Abstract
Background The increasing availability of molecular and clinical data of cancer patients combined with novel machine learning techniques has the potential to enhance clinical decision support, example, for assessing a patient's relapse risk. While these prediction models often produce promising results, a deployment in clinical settings is rarely pursued.
Objectives In this study, we demonstrate how prediction tools can be integrated generically into a clinical setting and provide an exemplary use case for predicting relapse risk in melanoma patients.
Methods To make the decision support architecture independent of the electronic health record (EHR) and transferable to different hospital environments, it was based on the widely used Observational Medical Outcomes Partnership (OMOP) common data model (CDM) rather than on a proprietary EHR data structure. The usability of our exemplary implementation was evaluated by means of conducting user interviews including the thinking-aloud protocol and the system usability scale (SUS) questionnaire.
Results An extract-transform-load process was developed to extract relevant clinical and molecular data from their original sources and map them to OMOP. Further, the OMOP WebAPI was adapted to retrieve all data for a single patient and transfer them into the decision support Web application for enabling physicians to easily consult the prediction service including monitoring of transferred data. The evaluation of the application resulted in a SUS score of 86.7.
Conclusion This work proposes an EHR-independent means of integrating prediction models for deployment in clinical settings, utilizing the OMOP CDM. The usability evaluation revealed that the application is generally suitable for routine use while also illustrating small aspects for improvement.
Keywords: decision support techniques, OMOP, data integration, prediction model
Background and Significance
With the rapidly growing volume and diversity of data in health care and biomedical research, traditional statistical methods are often complemented by modern machine learning techniques. 1 Such techniques are applied to gain valuable insights from ever-growing biomedical databases, leading, example, to patient stratification and personalized predictive models. 2 3 However, researchers in medical information systems development have pointed out, that applying predictive models for clinical decision support not only involves the model development process, but even more important the deployment in point-of-care settings. To achieve real clinical impact, researchers should thus also be concerned about the deployment and dissemination of their algorithms and tools into day-to-day clinical decision support. 4 This typically challenges the developers to integrate their model into proprietary commercial electronic health record (EHR) products.
This work was conducted within the MelEVIR project of the Erlangen Dermatology Department, which aims to develop, test, and deploy a diagnostics tool to assess the probability of a tumor relapse in melanoma patients. The tool uses a machine learning model to identify low- and high-risk patients based on a combination of clinical data elements as well as molecular markers. Molecular markers consist of gene expression in tumor samples, but especially also plasma-derived extracellular vesicles (pEVs) obtained from blood samples. The use of pEVs has the potential to make predictions more accurate and sample collection simpler and possible regardless of the presence of a solid tumor. 5 This particular predictive modeling project, with its model being currently in development and validation, was used to derive the general and generic concept of this work for the integration of machine learning-based decision support tools into clinical information technology environments.
To use the resulting model in the clinical environment of Erlangen University Hospital, it needed to be integrated both with (1) the hospital's EHR system and (2) a data source providing the diagnostic results of the molecular analyses. To keep this development generic and adaptable to other hospitals, the representation of required clinical and molecular data in a widely used common data model (CDM)—the Observational Medical Outcomes Partnership (OMOP) CDM 6 —was chosen. To further improve the universality of our approach, that is, the ability to exchange the underlying prediction model, the architecture makes use of REST interfaces for communication.
Objectives
The objective of this article is to illustrate the architectural design of this loosely EHR-coupled decision support tool and the modeling work required for mapping the respective data to OMOP. A major focus of the resulting application and architecture is broad generalizability, that is, the ability to not only be integrated with different EHRs, but also with various prediction models. The benefits of this approach are demonstrated by the prototypical development of such a system within a specific use case and the evaluation thereof by means of a usability analysis.
Methods
The first step of our work was to develop an extract-transform-load (ETL) process to map the clinical and demographic data from the EHR as well as the molecular data to the OMOP CDM. The ETL process consists of extracting the needed attributes from the data sources, mapping them to corresponding concepts of the standardized vocabulary Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT), which is used in OMOP, and loading them to the OMOP database. As of now OMOP does not use a standardized vocabulary for omics data. Therefore, we created a new vocabulary within the OMOP CDM using the HUGO Gene Nomenclature Committee (HGNC) 7 to enable the mapping of genomic data. For an in-depth overview of how to add custom vocabularies, refer to Maier et al. 8
To retrieve the input data for the prediction model from the OMOP database, we decided to not access the database directly via SQL statements, but rather use an abstraction layer, that is, the REST WebAPI, built on top of the OMOP CDM. For its application within the OHDSI ATLAS tool, the existing REST GET path /person/id only retrieves stored observation and measurement concepts with their respective start and end date, but not their actual stored values. Thus, we extended this WebAPI to also provide the actual observation and measurement values.
This extended WebAPI was then called from the newly developed Web application which first loads all relevant input data, display those to the physician, allows to revise and complete the data if necessary (accounting for data quality issues 9 ), and then consults the prediction model. The findings of the model—in this case the patient's relapse risk—is then presented to the physician, thus supporting his therapeutic decision-making process. All such steps are completely independent from the underlying EHR.
To evaluate the practicability of our application in the point-of-care setting, we performed a usability analysis with physicians. They were asked to perform a real-world use case consisting of loading the patient data, revising them where necessary, and querying the decision support tool. For this they were given a manual for the major steps involved and a mocked pathology report for data reconciliation. This was accompanied by a thinking-aloud protocol. Afterwards the participants were interviewed for additional feedback on the application and were requested to complete the system usability scale (SUS) 10 questionnaire.
Results
The demographic and clinical data of the patient as well as a reference to a molecular analysis are exported via a comma-separated values (CSV) file from the EHR. The gene expression values are provided via a second CSV file and can be merged with the patient data file using the aforementioned reference, creating a single combined file to be further processed.
The observation type of all observations is the OMOP standard concept “Observation recorded from EHR (38000280).” For an overview of the mapped data elements and their corresponding concepts see Table 1 . As it is a commonly used standardized vocabulary for gene names, we used the HGNC to map the analyzed genes to the corresponding HGNC-IDs. For storing the type of expression data measurement, we added a new concept called “RT-qPCR Measurement,” which represents the used technique for quantifying gene expression levels. The data are then loaded in the OMOP database. The demographic data are stored in the person table, clinical data in the observation table, and gene expression data in the measurement table.
Table 1. Overview of the data elements required by the prediction model and mapped to the OMOP CDM, including corresponding concepts in the SNOMED CT or applied transformation rules.
| Category | Attribute | Concept (SCTID) or transformation rule |
|---|---|---|
| Demographic data | Birth date | Split in year, month, and day |
| Gender | M and F expanded to MALE and FEMALE gender (263495000) |
|
| Clinical data | Date of primary diagnosis | Used as the date of all other attributes |
| Location | Tumor location after sectioning (396985003) | |
| Clark level | Clark level (260763001) | |
| pT | pT category finding (385385001) | |
| Breslow level | Breslow depth staging for melanoma (394648007) | |
| Omics data | Gene expression (770 genes) | Custom vocabulary and concepts based on HGNC |
Abbreviations: CDM, common data model; HGNC, HUGO Gene Nomenclature Committee; OMOP, Observational Medical Outcomes Partnership; SCTID, SNOMED CT Identifier; SNOMED CT, Systematized Nomenclature of Medicine – Clinical Terms.
There are three different ways to store values of observations and measurements in OMOP, namely valueAsString , valueAsNumber , and valueAsConceptId . While the first two ways are used for storing (α)numerical values, valueAsConceptId is used for categorical data, which can be represented using concept identifiers (e.g., the standard OMOP concepts for gender). The adapted WebAPI request getPerson 11 now additionally retrieves the three-value fields for all tables with relevant patient data. For our use case only the tables measurement and observation are used, but for a generalized approach all other tables can be read out as well (drug, drug_era, condition, condition_era, visit, death, device, procedure, specimen).
As measurements and observations generally do not have a start and end date, but just one date, we added the field timestamp for retrieving the stored timestamp in its original format. This also prevents an issue we encountered with the original implementation, which used the only date of those attributes both for the start and the end date and tried to parse them to the Java Datetime format. This was not guaranteed to be successful and led to multiple null values in the date fields of the generated JSON. The returned demographic data originally included only the gender and year of birth of the patient and was therefore extended to additionally retrieve the month and day of birth for a more precise calculation of the age at primary diagnosis, which is needed as an input for the prediction model.
The decision support user interface comprises three main components (see Fig. 1 ). At the top the user can input the patient ID (when the application is directly called from an EHR module, the ID would be provided automatically) which triggers the call of the REST GET path /person/id service to load the patient's data from OMOP. Another option is filling in the data fields manually. The middle part is split between the display of the patient's demographic and clinical data, as well as a table view for the expression data of all genes or in general the overview of levels of genomic markers. The bottom part consists of an HTML iframe which displays the findings of the prediction model to the user.
Fig. 1.
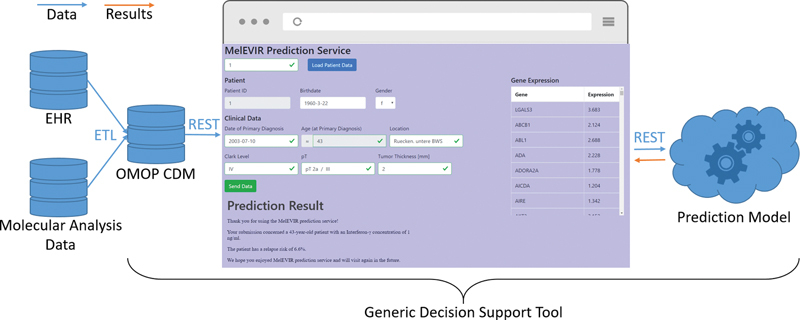
Architectural overview of the data sources and the integration.
All fields are validated after data loading and inputting new data to provide a consistent data format to the prediction model, which means that the physician may need to revise incorrect or missing data. Validation of fields includes checking format and correctness of date fields, validating numerical input format on number fields, and verifying that categorical input fields hold appropriate values (e.g., the Clark level must be a Roman or Arabic numeral between 1 and 5). Additionally, the form is checked for completeness before enabling the submit button to send the data via a REST POST request to the prediction model service.
For now, the findings of the prediction model are rendered as a textual message and displayed as they are provided by the model, but it would be also possible to display custom visualizations or statistics.
In total six physicians were recruited for the usability evaluation. While all participants could complete the given task without help, the thinking-aloud protocol revealed some small issues for improvement in the application. One example is the data field “age at primary diagnosis.” There may be occasions where based on transposition errors in data entry of the date fields (either originating from the EHR or by manual input) the age becomes negative or implausibly high. The fact that this field is not fetched from an external data source but instead calculated on the fly using the patient's birth date and date of primary diagnosis (and thus cannot by edited itself) was not obvious enough (gray background) for half of the participants. The dependency of this field from the two fields birth date and date of primary diagnosis (which both could be modified) needs to be illustrated more clearly. Other features of the application like the visual hints of the validation, that is, red and green borders around the field including a cross respectively a checkmark, were given a mixed reception ( Fig. 2 ). One half of the participants did not notice them at first or at all while the other half commended them greatly during the interview. All participants commented positively on the simple and responsive design. Five of the six physicians stated, however, that they would require some form of an explanatory component for the model or at least links to the backing literature or documentation. The evaluation of the SUS questionnaire resulted in an overall SUS score of 86.7 (number of participants 6, standard deviation 8.6, individual scores [70, 85, 85, 90, 92.5, 97.5]).
Fig. 2.

Display of the visual hints for revising input data. The depicted field represents the stage of primary tumor (pT) as an example.
Discussion
In many cases the statistical validation of a trained predictive model on a test data set marks the end of a machine learning project with no attempt to deploy those models into real practice. 12 To achieve real impact, researchers in the field of artificial intelligence should however be concerned about the deployment and dissemination of their algorithms and tools into day-to-day routine processes of clinicians and to directly apply such models as decision support tools at the point of care. This work proposes methods toward overcoming this issue by providing a simple and generic architecture for integrating prediction models into clinical settings. Therefore, the designing of the architecture was completely independent of developing the underlying predictive model. Validating the model in clinical routine and describing its implementation in detail remains subject of future work.
Using an EHR-independent integration based on OMOP has several advantages. First, it can be easily reintegrated when migrating to a different EHR, either as an embedded frame in the EHR or as a standalone application. Second, it can be easily transferred to another hospital with a different EHR. Third, there are already efforts to integrate prediction models using the OMOP CDM, 4 which could be transferred more easily to our approach. And finally, even the prediction model itself can be changed easily given the simple REST interface (although it may be necessary to adjust the provided data elements). Using techniques like OMOP, originating from a data warehousing background, has also disadvantages. Usually, these databases are updated via scheduled (e.g., nightly) ETL jobs, which can introduce substantial data latency for point-of-care decision support. However, when using molecular markers as input, the consequences of this delay are diminished by the prolonged process of molecular data acquisition. Additionally, the user interface of the tool was therefor designed to enable input of not yet mapped and revision of outdated data. Although, this introduces a limitation to the used approach, as there is no feasible technical solution for the generic tool to feed back inputted data into the EHR.
The exemplary implementation of our design presented in this article relies on a small subset of patient data, which although being limited, should illustrate the generic concept of providing EHR data in combination with molecular data (which are typically not yet included in the EHR) to prediction models utilizing the OMOP CDM. The ETL process of additional data items to OMOP can constitute a nontrivial task; however, there are multiple efforts to map EHR data to OMOP and institutions that already implement the CDM for other projects can start using the application without further work. 8 13 14 15 16 17 While other data models such as from i2b2 18 would have been conceivable, we decided for the use of the OMOP CDM for the integration of clinical and genomic data. We found this approach preferable because it naturally extended our previous local developments of established and well-maintained ETL processes to OMOP, as well as the standardized and yet easily extendable vocabularies and REST API. To further improve long-term interoperability, it is planned to introduce Health Level Seven Fast Healthcare Interoperability Resources (FHIR) 19 for exchanging clinical data with our application. While this would facilitate the integration on the EHR side (granted the EHR supports FHIR, which is not the case in our current setup), it would not replace OMOP as a means for integrating the data.
In terms of molecular markers, this study demonstrates how gene expression levels can be loaded to an OMOP database. The pEVs will also be integrated in OMOP when the set of used markers has been finalized. The pEVs could be loaded to OMOP the same way as the expression levels using a custom vocabulary, but it is also possible that some of them are already present in terminologies like the SNOMED CT which would further increase the generalizability of our approach.
There are currently efforts to integrate genomic data with the OMOP CDM using an extension called the genomic CDM (G-CDM). 20 While this approach would provide the HGNC as the standardized vocabulary for omics data required by us, it is still work in progress and does currently only provide a uniform solution for sequencing and not for gene expression data.
While our usability analysis conducted with six participants was not an extensive evaluation of a final application, it produced valuable feedback from real end-users on our prototype and passed the often cited threshold of five subjects to detect the most severe usability problems. 21 This also applies to the use of the SUS questionnaire, whose resulting SUS score of 86.7 is well beyond the limit of acceptable on the acceptability scale and translates to an adjective rating of “excellent.” 22 Although these, in relative terms, very positive findings are probably also due to the simplicity of the application, they provide a good indication of the usability of the EHR-integrated prototype. Even though this should be further confirmed using a more detailed analysis with the final application, we currently did not follow-up on this, since our current research focus was more on the generic EHR-independent application architecture, than on the decision support tool itself.
One major (usability) issue of the prototype was the missing of an explanatory component of the underlying prediction model. It is reported that one of the key factors in user acceptance of clinical decision support systems is providing an explanation on how the model internally computes its outcome, 23 which was supported by our evaluation. Providing such information is possible with our application; however, it needed to be supplied by the used prediction model. In favor of being able to generically support arbitrary models, the display of the findings and also possibly its detailed explanation is presented without further processing and in the case of our exemplary implementation the used prediction model did not yet supply an explanatory component.
Based on the users' feedback, implementing the user interface as simple as possible with a special focus on a responsive design was well received. This observation coincides with studies reporting that the speed of a decision support application determines a large portion of the users' perception. 24
Conclusion
This work proposes a method for integrating decision support tools generically, regardless of the underlying EHR, using the OMOP CDM. Together with the efforts on the G-CDM it can provide an approach to simplify the deployment and dissemination of prediction models in clinical environments. The evaluation of the application showed an excellent usability, while also revealing valuable user feedback for future refinement.
Clinical Relevance Statement
This study demonstrates a generic solution for the integration of prediction models in clinical settings. This facilitates the deployment of such decision support tools and therefore promotes their dissemination and use in clinical routine use.
Multiple Choice Questions
-
How is patient data loaded into the application?
Data loading is not possible and data can only be inputted manually.
The application sends SQL queries to an OMOP database.
The REST GET path from a WebAPI is called.
The data can be uploaded via a CSV file.
Correct Answer: The correct answer is option c. The application uses the REST GET path /person/id/ of the extended WebAPI. The WebAPI was extended to additionally provide the values of observations and measurements and acts as an abstraction layer on top of the OMOP database.
-
Which component(s) of the architecture can be easily replaced?
The EHR and the prediction model.
Only the EHR.
Only the underlying prediction model.
None of the integrated components can be easily replaced.
Correct Answer: The correct answer is option a. The architecture is designed generically. Using OMOP for storing the patient data allows for the use with different EHRs and given the simple REST interface the used prediction model can be exchanged as well.
Funding Statement
Funding This research has been conducted within the MelEVIR project. MelEVIR is funded by the German Federal Ministry of Education and Research (BMBF) under the Funding Number FKZ 031L0073A.
Conflict of Interest None declared.
Note
The present work was performed in (partial) fulfillment of the requirements for obtaining the degree “Dr. rer. biol. hum.” from the Friedrich-Alexander-Universität Erlangen-Nürnberg (P.U.).
Protection of Human and Animal Subjects
Ethical approval was not required.
References
- 1.Luo W, Phung D, Tran T et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res. 2016;18(12):e323. doi: 10.2196/jmir.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayaru L, Ypsilantis P P, Nanapragasam A et al. Prediction of outcome in acute lower gastrointestinal bleeding using gradient boosting. PLoS One. 2015;10(07):e0132485. doi: 10.1371/journal.pone.0132485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogutu J O, Schulz-Streeck T, Piepho H P. Genomic selection using regularized linear regression models: ridge regression, lasso, elastic net and their extensions. BMC Proc. 2012;6 02:S10. doi: 10.1186/1753-6561-6-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalilia M, Choi M, Henderson A, Iyengar S, Braunstein M, Sun J. Clinical predictive modeling development and deployment through FHIR web services. AMIA Annu Symp Proc. 2015;2015:717–726. [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J H, Dindorf J, Eberhardt M et al. Innate extracellular vesicles from melanoma patients suppress β-catenin in tumor cells by miRNA-34a. Life Sci Alliance. 2019;2(02):e201800205. doi: 10.26508/lsa.201800205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hripcsak G, Duke J D, Shah N H et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 7.Povey S, Lovering R, Bruford E, Wright M, Lush M, Wain H. The HUGO Gene Nomenclature Committee (HGNC) Hum Genet. 2001;109(06):678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 8.Maier C, Lang L, Storf H et al. Towards implementation of OMOP in a German University Hospital Consortium. Appl Clin Inform. 2018;9(01):54–61. doi: 10.1055/s-0037-1617452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCormack J L, Ash J S.Clinician perspectives on the quality of patient data used for clinical decision support: a qualitative studyIn, AMIA Annual Symposium Proceedings: American Medical Informatics Association;20121302. [PMC free article] [PubMed]
- 10.Brooke J. SUS-A quick and dirty usability scale. Usabil Eval Ind. 1996;189:4–7. [Google Scholar]
- 11.Unberath P.2019. Available at:https://github.com/Unberath/WebAPI/tree/v2.4.0-custom. Accessed April 16, 2020
- 12.Bellazzi R, Zupan B. Predictive data mining in clinical medicine: current issues and guidelines. Int J Med Inform. 2008;77(02):81–97. doi: 10.1016/j.ijmedinf.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Yoon D, Ahn E K, Park M Y et al. Conversion and data quality assessment of electronic health record data at a Korean tertiary teaching hospital to a common data model for distributed network research. Healthc Inform Res. 2016;22(01):54–58. doi: 10.4258/hir.2016.22.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Murugesan S, Bhullar H et al. An evaluation of the THIN database in the OMOP Common Data Model for active drug safety surveillance. Drug Saf. 2013;36(02):119–134. doi: 10.1007/s40264-012-0009-3. [DOI] [PubMed] [Google Scholar]
- 15.Lamer A, Depas N, Doutreligne M et al. Transforming French electronic health records into the Observational Medical Outcome Partnership's common data model: a feasibility study. Appl Clin Inform. 2020;11(01):13–22. doi: 10.1055/s-0039-3402754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch K E, Deppen S A, DuVall S L et al. Incrementally transforming electronic medical records into the Observational Medical Outcomes Partnership common data model: a multidimensional quality assurance approach. Appl Clin Inform. 2019;10(05):794–803. doi: 10.1055/s-0039-1697598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.FitzHenry F, Resnic F S, Robbins S L et al. Creating a common data model for comparative effectiveness with the observational medical outcomes partnership. Appl Clin Inform. 2015;6(03):536–547. doi: 10.4338/ACI-2014-12-CR-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy S N, Weber G, Mendis M et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17(02):124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bender D, Sartipi K.HL7 FHIR: an agile and RESTful approach to healthcare information exchangeIn, Proceedings of the 26th IEEE international symposium on computer-based medical systems: IEEE;2013326–331.
- 20.Shin S J, You S C, Park Y R et al. Genomic common data model for seamless interoperation of biomedical data in clinical practice: retrospective study. J Med Internet Res. 2019;21(03):e13249. doi: 10.2196/13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virzi R A. Refining the test phase of usability evaluation: how many subjects is enough? Hum Factors. 1992;34:457–468. [Google Scholar]
- 22.Bangor A, Kortum P, Miller J. Determining what individual SUS scores mean: adding an adjective rating scale. J Usability Stud. 2009;4:114–123. [Google Scholar]
- 23.O'Sullivan D, Fraccaro P, Carson E, Weller P. Decision time for clinical decision support systems. Clin Med (Lond) 2014;14(04):338–341. doi: 10.7861/clinmedicine.14-4-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bates D W, Kuperman G J, Wang S et al. Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality. J Am Med Inform Assoc. 2003;10(06):523–530. doi: 10.1197/jamia.M1370. [DOI] [PMC free article] [PubMed] [Google Scholar]


