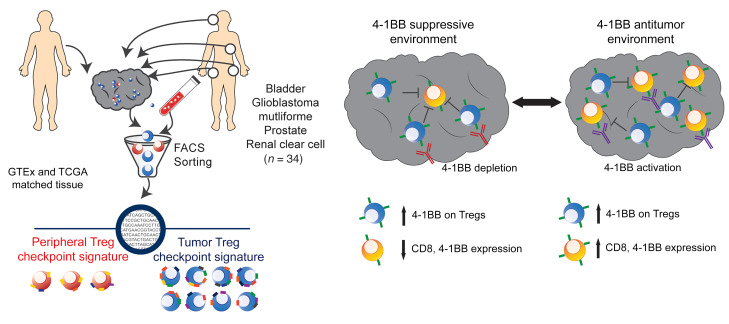
We sought to understand the global landscape of immune checkpoint and agonist molecule expression on Tregs across different tumor types. By generating RNA-Seq data from carefully sorted Tregs from multiple cancer types and mining publicly available sequencing data from human normal and malignant tissues, we identified a tumor Treg signature that was conserved across multiple cancer types and that was distinct from that of peripheral Tregs and other CD4 lineage T cells. Within this signature, 4-1BB best discriminated among intratumoral Tregs across cancer types. Accordingly, coexpression of 4-1BB and FOXP3 was associated with poorer survival outcomes, again across multiple cancer types. To corroborate these data, we treated murine tumors with a 4-1BB antibody of the IgG2a (depleting) isotype and found that treatment resulted in decreased tumor growth associated with depletion of intratumoral Tregs. Collectively, these data strongly support a conserved pattern of Treg checkpoint molecule expression and highlight 4-1BB as a target for modulating Treg function via depletion.
Results
A tumor Treg immune checkpoint signature associated with 4-1BB.
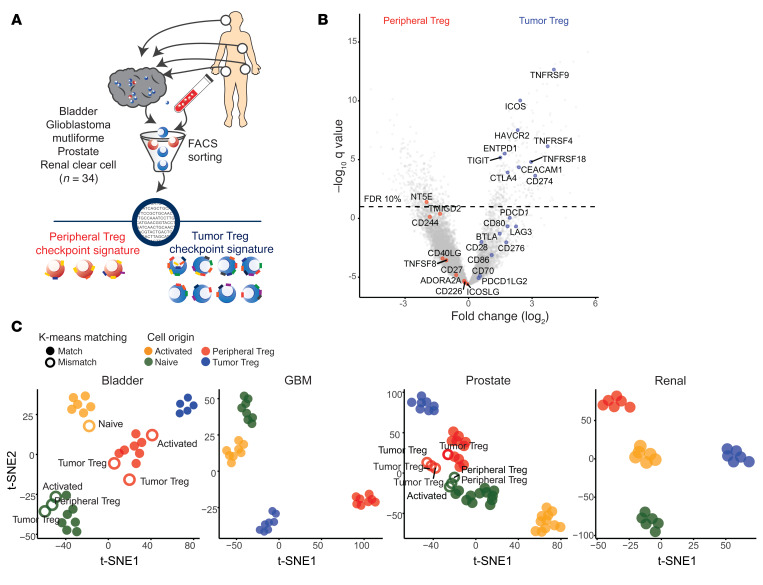
To comprehensively profile Treg-associated relevant immune checkpoint and agonist molecules in cancer, we performed RNA-Seq of matched FACS-sorted Tregs from the peripheral blood and tumors of treatment naive patients with 1 of 4 cancer types (urothelial bladder carcinoma [n = 8], glioblastoma multiforme [n = 8], prostate adenocarcinoma [n = 12], or clear cell renal carcinoma [n = 6]) (Figure 1A, Supplemental Figure 1, and Supplemental Data Set 1; supplemental material available online with this article; https://doi.org/10.1172/JCI128672DS1). Differential expression analysis focused on relevant molecules showed that peripheral blood Tregs were characterized by CD244, NT5E, and TMIGD2 expression, while tumor Tregs upregulated multiple molecules, with the greatest changes in the TNFR family members TNFRSF4 (OX40), TNFRS9 (4-1BB), and TNFRSF18 (GITR) (Figure 1B). Remarkably, tumor Tregs and peripheral Tregs as well as naive and activated CD4 T cells in all 4 tumor types clustered independently based on relevant immune molecule expression alone (Figure 1C and Supplemental Figure 2A). Only 14 of the total 140 purified cell samples demonstrated immune checkpoint signatures in which their k-means clustering designation did not match the original cell source and are marked with white circles as well as labels of the true cell type (Figure 1C). Unsupervised hierarchical clustering on immune molecule expression also distinguished peripheral versus tumor Treg populations, with a subset of 14 molecules clustering together to discriminate the subsets (Figure 2A, top annotation row with black boxes, green dendrogram on y axis). Peripheral and tumor Tregs could be characterized based on expression of 13 such molecules, while naive and activated CD4 T cells had more diffuse differences in expression (Figure 2A and Supplemental Figure 2B).
Figure 1. A conserved immune checkpoint signature differentiates peripheral and tumor Tregs across cancers.
(A) Treg immune checkpoint signatures were examined on peripheral and tumor Tregs isolated by FACS sorting from peripheral blood and tumor from patients with 1 of 4 cancer types (bladder carcinoma, n = 8; glioblastoma [GBM], n = 8; prostate carcinoma, n = 12; renal clear cell carcinoma, n = 6). (B) Differential expression analysis comparing gene expression for peripheral and tumor Tregs, with immune checkpoint genes highlighted. (C) Unsupervised clustering analysis based on immune checkpoint molecule expression in CD4 T cell subsets purified from patients with bladder cancer, glioblastoma, prostate cancer, or renal clear cell cancer. K-means clustering was used to assign T cell subtype labels based on immune checkpoint expression patterns, which were then compared with the true cell source origin. White circles represent mismatches between the k-means clustering assignment and the true cell identity; true cell identity is written adjacent to the circle.
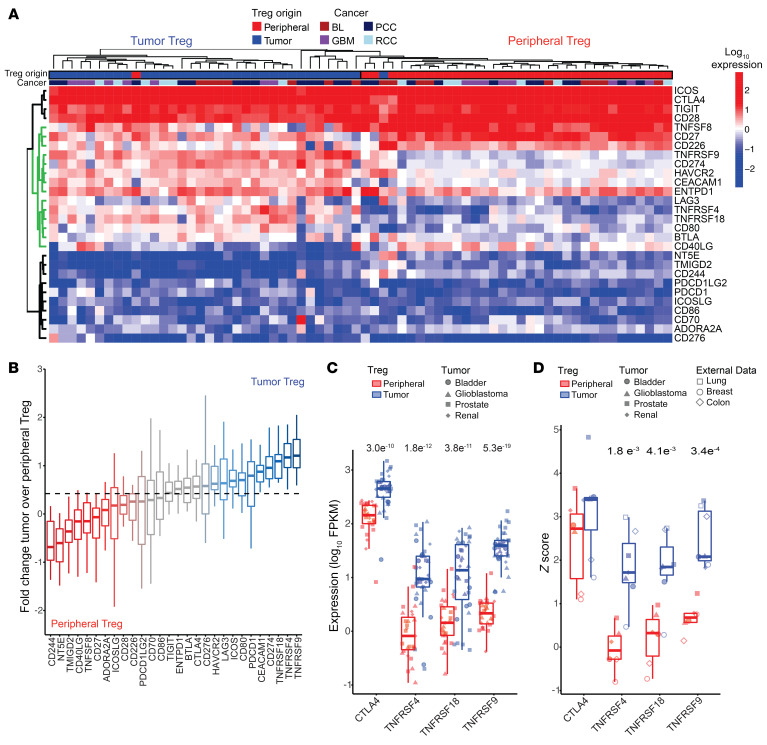
Figure 2. 4-1BB is a tumor Treg-specific immune checkpoint.
(A) Immune checkpoint expression in peripheral and tumor Tregs. The green dendrogram represents immune checkpoints important for differentiating Treg origin. The top annotation row designates Treg origin and the second annotation row identifies tumor origin. (B) Log2 fold change of the ratio of tumor to peripheral Treg expression of checkpoint genes. The dashed line represents the median log2 fold change ratio for all checkpoints. (C) Peripheral and tumor Treg expression of CTLA4, ICOS, TNFRSF4 (OX40), TNFRSF18 (GITR), and TNFRSF9 (4-1BB) expression across 4 cancer types. (D) Representative Z score comparison of CTLA4, ICOS, TNFRSF4, TNFRSF18, and TNFRSF9 expression across 7 cancer types from 4 cancers acquired as a part of this study and 3 published data sets (14, 15). Statistical comparisons were performed using paired t tests to compare peripheral and tumor Tregs for each genes’ expression. Values show in C and D are P values.
To more precisely determine which molecules are specific tumor Treg markers, we compared peripheral and intratumoral expression, examining the change in tumor versus peripheral Treg expression across immune checkpoint genes. These analyses showed which molecules were predominantly associated with peripheral versus tumor Tregs (Figure 2B). We found that, while CTLA-4 expression was enhanced on tumor Tregs, several TNFRSF members had relatively low expression in peripheral Tregs and effector CD4 T cells while showing increased expression on tumor Tregs (Figure 2C and Supplemental Figure 3A). These findings were consistent across the 4 cancers examined here as well as an additional 3 cancer types profiled in the literature: breast (14), lung (15), and colon cancer (ref. 15 and Figure 2D). We used cross-validated L1-norm constrained logistic regression (lasso) logistic regression to identify a limited set of molecules that could discriminate tumor Tregs from other CD4 T cells or from peripheral Tregs (Supplemental Figure 3, B and C). In both instances, tumor Tregs were best discriminated from other CD4 T cell subsets by TNFRSF9 expression. These data indicate that tumor 4-1BB discriminates tumor Tregs from other T cell subsets in multiple distinct cancers. Further, TNFRSF9 expression was enhanced on Tregs isolated from hepatocellular cancer (16) compared with peripheral Tregs in single-cell analyses (Supplemental Figure 4A). Additionally, tumor Tregs expressing FOXP3 and TNFRSF9 coexpressed a number of relevant molecules, including TNFRSF4, TNFRSF18, and CTLA-4 (Supplemental Figure 4B). 4-1BB levels were elevated on purified CD8 T cells from renal clear cell carcinoma, while tumor Tregs demonstrated increased TNFRSF9 across multiple cancers (Supplemental Figure 4C). Corroborating these data, we found that protein-level expression of 4-1BB was elevated on tumor Tregs as compared with tumor CD8 and CD4 T cells in hepatocellular cancer (Supplemental Figure 4D).
Tumor Treg immune checkpoint signature characterized by 4-1BB.
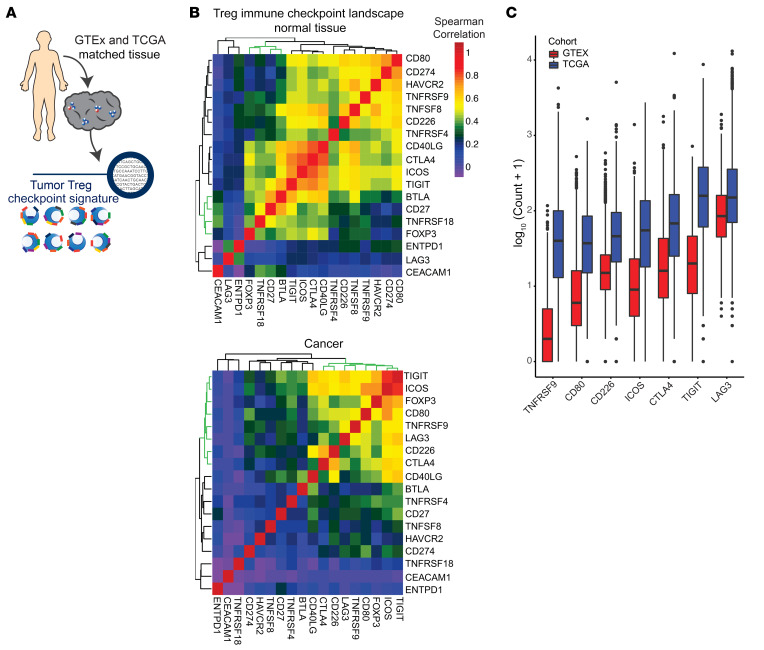
Given our findings using purified intratumoral and peripheral Treg populations, we sought to determine how Treg-associated immune checkpoint molecule expression varies between normal tissue and matched cancers. For these analyses, we capitalized on available RNA-Seq data from uniformly processed (17), matched noncancerous tissues and cancer tissues available through Genotype-Tissue Expression (GTEx) (18, 19) and The Cancer Genome Atlas (TCGA) (ref. 20 and Figure 3A). While FOXP3 expression was highly correlated with a subset of immune checkpoints in normal tissue, a different subset of immune checkpoints correlated with FOXP3 in tumors. This cancer immune landscape was characterized by TIGIT, ICOS, LAG-3, TNFRSF9, CD80, CD226, and CTLA-4 (Figure 3, B, green dendrograms, and C).
Figure 3. Conserved Treg checkpoint landscape present in bulk sequencing of multiple cancers.
(A) Treg immune checkpoint signatures were examined by analysis of data available through TCGA and RNA-Seq performed on purified Tregs. Normalized and batch effect controlled data from GTEx and TCGA was used to examine bulk tissue checkpoint signature across normal and cancer tissue. (B) Correlation matrix of immune checkpoints and FOXP3 expression in normal tissue or cancer. The green dendrogram represents FOXP3-associated checkpoints. (C) Box plots of log10-normalized expression of Treg-correlated immune checkpoints ordered by median expression in normal tissue and cancer. All immune checkpoints were significantly higher in TCGA versus GTEx samples (P < 2 × 10–16 for GTEx vs. TCGA for each checkpoint).
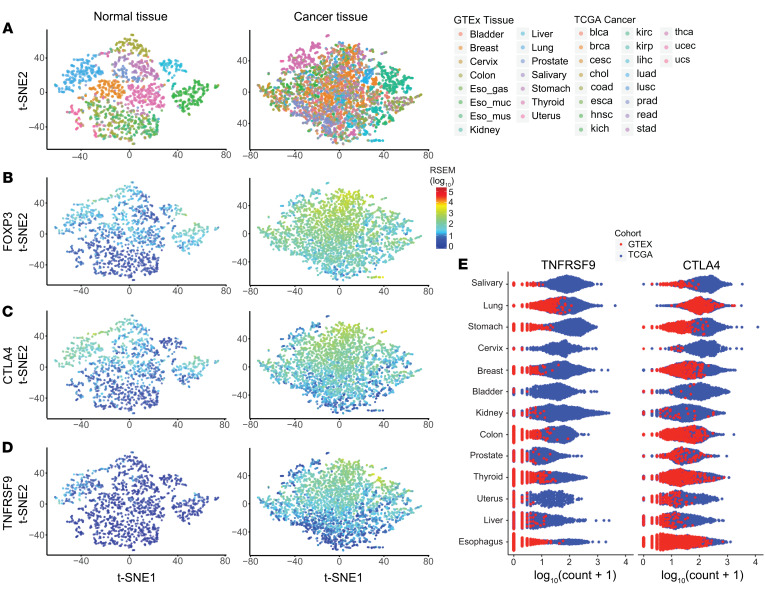
High-dimensional visualization using t-distributed stochastic neighbor embedding (t-SNE) identified a normal tissue-specific immune checkpoint landscape, which was substantially altered in cancer (Figure 4A). Expression of the Treg transcription factor FOXP3 was enriched across some tissues at low levels while other tissues had little expression. A gradient of FOXP3 expression was present in cancers irrespective of tissue origin, with most tumor tissues having greater expression as compared with normal tissues (Figure 4B). We next examined patterns of expression for each Treg checkpoint molecule in the signature, comparing expression in normal tissues and cancer (Figure 4C and Supplemental Figure 5A). TNFRSF9 and CTLA-4 had similar patterns of expression as related to FOXP3 in cancer while TNFRSF9 had lower levels of expression across normal tissues (Figure 4D and Supplemental Figure 5A). Furthermore, TNFRSF9 had increased enhancement in cancer versus normal tissue as compared with the other checkpoints. We next compared expression of the FOXP3-correlated immune checkpoints across each of the matched normal tissue and cancer samples by tissue origin (Figure 4E and Supplemental Figure 5B). Across all tissue types, TNFRSF9 generally had low levels on normal tissue as compared with levels that were enhanced in the corresponding cancer. By contrast, CTLA-4 expression was observed to be elevated in multiple normal tissues as well as in cancer. These data suggest that the conserved Treg immune checkpoint signature is present across multiple cancers and that 4-1BB expression is the signature member with the greatest differential expression between cancer and normal tissues.
Figure 4. 4-1BB displays tumor specificity across multiple tissues and cancer types.
(A) t-SNE clustering of FOXP3 and immune checkpoint gene expression in normal tissue and several different cancers. (B) Distribution of FOXP3 expression overlaid on cancer clustering analysis from D of immune checkpoint molecules and FOXP3 expression from 28 cancer types (n = 7608). (C and D) Expression of CLTA4 and TNFRSF9 in normal tissue and cancer. The color scale is the same as in B. (E) Expression of TNFRSF9 and CTLA4 across multiple normal and cancer tissue-matched samples demonstrating the normal and cancer landscape.
In vivo depletion of 4-1BB–expressing Tregs inhibits tumor growth.
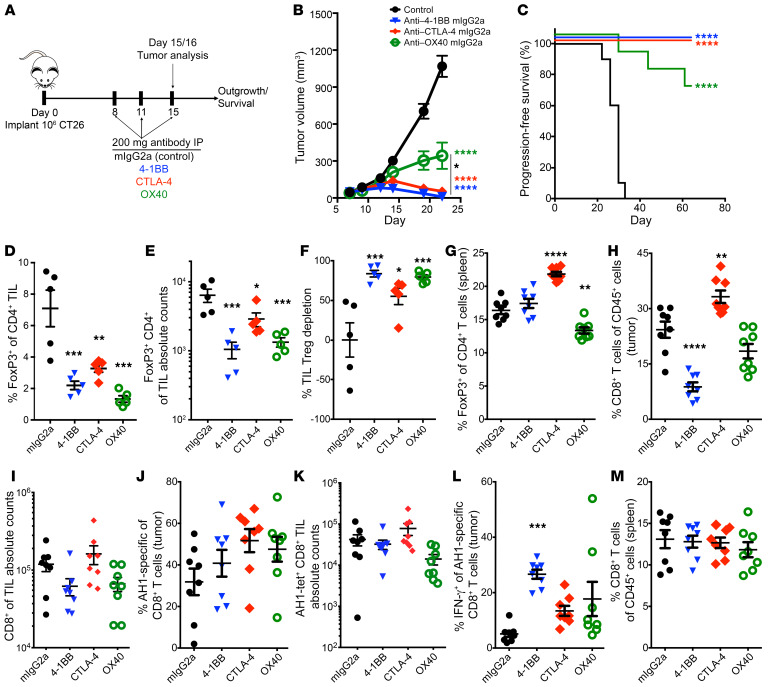
To determine whether depletion of 4-1BB–expressing Tregs alters tumor growth, we treated tumor-bearing mice with murine 4-1BB antibodies of the IgG2a subclass; murine IgG2a antibodies bind to activating FcγRs and mediate target depletion (Figure 5A and refs. 8, 21). We used the CT26 model to test these antibodies due to its increased numbers of effector CD8 T cells and Tregs as well as the ability to use tetramers to track the AH1 antigen-specific CD8 T cells. Additionally, this model was used in prior studies to demonstrate the efficacy of depleting anti–CTLA-4 antibodies (IgG2a). Although all 3 depleting antibodies led to significantly decreased tumor growth, 4-1BB showed the numerically largest treatment effect (Figure 5B). Mice treated with either anti–CTLA-4 or 4-1BB also had the greatest long-term survival (Figure 5C). Total numbers of tumor Tregs decreased similarly across the treatments, although anti–4-1BB treatment led to the greatest total reduction in total Treg number (Figure 5, D–F). Importantly, 4-1BB treatment did not appear to deplete Tregs or CD8 T cells in the spleen (Figure 5, G and M). As previously described (8), CTLA-4 treatment significantly increased the percentage of Tregs in the spleen while OX40 decreased splenic Tregs. As might be predicted by their activated phenotype (22, 23), anti–4-1BB did lead to an overall decrease in CD8 T cells in the tumor (Figure 5H); despite this decrease, the total percentage of CD8 T cells present in the tumor that were tumor antigen (AH1) specific was similar in the 4-1BB treatment group as compared with other treatment groups (Figure 5, I–K). Additionally, 4-1BB antibody treatment enhanced IFN-γ production by AH1-positive CD8 T cells (Figure 5L).
Figure 5. IgG2a antibody-mediated depletion of 4-1BB inhibits Tregs, leading to decreased tumor growth in mouse cancer.
(A) Schematic diagram of IgG2a treatment in murine CT26 tumor model. (B) Average median volume tumor growth (mm3) curves for mice treated as in A. (C) Kaplan-Meier survival curves for mice treated with depleting antibodies as in A. (D and E) Comparison of FOXP3+ CD4 T cells in tumor based on the percentage and absolute numbers of Tregs (n = 5/group). (F) The percentage of Treg depletion across antibody treatment conditions. (G) Splenic Treg numbers with different antibody treatments. (H and I) CD8 T cell frequency and absolute counts in tumor across treatment groups. (J and K) The percentage and absolute counts of AH1-specific CD8 T cells and (L) IFN-γ production by AH1 CD8 T cells across treatments. (M) Splenic CD8 T cell frequencies across treatment conditions. Representative example of 2 experiments. n = 10 for survival studies; n = 5–8 for flow cytometry studies. Statistical comparisons were performed using repeated-measures 2-way ANOVA with Tukey multiple comparisons test for tumor growth curve response to treatment and 1-way ANOVA with Dunnett multiple comparisons test for intratumoral analysis of different T cell populations. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Colored asterisks correspond to statistical comparison to control group.
To validate these findings, we tested whether targeting 4-1BB had similar antitumor effects in the MC38 colorectal cancer model. Treatment of MC38 tumors with either 4-1BB IgG1 or IgG2a subclass antibodies lead to decreased tumor growth and improved overall survival as compared with isotype or anti PD-1 treatment (Supplemental Figure 6, A–C). In this model, 4-1BB IgG2a treatment led to a significant decrease in intratumor Treg levels while slightly decreasing intratumoral CD8 T cells (Supplemental Figure 6, D–F). Given that the overall efficacy of the IgG1 and IgG2a subclass antibodies was similar, we examined Tregs and CD8 T cells for granzyme B and PD-L1 expression. 4-1BB IgG2a–mediated Treg depletion was accompanied by decreased expression of granzyme B and PD-1, while IgG1 treatment led to increased production of granzyme B by tumor Tregs (Supplemental Figure 6G). 4-1BB IgG1 also led to an increase in CD8 T cell granzyme B expression as anticipated (Supplemental Figure 6H), which was not present in 4-1BB IgG2A–treated mice (Supplemental Figure 6H).
Tumor T cell profiles characterized by low numbers of CD8 T cells and high numbers of 4-1BB–expressing Tregs signal poor survival outcomes across multiple human cancers.
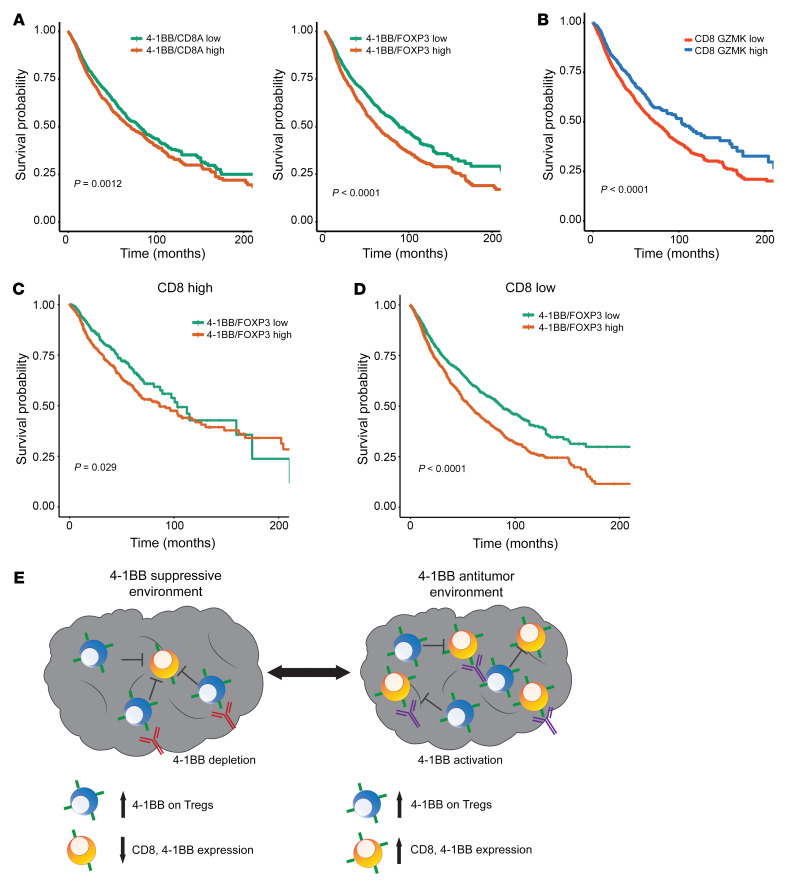
4-1BB is expressed on a population of CD8 tumor-infiltrating lymphocytes (TILs), where it serves as an activating coreceptor. Thus, 4-1BB–targeted agonist (cross-linking) antibodies are in development for cancer immunotherapy (24). This is in seeming contrast to the data above, demonstrating that 4-1BB is preferentially expressed on TIL Tregs in at least 8 different tumor types. To investigate the potential role of 4-1BB as an activating molecule in CD8 effector T cells versus in suppressive Tregs, we examined whether expression was associated with improved survival (as might be expected if a CD8 agonist role dominated) versus decreased survival (as might be expected if 4-1BB’s role on Tregs was more critical). As shown in Figure 6, an increased ratio of TNFRSF9/FOXP3 expression correlated with decreased overall survival, while the ratio of TNFRSF9/CD8A did not greatly effect survival across multiple cancer types from TCGA (Figure 6A). Furthermore, high coexpression of FOXP3 (above versus below the median) along with other Treg immune checkpoint molecules, including CTLA4/FOXP3, TNFRSF4/FOXP3, or TNFRSF18/FOXP3, did not correlate with differences in overall survival (Supplemental Figure 7A).
Figure 6. Elevated Treg 4-1BB in the absence of CD8 T cells leads to decreased survival across multiple human cancers.
(A) Kaplan-Meier curve for survival based on the TNFRSF9/CD8A and TNFRSF9/FOXP3 ratio for 13 cancers from TCGA. (B) Kaplan-Meier curves based on cohorts with high or low CD8A GZMK expression. (C) Kaplan-Meier curve for TNFRSF9/FOXP3 in the cohort with high CD8A GZMK expression. (D) Kaplan-Meier curve for TNFRSF9/FOXP3 in the cohort with low CD8A GZMK expression. (E) Diagram of the different functional relationship between 4-1BB with either CD8 or Treg expression in the TME.
Survival analyses further showed that increased levels of CD8A above the median were associated with improved survival, while FOXP3 expression did not have an effect (Supplemental Figure 7B). Given that tumors vary widely in their degree of CD8 T cell infiltration, we hypothesized that tumors with elevated levels of functional CD8 T cell infiltration would show relatively improved overall survival as a function of TNFRSF9/FOXP3. To test this, we stratified tumors into 4 quantile groups based on total CD8A and GZMK levels (Figure 6B and Supplemental Figure 7C). Tumors within the top quantile with the highest CD8A and granzyme K (GZMK) coexpression were associated with an increased survival outcome compared with those with low CD8A and GZMK coexpression (Supplemental Figure 7D). Only a few tumor types were characterized by having increased levels of CD8A and GZMK in the top quantile, the majority of which are known to respond to current checkpoint targeting immunotherapies (Supplemental Figure 7E).
Interestingly, we found that for those tumors with low CD8A and GZMK coexpression, high TNFRSF9/FOXP3 coexpression correlated with significantly worse survival compared with low TNFRSF9/FOXP3 coexpression (Figure 6D). This association between TNFRSF9/FOXP3 levels and survival was not seen in those tumors with high CD8A and GZMK levels (Figure 6C). We found that these differences in survival pattern were not the observed for CTLA4, TNFRSF4, or TNFRSF18 when normalized to FOXP3 (Supplemental Figure 8, A and B). In the cohort with high expression of CD8A and GZMK (CD8 high), TNFRSF9/FOXP3 expression changes were associated with increased expression of CD8A, GZMK, and perforin 1 (PRF1) (Supplemental Figure 8C). By contrast, TNFRSF9/FOXP3 levels were inversely correlated with CD8A, GZMK, and PRF1 in tumors characterized by elevated TNFRSF9/FOXP3 (Supplemental Figure 8C). Our data suggest that presence of 4-1BB–expressing Tregs in the absence of CD8A effector cells can signal an immune suppressive microenvironment that may be amenable to targeting with 4-1BB depletion (Figure 6E).
Loss of 4-1BB in murine Tregs impairs suppressive function in vitro.
Given our observations that increased Treg-associated 4-1BB was associated with impaired survival, we hypothesized that, in addition to serving as a tumor-specific Treg marker, 4-1BB might effect the overall suppressive function of Tregs. To test this, we performed Treg suppression assays with murine T cells in the presence of isotype- and 4-1BB–activating antibodies (25). Tregs were less able to suppress CD4 or CD8 responder cells in the presence of 4-1BB–agonizing antibodies as compared with isotype control (Supplemental Figure 9A). To rule out the possibility that the 4-1BB–agonizing antibodies were also acting on responder cells in these assays, we bred mice with a Treg-specific loss of 4-1BB. Using these cells, we found that Tregs lacking 4-1BB showed impaired suppressive function as compared with control littermates (Supplemental Figure 9, B and C). 4-1BB–deficient Tregs demonstrated increased expression of CTLA-4 and PD-1 as compared with littermate controls (Supplemental Figure 9, E and F). Taken together, these data using antibodies and Treg-specific knockout support the notion that 4-1BB expression on Tregs may be important in their function.
Discussion
In this comprehensive analysis of RNA-Seq data from flow-sorted Treg populations, we found that human Tregs have a conserved pattern of expression of relevant immune molecules in the TME, a pattern significantly different from that observed in the periphery. These changes were consistent across multiple tumor types, including those that are both sensitive and resistant to current immunotherapy treatments. We further showed that 4-1BB antibody–mediated depletion of Tregs leads to decreased tumor growth in mouse models, consistent with recent work that demonstrated depletion of Tregs with 4-1BB IgG2a antibodies (26). We found that 4-1BB represents a relatively specific tumor Treg marker, which is associated with impaired overall survival outcomes. 4-1BB was also identified as an important discriminator of tumor Tregs associated with worse patient outcomes in non–small cell lung cancer (27).
Although our approach identified 4-1BB as a Treg-specific target across multiple cancers, 4-1BB has been previously shown to be important in positively regulating effector T cell responses in cancer (28–30). 4-1BB agonism has been extensively studied in the context of anticancer immune responses, and agonistic therapies have demonstrated benefit in preclinical mouse models across multiple cancer types (24, 31). 4-1BB expression on tumor CD8 and CD4 T cells, as well as peripheral blood CD8 T cells, is also a predictor of response to ipilimumab or ipilimumab and pembrolizumab in high-grade melanoma (32). Consistent with this, our data highlight that, in tumor types heavily infiltrated with CD8 T cells, including melanoma, 4-1BB is associated with increased CD8 T cell effector function and improved patient survival.
4-1BB functions as a costimulatory molecule that potentiates TCR-mediated NF-κB signaling, leading to increased activation and proliferation of T cells and NK cells (33). 4-1BB likely uses similar signaling pathways in CD8 T cells and Tregs, but outcomes are likely related to the functional context of the cell and its contribution to the overall tumor immune environment. In support of this, previous work has demonstrated that FOXP3 binds to the 4-1BB promoter, increasing expression preferentially in activated Tregs (34). We found that 4-1BB on Tregs was associated with worse patient outcomes in multiple solid cancers and was inversely associated with antitumor effector responses.
Importantly, we confirmed in mouse cancer models that depleting anti–4-1BB antibodies of the IgG2a isotype mediated decreased tumor growth and Treg number as compared with either CTLA-4 or OX40. Anti–4-1BB–mediated decreases in tumor Tregs were accompanied by alteration in the phenotype of remaining Tregs, as they expressed less PD-L1 and secreted less granzyme B. This expression pattern is associated with decreased Treg suppressive function (35). We also found, in mice treated with IgG1 anti–4-1BB antibody, that Tregs had increased granzyme B production, suggestive of increased Treg suppressive activity when 4-1BB was activated. Of note, relevant recent studies also demonstrated that IgG2a-targeted depletion of 4-1BB led to tumor control in murine models (25). In those studies, IgG2a-mediated depletion was dependent on activating FcγR while the same was not true of IgG1. In the MC38 model, we demonstrated that IgG1 and IgG2a anti–4-1BB had an antitumor effect while Buchan et al. observed little efficacy of IgG1 anti–4-1BB in the CT26 tumor model (26). It remains unclear why IgG2a and IgG1 anti–4-1BB antibodies have similar efficacy in the MC38 model but not the CT26 model. This may stem from differences in the Fc receptors in the respective TME or in strain-specific differences that could play a role in the relative sensitivity to IgG1 treatment. In support of a genetic mechanism, SNP differences in various mouse strains have been demonstrated to affect FcγRIII levels and to modulate circulating levels of antigen-specific IgG1 (36).
A potential concern with antibodies against 4-1BB is the depletion of antitumor CD8 T cells in the process of removing Tregs. In keeping with this, in the CT26 mouse tumor model, we did observe a decrease in the overall frequency of tumor CD8 T cells in mice treated with depleting 4-1BB antibodies although it did not alter total number of tumor CD8 T cells. However, in spite of the decrease in CD8 T cells, 4-1BB depletion had significant antitumor activity and enhancement of IFN-γ production by antigen-specific CD8 T cells, suggesting that CD8 T cell tumor immune response may be preferentially selected. In fact, 4-1BB IgG2a treatment may select against bystander CD8 T cells, which have been recently demonstrated to be a significant portion of the tumor-infiltrating immune response (37). It remains unclear how this selection may occur. One hypothesis is that transient loss of Tregs may reverse the immunosuppressive environment and simultaneously affect CD8 T cells in the tumor while also allowing for new CD8 T cells to traffic to the TME. Antigen-specific CD8 T cells with elevated 4-1BB in the TME may also undergo ongoing proliferation that renders depletion more challenging to detect. Alternatively, antigen-specific CD8 T cells may upregulate antiapoptotic molecules, such as Bcl-2, that render them less susceptible to depletion (38, 39). Further investigation is warranted to determine the mechanisms that promote a differential increase in antigen-specific CD8 T cells in the TME after 4-1BB–mediated depletion.
Our studies on purified Tregs from 8 tumor types and bulk TCGA profiles identify 4-1BB as a marker of TIL Tregs with correspondingly low expression in normal tissue. Thus, 4-1BB–mediated depletion of Tregs may be broadly applicable as a therapeutic strategy in cancers resistant to current immunotherapies.
Methods
Patients.
Peripheral blood and tumor samples were collected for each patient on the day of surgery after the patient provided consent. A total of 34 previously untreated patients were sampled with 1 of 4 tumor types, including bladder transitional cell carcinoma (n = 8), glioblastoma multiforme (n = 8), prostate adenocarcinoma (n = 12), or renal clear cell carcinoma (n = 6). Tumor samples were obtained with a board-certified pathologist present to confirm initial stage and extent of disease. All tumor pathology was later confirmed via immunohistochemistry for tumor type and grade. All samples were processed and CD4 T cell subsets isolated by FACS sorting within 4 hours of original collection.
Mouse studies.
Animal experiments were performed in Association for Assessment and Accreditation of Laboratory Animal Care International–accredited housing facilities at Bristol-Myers Squibb or the University of Michigan.
C57BL/6N Tnfrsf9tm1a(EUCOMM)Wtsi mice (obtained from the Institut Clinique de la Souris, created by the International Mouse Phenotyping Consortium) were crossed to the Flpo deletor strain C57BL/6N-Tg(CAG-Flpo)1Afst/Mmucd (MMRRC:036512-UCD) to remove the FRT-flanked reporter sequence between exons 4 and 5 to generate Tnfrs9fl/fl mice. Tnfrsf9fl/fl mice were then crossed with B6.129 Foxp3tm4(YFP/icre)Ayr/J mice (obtained from The Jackson Laboratory) to generate Foxp3 Cre+ Tnfrsf9fl/fl mice. C57BL/6 or BALB/c mice (Charles Rivers Laboratories) were subcutaneously injected with 2 × 106 MC38 or 1 × 106 CT26 tumor cells (ATCC). Tumor volumes were measured 7 days after implantation, and mice were randomized into treatment groups to have similar mean tumor volumes. Tumor volumes were then measured every 2 to 3 days using electronic calipers and volume calculated using the formula (L12 × L2)/2, with L1 being the shortest diameter. Mice were treated intraperitoneally on days 8, 11, and 15 with 200 μg of corresponding antibodies. Tumors from mice used for TIL analysis were harvested at day 15 or 16. Mice were sacrificed with CO2 at either study termination or any of the following clinical endpoints: tumor volume ≥2000 mm3, tumor ulceration, body weight loss ≥20%, or moribund appearance.
Tumor-bearing mice were treated with monoclonal antibodies against mouse CTLA-4 (clone 9D9, mIgG2a isotype), OX40 (clone OX86, mIgG2a isotype), and CD137 (clone 1D8, mIgG1 and mIgG2a isotypes). A monoclonal antibody against keyhole limpet antigen (mIgG2a isotype) was used as a treatment control. Mice were treated on days 8, 11, and 15 with intraperitoneal injections of 200 μg of the indicated antibodies.
For TIL analysis, tumors were harvested and processed using GentleMacs cell disruptors (Miltenyi Biotec). Resulting cell suspensions were clarified through 70-μM filters, pelleted, resuspended in PBS or DMEM, and counted. Cells were incubated with anti-CD16/32 mAb 24G.2 (BioXCell or BD Biosciences) to reduce background FcγR binding and then stained with antibodies specific for CD8 (BioLegend, 53–6.7), CD4 (BioLegend, GK1.5), and CD45 (BioLegend, 30-F11). Cells were also stained with the LiveDead Aqua fixable viability dye (Thermo Fisher Scientific, L34597). For intracellular staining, samples were fixed, permeabilized, and stained with antibodies specific for FoxP3 (eBioscience, FJK-16s) and IFN-γ (eBioscience, XMG1.2). CT26 tumor antigen-specific CD8+ T cells were identified using AH-1 MHC class I tetramers (MBL MuLV gp70 SPSYVYHQF). Tumor and splenic cell suspensions were incubated with tetramer in DMEM and 10% FCS for 30 minutes at 37°C, washed, and stained with surface and intracellular antibodies, as above. Ex vivo AH-1 peptide stimulation was performed by culturing tumor or splenic cells with 2 μM AH-1 peptide (MBL) in the presence of brefeldin A for 4 hours at 37°C. Ex vivo cytokine staining was performed by fixing and staining cells as described above, directly after tissue harvest. Samples were analyzed on FACS Canto and Fortessa flow cytometers (BD Biosciences). TIL Treg depletion was calculated as the percentage reduction in Treg numbers relative to the mean Treg number in control mIgG2a-treated mice.
In vitro suppression assay.
Mice were euthanized to harvest lymph nodes and spleens for Treg isolation. Tregs were isolated from the spleens and lymph nodes of naive mice with the MojoSort Mouse CD4 T Cell Isolation Kit (Biolegend) followed by cell sorting to enrich for YFP+ and/or CD25+ Tregs. For responder cells, naive CD4+ or CD8 T cells from the spleens and lymph nodes of congenically distinct mice (CD90.1+) were isolated with the Pan T Cell Isolation Kit (Miltenyi Biotec) and then sorted based on CD62Lhi population. Sorted naive CD4+ or CD8 T cells were then labeled with proliferation dye eFluor450 (5 μM) (eBioscience) following the manufacturer’s protocol. Cells from the positive fraction from the Pan T cell isolation were irradiated with 30 Gy and were used as antigen-presenting cells (APCs). Different ratios of Tregs (suppressor) and naive CD4+ or Cd8 T cells (responder) were cocultured with soluble anti-CD3 (1 μg/ml) and/or rat IgG2a isotype or anti 4-1BB (10 μg/ml, 3H3 clone BioXCell) and irradiated APCs for 72 hours. The proliferation of the responder CD90.1+ T cells was measured by eFluor450 dilution by flow cytometry. The percentage of suppression was calculated by using the following formula: percentage suppression = (percentage of proliferated responders with no Tregs – percentage of proliferated responders)/percentage of proliferated responders with no Tregs × 100
Peripheral blood preparation.
Peripheral blood lymphocytes were isolated from whole blood using density gradient. Whole blood was diluted with HBSS (Corning Cellgro, catalog 21-022-CV) and underlaid with Ficoll-Paque Premium (GE Healthcare, catalog 17-5442-03). Gradients were centrifuged and the buffy coat was washed with 1× PBS (Corning Cellgro, catalog 21-040-CV) and resuspended in 1× PBS for downstream application.
Tumor dissociation.
Tumors were homogenized using the gentleMACS human tumor dissociation kit (Miltenyi Biotec, catalog 130-095-929) using a gentleMACS Octo Dissociator (Miltenyi Biotec, catalog 130-095-929) according to the manufacturer’s protocol. Samples were incubated at 37°C per Miltenyi Biotec’s protocol. Following this step, tumor homogenates were passed through a 100-μm filter prior to staining for FACS sorting.
FACS sorting.
PBMCs and tumor lymphocytes were enriched using Dynabeads FlowComp Human CD4 Kits (Thermo Fisher Scientific, catalog 11331D) according the manufacturer’s protocol prior to FACS sorting. Peripheral and tumor lymphocytes were stained with the following antibodies for sorting: Pacific Blue-CCR7 (BioLegend, catalog 353210), Brilliant Violet 570-CD45RO (BioLegend, catalog 304226), Alexa Fluor 488-CD127 (BioLegend, catalog 351314), PE-CD25 (BioLegend, catalog 302606), PerCP-Cy5.5-CD45RA (BioLegend, catalog 304122), PE-Cy5-CD4 (BioLegend, catalog 317412), PE-Cy7-CD8 (BioLegend, catalog 300914), APC-CD28 (BioLegend, catalog 302912), and APC-Cy7-CD27 (BioLegend, catalog 302816). For flow cytometry staining of human TILs for protein level expression, the following antibodies were used: BV-421 4-1bb (BioLegend, catalog 309819), PE/Cy7 FOXP3 (eBiosciences, catalog 25-4777-42), BV786 CD3 (BD Biosciences, catalog 563800), BV650 CD45 (BD Biosciences, catalog 563717), Alexa Fluor 700 CD8 (Biolegend, catalog 301028), and BV605 CD4 (Biolegend, catalog 317438).
Purified CD4 populations were defined as follows: peripheral naive, CD4+CD25loCD127+/–CCR7+CD45RA+CD27+CD28+; peripheral activated, peripheral naive cells that were activated ex vivo; peripheral Tregs, CD4+CD25hiCD127lo; tumor Tregs, CD4+CD25hiCD127lo. After sorting, cells were lysed with Trizol Reagent LS (Thermo Fisher Scientific, catalog 10296010) and stored at –80°C for later RNA isolation. A subset of naive cells were collected for later in vitro activation.
RNA-Seq and analysis.
All RNA-Seq on purified T cell populations was performed by the Johns Hopkins School of Medicine Sidney Kimmel Comprehensive Cancer Center Experimental and Computational Genomics Core. All RNA was extracted using Trizol reagent (Thermo Fisher Scientific, catalog 10296010) according to the manufacturer’s protocol. RNA yield was typically in the range of 100 pg to 200 ng for these sorted cell populations, with tumor-associated CD4 and CD8 populations having the smallest amounts. For samples with sufficient RNA input >1 ng total RNA, Bioanalyzer (Agilent Technologies) analysis was used to assess RNA integrity with values all exceeding 7.0. RNA-Seq barcoded libraries were prepared using the Nugen Ovation RNA-Seq System v2 kit (Nugen) according to the manufacturer’s instructions. Libraries were sequenced on an Illumina HiSeq 2500 with paired end 2 × 100 bp reads. Reads were aligned using RNA-Seq expression (RSEM) (v1.2.8–1.2.9) with bowtie2 to derive gene-level expression measures, represented as posterior fragments per thousand per million (FPKM), normalized using log2, log10 transformation, and Z score normalization for all analysis.
GTEx/TCGA data.
GTEx and TCGA expression values that were processed on the same bioinformatic pipeline and batch normalized were obtained from Wang et al. (17). Complete clinicopathological and normalized RSEM values for 74,149 TCGA samples were downloaded from cBioPortal (access date: 12/20/2017). Briefly, sample and survival data were downloaded individually for 16 separate TCGA cohorts, with normalized RSEM values for selected checkpoint markers retrieved via the cgdsr (v1.2.10) R-based cBioPortal API. For survival analysis, survival criteria were used from the clinicopathological data sets. All samples were stratified based on median expression or ratio of expression unless specified otherwise (CD8A and GZMK analysis). Renal clear cell carcinoma was excluded from the survival analysis based on previously published work demonstrating that increased CD8 T cells are associated with worse clinical outcomes (40).
High-dimensional visualization and clustering analysis.
t-SNE was performed on log10-transformed FPKM or TPM using the Rtsne package. For sorted T cell analysis of each independent tumor type, cluster assignment was performed independent of cell origin using k-means clustering with the 4 clusters corresponding with the number of original T cell types isolated. Cluster identity was then compared with original cell identity and mismatches were designated with white circles (Figure 1C).
Statistics.
All statistical analysis were performed in R (3.4.3, CRAN) and GraphPad Prism 7. Two-group comparisons were performed with 2-tailed Student’s t test or 2-tailed paired Student’s t test depending on relationship between the groups. Statistical comparisons for group comparisons were performed using 1-way ANOVA with Dunnett multiple comparisons test for individual group comparison. We used cross-validated LASSO (41) to identify a discriminatory subset of genes to identify tumor Tregs from either all other CD4 T cell types or peripheral Tregs. For tumor growth experiments, differences in average tumor growth were determined using repeated-measures 2-way ANOVA with Tukey multiple comparisons testing. For human flow cytometry, Friedman test with Dunn’s post test or 2-tailed paired t test was used to compare groups. Significance was determine to be at P ≤ 0.05. Numbers of samples and repeats of experiments are indicated in the figure legends where applicable.
Study approval.
All human tumors were collected with approval from the Johns Hopkins University IRB with the following protocol IDs: renal clear cell carcinoma (IRB00033839), prostate cancer (NA_00082175), bladder cancer (NA_00026693), and glioblastoma (IRB00049987). Human tumor samples for protein analysis were purchased from Conversant Bio. All animal experiments were reviewed and approved by local IACUCS as follows. In vivo antibody blockade experiments were reviewed and approved by the Bristol-Myers Squibb IACUC under protocol 1311-01. Generation of Foxp3 Cre+ Tnfrsf9fl/fl mice was reviewed and approved by the University of Michigan IACUC under protocol PRO00008577.
Author contributions
ZTF, LAS, AJK, SY, and CDG conceptualized and developed the project. ZTF, TRN, DHH, RJJ, JJE, MJS, CMK, RYL, A. Ghasemzadeh and A. Gupta, JZ, AMS, SJW, ATP, and SY contributed to acquisition and/or analysis of data. LAS, AJK, HJ, ATP, and SAT provided tools, resources and/or reagents, and key scientific input. ZTF, SY, and CGD wrote the manuscript. All authors edited and approved the manuscript.
Supplementary Material
Acknowledgments
This project was made possible through a Prostate Cancer Foundation Challenge Award to CGD and SY, a Commonwealth Foundation grant to SY, sponsored research funding from the Bristol-Myers Squibb International Immuno-Oncology Network and Janssen Inc, and supported by NIH/National Cancer Institute (NCI) Specialized Programs of Research Excellence (SPORE) in Prostate Cancer grant P50CA58236, NIH/NCI grant U01 CA196390, U.S. Department of Defense Prostate Cancer Research Program (PCRP) grant W81XWH-18-2-0015. We wish to thank the laboratories of CGD and SAT for helpful discussion. We also thank Jennifer Meyers and Michael Rongione from the Johns Hopkins School of Medicine Sidney Kimmel Comprehensive Cancer Center Experimental and Computational Genomics Core for their support of the RNA-Seq experiments. We wish to thank Im Hong Sun for guidance on performing suppression assays. We also wish to thank Ada Tam, Lee Blosser, and Raffaello Cimbro for maintenance of the flow cytometry core facilities. We wish to thank Amy Puffenberger for assistance with creation of visual abstracts.
Version 1. 02/04/2020
Electronic publication
Version 2. 03/02/2020
Print issue publication
Version 3. 06/24/2020
Additional support included in Acknowledgments.
Footnotes
CGD’s present address is: Division of Hematology and Oncology, Herbert Irving Comprehensive Cancer Center, Columbia University Medical Center, New York, New York, USA.
Conflict of interest: CGD is a coinventor on a patent for LAG-3, filing case C04255, licensed from Johns Hopkins to Bristol-Myers Squibb; has served as a paid consultant for Roche Genentech, Merck, and Novartis; and has received sponsored research funding from the Bristol-Myers Squibb International Immuno-Oncology Network and Janssen. SAT has served as a consultant for and received honoraria from Janssen, AbbVie, Sanofi, Almac Diagnostics, and Astellas/Medivation. SAT is a cofounder of, previous consultant for, equity holder in, and employee of Strata Oncology.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(3):1405–1416.https://doi.org/10.1172/JCI128672.
Contributor Information
Thomas R. Nirschl, Email: tnirsch1@jhmi.edu.
Jingyi Zhai, Email: jyzhai@umich.edu.
Ali Ghasemzadeh, Email: aghasemzadeh@jhmi.edu.
Anuj Gupta, Email: agupta52@jhu.edu.
Sarah J. Wheelan, Email: swheelan@jhmi.edu.
Hui Jiang, Email: jianghui@umich.edu.
Alexander T. Pearson, Email: apearson5@medicine.bsd.uchicago.edu.
Srinivasan Yegnasubramanian, Email: syegnasu@jhmi.edu.
References
- 1.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8(328):328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pauken KE, Wherry EJ. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015;36(4):265–276. doi: 10.1016/j.it.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binnewies M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curiel TJ, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10(9):942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 6.Arce Vargas F, et al. Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell. 2018;33(4):649–663.e4. doi: 10.1016/j.ccell.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen YC, et al. Combining intratumoral Treg depletion with androgen deprivation therapy (ADT): preclinical activity in the Myc-CaP model. Prostate Cancer Prostatic Dis. 2018;21(1):113–125. doi: 10.1038/s41391-017-0013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selby MJ, et al. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res. 2013;1(1):32–42. doi: 10.1158/2326-6066.CIR-13-0013. [DOI] [PubMed] [Google Scholar]
- 9.Wing K, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322(5899):271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 10.Gautron AS, Dominguez-Villar M, de Marcken M, Hafler DA. Enhanced suppressor function of TIM-3+ FoxP3+ regulatory T cells. Eur J Immunol. 2014;44(9):2703–2711. doi: 10.1002/eji.201344392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowther DE, et al. PD-1 marks dysfunctional regulatory T cells in malignant gliomas. JCI Insight. 2016;1(5):e85935. doi: 10.1172/jci.insight.85935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurtulus S, et al. TIGIT predominantly regulates the immune response via regulatory T cells. J Clin Invest. 2015;125(11):4053–4062. doi: 10.1172/JCI81187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ward-Kavanagh LK, Lin WW, Šedý JR, Ware CF. The TNF receptor superfamily in co-stimulating and co-inhibitory responses. Immunity. 2016;44(5):1005–1019. doi: 10.1016/j.immuni.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plitas G, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45(5):1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Simone M, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45(5):1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169(7):1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, et al. Unifying cancer and normal RNA sequencing data from different sources. Sci Data. 2018;5:180061. doi: 10.1038/sdata.2018.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GTEx Consortium. et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimmerjahn F, Ravetch JV. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 2005;310(5753):1510–1512. doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- 22.Lyford-Pike S, et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73(6):1733–1741. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams JB, Horton BL, Zheng Y, Duan Y, Powell JD, Gajewski TF. The EGR2 targets LAG-3 and 4-1BB describe and regulate dysfunctional antigen-specific CD8+ T cells in the tumor microenvironment. J Exp Med. 2017;214(2):381–400. doi: 10.1084/jem.20160485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melero I, et al. Monoclonal antibodies against the 4-1BB T-cell activation molecule eradicate established tumors. Nat Med. 1997;3(6):682–685. doi: 10.1038/nm0697-682. [DOI] [PubMed] [Google Scholar]
- 25.Menk AV, et al. 4-1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. J Exp Med. 2018;215(4):1091–1100. doi: 10.1084/jem.20171068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchan SL, et al. Antibodies to costimulatory receptor 4-1BB enhance anti-tumor immunity via T regulatory cell depletion and promotion of CD8 T cell effector function. Immunity. 2018;49(5):958–970.e7. doi: 10.1016/j.immuni.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24(7):978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 28.Palazón A, et al. The HIF-1α hypoxia response in tumor-infiltrating T lymphocytes induces functional CD137 (4-1BB) for immunotherapy. Cancer Discov. 2012;2(7):608–623. doi: 10.1158/2159-8290.CD-11-0314. [DOI] [PubMed] [Google Scholar]
- 29.Wolfl M, et al. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110(1):201–210. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi BK, et al. 4-1BB-based isolation and expansion of CD8+ T cells specific for self-tumor and non-self-tumor antigens for adoptive T-cell therapy. J Immunother. 2014;37(4):225–236. doi: 10.1097/CJI.0000000000000027. [DOI] [PubMed] [Google Scholar]
- 31.Chester C, Sanmamed MF, Wang J, Melero I. Immunotherapy targeting 4-1BB: mechanistic rationale, clinical results, and future strategies. Blood. 2018;131(1):49–57. doi: 10.1182/blood-2017-06-741041. [DOI] [PubMed] [Google Scholar]
- 32.Jacquelot N, et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat Commun. 2017;8(1):592. doi: 10.1038/s41467-017-00608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollok KE, et al. Inducible T cell antigen 4-1BB. Analysis of expression and function. J Immunol. 1993;150(3):771–781. [PubMed] [Google Scholar]
- 34.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445(7130):931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maj T, et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat Immunol. 2017;18(12):1332–1341. doi: 10.1038/ni.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrén M, Johanneson B, Alarcón-Riquelme ME, Kleinau S. IgG Fc receptor polymorphisms and association with autoimmune disease. Eur J Immunol. 2005;35(10):3020–3029. doi: 10.1002/eji.200526291. [DOI] [PubMed] [Google Scholar]
- 37.Simoni Y, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557(7706):575–579. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 38.Akondy RS, et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol. 2009;183(12):7919–7930. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamphorst AO, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci USA. 2017;114(19):4993–4998. doi: 10.1073/pnas.1705327114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giraldo NA, et al. Tumor-infiltrating and peripheral blood T-cell immunophenotypes predict early relapse in localized clear cell renal cell carcinoma. Clin Cancer Res. 2017;23(15):4416–4428. doi: 10.1158/1078-0432.CCR-16-2848. [DOI] [PubMed] [Google Scholar]
- 41.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.