A transcription factor gradient conveys positional information in the vertebrate limb bud.
Abstract
The positional information theory proposes that a coordinate system provides information to embryonic cells about their position and orientation along a patterning axis. Cells interpret this information to produce the appropriate pattern. During development, morphogens and interpreter transcription factors provide this information. We report a gradient of Meis homeodomain transcription factors along the mouse limb bud proximo-distal (PD) axis antiparallel to and shaped by the inhibitory action of distal fibroblast growth factor (FGF). Elimination of Meis results in premature limb distalization and HoxA expression, proximalization of PD segmental borders, and phocomelia. Our results show that Meis transcription factors interpret FGF signaling to convey positional information along the limb bud PD axis. These findings establish a new model for the generation of PD identities in the vertebrate limb and provide a molecular basis for the interpretation of FGF signal gradients during axial patterning.
INTRODUCTION
Since the proposal of the positional information theory 50 years ago (1, 2), several cases have been described in developing embryos of diffusible signals—or morphogens—that generate gradients to instruct cells on their positions along patterning axes (3). At the molecular level, morphogen concentration generally translates into gradients of transcription factor activity that interpret the morphogen gradient (4). Coupling between morphogens and interpreter transcription factors has been defined for most embryonic signaling pathways (5–9) but is lacking for the fibroblast growth factor (FGF) family (10), despite their well-characterized role as morphogens (11). Here, we investigate this question in developing mouse limbs, where FGFs have an instructive role in establishing proximo-distal (PD) identities (12).
The vertebrate limb PD dimension is a secondary embryonic axis that shows progressive development. The proximal-most identities represent the “ground” state, and progressively more distal identities appear sequentially at a distal mesenchymal region known as the progress zone (PZ) (13). The PZ underlies an ectodermal thickening called the apical ectodermal ridge (AER) (14), which produces FGF and Wnt signals, both of which are essential for maintaining the PZ undifferentiated and actively generating PD segments (15). As in the main embryonic axis, the progressive generation of PD identities in the limb involves the temporal colinear opening of Hox complexes, with hox9-10 paralogs involved in segments proximal to the elbow/knee, hox10-11 paralogs in the forearm/leg (zeugopod), and hox12-13 paralogs in the hand/foot (autopod) (16–21).
Meis1 and Meis2 encode highly similar homeodomain transcription factors expressed in proximal limb regions (22). Meis overexpression disrupts limb segments distal to the elbow/knee (23, 24), supporting the current view that they are segmentally regulated genes exclusively involved in the development of regions proximal to the stylopod/zeugopod transition. A conclusive study on the role of Meis in PD limb patterning has remained elusive, because complete elimination of Meis function from the limb field results in abrogation of limb bud outgrowth. Here, we recombined Meis1 and Meis2 floxed alleles with Dll1Cre (25), which bypasses Meis early requirements in limb induction. Contrary to the established notion that Meis is only required for the development of the stylopod and further proximal limb elements, we found that the zeugopod also requires appropriately dosed Meis activity. Meis-deficient limbs show phocomelia, a human congenital condition in which all limb segments are severely hypoplastic except the hand/foot. The mechanism underlying these phenotypes is the formation of a PD gradient of Meis protein under the control of AER-FGFs and the regulation of Hox gene segmental activation by different Meis concentration thresholds. Our findings show that Meis interprets distal FGF signaling to convey positional information along the vertebrate limb PD axis.
RESULTS
Elimination of Meis function produces phocomelia
To understand Meis roles in limb PD patterning, we eliminated Meis function with Dll1Cre (25), which recombines lateral plate precursors as they are being recruited to the limb field at the level of the presomitic mesoderm (Fig. 1, A to Ac). Meis1 and Meis2 elimination with Dll1Cre (M1M2DKO) does not affect limb induction and establishment of the PD and anterior-posterior signaling (n = 1/1 and 2/2; fig. S1). Recombination with Dll1Cre is incomplete, leaving, on average, 25% of cells that retain detectable Meis1/2 protein expression [range of % in mutant limbs: 15 to 37%; n = 3 wild type (WT) and 5 double knockouts (DKOs); fig. S2, A and B]. Despite the incomplete recombination, M1M2DKO hindlimbs showed severe phocomelia, with rudimentary skeletal elements in all limb segments except the foot, which was completely normal [n = 3/3 at birth + n = 3/5 at embryonic day 14.5 (E14.5); Fig. 1B]. Unexpectedly, forelimbs (FLs) were not affected at birth (n = 3/3; Fig. 1C); however, the frequency of M1M2DKO newborn animals was 37% of that predicted by Mendelian inheritance, indicating that the specimens analyzed had escaped from an earlier lethal phase. We therefore examined fetuses at 14.5 days of development, when M1M2DKO fetus survival was 67%. In these specimens, a proportion of FLs showed a phenotype similar to that of hindlimbs at birth (n = 3/5; Fig. 1B). We explain these results by variability in the anterior recombination border of Dll1Cre (n = 4; fig. S2C) and a likely collateral effect on heart development when recombination is anterior enough to completely affect the FLs. Examination of the early expression pattern of Sox9 indicated that the defects observed derive from mis-specification of skeletal elements during the patterning phase (n = 2/2; Fig. 1C). Furthermore, determination of the proliferation and cell death patterns showed no significant differences between control and mutant limb mesoderm, although mutant limbs showed a trend to lower proliferation (n = 3 WT and 5 DKOs; fig. S3). These results show that Meis deficiency provokes phocomelia by differentially affecting the patterning of PD limb skeletal elements.
Fig. 1. Elimination of Meis1 and Meis2 produces proximal skeletal element specification defects resulting in phocomelia.
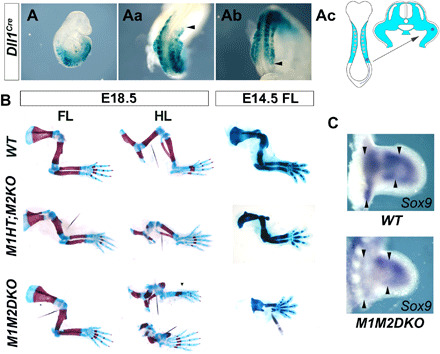
Recombination pattern of Dll1Cre (A to Ab) revealed by Rosa26RLacZ activation in whole-mount embryonic day 9.5 (E9.5) embryos (A), forelimbs (FL) (Aa), and hindlimbs (HL) (Ab). Black arrowheads point to the anterior border in the FLs and posterior in the HLs. (Ac) Schematic showing the recombination pattern of the Dll1Cre driver in limb bud precursors as they are recruited to the primordium. (B) Skeletal preparations of Meis mutants and WT embryos stained with Alcian Blue/Alizarin Red at E18.5 or Victorian Blue at E14.5. At E18.5, M1M2DKO FLs showed minor phenotypical defects (n = 3/3), while HLs showed severe phocomelia, with rudimentary skeletal elements in all limb segments except the autopod, which was completely normal (n = 3/3 at birth + n = 3/5 at E14.5). In Dll1Cre;Meis1+/f;Meis2f/f (M1HT;M2KO) and Dll1Cre;Meis1f/f;Meis2+/f (not shown) fetuses, FLs showed minor alterations, while HLs display smaller pelvis and severe specific stylopod reduction (n = 7/7). An extra anterior digit is observed in one specimen (n = 1/7). At E14.5, a proportion of FLs in M1M2DKO fetuses showed strong reductions or absence of all skeletal except the autopod (n = 3/5). (C) Sox9 mRNA whole-mount in situ hybridization in E11.5 WT and M1M2DKO HL buds, showing alterations of the chondrogenic precursor pattern in the presumptive stylopod and zeugopod (n = 2/2). Black arrowheads point to the proximal-most appendicular pre-condensations and to the prospective zeugopod-autopod boundary.
The requirement for Meis during zeugopod development shown by these results was unanticipated. Together with previous observations showing that Meis overexpression throughout the whole limb bud PD axis disrupts zeugopod development (23, 24, 26), these results suggested that zeugopod development requires a specific range of intermediate Meis levels.
A PD gradient of Meis proteins correlates with limb bud distalization
We then examined and quantified Meis protein expression at midbud stages, observing that it is distributed in a gradient along the limb bud PD axis (Fig. 2, A to Aa). Deletion of Meis genes using Hoxb6CreERT (27), which recombines the posterior half of the FL, provides an internal control within the limb bud, showing the reliability of endogenous Meis protein detection and signal quantification (Fig. 2, B and Ba). Because limb distalization is progressive, we studied Meis gradient formation over time in WT embryos. To examine the consistency of the Meis gradient across different specimens and between different regions of the limb, we developed a method for the regional quantification of Meis nuclear signal, which allowed us to statistically compare specimens for the PD gradient profile (fig. S4) and serially sectioned limbs at three different stages (figs. S5 to S7). In early limb buds, up to 20/21-somite stage (so), Meis protein is highly abundant and shows a flat profile along the whole PD axis (n = 3; Fig. 2, C to Cc). As the early limb bud grows, a linear gradient affecting all cells along the PD axis is established at 24so (n = 8; Fig. 2, D to Dc). Next, the proximal region containing the highest Meis levels expands, resulting in a plateau proximally and a persistent linear gradient in the rest of the limb bud at 30so (n = 9; Fig. 2, E to Ec). At 34so, a Meis-negative region is detected in the distal limb bud, restricting the gradient region to a medial segment of the developing limb (n = 9; Fig. 2, F to Fc). These results show that the Meis gradient dynamics parallel the limb distalization schedule.
Fig. 2. Meis protein forms a PD gradient that changes dynamically in correlation with the progressive distalization of the limb bud.
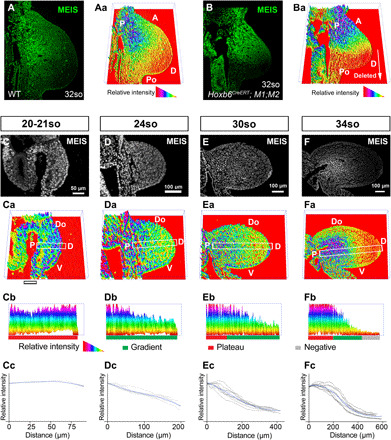
(A) Meis immunofluorescence in a WT FL bud from a 32-somite (so) embryo (~E10.5) and (Aa) its quantification represented in three dimensions (3D). (B and Ba) As a quantification control, the same analysis is shown in a HoxB6CreERT;M1KO;M2KO FL bud, in which only the posterior region lacks Meis proteins according to the reported HoxB6CreER recombination pattern after tamoxifen induction at E8.5 (27). (C to Fa) Temporal sequence of Meis distribution and corresponding 3D quantification plots showing individual representative examples of 20-21so to 34so stages (~E9.5 to E10.5). (Cb to Fb) PD cross sections of Meis abundance along the indicated rectangles in (Ca) to (Fa), with exclusion of the ectodermal signal. The rectangles align with the line of maximal Meis variation along the limb bud PD axis (see Materials and Methods). (Cc to Fc) Graphical representation of Meis gradient quantification at each stage. Black lines represent the individual replicates (see Materials and Methods and fig. S4 for a detailed explanation), and blue lines represent the loess curve fitted from the replicates. The gray shadow represents the 95% confidence interval of the loess curve. A, anterior; Po, posterior; P, proximal; D, distal; Do, dorsal; V, ventral. Scale bars, 50 μm (C) and 100 μm (D to F).
To study the contribution of Meis-expressing cells at different stages of PD limb development, we induced R26R-LacZ recombination from a Meis1CreERT2 allele (28) at different times (Fig. 3, A and B). The Meis1-expressing cell lineage initially contributed to all PD regions and was progressively restricted to more proximal regions (E8.5, n = 11; E10.5, n = 3; Fig. 3, A and B). At late stages, Meis expression border was established at the mid-humerus/femur region (Fig. 3B). These results agree with the observed dynamics of Meis protein expression and indicate that Meis expression does not adjust to limb segmental borders. In contrast, similar fate mapping experiments with a newly engineered Hoxa13CreERT2 allele (E9.5, n = 7; E10.5, n = 4; E11, n = 4; Fig. 3, C and D, and fig. S8), as well as previous results from Hoxa13Cre and Hoxa11GFP alleles (29, 30), show that Hox expression domains respect the main limb segmental borders at different developmental stages.
Fig. 3. Meis expression does not adjust to limb PD segmental borders.
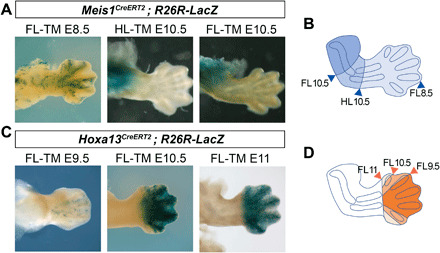
(A) Lineage tracing of Meis1CreERT2-labeled cells by tamoxifen (TM) injection at different stages (TM.E8.5, n = 11; TM.E10.5, n = 3). (B) Lineage tracing of Hoxa13CreERT2-labeled cells by tamoxifen injection at different stages (TM.E9.5, n = 7; TM.E10.5, n = 4; TM.E11, n = 4). (C and D) Schemes showing the boundaries of the regions colonized by Meis-expressing cells (C) and Hoxa13-expressing cells (D) at different labeling time points. Besides minor leakiness observed following injections at E9.5 (C), the lineage of Hoxa13-expressing cells respects the zeugopod-autopod boundary.
The Meis gradient controls HoxA gene activation timing and definitive expression domains
We next studied the correlation between the Meis gradient and distal Hox gene activation (Fig. 4). A detailed sequence of Hoxa11/a13 activation has been previously characterized and showed that Hoxa11 is first detected at the distal limb bud tip in 23so embryos and Hoxa13 in 32so embryos (24). Here, we confirmed the stage of onset of both Hoxa11 and Hoxa13 and studied the profile of Meis protein expression at these specific stages (Fig. 4). At the stage of Hoxa11 activation (23so), an incipient Meis gradient is detected (n = 3; Fig. 4, A to Ac, and C). At the stage of Hoxa13 activation at the distal limb bud tip (32so), Hoxa11 has expanded proximally (Fig. 4D) and Meis detection at the distal limb bud tip shows a plateau that cannot be distinguished from background signal (n = 3; Fig. 4, B to Bc, and F). As shown above, before Hoxa11 expression onset, at 20-21so, there is no evidence of gradient formation (n = 3; Fig. 2, C to Cc). These results show a correlation between the transition from zeugopod to autopod (Hoxa11 activation at 23so) and the initiation of the Meis gradient and a correlation between the transition from zeugopod to autopod (Hoxa13 activation at 32so) and the inactivation of Meis in the distal limb bud.
Fig. 4. Correlation between the Meis gradient and Hoxa11 and Hoxa13 activation schedule in FLs.
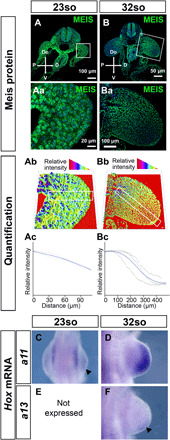
(A to Ba) Meis detection by immunofluorescence and (Ab and Bb) 3D representation of Meis level quantification in representative specimens at the stages in which Hoxa11 (so23) or Hoxa13 (so32) expression is first detected. (Ac and Bc) Graphical representation of Meis gradient quantification at so23 and so32. (C to F) Hoxa11 and Hoxa13 expression by whole-mount mRNA in situ hybridization at so23 and so32. At so23, when Hoxa11 mRNA is first detected (C), the Meis gradient is incipiently detected along the limb bud PD axis (A to Ac). At 32 somites, when Hoxa13 mRNA is first detected (F) and Hoxa11 has expanded proximally (D), the Meis gradient is mature (B to Bc), and Meis levels indistinguishable from background are detected in the distal limb bud (B to Ba). In (Ac) and (Bc), black lines represent the individual replicates (see Materials and Methods and fig. S4 for a detailed explanation), and blue lines represent the loess curve fitted from the replicates. The gray shadow represents the 95% confidence interval of the loess curve. Scale bars, 100 μm (A and Ba), 50 μm (B), and 20 μm (Aa).
In addition, the shape of Meis gradient, which shows a posterior bias, spatially matches the region of initial Hox activation (compare Figs. 2Aa and 4F), suggesting a relationship between the Meis gradient and the timing and/or patterning of Hox activation. In agreement with this view, elimination of Meis function from limb buds results in proximalized Hoxa11 and Hoxa13 expression domains (n = 3/4 and 4/5, respectively; Fig. 5, A and B). Furthermore, in 34so embryos, in which Hoxa13 is not yet expressed in control hindlimbs, a premature and misshaped Hoxa13 expression domain is found (Fig. 5B). These results indicate that Meis activity determines the timing and position of the expression domains of genes at the 5′ end of the HoxA complex in the limb bud. These results suggest that different thresholds of the Meis gradient are involved in this regulation. The control of Hox expression dynamics by Meis transcription factors could be mediated by direct interactions, given that Meis binds profusely to the HoxA complex DNA in limb buds (Fig. 5C). Meis binding profile to the HoxA complex is similar to that reported in E11 embryonic trunks (31). A comparison of the binding profile with that reported for epigenetic marks in E10.5 FLs shows that Meis binding does not correlate with enhancer or promoter activity in the HoxA complex (Fig. 5C) (32).
Fig. 5. Meis gradient controls the activation timing and positioning of HoxA expression domains along the limb bud PD axis.
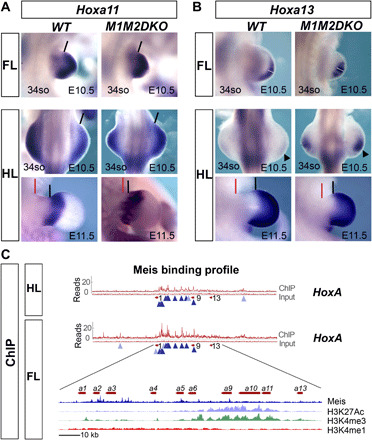
(A) Hoxa11 mRNA expression detected by whole-mount in situ hybridization in FLs (E10.5) and HLs (E10.5 and E11.5) of WT and M1M2DKO embryos. Black lines indicate the proximal border of Hoxa11 expression, and red lines indicate the point of limb insertion in the flank. Note the proximalization of the expression domain in the anterior limb bud (n = 3/4). (B) Hoxa13 expression in FLs (E10.5) and HLs (E10.5 and E11.5) of WT and M1M2DKO embryos reveals premature Hoxa13 activation and expression domain expansion in mutant limbs (n = 4/5). At E11.5, Hoxa13 proximal expression border (black lines) in mutant HLs is closer to the flank (red lines) than in controls. White lines on FL specimens indicate the maximal PD extension of the expression domains. Black arrowheads on E10.5 HLs mark the absence of expression in controls and the robust activation in mutants. (C) Chromatin immunoprecipitation sequencing (ChIPseq) profile of Meis binding to the HoxA complex in FL and HL buds at E10.5. Dark blue arrowheads, shared peaks between FL and HL; light blue arrowheads, peaks specific for either FL or HL. A detailed view of the HoxA cluster region for the FL together with ChIPseq profiles for chromatin marks (32) shows that there is no correlation between enhancer/promoter chromatin marks and Meis binding sites.
In contrast to the effect of Meis function on Hoxa11 and Hoxa13 expression, the expression domain and the mRNA level of the stylopod-related Hoxa9 gene are normal in M1M2DKO limbs (n = 2/2; fig. S9). These results indicate that Meis regulation of HoxA complex relates to regulating the timing and expression borders of distal limb genes but not to the transcriptional activity of proximal ones.
Meis gradient formation is regulated by AER-FGFs
To determine the mechanism of Meis protein gradient formation, we studied the PD distributions of Meis mRNA and a Meis1-CFP reporter (28) and compared them to Meis protein distribution in adjacent sections (Fig. 6). The mRNA and cyan fluorescent protein (CFP) signals both showed a PD-graded distribution, with the mRNA showing a more proximally retracted gradient, with a clear distal negative area, while the Meis protein in adjacent sections showed a sustained gradient across the whole section (n = 3; Fig. 6, A to Bb, E, and F). These results suggest that Meis protein gradient is shaped by mRNA regulation and that Meis protein stability contributes to shaping the gradient. Comparison of the CFP and Meis proteins showed a very similar distribution, with a slightly steeper profile of CFP (n = 3; Fig. 6, C to Db). These results show that the main mechanism controlling Meis protein gradient formation is PD regulation of mRNA abundance. The distribution of CFP and Meis suggests that the stability of the Meis proteins is similar or slightly over that of green fluorescent protein (GFP) family proteins, whose half-life is known to be around 24 hours (33).
Fig. 6. Transcriptional regulation of Meis gradient formation.
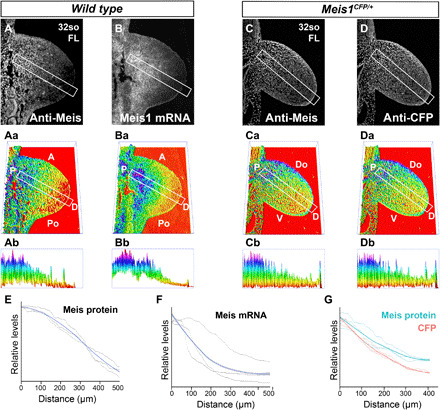
Detection of Meis protein distribution by immunofluorescence (A) and Meis1 mRNA by in situ hybridization (B) in adjacent sections of FL buds. (Aa and Ba) 3D representation of the signals detected in (A) and (B), respectively. (Ab and Bb) PD cross sections of Meis abundance along the indicated rectangles in (Aa) and (Ba). The rectangles align with the line of maximal Meis variation along the limb bud PD axis (see Materials and Methods). Detection of Meis protein (C) and CFP (D) detection by immunofluorescence in limb buds of mice heterozygous for a Meis1 knock-in allele in which the CFP reporter is inserted at the endogenous Meis ATG. Limb cells express CFP from this allele and endogenous Meis from the unaltered allele. (Ca and Da) 3D representation of the signals detected in (C) and (D), respectively. (Cb to Db) PD cross sections of Meis abundance along the indicated rectangles in (Ca) and (Da), with exclusion of the ectodermal signal. The rectangles align with the line of maximal Meis variation along the limb bud PD axis (see Materials and Methods). (E to G) Graphical representation of Meis protein, Meis mRNA, and CFP quantifications. Black lines represent the individual replicates (see Materials and Methods and fig. S4 for a detailed explanation), and colored lines represent the loess curve fitted from the replicates. The gray shadow represents the 95% confidence interval of the loess curve. (E and F) Graphs showing the quantification of MEIS protein and Meis mRNA, respectively, in adjacent sections. (G) Graph showing the quantification of Meis protein and CFP in adjacent sections.
We next sought to identify the upstream mechanism of Meis gradient formation. Previous results showed that Meis mRNA expression domain in the limb bud correlates with the PD distribution of retinoic acid (RA) and that RA manipulation affects PD limb patterning (26, 34–36). Furthermore, Meis mRNA in the distal limb is repressed by FGF, which has been demonstrated in pharmacological gain-of-function (36) and genetic loss-of-function (12) experiments. The effect of FGF on Meis is mediated by the transcriptional activation of the RA-degrading enzyme Cyp26b1 (37). The simplest model to explain Meis regulation thus involves RA distribution regulation by the FGF-Cyp26b1 pathway. We tested this hypothesis by determining Meis protein distribution in AER-FGF loss of function. Genetic elimination of several fgf alleles resulted in a flat distribution of Meis protein along the limb bud PD axis (n = 5/5; Fig. 7, A to Bb and C to Db), demonstrating that AER-FGF function is required for Meis protein gradient formation. Elimination of AER-FGF also leads to impairment in limb bud growth (12, 38), which raises the possibility that the Meis gradient depends on limb bud growth and that the effect of FGF on Meis protein gradient is due to an indirect effect on growth. To resolve this question, we compared control and mutant limbs of similar sizes irrespective of their stage (n = 3 fgf4−/−;8−/−;9−/−, 2 fgf4−/−;8−/−;9+/−, and 3 WT; Fig. 7E). This comparison showed that mutant limbs of a similar size, or even larger than control limbs, displayed a flatter Meis profile (Fig. 7E). These results do not discard a role for limb growth in the evolution of the Meis gradient but show that AER-FGFs do regulate Meis gradient formation independently of their effects on growth.
Fig. 7. Regulation of Meis gradient formation by AER-FGFs.
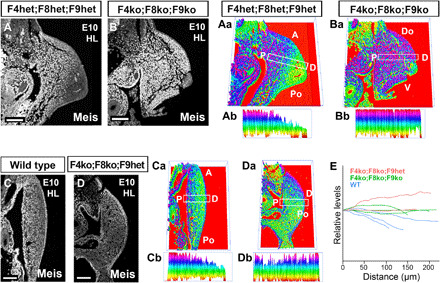
(A to Da) Meis distribution and corresponding 3D quantification plots showing individual representative examples of WT limbs and limbs with different combinations of AER-FGF mutant alleles. (Ab to Db) PD cross sections of Meis abundance along the indicated rectangles in (Aa) to (Da), with exclusion of the ectodermal signal. The rectangles align with the line of maximal Meis variation along the limb bud PD axis (see Materials and Methods). While the Meis PD gradient is detectable in controls (A to Ab and C to Cb), the gradient is swallower or completely undetectable in different combinations of AER-FGF mutants (B to Bb and D to Db) (n = 5/5). (E) Graphical representation of Meis expression profile against size in individual examples of WT limbs and limbs with different combinations of AER-FGF mutant alleles. While mutant limbs show no signs of Meis gradient formation, WT limbs of a similar or smaller size show a detectable PD Meis gradient. F4, FGF4; F8, FGF8; F9, FGF9. Scale bars, 100 μm.
DISCUSSION
Our results demonstrate a new mechanism for PD limb patterning in which FGF/RA antagonism is interpreted through a gradient of Meis protein abundance. The Meis gradient builds progressively and is temporally and spatially dynamic, correlating with the progressive development of the limb PD axis (fig. S10). Despite this dynamic, the shaping of the Meis gradient by FGF and the response of the segmental genes Hoxa11 and Hoxa13 to different Meis levels match the classical positional information mechanisms described in other developmental processes. PD limb patterning responds to levels of Meis expression. While Meis loss of function anticipates distal Hox activation and proximalizes Hox expression borders, Meis overexpression delays distal Hox activation and distalizes Hox expression borders (24, 26). The zeugopod is disrupted by both Meis overexpression and Meis elimination, which indicates that intermediate levels of Meis are required for proper zeugopod development, which agrees with the observed shape of the Meis gradient during zeugopod specification.
Regulation of HoxA complex activation might result from direct interpretation of Meis protein concentration through the abundant interactions between Meis and the Hox complex chromatin. A notable feature of Meis binding sites in the HoxA complex is their restriction to the region containing paralogs 1 to 9 and their exclusion from regions containing paralogs 10 to 13, which suggests roles related to the regulation of colinear activation of the complex. Meis binding sites do not correlate with enhancer or promoter regions within the Hox complex, which suggests that Meis activity is not involved in canonical enhancer-promoter interactions. The fact that the expression of the stylopod-associated gene Hoxa9 is not affected by Meis loss of function matches this view. Nonetheless, the possibility that Meis controls Hox transcriptional activity through its direct binding to the complex will need to be tested by functional analyses in the future. In addition, Meis may cooperate with Hoxa functions associated to stylopod patterning, given that Meis proteins are essential cofactors of Hox protein activity (39) and therefore can affect Hox function at a posttranslational level.
We previously proposed a two-phase regulation model for limb distalization (26), in which Meis transcriptional shut down and Hoxa11 activation were concomitantly triggered by a balance of signals, and marked the end of the first phase (i.e., the stylopod-zeugopod transition). This set in motion a second phase, in which we proposed that the absence of Meis led to a signal environment permissive for Hoxa13 activation, but this activation did not happen until certain time had elapsed, suggesting epigenetic control of distalization. We now show that Meis protein expression and Hoxa11 expression actually overlap for a while and that the end of second phase (Hoxa11-Hoxa13 or zeugopod-autopod transition) occurs when Meis protein is exhausted from the distal limb bud. Our new data suggest that the duration of the second phase is essentially controlled by the dynamics of Meis degradation, which could be related or not to the epigenetic mechanism we proposed before. The considerable stability of the Meis protein, suggested by comparison with CFP, predicts that persistence of Meis protein following experimental or natural Meis mRNA silencing would provide enough function for a delayed Hoxa13 activation. Meis degradation rate may also be relevant to the intrinsic control of distalization timing reported from experiments in the chicken embryo, in which early distal limb cells transplanted to a late distal limb bud environment retain the Hoxa13 activation timing of the donor (40, 41).
Beyond the role demonstrated here, the Meis gradient could also be directly interpreted for regulation of cellular properties that vary continuously along the limb PD axis (42). This possibility has important evolutionary implications, because Meis is also involved in PD patterning during urodele limb regeneration (42). PD positional information during limb regeneration is encoded in a continuous manner (36, 43, 44), which cannot be explained by segmental regionalization alone.
The mechanisms described here are relevant to the origin of genetic and pharmacological phocomelia. The recent identification of cereblon (CRBL) as a thalidomide-binding protein and of Meis2 as a CRBL degradation target (45) suggests that modification of the Meis gradient shape might underlie some of the limb birth defects associated with thalidomide.
MATERIALS AND METHODS
Mice
Mice were handled in accordance with Centro Nacional de Investigaciones Cardiovasculares (CNIC) Ethics Committee, Spanish laws, and the European Union (EU) Directive 2010/63/EU for the use of animals in research. All mouse experiments were approved by the CNIC and Universidad Autónoma de Madrid Committees for “Ética y Bienestar Animal” and the area of “Protección Animal” of the Community of Madrid with reference PROEX 220/15.
Generation of Hoxa13CreER mice
Cloning
The FRT-Neo-FRT-loxP cassette from the FRT-PGK-gb2-neo-FRT-loxP plasmid (GeneBridges) was cloned downstream of the CreERT2 complementary DNA (cDNA) in pBluescript by standard restriction-ligation methods. The ~10.12-kb region used to construct the targeting vector was first subcloned from a C57BL/6 BAC (bacterial artificial chromosome) clone (RPCI23:33N14) into pSP72 (Promega) using homologous recombination. The Cre-ERT2-FRT-Neo-FRT-LoxP cassette was then inserted into exon 1 just downstream of the endogenous ATG site, replacing 1864 base pairs of the coding region spanning exons 1 and 2. The short homology arm extends 2.22 kb downstream of the 3′ end of the FRT-Neo-FRT-LoxP cassette. The long homology arm ends immediately 5′ of the Cre-ERT2 gene and measures ~6.03 kb (fig. S8).
Screening
C57BL/6NTac × 129S6/SvEvTac embryonic stem (ES) cells (Taconic) were electroporated with the targeting construct. Neomycin-resistant ES cell clones were screened by Southern blot according to the depicted strategy. Of 288 clones, 26 were confirmed as positive.
Chimera generation
Targeted clones were expanded, confirmed, and injected into C57BL/6 blastocysts, and microinjected embryos were transferred to pseudopregnant females using standard procedures. Microinjection experiments were performed by the Transgenesis Unit at CNIC.
Deletion of the Neo cassette
After germline transmission, the Neo resistance cassette was eliminated by breeding with a Flpe deleter line.
Previously described mouse strains
Dll1Cre [Tg(Dll1-cre)33Dchp] (25), Meis1CreERT2 [Meis1tm3.1(cre/ERT2)Mtor] (28), Meis1flox (Meis1tm2Ngc) (46), HoxB6CreERT (47), and R26RlacZ [Gt(ROSA)26Sortm1Sho] (48) were as described. Msx2Cre;Fgf mutants are described in (12). The Meis2flox allele was generated by floxing exon 3. Deletion of this exon results in a premature stop codon, which eliminates all functional domains of the Meis2 protein.
Chromatin immunoprecipitation sequencing
Chromatin immunoprecipitation sequencing (ChIPseq) experiments for Meis have been deposited in the Gene Expression Omnibus (GEO) database with reference GSE134066. ChIPseq data for chromatin marks were obtained from Andrey et al. (32).
β-Galactosidase reporter analysis
HoxB6CreERT;R26RlacZ embryos were fixed for 30 min, rinsed in phosphate-buffered saline, and incubated in the presence of X-galactosidase as described (49).
Immunofluorescence
Immunofluorescence was performed according to standard protocols using antibodies targeting the C termini of Meis1 and Meis2 (42). To determine the recombination rate of Meis alleles by Dll1Cre, equivalent limb section from control and mutant cells was stained for Meis detection by immunofluorescence. Background fluorescence from blood cells was manually eliminated. Ectoderm was also eliminated from the quantification. Meis-positive cells were counted using nuclear detection and quantification with the ITCN plugin in ImageJ. All nuclei were identified using 4′,6-diamidino-2-phenylindole (DAPI) staining, and Meis-positive cells were detected from the immunofluorescence signal. Hindlimbs from three WT and five M1M2DKO embryos (three sections each) were quantified. Statistical comparisons were performed using an unpaired t test.
Cell death and proliferation assay
Rabbit polyclonal anti–phospho–histone 3 antibody (Millipore) and TUNEL (terminal deoxynucleotidyl transferase–mediated biotin–deoxyuridine triphosphate nick end labeling) detection kit (Roche) were used according to the manufacturer’s instructions. Quantification was performed manually, counting TUNEL- or H3-positive cells within the limb mesoderm excluding the ectoderm and determining the percentage of positive nuclei in each section by detection of DAPI signal using the ITCN plugin in ImageJ. Three WT limbs and five M1M2DKO were considered, and four sections of each limb were quantified. Statistical comparisons were performed using an unpaired t test.
Skeletal preparations
Skeletal preparations were stained with Victoria Blue and Alcian Blue/Alizarin Red according to standard procedures (50).
In situ hybridization
Whole-mount and on-section in situ hybridization was performed according to standard protocols (51).
Image analysis and quantification
Image quantification was performed by defining an envelope function that describes the distribution of Meis immunodetection in nuclei. Intensities were calculated using an image processing routine developed in MATLAB. First, the nuclear region is detected using an adaptive image threshold inside the region of interest, which resulted from excluding blood cells (autofluorescent) and the ectoderm. Regional intensity maxima for the Meis detection channel are then computed by identifying connected components of pixels with a constant intensity value, surrounded by pixels with a lower value within the nuclear region. A subsampling grid of 103 by 103 points is used to sample Meis detection channel intensity values. Background sample intensities are set to 0. Foreground intensities are subsampled assigning to every point in the grid the value of the closest regional maximum. The subsampled image is smoothed with a median filter using a 10-by-10 neighborhood. Last, previous subsampled image values are interpolated to estimate the envelopes of Meis channel nuclear intensities using spline function. For the RNA in situ hybridization images, signal regional maxima are computed inside the complete region of interest defined by authors and background is determined from the region of interest.
Envelope isolines and three-dimensional (3D) representations were generated using a 1/20 down-sampled version of originals to guarantee a proper visualization. This procedure generates the envelope disregarding interspersed Meis-negative cells, like endothelial cells. Topographic representations of the envelope values were used to determine the line of maximal variation of Meis levels along the PD axis for each specimen. The profile of Meis expression along this line was extracted from applying the ImageJ “Plot Profile” plugin to 255 gray-level 2D images displaying the intensities defined by the envelope function. The line thickness was programmed to 30 pixels to smooth local irregularities of the envelope function. The numerical datasets from the Plot Profile output were used for the fitting and statistical analysis of the results. For this, we plotted the data from all technical and biological replicates using the ggplot2 R package. Data were scaled to the minimum value from all replicates at X = 0. A loess curve was fitted for the technical/experimental replicates. Both the curve and its confidence interval (95%) are also represented in the figures.
Supplementary Material
Acknowledgments
We thank members of the Torres group for stimulating discussions and suggestions. We thank members of the Genomics, Bioinformatics, Transgenesis, and Animal Facility CNIC units for excellent support. We thank J. L. de la Pompa for providing RNA probes and S. Mackem for Hoxb6CreERT2 mice. Meis1 floxed mice were generated by K. Humphries and provided by H. Sadek. Funding: This study was supported by PGC2018-096486-B-I00 from the Spanish Ministry of Science, Innovation and Universities; RD16/0011/0019 from Instituto de Salud Carlos III; and grant S2017/BMD3875 from the Comunidad de Madrid. The CNIC is supported by the Ministerio de Ciencia, Innovación y Universidades and the Pro CNIC Foundation, and is a Severo Ochoa Center of Excellence (SEV-2015-0505). Author contributions: Conceptualization: M.T., I.D., and A.R.-D.; methodology: I.D., G.G., L.F.-d.-M., and F.S.-C.; formal analysis: L.F.-d.-M. and F.S.-C.; investigation: I.D., A.R.-D., G.G., V.C., A.C.L.-D., and M.J.A.; writing (original draft): M.T. and I.D.; writing (review and editing): M.L., A.R.-D., L.F.-d.-M., A.C.L.-D., and F.S.-C.; supervision: M.T. and M.L.; funding acquisition: M.T. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials and/or in “Mendeley Data” under DOI number 10.17632/tnj2drxwm6.1. The ChIp seq data is available from GEO with reference number GSE134066. The Meis1flox and Meis2flox mice can be provided by M.T. pending scientific review and a completed material transfer agreement. Requests for the fgf4flox, fgf8flox, fgf9flox, and fgf17flox should be submitted to M.L. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/23/eaaz0742/DC1
REFERENCES AND NOTES
- 1.Wolpert L., Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1–47 (1969). [DOI] [PubMed] [Google Scholar]
- 2.Wolpert L., Positional information and pattern formation. Curr. Top. Dev. Biol. 117, 597–608 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Rogers K. W., Schier A. F., Morphogen gradients: From generation to interpretation. Annu. Rev. Cell Dev. Biol. 27, 377–407 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Sagner A., Briscoe J., Morphogen interpretation: Concentration, time, competence, and signaling dynamics. Wiley Interdiscip. Rev. Dev. Biol. 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driever W., Nusslein-Volhard C., A gradient of bicoid protein in Drosophila embryos. Cell 54, 83–93 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Steward R., Zusman S. B., Huang L. H., Schedl P., The dorsal protein is distributed in a gradient in early Drosophila embryos. Cell 55, 487–495 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Wang B., Fallon J. F., Beachy P. A., Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100, 423–434 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Shimizu K., Gurdon J. B., A quantitative analysis of signal transduction from activin receptor to nucleus and its relevance to morphogen gradient interpretation. Proc. Natl. Acad. Sci. U.S.A. 96, 6791–6796 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiecker C., Niehrs C., A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development 128, 4189–4201 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Bökel C., Brand M., Generation and interpretation of FGF morphogen gradients in vertebrates. Curr. Opin. Genet. Dev. 23, 415–422 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Nowak M., Machate A., Yu S. R., Gupta M., Brand M., Interpretation of the FGF8 morphogen gradient is regulated by endocytic trafficking. Nat. Cell Biol. 13, 153–158 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Mariani F. V., Ahn C. P., Martin G. R., Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature 453, 401–405 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolpert L., The progress zone model for specifying positional information. Int. J. Dev. Biol. 46, 869–870 (2002). [PubMed] [Google Scholar]
- 14.Bell E., Saunders J. W. Jr., Zwilling E., Limb development in the absence of ectodermal ridge. Nature 184, 1736–1737 (1959). [DOI] [PubMed] [Google Scholar]
- 15.Towers M., Wolpert L., Tickle C., Gradients of signalling in the developing limb. Curr. Opin. Cell Biol. 24, 181–187 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Kmita M., Fraudeau N., Herault Y., Duboule D., Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature 420, 145–150 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Kmita M., Duboule D., Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331–333 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Davis A. P., Witte D. P., Hsieh-Li H. M., Potter S. S., Capecchi M. R., Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375, 791–795 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Davis A. P., Capecchi M. R., A mutational analysis of the 5' HoxD genes: Dissection of genetic interactions during limb development in the mouse. Development 122, 1175–1185 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Fromental-Ramain C., Warot X., Messadecq N., LeMeur M., Dollé P., Chambon P., Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122, 2997–3011 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Fromental-Ramain C., Fromental-Ramain C., Warot X., Lakkaraju S., Favier B., Haack H., Birling C., Dierich A., Doll e P., Chambon P., Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development 122, 461–472 (1996). [DOI] [PubMed] [Google Scholar]
- 22.González-Crespo S., Abu-Shaar M., Torres M., Martínez-A C., Mann R. S., Morata G., Antagonism between extradenticle function and Hedgehog signalling in the developing limb. Nature 394, 196–200 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Mercader N., Leonardo E., Azpiazu N., Serrano A., Morata G., Martínez-A C., Torres M., Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature 402, 425–429 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Mercader N., Selleri L., Criado L. M., Pallares P., Parras C., Cleary M. L., Torres M., Ectopic Meis1 expression in the mouse limb bud alters P-D patterning in a Pbx1-independent manner. Int. J. Dev. Biol. 53, 1483–1494 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Wehn A. K., Gallo P. H., Chapman D. L., Generation of transgenic mice expressing Cre recombinase under the control of the Dll1 mesoderm enhancer element. Genesis 47, 309–313 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Rosello-Diez A., Arques C. G., Delgado I., Giovinazzo G., Torres M., Diffusible signals and epigenetic timing cooperate in late proximo-distal limb patterning. Development 141, 1534–1543 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Nguyen M. T., Zhu J., Nakamura E., Bao X., Mackem S., Tamoxifen-dependent, inducible Hoxb6CreERT recombinase function in lateral plate and limb mesoderm, CNS isthmic organizer, posterior trunk neural crest, hindgut, and tailbud. Dev. Dyn. 238, 467–474 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Lazaro M., Roselló-Díez A., Delgado I., Carramolino L., Sanguino M. A., Giovinazzo G., Torres M., Two new targeted alleles for the comprehensive analysis of Meis1 functions in the mouse. Genesis 52, 967–975 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Scotti M., Kherdjemil Y., Roux M., Kmita M., A Hoxa13:Cre mouse strain for conditional gene manipulation in developing limb, hindgut, and urogenital system. Genesis 53, 366–376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson L. T., Rakshit S., Sun H., Wellik D. M., Generation and expression of a Hoxa11eGFP targeted allele in mice. Dev. Dyn. 237, 3410–3416 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penkov D., San Martín D. M., Fernandez-Díaz L. C., Rosselló C. A., Torroja C., Sánchez-Cabo F., Warnatz H. J., Sultan M., Yaspo M. L., Gabrieli A., Tkachuk V., Brendolan A., Blasi F., Torres M., Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Rep. 3, 1321–1333 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Andrey G., Schöpflin R., Jerković I., Heinrich V., Ibrahim D. M., Paliou C., Hochradel M., Timmermann B., Haas S., Vingron M., Mundlos S., Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Res. 27, 223–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corish P., Tyler-Smith C., Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 12, 1035–1040 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Cooper K. L., Hu J. K. H., ten Berge D., Fernandez-Teran M., Ros M. A., Tabin C. J., Initiation of proximal-distal patterning in the vertebrate limb by signals and growth. Science 332, 1083–1086 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosello-Diez A., Ros M. A., Torres M., Diffusible signals, not autonomous mechanisms, determine the main proximodistal limb subdivision. Science 332, 1086–1088 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Mercader N., Leonardo E., Piedra M. E., Martinez-A C., Ros M. A., Torres M., Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development 127, 3961–3970 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Yashiro K., Zhao X., Uehara M., Yamashita K., Nishijima M., Nishino J., Saijoh Y., Sakai Y., Hamada H., Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing mouse limb. Dev. Cell 6, 411–422 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Sun X., Lewandoski M., Meyers E. N., Liu Y. H., Maxson R. E. Jr., Martin G. R., Conditional inactivation of Fgf4 reveals complexity of signalling during limb bud development. Nat. Genet. 25, 83–86 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Mann R. S., Affolter M., Hox proteins meet more partners. Curr. Opin. Genet. Dev. 8, 423–429 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Saiz-Lopez P., Chinnaiya K., Towers M., Ros M. A., Intrinsic properties of limb bud cells can be differentially reset. Development 144, 479–486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saiz-Lopez P., Chinnaiya K., Campa V. M., Delgado I., Ros M. A., Towers M., An intrinsic timer specifies distal structures of the vertebrate limb. Nat. Commun. 6, 8108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mercader N., Tanaka E. M., Torres M., Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development 132, 4131–4142 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Crawford K., Stocum D. L., Retinoic acid coordinately proximalizes regenerate pattern and blastema differential affinity in axolotl limbs. Development 102, 687–698 (1988). [DOI] [PubMed] [Google Scholar]
- 44.da Silva S. M., Gates P. B., Brockes J. P., The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev. Cell 3, 547–555 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Fischer E. S., Böhm K., Lydeard J. R., Yang H., Stadler M. B., Cavadini S., Nagel J., Serluca F., Acker V., Lingaraju G. M., Tichkule R. B., Schebesta M., Forrester W. C., Schirle M., Hassiepen U., Ottl J., Hild M., Beckwith R. E. J., Harper J. W., Jenkins J. L., Thomä N. H., Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kocabas F., Zheng J., Thet S., Copeland N. G., Jenkins N. A., DeBerardinis R. J., Zhang C., Sadek H. A., Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood 120, 4963–4972 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu J., Nakamura E., Nguyen M. T., Bao X., Akiyama H., Mackem S., Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev. Cell 14, 624–632 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao X., Fujiwara Y., Orkin S. H., Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc. Natl. Acad. Sci. U.S.A. 96, 5037–5042 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whiting J., Marshall H., Cook M., Krumlauf R., Rigby P. W., Stott D., Allemann R. K., Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 5, 2048–2059 (1991). [DOI] [PubMed] [Google Scholar]
- 50.Carlson B. M., Simandl B. K., Stocker K. M., Connelly T. G., Fallon J. F., A method for combined gross skeletal staining and Feulgen staining of embryonic chick tissues. Stain Technol. 61, 27–31 (1986). [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson D. G., Nieto M. A., Detection of messenger RNA by in situ hybridization to tissue sections and whole mount. Methods Enzymol. 225, 361–373 (1993). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/23/eaaz0742/DC1


