Abstract
High levels of ecto-5′-nucleotidase (CD73) have been implicated in immune suppression and tumor progression, and have also been observed in cancer patients who progress on anti–PD-1 immunotherapy. Although regulatory T cells can express CD73 and inhibit T cell responses via the production of adenosine, less is known about CD73 expression in other immune cell populations. We found that tumor-infiltrating NK cells upregulate CD73 expression and the frequency of these CD73-positive NK cells correlated with larger tumor size in breast cancer patients. In addition, the expression of multiple alternative immune checkpoint receptors including LAG-3, VISTA, PD-1, and PD-L1 was significantly higher in CD73-positive NK cells than in CD73-negative NK cells. Mechanistically, NK cells transport CD73 in intracellular vesicles to the cell surface and the extracellular space via actin polymerization–dependent exocytosis upon engagement of 4-1BBL on tumor cells. These CD73-positive NK cells undergo transcriptional reprogramming and upregulate IL-10 production via STAT3 transcriptional activity, suppressing CD4-positive T cell proliferation and IFN-γ production. Taken together, our results support the notion that tumors can hijack NK cells as a means to escape immunity and that CD73 expression defines an inducible population of NK cells with immunoregulatory properties within the tumor microenvironment.
Keywords: Immunology
Keywords: NK cells
Introduction
The CD73 metabolic immune checkpoint orchestrates a crucial homeostatic balance of extracellular adenosine levels as part of a negative feedback mechanism to control inflammatory responses within a stressed or damaged tissue microenvironment (1). The lack of CD73 expression could implicate physiological wound healing and immunomodulation within the tissue microenvironment (2). Within the tumor microenvironment (TME), however, metabolic stress accumulates along with tumor progression, which leads to a dysregulated expression and activity of CD73 in cancers including breast cancer, metastatic melanoma, and ovarian cancer (3–6). Overexpression of CD73 within a tumor not only contributes to metastasis and anthracycline resistance, but also to immune evasion due to dysregulation of adenosine production (6, 7). For these reasons, inhibitors of CD73 are today used in cancer immunotherapy in combination with existing cancer treatments including anti–PD-1/anti–PD-L1 therapy (1, 8–11).
Tumor progression on immune checkpoint therapy can be associated with defects in tumor antigen presentation, which results in lack of tumor cell recognition by T cells (12). This has inspired the development of therapies based on activating natural killer (NK) cells (13). NK cells are characterized as granular lymphocytes capable of producing an abundance of inflammatory cytokines and induce cytotoxic responses against virus-infected and transformed cells. The regulation of their functions involves sophisticated mechanisms with a wide repertoire of signals from inhibitory and activating receptors, and at the same time is influenced by the type of cytokines and surrounding cells in the microenvironment (14–16). Although NK cells rarely infiltrate tumors, their presence in tumor biopsies has been shown to positively associate with increased survival (17). A recent clinical study investigated the clinical relevance of tumor-infiltrating NK cells in triple-negative breast cancer and reported a higher overall survival probability in patients with higher frequency of tumor-infiltrating NK cells (18). For these reasons, modulation of NK cell activity by tumor-targeting antibody therapies, immunomodulatory antibodies, cytokines, infusion of NK cells activated ex vivo, and bispecific killer engagers are currently being explored in clinical trials (19, 20).
Although NK cells are thought to contribute to tumor eradication, studies have established that NK cells can acquire a regulatory cell identity in acute infection and inflammation, producing adenosine and IL-10 (21, 22). More recently in the context of cancer, CD56+CD3– cells in patients with ovarian cancer suppressed the growth of T cells, as observed within an ex vivo expansion of tumor-infiltrating lymphocytes (TILs). Even though it was demonstrated that the suppression was mediated by NKp46 engagement, the underlying mechanisms of how NK cells suppress are still unclear (23). It is also still unclear how conventional NK cells can undergo a phenotypic switch to suppress other TIL populations and contribute to tumor immune escape.
We sought to investigate how peripheral blood and tumor-infiltrating NK cells differ in patients with breast cancer and sarcoma, and if tumor-infiltrating NK cells develop immunoregulatory functions. Compared with peripheral blood NK cells, tumor-infiltrating NK cells undergo phenotypic changes and acquire the expression of several immune checkpoint receptors. The expression of these immune checkpoint molecules was significantly higher on NK cells expressing CD73. Mechanistically, NK cells acquire CD73 surface expression via actin polymerization–dependent exocytosis upon engagement of 4-1BB ligand (4-1BBL). These CD73+ NK cells are reprogrammed to upregulate IL-10 and TGF-β production via STAT3 transcriptional activity, suppressing CD4+ T cell activity. Finally, we found that the NK cell signature influenced the predictive value of CD73 gene expression in the progression-free interval of sarcoma and breast cancer. Our results demonstrate that tumor cells can influence NK cells to acquire suppressive functions independently of CD73 activity.
Results
NK cell signature influences the predictive value of CD73 gene expression in progression-free interval of sarcoma and breast cancer.
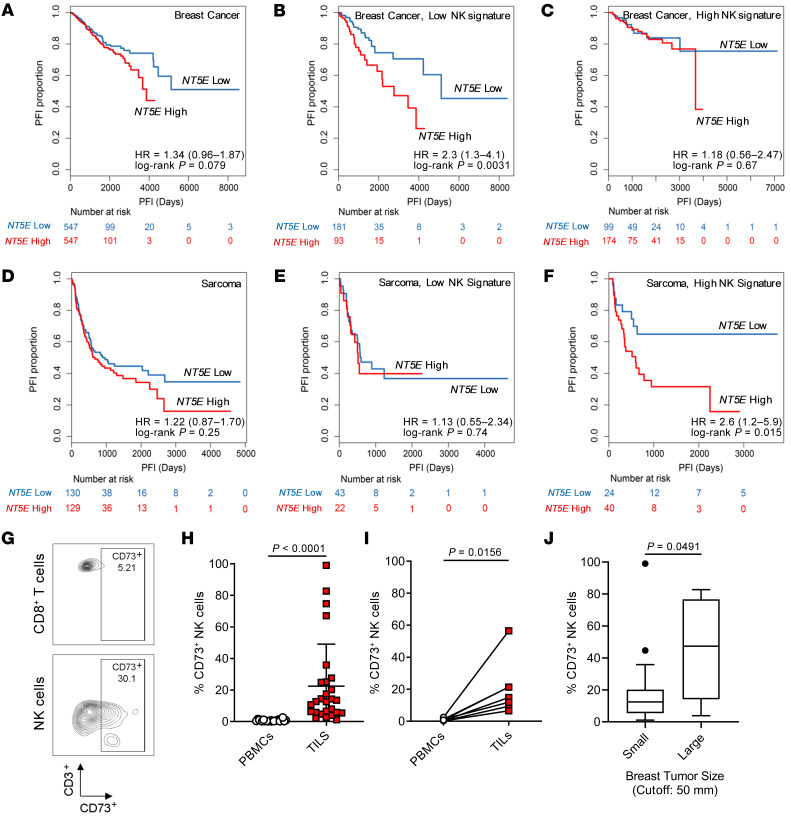
To determine the clinical relevance and relationship between NK cells and NT5E (encoding CD73) expression, we examined The Cancer Genome Atlas (TCGA) database, particularly focusing on breast and sarcoma patient cohorts. As reported earlier for several other solid tumors (24), a higher NT5E gene expression in breast cancer correlated with worse prognosis (Figure 1A). Using a 5-gene NK cell signature that was previously applied to analyze overall survival in solid tumors including breast cancer (25), progression-free survival comparing samples stratified by the top and bottom quartiles of the NK cell signature was analyzed in relation to NT5E gene expression. In breast cancer, the expression of NT5E had a greater influence on the progression-free survival (hazard ratio [HR] = 2.3, 95% confidence interval [CI] = 1.3–4.1) in patients with low NK cell gene signature (Figure 1, B and C). In sarcoma, however, NT5E expression alone did not correlate with poorer prognosis unless patients expressed a higher NK cell gene signature (HR = 2.6, 95% CI = 1.2–5.9) (Figure 1, D–F). In addition, the expression of NT5E correlated with NK cell gene signature in both sarcoma (r = 0.321) and breast cancer tissues (r = 0.326). In contrast, we did not observe that the regulatory T cell gene signature influenced the prognostic value of NT5E expression. Notably, in sarcoma but not in breast cancer, NT5E expression significantly influenced the prognosis in patients with high but not low CD8+ T cell signature (HR = 2.1, 95% CI = 1.1–4.3) (Table 1). Although the current understanding of CD73 as an immune checkpoint against tumor-infiltrating NK cells is not well understood, we show that the prognostic value of NT5E gene expression is influenced by the NK cell signature expressed by different types of tumors.
Figure 1. NT5E expression affects the prognostic value of NK cells in breast cancer and sarcoma patients.
(A) NT5E expression predicts progression-free interval (PFI) based on TCGA breast cancer cohort (n = 1094); (B) patients with low NK cell gene signature (n = 274) and (C) patients with high NK cell gene signature (n = 273). (D) NT5E expression predicts PFI based on TCGA sarcoma cohort (n = 259); (E) patients with low NK cell gene signature (n = 65) and (F) patients with high NK cell gene signature (n = 64). Log-rank Mantel-Cox test was used to assess significance. (G) Representative flow cytometric plot of breast tumor–infiltrating NK cells and CD8+ T cells based on CD3 versus CD73 expression (n = 25). (H and I) Differential expression of CD73 by NK cells from peripheral blood versus tumor resections for both breast cancer (n = 25) and sarcoma (n = 7), respectively. Mann-Whitney U test was used to determine significance in nonautologous comparison in H, while Wilcoxon’s signed-rank test was used for autologous comparison in I. (J) Correlation of percentage CD73+ tumor-infiltrating NK cells with breast cancer tumor size (n = 25) based on clinical measurement cutoff (>5 cm). Mann-Whitney U test was performed to assess significance.
Table 1. Prognostic value of CD73 gene expression influenced by immune gene signatures in TCGA sarcoma and breast cancer data sets.

Frequency of tumor-infiltrating CD73+NK cells correlates with larger tumor size in patients with breast cancer. To confirm the prognostic value of CD73 expression by tumor-infiltrating NK cells, peripheral blood and tumor resections were analyzed by flow cytometry from individual cohorts of breast cancer and sarcoma. Unlike peripheral blood NK cells and tumor-infiltrating T cells, tumor-infiltrating NK cells expressed significantly higher levels of cell surface CD73 (Figure 1, G–I and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI128895DS1). Upon analysis of clinical characteristics, it was observed that the frequency of CD73+ NK cells correlated with breast tumors of larger size based on clinical tumor, node, and metastasis (TNM) staging measurement cutoff (>5 cm) (Table 2 and Figure 1J). Although not significant, triple-negative breast cancer had a higher frequency of CD73+ NK cells, while the frequency of total NK cells among CD45+ TILs was significantly higher in the luminal B subtype (Supplemental Figure 1, B and C). Importantly, the presence of CD73+ NK cells was independent of tumor CD73 expression or frequency of tumor-infiltrating NK cells (Supplemental Figure 1, D and E). Taken together, these observations show that CD73+ NK cells were found only in the TME and that the frequency of these cells correlates with larger tumor size in patients with breast cancer.
Table 2. Characteristics of breast cancer patient cohort.
CD73+ NK cells show increased expression of multiple immune checkpoint receptors.
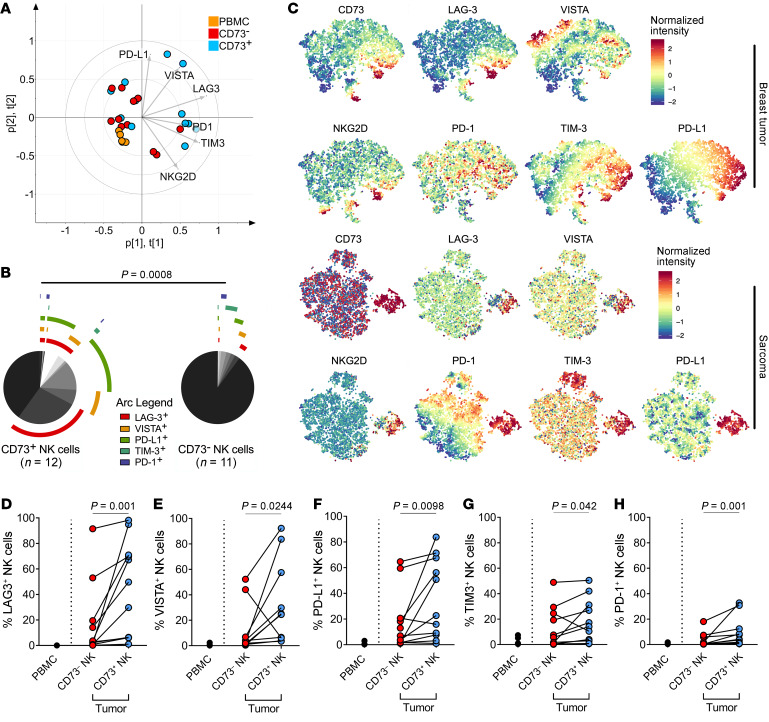
To investigate whether CD73 is upregulated as a bona fide immune checkpoint, flow cytometric analysis was performed to compare tumor-infiltrating NK cells and peripheral blood NK cells from patients with breast cancer and sarcoma for the multiple expression of other known immune checkpoints (gating strategy shown in Supplemental Figure 2A). Using principal components analysis (PCA), a greater phenotypic heterogeneity within CD73+ tumor-infiltrating NK cells in contrast with peripheral blood NK cells and CD73– tumor-infiltrating NK cells was observed (Figure 2A). The greatest proportion of the variability was attributed to surface expression of TIM-3 and LAG-3 (Supplemental Figure 2B). From analyzing the expression of different combinations of immune checkpoint receptors, we observed that the CD73– subset of tumor-infiltrating NK cells does not coexpress more than one immune checkpoint receptor (Figure 2B). Similarly, t-distributed stochastic neighbor embedding (t-SNE) analysis showed distinct populations of CD73+ NK cells expressing multiple immune checkpoints (Figure 2C). Unlike peripheral blood NK cells, tumor-infiltrating NK cells coexpressed other immune checkpoints including LAG-3, TIM-3, PD-1, VISTA, and PD-L1. Furthermore, the expression of these immune checkpoints was significantly higher in CD73+ compared with CD73– tumor-infiltrating NK cells (Figure 2, D–G and Supplemental Figure 2C). Although the expression of PD-1 on CD73+ NK cells was significantly higher compared with CD73– NK cells, it was generally lower compared with other immune checkpoint receptors on both CD73– (mean = 3.28%) and CD73+ (mean = 9.50%) tumor-infiltrating NK cells (Figure 2H). Analysis of the expression of activating receptors on tumor-infiltrating NK cells showed that the expression of NKG2C and NKp44 was significantly higher on CD73+ NK cells compared with CD73– NK cells. In contrast, the expression of NKG2D and NKp46 did not differ between CD73+ and CD73– tumor-infiltrating NK cells (Supplemental Figure 2, C–F). Collectively, NK cells from tumor resections, particularly the CD73+ cells, expressed a variable repertoire of immune checkpoints on their surface that were not observed on peripheral blood NK cells.
Figure 2. Characterization of immune checkpoint expression on CD73+ NK cells isolated from 11 individual tumor resections.
(A) Principal components analysis (PCA) showing an overview of heterogeneity in all samples based on their expression of 5 immune checkpoints and NKG2D. (B) Annotated pie chart showing proportions of cells expressing different combinations of immune checkpoints. Permutation test was performed to compare CD73+ NK cells (n = 12) and CD73– NK (n = 11) cells. (C) t-Distributed stochastic neighbor embedding analysis of tumor-infiltrating NK cell populations from the most representative sarcoma and breast tumor samples. (D–H) Differential expression of immune checkpoints (LAG-3, VISTA, PD-L1, TIM-3, and PD1) comparing CD73+ NK cells, CD73– NK cells, and total peripheral blood NK cells. Paired comparison was done with NK cells analyzed from 7 sarcoma and 4 breast tumor resections. Wilcoxon’s signed-rank test was used to assess significance in matching data points.
NK cells acquire CD73 surface expression upon engagement of 4-1BBL on tumor cells.
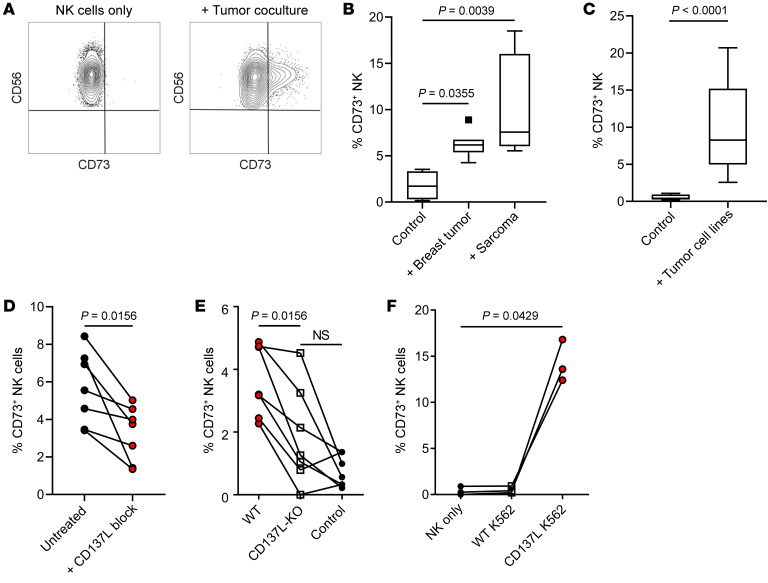
Based on our observations that tumor-infiltrating NK cells with CD73 expression also coexpressed higher levels of immune checkpoints, we hypothesized that CD73 acquisition was caused by an activation response by NK cells. To understand how NK cells acquired the expression of CD73, peripheral blood NK cells from healthy individuals were cocultured with fresh sarcoma and breast tumor resections and analyzed for their expression of CD73. Upon 4 hours of in vitro coculture with different tumors isolated from patient samples, these NK cells indeed upregulated the expression of CD73 (Figure 3, A and B). Similar results were reproduced upon coculture with a panel of tumor cell lines (Figure 3C). These observations were dependent on physical cell contact, as NK cells did not acquire the expression of CD73 when cocultured with tumor cells separated by Transwells or treated with tumor-conditioned culture media (data not shown).
Figure 3. 4-1BBL engagement promotes CD73 surface upregulation on NK cells within 4 hours of tumor coculture.
(A) Representative flow cytometric plot showing induction of CD73 expression by NK cells after 4 hours of tumor coculture. (B) Percentage of CD73+ CFSE-labeled NK cells after coculture with fresh patient tumor resections for 4 hours (n = 8 for breast cancer and n = 6 for sarcoma). (C) Percentage of CD73+ NK cells after coculture with ATCC tumor cell lines (MDA-MB-231, MDA-MB-436, 786O, and CAKI2) (n = 17). (D) Percentage of CD73+ NK cells after coculture with patient-derived sarcoma cell lines in the presence or absence of inhibitory recombinant 4-1BB protein (1 μg/mL) (n = 6). (E) Percentage of CD73+ NK cells after coculture with CD137L-knockout patient-derived sarcoma cell lines (n = 7). (F) Percentage of CD73+ NK cells after coculture with K562 and K562 transduced with 4-1BBL (n = 3). Friedman’s test was used to assess significance in matching data points for panels B and D–F.
Because it was recently shown that agonistic anti-CD137 (anti–4-1BB) induces CD73 expression on tumor-infiltrating T cells, we sought to investigate if 4-1BB engagement could play a role in the induction of CD73 on NK cells (26). To test this, recombinant 4-1BB soluble ligand was added to cocultures of NK cells and primary sarcoma cell lines. Following a 4-hour coculture, a significant reduction in CD73 expression was observed in the presence of recombinant 4-1BB ligand compared with untreated controls (Figure 3D). To further strengthen these observations, CRISPR/Cas9 was used to knock out 4-1BBL in these patient-derived cell lines. Similarly, upon coculture with 4-1BB ligand–edited cell lines, the expression of CD73 on NK cells was significantly reduced (Figure 3E). To exclude the possibility that the induction of CD73 was not due to general activation, NK cells were cultured with the MHC class I–deficient K562 cell line. Although coculture with K562 does activate NK cells, it did not induce CD73 upregulation unless modified to overexpress 4-1BBL (Figure 3F). These results demonstrated that 4-1BB engagement can promote the induction of CD73 expression, although we do not exclude the possibility that other costimulatory receptors could be involved in this process.
NK cells acquire CD73 surface expression via actin polymerization–dependent exocytosis.
Although we observed that 48 hours of IL-2 stimulation induced up to 1%–2% surface expression of CD73, confocal imaging and intracellular flow cytometry staining revealed the presence of CD73 even in tumor-experienced CD73– NK cells (Figure 4A and Supplemental Figure 3A). Based on quantification of CD73 fluorescence intensity, no significant differences in overall cellular CD73 fluorescence were observed upon coculture with 4-1BBL–transfected K562 cells, whereas significant differences in plasma membrane–associated CD73 were observed (Figure 4, B and C). Furthermore, confocal imaging showed that the expression of CD107a and CD73 did not colocalize. To test if exposure to tumor cells stimulated the transport of CD73 protein to the plasma membrane, inhibitors of actin polymerization were added during coculture of NK cells and tumor cells. Indeed, coculture of peripheral blood NK cells with 4-1BBL–expressing K562 target cells in the presence of cytochalasin D, latrunculin B, or jasplakinolide resulted in a dose-dependent reduction in CD73 surface expression (Figure 4, D–F). Because NK cells acquire higher surface CD73 expression in 4 hours of tumor coculture compared with overnight coculture (data not shown), we hypothesized that CD73 can be shed. Indeed, soluble CD73 was detected upon coculture with 4-1BBL–transduced K562 cells. Furthermore, cytochalasin D inhibited the secretion of soluble CD73, confirming that actin polymerization was essential also for the secretion of CD73 by NK cells (Figure 4G). To address if CD73 expression and shedding were due to NK cell degranulation or vesicular transport, NK cells were simultaneously stained for the degranulation and vesicle markers CD107a and CD63, respectively. Upon contact with 4-1BBL–transduced K562 cells, CD73 was mainly expressed on CD63+ NK cells. Degranulating NK cells (CD107a+) that lack surface CD63 expression did not express CD73. Although blocking actin dynamics can downregulate both CD107a and CD63 expression, cells that are still positive for CD63 and negative for CD107a had reduced expression of surface CD73 after treatment with either cytochalasin D or latrunculin B during 4 hours of coculture (Supplemental Figure 3, B and C). Collectively, these results show that NK cells express CD73 intracellularly and transport it to the cell surface, and to the extracellular space upon contact with tumor cells.
Figure 4. Tumor cells stimulate NK cells to translocate CD73 into the cell membrane and the extracellular space.
(A) Maximum intensity projection of confocal images showing localization of CD73 and CD107a expression in NK cells sorted after tumor coculture under 63× objective lens. Scale bar: 10 μm (n = 4). (B) Overall mean fluorescence intensity (MFI) of CD73 staining per cell comparing CD73+ and CD73– NK cells sorted after tumor coculture (n = 4). (C) MFI of CD73 on cell membrane of every cell comparing CD73+ and CD73– NK cells sorted after tumor coculture (n = 4). Significance was tested by Mann-Whitney U test for both B and C. (D–F) Dose-dependent inhibition of CD73 surface expression on NK cells after 4 hours of coculture with 4-1BBL–transduced K562 in the presence of latrunculin B (D), cytochalasin D (E), or jasplakinolide (F) (n = 4). (G) ELISA showing concentration of CD73 protein released after 24 hours of NK cells cocultured with 4-1BBL–transduced K562 (n = 4). Friedman’s test was used to assess significance in D–F.
CD73+ NK cells undergo transcriptional reprogramming to acquire suppressive functions.
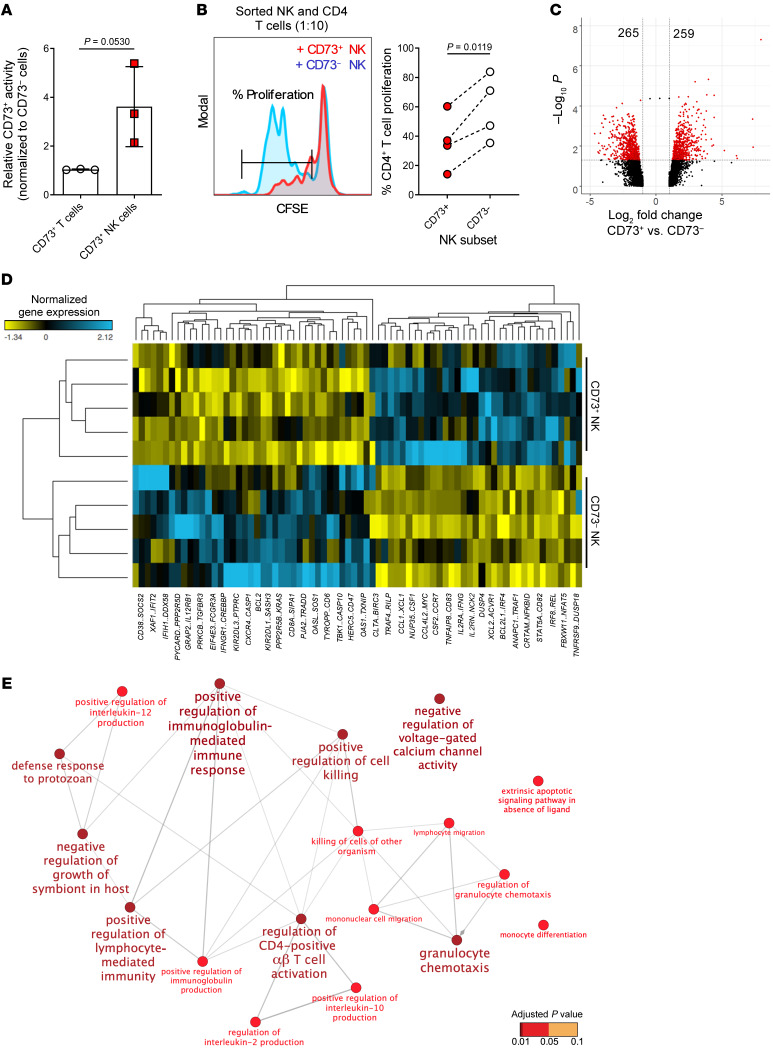
To investigate if the CD73 receptor expressed by NK cells is functional, the capacity of purified CD73+ NK cells to hydrolyze AMP was measured. CD73 enzymatic activity was only detected in CD73+ NK cells despite no significant differences in CD73 levels between tumor-experienced NK cells and CD73+ T cells (Figure 5A). To address if CD73+ NK cells differ in their suppressive activity, purified NK cells were added to proliferating autologous CD4+ T cells. At an NK to T cell responder ratio of 1:10, CD73+ NK cells significantly reduced the proliferation of CD4+ T cells compared with CD73– NK cells (Figure 5B). Importantly, these assays were performed without the addition of exogenous AMP, suggesting that CD73+ NK cells may inhibit CD4+ T cell proliferation through mechanisms other than the production of adenosine. To uncover such underlying mechanisms, CD73+ and CD73– NK cells were purified from tumor cocultures and subjected to RNA sequencing. From 5 independent tumor cocultures, results showed that a total of 524 genes (259 up and 265 down) were differentially expressed by more than 2-fold (Figure 5C, Supplemental Figure 4A, and Supplemental Table 2). When filtered for immune-related genes, CD73+ NK cells generally upregulated genes associated with immune activation, chemokines, CSF1 (encoding M-CSF), and CSF2 (encoding GM-CSF) (Figure 5D). In analyzing the top 100 upregulated genes, functional nodes mostly related to lymphocyte activation were identified (Figure 5E). Of interest, CD73+ NK cells upregulated genes that are related to IL-10 production and granulocyte chemotaxis, both of which could potentially play a role in immune suppression.
Figure 5. Characterization of CD73+ NK cells based on differential gene expression.
(A) CD73 enzymatic activity normalized to CD73+ T cells from PBMCs from healthy donors (n = 3). Student’s t test was used to assess significance. (B) Left: Representative histogram showing proliferation of CD4+ responder T cells after 2 days of coculture with different sorted populations on NK cells at a 10:1 ratio (n = 4). Right: Percentage of dividing CD4+ T cells in a 48-hour suppression assay. Significance was tested with Wilcoxon’s signed-rank test (n = 8). (C) Volcano plot generated to visualize the significance and magnitude of changes in gene expression comparing CD73+ NK cells versus CD73– NK cells. The x axis represents the fold change between the 2 groups and is on a log2 scale, and the y axis shows the negative log10 of the P values from mixed-model ANOVA. Genes with significant fold change less than 2 are represented in red (n = 5). (D) Gene expression heatmap generated to visualize immune-related genes that are differentially expressed in CD73+ NK cells based on Supplemental Table 1 (n = 5). (E) Gene ontology enrichment analysis showing functional pathways for the top 100 genes upregulated by CD73+ NK cells from the gene list in Supplemental Table 2. Each node has a minimum of 3 genes and the size of the nodes is proportional to the number of genes. Color of the nodes is determined by the significance of the enriched term assessed by 2-sided hypergeometric test with Bonferroni’s step-down method for P value correction (n = 5).
To validate and substantiate our findings from these coculture assays, we further isolated tumor-infiltrating NK cells from 2 breast tumors and 2 sarcomas for gene expression analysis. Not only did these experiments confirm the upregulation of IL-10 gene expression but also that several known targets that can be regulated by the STAT3 transcription factor. Notably, cytolytic genes such as granzyme B (GZMB) and perforin (PRF1) were downregulated (Supplemental Figure 4B). Similarly to the gene expression data generated from in vitro cultures, the presence of STAT3 binding motifs was examined 2000 base pairs upstream (5′) of the transcriptional start site of 259 significantly upregulated genes. Of these genes, 256 were recognized in the JASPAR database (http://jaspar.genereg.net/). Of the 256 genes identified, 61 genes with STAT3 binding motifs were identified. From these 61 genes we identified HIF1, CREB1, and NFKB, the protein products of which are all known to dimerize with STAT3 for transcriptional regulation (Supplemental Figure 4C). Together, gene expression analysis from both in vitro coculture and tumor-infiltrating NK cells suggested STAT3 as a key regulator for transcriptional reprogramming of these CD73+ NK cells to acquire a regulatory phenotype.
CD73+ NK cells upregulate IL-10 and TGF-β production via STAT3 transcriptional activity.
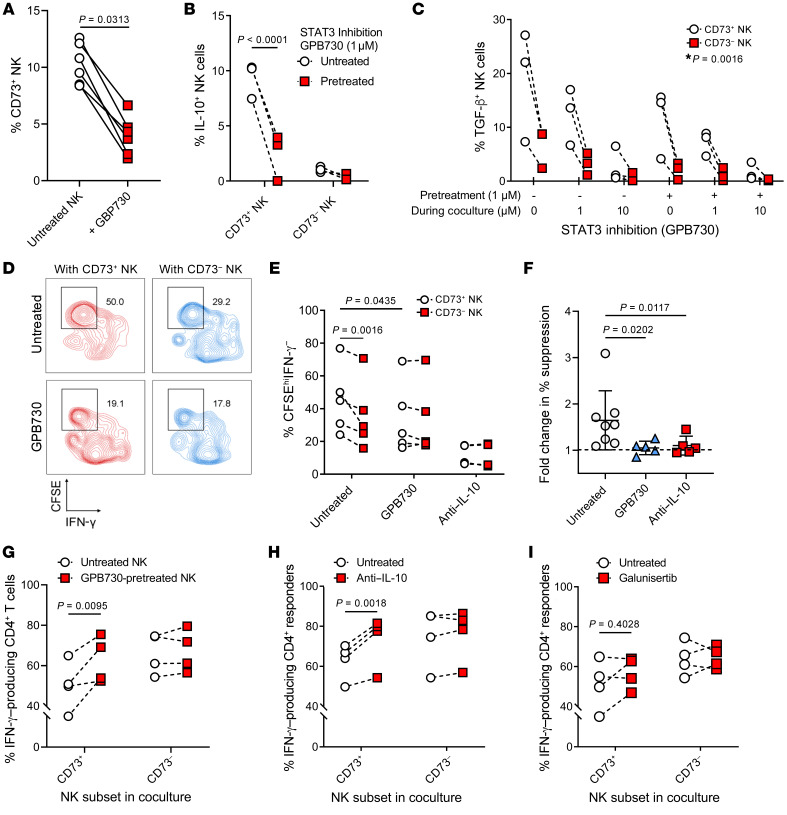
Because STAT3 is known to have immunoregulatory capacity and also regulates CD73 expression (27, 28), we sought to further investigate the role of STAT3 activity in these inducible CD73+ regulatory NK cells. We found that CD73+ NK cells displayed increased phosphorylation of serine residue 727 and tyrosine residue 705 of STAT3 as compared with CD73– NK cells (Supplemental Figure 5, A and B). To confirm if STAT3 is involved in inducing CD73+ immunoregulatory NK cells, the selective STAT3 inhibitor GPB730 was added to NK-tumor cocultures (Supplemental Figure 5C) (29, 30). During tumor coculture, GPB730 decreased STAT3-S727 phosphorylation in NK cells, while no significant changes in Y705 phosphorylation were observed (Supplemental Figure 5, D and E). Treatment with GPB730 before and during overnight tumor coculture resulted in an average 3-fold (SD = 1.19) reduction in the expression of CD73 on NK cells (Figure 6A). Because IL-10 was upregulated at both gene and protein levels by tumor-infiltrating CD73+ NK cells (Supplemental Figure 4B and Supplemental Figure 5F), NK cells were cocultured with tumors cells and assayed for their production of IL-10. Indeed, upon coculture with tumor cells, a significant upregulation of IL-10 production by CD73+ NK cells compared with CD73– NK cells was observed. In addition, CD73+ NK cells also produced higher levels of TGF-β compared with CD73– NK cells, and the capacity of CD73+ NK cells to produce these 2 cytokines was significantly reduced in the presence of GPB730. In addition, the production of TGF-β was significantly inhibited only at high doses of GPB730 during coculture, whereas IL-10 production was inhibited by pretreating NK cells with a low dose of GPB730 (Figure 6, B and C). Next, NK cells were treated with GPB730 before tumor coculture and tested for their ability to suppress CD4+ T cell production of IFN-γ and proliferation. Compared with CD73– NK cells, CD73+ NK cells significantly suppressed CD4+ T cell activity. Upon pretreatment with GPB730, a significantly reduced suppression by NK cells was observed, as evidenced by a reduced CFSEhiIFN-γ–CD4+ T cell population (Figure 6, D–F).
Figure 6. STAT3-dependent TGF-β and IL-10 production by CD73+ NK cells and suppressed proliferation and cytokine production of CD4+ T cells.
(A) Percentage of CD73+ NK cells after treatment with GPB730 (1 μM). Wilcoxon’s signed-rank test was used to assess significance (n = 6). (B) Percentage of IL-10–producing NK cells pretreated with GPB730 (1 μM for 48 hours) before overnight tumor coculture (n = 3). (C) TGF-β–producing NK cells with GPB730 treatment during tumor coculture (n = 3). *P value obtained for significant interaction between 2 variables based on 2-way ANOVA with Tukey’s post hoc test. (D) Representative flow cytometric plots showing percentage of suppressed CD4+ T cell activity based on CFSE labeling and IFN-γ production (n = 5). (E) Percentage of CFSEhiIFN-γ– CD4+ T cells after 48 hours of coculture with different subsets of NK cells at a 10:1 ratio (n = 5). Treatment conditions: NK cells were pretreated with GPB730 (1 μM) or cocultured with neutralizing antibody against IL-10 (10 μg/mL). (F) Fold change in percentage suppression (CD73+ vs. CD73–) calculated based on percentage of CFSEhiIFN-γ– responder CD4+ T cells in autologous coculture. A sample size of n = 5 was used in each treated group. Friedman’s test with multiple comparisons was used to assess significance. (G) IFN-γ+CD4+ T cells in a suppression assay comparing sorted CD73+ NK cells versus CD73– NK cells with or without GPB730 pretreatment (n = 4). (H and I) IFN-γ+CD4+ T cells in a suppression assay comparing sorted CD73+ NK cells versus CD73– NK cells in the presence of (I) neutralizing antibody against IL-10 (10 μg/mL) and (J) galunisertib (1 μM) (n = 4). Repeated-measures 2-way ANOVA with multiple comparisons was used to test for significant differences in C, F, and G–I.
Because GPB730-pretreated NK cells showed reduced suppression of CD4+ T cell IFN-γ production (Figure 6G), we further investigated if production of either IL-10 or TGF-β was the direct underlying suppressive mechanism of CD4+ T cells by CD73+ NK cells. Suppression assays performed in the presence of neutralizing antibodies against IL-10 restored the production of IFN-γ by CD4+ T cells, whereas the presence of the TGF-β1 receptor inhibitor galunisertib did not (Figure 6, H and I). These results show that CD73+ NK cells upregulate STAT3 activity due to immune activation, resulting in the production IL-10 that inhibits IFN-γ production by CD4+ T cells.
Discussion
Under physiological conditions, purinergic receptors play an important role in lineage commitment, tissue repair, and conferring immune tolerance (2, 31). However, extracellular adenosine could be dysregulated by CD73-overexpressing tumors, contributing to resistance against cytostatic drugs and immune therapies (7, 26, 32, 33). Despite extensive efforts to introduce CD73 inhibitors into the clinic, the complexity in the regulation of this ectonucleotidase is not well understood (34). We observed that the predictive value of tumor NT5E expression could be influenced by the magnitude of the NK cell gene signature in breast cancer and sarcoma. Notably, the expression of NT5E influenced the prognosis of breast cancer and sarcoma patients with low and high NK cell gene signatures, respectively. The underlying mechanisms of these findings are, however, unclear.
Since sarcomas are mesenchymal tumors and breast cancers are epithelial tumors, their respective influence on NK cell infiltration and activity may differ. Although a positive correlation between tumor NT5E expression and NK cell gene signature was observed, CD73 was also found to be expressed on tumor-infiltrating NK cells. The proportion of CD73+ tumor-infiltrating NK cells also positively correlated with larger breast tumors. Although regulatory T cells have been shown to express the ectonucleotidases CD39 and CD73 and inhibit T cell responses via the production of adenosine (35), less is known regarding the expression of these ectonucleotidases in conventional T cells as well as NK cells. Upon exposure to mesenchymal stem cells (MSCs), T cells upregulate CD39, resulting in suppression of activated T cells via increased production of extracellular adenosine (36). Similarly, studies reported that NK cells acquire the expression of CD73 after physical contact with human umbilical cord–derived MSCs or dental pulp stem cells (37, 38). In cancer, NK cells from gastrointestinal stromal tumors (GISTs) have been found to express higher levels of surface CD73 (39). Yet, the current understanding of tumor-infiltrating NK cells is unclear with regard to their robustness in phenotype and function. Here we extend these observations and demonstrate that NK cells not only upregulate the expression of CD73 but also several other immune checkpoint receptors implicating immune exhaustion. We found that these NK cells underwent transcriptional reprogramming to acquire noncanonical functions to suppress the immune environment.
The basis of our study was to coculture activated NK cells with tumor cells to model tumor-experienced NK cells within the TME. Upon physical contact with tumor cells, a rapid cell-surface expression and secretion of CD73 by NK cells via vesicular transport was observed. This observation was not restricted to tumor cell contact, as we also observed that endothelial cells and fibroblasts could also induce CD73 expression on NK cells (data not shown). Although agonizing 4-1BB activates both CD4+ and CD8+ T cells in vitro, engaging 4-1BB can result in an impaired NK cell development and induce cell death in resting NK cells (40–42). Mechanistically, 4-1BB stimulation was involved in the induction of CD73, which was also previously studied in tumor-infiltrating T cells (26). Given that there could be other chronic stimulation involved in the generation of CD73+ NK cells within the TME, there could be other potential underlying mechanisms left uncovered. Considering that prior studies have demonstrated that CTLA-4 and CD73 can be stored within intracellular vesicles, we hypothesized and demonstrated that exocytosis was involved in the underlying mechanism in which NK cells acquire CD73 expression (35, 43, 44). Although CD73 is an enzyme that hydrolyzes extracellular AMP on the cell surface, we showed that these tumor-experienced NK cells could also shed CD73 protein into the extracellular space — a finding that might have clinical relevance because soluble CD73 has been suggested to be a biomarker in patients with metastatic cancer (45). Another possibility that we did not explore was if these NK cells could secrete exosomes expressing CD73, which was previously found to be upregulated in serum from patients with head and neck squamous cell carcinoma (35, 46).
The concept of regulatory NK cells is controversial. An early study demonstrated that NK cells become immunosuppressive and produce IL-10 during acute infection (22). Later it was reported that CD56bright NK cells not only serve as regulatory cells secreting IL-10 but also synthesize adenosine via the CD38/PC1/CD73 pathway (21). A recent study by Crome et al. showed that CD56+CD3– regulatory innate lymphoid cells express higher levels of NKp46, with elevated IL-22 production (23). Without the assumption that these cells are NK cells, the group demonstrated that this unique population of innate lymphoid cells suppresses the ex vivo expansion of TILs from high-grade serous cancers.
By studying how tumor cells can influence normal NK cells to acquire CD73, we demonstrated that CD73+ NK cells suppress both proliferation and cytokine production of CD4+ T cells as compared with CD73– NK cells. Although the induced CD73 receptor on NK cells was functional as evidenced by the ability to hydrolyze AMP, the addition of exogenous AMP did not influence the immune suppression mediated by CD73+ NK cells (data not shown). Likewise, adenosine receptor inhibitors did not significantly change the ability of CD73+ NK cells to suppress CD4+ T cell responses (data not shown). Thus, we hypothesize that CD73+ NK cells may suppress CD4+ T cells via mechanisms other than the production of extracellular adenosine. Although studies have shown that NK cells can produce IL-10 via binding of Ly49H or IL-12 stimulation in viral and systemic infections respectively, we found that tumor-experienced NK cells produced IL-10 via STAT3 activation (22, 47). It was recently shown that STAT3 negatively regulates several NK cell functions and drives the expression of PD-L1 (48, 49). Our results support the idea that STAT3 drives the generation of CD73+ regulatory NK cells as well as the production of IL-10 by these NK cells.
In summary, NK cells undergo dynamic phenotypic and functional changes influenced by different cues within the heterogeneous TME. It is plausible to envision that future investigations on regulatory NK cells could potentially correlate with other undesirable clinical outcomes in a variety of solid tumors. Our results also support the use of STAT3 and IL-10 inhibitors by dampening the suppressive tumor immune landscape and thereby complement current immunotherapies.
Methods
Cell lines.
Commercial cell lines used for cocultures are 786O, CAKI2, MDAMB231, MDAMB436, and K562 (ATCC). K562 transduced with 4-1BB ligand were provided by Crystal Mackall (National Cancer Institute, Bethesda, Maryland, USA). Patient-derived sarcoma cell lines were established from surgical resections of patients 8 and 9 listed in Table 3. In brief, whole tumors were digested and processed (Tumor Dissociation Kit, Miltenyi Biotec). Tumor cells were then isolated by negative selection (Tumor Cell Isolation Kit, Miltenyi Biotec). Adherent cells were passaged at least 5 times before being used for experiments. All cell lines were maintained in RPMI1640 or in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific).
Table 3. Characteristics of sarcoma patient cohort.
Chemicals.
Cytochalasin D, latrunculin B, and jasplakinolide (Sigma-Aldrich) were used to inhibit actin polymerization in tumor cocultures. To inhibit STAT3, the selective small-molecule inhibitor GPB730 (N-[(4R,7R,9R,11S)-11-hydroxy-9-methyl-2-oxo-3-oxatricyclo [5.3.1.04,11] undec-l(10)-en-9-yl-4-methylbenzene-l-sulfonamide]) (Glactone Pharma) was used (see Supplemental Figure 5C for chemical structure). GPB730 and the inhibitor for TGF-β type 1 receptor, galunisertib (Selleckchem), were used for cocultures and suppression assays. Recombinant 4-1BB was used in cocultures to block 4-1BBL engagement (PeproTech, 310-15).
CRISPR knockout of CD137L in patient-derived cell lines.
To generate sarcoma cells with a knockout of CD137L, gRNAs were designed to bind within exon 1 of the gene using the CRISPOR algorithm (50). The gRNA (ATACGCCTCTGACGCTTCAC) was subcloned into the lentiviral expression vector lentiCRISPR v2 using Esp3I insertion sites (51). Plasmids were verified by sequencing. Lentivirus was produced as previously described (52). Briefly, 1 × 106 HEK293FT cells were plated into poly-D-lysine–coated 60-mm dishes (BD Biosciences). The following day, the cells were transfected with 4.8 μg of lentiCRISPR v2 containing the gRNA, 2.4 μg of pMDLg/pRRE (Addgene), 1.6 μg of pRSV-REV (Addgene), and 0.8 μg of phCMV-VSV-G (Addgene) using a calcium phosphate transfection kit (Sigma-Aldrich) in the presence of 25 μM chloroquine (Sigma-Aldrich). At 16 hours after transfection, the medium was changed, and the virus particles were collected after an additional 28 hours by filtering the supernatant through a 0.45-μm filter and stored at –80°C. Sarcoma cells (15,000) suspended in 2 mL of RPMI medium supplemented with 10% FBS were transduced with 500 μL of virus-containing supernatant for 8 hours in 8 μg/mL protamine sulfate (Sigma-Aldrich). After transduction, cells were cultured for 14 days and then sorted for absent staining with CD137L antibody (BioLegend, 311503) by using fluorescence-activated cell sorting (FACS) with a FACSAria Fusion (BD Biosciences). Sorted cells were seeded overnight in culture before coculture experiments.
NK cell isolation and culture.
Peripheral blood mononuclear cells (PBMCs) were collected after Ficoll gradient centrifugation (GE Healthcare). NK cells were isolated by negative selection based on the manufacturer’s protocol (Human NK Cell Isolation Kit, Miltenyi Biotec). Isolated NK cells were then cultured in X-VIVO 20 (Lonza) supplemented with 10% human AB serum (Karolinska University Hospital Blood Bank) and 1000 IU/mL IL-2 for 48 hours (Proleukin). In the absence of IL-2, NK cells were cocultured with tumor cell lines for 4 or 16 hours before cell sorting and FACS analysis.
CD4+ T cell isolation and suppression assay.
CD4+ T cells were isolated by negative selection based on the manufacturer’s protocol (Human CD4 T Cell Isolation Kit, Miltenyi Biotec). Isolated CD4+ T cells were labeled with 1 μM CFSE (BioLegend) and stimulated with 12 μL of CD3/CD28 beads (Thermo Fisher Scientific) per million cells and 50 IU/mL IL-2 for 48 hours before suppression assay. For the suppression assay, CD73+ and CD73– NK cells were isolated by FACS and cocultured with autologous activated CD4+ T cells at a suppressor to responder ratio of 1:10 in serum-free X-VIVO 20 media. FACS analysis was performed after 48 hours of coculture.
FACS analysis.
Single-cell suspensions of PBMCs and tissue samples were washed with FACS buffer (5% FBS in PBS) before staining with antibody mix (see Supplemental Table 3) in the presence of Human Fc Block (BD Biosciences). All samples were acquired on a Novocyte (ACEA Biosciences). All data were analyzed with FlowJo software (Tree Star). From FlowJo, FCS files with only NK cells were concatenated for downstream t-SNE analysis using the cytofkit R package (https://github.com/JinmiaoChenLab/cytofkit). PCA was performed using SIMCA 15 software (Umetrics). For staining of IFN-γ, cells were incubated for 1 hour at 37°C with 100 ng/mL phorbol myristate acetate (PMA) and 1 μg/mL ionomycin (Sigma-Aldrich) in X-VIVO 20 media supplemented with 10% human AB serum. After 1 hour, GolgiSTOP and GolgiPLUG (BD Biosciences) were used in combination according to the manufacturer’s protocol. Cells were then further incubated for 3 hours under the same conditions before cell surface staining. After fixation and permeabilization, antibodies against IFN-γ were added for intracellular staining at room temperature for 20 minutes before cells were washed and acquired on the Novocyte. See Supplemental Table 3 for antibody panel. For IL-10 and TGF-β staining, GolgiSTOP and GolgiPLUG were used in combination for 24 hours within the tumor coculture setting. Surface and intracellular cytokine staining was then performed as described above. For staining of phosphorylated STAT3, cells were fixated with Fix Buffer I (BD Biosciences) at 37°C for 10 minutes. Permeabilization of cells was done by resuspension in ice-cold Perm Buffer III (BD Biosciences) for 45 minutes at 4°C. After permeabilization and washing, cells were stained with anti–STAT3 p-Y705 and anti–STAT3 p-S727 (BD Biosciences) diluted 1:100 in FACS buffer for 1 hour at 4°C. Cells were washed twice in FACS buffer before acquisition on the flow cytometer.
CD73 ELISA and enzymatic activity assay.
NK cells (0.5 × 106) were cocultured with 0.1 × 106 4-1BBL–transduced K562 cells for 24 hours at an NK to K562 ratio of 5:1. Following coculture, supernatants were collected to detect CD73 by ELISA based on the manufacturer’s protocol (Nordic Biosite). To test the enzymatic activity of CD73, a modified AMP depletion assay was used (53). Briefly, 5000 sorted CD73+ and CD73– T and NK cells were seeded in each well of 96-well plate in serum-free X-VIVO 20 media with 0.4 mM AMP (Sigma-Aldrich). Cells were incubated for 90 minutes at 37°C and 5% CO2. After incubation, 25 μL of cell supernatant was collected and mixed with 25 μL of X-VIVO 20 media with 200 μM ATP (Sigma-Aldrich). The mixture was then added in a white-bottom plate with CellTiter-Glo reagent (Promega) at a 1:1 ratio. Luminescence readout was obtained with an integration time of 100 ms on a SPARK 10M plate reader (TECAN). Raw values from CD73– control cells were used to subtract background signals.
STAT3 reporter gene assay.
Dose-response activity to determine IC50 at 8 different doses in triplicate was used in a STAT3 reporter system. Briefly, the STAT3 reporter/HEK293 cell line was plated in 96-well white plates for 16 hours. Cells were pretreated with GPB730 for 1 hour. Cells were then treated with IL-6 to induce STAT3 activation for 16 hours. Luciferase activity was measured and analyzed.
Confocal image analysis.
Cells were seeded and incubated for 20 minutes on slides that had been coated with ICAM-1 (BioLegend, 552904) at 1 μg/mL for 1 hour at 37°C. Cells were then fixed using 4% paraformaldehyde for 20 minutes before being permeabilized with a 0.1% Triton solution for 10 minutes. Blocking was performed using 5% FBS in PBS for 1 hour. The cells were thereafter stained using mouse anti–human CD73 antibody (Invitrogen, 41-0200, 1:100) for 1 hour before incubation with the secondary antibody goat anti–mouse IgG conjugated with Alexa Fluor 647 (Invitrogen, A-21236, 1:500) for 1 hour. Cells were thereafter incubated with biotinylated mouse anti–human CD63 (Abcam, ab134331, 1:200) for 1 hour or biotinylated mouse anti–human CD107a (BioLegend, 328604, 1:100) for 1 hour. Finally, the cells were incubated with a solution containing DAPI (1:100), Alexa Fluor 488 phalloidin (Invitrogen, A12379, 1:200), and Alexa Fluor 555 streptavidin (Invitrogen, S21381, 1:500) for 30 minutes. The cells were then mounted in ProLong Diamond Antifade Mountant (Invitrogen, P36965). The cells were imaged using a Zeiss LSM800 confocal microscope equipped with Plan-Apochromat 63×/1.40 oil DIC M27 lens and analysis was performed using FIJI/ImageJ and CellProfiler.
Gene expression analysis of NK cells after tumor coculture.
Following 4 hours of coculture with the 786O tumor cell line and NK cells (n = 5), FACS-isolated CD73+ and CD73– NK cells were used to isolate RNA using an RNeasy Micro Kit based on the manufacturer’s protocol (Qiagen). Total RNA integrity was analyzed using the Agilent 2100 Bioanalyzer RNA 6000 Pico Kit and quantified on the Qubit 3.0 Fluorometer using the Qubit RNA High Sensitivity Assay Kit or by real-time PCR using Applied Biosystems’ RNA Quantification Kit on the Agilent AriaMx. Total RNA was normalized to 28 pg of input for full-length cDNA generation using Takara’s SMART-Seq v4 Ultra Low Input RNA Kit following the manufacturer’s recommendations. cDNA was normalized to 1 ng of input for sequencing library generation using Illumina’s Nextera XT DNA Library Preparation Kit following the manufacturer’s protocol. cDNA libraries were quantified by qPCR using Kapa Biosystems’ Library Quantification Kit on the Roche LightCycler 480 Instrument II following the manufacturer’s protocol. cDNA libraries were normalized to 1.5–2 nM, pooled, and denatured following Illumina’s NextSeq System Denature and Dilute Libraries Guide. Final pooled libraries (n = 5 samples) were spiked with 1% PhiX as an internal control and loaded at a final concentration of 1.6 pM onto the Illumina NextSeq 500 platform. cDNA libraries were sequenced on a 2 × 75-bp paired-end run using the NextSeq 500 High Output v2 Kit (150 cycles) for n = 2 sequencing runs. Approximately 763 million indexed pass-filter paired-end reads were generated during sequencing run 1 and approximately 760 million indexed, pass-filter paired-end reads were generated during sequencing run 2.
Raw data were uploaded into Partek Flow for data analysis and NCBI’s Gene Expression Omnibus database (GEO GSE125119). Raw reads were quality trimmed based on a minimum Phred quality score of 20 and aligned to human genome assembly GRCh38 (hg38) using STAR v2.5.3a with default parameters. Gene level counts were normalized using counts per million (CPM) and quantified using a mixed-model ANOVA to account for donor ID as a random effect and CD73+/– expression status as a fixed factor. A total of 8456 differentially expressed genes were identified (see Supplemental Table 1), while 524 were significantly differentially expressed (P value < 0.05, fold change ±2; see Supplemental Table 2). A volcano plot (Figure 5C) was generated to visualize the significance and the magnitude of changes in gene expression. From the filtered 524 genes (Supplemental Table 2), 74 immune-related genes were identified from the Gene Ontology (GO) database; a heatmap was generated using (unsupervised) hierarchical clustering (average linkage distance metric and Euclidean point distance metric) (Figure 5D). Note binning of genes in the x axis for display purposes.
GO enrichment analysis was done based on the top 100 genes upregulated (see Supplemental Table 2) with the ClueGO plugin in Cytoscape. Using default ClueGO settings and GO term fusion, the 100 gene IDs were submitted to query the GO Biological Process database (EBI, QuickGO, 15,783 terms associated with 17,268 unique genes; updated on November 20, 2017). GO terms are presented as nodes and clustered together based on the term similarity (Figure 5E). From Supplemental Table 2, 266 significantly upregulated genes were uploaded onto oPOSSUM-3 to query for the STAT3 transcription binding site up to 2000 bp upstream.
Gene expression analysis of tumor-infiltrating NK cells.
Before FACS isolation, immune cells were enriched from freshly processed tumor samples (CD45 Isolation Kit, Miltenyi Biotec). Based on CD73 expression, NK cells were sorted directly into lysis buffer for RNA extraction (RNeasy Micro Kit, Qiagen). A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using a NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample. In order to select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with an AMPure XP system (Beckman Coulter). Then, 3 μL of USER Enzyme (New England Biolabs) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 minutes followed by 5 minutes at 95°C before PCR. PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform and paired-end reads were generated. Reference genome and gene model annotation files were downloaded directly from the UCSC genome browser (https://genome.ucsc.edu/). Paired-end clean reads were mapped to the reference genome using HISAT2 software. Before differential gene expression analysis, for each sequenced library the read counts were adjusted by the edgeR program through one scaling-normalized factor. Differential expression analysis of 2 conditions was performed using the DEGseq R package. P values were adjusted using the Benjamini and Hochberg method.
Application of an NK cell signature to public data sets.
Normalized, batch corrected, RNA sequencing data and patient/tumor clinico-pathological characteristics from the Pan-Cancer Genome Atlas project (Pan-Can) were accessed from the NIH genomic data commons (GDC) database (https://gdc.cancer.gov). Genes representing the NK signature (KLRB1, CD160, NCR1, NCR3, and PRF1) were extracted from the data set and their expression summed on a per tumor (column-wise) basis to generate an NK signature score. Signatures for CD8+ cytotoxic T cells (CD3E, CD8A) and regulatory T cells (FOXP3) were derived using the same scoring method (25). For breast tumor– and sarcoma-specific analyses this score was divided into quartiles within each cancer type and the expression of the NT5E gene was examined within the highest and lowest NK signature quartiles. Results are tabulated in Table 1.
Statistics.
Kaplan-Meier analysis was performed using the survival and survplot R packages with NT5E split into a binary (low/high) variable based on the median value and with progression-free interval as the survival endpoint. The permutation test shown in Supplemental Figure 2 was performed using SPICE version 6.0 software (NIH). All other experimental data were plotted and tested for significance using Prism 8.0 (GraphPad Software) as described in figure legends unless stated otherwise. P values below 0.05 were considered significant. All error bars represent SD of the mean.
Study approval.
Tumor resections were obtained from breast cancer patients at Karolinska University Hospital, St. Göran Hospital, and Stockholm South General Hospital (Table 2). All cases except case 17 are untreated primary tumors. Tumor resections and peripheral blood were collected from sarcoma patients at Karolinska University Hospital (Table 3). Before resection, informed consent was given and the collection of patient samples was approved by the ethical review board of Karolinska Institutet (2012/90-31/2, 2013/1979-31, 2016/957-31 and 2017/742-32) and in accordance with the Declaration of Helsinki. Peripheral blood samples were obtained from purchased anonymized by-products of blood donations from healthy adult donors at the Karolinska University Hospital Blood Bank.
Author contributions
SYN, JR, NPT, EB, JA, JH, and AL contributed to the study conception and design. SYN, YY, JR, RM, XC, ZC, AKW, SM, EA, MJ, LSW, FH, JH, and AL contributed to the development of research methodology. SYN, YY, JR, RM, XC, ZC, NPT, EB, CS, RT, and AKW contributed to the acquisition of data and conducted experiments. SYN, YY, NPT, EB, JA, JDB, JB, RC, MJ, LSW, FH, JH, and AL contributed to data analysis and interpretation, and essential reagents. SYN, JR, AKW, NPT, and AL contributed to the writing the manuscript.
Supplementary Material
Acknowledgments
We thank Maria Johansson and Juan Basile at the Biomedicum Flow Cytometry Core Facility, Karolinska Institute. We thank Marissa Brooks at the Genomics Core Facility and Kim Kusser at the Flow Cytometry Facility at the Center for Collaborative Research, Nova Southeastern University. We thank Chris Tibitt, Apple Tay Hui Min, and Anna Malmerfelt, Karolinska Institute, for technical assistance. This work was supported by the Swedish Cancer Society (CAN 2015/421 and CAN 2018/451), the Swedish Childhood Cancer Foundation (PR2014-0093 and PR2017-0049), the Cancer Research Foundations of Radiumhemmet (161192 and 181183), Vinnova Swelife, and Medtech4Health (2018-00262), and The Sagen Foundation.
Version 1. 11/26/2019
In-Press Preview
Version 2. 02/04/2020
Electronic publication
Version 3. 03/02/2020
Print issue publication
Funding Statement
This foundation does not provide a grant or fund number
Footnotes
Conflict of interest: MJ is an employee at Glactone Pharma Development.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(3):1185–1198.https://doi.org/10.1172/JCI128895.
Contributor Information
Shi Yong Neo, Email: shiyong.neo@ki.se.
Ying Yang, Email: ying.yang@ki.se.
Ran Ma, Email: ran.ma@ki.se.
Xinsong Chen, Email: xinsong.chen@ki.se.
Ziqing Chen, Email: chen.ziqing@ki.se.
Nicholas P. Tobin, Email: nick.tobin@ki.se.
Emily Blake, Email: Blake.Emily@mayo.edu.
Christina Seitz, Email: christina.seitz@ki.se.
Ron Thomas, Email: rthomas2@nova.edu.
Arnika Kathleen Wagner, Email: arnika.wagner@ki.se.
John Andersson, Email: john.andersson@ki.se.
Jana de Boniface, Email: janamarit@yahoo.com.
Jonas Bergh, Email: jonas.bergh@ki.se.
Shannon Murray, Email: smurray@fredhutch.org.
Evren Alici, Email: evren.alici@ki.se.
Richard Childs, Email: childsr@nhlbi.nih.gov.
Martin Johansson, Email: martin.johansson@glactone.com.
Lisa S. Westerberg, Email: Lisa.Westerberg@ki.se.
Felix Haglund, Email: Felix.Haglund@ki.se.
Johan Hartman, Email: johan.hartman@ki.se.
Andreas Lundqvist, Email: Andreas.Lundqvist@ki.se.
References
- 1.Allard B, Longhi MS, Robson SC, Stagg J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol Rev. 2017;276(1):121–144. doi: 10.1111/imr.12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scarfì S. Purinergic receptors and nucleotide processing ectoenzymes: Their roles in regulating mesenchymal stem cell functions. World J Stem Cells. 2014;6(2):153–162. doi: 10.4252/wjsc.v6.i2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard D, Allard B, Gaudreau PO, Chrobak P, Stagg J. CD73-adenosine: a next-generation target in immuno-oncology. Immunotherapy. 2016;8(2):145–163. doi: 10.2217/imt.15.106. [DOI] [PubMed] [Google Scholar]
- 4.Häusler SF, et al. Anti-CD39 and anti-CD73 antibodies A1 and 7G2 improve targeted therapy in ovarian cancer by blocking adenosine-dependent immune evasion. Am J Transl Res. 2014;6(2):129–139. [PMC free article] [PubMed] [Google Scholar]
- 5.Antonioli L, Yegutkin GG, Pacher P, Blandizzi C, Haskó G. Anti-CD73 in cancer immunotherapy: awakening new opportunities. Trends Cancer. 2016;2(2):95–109. doi: 10.1016/j.trecan.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stagg J, et al. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc Natl Acad Sci USA. 2010;107(4):1547–1552. doi: 10.1073/pnas.0908801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loi S, et al. CD73 promotes anthracycline resistance and poor prognosis in triple negative breast cancer. Proc Natl Acad Sci USA. 2013;110(27):11091–11096. doi: 10.1073/pnas.1222251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Astra Zeneca. Phase II umbrella study of novel anti-cancer agents in patients with NSCLC who progressed on an anti-PD-1/PD-L1 containing therapy (HUDSON). NIH. https://ClinicalTrials.gov/show/NCT03334617 Published November 7, 2017. Updated October 25, 2017. Accessed December 17, 2019.
- 9. MedImmune LLC. Oleclumab (MEDI9447) EGFRm NSCLC novel combination study. NIH. https://ClinicalTrials.gov/show/NCT03381274 Published December 21, 2017. Updated December 9, 2019. Accessed December 17, 2019.
- 10. MedImmune LLC. MEDI9447 (Oleclumab) pancreatic chemotherapy combination study. NIH. https://ClinicalTrials.gov/show/NCT03611556 Published August 2, 2018. Updated December 5, 2019. Accessed December 17, 2019.
- 11. MedImmune LLC. MEDI9447 alone and in combination with MEDI4736 in adult subjects with select advanced solid tumors. NIH. https://ClinicalTrials.gov/show/NCT02503774 Published July 21, 2015. Updated December 9, 2019. Accessed December 17, 2019.
- 12.Gettinger S, et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 2017;7(12):1420–1435. doi: 10.1158/2159-8290.CD-17-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlberg CI, Sarhan D, Chrobok M, Duru AD, Alici E. Natural killer cell-based therapies targeting cancer: possible strategies to gain and sustain anti-tumor activity. Front Immunol. 2015;6:605. doi: 10.3389/fimmu.2015.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kärre K. Natural killer cell recognition of missing self. Nat Immunol. 2008;9(5):477–480. doi: 10.1038/ni0508-477. [DOI] [PubMed] [Google Scholar]
- 15.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 16.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen SK, Gao Y, Basse PH. NK cells in the tumor microenvironment. Crit Rev Oncog. 2014;19(1–2):91–105. doi: 10.1615/critrevoncog.2014011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian W, et al. A prognostic risk model for patients with triple negative breast cancer based on stromal natural killer cells, tumor-associated macrophages and growth-arrest specific protein 6. Cancer Sci. 2016;107(7):882–889. doi: 10.1111/cas.12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17(9):1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 20.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16(1):7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 21.Morandi F, et al. CD56brightCD16- NK cells produce adenosine through a CD38-mediated pathway and act as regulatory cells inhibiting autologous CD4+ T cell proliferation. J Immunol. 2015;195(3):965–972. doi: 10.4049/jimmunol.1500591. [DOI] [PubMed] [Google Scholar]
- 22.Vivier E, Ugolini S. Regulatory natural killer cells: new players in the IL-10 anti-inflammatory response. Cell Host Microbe. 2009;6(6):493–495. doi: 10.1016/j.chom.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Crome SQ, et al. A distinct innate lymphoid cell population regulates tumor-associated T cells. Nat Med. 2017;23(3):368–375. doi: 10.1038/nm.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang T, et al. Comprehensive evaluation of NT5E/CD73 expression and its prognostic significance in distinct types of cancers. BMC Cancer. 2018;18(1):267. doi: 10.1186/s12885-018-4073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Böttcher JP, et al. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell. 2018;172(5):1022–1037.e14. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, et al. CD73 expression on effector T cells sustained by TGF-β facilitates tumor resistance to anti-4-1BB/CD137 therapy. Nat Commun. 2019;10(1):150. doi: 10.1038/s41467-018-08123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalmin F, et al. Stat3 and Gfi-1 transcription factors control Th17 cell immunosuppressive activity via the regulation of ectonucleotidase expression. Immunity. 2012;36(3):362–373. doi: 10.1016/j.immuni.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Hedrich CM, et al. Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc Natl Acad Sci USA. 2014;111(37):13457–13462. doi: 10.1073/pnas.1408023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Don-Doncow N, et al. Galiellalactone is a direct inhibitor of the transcription factor STAT3 in prostate cancer cells. J Biol Chem. 2014;289(23):15969–15978. doi: 10.1074/jbc.M114.564252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Escobar Z, et al. Preclinical characterization of 3β-(N-acetyl l-cysteine methyl ester)-2aβ,3-dihydrogaliellalactone (GPA512), a prodrug of a direct STAT3 inhibitor for the treatment of prostate cancer. J Med Chem. 2016;59(10):4551–4562. doi: 10.1021/acs.jmedchem.5b01814. [DOI] [PubMed] [Google Scholar]
- 31.MacDonald GI, Augello A, De Bari C. Role of mesenchymal stem cells in reestablishing immunologic tolerance in autoimmune rheumatic diseases. Arthritis Rheum. 2011;63(9):2547–2557. doi: 10.1002/art.30474. [DOI] [PubMed] [Google Scholar]
- 32.Streicher K, et al. Increased CD73 and reduced IFNG signature expression in relation to response rates to anti-PD-1(L1) therapies in EGFR-mutant NSCLC. J Clin Oncol. 2017;35(15_suppl):11505. doi: 10.1200/JCO.2017.35.15_suppl.11505. [DOI] [Google Scholar]
- 33.Inoue Y, et al. Prognostic impact of CD73 and A2A adenosine receptor expression in non-small-cell lung cancer. Oncotarget. 2017;8(5):8738–8751. doi: 10.18632/oncotarget.14434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garber K. Adenosine checkpoint agent blazes a trail, joins immunotherapy roster. Nat Biotechnol. 2017;35(9):805–807. doi: 10.1038/nbt0917-805. [DOI] [PubMed] [Google Scholar]
- 35.Schuler PJ, et al. Human CD4+ CD39+ regulatory T cells produce adenosine upon co-expression of surface CD73 or contact with CD73+ exosomes or CD73+ cells. Clin Exp Immunol. 2014;177(2):531–543. doi: 10.1111/cei.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saldanha-Araujo F, et al. Mesenchymal stromal cells up-regulate CD39 and increase adenosine production to suppress activated T-lymphocytes. Stem Cell Res. 2011;7(1):66–74. doi: 10.1016/j.scr.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Chatterjee D, Tufa DM, Baehre H, Hass R, Schmidt RE, Jacobs R. Natural killer cells acquire CD73 expression upon exposure to mesenchymal stem cells. Blood. 2014;123(4):594–595. doi: 10.1182/blood-2013-09-524827. [DOI] [PubMed] [Google Scholar]
- 38.Yan F, et al. Human dental pulp stem cells regulate allogeneic NK cells’ function via induction of anti-inflammatory purinergic signalling in activated NK cells. Cell Prolif. 2019;52(3):e12595. doi: 10.1111/cpr.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vijayan D, et al. Selective activation of anti-CD73 mechanisms in control of primary tumors and metastases. Oncoimmunology. 2017;6(5):e1312044. doi: 10.1080/2162402X.2017.1312044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu L, et al. Cytokine-mediated disruption of lymphocyte trafficking, hemopoiesis, and induction of lymphopenia, anemia, and thrombocytopenia in anti-CD137-treated mice. J Immunol. 2007;178(7):4194–4213. doi: 10.4049/jimmunol.178.7.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi BK, et al. Peripheral 4-1BB signaling negatively regulates NK cell development through IFN-gamma. J Immunol. 2010;185(3):1404–1411. doi: 10.4049/jimmunol.1000850. [DOI] [PubMed] [Google Scholar]
- 42.Vinay DS, Cha K, Kwon BS. Dual immunoregulatory pathways of 4-1BB signaling. J Mol Med. 2006;84(9):726–736. doi: 10.1007/s00109-006-0072-2. [DOI] [PubMed] [Google Scholar]
- 43.Schneider H, Rudd CE. Diverse mechanisms regulate the surface expression of immunotherapeutic target CTLA-4. Front Immunol. 2014;5:619. doi: 10.3389/fimmu.2014.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smyth LA, et al. CD73 expression on extracellular vesicles derived from CD4+ CD25+ Foxp3+ T cells contributes to their regulatory function. Eur J Immunol. 2013;43(9):2430–2440. doi: 10.1002/eji.201242909. [DOI] [PubMed] [Google Scholar]
- 45.Morello S, et al. Soluble CD73 as biomarker in patients with metastatic melanoma patients treated with nivolumab. J Transl Med. 2017;15(1):244. doi: 10.1186/s12967-017-1348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theodoraki MN, Hoffmann TK, Jackson EK, Whiteside TL. Exosomes in HNSCC plasma as surrogate markers of tumour progression and immune competence. Clin Exp Immunol. 2018;194(1):67–78. doi: 10.1111/cei.13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SH, Kim KS, Fodil-Cornu N, Vidal SM, Biron CA. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J Exp Med. 2009;206(10):2235–2251. doi: 10.1084/jem.20082387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song TL, et al. Oncogenic activation of the STAT3 pathway drives PD-L1 expression in natural killer/T-cell lymphoma. Blood. 2018;132(11):1146–1158. doi: 10.1182/blood-2018-01-829424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cacalano NA. Regulation of natural killer cell function by STAT3. Front Immunol. 2016;7:128. doi: 10.3389/fimmu.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haeussler M, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17(1):148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sutlu T, Nyström S, Gilljam M, Stellan B, Applequist SE, Alici E. Inhibition of intracellular antiviral defense mechanisms augments lentiviral transduction of human natural killer cells: implications for gene therapy. Hum Gene Ther. 2012;23(10):1090–1100. doi: 10.1089/hum.2012.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sachsenmeier KF, et al. Development of a novel ectonucleotidase assay suitable for high-throughput screening. J Biomol Screen. 2012;17(7):993–998. doi: 10.1177/1087057112443987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.