Abstract
Posttranslational modifications (PTMs) are common among proteins that aggregate in neurodegenerative disease, yet how PTMs impact the aggregate conformation and disease progression remains unclear. By engineering knockin mice expressing prion protein (PrP) lacking 2 N-linked glycans (Prnp180Q/196Q), we provide evidence that glycans reduce spongiform degeneration and hinder plaque formation in prion disease. Prnp180Q/196Q mice challenged with 2 subfibrillar, non–plaque-forming prion strains instead developed plaques highly enriched in ADAM10-cleaved PrP and heparan sulfate (HS). Intriguingly, a third strain composed of intact, glycophosphatidylinositol-anchored (GPI-anchored) PrP was relatively unchanged, forming diffuse, HS-deficient deposits in both the Prnp180Q/196Q and WT mice, underscoring the pivotal role of the GPI-anchor in driving the aggregate conformation and disease phenotype. Finally, knockin mice expressing triglycosylated PrP (Prnp187N) challenged with a plaque-forming prion strain showed a phenotype reversal, with a striking disease acceleration and switch from plaques to predominantly diffuse, subfibrillar deposits. Our findings suggest that the dominance of subfibrillar aggregates in prion disease is due to the replication of GPI-anchored prions, with fibrillar plaques forming from poorly glycosylated, GPI-anchorless prions that interact with extracellular HS. These studies provide insight into how PTMs impact PrP interactions with polyanionic cofactors, and highlight PTMs as a major force driving the prion disease phenotype.
Keywords: Infectious disease, Neuroscience
Keywords: Glycobiology, Neurodegeneration, Prions
Introduction
Amyloid plaques accumulate in the brain of patients with Alzheimer’s disease and certain familial prion diseases, and are often associated with a clinical course that progresses for more than 3 years (1–3). In contrast, in sporadic Creutzfeldt-Jakob disease (sCJD), prion aggregates more commonly form diffuse, synaptic, or plaque-like deposits in the brain and symptoms advance with extraordinary rapidity, with a median of 6 months from clinical onset to terminal disease (4–6). Similar to sCJD, in animal prion diseases, such as chronic wasting disease of cervids, prion aggregates frequently form diffuse or punctate deposits and the clinical phase is brief (7–11). Although the histopathology has been extensively characterized, the molecular mechanisms that drive the prion fold into fibrillar plaques or subfibrillar diffuse deposits are unclear.
Prion aggregates accumulate during the conversion of the cellular prion protein, PrPC, into a β-sheet rich multimer known as PrPSc (12, 13). Although the PrPSc structure has not yet been solved, several lines of evidence suggest that PrP posttranslational modifications (PTMs), a glycophosphatidylinositol (GPI) anchor and 2 N-linked glycans (14–18), impact the prion conformation (19–21), species barriers (22, 23), and disease progression (24, 25). Previous studies have demonstrated that PrPSc maintains the sialylated glycans (α2,6-linked) (17, 26) and that sialylation influences prion conversion and intracellular trafficking (27–31), yet how these bulky, anionic glycans modify prion assembly and the disease phenotype remains unresolved (32). Additionally, while cell and mouse models have revealed that unglycosylated PrPC can be converted into PrPSc (24, 33), further study has been hampered by the intracellular retention of unglycosylated PrPC (34, 35), which normally traffics to the cell surface (36, 37).
To address the question of how glycans impact prion protein aggregation and ultimately disease progression in vivo, here we employed 2 new knockin mouse models expressing cell surface PrPC with 0 or 3 glycans and 5 complementary approaches. First, using 4 conformationally distinct prion strains, we show that unglycosylated PrP favors extracellular plaque formation and induces severe spongiform degeneration in the brain. Second, using an antibody specific for shed PrP (38), we show that the plaques are highly enriched in ADAM10-cleaved PrP. Third, using mass spectrometry and immunohistochemical labelling, we show that these unglycosylated, ADAM10-cleaved prions tightly bind heparan sulfate (HS) and HS colocalizes to plaques. Fourth, using heparin affinity chromatography, we directly show that unglycosylated PrP has the highest affinity to heparin and that each additional glycan decreases the binding affinity. Finally, using knockin mice that express PrP having a third N-linked glycan, we show a complete reversal of a typical plaque-forming disease phenotype, as mice instead developed primarily subfibrillar aggregates that lack HS binding and showed a rapid disease progression. Collectively, these studies support the conclusion that PrP glycans can impede fibril formation, potentially through reduced binding of shed, glycosylated PrP to extracellular HS, and suggest that the PTMs on PrP contribute to the profoundly rapid clinical progression observed in the majority of prion diseases.
Results
Prnp180Q/196Q mice generated using the CRISPR-Cas system show normal PrPC expression and trafficking.
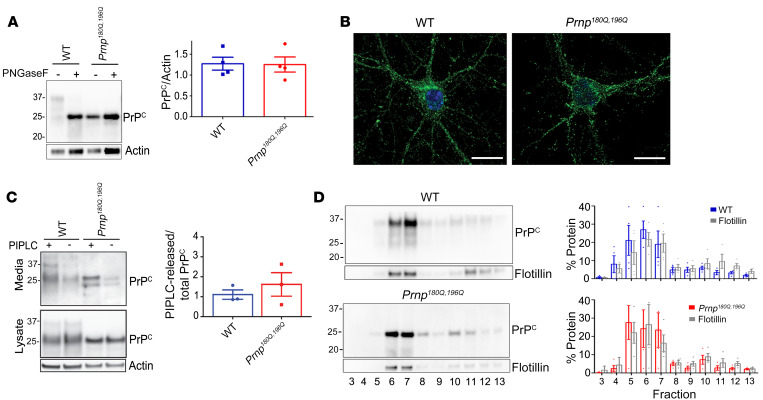
Mice expressing unglycosylated PrPC were generated using a single guide RNA to create 2 point mutations in the endogenous Prnp locus, corresponding to asparagine to glutamine substitutions at positions 180 and 196 (mouse PrP numbering) thereby altering the N-glycosylation sequons. Six founder lines were generated, 2 were sequenced, and 1 (line 191) was selected and bred to homozygosity on a C57BL/6 background. Prnp180Q/196Q mice developed normally with no clinical signs or histologic lesions observed in the brain (n = 12 mice, 150–639 days of age) (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI131564DS1). PrPC was expressed in the brain at levels similar to WT mice, and the unglycosylated forms showed an equivalent electrophoretic mobility (Figure 1A). To assess the PrPC distribution in neurons, cortical neurons were isolated from Prnp180Q/196Q and WT mice, immunolabelled for PrP, and evaluated by confocal microscopy, revealing an indistinguishable PrP distribution throughout the neuronal cell body and neurites (Figure 1B).
Figure 1. PrP180Q/196Q traffics similarly to WT PrPC in primary neurons and in mice.
(A) Representative Western blot of PNGase-F–treated brain extracts from age-matched Prnp180Q/196Q and WT mice reveal similar PrPC expression levels (quantified in right panel) (100–250 day old mice); n = 4/group. (B) PrP immunocytochemistry shows that unglycosylated PrP180Q/196Q traffics to neuronal processes in primary cortical neurons, as does PrP in WT neurons; n = 3 experiments. Scale bars: 10 μm. (C) Representative Western blots of phospholipase C–cleaved (PIPLC-cleaved) PrP180Q/196Q and WT PrP from the surface of cortical neurons show that surface PrPC levels are similar (media); n = 3 experiments. The additional band in the media (~23 kDa) may be a cleaved form of PrP. (D) PrP180Q/196Q and WT PrPC, together with flotillin, localize to detergent-resistant membranes in the brain; n = 3/group. Unpaired, 2-tailed Student’s t test, no significant differences (A and C).
PrPC is GPI-anchored in the outer leaflet of the cell membrane, yet whether unglycosylated PrP traffics to the cell membrane has been controversial and may depend on the specific amino acids substituted into the N-linked glycan consensus sequence, Asn-X-Ser/Thr (39). Asparagine-to-glutamine substitutions were chosen here due to their structural similarity, differing only by a single methylene. To quantify surface expression in primary cortical neurons isolated from WT and Prnp180Q/196Q mice, phosphatidylinositol-specific phospholipase C (PIPLC) hydrolysis was performed on live primary neurons to cleave the GPI-anchor. Similar levels of cleaved PrP were detected in the media, indicating that unglycosylated PrP180Q/196Q localized to the cell surface (Figure 1C). To further confirm that PrP180Q/196Q traffics to the cell surface, PrP-deficient RK13 cells were transfected with PrnpWT, Prnp180Q/196Q, or Prnp180A/196A, a mutant reported to show impaired PrP trafficking (40). Again PrP180Q/196Q was released into the media by PIPLC similar to WT PrPC, whereas only approximately 40% of PrP180A/196A was released, supporting that PrP with asparagine-to-glutamine substitutions traffics to the cell surface (Supplemental Figure 1B). To assess whether unglycosylated PrP localizes to lipid rafts in vivo, detergent-resistant membranes from WT and Prnp180Q/196Q brains were collected following equilibrium density gradient centrifugation, and revealed WT and unglycosylated PrPC localized in lipid raft fractions together with flotillin (Figure 1D), further confirming surface localization of PrP180Q/196Q. Thus by multiple measures, PrP180Q/196Q traffics to the plasma membrane.
Aged Prnp180Q/196Q mice show no evidence of spontaneous prion disease.
To determine whether unglycosylated PrPC spontaneously aggregates, we histologically and biochemically investigated 12 aged Prnp180Q/196Q mice. Brain was immunolabelled for PrP and showed no evidence of prion aggregation (Supplemental Figure 1A). To test for the presence of proteinase K–resistant (PK-resistant) PrP, WT and PrP180Q/196Q brain extracts were treated with low PK concentrations, which revealed no differences in PrPC digestion (Supplemental Figure 1C). Last, to test for insoluble PrP, brain homogenates were ultracentrifuged, which revealed that PrP180Q/196Q was highly soluble, similar to WT PrPC (approximately 92% and 95% soluble, respectively), and differed from a prion-infected brain (approximately 22% soluble) (Supplemental Figure 1D). Thus, Prnp180Q/196Q mice show no evidence of aggregated PrP, consistent with their normal lifespan and lack of neurologic disease.
Prnp180Q/196Q mice infected with 4 prion strains develop plaque and plaque-like deposits.
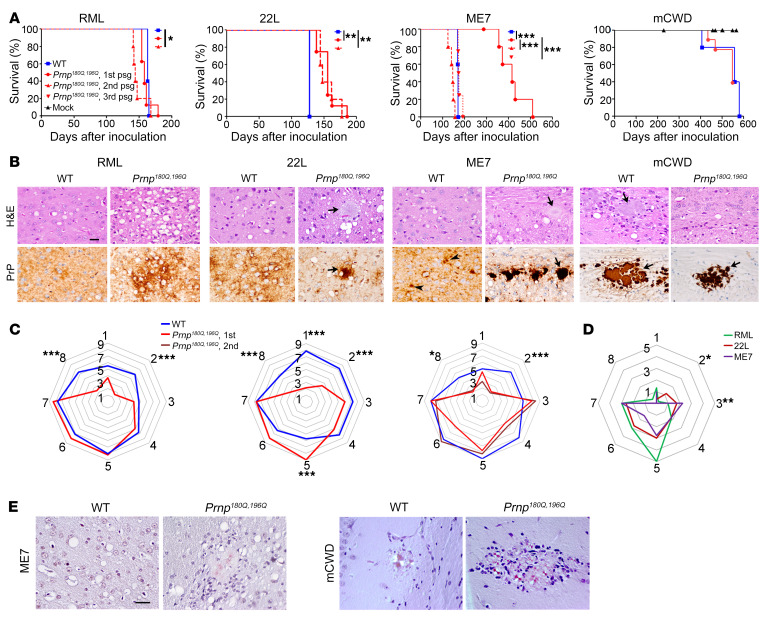
We next assessed the susceptibility of the Prnp180Q/196Q mice to prion infection. Prnp180Q/196Q and WT mice were inoculated intracerebrally with 4 mouse-adapted prion strains known as RML, 22L, ME7, and mCWD. RML, 22L, and ME7 are subfibrillar strains derived from sheep scrapie and show no fibrils ultrastructurally in situ (41–43), whereas mCWD is a mouse-adapted fibrillar strain derived from deer chronic wasting disease that is characterized by dense plaques composed of bundles of long, extracellular fibrils (44). All RML- and 22L-inoculated Prnp180Q/196Q mice developed prion disease. Following inoculation with RML prions, WT and Prnp180Q/196Q mice showed no significant difference in survival times on first passage (WT: 164 ± 1 days after inoculation [dpi]; Prnp180Q/196Q: 160 ± 3 dpi [mean ± SEM]), and a modest decrease in survival on second passage in Prnp180Q/196Q mice (148 ± 5 dpi) (Figure 2A). Following inoculation with 22L prions, Prnp180Q/196Q mice showed a prolonged survival (WT: 127 ± 0 dpi; Prnp180Q/196Q: 155 ± 5 dpi), which was unchanged on second passage in Prnp180Q/196Q mice (155 ± 6 dpi) (Figure 2A). In contrast, there was a significant transmission barrier following inoculation with ME7 prions, as there was an incomplete attack rate (6/7 mice, 86%) and a highly variable and significantly prolonged incubation period (WT: 169 ± 1 dpi; Prnp180Q/196Q: 389 ± 27 dpi). Yet by the second passage, the attack rate was 100% and the incubation period decreased by 60% (142 ± 5 dpi), and by third passage the incubation period was similar to ME7 in WT mice (177 ± 5 dpi), suggestive of strain adaptation. Similarly, the fibrillar mCWD prions led to an incomplete attack rate on first passage (5/9, 56%), suggestive of a transmission barrier, although the incubation period in WT mice was longer (WT: 569 ± 7 dpi; Prnp180Q/196Q: 508 ± 24 dpi) (Figure 2A). Notably, most Prnp180Q/196Q mice positive for mCWD prions (4 of 5 positive mice)had low PrPSc levels in brain and were euthanized due to age (n = 3) or concurrent disease (n = 1), thus were not likely at terminal disease stages.
Figure 2. Survival and histopathologic lesions in prion-infected Prnp180Q/196Q and WT mice.
(A) For mCWD-infected mice, there was no difference in survival times. In contrast, 22L- and ME7-infected Prnp180Q/196Q mice showed prolonged survival compared with WT mice. By the second and third passage, ME7-infected Prnp180Q/196Q mice showed a 100% attack rate and similar survival times as the WT mice. (B) Brain sections stained with H&E or immunolabelled for PrP revealed severe spongiform degeneration and plaque-like deposits (RML) or plaques (22L, ME7, and mCWD, arrows) in prion-infected Prnp180Q/196Q mice. Less spongiform degeneration was noted in the mCWD-infected mice. Scale bar: 50 μm. (C) Lesion scores of spongiform change, gliosis, and PrPSc in 8 brain regions differed significantly in RML-, 22L-, and ME7-infected Prnp180Q/196Q mice compared with WT mice. Cerebellum was consistently less severely affected in the Prnp180Q/196Q mice. 1-dorsal medulla, 2-cerebellum, 3-hypothalamus, 4-medial thalamus, 5-hippocampus, 6-septum, 7-cerebral cortex, and 8-cerebral peduncle. (D) Right panel shows that PrPSc levels differed in the cerebellum (RML versus 22L; 0.2 ± 0.2 versus 1.2 ± 0.2, respectively) and hypothalamus (22L versus ME7; 1.8 ± 0.5 versus 2.3 ± 0.7, respectively) of the Prnp180Q/196Q mice. (E) ME7 and mCWD plaques bind Congo red in Prnp180Q/196Q brains. Scale bar: 50 μm. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; 1-way ANOVA with Tukey’s test (A), 2-way ANOVA with Bonferroni’s post hoc test (C and D). RML: n = 5–8 mice per group; ME7: n = 5–7 mice per group; 22L: n = 5–8 mice per group, mCWD: n = 5–9 mice per group, mock: n = 4–8 mice per group.
A comparison of the histopathologic lesions in RML-, 22L-, and ME7-infected WT and Prnp180Q/196Q mice revealed intriguing differences in spongiform degeneration, gliosis, and PrPSc distribution (Figure 2, B and C). First, the spongiform degeneration was consistently scored as more severe for all 3 strains in the Prnp180Q/196Q mice as compared with the WT mice (Figure 2B). Additionally, in the Prnp180Q/196Q mice infected with ME7 prions, the hippocampus was consistently markedly atrophied due to severe neuronal loss, with complete loss of the CA1 pyramidal neurons and numerous gemistocytic astrocytes, which persisted upon second passage, whereas WT mice showed a moderate loss of hippocampal neurons (Supplemental Figure 2A). Finally, the cerebellum lacked PrPSc in all infected Prnp180Q/196Q mice, a striking difference from WT mice in which all 3 strains were present in the cerebellum (Figure 2, C and D, region 2). This was not due to a lack of PrPC expression, as PrP180Q/196Q was expressed in the cerebellum at levels similar to WT PrP (Supplemental Figure 2B).
PrPSc morphology also differed from WT mice in that most Prnp180Q/196Q brains showed some plaque-like deposits or dense parenchymal plaques. In WT mice, RML and 22L prions typically form diffuse aggregates, ME7 prions form diffuse and small plaque-like aggregates (15–30 μm), and mCWD prions form large plaques (>100 μm) (Figure 2B). In the Prnp180Q/196Q mouse brain, the RML prion deposits remained mostly diffuse and occasionally formed plaque-like clusters (Figure 2B). In contrast, 22L and ME7 prions formed both diffuse and significantly larger plaques (up to 100 μm for ME7) that were congophilic (Figure 2, B and E; Supplemental Figure 2, C and D). There were also numerous florid ME7 plaques that were surrounded by small vacuoles, similar to those described in variant CJD-infected human brains (45) (Supplemental Figure 2E). Notably, large mCWD plaques were morphologically identical in the WT and Prnp180Q/196Q mice (Figure 2B). Thus the Prnp180Q/196Q mice challenged with the 3 subfibrillar prion strains shared certain newly acquired disease features, including an increase in plaques and plaque-like structures, more severe cortical spongiosis, and a notable lack of prions in the cerebellum.
PrPSc biochemical properties were altered in Prnp180Q/196Q mice.
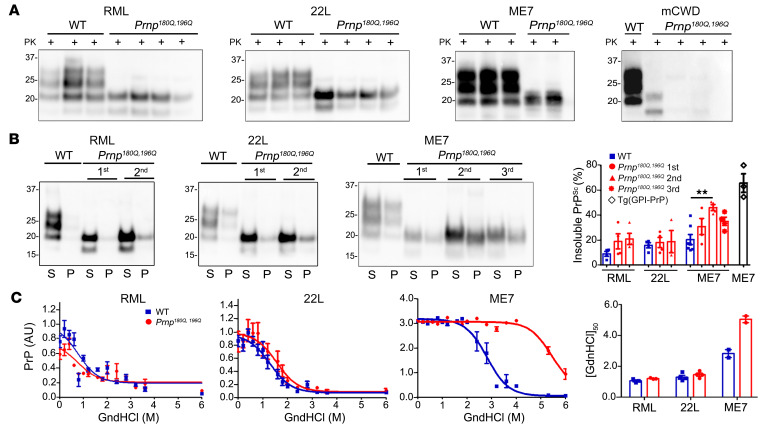
To assess the differences in the biochemical properties of the unglycosylated prions, we first assessed the presence of PK-resistant PrPSc in all Prnp180Q/196Q mice. All RML- and 22L-challenged and some ME7- and mCWD-challenged mice showed PK-resistant PrPSc (Figure 3A). The solubility and stability of RML, 22L, and ME7 PrPSc were assessed (mCWD prion levels were too low to assess). Notably, the insoluble PrP levels were similar for RML- and 22L-infected WT and Prnp180Q/196Q mice, yet for ME7, we found that further passage in the Prnp180Q/196Q mice was associated with significantly increased levels of insoluble PrPSc (Figure 3B, second passage).
Figure 3. Comparison of PK resistance, solubility, and stability of RML, 22L, ME7, and mCWD prions in Prnp180Q/196Q and WT brain samples.
(A) Western blots of brain homogenates reveal PK-resistant unglycosylated PrPSc in RML-, 22L-, ME7-, and mCWD-infected Prnp180Q/196Q mice. (B) Western blots of soluble (S, supernatant) and insoluble (P, pellet) PrPSc show similar levels of insoluble PrPSc in Prnp180Q/196Q and WT brains on first passage, but significantly higher levels in ME7-infected Prnp180Q/196Q brains on further passage. The GPI-ME7 was included to compare samples with a known highly fibrillar prion; n = 2 (ME7, first passage only) and n = 3–7/strain (all other strains, including ME7, second and third passage). (C) Representative PrPSc stability curves from single mouse brain samples run in triplicate. The right graph shows the guanidine hydrochloride concentration at which half the PrPSc remains ([GndHCl]2) and reveals no significant differences in the stability of RML or 22L, but a significantly more stable ME7 prion after passage in Prnp180Q/196Q mice. Plotted (right graph) are the mean and SEM for n = 2 (ME7-infected Prnp180Q/196Q), n = 3 (WT), or n = 4 (RML- and 22L-infected Prnp180Q/196Q) mice per strain, each run in triplicate. **P ≤ 0.01, 1-way ANOVA with Tukey’s test (B).
To next measure the aggregate stability, aliquots of brain homogenate were first denatured with guanidine hydrochloride (GndHCl) ranging from 0 to 6 M, and then diluted and digested with PK. The PrP was measured by ELISA to determine the [GndHCl]1/2 concentration at which half the PrPSc remained. While there were no stability differences for RML and 22L prions in the WT and Prnp180Q/196Q brains, ME7 prions were more stable in the Prnp180Q/196Q brain (Figure 3C). Therefore, the aggregate stability of the 3 unglycosylated prions correlated with plaque formation, with ME7 showing the highest stability.
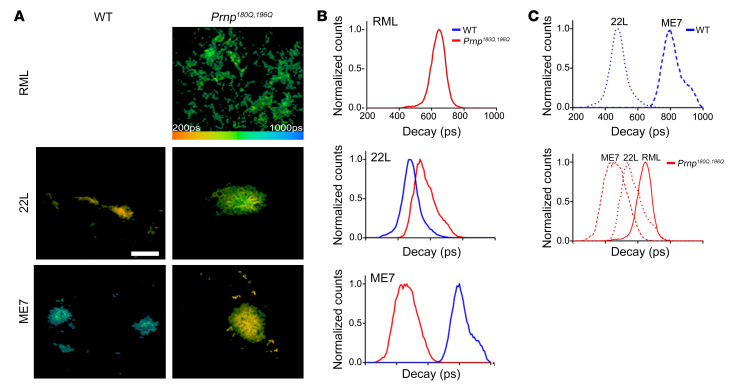
To further probe the conformation of unglycosylated PrPSc, we next applied a conformationally sensitive amyloid probe, heptameric formic thiophene acetic acid (h-FTAA), to brain cryosections and performed fluorescence lifetime imaging (FLIM) to measure the decay of emitted light from prion-bound h-FTAA. The decay of light depends on the immediate chemical environment of the h-FTAA, and thus is useful to distinguish differential binding modes of the probe to distinct aggregates. Variations in the decay times have previously been observed for h-FTAA bound to different aggregated morphotypes of amyloid-β (46), as well as prion deposits associated with distinct prion strains (47). Here we found that for every strain compared, the FLIM of prion-bound h-FTAA in the Prnp180Q/196Q mice differed from WT mice. Whereas h-FTAA does not bind RML in WT mice, h-FTAA binds RML in the Prnp180Q/196Q mice (Figure 4). For 22L and ME7 prions, the lifetime distribution was shifted in the Prnp180Q/196Q mice as compared with the WT mice (Figure 4). Since theoretical calculations and solid-state NMR experiments have verified that the binding mode and the optical properties of the oligothiophene are determined by regularly spaced surface charge patterns and highly accessible grooves on the fibril surface (48–50), the observed differences in life time distribution are most likely due to PrP structural variations in Prnp180Q/196Q mice.
Figure 4. Fluorescence lifetime (FLIM) decay of h-FTAA bound to prion aggregates.
(A) Fluorescence lifetime images and (B and C) intensity-weighted mean lifetime (ti) distributions of h-FTAA–stained RML, 22L, and ME7 prion plaques in Prnp180Q/196Q (red) or WT (blue) brain show that the FLIM decay curves in Prnp180Q/196Q brain differ from those in WT brain and reveal 3 different aggregate conformers in prion-infected Prnp180Q/196Q mice. The decay data were collected with the excitation wavelength set at 490 nm. The color bar represents lifetimes from 200 ps (orange) to 1000 ps (blue) and the images are color coded according to the representative lifetime. Decay curves were collected from 5–10 prion deposits from a minimum of n = 3 mice/strain. Scale bar: 20 µm.
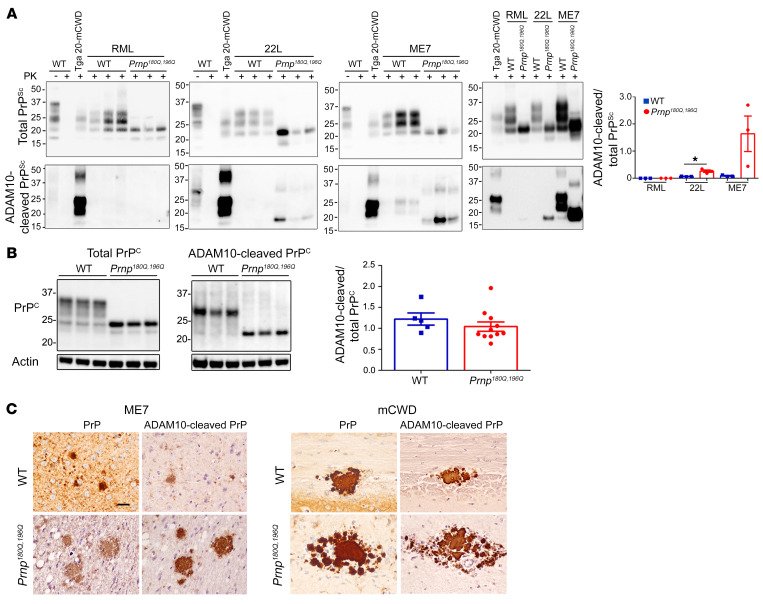
ME7 and mCWD prions in Prnp180Q/196Q mice largely consist of ADAM10-cleaved PrP.
Unglycosylated PrP showed an increased propensity for fibril formation in a strain-dependent manner. How does the lack of PrP glycans promote fibril and plaque formation? Previously we found that extracellular prion plaques (mCWD strain) were largely, if not entirely, composed of ADAM10-cleaved, GPI-anchorless PrP (Figure 5A) (51), which comprises 10%–15% of PrPC in the brain (52, 53).
Figure 5. Increased ADAM10-cleaved PrPSc in prion-infected Prnp180Q/196Q mice.
(A) Western blots reveal 22L- and ME7-infected Prnp180Q/196Q mice harbor higher levels of ADAM10-cleaved PrPSc in the brain compared with WT mice; n = 3/group. Note that the PK-digested ADAM10-cleaved PrP runs at a lower molecular weight due to lack of the GPI-anchor and terminal 3 amino acids. *P ≤ 0.05, unpaired, 2-tailed Student’s t test. (B) Western blots show that uninfected Prnp180Q/196Q and WT mice have similar levels of ADAM10-cleaved/total PrPC in the brain; n = 5 WT and 11 Prnp180Q/196Q mice. (C) Brain sections immunolabeled for PrP with SAF84 (labelled PrP) or with sPrPG228 antibody (labelled ADAM10-cleaved PrP) reveal ADAM10-cleaved PrP localizes to plaque-like and plaque deposits in ME7-infected Prnp180Q/196Q, mCWD-infected Prnp180Q/196Q, and WT brain sections. Note that the diffuse aggregates are not labelled by the sPrPG228 antibody. Cortex (ME7-infected WT), thalamus (ME7-infected Prnp180Q/196Q), hippocampus (mCWD-infected Prnp180Q/196Q), and corpus callosum (WT-infected mCWD) are shown. Scale bar: 50 μm.
The plaques and plaque-like structures prompted us to investigate the level of ADAM10-cleaved PrPSc in Prnp180Q/196Q mice. Using antibodies targeting ADAM10-cleaved PrP (sPrPG228) (38) or total PrP (POM1) (54), we found that ADAM10-cleaved PrP levels were very low for RML, yet substantially higher for 22L and ME7, showing a 4-fold and an 18-fold increase as compared with WT mice (Figure 5A). The increase in ADAM10-cleaved PrPSc was not simply due to an increase in cleaved PrPC in the Prnp180Q/196Q mice, as the levels were similar to those in the WT mice (Figure 5B).
To next localize the ADAM10-cleaved PrP in the 22L- and ME7-infected brain, we used the sPrPG228 and SAF84 antibodies specifically against ADAM10-cleaved PrP and the PrP epitope at residues 166-172 (55, 56), respectively. Remarkably, the ADAM10-cleaved PrP localized to plaques, but not diffuse deposits (Figure 5C and Supplemental Figure 3A). Thus, the relative levels of ADAM10-cleaved PrPSc in the Prnp180Q/196Q mice correlated with those in WT mice, with ME7 exceeding 22L and RML, yet the absolute levels of cleaved PrPSc were higher in the Prnp180Q/196Q mice and were localized to extracellular plaques in situ. For mCWD, the prion plaques were composed of ADAM10-cleaved PrP in both the WT and Prnp180Q/196Q mice (Figure 5C).
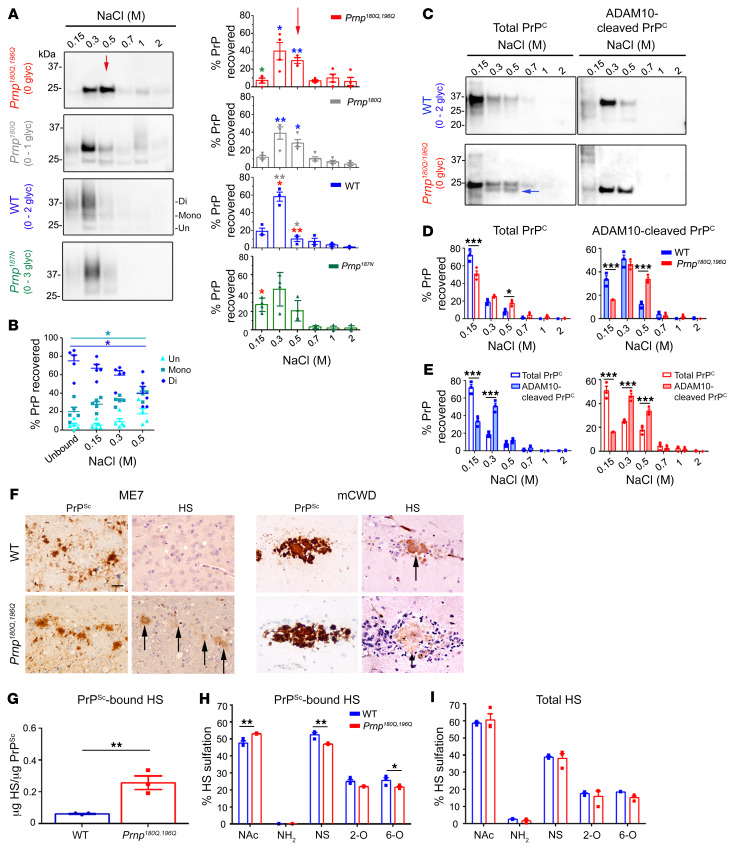
PrP glycans decrease the heparin-binding affinity.
Why does ADAM10-cleaved, unglycosylated PrP have a higher tendency to form fibrils than glycosylated PrP? We previously found high levels of HS bound to ADAM10-cleaved PrPSc having 0 to 1 glycan (51). We reasoned that unglycosylated PrP may bind with higher affinity to extracellular HS due to a lack in electrostatic repulsion between the anionic glycans and HS. To test this hypothesis, we assessed how PrP glycans impact the binding affinity to heparin, a highly sulfated surrogate of HS, using affinity chromatography and PrP with 0, 1, 2, or 3 glycans from transfected RK13 cells. PrPC was cleaved from the cell surface by PIPLC hydrolysis and applied to the heparin sepharose column. While PrPC with 2 to 3 glycans showed a low affinity to heparin, unglycosylated PrPC showed a high affinity to heparin; the affinity progressively decreased with each additional glycan (Figure 6A). Notably, even within the glycoform mixtures of WT PrPC (0–2 glycans), the unglycosylated PrPC showed the highest affinity to heparin (Figure 6B).
Figure 6. PrP glycans hinder binding to HS.
(A) Immunoblots of heparin affinity chromatography experiments assessing variably glycosylated PrP. Relative levels of PrP from each elution are shown in the graphs. Differences in the binding affinity are most notable in the 0.5 M NaCl elution (red arrow). Asterisk color indicates the mutants with significant differences. The triglycosylated PrP isoform level was low in RK13 cells; n = 3–4 experiments. (B) Among the WT PrP glycoforms, diglycosylated PrP has a lower heparin affinity than mono- or unglycosylated PrP (unbound is PrP in the flow-through); n = 5 experiments, 4 also included in A. (C) Affinity chromatography of the soluble brain fraction reveals that total and ADAM10-cleaved PrP180Q/196Q have significantly higher heparin affinity than the corresponding WT PrP. PrP180Q/196Q shows a second band (blue arrow) that corresponds to ADAM10-cleaved PrPC. (D) Quantification of PrP shown in C; n = 3/strain. (E) ADAM10-cleaved PrP has a higher heparin-binding affinity than total PrP for both WT PrP and PrP180Q/196Q. (F) Immunolabelling reveals HS colocalizes to ME7 plaques in the Prnp180Q/196Q brain only and to mCWD plaques in both the WT and Prnp180Q/196Q brain; n = 4/strain. Scale bar: 50 μm. (G) LC-MS reveals approximately 6-fold more HS bound to unglycosylated ME7 PrPSc than to highly glycosylated ME7 PrPSc (WT); n = 3/strain. (H) Composition analysis of HS bound to purified PrPSc (ME7) reveals less N-sulfated (NS) and 6-O sulfated (6-O) HS bound to unglycosylated (Prnp180Q/196Q) as compared with glycosylated (WT) PrPSc; n = 3/group. (I) The overall HS composition in ME7-infected Prnp180Q/196 and WT whole-brain lysates are similar; n = 3/group. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; 2-way ANOVA with Bonferroni’s post hoc test (A, D, E, and H). *P < 0.05; 1-way ANOVA with Tukey’s test (B). **P ≤ 0.01, unpaired, 2-tailed Student’s t test (G).
To determine whether brain-derived unglycosylated PrPC also shows a higher affinity to heparin, we next tested the heparin-binding affinity of PrPC from age-matched WT and Prnp180Q/196Q mouse brain homogenates (Figure 6C). Again we found that unglycosylated PrPC had a higher heparin-binding affinity than glycosylated WT PrPC (Figure 6, C and D), which also held true for their ADAM10-cleaved counterparts. Interestingly, for both genotypes, ADAM10-cleaved PrP showed a higher heparin-binding affinity than full-length PrP (Figure 6E), suggesting that the GPI anchor with the 3 C-terminal amino acids markedly reduces heparin binding. Collectively, these data suggest that N-linked glycans and the GPI-anchor hinder PrPC binding to HS.
Unglycosylated, ADAM10-cleaved PrP binds HS in vivo.
Sulfated glycosaminoglycans (GAGs) promote fibril assembly in vitro (57, 58) and the sulfated GAG, HS, codeposits with amyloid beta and prion plaques in vivo (59, 60). However, whether HS preferentially binds to unglycosylated prions is unknown. To determine whether the 22L, ME7, and mCWD plaques harbor HS in the Prnp180Q/196Q mice, we immunolabelled prion-infected brain sections for HS using the 10E4 antibody, which recognizes an N-sulfated glucosamine residue and does not react with hyaluronan, chondroitin sulfate, or dermatan sulfate. We found that HS clearly localized to prion plaques in Prnp180Q/196Q mice, as compared with only a low level of diffuse HS stain in RML-, 22L-, and ME7-infected WT mice (Figure 6F, Supplemental Figure 3B) and RML-infected Prnp180Q/196Q mice (Supplemental Figure 3B). Pretreating brain sections with heparin lyases abolished the HS immunolabelling (Supplemental Figure 3C), supporting specificity of the antibody binding.
Mass spectrometry was next used to quantify HS bound to ME7 prions from Prnp180Q/196Q and WT brain. ME7 was selected due to the abundance of plaques in the Prnp180Q/196Q mice. PrPSc was purified by solubilization and multiple rounds of ultracentrifugation (61), levels were quantified against a recombinant PrP standard, and the sample was denatured with sodium hydroxide. HS chains were then depolymerized with a cocktail of heparin lyases, the resulting disaccharides were [12C6] aniline-tagged, and disaccharides were identified by liquid chromatography-mass spectrometry (LC-MS) using [13C6] aniline-tagged disaccharide standards (62, 63). The HS relative to PrP level was calculated for each sample (HS/PrP ratio [μg/μg]). Consistent with the HS immunostaining results, we found significantly higher levels of HS bound to unglycosylated PrPSc compared with WT PrPSc (Figure 6G). In addition, the composition of HS bound to ME7 in the WT versus Prnp180Q/196Q brain differed, as HS was generally less sulfated in the Prnp180Q/196Q brain, with lower levels of N-sulfated and 6-O sulfated HS (Figure 6H). Interestingly, the HS composition in the whole brain lysates from prion-infected WT and Prnp180Q/196Q mice was similar. This finding suggests that the observed composition differences in PrPSc-bound HS were not due to overall differences in HS biosynthesis, but were instead due to PrPSc selectively binding to specific HS molecules (Figure 6I).
A third glycan on PrP reverses the prion disease phenotype.
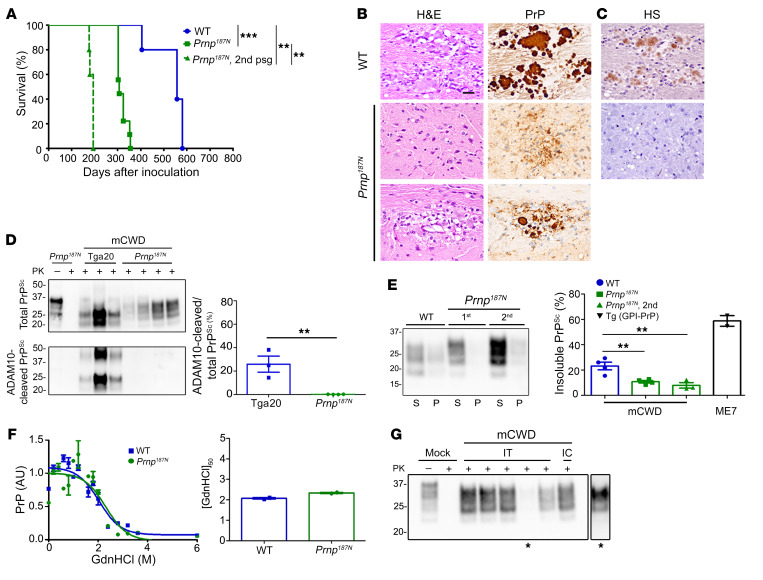
We reasoned that since unglycosylated, ADAM10-cleaved PrP binds HS and favors plaque formation, the addition of a third glycan may correspondingly decrease plaque formation. To directly test this hypothesis, we developed a new knockin mouse model having a third glycan at position 187 of PrP through a single amino acid mutation (T187N) (mouse numbering). We confirmed the addition of the third glycan and characterized the PrP expression and cell membrane localization in Prnp187N mice (Supplemental Figure 4, A and B), which did not show clinical signs or spontaneous disease development. We next challenged mice with a strain known to form long fibrils, mCWD (44). Strikingly, despite an amino acid substitution expected to lead to a transmission barrier and prolong survival, the Prnp187N mice developed a profoundly accelerated disease (326 ± 9 dpi), which was approximately 60% of the incubation period of mCWD in WT mice (Figure 7A). Remarkably, on second passage, the incubation period decreased even further to 194 ± 7 dpi, approximately 35% of the WT incubation period (Figure 7A). This disease acceleration was likely due to the additional glycans and not to a difference in the amino acid side chain, as PrPC-PrPSc sequence differences typically prolong survival.
Figure 7. Knockin mice expressing triglycosylated PrP show a reversal in the clinical, histologic, and biochemical phenotype when infected with the plaque-forming prion strain, mCWD.
(A) Survival curves of Prnp187N mice inoculated with mCWD prions show a profound decrease in the survival time as compared with WT mice; n = 5–9/group. (B) The spongiform degeneration is similar in Prnp187N mice as compared with WT mice; however, PrP deposits switched from the typical large, dense plaques in WT mice to primarily diffuse aggregates, with occasional rare small plaques in the corpus callosum (lower panel). (C) HS immunolabelling of the diffuse aggregates or plaque-like structures was not evident in the Prnp187N brain. (D) Western blot reveals that the PrPSc in the Prnp187N brain is no longer composed of ADAM10-cleaved PrPSc that predominated in the original mCWD strain in tga20 mice (overexpressed WT mouse PrP); n = 3–5/mouse strain. (E) Western blot of soluble versus insoluble PK-resistant PrPSc reveals a decrease in insoluble PrPSc in the Prnp187N mice; n = 4–7 WT or Prnp187N mice and n = 2 Tg (GPI-PrP) positive control brain samples. (F) Representative PrPSc stability assay and quantification show that the resistance to unfolding in chaotropes is similar for PrPSc aggregates in the mCWD-infected Prnp187N and WT brain; n = 2/group. (G) Western blot of Prnp187N mice inoculated intratongue (IT) or intracerebrally (IC) with mCWD-passaged Prnp187N brain homogenate shows that all clinically terminal mice (5/5 mice) harbored PrPSc in the brain. *A longer exposure of one sample is shown on the right. **P ≤ 0.01, ***P ≤ 0.001; 1-way ANOVA with Tukey’s test (A and E), unpaired, 2-tailed Student’s t test (D). Scale bar: 100 μm (B and C).
To investigate whether the Prnp187N mice harbored the large plaques characteristic of mCWD in WT mice, we assessed the histopathological phenotype. We found a notable lack of large plaques in the Prnp187N mice, and instead only diffuse aggregates and rare small plaques in the corpus callosum (Figure 7B). In addition to the rapidly progressive disease and small aggregates, the aggregates no longer bound HS (Figure 7C). Moreover, the prion aggregates were no longer composed of ADAM10-cleaved PrP (Figure 7D), were significantly more soluble (Figure 7E), and showed a similar stability in chaotropes (Figure 7F).
We previously found that mCWD prions do not spread from the tongue into the CNS, thus were nonneuroinvasive, whereas the subfibrillar strains were neuroinvasive (64). To test for neuroinvasion of the new subfibrillar mCWD prions in the Prnp187N mice, we inoculated these prions into the tongue. Notably, all Prnp187N mice developed prion disease (5/5 mice, 312 ± 10 dpi), indicating that the prions had switched from a nonneuroinvasive to a highly neuroinvasive prion (Figure 7G). Therefore, by multiple histological and biochemical measures, the mCWD prion had undergone a profound transformation to a GPI-anchored, non–HS binding, highly soluble, neuroinvasive prion causing a rapidly progressive disease. Collectively, these data support the conclusion that PrP glycans are linked to a decreased tendency for plaque formation.
Discussion
In contrast to Alzheimer’s disease, amyloid plaques in the brain are rare in prion disease and are often associated with longer disease durations, as observed in Gerstmann–Sträussler–Scheinker disease, for example (3). Prions more commonly form diffuse, synaptic, or punctate aggregates associated with an extraordinarily rapid disease progression, and the molecular determinants that underlie these disease differences are unclear. Our work here indicates that the PrP glycans and GPI-anchor, together with HS, orchestrate major components of the disease phenotype in prion disease. The absence of glycans was linked to the formation of parenchymal plaques, particularly for prions lacking a GPI-anchor. Therefore, our results support a model in which the PrP posttranslational state markedly influences the fate of a prion aggregate, with cleaved, unglycosylated prions efficiently binding to extracellular HS to form parenchymal plaques.
The molecular mechanisms that underlie extracellular fibril formation in prion disease have not been previously identified and tested. Our approach to determining these mechanisms was prompted by studies of knockin mice expressing unglycosylated PrP, known as G3 mice, which revealed that glycosylation is not essential for prion conversion or transmission of infectivity (21, 24). G3 mice also showed a tendency to form thioflavin-positive plaques, suggestive of fibrillar prions (24). Similar to the G3 mice, transgenic mice expressing GPI-anchorless PrPC also develop fibrillar, thioflavin-positive prion aggregates, but fibrils accumulate perivascularly (19). Our use of prion-infected knockin mice expressing unglycosylated and triglycosylated PrP, together with WT mice, also reveal a correlation between a lack of PTMs and fibrillar prion formation. Using recently developed antibodies that recognize ADAM10-cleaved PrP (38), we now identify ADAM10-cleaved PrP as a major component of the unglycosylated PrPSc in plaques. Thus, prions with the capacity to replicate ADAM10-cleaved PrPC, such as ME7, preferentially recruit unglycosylated, cleaved PrPC and form large plaques. In contrast, largely GPI-anchored prions, such as RML, recruit unglycosylated, GPI-anchored PrPC and show a minimally altered disease phenotype. Collectively, these findings suggest that the presence or absence of glycans is relevant for prion conformers that can convert ADAM10-cleaved PrPC, in which case the unglycosylated PrPC is recruited by cleaved, GPI-anchorless PrPSc.
Glycans stabilize the prion protein structure (65, 66) and impede prion conversion and fibril formation in vitro (66–68), potentially through modifying or blocking PrPC/PrPSc recognition domains (69). The diverse glycans on PrP are bi-, tri-, and tetra-antennary and are highly sialated (14, 17, 70, 71), with high sialation levels negatively impacting the prion replication rate (27). In the GPI-anchored prions RML and 22L, we found that the glycosylation state did not seem to markedly affect prion conversion, as the incubation period was only modestly affected. Additionally, for RML prions, the aggregate morphology, aggregate stability, and solubility were remarkably similar in the unglycosylated state. For 22L, however, the prion aggregate morphology was subtly altered, as PrPSc recruited ADAM10-cleaved PrPC and formed some plaques. Consistent with a morphological change, alterations in the h-FTAA binding environment also suggested differences in the structure of unglycosylated PrPSc, in agreement with reports that suggest the glycans may affect the tertiary and quaternary structure, but do not significantly affect the secondary structure (72). Thus, in contrast with in vitro reports (67, 68), our data suggest that the glycans do not impede conversion in vivo, as switching from glycosylated to unglycosylated PrPC did not accelerate prion conversion. Moreover, triglycosylated PrP was efficiently converted and led to a rapidly progressive prion disease.
In contrast to RML, ME7 prions were markedly altered by the glycosylation state, as unglycosylated ME7 PrPSc formed large extracellular plaques, recruited ADAM10-cleaved PrPC, was more stable in chaotropes, had more insoluble PrPSc, and showed an altered h-FTAA decay profile, suggestive of an altered structure due to either adaptation or a switch in the dominant PrPSc structure. Why did unglycosylated ME7 prion aggregates assemble as highly stable, large parenchymal plaques? HS serves as an extracellular scaffold (73–75) and is widely recognized to bind prion aggregates, both in vitro and in vivo (76–79). In accordance with these observations, we found that HS localizes to parenchymal plaques, but not to diffuse aggregates within the same brain. Additionally, mass spectrometry quantification revealed 4 times more HS bound to unglycosylated versus glycosylated ME7 prions. This inverse correlation of PrP glycosylation state with HS binding and plaque formation may be explained by the effect of PrP glycans on HS affinity, potentially decreased by electrostatic repulsion between the anionic glycans of PrP and HS. Our experiments indicating that unglycosylated PrP has the highest affinity for heparin support this idea. Another possible explanation is that PrP glycans sterically block heparin and HS binding sites. Further research will be required to distinguish among these possibilities.
Is highly glycosylated PrP compatible with parenchymal plaque formation? We tested this possibility by challenging mice that express triglycosylated PrP with mCWD prions, which are ADAM10-cleaved, monoglycosylated, and form large plaques. Notably, the first mCWD passage in Prnp187N mice resulted in a near abrogation of plaques. Instead, prions formed mostly diffuse aggregates concurrent with a sharp reduction in the survival period. Although it remains a possibility that the change in the amino acid side chain and not the third glycans caused a switch in the strain phenotype, this would more likely have led to a lengthening of the incubation period. Instead, these findings suggest that highly glycosylated PrP may not form large, fibrillar parenchymal plaques, at least for certain strains, further supporting a link between PTM-deficient PrP and fibrillar parenchymal plaque formation.
We propose that a triad of factors act in concert to determine parenchymal prion plaque formation: (a) lack of a GPI-anchor, (b) one or no glycans, and (c) extracellular HS. Therefore, prions that replicate ADAM10-cleaved, unglycosylated PrP can bind HS and assemble into fibrils, forming extracellular parenchymal plaques. A key role for HS in parenchymal plaque formation is further supported by the finding that shortening HS chains in mice haploinsufficient in HS polymerase exostosin 1 decreases parenchymal plaques and increases vascular deposition of ADAM10-cleaved PrPSc (51). Similarly, in an AD mouse model, depleting neuronal HS chains leads to a reduction in parenchymal plaques and an increase in vascular plaques (80), also indicating a role for HS in amyloid-β parenchymal plaque formation. Future studies will be necessary to understand the exact role of HS in prion parenchymal plaque formation.
Our findings support and provide a possible explanation for the parenchymal plaques observed histologically in certain familial prion diseases, for example those caused by the F198S mutation in which the plaque core consists of an unglycosylated PrP fragment (81). Additional familial mutations associated with parenchymal plaque formation are also composed of mono- or unglycosylated PrP, including V180I (fCJD) (82) and T183A (1, 83, 84). Whether these prions are composed of GPI-anchored or ADAM10-cleaved PrP remains unclear, and antibodies that can distinguish the posttranslationally modified forms of PrP will be useful for future studies. Since most PrPC in mammals is posttranslationally modified with a GPI-anchor and N-linked glycans, these results may help explain the rarity of fibrillar plaques in prion disease and the predominance of small aggregates associated with the highest levels of infectivity.
Methods
Additional Methods are included in the Supplemental Material.
Prnp180Q/196Q and Prnp187N mouse generation using the CRISPR/CAS9 system.
The Prnp180Q/196Q and Prnp187N knockin mice were generated by the UC Irvine Mouse Genetics core by microinjection of Cas9 ribonucleoprotein (PNA Bio) into B6SJLF1 × C57BL/6NJ zygotes. Briefly, Cas9 (20 ng/μL), gRNA-1 (20 ng/μL), and ssDNA HDR template (10 ng/μL) were mixed in injection buffer (10 mM Tris, 0.1 mM EDTA) and incubated on ice for 10 minutes, as per the manufacturer’s instructions. The Cas9 mixture was microinjected into the pronucleus of single-cell zygotes isolated from super-ovulated females. All founders and select progeny were genotyped by Sanger sequencing genomic DNA from proteinase K digested tail tissue. Six founder mice carried the mutated Prnp180Q/196Q allele and 10 founders carried the Prnp187N. Select lines were backcrossed and bred to homozygosity. Aged mice investigated for spontaneous aggregation were 100–600 days old.
Statistics.
A Student’s t test (2-tailed, unpaired) was used to determine the statistical significance between the Prnp180Q/196Q and Prnp187N versus WT mouse brain samples for the PrPC level of expression, PK digestion of PrPC, PrPSc conformational stability, PrPC solubility, levels of ADAM10-cleaved PrPC and PrPSc, and levels of HS bound to PrPSc in ME7-infected WT and Prnp180Q/196Q brains. Student’s t test (2-tailed, unpaired) was also used to compare numbers of PrPSc plaques found in ME7-infected WT and Prnp180Q/196Q mice, PrPC cell surface levels from WT and Prnp180Q/196Q cortical neurons, and cerebellar PrPC levels from WT and Prnp180Q/196Q mice.
One-way ANOVA with Tukey’s post test was performed to assess survival differences between mouse groups, PrPSc solubility differences in the prion-infected mouse brain over multiple passages (Prnp180Q/196Q and Prnp187N as compared with WT mice), and to compare the binding affinity of unglycosylated, monoglycosylated, and diglycosylated isoforms of WT PrPC to heparin.
Two-way ANOVA with Bonferroni’s post hoc test was performed to assess differences in the lesion profiles, PrPSc levels in each brain region, plaque size of the prion-infected WT and Prnp180Q/196Q brain, the heparin-binding affinity of RK13-derived PrP180Q/196Q, PrP180Q, PrP187N, and WT PrPC, the heparin-binding affinity of full-length and ADAM10-cleaved PrPC from Prnp180Q/196Q and WT mouse brains, and the composition of HS bound to PrPSc in ME7-infected Prnp180Q/196Q and WT mouse brain. GraphPad Prism 5 software was used for statistical analyses. No measurement was excluded for statistical analysis. For all analyses, P less than or equal to 0.05 was considered significant. Data displayed in graphs represent mean ± SEM.
Study approval.
All animal studies were performed following procedures to minimize suffering and were approved by the Institutional Animal Care and Use Committee at UC San Diego. Protocols were performed in strict accordance with good animal practices, as described in the Guide for the Use and Care of Laboratory Animals published by the National Institutes of Health.
Author contributions
AMS, PAC, TDK, JAL, KPRN, CJS designed experiments, HA and MG provided reagents, AMS, PAC, TDK, JAL, KS, THN, BC, TS, DPP, SN performed the experiments, AMS, PAC, TDK, JAL, THN, TS, DPP, HA, JDE, MG, KPRN, and CJS analyzed the experiments, AMS, PAC and CJS wrote the manuscript. The order of the co–first authors was determined by the study design, data, and experimental interpretation contributed by each co-author.
Supplementary Material
Acknowledgments
We thank Jonathan Neumann, Taylor Winrow, and Nazilla Alderson for outstanding technical support, and the animal care staff at UC San Diego for excellent animal care. We also thank the UC Irvine Mouse Genetics core for their help generating the knockin mice and the UC San Diego GlycoAnalytics Core for the mass spectrometry analysis. This study was supported by the National Institutes of Health grants NS069566 (to CJS), NS076896 (to CJS), NS103848 (to JGS), NS103588 (to JAL), AG061251 (to PAC), CJD Foundation (to CJS and HCA), the Werner-Otto-Stiftung (to HCA), and the Ramón Areces Foundation (to PAC). The UC Irvine Transgenic Mouse Facility is funded in part by the Chao Family Comprehensive Cancer Center Support Grant (P30CA062203) from the National Cancer Institute.
Version 1. 01/27/2020
Electronic publication
Version 2. 03/02/2020
Print issue publication
Footnotes
AMS’s present address is: Department of Clinical Cancer Prevention, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
TDK’s present address is: Foundation for Food and Agriculture Research, Washington, DC, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(3):1350–1362.https://doi.org/10.1172/JCI131564.
See the related Commentary at Underglycosylated prion protein modulates plaque formation in the brain.
Contributor Information
Alejandro M. Sevillano, Email: a1sevillanomantas@ucsd.edu.
Patricia Aguilar-Calvo, Email: p2aguilar@ucsd.edu.
Katrin Soldau, Email: katrin.soldau@gmail.com.
Taylor Schumann, Email: tschumann@ucsd.edu.
Sofie Nyström, Email: Sofie.nystrom@liu.se.
Biswa Choudhury, Email: bchoudhury@ucsd.edu.
Jeffrey D. Esko, Email: jesko@ucsd.edu.
Markus Glatzel, Email: m.glatzel@uke.de.
K. Peter R. Nilsson, Email: petni@ifm.liu.se.
References
- 1.Ghetti B, Piccardo P, Zanusso G. Dominantly inherited prion protein cerebral amyloidoses - a modern view of Gerstmann-Sträussler-Scheinker. Handb Clin Neurol. 2018;153:243–269. doi: 10.1016/B978-0-444-63945-5.00014-3. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ. The molecular pathology of Alzheimer’s disease. Neuron. 1991;6(4):487–498. doi: 10.1016/0896-6273(91)90052-2. [DOI] [PubMed] [Google Scholar]
- 3.Kim MO, Takada LT, Wong K, Forner SA, Geschwind MD. Genetic PrP prion diseases. Cold Spring Harb Perspect Biol. 2018;10(5):a033134. doi: 10.1101/cshperspect.a033134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zerr I, et al. Current clinical diagnosis in Creutzfeldt-Jakob disease: identification of uncommon variants. Ann Neurol. 2000;48(3):323–329. doi: 10.1002/1531-8249(200009)48:3<323::AID-ANA6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki Y, Kato H, Ando T, Mimuro M, Kitamoto T, Yoshida M. MM1-type sporadic Creutzfeldt-Jakob disease with 1-month total disease duration and early pathologic indicators. Neuropathology. 2017;37(5):420–425. doi: 10.1111/neup.12379. [DOI] [PubMed] [Google Scholar]
- 6.Zerr I, Parchi P. Sporadic Creutzfeldt-Jakob disease. Handb Clin Neurol. 2018;153:155–174. doi: 10.1016/B978-0-444-63945-5.00009-X. [DOI] [PubMed] [Google Scholar]
- 7.Wells GA, Wilesmith JW. The neuropathology and epidemiology of bovine spongiform encephalopathy. Brain Pathol. 1995;5(1):91–103. doi: 10.1111/j.1750-3639.1995.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 8.Spraker TR, Zink RR, Cummings BA, Sigurdson CJ, Miller MW, O’Rourke KI. Distribution of protease-resistant prion protein and spongiform encephalopathy in free-ranging mule deer (Odocoileus hemionus) with chronic wasting disease. Vet Pathol. 2002;39(5):546–556. doi: 10.1354/vp.39-5-546. [DOI] [PubMed] [Google Scholar]
- 9.Spraker TR, Balachandran A, Zhuang D, O’Rourke KI. Variable patterns of distribution of PrP (CWD) in the obex and cranial lymphoid tissues of Rocky Mountain elk (Cervus elaphus nelsoni) with subclinical chronic wasting disease. Vet Rec. 2004;155(10):295–302. doi: 10.1136/vr.155.10.295. [DOI] [PubMed] [Google Scholar]
- 10.Spiropoulos J, Casalone C, Caramelli M, Simmons MM. Immunohistochemistry for PrPSc in natural scrapie reveals patterns which are associated with the PrP genotype. Neuropathol Appl Neurobiol. 2007;33(4):398–409. doi: 10.1111/j.1365-2990.2007.00800.x. [DOI] [PubMed] [Google Scholar]
- 11.Wemheuer WM, et al. Similarities between forms of sheep scrapie and Creutzfeldt-Jakob disease are encoded by distinct prion types. Am J Pathol. 2009;175(6):2566–2573. doi: 10.2353/ajpath.2009.090623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216(4542):136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 13.Prusiner SB, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63(4):673–686. doi: 10.1016/0092-8674(90)90134-Z. [DOI] [PubMed] [Google Scholar]
- 14.Endo T, Groth D, Prusiner SB, Kobata A. Diversity of oligosaccharide structures linked to asparagines of the scrapie prion protein. Biochemistry. 1989;28(21):8380–8388. doi: 10.1021/bi00447a017. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin MA, Stahl N, Reinders LG, Gibson BW, Prusiner SB, Burlingame AL. Permethylation and tandem mass spectrometry of oligosaccharides having free hexosamine: analysis of the glycoinositol phospholipid anchor glycan from the scrapie prion protein. Anal Biochem. 1990;191(1):174–182. doi: 10.1016/0003-2697(90)90405-X. [DOI] [PubMed] [Google Scholar]
- 16.Stahl N, Baldwin MA, Prusiner SB. Electrospray mass spectrometry of the glycosylinositol phospholipid of the scrapie prion protein. Cell Biol Int Rep. 1991;15(9):853–862. doi: 10.1016/0309-1651(91)90037-J. [DOI] [PubMed] [Google Scholar]
- 17.Rudd PM, et al. Glycosylation differences between the normal and pathogenic prion protein isoforms. Proc Natl Acad Sci USA. 1999;96(23):13044–13049. doi: 10.1073/pnas.96.23.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudd PM, Wormald MR, Wing DR, Prusiner SB, Dwek RA. Prion glycoprotein: structure, dynamics, and roles for the sugars. Biochemistry. 2001;40(13):3759–3766. doi: 10.1021/bi002625f. [DOI] [PubMed] [Google Scholar]
- 19.Chesebro B, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308(5727):1435–1439. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 20.Cancellotti E, et al. Glycosylation of PrPC determines timing of neuroinvasion and targeting in the brain following transmissible spongiform encephalopathy infection by a peripheral route. J Virol. 2010;84(7):3464–3475. doi: 10.1128/JVI.02374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancellotti E, et al. Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties. EMBO J. 2013;32(5):756–769. doi: 10.1038/emboj.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Race B, Phillips K, Meade-White K, Striebel J, Chesebro B. Increased infectivity of anchorless mouse scrapie prions in transgenic mice overexpressing human prion protein. J Virol. 2015;89(11):6022–6032. doi: 10.1128/JVI.00362-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiseman FK, et al. The glycosylation status of PrPC is a key factor in determining transmissible spongiform encephalopathy transmission between species. J Virol. 2015;89(9):4738–4747. doi: 10.1128/JVI.02296-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuzi NL, et al. Host PrP glycosylation: a major factor determining the outcome of prion infection. PLoS Biol. 2008;6(4):e100. doi: 10.1371/journal.pbio.0060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguilar-Calvo P, et al. Post-translational modifications in PrP expand the conformational diversity of prions in vivo. Sci Rep. 2017;7:43295. doi: 10.1038/srep43295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katorcha E, Baskakov Analyses of N-linked glycans of PrPSc revealed predominantly 2,6-linked sialic acid residues. FEBS J. 2017;284(21):3727–3738. doi: 10.1111/febs.14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katorcha E, Makarava N, Savtchenko R, D’Azzo A, Baskakov Sialylation of prion protein controls the rate of prion amplification, the cross-species barrier, the ratio of PrPSc glycoform and prion infectivity. PLoS Pathog. 2014;10(9):e1004366. doi: 10.1371/journal.ppat.1004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katorcha E, Makarava N, Savtchenko R, Baskakov Sialylation of the prion protein glycans controls prion replication rate and glycoform ratio. Sci Rep. 2015;5:16912. doi: 10.1038/srep16912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bate C, Nolan W, Williams A. Sialic acid on the glycosylphosphatidylinositol anchor regulates PrP-mediated cell signaling and prion formation. J Biol Chem. 2016;291(1):160–170. doi: 10.1074/jbc.M115.672394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katorcha E, et al. Reversible off and on switching of prion infectivity via removing and reinstalling prion sialylation. Sci Rep. 2016;6:33119. doi: 10.1038/srep33119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava S, Katorcha E, Daus ML, Lasch P, Beekes M, Baskakov Sialylation controls prion fate in vivo. J Biol Chem. 2017;292(6):2359–2368. doi: 10.1074/jbc.M116.768010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baskakov Limited understanding of the functional diversity of N-linked glycans as a major gap of prion biology. Prion. 2017;11(2):82–88. doi: 10.1080/19336896.2017.1301338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taraboulos A, et al. Acquisition of protease resistance by prion proteins in scrapie-infected cells does not require asparagine-linked glycosylation. Proc Natl Acad Sci USA. 1990;87(21):8262–8266. doi: 10.1073/pnas.87.21.8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancellotti E, et al. Altered glycosylated PrP proteins can have different neuronal trafficking in brain but do not acquire scrapie-like properties. J Biol Chem. 2005;280(52):42909–42918. doi: 10.1074/jbc.M509557200. [DOI] [PubMed] [Google Scholar]
- 35.Salamat MK, Dron M, Chapuis J, Langevin C, Laude H. Prion propagation in cells expressing PrP glycosylation mutants. J Virol. 2011;85(7):3077–3085. doi: 10.1128/JVI.02257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caughey B, Race RE, Ernst D, Buchmeier MJ, Chesebro B. Prion protein biosynthesis in scrapie-infected and uninfected neuroblastoma cells. J Virol. 1989;63(1):175–181. doi: 10.1128/JVI.63.1.175-181.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borchelt DR, Scott M, Taraboulos A, Stahl N, Prusiner SB. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J Cell Biol. 1990;110(3):743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linsenmeier L, et al. Structural and mechanistic aspects influencing the ADAM10-mediated shedding of the prion protein. Mol Neurodegener. 2018;13(1):18. doi: 10.1186/s13024-018-0248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuendorf E, et al. Glycosylation deficiency at either one of the two glycan attachment sites of cellular prion protein preserves susceptibility to bovine spongiform encephalopathy and scrapie infections. J Biol Chem. 2004;279(51):53306–53316. doi: 10.1074/jbc.M410796200. [DOI] [PubMed] [Google Scholar]
- 40.Rogers M, Taraboulos A, Scott M, Groth D, Prusiner SB. Intracellular accumulation of the cellular prion protein after mutagenesis of its Asn-linked glycosylation sites. Glycobiology. 1990;1(1):101–109. doi: 10.1093/glycob/1.1.101. [DOI] [PubMed] [Google Scholar]
- 41.Bett C, et al. Biochemical properties of highly neuroinvasive prion strains. PLoS Pathog. 2012;8(2):e1002522. doi: 10.1371/journal.ppat.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Godsave SF, et al. Cryo-immunogold electron microscopy for prions: toward identification of a conversion site. J Neurosci. 2008;28(47):12489–12499. doi: 10.1523/JNEUROSCI.4474-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeffrey M, Goodsir CM, Bruce ME, McBride PA, Fraser JR. In vivo toxicity of prion protein in murine scrapie: ultrastructural and immunogold studies. Neuropathol Appl Neurobiol. 1997;23(2):93–101. doi: 10.1111/j.1365-2990.1997.tb01191.x. [DOI] [PubMed] [Google Scholar]
- 44.Sigurdson CJ, et al. Strain fidelity of chronic wasting disease upon murine adaptation. J Virol. 2006;80(24):12303–12311. doi: 10.1128/JVI.01120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Will RG, et al. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996;347(9006):921–925. doi: 10.1016/S0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 46.Ulrich JD, et al. ApoE facilitates the microglial response to amyloid plaque pathology. J Exp Med. 2018;215(4):1047–1058. doi: 10.1084/jem.20171265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magnusson K, Simon R, Sjölander D, Sigurdson CJ, Hammarström P, Nilsson KP. Multimodal fluorescence microscopy of prion strain specific PrP deposits stained by thiophene-based amyloid ligands. Prion. 2014;8(4):319–329. doi: 10.4161/pri.29239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herrmann US, et al. Structure-based drug design identifies polythiophenes as antiprion compounds. Sci Transl Med. 2015;7(299):299ra123. doi: 10.1126/scitranslmed.aab1923. [DOI] [PubMed] [Google Scholar]
- 49.König C, Skånberg R, Hotz I, Ynnerman A, Norman P, Linares M. Binding sites for luminescent amyloid biomarkers from non-biased molecular dynamics simulations. Chem Commun (Camb) 2018;54(24):3030–3033. doi: 10.1039/C8CC00105G. [DOI] [PubMed] [Google Scholar]
- 50.Schütz AK, et al. Binding of polythiophenes to amyloids: structural mapping of the pharmacophore. ACS Chem Neurosci. 2018;9(3):475–481. doi: 10.1021/acschemneuro.7b00397. [DOI] [PubMed] [Google Scholar]
- 51.Aguilar-Calvo P, et al. Shortening heparan sulfate chains prolongs survival and reduces parenchymal plaques in prion disease caused by mobile, ADAM10-cleaved prions [published online ahead of print October 31, 2019] Acta Neuropathol. doi: 10.1007/s00401-019-02085-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stahl N, Baldwin MA, Burlingame AL, Prusiner SB. Identification of glycoinositol phospholipid linked and truncated forms of the scrapie prion protein. Biochemistry. 1990;29(38):8879–8884. doi: 10.1021/bi00490a001. [DOI] [PubMed] [Google Scholar]
- 53.Taylor DR, et al. Role of ADAMs in the ectodomain shedding and conformational conversion of the prion protein. J Biol Chem. 2009;284(34):22590–22600. doi: 10.1074/jbc.M109.032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polymenidou M, et al. The POM monoclonals: a comprehensive set of antibodies to non-overlapping prion protein epitopes. PLoS One. 2008;3(12):e3872. doi: 10.1371/journal.pone.0003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacobs JG, et al. Proteinase K-resistant material in ARR/VRQ sheep brain affected with classical scrapie is composed mainly of VRQ prion protein. J Virol. 2011;85(23):12537–12546. doi: 10.1128/JVI.00448-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sevillano AM, et al. Recombinant PrPSc shares structural features with brain-derived PrPSc: insights from limited proteolysis. PLoS Pathog. 2018;14(1):e1006797. doi: 10.1371/journal.ppat.1006797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corlin DB, Johnsen CK, Nissen MH, Heegaard NH. Glycosaminoglycans enhance the fibrillation propensity of the β2-microglobulin cleavage variant--ΔK58-β2m. Biochem Biophys Res Commun. 2010;402(2):247–251. doi: 10.1016/j.bbrc.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Bourgault S, Solomon JP, Reixach N, Kelly JW. Sulfated glycosaminoglycans accelerate transthyretin amyloidogenesis by quaternary structural conversion. Biochemistry. 2011;50(6):1001–1015. doi: 10.1021/bi101822y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snow AD, et al. Immunolocalization of heparan sulfate proteoglycans to the prion protein amyloid plaques of Gerstmann-Straussler syndrome, Creutzfeldt-Jakob disease and scrapie. Lab Invest. 1990;63(5):601–611. [PubMed] [Google Scholar]
- 60.Snow AD, et al. The presence of heparan sulfate proteoglycans in the neuritic plaques and congophilic angiopathy in Alzheimer’s disease. Am J Pathol. 1988;133(3):456–463. [PMC free article] [PubMed] [Google Scholar]
- 61. Raymond GJ, Chabry J. Purification of the pathological isoform of prion protein (PrPSc or PrPres) from transmissible spongiform encephalopathy-affected brain tissue. In: Lehmann S, Grassi J, eds. Techniques in Prion Research. Birkhäuser, Basel; 2004:16–26. [Google Scholar]
- 62.Lawrence R, et al. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J Biol Chem. 2008;283(48):33674–33684. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawrence R, et al. Disease-specific non-reducing end carbohydrate biomarkers for mucopolysaccharidoses. Nat Chem Biol. 2012;8(2):197–204. doi: 10.1038/nchembio.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bett C, et al. Enhanced neuroinvasion by smaller, soluble prions. Acta Neuropathol Commun. 2017;5(1):32. doi: 10.1186/s40478-017-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu EL, et al. Insight into early-stage unfolding of GPI-anchored human prion protein. Biophys J. 2015;109(10):2090–2100. doi: 10.1016/j.bpj.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mishra R, et al. Impact of N-glycosylation site variants during human PrP aggregation and fibril nucleation. Biochim Biophys Acta Proteins Proteom. 2019;1867(10):909–921. doi: 10.1016/j.bbapap.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 67.Bosques CJ, Imperiali B. The interplay of glycosylation and disulfide formation influences fibrillization in a prion protein fragment. Proc Natl Acad Sci USA. 2003;100(13):7593–7598. doi: 10.1073/pnas.1232504100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Araman C, Thompson RE, Wang S, Hackl S, Payne RJ, Becker CFW. Semisynthetic prion protein (PrP) variants carrying glycan mimics at position 181 and 197 do not form fibrils. Chem Sci. 2017;8(9):6626–6632. doi: 10.1039/C7SC02719B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeMarco ML, Daggett V. Characterization of cell-surface prion protein relative to its recombinant analogue: insights from molecular dynamics simulations of diglycosylated, membrane-bound human prion protein. J Neurochem. 2009;109(1):60–73. doi: 10.1111/j.1471-4159.2009.05892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stahl N, et al. Structural studies of the scrapie prion protein using mass spectrometry and amino acid sequencing. Biochemistry. 1993;32(8):1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 71.Stimson E, Hope J, Chong A, Burlingame AL. Site-specific characterization of the N-linked glycans of murine prion protein by high-performance liquid chromatography/electrospray mass spectrometry and exoglycosidase digestions. Biochemistry. 1999;38(15):4885–4895. doi: 10.1021/bi982330q. [DOI] [PubMed] [Google Scholar]
- 72.Baron GS, et al. Effect of glycans and the glycophosphatidylinositol anchor on strain dependent conformations of scrapie prion protein: improved purifications and infrared spectra. Biochemistry. 2011;50(21):4479–4490. doi: 10.1021/bi2003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3(7):a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stoler-Barak L, Moussion C, Shezen E, Hatzav M, Sixt M, Alon R. Blood vessels pattern heparan sulfate gradients between their apical and basolateral aspects. PLoS One. 2014;9(1):e85699. doi: 10.1371/journal.pone.0085699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ortmann C, et al. Sonic hedgehog processing and release are regulated by glypican heparan sulfate proteoglycans. J Cell Sci. 2015;128(12):2374–2385. doi: 10.1242/jcs.170670. [DOI] [PubMed] [Google Scholar]
- 76.Snow AD, Kisilevsky R, Willmer J, Prusiner SB, DeArmond SJ. Sulfated glycosaminoglycans in amyloid plaques of prion diseases. Acta Neuropathol. 1989;77(4):337–342. doi: 10.1007/BF00687367. [DOI] [PubMed] [Google Scholar]
- 77.McBride PA, Wilson MI, Eikelenboom P, Tunstall A, Bruce ME. Heparan sulfate proteoglycan is associated with amyloid plaques and neuroanatomically targeted PrP pathology throughout the incubation period of scrapie-infected mice. Exp Neurol. 1998;149(2):447–454. doi: 10.1006/exnr.1997.6740. [DOI] [PubMed] [Google Scholar]
- 78.Wong C, et al. Sulfated glycans and elevated temperature stimulate PrP(Sc)-dependent cell-free formation of protease-resistant prion protein. EMBO J. 2001;20(3):377–386. doi: 10.1093/emboj/20.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Imamura M, et al. Heparan sulfate and heparin promote faithful prion replication in vitro by binding to normal and abnormal prion proteins in protein misfolding cyclic amplification. J Biol Chem. 2016;291(51):26478–26486. doi: 10.1074/jbc.M116.745851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu CC, et al. Neuronal heparan sulfates promote amyloid pathology by modulating brain amyloid-β clearance and aggregation in Alzheimer’s disease. Sci Transl Med. 2016;8(332):332ra44. doi: 10.1126/scitranslmed.aad3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giaccone G, et al. Prion protein preamyloid and amyloid deposits in Gerstmann-Sträussler-Scheinker disease, Indiana kindred. Proc Natl Acad Sci USA. 1992;89(19):9349–9353. doi: 10.1073/pnas.89.19.9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chasseigneaux S, et al. V180I mutation of the prion protein gene associated with atypical PrPSc glycosylation. Neurosci Lett. 2006;408(3):165–169. doi: 10.1016/j.neulet.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 83.Grasbon-Frodl E, Lorenz H, Mann U, Nitsch RM, Windl O, Kretzschmar HA. Loss of glycosylation associated with the T183A mutation in human prion disease. Acta Neuropathol. 2004;108(6):476–484. doi: 10.1007/s00401-004-0913-4. [DOI] [PubMed] [Google Scholar]
- 84.Xiao X, et al. Glycoform-selective prion formation in sporadic and familial forms of prion disease. PLoS One. 2013;8(3):e58786. doi: 10.1371/journal.pone.0058786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.