Fusion, fission, and formation of complex shapes of droplets made of DNA were controlled by a sequence design approach.
Abstract
DNA has the potential to achieve a controllable macromolecular structure, such as hydrogels or droplets formed through liquid-liquid phase separation (LLPS), as the design of its base sequence can result in programmable interactions. Here, we constructed “DNA droplets” via LLPS of sequence-designed DNA nanostructures and controlled their dynamic functions by designing their sequences. Specifically, we were able to adjust the temperature required for the formation of DNA droplets by designing the sequences. In addition, the fusion, fission, and formation of Janus-shaped droplets were controlled by sequence design and enzymatic reactions. Furthermore, modifications of proteins with sequence-designed DNAs allowed for their capture into specific droplets. Overall, our results provide a platform for designing and controlling macromolecular droplets via the information encoded in component molecules and pave the way for various applications of sequence-designed DNA such as cell mimics, synthetic membraneless organelles, and artificial molecular systems.
INTRODUCTION
Liquid-liquid phase separation (LLPS) of water-soluble molecules induces the formation of macromolecular droplets in water. In polymer chemistry and soft matter physics, LLPS has been studied to elucidate the underlying mechanisms of phase separation (1–3) and to construct cell-like structures (4) or bioreactors (5). With recent advances in LLPS in biological research fields, studies have found that macromolecular droplets composed of proteins and RNAs are formed within living cells via moderate interactions among the biomolecules (6–10). LLPS of biomolecules has garnered considerable attention, as the droplets have an important role in regulating various biological functions (11). Although several in vitro studies have reproduced droplet formation by changing the concentrations of proteins/RNAs and salts (8, 9, 12, 13), no study has demonstrated the control of dynamic behavior of macromolecular droplets by varying the biological information encoded in the biopolymer sequence. The sequence of biopolymers plays an essential role in determining intra- and intermolecular interaction strength (14). However, the potential to regulate the formation and behavior of macromolecular droplets via the sequence design of biopolymers remains less explored. It is expected that such a technology would provide insights into not only mechanisms of droplet formation via LLPS but also applications for droplets made of biopolymer molecules.
DNA can be used as a programmable material by designing the base sequences because the self-assembly of DNA is precisely predictable from sequence information; however, this controllability has not been translated into the ability to control the dynamic behaviors and functions of droplets formed via LLPS. In the field of DNA nanotechnology, arbitrarily shaped nanostructures have been constructed using sequence-designed DNA (15–20). The nanostructures can be further assembled into micrometer-sized structures. For example, a DNA hydrogel can be formed via the self-assembly of branched-DNA nanostructures (called motifs or nanostars) (21–25), and it can serve as a substrate for cell-free protein expression (21), cytoskeleton for artificial cells (22), or carrier for drug delivery (23). Moreover, several studies have reported the phase behavior of DNA nanostructures, such as the phase separation of DNA nanostructures into DNA-poor and DNA-rich phases (26), formation of a dense DNA liquid or droplets (27, 28), and reentrant phase dynamics (29). These studies focused on the effects of temperature, DNA concentration, and ionic strength on the LLPS of DNA nanostructures; however, control of DNA-based droplets, formed via phase separation, by designing base sequences has not yet been achieved.
We herein report the control of a “DNA droplet” made of sequence-designed DNAs (Fig. 1). We exploit DNA nanostructures (Fig. 1A) to control the droplet formation via LLPS depending on the surrounding temperature (Fig. 1B) and the designed sequence. We demonstrate that control of the dynamic functions of DNA droplets, including fusion, fission, and segregation of DNA droplets, can be achieved via rational design of DNA sequences (Fig. 1C). Furthermore, as a potential application of our DNA droplet technology, we demonstrate the capture and partitioning of protein cargos in the DNA droplets in a sequence-specific manner (Fig. 1D).
Fig. 1. Droplet formation of DNA nanostructures and sequence-dependent control of DNA droplets.
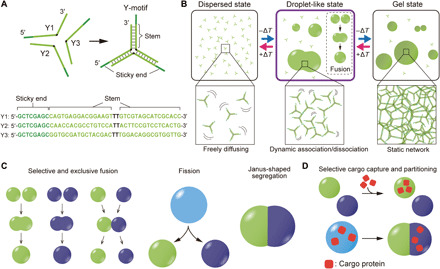
(A) Schematic of the Y-shaped DNA nanostructure (Y-motif) and strand sequences of the Y-motif. (B) Temperature-dependent behavior of the Y-motifs. The Y-motifs are dispersed in solution at a high temperature (“dispersed state”). Decreasing the temperature induces the LLPS of Y-motifs into a “droplet-like state” in which the assembled Y-motifs exhibit liquid droplet-like behavior such as fusion. By further decreasing the solution temperature, the droplet-like state changes to a “gel state.” The temperature for the LLPS is dominated by the sequences of the DNAs. (C) Schematic representations for the control of DNA droplet behavior. (D) Selective cargo capture and partitioning into DNA droplets in a sequence-specific manner.
RESULTS
Formation of DNA droplets via phase separation of DNA nanostructures
We designed a Y-shaped DNA nanostructure (hereafter referred to as a Y-motif) with three “sticky ends” (SEs) (Fig. 1A) (22). The Y-motif is composed of three single-stranded DNAs (ssDNAs): Y1, Y2, and Y3 (Fig. 1A). The “stems” of the Y-motif have 16 base pairs that hybridized at 75°C in a buffer condition [20 mM Tris-HCl (pH 8.0), 350 mM NaCl] (fig. S1); the SE was a palindromic 8–nucleotide (nt) sequence, and the same was true for the three branches of the Y-motif, allowing for connections among the Y-motifs.
We observed the Y-motif solution using a confocal laser scanning microscope and a stage heater. The Y-motifs were stained with a dye molecule (SYBR Gold). At 70°C, the Y-motifs were dispersed in a buffer solution (“dispersed state”) (Fig. 1B), and no micrometer-sized structures were observed owing to dehybridization of the SE at this temperature (Fig. 2A, a). When the temperature was lowered to 62°C, spherical microparticles (from several to several tens of micrometers) were observed in the solution, although the temperature was still much higher than the melting temperature (Tm) of the 8-nt SE (Tm, 45.9°C) (Fig. 2A, b). These particles may have been formed through a nucleation and growth process (1). Fusion of the two particles occurred when they collided with each other (Fig. 2, B and C, and movie S1); the fusion behavior is reminiscent of that of the droplets formed via LLPS in living cells (6). We called the particles that exhibit such fusion “DNA droplets,” where Y-motifs spontaneously phase separate into Y-motif–rich liquid droplets in equilibrium with a Y-motif–poor phase (26) (“droplet-like state”) (Fig. 1B). The DNA droplets grew in size via collision with one another (figs. S2 and S3). The fusion is caused by the complementation of the SE and the surface tension of the DNA droplets (13). Fluorescence recovery after photobleaching (FRAP) experiments (Fig. 2, D and E) showed immediate recovery of the fluorescence intensity, suggesting that the Y-motifs may diffuse inside droplets (do not form static networks) and that the dynamic dissociation and association of the Y-motifs result in the formation of the droplet. This recovery degree, representing the dissociation/association, gradually decreased with temperature (fig. S4). Two types of LLPS have mainly been reported: LLPS of high-concentration polymer blends such as polyethylene glycol/dextran mixture, and LLPS by aggregation of low-concentration macromolecules such as protein droplets. The phase separation of DNA nanostructures in our system would be considered to be the latter phenomenon because the hybridization of SEs of low-concentration DNA nanostructures induced the DNA droplet formation.
Fig. 2. Microscopic observation of DNA droplets and hydrogels.
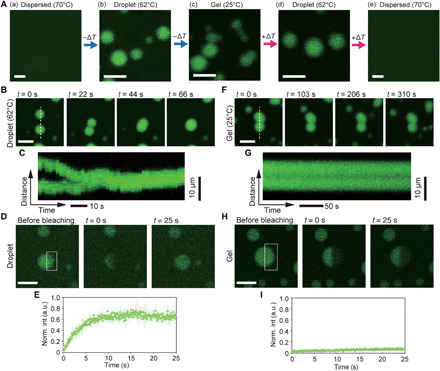
(A) Microscopic images for reversible state changes in Y-motifs with 8-nt SEs between the dispersed, droplet-like, and gel states, in response to the surrounding temperature. (B and C) Sequential images for DNA droplets (B) at 62°C, and kymographs along the dashed line shown in the image at t = 0 s (C). (D and E) Image sequences of the FRAP experiments (D) and time series of the fluorescence intensity (E) of the DNA droplets at 60°C. The bleached region is indicated by a white box in the image. Bars show the mean ± SD. The number of measurements (n) = 3. a.u., arbitrary unit. (F and G) Sequential images for DNA hydrogels (B) at 25°C, and kymographs along the dashed line shown in the image at t = 0 s (G). (H and I) Image sequences of FRAP experiments (H) and time series of the fluorescence intensity (I) of DNA hydrogels at 30°C. Bars show the mean ± SD. The number of measurements (n) = 3. Scale bars, 100 μm (A, a and e), 10 μm (other images).
When the temperature was further decreased to 25°C (Fig. 2A, c), the microparticles did not fuse (Fig. 2, D and E) during the observation period (~5 min). In addition, in the FRAP experiments, the fluorescence intensity did not immediately recover at around this temperature (Fig. 2, H and I). Thus, the Y-motifs formed static networks, and the DNA droplets became Y-motif–rich hydrogels (“gel state”) (Fig. 1B).
For the formation of DNA droplets, the range of the concentrations of DNA nanostructures is important. In general, the phase separation into the Y-motif–rich droplet/gel phase and the Y-motif–poor phase does not occur at very high concentrations of Y-motifs, even with decreasing temperatures, because the entire solution of Y-motifs homogeneously gelates; for instance, a previous microrheological study using 500 μM DNA nanostructures reported the gradual transition from a dilute DNA nanostructure solution into a hydrogel (25). In contrast, a very dilute Y-motif solution, of course, does not transition into a droplet/hydrogel. In the case of the middle range of the concentrations of DNA nanostructures, by decreasing the temperature, DNA droplets are formed in the DNA nanostructure solution in a discontinuous manner (i.e., the phase separation occurs), and, lastly, the DNA droplet gradually transitions into a hydrogel by losing its fluidity; for example, our study observed such phase behavior using a 5 μM concentration of DNA nanostructures (fig. S5), and Saleh and co-workers (28) have reported a 10 μM concentration of DNA nanostructures that phase separated into spherical liquid droplets after incubation at room temperature for 1 hour.
We further found that the droplet formation was reversible. The hydrogels changed into droplets when temperatures increased to 62°C (Fig. 2A, d), and the Y-motifs dispersed at 70°C (Fig. 2A, e). This temperature-dependent droplet formation was confirmed with further continuous temperature changes (fig. S6).
Effects of sequence and number of branches on the state changes
We next investigated the effects of the SE sequences on the “state-change temperatures” between the dispersed and droplet-like states (Td) and between the droplet-like and gel states (Tg). We defined Td as the temperature at which droplet formation was observed (fig. S5), and Tg as the temperature at which a reduced recovery of fluorescence was detected in the FRAP experiment (fig. S4). Td and Tg were identified by decreasing the temperature from 85°C at a rate of −1°C/min. The state diagram (Fig. 3A) was experimentally constructed for the Y-motifs with 4-, 6-, 8-, 10-, and 12-nt SEs (Fig. 3B and table S1). Enthalpy changes in the hybridization of SEs and Tm were obtained by numerical calculation using the web-based software NUPACK (30) and DINAMelt (31). The state diagram showed that a larger –ΔH (i.e., longer sticky-end length) tended to result in increases in both Td (solid line in Fig. 3A) and Tg (dashed line in Fig. 3A). This means that the interaction strength among the motifs, i.e., the sequence of the SEs, has an important role in the assembly of the motifs, resulting in the formation of the droplets/hydrogels. Note that droplet formation was observed for the Y-motifs whose SEs were 4 to 12 nt long (fig. S7), but not for those with 2-nt SEs (ΔH = −9.6 kcal/mol) (fig. S8). Considering the formation process of the droplets, SEs should interact after the formation of a motif. Thus, to achieve a droplet formation in the Y-motif under our experimental conditions, SE sequences ought to have parameters within the range of 9.3°C (Tm of the SE for 4 nt) < Tm < 75.0°C (folded temperature of the Y-motif).
Fig. 3. SE-dependent state change of DNA nanostructures.
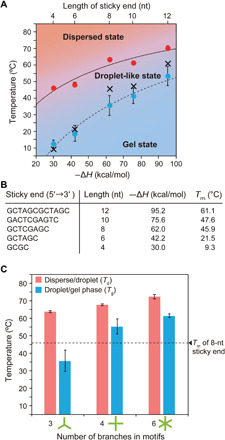
(A) State diagram of Y-motifs with different lengths of SEs. Red and blue dots represent mean values of the state-change temperatures between the dispersed and droplet-like states (Td), and between the droplet and gel states (Tg), respectively. Cross-marks in the diagram show melting temperatures (Tm) of each SE calculated under conditions of 15 μM Y-motifs and 350 mM Na+ ions (corresponds to the experimental condition). Error bars indicate SD (mean ± SD, n = 3). Solid and dashed lines are guides. (B) List of SE sequences, lengths, enthalpy changes in the hybridization of SEs (–ΔH), and Tm. (C) Td and Tg of a motif with different numbers of branches. Error bars indicate SD (mean ± SD, n = 3). Length of the SE was fixed at 8 nt (–ΔH, 62.0 kcal/mol, Tm, 45.9°C). Horizontal dashed line indicates the Tm of 8-nt SE.
We also investigated the effect of the number of branches in the motifs on the Td and Tg. We additionally designed two motifs that had four or six branches with 8-nt SEs (table S2). The experimental results showed that the Td and Tg increased with the number of branches in the motifs (Fig. 3C and figs. S9 to S11).
The fact that the DNA droplets were formed at higher temperatures than the Tm of the SEs suggests that unstable hybridization between the SEs plays a key role in the formation of the DNA droplets. Such weak interactions of the SEs would allow for the dynamic association/dissociation of the motifs, which leads to the fluidic behavior of DNA droplets, e.g., fusion of the droplets (Fig. 2, B and C). In contrast, an increase in the number of branches in a motif resulted in an increase in Td (Fig. 3C). Thus, both the stability of hybridization in the SEs and the number of branches in a motif determine Td.
We considered that the Tg was attributed to not only the stability of the SEs but also to the multiple bonding points of the SEs. Previous studies have reported that the gelation process in the branched-DNA nanostructures occurs at around the Tm of SEs (25) and the concentration of the DNA changes the Tm. We confirmed that Y-motifs were concentrated into the hydrogels by phase separation (figs. S12 and S13), which resulted in an increase in the Tm of the SEs. In addition, an increase in the number of branches in the motifs increased the structural stability due to an increase in the number of binding points among motifs, which also decreased the mobility of the motifs and increased the Tg (Fig. 3C). Further discussion regarding the Td and Tg can be found in the Supplementary Discussion (see also figs. S12 to S20).
Orthogonality in DNA droplets resulting in selective and exclusive fusion
We expected that the rational design of the motif sequence would allow for the control of the droplet-droplet interactions. Because DNA is an anionic polymer, two different motifs with noncomplementary SEs cannot hybridize due to an electrical repulsion. This will induce the formation of two immiscible types of droplets. We designed an “orthogonal Y-motif” with an 8-nt SE (orthY-motif); the sequence of this motif was not complementary to the Y-motif with the 8-nt SE, but they had almost identical thermodynamic parameters (ΔH and Tm) in their stem and SEs (tables S3 and S4). The orthogonality of the SE sequences between the Y- and orthY-motifs enabled us to assess the sequence specificity of the fusion of DNA droplets (Fig. 4A). The Y- and orthY-motifs were labeled with fluorescein amidite (FAM) (green) and Alexa405 (blue), respectively. Then, all strands for both of the motifs were mixed in a solution, and the motifs were annealed on the stage heater from 85°C at a rate of −1°C/min to 60°C.
Fig. 4. Sequence-dependent fusion control in DNA droplets.
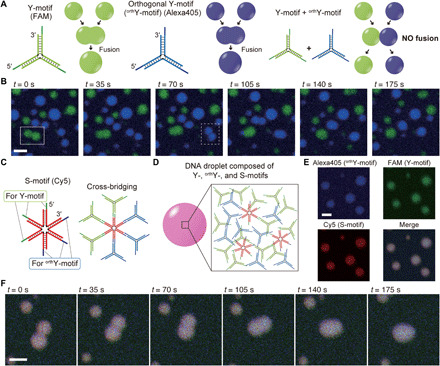
(A) Schematic illustrations of the selective fusion in the Y- and orthogonal Y-motifs (orthY-motif). (B) Time series of microscopic images representing fusion events for the droplets composed of the Y-motifs (box with solid line at t = 0 s) and orthY-motifs (box with dashed line at t = 70 s). Blue and green channels indicate orthY- (Alexa405) and Y-motifs (FAM), respectively. (C) Six-junction motif (S-motif) having SEs for the Y- and orthY-motifs. (D) Schematic of DNA droplets composed of the Y-, orthY-, and S-motifs. (E and F) Microscopic images and time series of the fusion of DNA droplets composed of the Y-, orthY-, and S-motifs. Scale bars, 10 μm.
As expected, the Y- and orthY-motifs were individually phase separated into droplets. Fusion events were observed between the droplets composed of the same motifs but not between the droplets formed by different motifs (Fig. 4B and movie S2). To confirm that the selective and exclusive fusion was derived from the sequence orthogonality, we further designed a six-junction motif (S-motif) with two types of SEs corresponding to the SEs of the Y- and orthY-motifs (Fig. 4C and table S5). The S-motifs are involved in cross-bridging between the Y- and orthY-motifs and can eliminate the orthogonality between the two types of motifs, resulting in the formation of DNA droplets composed of Y-, orthY-, and S-motifs (Fig. 4D). The Cy5-labeled S-motif, FAM-labeled Y-motif, and Alexa405-labeled orthY-motif were mixed in a 1:3:3 molar ratio and visualized at 65°C. In the experiment, fluorescence derived from Cy5, FAM, and Alexa405 was observed in a single droplet, suggesting that the Y- and orthY-motifs were mixed via the S-motifs in the droplets (Fig. 4E). The elimination of orthogonality was also confirmed in a two-dimensional histogram of FAM/Alexa405 fluorescence intensities (fig. S21). In addition, the droplets composed of Y-, orthY-, and S-motifs exhibited fusion (Fig. 4F), similar to the droplets composed of a single motif. Thus, selective fusion of the DNA droplets was achieved in two motifs that were orthogonal, and the addition of a motif capable of cross-bridging eliminated the selectivity.
Fission and segregation of DNA droplets
To further demonstrate the controllability of the DNA droplets, we designed a mechanism for their fission (Fig. 5, A and B). DNA-RNA chimera strands were introduced into an S-motif, termed “chimerized-S-motif (CS-motif),” that also has SEs for both the Y- and orthY-motifs (Fig. 5A and table S5). Two RNA regions were located at around the center of the CS-motif, and ribonuclease A (RNase A) was used to degrade the RNA parts. The enzymatic reaction resulted in splitting of the CS-motif into two three-branched portions with affinity to the Y-motif (“Y-portion”) and the orthY-motif (“orthY-portion”) (Fig. 5A and fig. S22). After splitting, the cross-bridging ability of the CS-motif disappeared, and the Y- and orthY-motifs could not be mixed owing to their intrinsic orthogonality. As a result, the Y- and orthY-motifs gradually separated inside the droplets via a mechanism probably based on spinodal decomposition (32), and, lastly, we observed fission into multiple droplets composed of Y-motif/Y-portion or orthY-motif/orthY-portion (Fig. 5B).
Fig. 5. Fission and segregation of DNA droplets based on sequence design and enzymatic reaction.
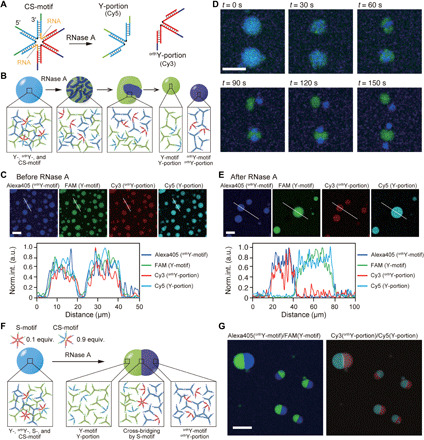
(A) S-motif containing DNA-RNA chimera strands (CS-motif). RNA sequences are introduced at around the center of the CS-motif. Enzymatic reaction of ribonuclease A (RNase A) results splits the CS-motif into two portions, termed Y- and orthY-portions. Y- and orthY-portions were labeled with Cy5 and Cy3, respectively. (B) Fission process of DNA droplets and the motifs that comprise the droplets. (C) Microscopic images of DNA droplets before the addition of RNase A for Alexa405 (orthY-motif), FAM (Y-motif), Cy3 (orthY-portion), and Cy5 (Y-portion). The bottom graph shows the normalized intensity profile of each fluorophore on the line shown in the above images. (D) Time series images of fission of DNA droplets. Channels for Alexa405 (blue) and FAM (green) were merged. (E) Microscopic images of DNA droplets and line profile for each fluorophore after the addition of RNase A. (F) Schematic of Janus-shaped DNA droplet. At the middle of the Janus DNA droplet, a few S-motifs cross-bridge the immiscible motif groups (Y-motifs/Y-portions and orthY-motifs/orthY-portions). (G) Microscopic images for Alexa405/FAM (blue/green) and Cy3/Cy5 (red/cyan) channels in the Janus-shaped DNA droplets. Scale bars, 20 μm (D), 30 μm (other images).
The Y-, orthY-, and CS-motifs formed droplets, representing the mixing of the Y- and orthY-motifs via the CS-motifs (Fig. 5C). Real-time monitoring of the RNase-induced split of the CS-motif allowed visualization of the fission of the DNA droplets (Fig. 5D and movie S3). After the addition of RNase A to the sample solution during the experiment, the Y- and orthY-motifs were gradually separated inside the droplets (t = 30 s in Fig. 5D and fig. S23), and we, lastly, observed complete fission into two or three droplets (t = 150 s in Fig. 5D and fig. S24). Line profiles for each fluorophore showed the colocalization of Y-motifs and Y-portions, and orthY-motifs and orthY-portions (Fig. 5E), indicating the successful split of the CS-motifs as designed. The addition of only the buffer solution did not induce such fission behavior (fig. S25).
By using S-motifs (noncleavable by RNase A) in addition to CS-motifs (cleavable by RNase A), we demonstrated the formation of complex-shaped droplets. When the proportion of CS-motifs was high but that of S-motifs was low, addition of RNase A did not induce fission but caused the segregation of the droplets, owing to the immiscibility and the partial cross-bridging by a few S-motifs (Fig. 5F). Experimental results showed that at a 90% ratio of CS-motifs, the DNA droplets segregated and, lastly, formed Janus-shaped droplets with both Y- and orthY-motifs in the droplets (Fig. 5G and figs. S26 and S27). Fluorescence of Cy3 (orthY-portions) and Cy5 (Y-portions) colocalized to Alexa405 (orthY-motif) and FAM (Y-motif), respectively. Furthermore, a 50% ratio of CS-motifs resulted in a patchy-like pattern formation in the DNA droplets (figs. S28 and S29). These results showed that the ratio of the S- and CS-motifs determined the shape of the DNA droplets after the addition of the RNase A and indicated that the S-motifs played a role not only by cross-bridging but also for a “surfactant” between the Y- and orthY-motifs.
Selective cargo capture and partitioning in DNA droplets
Combining our sequence-designed DNA droplets with other molecules could provide a basis for additional applications and increase the functionality of the DNA droplets. We focused on the capture of cargo into the DNA droplets via DNA modification. DNA-streptavidin conjugates that had SEs for Y- or orthY-motifs were prepared (Fig. 6A), and the selective capture of streptavidin in the DNA droplets was visualized (Fig. 6B).
Fig. 6. Protein capture and partitioning in DNA droplets.
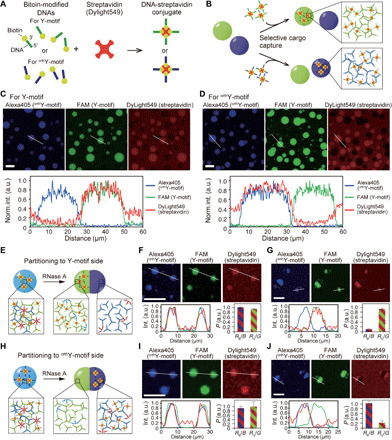
(A) Streptavidin modified with biotinylated-DNAs that have SE sequences for Y- and orthogonal Y-motifs (orthY-motifs). (B) The DNA-modified streptavidin can be captured in DNA droplets in a sequence-dependent manner. (C and D) Microscopic images and line profiles representing localization of streptavidin modified with SEs for Y- (C) or orthY-motifs (D). The bottom graphs show normalized intensity profile of each fluorophore on the line shown in the above images. (E and H) Schematic of partitioning DNA-modified streptavidin to the portion composed of Y- (E) or orthY-motifs (H). (F and G) Microscopic images of DNA droplets and streptavidin modified with SEs for Y-motifs before (F) and after (G) the addition of RNase A. The bottom left graphs show normalized intensity profile of each fluorophore on the line shown in the above images. The bottom right graphs show the partitioning coefficient value, P. RB/B and RG/G indicates the fluorescence intensity ratio of Dylight549 fluorescence (streptavidin) on Alexa405 fluorescence (orthY-motif) and on FAM fluorescence (Y-motif), respectively, on the line profile (detailed methods on how to obtain the P values have been described in Materials and Methods). Error bars indicate SD (mean ± SD, n = 5). (I and J) Microscopic images of DNA droplets and streptavidin modified with SEs for orthY-motifs before (I) and after (J) the addition of RNase A. The bottom left graphs show normalized intensity profile of each fluorophore on the line shown in the above images. The bottom right graphs show P. Error bars indicate SD (mean ± SD, n = 5). Scale bars, 30 μm (C and D), 20 μm, (other images).
DNA-modified streptavidin, labeled with DyLight549 fluorophores, was added during the visualization of DNA droplets composed of Y- or orthY-motifs. The streptavidin was successfully localized in specific DNA droplets, depending on the modified DNA sequence (Fig. 6, C and D). In addition, the accumulation process of the streptavidin into DNA droplets was also visualized (fig. S30).
Moreover, by adopting the technique used to prepare Janus-shaped DNA droplets (Fig. 5F), partitioning of the cargo in DNA droplets was achieved (Fig. 6, E to J). Streptavidin, modified with SE sequences, was homogeneously distributed in DNA droplets in which Y- and orthY-motifs were mixed by the S- and CS-motifs (Fig. 6, F and I). After the addition of RNase A, DNA droplets segregated into the Janus shape, and streptavidin was partitioned into the complementary sides depending on the modified sequence (Fig. 6, G and J). As we have demonstrated here, modification of targets with SE sequences that encode interaction information allows us to capture them in DNA droplets and to control their partitioning in a sequence-dependent manner.
DISCUSSION
Here, we demonstrated the control of dynamic behaviors and functions of DNA droplets according to the information encoded in the SE sequences. Typically, LLPS of biomolecules (droplet formation) is induced via a hydrophobic (7) and/or an electrostatic interaction (10). In contrast, our system achieved the droplet formation based on a sequence design (hybridization of the base pairs), which allowed for an increased controllability of the droplet behaviors. Because we focused on the sequence design to control the DNA droplets, the detailed effects of the motif concentration were not addressed in this study. LLPS is a phenomenon that depends on the concentration of components. Therefore, revealing the mechanism by which the motif concentration influences the formation/behavior of DNA droplets may allow widening of our understanding of LLPS in biomolecules.
Sequence-dependent state changes were reversible between the dispersed, droplet-like, and gel states in response to the surrounding temperature (Fig. 2A). In addition, the temperature range of each state was altered by changing the SE sequence and the number of SE branches (Fig. 3). Longer SEs and more branches narrowed the temperature range for the droplet-like state. We expect that these results will be applicable to other nanostructures such as DNA origami (16), DNA-modified proteins (33), or nanoparticles covered with DNA strands (34). In other words, the state changes shown in the present study could be expanded to matter that can only be modified with a designed DNA molecule. This may enable us to design the state and create droplets for any material under selected conditions. Selective fusion, fission, and segregation of the DNA droplets were demonstrated by the rational design of the SE sequences. Although the present design cannot control the number of droplets after fission, this can be achieved by encapsulating the DNA droplets in a confined space and designing multiple orthogonal motifs.
The controllability of the interaction and dynamic functions of the droplets (Figs. 3 to 5) enables various applications of our DNA droplet system. As living systems are well-organized, dynamic structures whose behavior is regulated by the information encoded in biopolymers (35), our DNA-based LLPS system provides a basis for the development of artificial cell engineering. In addition, our demonstration of the capture of proteins in DNA droplets with sequence specificity (Fig. 6) may make a notable contribution to synthetic biology and/or artificial cell engineering by enabling the generation of a biochemical reaction environment, such as artificial membraneless organelles, composed of designed molecules. Furthermore, DNA droplets that capture arbitrary cargos would be useful as a carrier for drug delivery.
From the viewpoint of material science, the successful creation of the Janus-shaped DNA droplets (Fig. 5G) provides a fundamental technique for constructing water-soluble complex-shaped droplets in an aqueous phase; this is intrinsically different from the conventional creation of Janus droplets (36), which is based on water/oil immiscibility. Our DNA-Janus droplets may be used as a chassis to achieve a chemotaxis-like motion by incorporating metal particles (37). Furthermore, the Janus DNA droplets were created by taking advantage of the surfactant-like role of DNA nanostructures in the cross-bridging of the two types of Y-motifs. This suggests that multiaffinity surfactants could be achieved by the modification of designated molecules with DNAs.
Last, we envision that a combination of our DNA droplet technology and biochemical circuits that can be programmed to produce and decompose specific DNA strands (38–40) will lead to the design and control of autonomous dynamic functions of DNA droplets as in living organisms. Ultimately, such autonomous functions of macromolecular structures could serve as the basis for the development of molecular robotic systems (41, 42), which could become comparable with living cellular systems in the future. We believe that this study provides the basis for a programmable LLPS system that can be controlled by information encoded in component molecules.
MATERIALS AND METHODS
Oligonucleotide preparation
Sequences for the DNA and DNA-RNA chimera strands are provided in tables S1 to S3 and S5. The oligonucleotides were purchased from Eurofins Genomics (Tokyo, Japan) and were of oligonucleotide purification cartridge (OPC)–grade purification, except for the fluorophore-labeled and chimerized strands, which were purified to high-performance liquid chromatography (HPLC) grade. The oligonucleotides were dissolved in ultrapure water (18 megaohms⋅cm in resistance) at 100 μM concentrations and stored at −20°C until use.
Measurement of Tm for the Y-motif
DNA strands for the Y-motifs without SEs were mixed in a test tube at 5 μM of each strand with a buffer consisting of 20 mM Tris-HCl (pH 8.0) and 350 mM NaCl. SYBR Green I (Takara Bio, Kusatsu, Japan) was added at 1× concentration to the test tube to detect the formation of the double helix. The test tubes were heated using a real-time polymerase chain reaction (PCR) detection system (CFX Connect, Bio-Rad, Hercules, CA, USA). The sample was heated at 85°C for 3 min and then cooled to 5°C at a rate of −1°C/min. Subsequently, the temperature was increased from 5° to 85°C at the same rate. The fluorescence intensity was measured at 1°C intervals during the decrease and increase in temperature (fig. S1).
Preparation of the observation chamber
Glasses and coverslips, measuring 30 mm by 40 mm and 18 mm by 18 mm, respectively, with a thickness of 0.17 mm, were purchased from Matsunami Glass (Kishiwada, Japan). The glasses (30 × 40) were treated with oxygen plasma using a plasma cleaning machine (PIB-20, Vacuum Device, Mito, Japan) and were then soaked in 5% (w/v) bovine serum albumin (BSA) and dissolved in 20 mM Tris-HCl (pH 8.0) for over 30 min to prevent nonspecific interactions of the DNA on the glass surface. After the BSA coating, the glasses were washed with distilled water and dried under an airflow. The BSA-coated glasses and the cover slips (18 mm by 18 mm) were assembled using double-sided tape. The sample solutions were immersed into the slit between the coated glass and the cover slips, and the edges were sealed with manicure paste. To avoid evaporation of the sample solution during the observation period, the chamber was further covered with a mineral oil (Nacalai Tesque, Kyoto, Japan) using a bank made of a silicone sheet.
Visualization
Samples in the observation chamber were visualized using a confocal laser scanning microscope (FV1000, Olympus, Tokyo, Japan) and a stage heater [10021-PE120 system, Japan high tech (Linkam), Hukuoka, Japan]. SYBR Gold and FAM were visualized at an excitation wavelength of 473 nm. Alexa405, Cy3, and Cy5 were visualized at 405, 559, and 635 nm, respectively.
Repetitive state change of Y-motifs
DNA strands for the Y-motifs with 8-nt-long SEs were mixed in a test tube with the buffer at 5 μM for each strand. SYBR Gold was also added at 1× concentration to the test tube to stain the DNA. The test tube was heated at 85°C for 3 min and then gradually cooled to 25°C at a rate of −1°C/min using a thermal cycler (Mastercycler Nexus X2, Eppendorf, Hamburg, Germany). The annealed sample was first visualized at 25°C, and then the temperature was changed to 70°C at a rate of 20°C/min. Fluorescence images were then obtained. The temperature was decreased to 62°C, and droplet formation via the phase separation was sequentially visualized at a scanning velocity of 2.2 s per frame. Subsequently, the temperature was further changed to 25°, 62°, 70°, 62°, and 25°C, in this order, at a rate of 20°C/min. The samples were visualized at each temperature. A detailed protocol for the repetitive state change and representative microscopic images for each temperature are shown in fig. S6.
Measurement of the state-change temperatures and FRAP experiments
Each motif was labeled with dye-modified DNA without SEs, which was mixed at a 10% molar ratio in the solution (e.g., 5 μM for Y-1 and Y-3, 4.5 μM for Y-2, and 0.5 μM for Y2_0_FAM). The sample solution was poured into the chamber on the stage heater and incubated at 85°C for 3 min. Subsequently, the temperature was decreased to a designated temperature at a rate of −1°C/min. To determine the temperature at which the DNA droplets were formed, the sample was visualized at each 1°C interval. To evaluate the state change from the droplet-like to the gel state, the fluidity of the motifs in the particles formed by the phase separation was analyzed using FRAP experiments. A rectangular-shaped region in the DNA droplets or hydrogels was photobleached at the respective wavelength for each fluorophore. Sequential images were obtained at a scanning velocity of 65 ms per frame. The average fluorescence intensity values for the fluorescence in the bleached region were measured at designated temperatures.
Concentration estimation
The concentrations of the Y-motifs in the droplets and gel states were estimated by measuring the fluorescence intensity of the supernatant, after centrifugation, that precipitated the DNA droplets and hydrogels. First, the calibration curve for the intensity-based concentration values was prepared using Y-motifs without SEs [composed of Y-1_0, Y-2_0, Y-3_0, and 10% (mol) Y-2_0_FAM]. The Y-motif without an SE was annealed at 5 μM in a 20-μl volume in the PCR test tube with the buffer, using the thermal cycler from 85° to 4°C, at a rate of −1°C/min. The annealed Y-motifs without SEs were then diluted to 100, 10, and 1 nM for a 20-μl volume using the buffer, and the fluorescence intensity was measured using a spectrometer (FP-6500, JASCO, Hachioji, Japan).
Next, the Y-motifs with 4-, 6-, 8-, 10-, and 12-nt SEs were also annealed using the same procedure. After annealing, to precipitate the DNA droplets and hydrogels at the bottom of the test tubes, the PCR tube was placed in a larger test tube as a support during centrifugation (fig. S13A). Then, the tubes were centrifuged at 2000g at 4°C for 20 min. The precipitation of the phase-separated Y-motifs (Y-motif–rich and Y-motif–poor phases) was confirmed using a gel imager (Gel Doc EZ system, Bio-Rad, Hercules, CA, USA) (fig. S13B). The PCR tubes containing the precipitate were set on the thermal cycler. For measuring the concentrations in the droplet and gel states, the temperatures were set at 1°C lower than the Td for each length of the SE or 4°C, respectively. The test tubes were incubated for 10 min at each temperature, and the supernatant was collected using a gel loading tip (BM Equipment, Tokyo, Japan). Ten microliters of supernatant was diluted to 200-μl using the buffer, and the fluorescence intensity was measured. The concentration of Y-motifs was estimated using the calibration curve.
Observations of orthogonality in DNA droplets and its elimination
For the orthogonality experiments, DNA strands were mixed in a test tube with the buffer at the concentrations shown in table S6. This mixture was poured in the observation chamber and set on the stage heater. Then, the temperature was kept at 85°C for 3 min and decreased at a rate of −1°C/min. The fusion events of the DNA droplets were visualized at 60°C using the confocal microscope at a scanning velocity of 7 s per frame.
To confirm the elimination of the orthogonality, the Y-, orthY-, and S-motifs were mixed in a test tube with the buffer at the concentrations shown in table S7. The mixture was then visualized using the same method as described above.
Confirmation of the cleavage of DNA-RNA chimera strands by RNase A
Cleavage of the DNA-RNA chimera strands was confirmed via denaturing urea polyacrylamide gel electrophoresis. A denaturing 10% polyacrylamide gel containing 8 M urea was prepared by mixing acrylamide/Bis (29:1) solution (Nacarai Tesque), urea (FUJIFILM Wako Pure Chemical, Osaka, Japan), Tris-borate EDTA buffer (Nippon Gene, Toyama, Japan), ammonium peroxodisulfate (FUJIFILM Wako Pure Chemical), and N,N,N′,N′-tetramethylethylenediamine (FUJIFILM Wako Pure Chemical). The DNAs, DNA-RNA chimera strands, CS-motifs, and S-motifs (all of them were annealed) were mixed with 20 μg/ml of RNase A (Nippon Gene, Toyama, Japan) at 5 μM concentrations in test tubes. They were incubated at 37°C for 15 min, and then the sample solutions were mixed with a loading buffer composed of 0.05% (w/v) bromophenol blue, 1 mM EDTA (Nippon Gene), and ~100% (v/v) deionized formamide (FUJIFILM Wako Pure Chemical) and further incubated at 95°C for 10 min. Electrophoresis was performed at 200 V for 30 min at 25°C. The gel was stained with SYBR Gold and imaged using the gel imager (fig. S22).
Fission of DNA droplets
To achieve fission of the DNA droplets, the part of the DNA strands constituting the S-motif was replaced with DNA-RNA chimera strands, considering the motif geometry for the split into the Y- and orthY- portions. DNA and DNA-RNA chimera strands were mixed at the concentrations given in table S8. The samples were then annealed from 85° to 25°C at a rate of −1°C/min using the thermal cycler. Twenty microliters of the annealed samples was put on BSA-coated glass, on which a silicone sheet with a punch hole 8 mm in diameter was placed. Mineral oil was poured on the sample in the pore to avoid evaporation of the sample during the observations. The glass was put on the stage heater and incubated at 60°C for 30 min so that the sample solution was fully in the droplet states. RNase A was dissolved in the buffer and was added to the sample solution at a final concentration of 20 μg/ml during the observations at 65°C. As a control experiment, buffer solution was added to the sample instead of the RNase A solution.
Segregation of DNA droplets
The Janus-shaped droplets and patchy-like patterns in the DNA droplets were created by adjusting the concentrations of DNA-RNA chimera strands. For the Janus-shaped and patchy-like droplets, 90 and 50 mol % of the two DNA strands in the motif were replaced with the chimera strands, respectively (tables S9 and S10). Sample solutions in the test tubes were annealed from 85° to 25°C at a rate of −1°C/min using the thermal cycler; then, 1 μl of the RNase A solution was added to the solutions for a 20 μg/ml final concentration. These mixtures were poured into the observation chamber on the stage heater, the temperature was increased to 62°C, and the mixtures were visualized using the confocal microscope.
Capture and partitioning of DNA-modified streptavidin
DyLight549-labeled streptavidin was purchased from Vector Laboratories (CA, USA). Biotin molecules were modified to the 3′ end in DNA with the SE sequence. The streptavidin and the biotinylated DNA were mixed in a sample solution for a final concentration of 20 μg/ml and 1.5 μM. To evaluate the partitioning of streptavidin in the Janus droplets, the partitioning coefficient, P, was calculated by analyzing the line profile of the distribution of the dye molecules. The analysis was performed by obtaining the sum values of the fluorescence intensity of Alexa405 (defined as B) or FAM (defined as G) in the full width at half maximum (FWHM) range. Then, the sum values of Dylight549 fluorescence intensity in the FWHM range of Alexa405 or FAM (RB or RG) were obtained. Last, P was calculated by dividing RB by B or RG by G.
Supplementary Material
Acknowledgments
We would like to thank H. Watanabe for support in the data analysis. Funding: This research was supported by JSPS KAKENHI to M.T. (nos. JP17H01813 and JP18K19834) and Y.S. (no. JP18J00720), Research Encouragement Grants from The Asahi Glass Foundation to M.T., and Support for Tokyo Tech Advanced Researchers (STAR) to M.T. Y.S. is a JSPS Research Fellow (SPD). Author contributions: M.T. provided the original concept. Y.S. and M.T. planned the experiments. Y.S. performed all experiments. T.S. aided in data analysis. Y.S. and M.T. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The original manuscript was uploaded in an arXiv preprint server (accession number: 1907.10252). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Contact M.T. for addition information.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/23/eaba3471/DC1
REFERENCES AND NOTES
- 1.Bates F. S., Polymer-polymer phase behavior. Science 251, 898–905 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Ianiro A., Wu H., van Rijt M. M. J., Vena M. P., Keizer A. D. A., Esteves A. C. C., Tuinier R., Friedrich H., Sommerdijk N. A. J. M., Patterson J. P., Liquid-liquid phase separation during amphiphilic self-assembly. Nat. Chem. 11, 320–328 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Yanagisawa M., Nigorikawa S., Sakaue T., Fujiwara K., Tokita M., Multiple patterns of polymer gels in microspheres due to the interplay among phase separation, wetting, and gelation. Proc. Natl. Acad. Sci. U.S.A. 111, 15894–15899 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakatani N., Sakuta H., Hayashi M., Tanaka S., Takiguchi K., Tsumoto K., Yoshikawa K., Specific spatial localization of actin and DNA in a water/water microdroplet: Self-emergence of a cell-like structure. Chembiochem 19, 1370–1374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewey D. C., Strulson C. A., Cacace D. N., Bevilacqua P. C., Keating C. D., Bioreactor droplets from liposome-stabilized all-aqueous emulsions. Nat. Commun. 5, 4670 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A. A., Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G. M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., Tjian R., Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C. C.-H., Eckmann C. R., Myong S., Brangwynne C. P., The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franzmann T. M., Jahnel M., Pozniakovsky A., Mahamid J., Holehouse A. S., Nüske E., Richter D., Baumeister W., Grill S. W., Pappu R. V., Hyman A. A., Alberti S., Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., Baldwin A. J., Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berry J., Weber S. C., Vaidya N., Haataja M., Brangwynne C. P., RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E5237–E5245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., Kriwacki R. W., Pappu R. V., Brangwynne C. P., Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Runnels C. M., Lanier K. A., Williams J. K., Bowman J. C., Petrov A. S., Hud N. V., Williams L. D., Folding, assembly, and persistence: The essential nature and origins of biopolymers. J. Mol. Evol. 86, 598–610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson E., Mohammed A., Gardell J., Masich S., Czeizler E., Orponen P., Högberg B., DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Douglas S. M., Dietz H., Liedl T., Högberg B., Graf F., Shih W. M., Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y., Ye T., Su M., Zhang C., Ribbe A. E., Jiang W., Mao C., Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452, 198–201 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Ong L. L., Hanikel N., Yaghi O. K., Grun C., Strauss M. T., Bron P., Lai-Kee-Him J., Schueder F., Wang B., Wang P., Kishi J. Y., Myhrvold C., Zhu A., Jungmann R., Bellot G., Ke Y., Yin P., Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 552, 72–77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothemund P. W. K., Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Seeman N. C., Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982). [DOI] [PubMed] [Google Scholar]
- 21.Park N., Um S. H., Funabashi H., Xu J., Luo D., A cell-free protein-producing gel. Nat. Mater. 8, 432–437 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Kurokawa C., Fujiwara K., Morita M., Kawamata I., Kawagishi Y., Sakai A., Murayama Y., Nomura S.-I. M., Murata S., Takinoue M., Yanagisawa M., DNA cytoskeleton for stabilizing artificial cells. Proc. Natl. Acad. Sci. U.S.A. 114, 7228–7233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikawa M., Mizuno Y., Mohri K., Matsuoka N., Rattanakiat S., Takahashi Y., Funabashi H., Luo D., Takakura Y., Biodegradable CpG DNA hydrogels for sustained delivery of doxorubicin and immunostimulatory signals in tumor-bearing mice. Biomaterials 32, 488–494 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Um S. H., Lee J. B., Park N., Kwon S. Y., Umbach C. C., Luo D., Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 5, 797–801 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Xing Z., Caciagli A., Cao T., Stoev I., Zupkauskas M., O'Neill T., Wenzel T., Lamboll R., Liu D., Eiser E., Microrheology of DNA hydrogels. Proc. Natl. Acad. Sci. U.S.A. 115, 8137–8142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biffi S., Cerbino R., Bomboi F., Paraboschi E. M., Asselta R., Sciortino F., Bellini T., Phase behavior and critical activated dynamics of limited-valence DNA nanostars. Proc. Natl. Acad. Sci. U.S.A. 110, 15633–15637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon B.-J., Nguyen D. T., Abraham G. R., Conrad N., Fygenson D. K., Saleh O. A., Salt-dependent properties of a coacervate-like, self-assembled DNA liquid. Soft Matter 14, 7009–7015 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Nguyen D. T., Saleh O. A., Tuning phase and aging of DNA hydrogels through molecular design. Soft Matter 13, 5421–5427 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Bomboi F., Romano F., Leo M., Fernandez-Castanon J., Cerbino R., Bellini T., Bordi F., Filetici P., Sciortino F., Re-entrant DNA gels. Nat. Commun. 7, 13191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zadeh J. N., Steenberg C. D., Bois J. S., Wolfe B. R., Pierce M. B., Khan A. R., Dirks R. M., Pierce N. A., NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Markham N. R., Zuker M., DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 33, W577–W581 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen D. T., Jeon B.-J., Abraham G. R., Saleh O. A., Length-dependent and spatial structure of DNA partitioning into a DNA liquid. Langmuir 35, 14849–14854 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Saccà B., Niemeyer C. M., Functionalization of DNA nanostructures with proteins. Chem. Soc. Rev. 40, 5910–5921 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Rogers W. B., Manoharan V. N., DNA nanotechnology. Programming colloidal phase transitions with DNA strand displacement. Science 347, 639–642 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Thieffry D., Sarkar S., Forty years under the central dogma. Trends Biochem. Sci. 23, 312–316 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Nisisako T., Recent advances in microfluidic production of Janus droplets and particles. Curr. Opin. Colloid Interface Sci. 25, 1–12 (2016). [Google Scholar]
- 37.Hayakawa M., Onoe H., Nagai K. H., Takinoue M., Influence of asymmetry and driving forces on the propulsion of bubble-propelled catalytic micromotors. Micromachines. 7, 229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kishi J. Y., Schaus T. E., Gopalkrishnan N., Xuan F., Yin P., Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. 10, 155–164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montagne K., Plasson R., Sakai Y., Fujii T., Rondelez Y., Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soloveichik D., Seelig G., Winfree E., DNA as a universal substrate for chemical kinetics. Proc. Natl. Acad. Sci. U.S.A. 107, 5393–5398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato Y., Hiratsuka Y., Kawamata I., Murata S., Nomura S.-I. M., Micrometer-sized molecular robot changes its shape in response to signal molecules. Sci. Robot. 2, eaal3735 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Hagiya M., Konagaya A., Kobayashi S., Saito H., Murata S., Molecular robots with sensors and intelligence. Acc. Chem. Res. 47, 1681–1690 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/23/eaba3471/DC1


