Abstract
MS-assisted lipidomic tissue analysis is a valuable tool to assess sphingolipid metabolism dysfunction in disease. These analyses can reveal potential pharmacological targets or direct mechanistic studies to better understand the molecular underpinnings and influence of sphingolipid metabolism alterations on disease etiology. But procuring sufficient human tissues for adequately powered studies can be challenging. Therefore, biorepositories, which hold large collections of cryopreserved human tissues, are an ideal retrospective source of specimens. However, this resource has been vastly underutilized by lipid biologists, as the components of OCT compound used in cryopreservation are incompatible with MS analyses. Here, we report results indicating that OCT compound also interferes with protein quantification assays, and that the presence of OCT compound impacts the quantification of extracted sphingolipids by LC-ESI-MS/MS. We developed and validated a simple and inexpensive method that removes OCT compound from OCT compound-embedded tissues. Our results indicate that removal of OCT compound from cryopreserved tissues does not significantly affect the accuracy of sphingolipid measurements with LC-ESI-MS/MS. We used the validated method to analyze sphingolipid alterations in tumors compared with normal adjacent uninvolved lung tissues from individuals with lung cancer and to determine the long-term stability of sphingolipids in OCT compound-cryopreserved normal lung tissues. We show that lung cancer tumors have significantly altered sphingolipid profiles and that sphingolipids are stable for up to 16 years in OCT compound-cryopreserved normal lung tissues. This validated sphingolipidomic OCT compound-removal protocol should be a valuable addition to the lipid biologist’s toolbox.
Keywords: lipidomics; mass spectrometry; cancer, ceramide; lung adenocarcinoma; lung squamous cell carcinoma; biorepository; non-small cell lung cancer
Sphingolipids are structurally related lipid metabolites with well-established roles in human physiology and disease (1, 2). Many are highly bioactive, and regulate complex biological programs like cellular migration, survival, motility, intracellular vesicular trafficking, metastasis, angiogenesis, invasion, and cytokine production (1–9). Sphingolipid dysregulation is known to significantly contribute to cancer initiation, progression, response to therapy, and the development of multi-drug resistance (3, 10). Given this broad impact on the etiology of disease, there is great interest in precisely establishing disease-specific sphingolipid alterations.
A popular MS platform to quantify sphingolipids in cells and tissues is LC-ESI-MS/MS (2, 3, 11–13). However, there are limitations to the types of tissue specimens that can be analyzed. For example, it has not previously been shown that LC-ESI-MS/MS can be used for the analysis of sphingolipids in tissues embedded in OCT compound. OCT compound contains polyvinyl alcohol and polyethylene glycol that readily ionize and result in ion suppression and loss of signal in most MS platforms (14–17). Therefore, as embedding tissues in OCT compound is one of the primary cryopreservation methods used by tissue biorepositories (18), the vast numbers of tissue specimens held by these facilities have mostly remained untouched by sphingolipid biologists. In stark contrast, there are various methods developed for analyzing DNA, RNA, and proteins from OCT compound-embedded tissues (14, 16, 19–22). However, most of these methods are unsuitable for the extraction and analysis of lipids, as many rely on organic solvents or detergents, which would deplete sphingolipids during the OCT compound-removal stages. Given that tissue biorepositories perform front-end quality control to confirm histology and pathology, follow standard operating procedures to cryogenically store human tissue specimens, and can access subjects’ clinical data that can be used to find clinically relevant correlations between lipidomic data and clinicopathological variables, biorepositories are ideal specimen sources for molecular lipid research. Thus, it is critical that methods are developed for sphingolipid biologists to take advantage of the excellent research materials and resources that can be provided by tissue biorepositories. Here, we present a sphingolipidomic OCT compound-removal protocol (sOCTrP) that makes it possible to analyze sphingolipids in OCT compound-embedded tissues using LC-ESI-MS/MS.
MATERIALS AND METHODS
Human tissue specimens
De-identified human lung tissues were procured from the Virginia Commonwealth University (VCU) Tissue and Data Acquisition and Analysis Core (TDAAC) under a VCU IRB-approved protocol (#HM2471). Specimens were obtained during standard-of-care procedures and banked when judged to be in excess of that required for patient diagnosis and treatment. Fresh samples procured during routine gross examination in surgical pathology were deposited into 2 ml cryovials, snap-frozen in liquid nitrogen, and then stored at −80°C. These tissues were then carefully evaluated for histological type, and then tissues were placed in plastic 24 × 5 mm round disposable base molds (TissueTek, #NC0548726) and filled with OCT compound (Fisher, #23-730-571). OCT compound-embedded specimens were then sectioned, and representative hematoxylin and eosin-stained sections were prepared for histological review by a pathologist to determine the presence or lack of tumor. Only tumor sections with greater than 35% tumor enrichment were selected for lipidomics studies. Suitable samples were prepared as 10 μm frozen sections as needed. On average, 21.4 ± 8.9 mg of tissue sections were prepared for lipidomic analysis. Frozen tissue sections were placed into labeled 15 ml polypropylene conical tubes and stored at −80°C. All associated anonymized sample annotation was generated by the TDAAC SQL2000 secure biorepository database.
Mouse lung and liver specimens
Liver.
Mice were euthanized by CO2 inhalation followed by cervical dislocation. Livers were removed en toto and dipped in PBS to wash off excess blood. Livers were then mechanically minced into six to eight pieces and rinsed with PBS. Minced liver tissues were then placed in 2 ml cryovials and snap-frozen in liquid nitrogen or placed in disposable plastic pathology base molds (Fisher, #22-363-554) containing 2 ml of OCT compound (Fisher, #23-730-571), and then placed over a bed of dry ice until frozen.
Lung.
Mice were euthanized by CO2 inhalation followed by cervical dislocation. Lungs were removed en toto and dipped in PBS to wash off excess blood. Lungs were then separated into right and left lobes, minced, washed with PBS, and placed into 2 ml cryovials and snap-frozen in liquid nitrogen, or placed in disposable plastic pathology base molds (Fisher, #22-363-554) containing 2 ml of OCT compound (Fisher, #23-730-571) and then placed over a bed of dry ice until frozen. For each euthanized mouse, one snap-frozen lung control cryovial and one OCT compound-embedded lung block were prepared. All samples were stored at −80°C until they were processed for lipidomics.
sOCTrP
Human lung specimens.
Human tissues were processed in a class II type A2 biosafety cabinet following safety protocols for specimens of human origin. All steps were performed at 4°C or on ice. Frozen sections of OCT compound-embedded lung tumors or adjacent uninvolved tissues (AUTs) in 15 ml polypropylene tubes were thawed on ice, and then 10 ml of ice-cold ultra-pure deionized Milli-Q water (Millipore) were added. No difference in sphingolipid quantification was observed if PBS or water was used for washing tissues (data not shown). Tubes containing tissues were then vigorously vortexed (typically 20 s). Samples were then centrifuged at 5,000 g and 4°C in a swinging bucket rotor for 10 min. Importantly, 15 ml conical tubes containing human specimens were centrifuged in swinging buckets equipped with ClickSeal biocontainment lids (Thermo Fisher, #75007309) to avoid potential exposure to biohazardous aerosols generated during centrifugation. Following centrifugation, the supernatant was aspirated with great care to avoid taking up any tissue. This was considered one standard sOCTrP cycle. If more sOCTrP cycles were required, 10 ml of ice-cold ultra-pure deionized Milli-Q water were added, the samples vortexed, centrifuged, the supernatant aspirated, and the process repeated until all cycles had been completed. After the final aspiration step, the tissues were resuspended in 500 μl of ice-cold potassium phosphate buffer (Gibco, #20012027). Milli-Q water could also be used, but in practice PBS maximized the amount of tissue that could be retrieved from the 15 ml conical tube. Moreover, to assist in the transfer of lung sections from the 15 ml tubes, 4–5 mm of the tip of a 1,000 μl pipette tip (TipOne, #1111-2721) were cut off using clean surgical scissors prerinsed with methanol, and the shortened tip used to resuspend and uptake the finely sectioned washed lung tissues. Tissues were then placed into preweighed and labeled 1.5 ml polypropylene micro-centrifuge tubes (USA Scientific, #1615-5500). Tube preweighing was done in an analytical balance and the weights recorded in an electronic spreadsheet. The preweighed 1.5 ml tubes containing the samples in PBS were then centrifuged at 7,500 g and 4°C for 10 min. The tubes were then retrieved and the supernatant carefully aspirated. Prior to reweighing the tubes, a fine wick formed by twisting the corner of a Kimwipe tissue (Kimtech, #34120) was used to carefully remove all remaining aqueous solution. Touching specimens with the wick was avoided to prevent contamination and tissues adhering to the wick. However, in control experiments, Kimwipes were processed with the lipid extraction protocol and analyzed with LC-ESI-MS/MS. Kimwipes inherently are low shedding, but at theoretical shedding quantities of 0.1 to 0.5 mm2, which is likely hundreds of times the amount of potential Kimwipe contamination, there were negligible amounts of sphingolipids (data not shown). After aspirating and wicking as much aqueous solution as possible, tubes were reweighed in an analytical balance and weights recorded in an electronic spreadsheet. Tissue weight was calculated by subtracting the weight of tube with tissue specimen from the weight of the tube without tissue specimen and considered “wet weight”. In control experiments, wet weight and initial tissue weights were highly and significantly correlated (see Fig. 1A), as were wet weight and protein concentration from tissues (see Fig. 1B), indicating that wet weight is an accurate measure of original tissue weights. After weighing, 300 μl of ice-cold PBS were added to the tube, and a shortened pipette was used to retrieve the samples and place them in 1 ml of LC-MS grade ice-cold methanol in a 13 × 100 mm screw-top glass tube (VWR, #53283-800) that was then capped (Kimble, #45066C-13). Tissues in methanol were stored at −80°C until processed for sphingolipidomic analyses as described below.
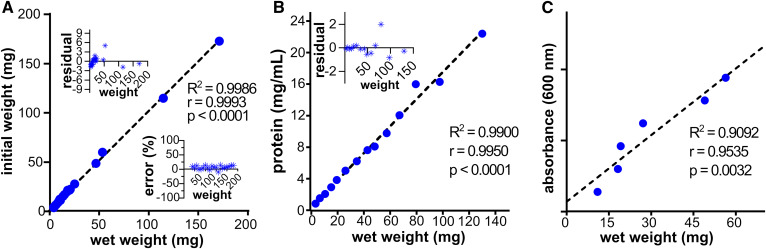
Fig. 1.
Correlation between wet weight and common data normalization measures. A–C: XY-scatterplots and Pearson correlation analysis between initial weights and wet weights (A), protein concentrations and wet weights (B), and lipid phosphates and wet weights (C). For A, mouse livers were weighed after removal (called initial weight), processed with sOCTrP, and weighed (called wet weight). In B, mouse livers were processed with sOCTrP, wet weights were recorded, and then the tissues were resuspended in buffer, sonicated, centrifuged, and protein concentrations of the supernatants determined using a Bio-Rad protein assay. In C, mouse livers were processed by sOCTrP; wet weights were recorded; and then total lipid phosphates were extracted, resuspended, and absorbances recorded. Dotted lines are linear correlation fits of variables, and the linear fit coefficients (R2), Pearson correlation coefficients (r), and P-values for Pearson correlation analysis (p) are shown in each plot. In A, the bottom right inset shows the relative percent error across all weights measured. In A and B, residuals (milligrams) of linear correlation fits are shown in the top left insets. Data were fit with Prism. P ≤ 0.05 was considered significant.
Mouse lung and liver tissues.
OCT compound-embedded and frozen mouse tissues were thawed by placing base molds and tubes over a bed of ice. Upon thawing, tissues were retrieved from the OCT compound medium using forceps or a small spatula and placed in 15 ml polypropylene conical tubes (USA Scientific #14752610) containing 10 ml of prechilled ice-cold ultra-pure deionized water (Milli-Q; Millipore). Retrieving tissues out of the OCT compound-embedding medium minimized the amount of OCT compound carried over. All tissues were maintained at 4°C post-thawing. All other sOCTrP steps were performed as described for human tissues above.
Lipid extraction
Tissues stored in methanol at −80°C were equilibrated to room temperature (25°C) prior to processing, and 10 μl of internal standard solution (see below) were added to each tube with a mechanical repeating pipette (Eppendorf, Hauppauge, NY). Tissues were then homogenized (Homogenizer 150, Fisher Scientific) at maximum power until fully triturated (5–10 s). To ensure that no tissue clumps remained, tubes were sonicated in a Branson 2800 series ultrasonic bath (Branson Ultrasonics, Danbury, CT). If any tissue aggregates formed upon sonication, the samples were rehomogenized and the process repeated iteratively until no more clumps formed upon sonication. Methanol and chloroform were then added to achieve a 2:1:0.1 ratio of methanol:chloroform:water. Typically, an extraction volume of 3–4 ml was used for specimens weighing 50 mg or less and adjusted accordingly for larger specimens. Tubes were then tightly recapped, and the single-phase Bligh-Dyer mixture was incubated at 48°C overnight. The next morning, tubes were centrifuged for 10 min at 4°C and 5,000 g to remove insoluble debris, and the supernatant decanted or pipetted into clean 13 × 100 mm borosilicate glass tubes. Solvent was then evaporated under vacuum at 45°C in Speed-vac equipped with a solvent condenser (Thermo Scientific). Extracted lipids were then reconstituted in 0.5 ml of the starting mobile phase solvent for LC-ESI-MS/MS analysis (see below), and extensively sonicated, centrifuged for 5 min at 5,000 g and 4°C, and the supernatant decanted or pipetted into an autoinjector vial (VWR, #46610-724) and capped (VWR, #89239-020) for analysis.
Internal standards
Internal standards were from Avanti Polar Lipids (Alabaster, AL) and were added to samples as a cocktail containing 250 pmol each in 10 µl ethanol:methanol:water (7:2:1). Sphingoid base and sphingoid base 1-phosphate standards were 17-carbon chain-length analogs: C17-sphingosine, (2S,3R,4E)-2-aminoheptadec-4-ene-1,3-diol (d17:1-So); C17-sphingosine 1-phosphate, heptadecasphing-4-enine-1-phosphate (d17:1-So1P). Standards for N-acyl sphingolipids were C12-fatty acid analogs: C12-Cer, N-(dodecanoyl)-sphing-4-enine (d18:1/C12:0); C12-Cer 1-phosphate, N-(dodecanoyl)-sphing-4-enine-1-phosphate (d18:1/C12:0-Cer1P); C12-SM, N-(dodecanoyl)-sphing-4-enine-1-phosphocholine (d18:1/C12:0-SM); and C12-glucosylceramide, N-(dodecanoyl)-1-β-glucosyl-sphing-4-eine.
MS
Analysis was performed as described (13, 23). Briefly, for LC-ESI-MS/MS analyses, a Shimadzu Nexera LC-30 AD binary pump system coupled to a SIL-30AC autoinjector and DGU20A5R degasser coupled to an AB Sciex 5500 quadrupole/linear ion trap (QTrap) (SCIEX, Framingham, MA) operating in a triple quadrupole mode was used. Q1 and Q3 were set to pass molecularly distinctive precursor and product ions (or a scan across multiple m/z in Q1 or Q3), using N2 to collisionally induce dissociations in Q2 (which was offset from Q1 by 30–120 eV); the ion source temperature set to 500°C. Q1 and Q3 precursor and product ion transitions are shown in Table 1.
TABLE 1.
MRM pairs, ionization parameters, and collision energies utilized for the LC-ESI-MS/MS of sphingolipids
| Q1 Mass (Da) | Q3 Mass (Da) | DP | CE | |
| Sphingoid bases | ||||
| d18:1 Sph | 300.5 | 282.3 | 100 | 21 |
| d18:0 dihydro- Sph | 302.5 | 284.3 | 100 | 21 |
| d18:1 S1P | 380.4 | 264.4 | 90 | 25 |
| d18:0 dihydro-S1P | 382.4 | 266.4 | 90 | 25 |
| Ceramide acyl chain-length | ||||
| C14:0 | 510.7 | 264.4 | 80 | 43.5 |
| C16:0 | 538.7 | 264.4 | 80 | 46 |
| C18:1 | 564.7 | 264.4 | 80 | 48.5 |
| C18:0 | 566.7 | 264.4 | 80 | 48.5 |
| C20:0 | 594.7 | 264.4 | 80 | 51 |
| C22:0 | 622.8 | 264.4 | 80 | 53.5 |
| C24:1 | 648.9 | 264.4 | 80 | 56 |
| C24:0 | 650.9 | 264.4 | 80 | 56 |
| C26:1 | 676.9 | 264.4 | 80 | 58.5 |
| C26:0 | 678.9 | 264.4 | 80 | 58.5 |
| Monohexosylceramide acyl chain-length | ||||
| C14:0 | 672.6 | 264.4 | 80 | 43.5 |
| C16:0 | 700.7 | 264.4 | 80 | 46 |
| C18:1 | 726.7 | 264.4 | 80 | 48.5 |
| C18:0 | 728.7 | 264.4 | 80 | 48.5 |
| C20:0 | 756.7 | 264.4 | 80 | 51 |
| C22:0 | 784.8 | 264.4 | 80 | 53.5 |
| C24:1 | 810.9 | 264.4 | 80 | 56 |
| C24:0 | 812.9 | 264.4 | 80 | 56 |
| C26:1 | 838.9 | 264.4 | 80 | 58 |
| C26:0 | 840.9 | 264.4 | 80 | 58 |
| SM acyl chain-length | ||||
| C14:0 | 675.7 | 184.4 | 80 | 43.5 |
| C16:0 | 703.8 | 184.4 | 80 | 46 |
| C18:1 | 729.8 | 184.4 | 80 | 48.5 |
| C18:0 | 731.8 | 184.4 | 80 | 48.5 |
| C20:0 | 759.9 | 184.4 | 80 | 51 |
| C22:0 | 787.9 | 184.4 | 80 | 53.5 |
| C24:1 | 813.9 | 184.4 | 80 | 56 |
| C24:0 | 815.9 | 184.4 | 80 | 56 |
| C26:1 | 841.9 | 184.4 | 80 | 58 |
| C26:0 | 843.9 | 184.4 | 80 | 58 |
DP, declustering potential; CE, collision energy.
Sphingolipids were separated by reverse-phase LC on a Supelco 2.1 × 50 mm Ascentis Express C18 column (Sigma, St. Louis, MO) at 35°C using a binary solvent system (flow rate of 0.5 ml/min). Prior to injection, the column was equilibrated for 0.5 min with a solvent mixture of 95% mobile phase A1 (CH3OH:water:HCOOH, 58:41:1, v:v:v, with 5 mM ammonium formate) and 5% mobile phase B1 (CH3OH:HCOOH, 99:1, v:v, with 5 mM ammonium formate), and after sample injection (typically 5 μl), the A1/B1 ratio was maintained at 95/5 for 2.25 min, followed by a linear gradient to 100% B1 over 1.5 min, which was held at 100% B1 for 5.5 min, followed by a 0.5 min gradient return to 95/5 A1/B1. The column was re-equilibrated with 95/5 A1/B1 for 0.5 min before the next run.
Lipid phosphate quantification
Total lipid phosphates were extracted and quantified as described (24). Briefly, lipids were extracted using a single-phase Bligh-Dyer extraction as described above, and the extracts dried down in a Speed-vac. Lipids were then resuspended in 3 ml of a 1:2 chloroform:methanol mixture and vortexed, 0.8 ml of water added to form a single-phase solution, vortexed, and then 1 ml of chloroform and 1 ml of water sequentially added and vortexed to obtain a two-phase solution. Samples were then centrifuged at 3,000 g and the upper aqueous layer aspirated and discarded. A fraction of the organic layer was then removed (typically 1.6 ml), placed in a clean 13 × 100 mm borosilicate glass tube, and the solvent evaporated in a Speed-vac. Once dried, 600 μl of ashing solution (1.8 N H2SO4, 1.4% HClO4 in water) were added to each sample and to sodium phosphate standards (solutions containing 0.1–1 mM NaH2PO4). Samples and standards were then vortexed and placed on a 160°C heating block overnight. The next day, samples and standards were allowed to cool and 0.9 ml of water, 0.5 ml of 0.9% ammonium molybdate, and 200 μl of 9.0% ascorbic acid solution added sequentially with intermittent vortexing. Samples and standards were then incubated at 45°C for 30 min. Absorbance of a 200 μl aliquot of each sample and standard was measured at 600 nm with a microplate reader (Bio-Tek).
RESULTS
Data normalization strategies
Robust analysis strategies rely on the ability to compare relative levels of sphingolipids across different experimental groups, which might have vastly different inputs. For example, one might have large amounts of normal tissue, but have a very limited amount of a tumor. Therefore, it is critical that MS data be internally (with internal standards) and externally normalized. Tissue weight, protein concentration, and total lipid-phosphate content are routinely used to externally normalize data with different amounts of specimen input, and produce data of sphingolipid levels relative to milligrams of tissue (if by weight), per microgram of protein (if by protein concentration), or per total lipid phosphate content. In sOCTrP, tissues are washed several times prior to MS analysis, and we have suggested that tissues can be “wet-weighed” as a method to externally normalize data. However, it was important to assess the accuracy of this strategy in faithfully reflecting initial tissue weights. For these experiments, mouse livers were weighed and recorded following surgical extraction, placed in preweighed tubes, washed with sOCTrP, and the tubes reweighed and recorded after washing. For analysis, data were plotted as an XY-scatterplot with initial- and wet weights as axes. For specimens weighing between 4.1 and 171 mg, as shown in Fig. 1A, there was a strong linear relationship (R2 = 0.9986) between initial and wet weights, which were statistically correlated by a significant (P < 0.0001) Pearson product-moment correlation coefficient of 0.9993 and a mean residual error of 0.3 mg (Fig. 1A, inset). These results show that wet weight is strongly correlated with tissue weight and should be an acceptable method to obtain external normalization values for tissues.
A common method for normalizing sphingolipidomic data involves measuring the protein concentration of lysates prepared from tissues being analyzed, and lipids are normalized per milligram or microgram of protein. To examine the relationship between protein concentration and wet weight, mouse liver specimens ranging in size from 3.3 to 130.0 mg were also wet weighed after sOCTrP, and then resuspended in 1 ml of buffer containing 30 mM HEPES, 150 mM NaCl, 10% glycerol, 1 mM EDTA, and 0.1% Triton X-100, homogenized, and sonicated on ice for three cycles of 60 s sonication (60% power; microtip) with a Branson probe sonicator (Branson Ultrasonics) and a 1 min rest on ice. The resulting homogenates were centrifuged at 12,000 g at 4°C, and protein concentrations of the supernatants determined using a Bradford protein assay (Bio-Rad). As shown in Fig. 1B, an XY-scatterplot with wet weight and protein concentration axes showed a strongly linear relationship (R2 = 0.9900) between variables and had a significant (P < 0.0001) Pearson product-moment correlation coefficient of 0.995 and a mean residual error of 0.1 mg (Fig. 1B, inset). These data suggest that wet weights and protein concentrations of tissue lysates are equally suitable and comparatively accurate methods to normalize data, even from tissue specimens that do not contain OCT compound.
Lastly, we assessed the relationship between wet weight and total lipid phosphates extracted and quantified from mouse livers ranging from 11 to 56 mg. As shown in Fig. 1C, under the conditions used to extract lipid phosphates (see the Materials and Methods), quantified levels were strongly correlated (r = 0.9535, P = 0.0032) and linear (R2 = 0.9092) with wet weight.
Interference of OCT compound with data normalization strategies
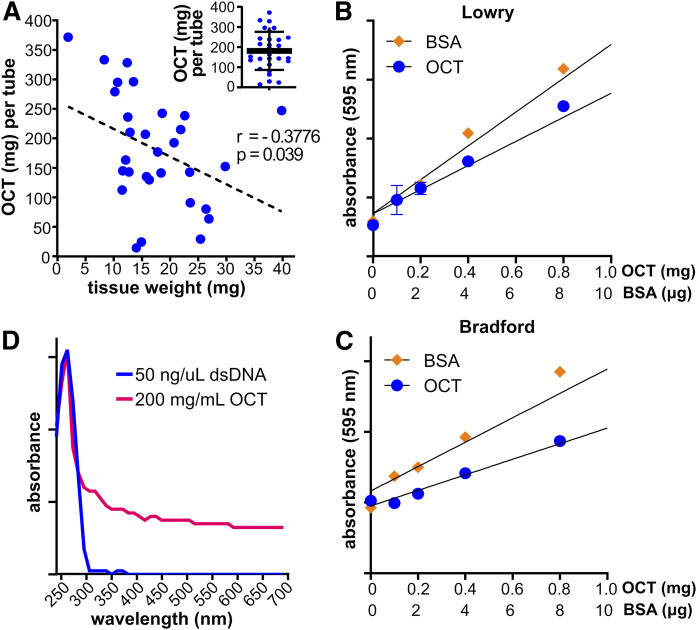
To accurately assess whether OCT compound interferes with data normalization or MS, it was first important to estimate how much OCT compound ordinarily accompanies biorepository tissue specimens. Tubes containing tissue sections embedded in OCT compound were preweighed, the tissues washed with sOCTrP, and the tubes reweighed after the OCT compound had been removed. The difference between initial and final tube weight was assumed to be an estimate of the amount of OCT compound contained in each tube. On average, there were 181.4 mg of OCT compound (Fig. 2A, inset) with specimens weighing between 1.8 and 39.7 mg (Fig. 2A), but OCT compound content per tube ranged between 14.7 and 375.7 mg (Fig. 2A). As expected, there was a statistically significant negative correlation between tissue weight and OCT compound content, with the smallest specimens containing higher amounts of OCT compound (Fig. 2A). This is likely because tissues are embedded in standard size blocks and then backfilled with OCT compound. Therefore, the proportion of OCT compound per tissue weight increases almost proportionally with smaller tissue specimens. These results suggest that using raw tissue weight could dramatically skew external data normalization.
Fig. 2.
Ratios of OCT compound to tissue weights in biorepository tissues sections and interference of OCT compound with protein quantification and absorption spectroscopy analysis. A: Lung tissues (n = 30) were procured from the VCU TDAAC. Tubes were weighed, the tissues were washed with sOCTrP, and then the tubes were reweighed to estimate OCT compound in each tube. Tissue weights were determined by weighing the empty tubes after OCT compound removal and then verified using the wet weighing protocol. The dashed line is a linear correlation fit analysis between OCT compound content and tissue weights, and the Pearson correlation coefficient (r) and P-value (p) are shown. Inset: OCT compound weight distribution across 30 lung tissue specimens. B, C: Solutions (n = 3) were prepared containing BSA, OCT amounts based on the data in A, and protein concentrations measured with Lowry (B) or Bradford (C) assays. Absorbance was read on a plate reader at 595 nm. D: Absorption spectra of solutions containing dsDNA or OCT compound. The quantity of dsDNA used is the expected yield of DNA from 25 mg of lung tissue (Qiagen), and OCT compound amount was based on the data in A.
As protein concentration of tissue lysates was strongly correlated with tissue weight (Fig. 1B), it was important to establish whether OCT compound interfered with the standard colorimetric or spectrophotometric assays used to determine protein concentration. Therefore, a series of solutions containing OCT compound, at concentrations that were expected based on the quantification of OCT compound contained by biorepository specimens (Fig. 2A), were prepared, along with standard solutions containing BSA. As shown in Fig. 2B and C, OCT compound solutions had concentration-dependent absorbance changes with Lowry and Bradford colorimetric protein assays, respectively. Moreover, OCT compound solutions had an absorbance spectrum in water below 300 nm like those of double stranded nucleic acids and proteins (Fig. 2D). Therefore, these results show that OCT compound interferes with standard assays used to estimate protein and nucleic acid concentration, and these assays will likely be unreliable when using samples containing large amounts OCT compound relative to specimen size.
OCT compound interference with sphingolipid quantification by LC-ESI-MS/MS
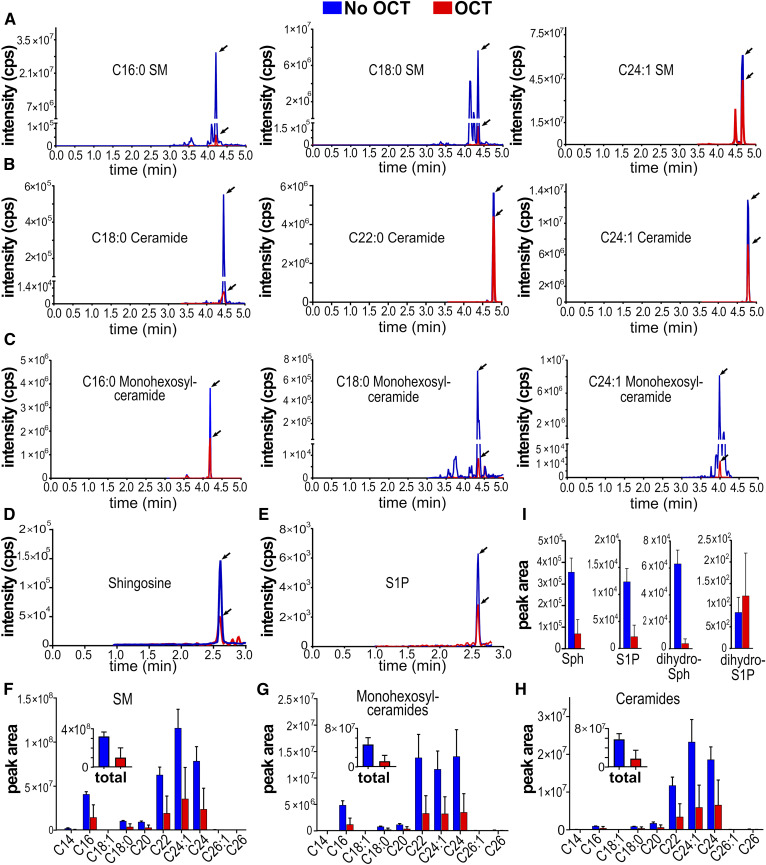
We have shown it is critical that OCT compound be removed from tissue sections because of its interference with external data normalization strategies. However, it was also important to examine whether OCT compound impacted established LC-ESI-MS/MS sphingolipid quantification methods. Thus, 200 mg of mouse liver were resuspended in 2 ml of PBS, homogenized until fully dispersed, and sonicated with a Branson probe sonicator (three rounds of 1 min sonication on ice, 1 min rest on ice). From this solution, six identical 300 μl liver-homogenate replicates were prepared; to three replicates, 200 mg of OCT compound were added (200 mg was based on data shown in Fig. 2A), and 200 µl of water were added to the other three replicates. One milliliter of methanol was then added, and samples were stored at −80°C. Replicates were then processed in parallel and analyzed following lipid extraction and MS protocols as described in the Materials and Methods. It was noted that, unlike control samples with water, OCT compound-containing replicates were cloudy in the single-phase lipid extraction solution, which could not be clarified with centrifugation. After overnight extraction, centrifugation, and solvent evaporation, OCT compound-containing replicates had large insoluble pellets that could not be reconstituted in methanol even under vigorous and extensive sonication. As shown in Fig. 3, OCT compound-containing samples had dramatic reductions in analyte peak heights (Fig. 3A–E), total peak areas (Fig. 3F–I, insets), and acyl-chain length peak areas (Fig. 3F–H). The effect was not systematic or species- or acyl chain-length-specific, and affected SMs (Fig. 3A), ceramides (Fig. 3B), monohexosylceramides (Fig. 3C), and sphingoid bases (Fig. 3D, E). These results show that failure to remove OCT compound from tissue specimens can impact the reliable quantification of sphingolipid levels by LC-ESI-MS/MS.
Fig. 3.
Quantification of sphingolipids by LC-ESI-MS/MS in the presence or absence of OCT compound. LC-ESI-MS/MS multiple reaction monitoring pairs of indicated lipids (A–E) and corresponding integrated peak areas (F–I). Mouse livers were homogenized and sonicated as described in the Materials and Methods, split into six identical samples, and 200 mg of OCT compound were added to three samples (red traces and bars) and water to the other three (blue traces and bars). All six samples were processed identically and analyzed by LC-ESI-MS/MS as described in the Materials and Methods.
sOCTrP validation
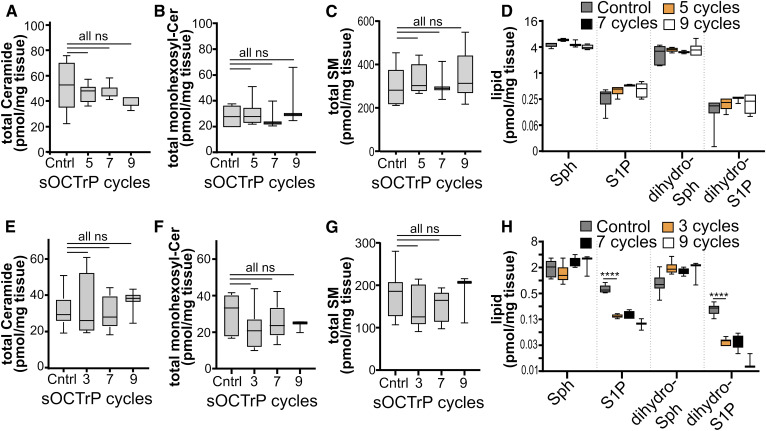
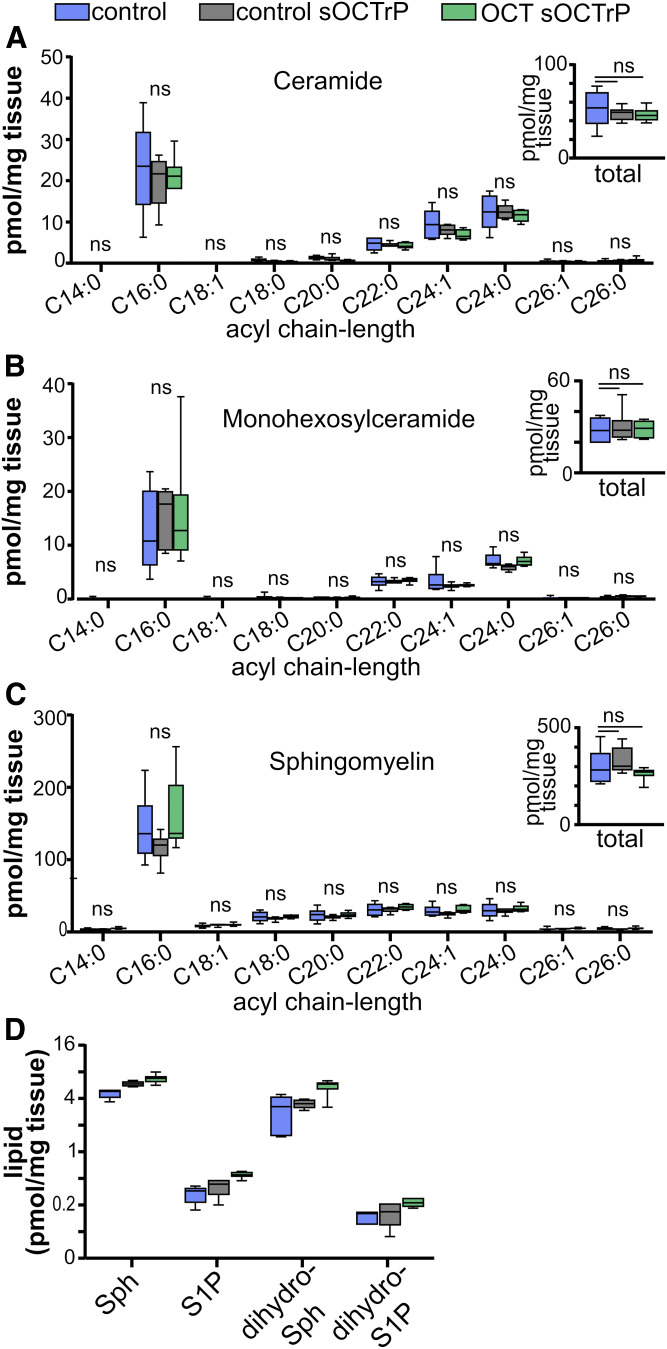
Because OCT compound interferes with multiple elements in the sphingolipidomic LC-ESI-MS/MS analysis workflow, it was important to establish that the protocol used to remove OCT compound did not affect determination of sphingolipid levels in tissues. Thus, mouse livers and lungs were used for validation experiments. First, the effect of continuous sOCTrP cycles on signal decay was examined using liver or lung tissues that had been embedded in OCT compound. Tissues not embedded in OCT compound or processed with sOCTrP were used as controls. Given that large pellets remained in lipid extractions performed with OCT compound, a minimum of three sOCTrP cycles were used to avoid damaging LC columns or contaminating MS equipment with OCT compound. Sample weights were used to normalize MS data. Total levels of individual sphingolipids were assessed. As shown in Fig. 4, successive sOCTrP cycles with lung tissues did not lead to significant reductions in the levels of complex sphingolipids, including ceramides (Fig. 4A), monohexosylceramides (Fig. 4B), and SMs (Fig. 4C), or of the sphingoid bases, sphingosine (Sph) (Fig. 4D), S1P (Fig. 4D), dihydrosphingosine (dihydro-Sph; Fig. 4D), or dihydrosphingosine-1-phosphate (dihydro-S1P; Fig. 4D). Similarly, successive sOCTrP cycles in liver tissues did not significantly deplete ceramides (Fig. 4E), monohexosylceramides (Fig. 4F), SMs (Fig. 4G), Sph (Fig. 4H), or dihydro-Sph (Fig. 4H). However, a significant loss of S1P and dihydro-S1P was observed in liver tissues with as few as three sOCTrP cycles (Fig. 4H). Yet, after the initial loss of signal of these lipids, there were no significant differences in S1P or dihydro-S1P levels between control-sOCTrP and OCT-sOCTrP tissues (Fig. 4H), suggesting that there is a stable pool of these lipids within the tissues that remains after the initial washes. Based on these data, three sOCTrP cycles are sufficient to produce reliable results.
Fig. 4.
Effect of sOCTrP cycles on total sphingolipid levels. Mouse lungs (A–D) and livers (E–H) were processed with the indicated number of sOCTrP cycles and the indicated sphingolipids quantified by LC-ESI-MS/MS. Data are expressed as picomoles of lipid per milligram of tissue. In all groups, control (Cntrl) samples were snap-frozen tissues not stored in OCT compound. sOCTrP-processed samples were embedded in OCT compound. In D and H, the y axis scale is log2 base. Total levels of ceramide, monohexosylceramide (monohexosyl-Cer), and SM are the sum of their C14:0, C16:0, C18:1, C20:0, C22:0, C24:1, C24:0, C26:1, and C26:0 acyl chain-length species. In A–H, Cntrl n = 10. In A–D, five and seven cycles n = 6; nine cycles n = 3. In E and F, five and seven cycles n = 6; nine cycles n = 3. Box plots show medians (black line) and whiskers of minimum to maximum. ****P ≤ 0.0005. ns, not significant.
The effects of short-term storage in OCT compound and processing with sOCTrP were examined. Lung and liver tissues were stored for 4 months at −80°C. Experimental conditions were: 1) specimens snap-frozen and not processed with sOCTrP (labeled “control”); 2) specimens that were snap-frozen, thawed, and processed with sOCTrP (labeled “control-sOCTrP”); and 3) specimens that were embedded in OCT compound, frozen, thawed, and then processed with sOCTrP (labeled “OCT-sOCTrP”). Upon thawing, “control” tissues were first weighed, and then immersed in 1ml of ice-cold LC-MS grade methanol; control-sOCTrP tissues were thawed, processed with sOCTrP, wet weighed, and then placed in 1 ml of ice-cold LC-MS grade methanol; “OCT-sOCTrP” tissues were thawed, processed with sOCTrP, wet weighed, and placed in 1ml of ice-cold LC-MS grade methanol. An analytical balance was used for weighing, and it was critical to minimize errors in tissue weighing by removing as much washing solution as possible from each sample prior to weighing using wicks.
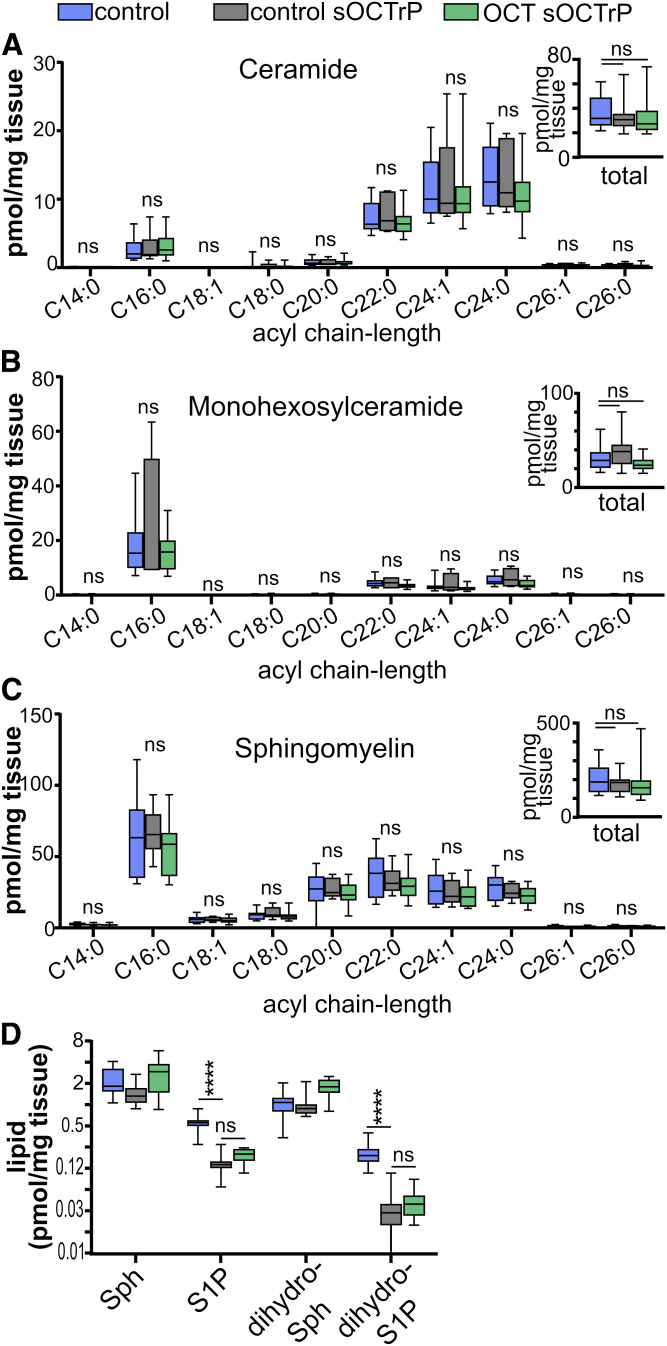
As shown in Figs. 5 and 6, respectively, short-term storage in OCT compound or processing with sOCTrP did not result in significant deterioration of total sphingolipid levels in lung or liver tissues. Furthermore, when acyl chain-length-specific fluctuations were evaluated, there were no significant changes between lung control, lung control-sOCTrP, and lung OCT-sOCTrP ceramides (Fig. 5A), monohexosylceramides (Fig. 5B), SMs (Fig. 5C), Sph (Fig. 5D), S1P (Fig. 5D), dihydro-Sph (Fig. 5D), or dihydro-S1P (Fig. 5D). Similar observations were made with most sphingolipids from liver tissues (Fig. 6). However, unlike lung tissues, sOCTrP processing of control liver tissues, which contain relatively large volumes of blood compared with lung (27), resulted in a significant decrease in the levels of S1P and dihydro-S1P (Fig. 6D).
Fig. 5.
Effect of sOCTrP on sphingolipid acyl chain-length species in mouse lung tissues. Mouse lung tissue snap-frozen (blue bars), snap-frozen and processed with sOCTrP (gray bars), or embedded in OCT compound, frozen, and processed with sOCTrP (green bars) were analyzed by LC-ESI-MS/MS and the indicated acyl chain-length species of ceramides (A), monohexosylceramides (B), SMs (C), and Sph (D), S1P (D), dihydro-Sph (D), and dihydro-S1P (D) quantified. Insets: Total levels of the indicated sphingolipids are the sum of the acyl chain-length species and are expressed as picomoles per milligram of tissue. In D, the y axis scale is log2 base. Box plots show medians (black line) and whiskers of minimum to maximum. ns, not significant. In A–D: Cntrl (blue bars), n = 6; control sOCTrP (gray bars), n = 6; OCT-sOCTrP (green bars), n = 6.
Fig. 6.
Effect of sOCTrP on sphingolipid acyl chain-length species in mouse liver tissues. Mouse livers snap-frozen (blue bars), snap-frozen and processed with sOCTrP (gray bars), or embedded in OCT compound, frozen, and processed with sOCTrP (green bars) were analyzed by LC-ESI-MS/MS and the indicated chain-length of ceramides (A), monohexosylceramides (B), SMs (C), and Sph (D), S1P (D), dihydro-Sph (D), and dihydro-S1P (D) were quantified. Insets: Total levels of the indicated sphingolipids are the sum of their acyl chain-length species and expressed as picomoles per milligram of tissue. In D, the y axis scale is log2 base. Box plots show medians (black line) and whiskers of minimum to maximum. ns, not significant. ****P ≤ 0.0005. In A–D: Cntrl (blue bars), n = 10; control sOCTrP (gray bars), n = 10; OCT-sOCTrP (green bars), n = 10.
Analysis of human non-small cell lung cancer tumors
Two small cohorts of human lung adenocarcinomas (LUADs; n = 5) and lung squamous cell carcinomas (LSCCs; n = 5) stored in OCT compound were procured from the VCU TDAAC. All tumors had matched adjacent uninvolved tissues (AUTs) surgically removed at the time of tumor resection. All tissues had been cryopreserved in OCT compound and were stored at −80°C. Specimens were received as fine sections taken from OCT compound-embedded tumor blocks. Upon thawing, tissues were processed with five cycles of sOCTrP, weighed, placed in LC-MS grade ice-cold methanol, lipids extracted, and then analyzed by LC-ESI-MS/MS as described in the Materials and Methods.
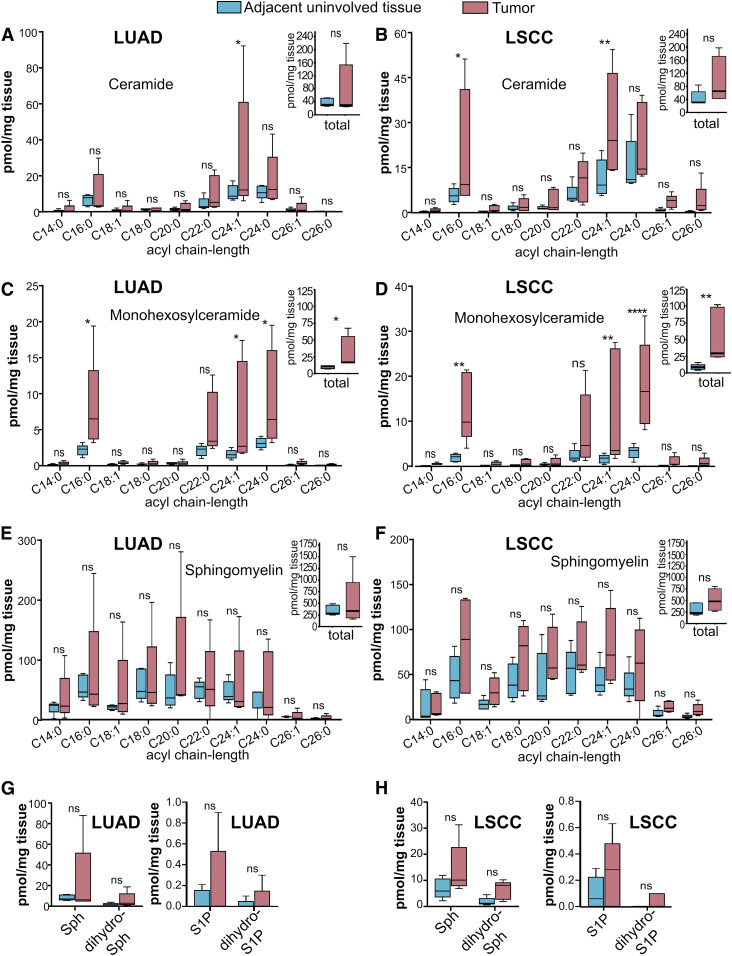
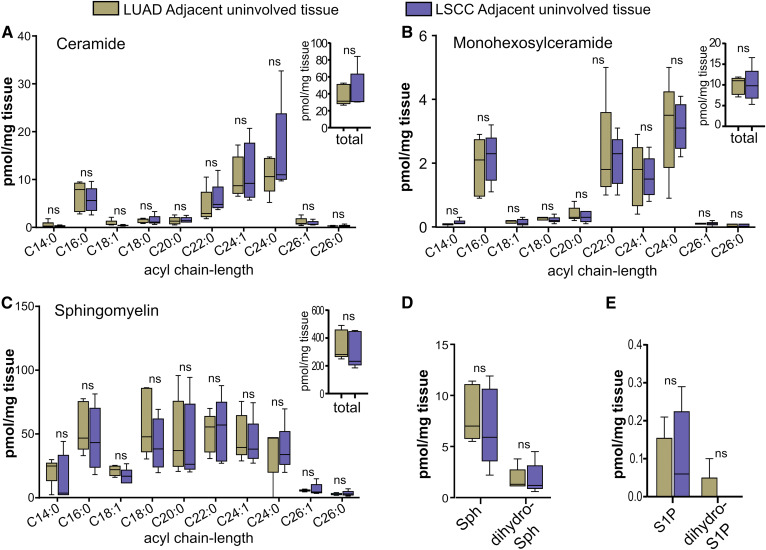
Levels of sphingolipids were altered in LUAD and LSCC tumors compared with AUTs. Specifically, C24:1 ceramide was significantly higher in LUADs (Fig. 7A) and LSCCs (Fig. 7B). Importantly, however, ceramide changes were not identical between LUADs and LSCCs, as only LSCC tumors had higher C16:0 ceramide (Fig. 7B). Interestingly, although total ceramide levels were not significantly greater in either disease compared with AUTs (Fig. 7A, B; insets), total monohexosylceramides were significantly higher in both diseases (Fig. 7C, D; insets). Moreover, there were dramatic differences between LUAD and LSCC tumors and AUTs with regard to various monohexosylceramide acyl chain-length species, such as C16:0 (4-fold higher in LUADs; 6.8-fold higher in LSCCs), C24:1 (4.5-fold in LUADs; 7.5-fold higher in LSCCs), and C24:0 (2.9-fold higher in LUADs; 5.6-fold in LSCCs) (Fig. 7C, D). In stark contrast to the significant changes in ceramides and monohexosylceramides, there were no alterations in total or acyl chain-length-specific SMs (Fig. 7E, F and insets) in either disease. Similarly, no tumor/AUT differences were observed for Sph (Fig. 7G), dihydro-S1P (Fig. 7G), S1P (Fig. 7H), or dihydro-Sph (Fig. 7H). Data from AUTs were further analyzed to assess differences in the AUT sphingolipid profiles of patients with LUAD and LSCC. As shown in Fig. 8, uninvolved lung tissues of subjects with LUAD or LSCC did not show significant differences in total or acyl chain-length-specific levels of ceramide (Fig. 8A and inset), monohexosylceramides (Fig. 8B and inset), SMs (Fig. 8C and inset), Sph (Fig. 8D), dihydro-Sph (Fig. 8D), S1P (Fig. 8E), or dihydro-S1P (Fig. 8E).
Fig. 7.
Sphingolipid profiles of LUADs, LSCCs, and AUTs banked in OCT compound and processed with sOCTrP. LUADs (A, C, E, G), LSCCs (B, D, F, H), and paired AUTs were obtained from the VCU TDAAC. Specimens had been embedded in OCT compound and cryogenically stored at −80°C. Tissues were processed with sOCTrP, weighed, and analyzed at the VCU Lipidomics Core. Lipid levels are in picomoles per milligram of tissue. The indicated acyl chain-length species of ceramide (A, B), monohexosylceramide (C, D), SM (E, F), and Sph (G, H), S1P (G, H), dihydro-Sph (G, H), and dihydro-S1P (G, H) were quantified. Insets: Total levels of the indicated sphingolipids are the sum of their acyl chain-length species. For LUADs (A, C, E,G), n = 5 tumors and n = 5 AUTs; LSCCs (B, D, F, H), n = 5 tumors and n = 5 AUTs. Box plots show medians (black line) and whiskers of minimum to maximum. ns, not significant. *P ≤ 0.05; **P ≤ 0.01; **** P ≤ 0.0005.
Fig. 8.
Sphingolipid levels in AUTs of subjects with LUAD or LSCC. A–E: Secondary analysis of sphingolipid data from AUTs shown in Fig. 7. Comparison of total (insets) and acyl chain-length ceramides (A), monohexosylceramides (B), SMs (C), and Sph (D), S1P (E), dihydro-Sph (D), and dihydro-S1P (E) in the AUTs from patients with LUAD and LSCC. Insets: Total lipids are the sum of C14:0, C16:0, C18:0, C18:1, C20:0, C22:0, C24:1, C24:0, C26:1, and C26:0 acyl chain-length species. All lipids are expressed in picomoles per milligram of tissue. Box plots show medians (black line) and whiskers of minimum to maximum. ns, not significant.
Storage-related sphingolipid deterioration in OCT compound-embedded lung tissues
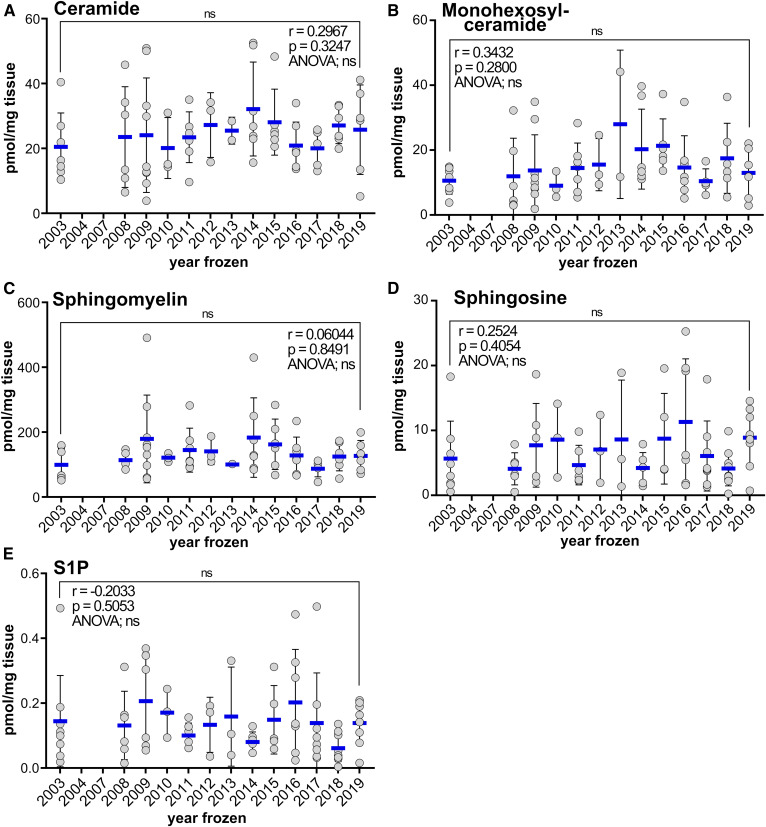
To assess the decay of sphingolipids in long-term cryogenic storage, 73 normal lung tissue specimens that had been frozen between 2003 and 2019 were processed with sOCTrP and analyzed by LC-ESI-MS/MS. Data were plotted in an XY-scatterplot with total lipid levels and year frozen as the axes. A Pearson moment correlation analysis revealed that there was not a significant age-related decay in ceramides (Fig. 9A), monohexosylceramides (Fig. 9B), SMs (Fig. 9C), Sph (Fig. 9D), SiP (Fig. 9E), dihydro-Sph (data not shown), or dihydro-S1P (data not shown). Remarkably, a multiple-comparison ANOVA analysis that examined differences between lipid levels from patients one year and any other year revealed no significant differences (Fig. 9). Finally, a paired analysis between the oldest (2003) specimens and most recently acquired specimens (2019) did not reveal any significant differences (Fig. 9).
Fig. 9.
Analysis of time-dependent effects on sphingolipid levels in tissues cryogenically stored in OCT compound for up to 16 years. XY-scatterplots of total (sum of C14:0, C16:0, C18:1, C18:0, C20:0, C22:0, C24:1, C24:0, C26:1, and C26:0 acyl chain-length species) lipid levels and the year tissues were cryopreserved in OCT compound. Each filled circle represents data from a single subject (n = 73). The x axis is the year that tissues were collected, embedded in OCT compound, and frozen; the y axis is the total levels of ceramide (A), monohexosylceramide (B), SM (C), Sph (D), and S1P (E) in normal lung tissues of subjects. Pearson correlation analysis (Prism) revealed no significant relationship between the year frozen and any of the lipids measured. ANOVA multi-comparison analysis (Prism) revealed no significant differences between the lipid levels of any given year and any other year (78 comparisons). ns, not significant. All lipids are expressed in picomoles per milligram of tissue. Blue lines are means with standard deviations.
DISCUSSION
Much is to be learned of the roles that sphingolipids play in human cancers, and large-scale studies that characterize sphingolipid metabolism alterations in tissues can greatly enhance our ability to probe the mechanistic underpinnings of these diseases. Yet, given current limitations in our ability to analyze tissues cryopreserved in OCT compound, obtaining sufficient human specimens for adequately powered studies can be challenging, oftentimes requiring prospective studies to acquire tissues directly from the surgical suite. In contrast, biorepositories already hold extensive tissue collections that can be procured for retrospective studies. However, the broad use of OCT compound for freezing specimens has been a barrier for MS analysis, given that polyvinyl alcohol and polyethylene glycol in OCT compound readily ionize and result in ion suppression and loss of signal (14–17, 21, 22). Here, we show that OCT compound is carried over through all steps of single-phase Bligh-Dyer extractions used for the extraction of sphingolipids from tissues, which dramatically impacted their quantification with LC-ESI-MS/MS (Fig. 3).
In contrast to the status quo for lipidomic analysis, there are established methods for DNA, RNA, and proteomic analysis of tissues embedded in OCT compound. For instance, laser capture microdissection is effective for extraction of RNA and DNA (19). Ether-methanol precipitation, SDS-PAGE-based isolation, trichloroacetic acid precipitation, solid-phase extraction of glycopeptides, immobilization of proteins followed by washing, and washing samples with ethanol are all methods that have been developed for proteomics (14, 16, 20–22). OCT compound-embedded tissues are also suitable for semi-quantitative analytical methods like immunostaining or Western blotting following protein extraction by mechanical or chemical methods. Importantly, the accuracy and reproducibility of these methods have led to the identification of numerous tumor-specific proteins and various potential druggable targets in lung and prostate cancers (20, 22). However, these methods are unsuitable for lipidomic analysis because the physicochemical properties of sphingolipids and proteins are vastly different, and any method that utilizes detergents or organic solvents to remove OCT compound will result in stripping or the extraction of sphingolipids into the organic phase, leading to lipid-specific depletion and introducing methodology-related artifacts. Given the demonstrated importance of sphingolipids in the etiology of human diseases, developing methodology for MS sphingolipid profiling of OCT compound-cryopreserved tissues is warranted. Therefore, we have developed a method for LC-ESI-MS/MS sphingolipidomic analysis of human specimens cryopreserved in OCT compound. Our method is simple, inexpensive, and 40 samples in 1 day can be routinely processed.
In addition to previously shown experimental barriers in the analysis of OCT compound-embedded specimens (14–17, 21, 22), here we show that OCT compound interfered with assays used for normalization of MS data. First, we found an inversely proportional relationship between the amount of OCT compound and tissue size in biorepository-procured tissue specimens (Fig. 2A). This was not unexpected, as specimens of different sizes are placed in similarly sized cryopreservation trays. Thus, smaller specimens occupy a smaller proportion of a frozen OCT compound-tissue block. Consequently, if OCT compound is not removed prior to weighing tissues, the net weight cannot be used for data normalization as their mass is proportionally mostly OCT compound. In our experiments, we found that this is particularly critical for tissue OCT compound specimens weighing 10 mg or less, as these had 275–375 mg of OCT compound (Fig. 2A). Failure to remove OCT compound would result in significant errors in data normalization. Second, tissue extract protein concentrations are commonly used to normalize MS data (23, 26–31). However, we found that OCT compound interferes in a concentration-dependent manner with Bradford and Lowry reagents (Fig. 2B, C). The concentrations of OCT compound tested in these assays were based on the quantities of OCT compound found in tissue sections (Fig. 2A). Third, OCT compound has an absorbance spectrum that overlaps with DNA and protein spectra (Fig. 2D). Therefore, in combination, these results indicate that OCT compound must be removed from tissue sections if weights or protein concentrations of tissue extracts will be used to externally normalize MS data. However, this can be easily remedied if OCT compound is removed from tissues with sOCTrP, and we demonstrate that tissue weights prior to sOCTrP washing closely correlated with post-sOCTrP wet weights (Fig. 1A). Moreover, wet weights closely correlated with the protein concentration of extracts (Fig. 1B) and total lipid phosphates prepared from tissues (Fig. 1C). We also demonstrate that it is safe to use Kimwipe wicks to fully remove all wash solution from tissues in the final steps of sOCTrP, which can increase the accuracy of tissue weighing. Therefore, tissue wet weights are a suitable normalization strategy for OCT compound-embedded specimens.
Validation experiments with mouse tissues demonstrated that storage in OCT compound and sOCTrP processing did not deplete most sphingolipids analyzed. Specifically, in mouse lung tissues, there were no changes in ceramides, monohexosylceramides, or SMs with C14:0, C16:0, C18:1, C20:0, C22:0, C24:1, C24:0, C26:1, and C26:0 chain-lengths, or of Sph, dihydro-Sph, S1P, and dihydro-S1P. In mouse liver tissues, ceramides, monohexosylceramides, SMs, Sph, and dihydro-Sph were also unchanged by sOCTrP. However, washing fresh mouse liver tissues with sOCTrP did result in a significant depletion of S1P and dihydro-S1P. One possibility for this depletion is that as there is a much larger blood volume in livers than in lungs (25), and blood contains some of the highest physiological levels of S1P (32, 33). Therefore, reduction in S1P is likely due to removal of blood from the liver tissues by sOCTrP. This is consistent with the observation that S1P levels were reduced after the initial three sOCTrP cycles but remained constant up to seven sOCTrP cycles (Fig. 4H). We suggest that S1P quantified in sOCTrP-washed livers is tissue-associated and likely membrane embedded. Nevertheless, in contrast to these observations in liver, there was no reduction in S1P or dihydro-S1P in lung tissues. Given these data, sOCTrP should be adequate for the analysis of sphingolipids such as ceramides, monohexosylceramides, SMs, Sph, and dihydro-Sph from lung and liver tissues. For lung tissues, our data suggest that sOCTrP is an amenable method for S1P analysis, but evaluation of S1P levels in tissues processed with sOCTrP should be interpreted with caution. However, in validation experiments, there were no significant differences in S1P or dihydro-S1P between control-sOCTrP- and OCT-sOCTrP-processed tissues. Thus, in cases where biorepository specimens have paired normal tissues, we suggest that so long as tumor and normal tissues are processed using the same number of sOCTrP cycles, S1P data might be informative.
Upon validation of sOCTrP, a small scale lipidomic analysis of tissues from patients with the two most common non-small cell lung cancer histological subtypes, LUAD and LSCC, was conducted. Data from five human subjects per disease with matched AUTs showed tumors had significant alterations in ceramides (C16:0 and C24:1 acyl chain-lengths) and monohexosylceramides (C16:0, C24:1, and C24:0 acyl chain-lengths). A disease-specific alteration was also observed, as C16:0 acyl chain-length ceramides were elevated only in LSCC and not in LUAD. Currently, there are no LSCC-specific molecular targets. Importantly, however, not all sphingolipids were higher in tumors, as SMs of all acyl chain-lengths and Sph were unaltered in either disease. Furthermore, a secondary analysis revealed that there were no significant differences between adjacent uninvolved lung tissues of subjects with adenocarcinomas or squamous cell carcinomas. These data suggest there are specific sphingolipid metabolism alterations in the tumors of patients with non-small cell lung cancer. This is consistent with the expanding body of evidence demonstrating that sphingolipid alterations are observed in many cancers. As these changes are thought to influence various steps along the carcinogenic pathway, and in progression, metastasis, and the development of multi-drug resistance (2, 3, 10), new tools, such as sOCTrP, that permit medium to large scale analysis of specimens from human subjects should grant further insight into the role that sphingolipid alterations play in the etiology these diseases. Moreover, as biorepositories already hold large numbers of specimens, these studies can be retrospective in nature, which should accelerate the collection of substantial volumes of data without the design and approval of prospective studies. Importantly, using 73 normal tissue specimens, we have shown that complex sphingolipids and sphingoid bases are stable when cryopreserved for 16 years. Moreover, this analysis showed that the normal lung levels of sphingolipids in many individuals are remarkably similar.
Few studies have reported measurements of sphingolipid levels in human lung specimens. However, C16:0, C18:0, C20:0, and C22:0 acyl chain-length ceramides have been measured in biopsies from freshly explanted lungs of subjects with pulmonary hypertension or with cystic fibrosis (34), where patients with pulmonary hypertension were considered “normal” subjects. Ceramide levels between these normal subjects and the adjacent uninvolved lung tissues in our study are comparable. These results suggest that tissues stored in OCT compound and processed with sOCTrP have ceramide levels similar to those of fresh human lung tissues. Moreover, it is reasonable to assume that any differences between our normal subjects could be biological and not of experimental origin. Nevertheless, these results further validate the utility of our protocol, as comparative studies between several diseases could be used to establish the molecular origins of sphingolipid alterations and define common mechanistic themes that might provide druggable targets. These types of studies could be highly insightful in cancers and cardiovascular and inflammatory diseases where sphingolipids are known to play important roles (2, 3, 10).
Data availability
All data are contained within the article.
Footnotes
Abbreviations:
- AUT
- adjacent uninvolved tissue
- dihydro-S1P
- dihydrosphingosine-1-phosphate
- dihydro-Sph
- dihydrosphingosine
- LSCC
- lung squamous cell carcinoma
- LUAD
- lung adenocarcinoma
- sOCTrP
- sphingolipidomic OCT compound-removal protocol
- Sph
- sphingosine
- TDAAC
- Tissue and Data Acquisition and Analysis Core
- VCU
- Virginia Commonwealth University
Services and support of this research project were provided by the Virginia Commonwealth University (VCU) Massey Cancer Center Tissue and Data Acquisition and Analysis Core and the VCU Lipidomics Core, which are supported in part with funding from National Institutes of Health-National Cancer Institute Cancer Center Support Grant P30CA016059. This work was supported by National Institutes of Health Grants K22CA187314 and R21CA232234 (S.L.), R01GM043880 (S.S.), and T32DK007150 (T.D.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Maceyka M., and Spiegel S.. 2014. Sphingolipid metabolites in inflammatory disease. Nature. 510: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogretmen B. 2018. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer. 18: 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannun Y. A., and Obeid L. M.. 2018. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 19: 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hla T., and Dannenberg A. J.. 2012. Sphingolipid signaling in metabolic disorders. Cell Metab. 16: 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima S., Milstien S., and Spiegel S.. 2017. Sphingosine and sphingosine kinase 1 involvement in endocytic membrane trafficking. J. Biol. Chem. 292: 3074–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lima S., Takabe K., Newton J., Saurabh K., Young M. M., Leopoldino A. M., Hait N. C., Roberts J. L., Wang H. G., Dent P., et al. 2018. TP53 is required for BECN1- and ATG5-dependent cell death induced by sphingosine kinase 1 inhibition. Autophagy. 14: 942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young M. M., Takahashi Y., Fox T. E., Yun J. K., Kester M., and Wang H. G.. 2016. Sphingosine kinase 1 cooperates with autophagy to maintain endocytic membrane trafficking. Cell Rep. 17: 1532–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young M. M., and Wang H. G.. 2018. Sphingolipids as regulators of autophagy and endocytic trafficking. Adv. Cancer Res. 140: 27–60. [DOI] [PubMed] [Google Scholar]
- 9.Shen H., Giordano F., Wu Y., Chan J., Zhu C., Milosevic I., Wu X., Yao K., Chen B., Baumgart T., et al. 2014. Coupling between endocytosis and sphingosine kinase 1 recruitment. Nat. Cell Biol. 16: 652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morad S. A. F., and Cabot M. C.. 2018. Chapter nine - The onus of sphingolipid enzymes in cancer drug resistance. In Advances in Cancer Research. C. E. Chalfant and P. B. Fisher, editors. Academic Press. 235–263. [DOI] [PubMed] [Google Scholar]
- 11.Haynes C. A., Allegood J. C., Park H., and Sullards M. C.. 2009. Sphingolipidomics: methods for the comprehensive analysis of sphingolipids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merrill A. H. Jr., Sullards M. C., Allegood J. C., Kelly S., and Wang E.. 2005. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 36: 207–224. [DOI] [PubMed] [Google Scholar]
- 13.Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., Sullards M. C., and Merrill A. H. Jr.. 2009. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50: 1692–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weston L. A., and Hummon A. B.. 2013. Comparative LC-MS/MS analysis of optimal cutting temperature (OCT) compound removal for the study of mammalian proteomes. Analyst. 138: 6380–6384. [DOI] [PubMed] [Google Scholar]
- 15.Holfeld A., Valdés A., Malmström P-U., Segersten U., and Lind S. B.. 2018. Parallel proteomic workflow for mass spectrometric analysis of tissue samples preserved by different methods. Anal. Chem. 90: 5841–5849. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W., Sakashita S., Taylor P., Tsao M. S., and Moran M. F.. 2015. Comprehensive proteome analysis of fresh frozen and optimal cutting temperature (OCT) embedded primary non-small cell lung carcinoma by LC–MS/MS. Methods. 81: 50–55. [DOI] [PubMed] [Google Scholar]
- 17.Shah P., Zhang B., Choi C., Yang S., Zhou J., Harlan R., Tian Y., Zhang Z., Chan D. W., and Zhang H.. 2015. Tissue proteomics using chemical immobilization and mass spectrometry. Anal. Biochem. 469: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steu S., Baucamp M., von Dach G., Bawohl M., Dettwiler S., Storz M., Moch H., and Schraml P.. 2008. A procedure for tissue freezing and processing applicable to both intra-operative frozen section diagnosis and tissue banking in surgical pathology. Virchows Arch. 452: 305–312. [DOI] [PubMed] [Google Scholar]
- 19.Espina V., Wulfkuhle J. D., Calvert V. S., VanMeter A., Zhou W., Coukos G., Geho D. H., Petricoin Iii E. F., and Liotta L. A.. 2006. Laser-capture microdissection. Nat. Protoc. 1: 586–603. [DOI] [PubMed] [Google Scholar]
- 20.Tian Y., Bova G. S., and Zhang H.. 2011. Quantitative glycoproteomic analysis of optimal cutting temperature-embedded frozen tissues identifying glycoproteins associated with aggressive prostate cancer. Anal. Chem. 83: 7013–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vrana M., Goodling A., Afkarian M., and Prasad B.. 2016. An optimized method for protein extraction from OCT-embedded human kidney tissue for protein quantification by LC-MS/MS proteomics. Drug Metab. Dispos. 44: 1692–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X., Huffman K. E., Fujimoto J., Canales J. R., Girard L., Nie G., Heymach J. V., Wistuba I. I., Minna J. D., and Yu Y.. 2017. Quantitative proteomic analysis of optimal cutting temperature (OCT) embedded core-needle biopsy of lung cancer. J. Am. Soc. Mass Spectrom. 28: 2078–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hait N. C., Allegood J., Maceyka M., Strub G. M., Harikumar K. B., Singh S. K., Luo C., Marmorstein R., Kordula T., Milstien S., et al. 2009. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 325: 1254–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Veldhoven P. P., and Bell R. M.. 1988. Effect of harvesting methods, growth conditions and growth phase on diacylglycerol levels in cultured human adherent cells. Biochim. Biophys. Acta. 959: 185–196. [DOI] [PubMed] [Google Scholar]
- 25.Hall C., Lueshen E., Mosat A., and Linninger A. A.. 2012. Interspecies scaling in pharmacokinetics: a novel whole-body physiologically based modeling framework to discover drug biodistribution mechanisms in vivo. J. Pharm. Sci. 101: 1221–1241. [DOI] [PubMed] [Google Scholar]
- 26.Tidhar R., Zelnik I. D., Volpert G., Ben-Dor S., Kelly S., Merrill A. H. Jr., and Futerman A. H.. 2018. Eleven residues determine the acyl chain specificity of ceramide synthases. J. Biol. Chem. 293: 9912–9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai A., Liu X., Bielawski J., and Hannun Y. A.. 2019. Bioactive sphingolipid profile in a xenograft mouse model of head and neck squamous cell carcinoma. PLoS One. 14: e0215770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullards M. C., Liu Y., Chen Y., and Merrill A. H. Jr.. 2011. Analysis of mammalian sphingolipids by liquid chromatography tandem mass spectrometry (LC-MS/MS) and tissue imaging mass spectrometry (TIMS). Biochim. Biophys. Acta. 1811: 838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coen P. M., Dubé J. J., Amati F., Stefanovic-Racic M., Ferrell R. E., Toledo F. G. S., and Goodpaster B. H.. 2010. Insulin resistance is associated with higher intramyocellular triglycerides in type I but not type II myocytes concomitant with higher ceramide content. Diabetes. 59: 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oyeniran C., Sturgill J. L., Hait N. C., Huang W-C., Avni D., Maceyka M., Newton J., Allegood J. C., Montpetit A., Conrad D. H., et al. 2015. Aberrant ORM (yeast)-like protein isoform 3 (ORMDL3) expression dysregulates ceramide homeostasis in cells and ceramide exacerbates allergic asthma in mice. J. Allergy Clin. Immunol. 136: 1035–1046.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alvarez S. E., Harikumar K. B., Hait N. C., Allegood J., Strub G. M., Kim E. Y., Maceyka M., Jiang H., Luo C., Kordula T., et al. 2010. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 465: 1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thuy A. V., Reimann C. M., Hemdan N. Y., and Graler M. H.. 2014. Sphingosine 1-phosphate in blood: function, metabolism, and fate. Cell. Physiol. Biochem. 34: 158–171. [DOI] [PubMed] [Google Scholar]
- 33.Nagahashi M., Yamada A., Aoyagi T., Allegood J., Wakai T., Spiegel S., and Takabe K.. 2016. Sphingosine-1-phosphate in the lymphatic fluid determined by novel methods. Heliyon. 2: e00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brodlie M., McKean M. C., Johnson G. E., Gray J., Fisher A. J., Corris P. A., Lordan J. L., and Ward C.. 2010. Ceramide is increased in the lower airway epithelium of people with advanced cystic fibrosis lung disease. Am. J. Respir. Crit. Care Med. 182: 369–375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.