Abstract
The transition to herbivory by insects is associated with distinct genomic signatures. Sequenced genomes of extant herbivore species reveal the result of these transitions, but in lieu of comparisons between herbivorous and non-herbivorous lineages that diverged recently, such datasets have shed less light on the evolutionary genomic processes involved in diet shifts to or from herbivory. Here we propose that the comparative genomics of diet shifts between closely related insect herbivores and non-herbivores, and within densely-sampled clades of herbivores, will help reveal the extent to which herbivory evolves through the co-option and subtle remodeling of widely-conserved gene families with functions ancestrally distinct from phytophagy.
Introduction
The genomes of arthropod herbivores bear the distinct signatures of phytophagy. Herbivore genomes tend to be enriched, albeit idiosyncratically, in gene functions involved in digesting plant primary metabolites and detoxifying plant secondary compounds, as well as in chemosensory functions for locating and identifying hosts (1). Beyond these general observations, there is much we do not know about how herbivory shapes the evolution of arthropod genomes. This is because many studies of arthropod herbivore genomes, including in the phytophagan (Curculionoidea and Chrymeloidea) beetles, Lepidoptera and Acari, although interesting, do not necessarily illuminate the process of transitioning to a diet of living plant tissues, but rather subsequent evolution in these lineages after they transitioned to herbivory hundreds of millions of years ago [2**, 3**].
Relying on comparisons between more closely-related taxa, a handful of studies are emerging that offer new insight into evolutionary processes underpinning diet shifts in insects. Drosophilid fly lineages that shifted from detritivory (microbe-feeding) to herbivory exhibit a sequential loss of yeast-tuned olfactory receptors (3), and those with diets that vary in toxicity exhibit predictable differences in the sizes of gene families encoding detoxifying enzymes (4*; 5**) (Figure 1). Based on a combination of transcriptome and genome sequences, herbaceous beetles exhibit lineage-specific expansions of detoxification and digestion-related gene families relative to predaceous suborders, and further display higher gene gain rates, lower loss rates, and higher absolute numbers of genes overall (6**). These results, coupled with comparable results on diet shifts in other invertebrates and mammals (8; 7), lend support to the general hypothesis that diet shifts in insects are associated with predictable evolutionary changes in genome architecture, most notably gene duplications, losses, and even parallel amino acid substitutions.
Figure 1.
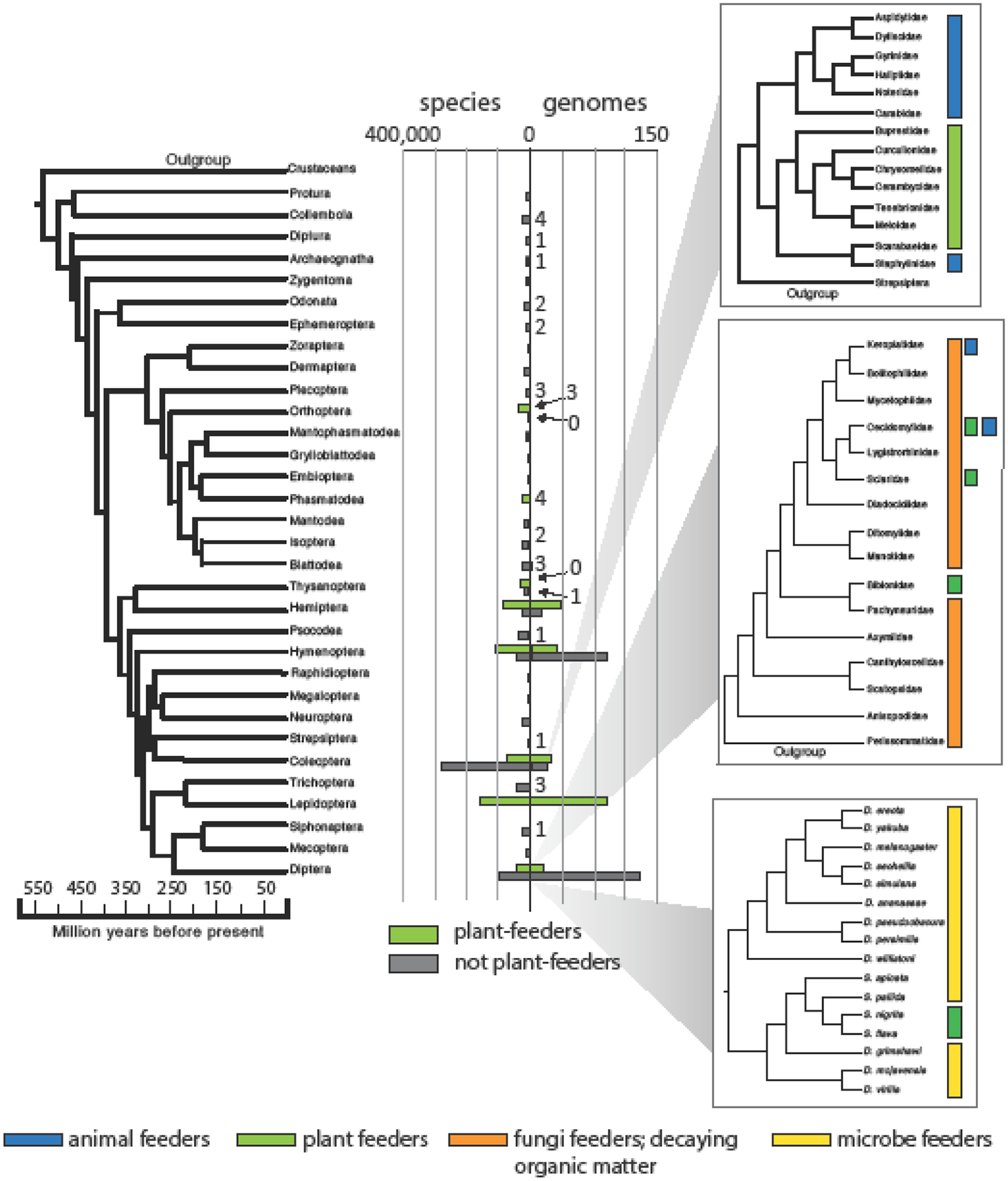
Phylogeny of the insect orders, after [55]. Indicated are the number of herbivorous and non-herbivorous species in each order, from [56], and the number of annotated genomes in each group, from the NCBI database in September 2019. Insets display diet variation in families of polyphagan beetles (top), modified from [6]; families of Bibionomorpha flies (middle), modified from [47]; and Drosophila and Scaptomyza flies (bottom), modified from [14]. Primary feeding modes are indicated in color to the right of each, from [6], [57] [51] and [56], respectively. Notable cases where there is exceptional diversity of feeding modes within groups are indicated by additional colored squares.
A challenge with broad-scale comparisons is that the passage of time tends to obscure more subtle changes in the pace and dynamics of genome evolution, especially because inferring events deep within phylogenies or along long branches is rife with uncertainty. Diet shifts may be correlated not just with changes in gene family size, which lend to straightforward comparisons among taxa, but with shifts in gene birth and death rates that require dense taxon sampling and more complex models in order to reliably infer. Depending on the ancestral repertoires, gene loss may actually outpace gene gain, for example (7, 9*, 10*, 12). Key unresolved questions include: What are the functional underpinnings of these diet shifts? What is the relative balance of selective, demographic and neutral processes acting on key genes/functions that drive highly uneven patterns of birth/death within and across gene families (4, 13, 14*)? Our aim here is to address some of the emerging genomic patterns associated with herbivory in insects, and going forward, to identify the types of studies needed to more fully understand how the process of diet shifts shapes the evolution of insect genomes.
Gene family dynamics and dietary shifts
The pervading idea on gene family dynamics and the role of diet shifts from the literature is that the chemical complexity of an organism’s dietary niche favors (or is enabled by) complexity of detoxification and chemosensory gene repertoires (1; 15). One general prediction from this literature is that plants are a chemically complex resource, so herbivores will have larger chemosensory and detoxification repertoires than animals with other diet types. Notably, such ideas could be confounded with those associated with diet breadth (e.g., specialization vs. generalization): generalists encounter more chemical diversity and will have larger repertoires than specialists (16; 17). However, these insights are derived from shifts in hosts or changes in host breadth among plant lineages in already-herbivorous lineages, from which the vast majority of studies derive. To what extent do changes within herbivorous lineages recapitulate the genomic dynamics of transitions to/from herbivory in insects? Rane et al. (18**) surveyed 160 species with sequenced genomes and found that omnivorous insect species tend to have relatively more detoxification genes than carnivores and herbivores, as do species that feed on tissues with more complex defensive chemistry. As compelling as this is, raw gene counts such as these don’t reveal the evolutionary processes that generated the gene family repertoire differences, or the order of the genetic changes involved in the evolution of herbivorous insects (4).
Consider dietary shifts in the relatively recently-derived mammals. Transitions associated with herbivory and insectivory reveal the need to digest recalcitrant carbohydrates such as chitin or cellulose for nutrient acquisition (19). Foregut fermenting and herbivorous ungulates and langur monkeys convergently evolved lysozyme enzymes able to function at low pH in the stomach, allowing them to lyse bacterial cells being cultivated on plant material in their stomachs (20). In turn, these herbivores have also repeatedly evolved duplicated and neofunctionalized gastrointestinal RNases to efficiently degrade bacterial RNA (21), a rich source of nitrogen. Similarly, three serine protease genes (PRSS1, PRSS36 and CPB1) encoded in giant and red panda genomes have convergently evolved to facilitate uptake of essential amino acids lysine and arginine (22). Finally, ancestral mammals are hypothesized to have been insectivorous, and new genomic evidence suggests that five copies of a gastrointestinal, acidic chitinase gene (CHIA) were present in the common ancestor of all placental mammals (23). The number of CHIA genes is associated with the proportion of arthropods in the diet, and were lost entirely in many herbivorous lineages. There are clear parallels with the few studies that have focused on this question in insects. This includes the origin, via horizontal gene transfer (HGT), of plant cell wall-degrading enzymes (PCWDEs) several times independently in herbivorous beetles (24).
Mammals also illustrate the dramatic changes that can occur in chemosensory gene repertoires with diet shifts. Since their split from terrestrial artiodactyls, cetaceans have experienced remarkable erosion of both olfactory receptor genes and the attendant peripheral and central nervous system components. This is most extreme in toothed whales, which lack olfactory nerves, the olfactory bulb, have few intact olfactory receptor genes (25) and have lost all gustatory receptor genes required for sweet, umami and bitter taste (26). The umami receptor gene TAS1R1 was also lost independently in the ancestors of the bamboo-specialist (and distantly related) giant panda and red panda (22).
Finally, an illustrative example of the genomic underpinnings of a diet shift can be found in the plant parasites in the genera Striga and Triphysaria. The origin of plant parasitism principally involves endogenous changes in the regulation of plant genes ancestrally involved in non-parasitic functions, rather than genes acquired by HGT or functions acquired by symbiosis (28). Most core parasitism genes have clear orthologs in non-parasitic relatives, such as those encoding subtilisin-like serine proteases that have roles in protein degradation/processing and signal transduction in non-parasitic plants, as well as genes involved in symbiotic nodulation and in root and flower biology (29).
Altogether, phylogenetically-informed studies from mammals and plants bolster the hypothesis that trophic shifts to living plants are associated with distinct evolutionary genomic signatures. Genic novelty, convergence and changes in the dynamics of gene gain and loss are notable features of insect herbivore genomes. But less obvious is the degree to which transitions to herbivory involve co-option and remodeling of ancestral genes and gene networks with roles not previously associated with herbivory (2; 14; 30*). Comparative genomics studies from more recently-derived mammals suggest that the focus should be on relatively recently derived (<50 million years) herbivorous insect lineages to have the best opportunity for mechanistic insight.
Gains, losses and remodeling in insect herbivores
Diet shifts represent radical changes in physiology, requiring novel gene functions. True & Carroll (31) describe four ways in which genes acquire new functions: changes in amino acid sequence, changes in gene regulation, the assembly of new genes out of the fragments of preexisting gene structures, and principally, gene duplication (Box 1). The genomic signatures of dietary shifts in herbivorous insects should include each of these processes (32*), which can be identified in three functional categories implicated in herbivory.
Box I. Gene duplication and the evolution of gene families.
Gene duplication and loss are a source of novel phenotypes in plants and animals (52). Initially, new gene copies replicate the functions of their original copies or else change optimal gene dosage, and therefore are selectively neutral or unfavorable (53). To persist, they must acquire either novel functions (neofunctionalization) or else partition ancestral functions between copies (subfunctionalization). Similarly, gene losses can change or be associated with a change the nature of the behavioral response or trophic relationship with an ancestral diet (3). Many variations exist on these themes. For example, multi-functional genes might go to fixation in the absence of environmental change if new mutations allow for functional subdivision between gene copies (subfunctionalization). If new functions interfere with old ones (antagonistic pleiotropy), then gene duplicates are potentially targets for new mutations that allow for selective resolution of pleiotropic conflicts (escape from adaptive conflict; 54). The persistence of gene duplicates that occur before dietary shifts might depend on the complementary subdivision and acquisition of novel functions in both gene copies (neo-subfunctionalization; 32). Gene family size following duplication can be governed by entirely neutral, stochastic processes. Global processes governing total genome size also affect the fate of gene families involved in chemical adaptation to host plants.
Chemosensory.
Insects use various chemosensory organs, which express odorant-binding proteins, olfactory receptors, and gustatory receptors, to select suitable hosts for oviposition and feeding. Studies of chemosensory receptors in well-sampled clades provide some of the most informative pictures of gene family dynamics in insect herbivores (33; 34). Expansions are often targeted to a few sub-families (in tandem arrays) and coupled with substitutions that change function and/or are predicted to be positively selected, which suggests selection to elaborate the functions of subsets of key genes. Some studies focus on gene expansions, others on losses, and others on total gene family size (9; 35; 36*). Genomic studies of turnover rate (i.e., cumulative rate of gene gain and loss events) are lacking, as are datasets to conduct them (but see 2, 37). Although the Drosophilidae are largely not herbivorous, most species tend to feed on decaying plant and microbial material and encounter plant and microbial toxins as a result. Comparisons of chemosensory gene turnover rate are intriguing (38), although by necessity are focused on a small number of recently diverged taxa and potentially confounded by demography (33). A new study on chemosensory gene evolution in a truly herbivorous drosophilid (Scaptomyza flava) and its close relatives found differences in turnover rate on the lineages leading to herbivorous taxa in olfactory receptor and odorant binding, but not gustatory receptor or ionotropic receptor gene families (2). Comparisons of overall gene family size would have missed these potentially important evolutionary patterns.
Digestion.
Herbivory requires unique feeding behaviors and physiology that differs from carnivory, insectivory or other diets. Insect herbivores ingest macromolecules such as proteins, polysaccharides, and lipids, but they also ingest defensive chemicals that target digestion itself. A principal means of defense against herbivory by plants is the production of proteolytic inhibitors (PIs) and molecular competitors that diminish the activity of enzymes involved in digestion. Insects respond to plant PIs by several mechanisms, including increasing copy number and expression of digestive or insensitive proteases and hydrolysis of plant PIs. Among the expanded orthologous groups in herbivorous beetles relative to predatory ones, for example, are serine and cysteine proteases with functions assumed to be related to digestion (6).
Detoxification.
Plants express defensive chemicals, which require defence mechanisms on the part of insects, including detoxification, sequestration, and/or excretion. Some of the most thorough work has concentrated on the classical case of the heme-binding cytochrome P450 monooxygenases (CYPs). CYPs can be roughly partitioned into two types, those involved in the biosynthesis of physiologically-important chemicals such as steroids and cholesterol, and those involved in the detoxification of xenobiotics (40). In both vertebrates and invertebrates, the detoxification-types have evolved from the biosynthesis-types and diversified following ancient duplication of biosynthesis-types. As Feyereisen describes, CYPs subsequently diversified and “bloomed”, probably due to a combination of selective and neutral processes acting on gene birth and death rates (41). In each of these cases, gene duplication fuels the evolution of novel functions necessary for niche adaptation, but coupled with complex patterns of genome dynamics that effectively remodel gene families. Johnson et al. (42**) compared CYP diversity in florivorous Bombus and Apis species to eusocial carnivorous Polistes species, and found evidence for positive selection on and expansion of genes encoding CYP6AS, which metabolize flavonols in nectar and honey. They hypothesized that these changes associated with the shift from carnivory to florivory in bees. Island populations of Drosophila yakuba have specialized on the toxic fruits of noni, much like its island-endemic and relative D. sechellia. In D. sechellia and the herbivorous Scaptomyza spp., shifts from microbe-feeding/detritivory to folivory and frugivory are associated with an increase in toxin resistance, respectively (2, 14; 43). In D. yakuba, candidate genes include genes involved in juvenile hormone biosynthesis and ecdysteroid signaling, as well as genes in the enigmatic Osiris cluster with poorly known functions that may be directly involved in resistance to octanoic acid, the latter mirrored in D. sechellia as well as Scaptomyza (4, 30). When known, Osiris gene products have diverse roles and may play an especially important one in adaptation to toxic diets (44). In the evolution of herbivorous Scaptomyza flava, Gloss et al. (2) found a small net loss in genes encoding CYPs and no net change in glutathione S-transferases, compared to the closest, non-herbivorous relatives. However, turnover rates were higher for both of these gene families in herbivorous Scaptomyza compared to non-herbivorous relatives (Fig. 2). The CYP gene cyp6g1 expanded from two copies in ancestral Drosophila and Scaptomyza (45) to six copies in S. flava. This gene is frequently involved in the evolution of insecticide resistance in other dipterans through an increase in gene dosage, via cis-regularly mutations or gene duplication events (45). The GstE5–8 gene expanded from one to five copies in S. flava since its divergence from non-herbivorous species. Remarkably, three of these GstE5–8 paralogs in S. flava encode enzymes that are the most efficient at detoxifying isothiocyanates (mustard oils), the principal toxins of their Brassicales host plants, among all known animal GSTs (2). It is possible, although speculative, that over longer periods of time, turnover rates could translate into differences in absolute numbers of genes encoding detoxification enzymes (e.g., 18), but clearly increases in the size of entire gene families (e.g, CYPs, GSTs) are not necessary for obligate herbivory to evolve in insects. Rather these gene family expansions may be more associated with changes in host breadth (e.g., in the evolution of host generalism).
Figure 2.
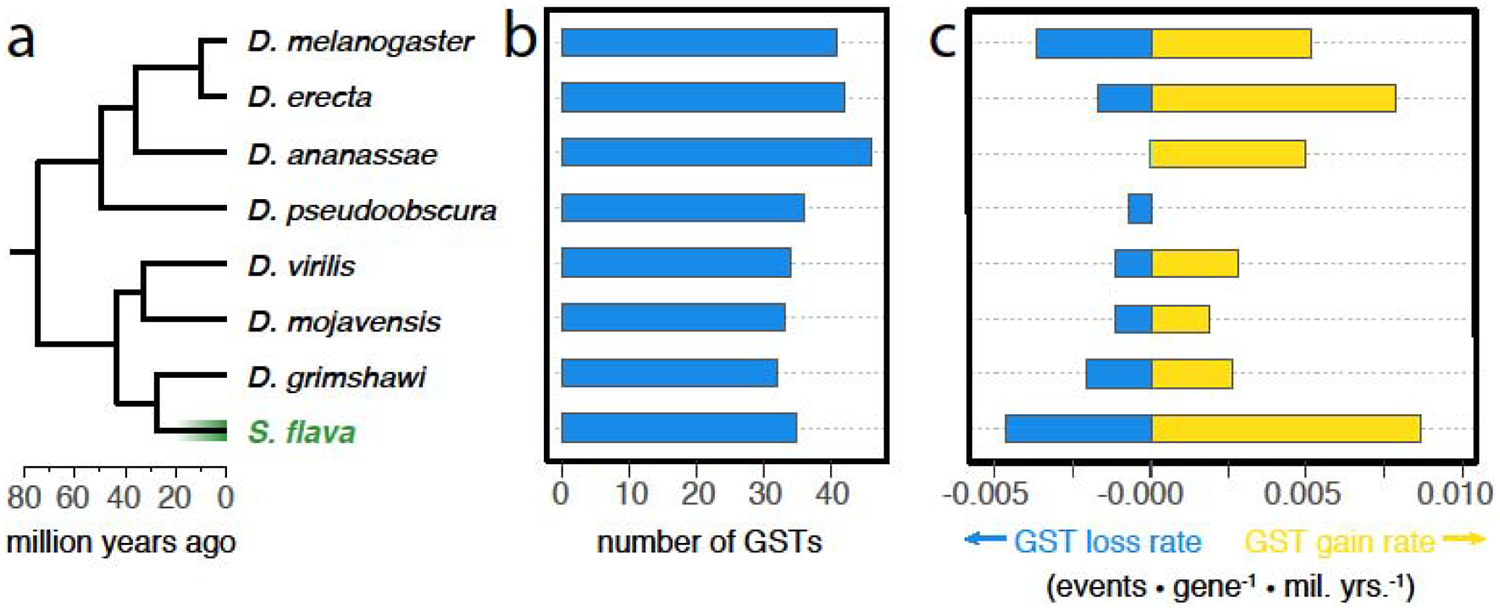
Accelerated turnover of genes encoding glutathione S-transferases (GSTs) coupled with a transition from microbe-feeding to herbivory in Scaptomyza. This figure was reproduced from [2], wherein full results and methods can be found. (a) Genes encoding GSTs were annotated in the genomes of eight drosophilids with diverse ecologies. Among these species, only S. flava is herbivorous. Divergence dates are taken from [30]. (b) Total number of GST genes in each genome. (c) Maximum likelihood estimates of GST gene gain and loss rates in each taxon. Of the eight taxa, only S. flava exhibits a rate of GST turnover (cumulative gains + losses) that is significantly higher than the background rate across the phylogeny as a whole. Yet because the total number of GST genes in S. flava falls within the range observed across the non-herbivorous Drosophila, a focus on overall gene family size would fail to detect this gene family “remodeling” coupled with a transition to herbivory.
Next steps
Outside of acquisition of functions by HGT or symbiosis, the transition to herbivory is hypothesized to derive from endogenous mechanisms not previously associated with herbivory. If so, then the question is how to identify these mechanisms? A central challenge is identifying phylogenetically and statistically robust sets of taxa. Most evidence supporting the prediction that the complexity of plant chemical defenses requires diverse detoxification strategies to enable dietary transitions to herbivory is noisy, lacks phylogenetic robustness, and/or is anecdotal. This is largely due to the uneven (and still relatively sparse) sampling of different diets across the insect phylogeny. The non-independence of different factors [diet type (e.g., carnivore vs. herbivore), diet sub-type (e.g., leaves are more complex than sap), and diet breadth across the phylogeny also confounds inferences (18).
While evidence for dramatic changes in gene family size is largely lacking, evidence is emerging that rates of gene turnover in detoxification and chemosensory gene families are accelerated in herbivores, particularly in genes that interact with key defensive secondary compounds. It would be unsurprising that rates of gene birth and death are relatively balanced: Important genes for plant chemical interactions are duplicated but genes important for a non-herbivorous lifestyle are lost. This means that analyses that model changes in total gene family size among distantly related taxa, rather than comparing closely related taxa to infer individual gain/loss events and model rate changes across taxa, may miss effects of herbivory on gene family evolution when gene birth and death rates are both accelerated.
Phylogenetic frameworks that consider multiple transitions to herbivory within a relatively recent radiation may offer more power to uncover patterns due to dietary shifts, as demonstrated with mammals. Yet even with dense sampling, biases in estimates of gene gains and losses could arise from rate heterogeneity, misassembled paralogs and gene conversion (46), requiring close attention to appropriate model selection and quality controls when estimating gene birth and death processes (9).
What groups are best suited to these constraints? Herbivory has evolved more often than any other diet shift in Diptera (at least 25 times among extant lineages, and probably much more; 47). Given the genomic resources already existing for dipterans, these are an obvious group in which to conduct detailed, phylogenetically-robust studies of the evolutionary consequences of transitions to herbivory (3; Figure 1). For example, a species of Scaptomyza in a different subgenus than the Scaptomyza leaf-miners mentioned above is especially intriguing because it is a facultative leaf-miner of New Zealand celery (Apiaceae: Apium prostraum). Although adults cannot oviposit directly into leaf tissue, second and third instar larvae move from decaying leaves, where they eclose from eggs, to living leaves where they become leaf-miners and complete development (48). There could be standing genetic variation for this trait segregating across its range, providing opportunity for population and quantitative genomics (49) as well as experimental approaches to identifying the genomic architecture of herbivory (50). Further, transcriptomics of flies reared on decaying vs. living leaves may reveal what alleles are involved prior to the full ecological transition to herbivory. The plant species on which they feed contain furanocoumarins, which have played an important role in our understanding of the evolutionary genomic basis of phytochemical detoxification in herbivores (37), further enhancing its comparative potential.
Diptera are not the only promising group. Basally-derived cecidomyiids are mycophagous, but herbivory, gall-forming, and predation have evolved more recently (51, 57). Detailed genomic comparisons within the mostly herbivorous Polyphaga clade of beetles can also provide insights into derived losses of herbivory. The resulting change in evolutionary constraint should help reveal genes necessary for herbivory in these taxa, and can complement studies of shifts to herbivory (6). Similar opportunities are available in the Orthoptera, Hemiptera and Hymenoptera (Fig. 1). As comparative genomics of arthropods blossoms, a clearer picture of the evolutionary genomic roots of herbivory will emerge.
Highlights.
Plant chemical complexity drives detoxification repertoire evolution in herbivores
Herbivore genomes are enriched for chemosensation, digestion, and detoxification genes
Unique genomic signatures are shaped by major transitions in diet
Co-option and gene family remodeling are major evolutionary feature of herbivory
An important goal is to identify relatively recently derived diet shifts in insects
Acknowledgements
This research was supported by the National Institute of General Medical Science of the National Institutes of Health award number R35GM119816 to NKW. This manuscript is based on work done by PA while serving at the U.S. National Science Foundation. The views expressed in this paper do not necessarily reflect those of the National Science Foundation or the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew D. Gloss, Department of Ecology and Evolution, University of Chicago, Chicago, IL, USA
Patrick Abbot, Department of Biological Sciences, Vanderbilt University, Nashville, TN, USA.
Noah K. Whiteman, Department of Integrative Biology, University of California, Berkeley, CA, USA
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Simon J-C, d’Alencon E, Guy E, Jacquin-Joly E, Jaquiery J, Nouhaud P, Peccoud J, Sugio A, Streiff R: Genomics of adaptation to host-plants in herbivorous insects. Briefings Func Gen 2015, 14:413–423. [DOI] [PubMed] [Google Scholar]
- 2. **.Gloss AD, Nelson Dittrich AC, Lapoint RT, Goldman Huertas B, Verster KI, Pelaez JL, Nelson ADL, Aguilar J, Armstrong E, Charboneau JLM, Groen SC, Hembry DH, Ochoa C, O’Connor TK, Prost S, Suzuki HC, Zaaijer S, Nabity PD, Whiteman NK. Evolution of herbivory remodels a Drosophila genome. bioRxiv 2019, doi: 10.1101/767160. [DOI] [Google Scholar]; A high-quality genome annotation of the herbivorous S. flava allowed a fine-grained study on the association between the relatively recent evolution of herbivory in an insect and the evolution of its chemosensory and detoxification gene families. Functionally important genes bloomed through recurrent duplications against a backdrop of accelerated gene loss. Functional validation of GSTs in vitro revealed that five recently duplicated paralogs were neofunctionalized to detoxify mustard oils.
- 3. **.Goldman-Huertas B, Mitchell RF, Lapoint RT, Faucher CP, Hildebrand JG, Whiteman NK. Evolution of herbivory in Drosophilidae linked to loss of behaviors, antennal responses, odorant receptors, and ancestral diet. Proc Natl Acad Sci 2015, 112:3026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]; A loss of attraction to yeast volatiles was associated with a loss of peripheral perception of yeast volatiles and a stepwise loss of canonical yeast-activated odorant receptors in Scaptomyza, across the phylogeny, leading to herbivory in S. flava.
- 4. *.Yassin A, Debat V, Bastide H, Gidaszewski N, David JR, Pool JE: Recurrent specialization on a toxic fruit in an island Drosophila population. Proc Natl Acad Sci 2016, 113:4771–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]; Parallel evolutionary genetic changes in many of the same genes involved in chemosensation and detoxification were discovered in an island population of D. yakuba that became independently adapted to the toxic noni fruit, when compared to the other independently adapted D. sechellia. At another level of convergence, Osiris genes were found to be involved in the response to toxins in the noni in these two species, as well as to mustard oils in S. flava. This suggests that Osiris genes may be commonly recruited during host shifts and the evolution of herbivory.
- 5. **.Rane RV, Clarke DF, Pearce SL, Zhang G, Hoffmann AA, Oakeshott JG: Detoxification genes differ between cactus-, fruit-, and flower-feeding Drosophila. J. Hered 2019, 110:80–91. [DOI] [PubMed] [Google Scholar]; Detailed gene loss/gain and positive selection analyses across Phase I, II and III detoxification gene families revealed a potential role of herbivory, in the case of D. suzukii, which feeds on living fruits, and toxin adaptation, in the case of D. sechellia in increasing overall detoxification gene number. However, contra to the high levels of toxins found in cacti, the cactophilic Drosophila were relatively depauperate in this regard. This illustrates the challenges of comparing absolute numbers vs. turnover rates in illuminating evolutionary genomic changes associated with trophic shifts and specialization towards toxic diets.
- 6. **.Seppey M, Ioannidis P, Emerson BC, Pitteloud C, Robinson-Rechavi M, Roux, Escalona HE, McKenna DD, Misof B, Shin S, et al. : Genomic signatures accompanying the dietary shift to phytophagy in polyphagan beetles. 2019, doi: 10.1186/s13059-019-1704-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ten adephagan (mostly predaceous) transcriptomes and eight polyphagan genomes and one polyphagan transcriptome (mostly herbivorous) were sequenced and annotated, along with a transcriptome of a strepsipteran outgroup. Gene family expansions, particularly those involved in detoxification and nutrient acquisition were detected in a phylogenetic context during the diversification of the polyphagan species, especially in curculionids and chrysomelids. The evolution of herbivory per se was not isolated as a variable, although its signatures could contribute to the patterns observed.
- 7.Hughes GM, Boston ESM, Finarelli JA, Murphy WJ, Higgins DG, Teeling EC: The birth and death of olfactory receptor gene families in mammalian niche adaptation. Mol. Biol. Evol 2018, 35:1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poore AGB, Ahyong ST, Lowry JK, Sotka EE: Plant feeding promotes diversification in the Crustacea. Proc Natl Acad Sci 2017, 114:8829–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. *.Almeida FC, Sánchez-Gracia A, Campos JL, Rozas J: Family size evolution in Drosophila chemosensory gene families: A comparative analysis with a critical appraisal of methods. Genome Biol Evol 2014, 6:1669–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chemosensory gene birth and death analyses are found to be sensitive to the method used and to rate heterogeneity. A particular outlier is D. sechellia. This suggests care must be taken to achieve robust analyses, including sampling and analytical methodology.
- 10. *.Hecker N, Sharma V, Hiller M: Convergent gene losses illuminate metabolic and physiological changes in herbivores and carnivores. Proc Natl Acad Sci 2019, 116:3036–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed genomic studies of herbivorous and carnivorous mammals revealed convergent losses within each guild, of genes with function in nutrient acquisition. Genes associated with detection of xenobiotics (from plant compounds) were lost independently in carnivores. This suggests that loss of function is an important locus of adaptation in the evolution of diet shifts in both vertebrates and arthropods.
- 11.Hughes GM, Boston ESM, Finarelli JA, Murphy WJ, Higgins DG, Teeling EC. The birth and death of olfactory receptor gene families in mammalian niche adaptation. Mol Biol Evol 2018, 35:1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang P, Josue J, Li X, Glaser D, Li W, Brand JG, Margolskee RF, Reed DR, Beauchamp GK: Major taste loss in carnivorous mammals. Proc Natl Acad Sci 2012, 109:4956–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res 2010, 20:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. *.Gloss AD, Vassão DG, Hailey AL, Nelson Dittrich AC, Schramm K, Reichelt M, et al. Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the Drosophilidae. Mol Bio Evol 2014, 31:2441–56. [DOI] [PMC free article] [PubMed] [Google Scholar]; The evolution of herbivory in Scaptomyza was associated with a shift to mustard oil producing plants in the Brassicales. Drosophilids were found, for the first time (including D. melanogaster) to use the mercapturic acid pathway to detoxify isothiocyanates. Adaptive evolution occurred in GST enzymes catalyzing the first step of this pathway in herbivorous Scaptomyza. Adaptation involved both amino acid substitutions and gene duplication of a candidate gene, GstD1. In vitro assays of GSTD1 showed that enzymes from herbivorous species outperformed those from microbe-feeding species. A handful of key amino acid residues were identified by solving the crystal structure from herbivorous species and this information was used, with site directed mutagenesis followed by functional studies, to swap activities between the D. melanogaster and Scaptomyza orthologs.
- 15.Zunjarrao SS, Tellis MB, Joshi SN, Joshi RS. Plant-Insect Interaction: The Saga of Molecular Coevolution In: Merillon JM, Ramawat K. (eds) Co-Evolution of Secondary Metabolites. 2019. Reference Series in Phytochemistry. Springer, Cham [Google Scholar]
- 16.Cheng T, Wu J, Wu Y, Chilukuri RV, Huang L, Yamamoto K, Feng L, Li W, Chen Z, Guo H, et al. : Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol 2017, 1:1–10. [DOI] [PubMed] [Google Scholar]
- 17.Gouin AXS, Bretaudeau A, Nam K, Gimenez S, Aury J-M, Duvic B, Hilliou FXDXR, Durand N, x000E9 NM, Darboux I, et al. : Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci Rep 2017, doi: 10.1038/s41598-017-10461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. **.Rane RV, Ghodke AB, Hoffmann AA, Edwards OR, Walsh TK, Oakeshott JG: Detoxifying enzyme complements and host use phenotypes in 160 insect species. Curr Opin Ins Sci 2019, doi: 10.1016/j.cois.2018.12.008. [DOI] [PubMed] [Google Scholar]; 160 insect species with genome sequences were screened for genes encoding esterases, CYP450s and GSTs. Insects feeding on complex plant diets and specialists tended to have more of these gene copies, and insects feeding on nutritionally poor diets tended to have fewer, as did generalist species.
- 19.Luca F, Perry GH, Di Rienzo A: Evolutionary adaptations to dietary changes. Annu. Rev. Nutr 2010, 30:291–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart CB, Schilling JW, Wilson AC: Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature 1987, 330:401–404. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J: Parallel functional changes in the digestive RNases of ruminants and colobines by divergent amino acid substitutions. Mol. Biol. Evol 2003, 20:1310–1317. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y, Wu Q, Ma S, Ma T, Shan L, Wang X, Nie Y, Ning Z, Yan L, Xiu Y, et al. : Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc Natl Acad Sci 2017, 114:1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emerling CA, Delsuc F, Nachman MW: Chitinase genes (CHIAs) provide genomic footprints of a post-Cretaceous dietary radiation in placental mammals. Sci Adv 2018, 4:eaar6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pauchet Y, Heckel DG: The genome of the mustard leaf beetle encodes two active xylanases originally acquired from bacteria through horizontal gene transfer. Proc the Roy Soc B 2013, 280:20131021–20131021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGowen MR, Clark C, Gatesy J: The vestigial olfactory receptor subgenome of odontocete whales: phylogenetic congruence between gene-tree reconciliation and supermatrix methods. Syst Biol 2008, 57:574–590. [DOI] [PubMed] [Google Scholar]
- 26.Feng P, Zheng J, Rossiter SJ, Wang D, Zhao H. Massive losses of taste receptor genes in toothed and baleen whales. Gen Biol Evol 2014, 6: 1254–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y, Wu Q, Ma S, Ma T, Shan L, Wang X, et al. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc Natl Acad Sci 2017, 114:1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandaranayake PCG, Yoder J. 2013. Evolutionary origins In: Joel DM, Gressel J, Musselman LJ, editors. Parasitic Orobanchaceae—parasitic mechanisms and control strategies. Heidelberg (Germany): Springer; p. 69–70. [Google Scholar]
- 29.Yang Z, Wafula EK, Honaas LA, Zhang H, Das M, Fernandez-Aparicio M, Huang K, Bandaranayake PCG, Wu B, Der JP, et al. : Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol. Biol. Evol 2014, 32:767–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. *.Whiteman NK, Gloss AD, Sackton TB, Groen SC, Humphrey PT, Lapoint RT, Sønderby IE, Halkier BA, Kocks C, Ausubel FM, et al. : Genes involved in the evolution of herbivory by a leaf-mining, Drosophilid fly. Genome Biol Evol 2012, 4:900–916. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gene expression for hundreds of loci is differentially regulated by dietary mustard oils in the mustard specialist S. flava. These loci are also evolving more rapidly, are enriched for novel genes, and include Osiris genes involved in resistance to plant toxins in distantly related D. yakuba and D. sechellia.
- 31.True JR, Carroll SB: Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol 2002, 18:53–80. [DOI] [PubMed] [Google Scholar]
- 32. *.Heidel-Fischer HM, Kirsch R, Reichelt M, Ahn S-J, Wielsch N, Baxter SW, Heckel DG, Vogel H, Kroymann J: An insect counteradaptation against host plant defenses evolved through concerted neofunctionalization. Mol. Biol. Evol 2019, 77:16–12. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gene duplication and neofunctionalization was studied from an evolutionary and functional perspective in Plutella xylostella and relatives, which are moth specialists on mustards. Genes encoding desulfating enzyme genes have been duplicated and diverged multiple times in this lineage and now have overlapping but also distinct glucosinolate substrates. A potentially new fate for gene duplication is proposed.
- 33.Gardiner A, Barker D, Butlin RK, Jordan WC, Ritchie MG: Drosophila chemoreceptor gene evolution: selection, specialization and genome size. Mol Ecol 2008, 17:1648–1657. [DOI] [PubMed] [Google Scholar]
- 34.Robertson HM: Molecular evolution of the major arthropod chemoreceptor gene families. Ann Rev Entomol 2019, 64:227–242. [DOI] [PubMed] [Google Scholar]
- 35.Engsontia P, Sangket U, Chotigeat W, Satasook C: Molecular evolution of the odorant and gustatory receptor genes in lepidopteran insects: Implications for their adaptation and speciation. J Mol Evol 2014, 79:21–39. [DOI] [PubMed] [Google Scholar]
- 36. *.Ramasamy S, Ometto L, Crava CM, Revadi S, Kaur R, Horner DS, Pisani D, Dekker T, Anfora G, Rota-Stabelli O: The evolution of olfactory gene families in Drosophila and the genomic basis of chemical-ecological adaptation in Drosophila suzukii. Genome Biol Evol 2016, 8:2297–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]; Combined a very detailed evolutionary genomics with neurophysiology to understand subtle patterns of odorant receptor evolution in the recently-derived frugivore D. suzukii. Remarkably, Or22a function was lost in this lineage, as in Scaptomyza, in association with the evolution of frugivory. Or22a is tuned to yeast-associated aliphatic esters, present in rotting, but not ripe fruit.
- 37.Calla B, Noble K, Johnson RM, Walden KKO, Schuler MA, Robertson HM et al. Cytochrome P450 diversification and host plant utilization patterns in specialist and generalist moths: Birth, death and adaptation. Mol Ecol. 2017, 26:6021–6035. [DOI] [PubMed] [Google Scholar]
- 38.McBride CS: Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proceedings of the National Academy of Sciences of the United States of America 2007, 104:4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardiner A, Barker D, Butlin RK, Jordan WC, Ritchie MG. Drosophila chemoreceptor gene evolution: selection, specialization and genome size. Mol Ecol 2008, 17:1648–1657. [DOI] [PubMed] [Google Scholar]
- 40.Feyereisen R. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta 2011, 1814:19–28. [DOI] [PubMed] [Google Scholar]
- 41.Kawashima A, Satta Y. Substrate-dependent evolution of cytochrome P450: rapid turnover of the detoxification-type and conservation of the biosynthesis-type. PLoS One 2014, 9:e100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. **.Johnson RM, Harpur BA, Dogantzis KA, Zayed A, Berenbaum MR. Genomic footprint of evolution of eusociality in bees: floral food use and CYPome “blooms”. Insectes Soc. 2018, 65:445–454. [Google Scholar]; Comparing CYP450 repertoires across 10 bee species, including polymorphism and divergence based tests for positive selection, revealed the CYP6AS subfamily expanded and was involved in the shift from predation to florivory and exposure to plant chemicals, including flavonoids.
- 43.Jones CD. The genetic basis of Drosophila sechellia’s resistance to a host plant toxin. Genetics 1998, 149: 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith CR, Morandin CM, Noureddine M, Pant S. Conserved roles of Osiris genes in insect development, polymorphism, and protection. J Evol Biol 2018, 31: 516–529. [DOI] [PubMed] [Google Scholar]
- 45.Le Goff G, Hilliou F. Resistance evolution in Drosophila: the case of CYP6G1. Pest Manag Sci 2017, 73:493–499. [DOI] [PubMed] [Google Scholar]
- 46.Demuth JP, Hahn MW: The life and death of gene families. BioEssays 2009, 31:29–39. [DOI] [PubMed] [Google Scholar]
- 47.Wiegmann B, Trautwein M, Winkler I, Barr N, Kim JW, Lambkin C, Yeates D. Episodic radiations in the fly tree of life. Proc Natl Acad Sci. 2011, 108:5690–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin NA. Scaptomyza (Bunostoma) flavella (Diptera: Drosophilidae) and the evolution of leaf mining. The Weta 2014, 47:8–12. [Google Scholar]
- 49.Gompert Z, Mandeville L, Buerkle CA. Analysis of population genomic data from hybrid zones. Ann Rev Ecol Syst 2017, 48:207–229. [Google Scholar]
- 50.Nallu S, Hill J, Don K, Sahagun C, Zhang W, Meslin C, Snell-Rood E, Clark N, Morehouse N, Bergelson J, Wheat C, Kronforst MR. The molecular genetic basis of herbivory between butterflies and their host-plants. Nature Ecol Evol 2018, 2: 1418–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gagné RJ. The Plant-feeding Gall midges of North America. 1989. Cornell University Press, Ithaca, NY. [Google Scholar]
- 52.Conant GC, Wolfe KH: Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet 2008, 9:938–950. [DOI] [PubMed] [Google Scholar]
- 53.Konrad A, Flibotte S, Taylor J, Waterston RH, Moerman DG, Bergthorsson U, Katju V: Mutational and transcriptional landscape of spontaneous gene duplications and deletions in Caenorhabditis elegans. Proceedings of the National Academy of Sciences 2018, 115:7386–7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storz JF. Gene duplication and the resolution of adaptive conflict. J. Hered 2009, 102:99–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Misof B, Liu SL, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, et al. : Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346:763–767. [DOI] [PubMed] [Google Scholar]
- 56.Wiens JJ, Lapoint RT, Whiteman NK: Herbivory increases diversification across insect clades. Nature Communications 2015, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gagné RJ: Chapter 16: Cecidomyiidae. In Manual of Nearctic Diptera. Volume 1 Edited by McAlpine JF, Peterson BV, Shewell GE, Teskey HJ, Vockeroth JR, Wood DM. Agriculture Canada Monograph, 27: 1981: 257–292. [Google Scholar]


