Abstract
Nasopharyngeal carcinoma (NPC) is a major health problem in the East and Southeast Asia, and the intensity modulated radiotherapy (IMRT) is the current preferred treatment method of NPC, but radioresistance-induced residual and recurrent tumors are the main cause of treatment failure. Till now, the mechanism of radioresistance and prognostic biomarkers of NPC are still unrevealed. In this study, we collected clinical NPC samples and established radioresistant NPC-R cell lines by irradiating NPC cells with fractionation doses of γ-rays. Using genechip assay between radioresistance and radiosensitive clinical samples and TMT assay between NPC and NPC-R cells, differential expressed genes were examined and the potential biomarker of radioresistance was screened. Immunohistochemical assay of NPC clinical specimens showed that CLIC4 was significantly up-regulated in radioresistance tumor tissues. In vitro studies confirmed that up-regulation of CLIC4 gene enhanced radioresistance in comparison with the alterations of intracellular oxidative metabolism of reactive oxygen species (ROS) and nitric oxide (NO) in an opposite way. Correspondingly, inhibition of CLIC4 sensitized NPC cells to irradiation and decreased nuclear translocation of iNOS and intracellular level of NO in NPC cells. Interestingly, the capacity for DNA repair had no difference between NPC and NPC-R cells. Moreover, because of great interests in using carbon ion irradiation to treat NPC effectively, we demonstrated that, after carbon ion irradiation, NPC-R and NPC cells had similar survival even under the status of up- or down-regulation of CLIC4. Conclusively, CLIC4 contributes to radioresistance of NPC to γ-rays but not carbon ions by regulating intracellular oxidative metabolism of nuclear translocation of iNOS.
Keywords: Nasopharyngeal carcinoma, ionizing radiation, oxidative stress, CLIC4, iNOS
Introduction
Nasopharyngeal carcinoma is characterized by distinct geographical distribution and is particularly prevalent in the East and Southeast Asia [1]. Because of the highly sensitive to ionizing radiation, the intensity modulated radiotherapy (IMRT) is the mainstay treatment modality for non-metastatic NPC with a 5-year recurrence rate as low as 7.4% [2]. But the development of radioresistance still limits the therapeutic efficacy and prognosis of NPC patients. Tumors recurring within one year are considered as radioresistant. It was reported that about 10% of patients have residual disease or develop a recurrent disease at the primary and/or regional site after IMRT [3]. In current, the advanced techniques of high energy particle therapy, especially the intensity modulated carbon ion therapy (IMCT), has been applied to treat NPC with fascinating therapeutic effect [4,5]. Understanding the mechanism of NPC radiosensitivity to different irradiations will be conducive to develop novel therapeutic strategies.
Over the past decades, enormous efforts have been made to identify the biomarkers of the poor prognosis of NPC after radiotherapy using genechip, microarray, and quantitative proteomic analyses [6-9]. In general, tumor radioresistance is related to multiple signaling pathways (e.g., the PI3K/Akt pathway), regulating genes (e.g., microRNA), anoxic condition, angiogenesis, cancer stem cells, autophagy and so on [10]. However, almost no overlap genes of radioresistance regulation was reported by different literatures, which has promoted us to study in-depth.
Chloride intracellular channel (CLIC) proteins are redox-regulated metamorphic proteins, where CLIC4 is a member of CLIC family. The CLIC family regulates ionic homeostasis, organellar volume and electro-neutrality [11], cell-cycle, cytoskeletal function, mitosis and differentiation [12]. CLIC4 protein has a low level in multiple human tumor tissues [13]. But it is abundant in the cytoplasm including endoplasmic reticulum, mitochondrial and nuclear membranes [14-16]. During cell response to various stresses, CLIC4 can be translocated to nuclear and activates p53 signaling pathway and thus contributes to apoptosis mediated by c-Myc and p53 [17,18] or by tumour necrosis factor (TNF)-α independent of nuclear factor-kappa B (NF-κB) [13]. Recently, it was found that chloride efflux could be triggered by reactive oxygen species (ROS)-dependent translocation of CLIC4 on plasma membrane [19]. Cytoplasmic CLIC4 has multiple functions in apoptosis induction, metabolic stress, growth inhibition and inflammatory response, but the role of CLIC4 in radiation response has not been reported yet.
Our previous studies found that intracellular homeostasis plays a key role in cell radiosensitivity. Complex interactions between pro-oxidants and antioxidants are crucial in maintaining the normal status of intracellular homeostasis. Radiation-induced ROS and reactive nitrogen species (RNS) could cause oxidative damage to proteins, lipids and DNA and even influence cell microenvironment that is closely related to radioresistance [20]. For example, the increase of ROS level in the irradiated human hepatocyte carcinoma HepG2 cells sensitized cellular radiation responses by inhibiting cell proliferation [21].
In this study, we demonstrated that the high expression and cluster distribution of CLIC4 in tumor cells regulated microenvironment of intracellular RNS and then contributed to radioresistance of NPC. It is speculated that CLIC4 has potential to become a biomarker of predicting the prognosis and radioresistance of NPC patients.
Material and methods
NPC patient specimens and online expression data
Patients with diagnosed NPC were consecutively recruited from 2007 to 2018 at the Southern Hospital, Southern Medical University (Guangzhou, China). All patients were treated with IMRT with a total dose of 68 to 70 Gy in tumor planning target volume (PTV), 5 times a week for 30 to 33 times in all. Paraffin-embedded NPC specimens (tumour tissues) used for gene expression analysis and immunohistochemistry (IHC) were obtained from 25 patients who had undergone conventional radiotherapy. Tumour regression status was evaluated by tumour enhanced magnetic resonance imaging (MRI) or computed tomography (CT) of head and neck according to the Response Evaluation Criteria in Solid Tumours (RECIST) in three months after treatment completion. Patients with complete remission (CR) and partial response (PR) were classified as radiosensitive group (RS), while those with stable diseases (SD) and progressive diseases (PD) were classified as radioresistant group (RR). Detailed clinicopathological parameters of patients are listed in Table S1. Ethical approval for the study of human subjects was obtained from the research and ethics committee of Nanfang Hospital, and the informed consent was obtained from each patient. Paired expression data of head and neck squamous cell carcinoma (HNSC) cases were obtained from the Cancer Genome Atlas (TCGA) Research Network. The data of the gene expression profile across all tumor samples and paired normal tissues were obtained from GEPIA2, a web server for analyzing the RNA sequence data from TCGA and GTEx projects, using a standard processing pipeline.
Microarray assay and gene expression analysis
Three formalin-fixed paraffin-embedded (FFPE) NPC samples with CR and 3 FFPE samples before radiotherapy from RR group were used for genechip screen. Briefly, total RNA of these samples was extracted and purified using RecoverAllTM Total Nucleic Acid Isolation kit (Ambion, Austin, TX, US). RNA was amplified, labeled and purified by using Ovation FFPE WTA system (NuGEN, San Carlos, CA, US) and FL-OvationTM cDNA Biotin Module V2 (NuGEN) to obtain the biotin-labeled cRNA. Array hybridization was performed by using GeneChip® Hybridization, wash and stain kit (Affymetrix, Santa Clara, CA, US) in Hybridization Oven 645 and Fluidics Station 450 (Affymetrix) followed the manufacturer’s instructions. Slides were scanned by GeneChip® Scanner 3000 and analyzed by Command Console Software 4.0 (Affymetrix) with default settings. Raw data were normalized by MAS 5.0 algorithm, GeneSpring Software12.6.1 (Agilent technologies, Santa Clara, CA, US).
Tumour immunohistochemistry assay
Immunohistochemical (IHC) assay of CLIC4 protein expression was performed in 10 radioresistant and 9 radiosensitive NPC patients’ tissue samples. The tumour sections were incubated with primary antibodies of anti-CLIC4 (Santa Cruz Biotechnology Inc., Dallas, TX, USA; 1:200) for 12 h at 4°C, and incubated with HRP-labeled secondary antibodies, then visualized with DAB kit (Dako, Glostrup, Denmark). Each tissue section was photographed in at least 6 fields randomly and analyzed with the ImageJ software (National Institutes of Health, USA). The average optical density of positive areas in each field was counted from 9-10 independent tumour samples.
Cell culture
The human NPC cell lines, CNE1 and CNE2, were obtained from the Cancer Center of Fudan University (Shanghai, China) and authenticated by Short Tandem Repeat (STR) analysis (Genesky Biotechnologies Inc., Shanghai). These cells were cultured with RPMI 1640 medium (Gibco Invitrogen, Grand Island, NY, USA) supplemented with 10% fetal calf serum (FCS, Gibco) and 1% penicillin-streptomycin (Gibco) and maintained in an atmosphere of 5% CO2 at 37°C.
Establishment of radioresistant NPC cell lines
NPC cell lines of CNE1 and CNE2 were seeded at a density of 1 × 106 per 10 cm dish. After 24 h of culture, cells received fractionated doses (2, 2, 4, 4, 4, 4, 6, 6, 6, 6, 8, 8 Gy) of γ-rays at a dose rate of 0.73 Gy/min. The γ-rays with a LET of 0.25 keV/µm were generated from a 137Cs irradiator (Nordion International Inc., Kanata, Ontario, Canada). After each radiation, cells were cultured for a few days to allow recovery and then the survived cells were exposed to next irradiation. After the final irradiation, cells became more radioresistant than their parent cells. The radioresistant cells were named as CNE1-R and CNE2-R cells (NPC-R cells collectively) and their parent cells were classified into radiosensitive group (NPC cells collectively). Further experiments, NPC-R cells were used within 4 to 10 passages after establishment.
TMT (Tandem mass tags) assay
Total protein samples were extracted from NPC and NPC-R cells using lysis buffer (8 M urea, 1% protease inhibitor cocktail) and centrifuged at 12,000 g for 10 min at 4°C. After trypsin digestion, peptides were desalted by Strata X C18 SPE column (Phenomenex) and dried in vacuum. Peptide was reconstituted in 0.5 M tetraethyl-ammonium bromide (TEAB) and processed according to the manufacturer’s protocol for TMT kit. Peptide mixtures were then incubated for 2 h at room temperature and pooled, desalted and dried by vacuum centrifugation. The tryptic peptides were dissolved in 0.1% formic acid, directly loaded onto a home-made reversed-phase analytical column for LC-MS/MS analysis. The MS/MS data were analyzed with Maxquant search engine (v.1.5.2.8).
Cell irradiation
The doubling time of NPC and NPC-R cells was approximately 20 to 22 h. At 18 h before irradiation, exponentially growing cells were trypsinized to generate single-cell suspension and seeded in six-well culture plates, Mylar-film based dishes and T-25 bottles, respectively. Keeping cells in the same cell cycle phases, the NPC and NPC-R cells were exposed to γ-rays, carbon ions (LETs of 31.6 and 70 keV/μm) and α-particles, respectively. The α-particles were generated from a 241Am plate source (Atom High Tech Co. Ltd., Beijing, China). The characteristic of α-particles with a LET of 100 keV/μm has been described before [22]. 290 MeV/u carbon ions with a LET of 70 keV/µm were accelerated by the HIMAC facility of NIRS (National Institute of Radiological Sciences, Japan) [23]. 80 MeV/u carbon ions with a LET of 31.6 keV/µm were accelerated by the Heavy Ion Research Facility in Lanzhou (HLRFL, Institute of Modern Physics, Chinese Academy of Sciences, China).
Clonogenic survival assay
After irradiation, cells were reseeded and cultured for 9-12 days to form colonies and then fixed and stained with crystal violet. The number of surviving colonies with >50 cells was counted and the survival fractions (SF) of irradiated cells was normalized to that of nonirradiated control cells. The dose response curve of survival was fitted using the single-hit multi-target model SF = 1-(1-exp (-k*D))N.
Transfection of lentiviral vector and chemical treatment
The cells were transfected with the recombinant lentiviral vector containing the full coding region of CLIC4 (Lv-CLIC4) or the shRNA against CLIC4 and their negative control vectors purchased from Hanyin Biotechnology Co. Ltd (Shanghai, China). Two target sequences from the RNA interference Consortium shRNA Library were used i.e., shRNA-1: GAT GGC AAT GAA ATG ACA TTA, shRNA-2: TAT GCC CTC CCA AGT ACT TAA. Cells were transduced using shRNA-1, shRNA-2 or their combination to knock down CLIC4, respectively. For transfection, NPC cells were cultured with lentivirus at a multiplicity of infection (MOI) of 20, and puromycin (Sigma-Aldrich, St. Louis, MO, USA) at a final concentration of 1 μg/ml was added into the culture medium to select the transfected cells. The efficiencies of CLIC4 knockdown and overexpression were monitored by Western blot assay. In some experiments, NPC-R cells were treated with 0.1 mM aminoguanidine (AG, SelleckChem. Shanghai, China) for 12 h to inhibit iNOS activity [24].
Immunofluorescence (IF) assay
At indicated time point after 4 Gy γ-ray irradiation, the formation of γ-H2AX, CLIC4 and iNOS in cells were detected. In brief, the exponentially growing cells on glass coverslip were washed with phosphate buffered saline (PBS) triply and permeabilized with 0.5% Triton X-100 solution for 15 min at room temperature. After blocking with 10% normal goat serum for 1 h, cells were incubated with primary antibodies at 4°C overnight. The primary antibodies included anti-CLIC4 (Santa Cruz, 1:200), anti-iNOS (Abcam, Cambridge, MA, USA; 1:200), and anti-phospho-H2AX (γ-H2AX) (Cell Signaling Technology, Inc., Danvers, MA, USA; 1:200). Then the cells were washed with PBS and probed with FITC/PI-conjugated secondary antibodies (Cell Signaling Technology, 1:400) at room temperature for 1 h. Cell nuclei were counterstained with DAPI for 5 min. The images in at least 6 fields were randomly captured using a fluorescence microscope (Zeiss Axioplan, Germany) and analyzed with the ImageJ software.
Western blot assay
The total cell protein was extracted using RIPA lysis buffer, and the nuclear protein was extracted according to instructions of nuclear and cytoplasmic extraction reagent kit (Beyotime Biotech Ltd., Haimen, China). All proteins were measured by Western blot assay using the following antibodies: anti-p-ATM (Ser1981), anti-p-ATR (Thr1989), anti-Rad51 (Cell Signaling Technology, 1:1000), anti-CLIC4 (Santa Cruz, 1:1000), anti-iNOS (Abcam, 1:500), anti-lamin A/C (Abclonal Technology, Rocky Hill, CT, USA) and anti-β-tubulin (Beyotime, 1:1000). Proteins transferred on a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA) were incubated with the primary antibodies overnight at 4°C and then labeled with HRP-conjugated antibodies (Beyotime; 1:1000) for 2 h at room temperature. The protein bands were visualized using the ChemiDoc XRS system (Bio-Rad Laboratories, Hercules, CA, USA) and their densities were measured using the Quantity One software (Bio-Rad Laboratories).
Intracellular ROS measurement
Intracellular ROS was detected by cell permeable molecular probe of 2’,7’-dichlorofluorescein diacetate (DCFH-DA). Cells were incubated in serum-free medium containing 3 μM DCFH-DA (Beyotime) for 20 min at 37°C and then washed with PBS twice to remove any residual DCFH-DA. DCF fluorescence intensity was immediately detected by a microplate reader (Synergy H1, BioTek Instruments, Vermont, USA) with an excitation wavelength at 488 nm and an emission wavelength at 535 nm. The relative level of ROS in the irradiated cells was normalized to the control without irradiation.
Nitric oxide measurement
NO free radical has a very short half-life and can be oxidized to form two stable NO products of nitrate (NO3 -) and nitrite (NO2 -) [25]. The NO free radical was quantified indirectly by measuring the concentrations of nitrate and nitrite in the irradiated cells and their culture media with a NO assay kit (Beyotime) according to the manufacturer’s protocol. After reaction, the absorbance of sample was measured at 540 nm using a microplate reader.
Functional enrichment analysis
Differential expressions of proteins and mRNAs were subjected to Gene Ontology (GO) and biological pathway enrichment analysis using the FunRich tool against human background database. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were conducted using the database DAVID Bioinformation Resources 6.8 (the Database for Annotation, Visualization and Integration Discovery).
Statistical analysis
Data from more than 3 independent experiments are presented as means ± SEM. Student’s t-tests are used for statistical analysis using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The dose response curves of cell survival were compared using the two-way ANOVA. P<0.05 was considered as significant difference between indicated groups.
Results
High expression of CLIC4 gene in HNSC and NPC
To identify genes involved in radioresistance of NPC, we conducted genechip analysis of tumour samples from radioresistant NPC patients (n=3) and radiosensitive NPC patients (n=3). We also analyzed the gene expression data of HNSC cases from TCGA database. After comprehensive analysis of the differentially expressed gene data from the genechip assay and TCGA database, we found that the expression level of CLIC4 in radioresistant NPC tumor samples was 1.36-times higher than that in radiosensitive NPC tumor samples (Figure 1A). CLIC4 expression in the HNSC tissues (n=520) was even 3.43-fold of that in normal tissues (n=44) (Figure 1B). To further identify the clinical implication of CLIC4, we measured the protein expression of CLIC4 in tumour tissues of other radiosensitive NPC patients (n=9) and radioresistant NPC patients (n=10) with immunohistochemistry assay. Figure 1C showed that the CLIC4 protein had a higher level in radioresistant NPC tumour tissue and localized mainly on the cell membrane and cytoplasm. Interestingly, the cells with high level of CLIC4 protein were distributed in cluster inside of radioresistant tumour but they were scattered in the radiosensitive tumour.
Figure 1.

Expression of CLIC4 in head and neck squamous cell carcinoma (HNSC) and nasopharyngeal carcinoma (NPC). A. Relative expression of CLIC4 in radioresistant (RR) NPC (n=3) and radiosensitive (RS) NPC (n=3) tumor tissues analyzed by genechip technology (*P<0.05). B. Expression of CLIC4 in primary HNSC and normal epithelium tissues (data from TCGA; raw data was normalized with the TMM method and transformed to count per million reads). C. Expression and distribution of CLIC4 in RR (n=10) and RS (n=9) NPC tumor tissues. Scale bars, 100 μm, 20 μm. CLIC4 IHC staining score showed the numbers of positive CLIC4 cells per 100 cells (***P<0.001). D. Ratio of the relative expression of CLIC4 in NPC and NPC-R cell lines examined by TMT technology. E. Protein expression of CLIC4 in NPC and NPC-R cell lines. β-tubulin was used as the internal control (***P<0.001 NPC-R versus NPC, ###P<0.01 CNE1 versus CNE2). Results correspond to the means ± SEM of three independent experiments with three replicates in each case.
To further investigate the role of CLIC4 in NPC radioresistance, we established radioresistant cell lines (NPC-R, including CNE1-R and CNE2-R) from NPC cell lines (CNE1 and CNE2) by fractionation irradiation of γ-rays, and then detected the differentially expressed proteins in these cell lines by TMT assay. It was found that CLIC4 protein level in NPC-R cells was about 1.6-fold of that in NPC cell lines (Figure 1D), in consistent with the status of clinical samples. In addition, the Western blot assay also showed that the expression of CLIC4 in NPC-R cells was higher than that in corresponding NPC cells (Figure 1E). These results manifest that CLIC4 contributes to the radioresistance of both NPC cells and patients, accordingly, it may be applicable as a potential biomarker of radioresistance.
Effect of CLIC4 on radiation sensitivity
Figure 2A demonstrated that, after γ-ray irradiation, the survival of NPC-R cells was higher than that of NPC cells. To understand the role of CLIC4 in cell radiation response, we decreased CLIC4 expression in NPC and NPC-R cells using shRNA-1, shRNA-2, or their combination (shRNA or sh-CLIC4 hereafter). Western blot assay verified that all of these shRNA transfection were effective in inhibiting the expression of CLIC4 (Figures 2B, S1A). In addition, we also overexpressed CLIC4 in NPC cells using a lentiviral vector (Figure 2C). It was found that the interfering of CLIC4 sensitized NPC and NPC-R cells to γ-ray irradiation (Figures 2D, S1B), whereas the overexpression of CLIC4 further enhanced the radioresistance of NPC cells (Figure 2E).
Figure 2.

CLIC4 regulated radiation sensitivity of NPC cells in γ-ray irradiation. A. Survival curves of CNE-1, CNE1-R, CNE-2 and CNE2-R cells exposed to γ-rays (**P<0.01, ***P<0.001 NPC-R versus NPC; ##P<0.01 CNE1 versus CNE2). B. Knockdown of CLIC4 using shRNA in NPC and NPC-R cells. C. CLIC4 in NPC cells overexpressed by the recombinant lentiviral vector with the full coding region of CLIC4. D. CLIC4 knockdown decreased radioresistance of NPC and NPC-R cells. E. CLIC4 overexpression increased radioresistance of CNE1, CNE2 cells. The dose-survival curves were fitted using the single-hit multi-target model. Results correspond to the means ± SEM of three independent experiments with three replicates in each case.
DNA damage response of NPC cells
The capacity of DNA damage response (DDR) has been considered a key factor in regulating cell radiosensitivity [26]. To compare DDR capacity between NPC and NPC-R cells, we conducted immunofluorescence assays of γ-H2AX foci at 0.5 h, 2 h and 24 h after 4 Gy of γ-ray irradiation (Figure 3A). It was found that the formations of γ-H2AX foci in NPC cells were slightly lower than in NPC-R cells from 0.5 h to 24 h after irradiation, except a significant difference at 2 h after irradiation (Figure 3B, 3C). These results indicate that the capability of DNA damage response may not be the main cause of the radiosensitivity difference between NPC and NPC-R cells.
Figure 3.
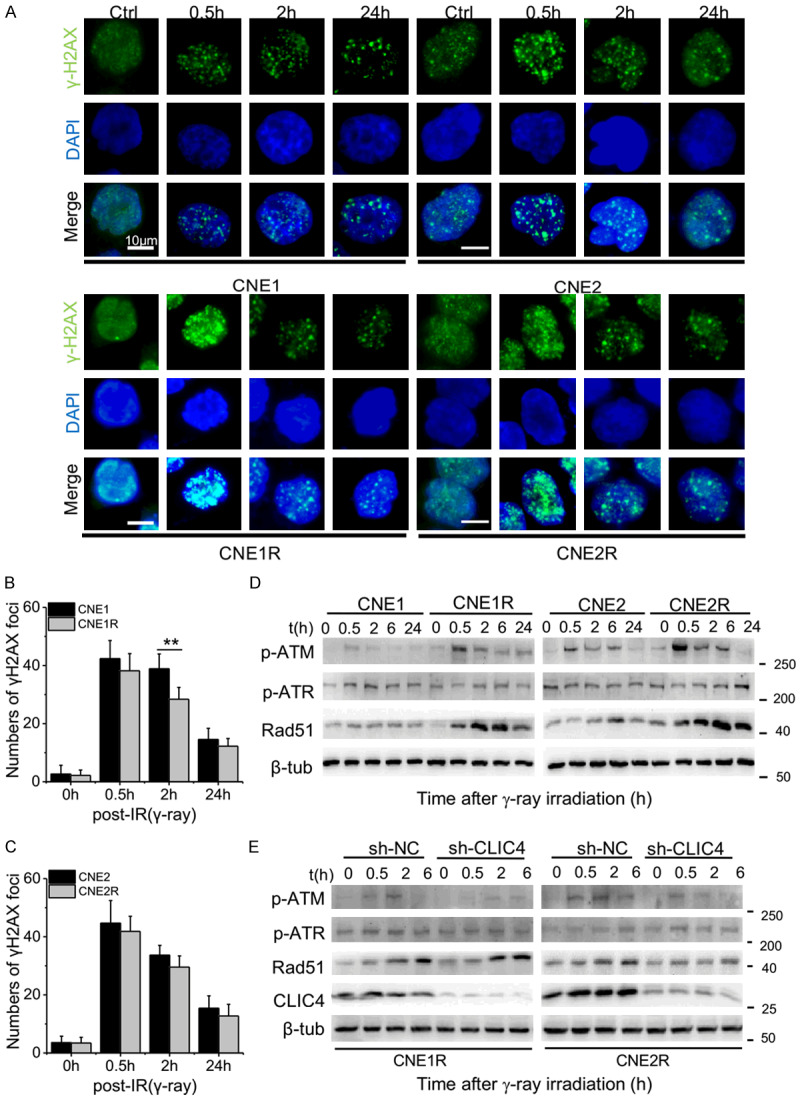
CLIC4 affected the DNA damage response of NPC cells. (A) Representative immunofluorescence images and graphs showing cells immunostained with antibodies against γ-H2AX. The indicated cells were exposed to 4 Gy of γ-rays and stained at 0.5, 2 and 24 h after irradiation. Untreated cells (0 h) were also stained and served as a control. The nuclei were stained with DAPI. Scale bars, 10 μm. Numbers of γ-H2AX foci per positive CNE1/CNE1-R cells (B) and CNE2/CNE2-R cells (C) (**P<0.01). (D) Expressions of DNA repair response proteins in the NPC and NPC-R cells at 0.5, 2, 6 and 24 h after irradiation. (E) Expressions of DNA repair response proteins and CLIC4 in the CLIC4-knockdown cells at 0.5, 2 and 6 h after irradiation. β-tubulin was used as the internal control. Western blot analysis was representative of three independent experiments.
Next, we detected the time response of some DDR proteins. Expressions of p-ATM (Ser1981), p-ATR (Thr1989) and Rad51 were detected in NPC and NPC-R cells after γ-ray irradiation (Figure 3D). Results showed that the expression of p-ATR had no significant difference between NPC and NPC-R cells. Expressions of Rad51 and p-ATM in NPC-R cells were higher than that in NPC cells and they were enhanced after irradiation. The expression of p-ATM in both cell lines increased to a high level at 0.5 after irradiation and then declined after 2 h after irradiation.
To explore whether CLIC4 is involved in DDR, we detected the expression of DDR proteins in CLIC4 knockdown NPC-R cells after γ-ray irradiation (Figure 3E). It could be seen that CLIC4 knockdown treatment had no significant influence in the expressions of p-ATR and Rad51 but it weakened the expressions of p-ATM in NPC-R cells at 0.5 h and 2 h after irradiation.
Effects of CLIC4 on the cellular oxidative stress after irradiation
It has been reported that the regulation of intracellular ROS and RNS are efficient antioxidant defenses against irradiation [27]. To know the relationship between CLIC4 and ROS/RNS in regulating radiosensitivity, we measured these free radicals in NPC and NPC-R cells after γ-ray irradiation. Figure 4A illustrates that the intracellular ROS in both NPC and NPC-R cells increased until 2 h after irradiation and then decreased quickly to control level at 6 h after irradiation. At 2 h after irradiation, the ROS level in NPC cells was about 1.35 times of NPC-R cells. When CLIC4 was knocked down, the intracellular ROS in all cell lines were significantly increased to 1.31-1.34 times of shRNA negative control cells at 2 h after irradiation. When CLIC4 was overexpressed, the intracellular ROS were significantly reduced by 1.3-1.4 times of the negative control cells at 2 h after irradiation (Figure 4B). Therefore, CLIC4 could negatively regulate radiation-induced generation of ROS.
Figure 4.

Time responses of intracellular ROS and NO in the medium of cells irradiated by 4 Gy irradiation of γ-rays. (A) Time response of intracellular ROS in NPC-R and NPC cells at 0.5, 2, 6 and 24 h after irradiation. (B) Time responses of intracellular ROS in CLIC4-knockdown and overexpression cells at 0.5, 2, 6 and 24 h after irradiation. (C) Time responses of the concentration of NO in the medium from NPC-R and NPC cells at 0.5, 2 and 6 h after irradiation. (D) Time responses of the concentration of NO in the medium from CLIC4-knockdown and overexpression cells at 0.5, 2 and 6 h after irradiation. Results correspond to the means ± SEM of three independent experiments with three replicates in each case. In (A) and (C), ***P<0.001 CNE1-R versus CNE1; ###P<0.001 CNE2-R versus CNE2. In (B) and (D), *P<0.05, **P<0.01, ***P<0.001 sh-CLIC4 versus negative control (NC); ##P<0.01, ###P<0.001 lv-CLIC4 versus NC.
In contrast to ROS, the concentration of NO in the medium of irradiated NPC-R cells were higher than that of NPC cells and it was maintained at a higher concentration for a long time after irradiation (Figure 4C). At 2 h after irradiation, the concentration of NO in NPC-R culture medium approached to about 2.35-times of NPC cells. When CLIC4 was knocked down, the concentration of NO in the cell culture medium decreased to 0.62-0.74 of shRNA control cells. When CLIC4 was overexpressed, the concentration of NO increased to about 2.1-2.6 times of the negative control cells at 2 h after irradiation (Figure 4D). Accordingly, CLIC4 promoted the generation of NO after irradiation.
CLIC4 enhances the nuclear translocation of iNOS
NO in the medium should be released from irradiated cells. Figure 5A shows that intracellular NO in NPC-R cells was much higher than NPC cells, and the intracellular NO was reduced by CLIC4 knockdown. Further immunofluorescence assay showed that the expressions of CLIC4 and iNOS in NPC and NPC-R cells were increased after irradiation and they had much higher level in NPC-R cells (Figure 5B). It was found that radiation-induced iNOS protein was diffused in the whole cell at 2 h after irradiation and then mainly located in nucleus at 6 h after irradiation, indicating nuclear translocation of iNOS. Meanwhile, CLIC4 was mainly located in the cell membrane when no stress. After 2 h of γ-ray irradiation, the protein level of CLIC4 in cytoplasm became much higher than non-irradiated control, indicating that radiation stimulated the movement of CLIC4 from membrane to cytoplasm.
Figure 5.

CLIC4 enhanced nuclear translocation of iNOS resulting in radioresistance. (A) Relative concentration of intracellular NO in the CLIC4-knockdown NPC-R and NPC cells (**P<0.01, ***P<0.001 versus NC; ###P<0.001 NPC-R versus NPC). (B) Representative immunofluorescence staining for CLIC4 (green) and iNOS (red) in NPC and NPC-R cells. Nuclei were stained with DAPI. Scale bars, 20 μm. (C) The protein expressions of CLIC4 and iNOS in the nucleus of NPC and NPC-R cells. Lamin A/C was used as the internal control. (D) Expressions of iNOS and CLIC4 in the CLIC4-knockdown cells. (E) Expressions of CLIC4 and iNOS in the nucleus of CLIC4-knockdown cells at 6 h after irradiation. (F) Radiosensitivity of NPC-R cells treated with Aminoguanidine (AG) and exposed to γ-ray (**P<0.01, ***P<0.001 AG versus Ctrl). In (B-E), cells were exposed to 4 Gy of γ-rays and detected at indicated time points. Untreated cells (0 h) were also stained and served as control. Results correspond to the means ± SEM of three independent experiments with three replicates in each case.
Western blot assay of nuclear proteins further confirmed the nuclear translocation of iNOS (Figure 5C). At 6 h after irradiation, nuclear iNOS protein approached to a high level in nucleus, especially in NPC-R cells. To explore the relationship between CLIC4 and iNOS, we detected the expression of iNOS protein in CLIC4-knockdown cells and found that knockdown of CLIC4 decreased the expression of iNOS at different time-points after irradiation (Figure 5D). Moreover, after knocking down CLIC4, nuclear iNOS expression was decreased at 6 h after irradiation (Figure 5E), indicating that the nuclear translocation of iNOS was also inhibited by CLIC4 suppression.
To verify the effect of NO in radioresistance, we treated CNE1-R and CNE2-R cells with aminoguanidine (AG) to inhibit iNOS (Figure S2). It was found that AG treatment increased the radiosensitivity of NPC-R cell lines (Figure 5F), indicating the radioresistance was associated with the level of intracellular NO in cells.
Radiosensitivity of NPC and NPC-R cells exposed to carbon ions irradiation
Due to the advantageous physical and biological characteristics of carbon ion beam, IMCT has been used to improve the outcome of locally recurrent NPC [28,29]. We wondered whether regulation of CLIC4 in radioresistance worked on heavy ion irradiation and then measured the survival curves of NPC and NPC-R cell lines with or without CLIC4 regulation after high-LET irradiations. When cells were exposed to 70 keV/μm (Figure 6A) and 31.6 keV/μm carbon ions (Figure 6B), the survival fractions of NPC and NPC-R cells had no significant difference, and so do the difference of survival curves between CNE1 and CNE2 cells. In addition, when cells were exposed to α-particles with a LET of 100 keV/μm, the survival of NPC and NPC-R cells were also similar (Figure S3). These results indicate that, in comparison to NPC cells, NPC-R cells are radioresistant to γ-rays but not to high-LET irradiations.
Figure 6.

CLIC4 had no effect on the radiosensitivity of high-LET irradiation. A. Survival curves of CNE-1, CNE1-R, CNE-2 and CNE2-R cells exposed to carbon ions (70 keV/µm). B. Survival curves of CNE-1, CNE1-R, CNE-2 and CNE2-R cells exposed to carbon ions (31.6 keV/µm). C. CLIC4 knockdown in CNE1, CNE1-R, CNE2, CNE2-R cells did not influence the radiosensitivity of CNE1 and CNE2 cells to carbon ion irradiation. D. CLIC4 overexpression did not influence the radiosensitivity of CNE1 and CNE2 cells to carbon ion irradiation. The dose-survival curves were fitted using the single-hit multi-target model. Results correspond to the means ± SEM of three independent experiments with three replicates in each case.
Furthermore, we measured the survival curves of CLIC4 regulated-cells exposed to 70 keV/μm carbon ions irradiation. It was found that, the radiation sensitivity of these NPC cells were not influenced by CLIC4 regulation of either knockdown (Figure 6C) or overexpression (Figure 6D). Therefore, CLIC4 has no influence in the sensitivity of NPC cells exposed to high-LET carbon ion irradiation.
Discussion
Radioresistance-induced treatment failure and local recurrence continue to be a major problem in the radiotherapy of NPC. The mechanism of tumor radioresistance has been a long research topic. Identification of radioresistance associated genes will be helpful for finding biomarkers to predict NPC radiosensitivity and tailoring individualized therapies for NPC patients. In this study, we analyzed NPC samples from RS/RR patients and NPC/NPC-R cells using genechip and TMT assays to identify differential expressed genes and proteins between radioresistant and radiosensitive samples. GO pathway enrichment analysis disclosed that the genes concerning immune system, oxidation-reduction and metabolic process, and cell death regulation were mainly enriched for biological process. Genes of extracellular region part, nucleoplasm and membrane-enclosed lumen were mainly enriched for cell component. Genes correlated with oxidoreductase activity, adhesion and ion binding, endopeptidase and DNA topoisomerase activity were mainly enriched for molecular functions (Figure S4). Enrichment analysis of KEGG pathway revealed that the differential expressed genes might be associated with many pathways including glutathione metabolism, carbon metabolism, ECM-receptor interaction, cytokine receptor interaction, cell adhesion molecules, sphingolipid metabolism and NF-κB pathway (Figure S5). Taking these analysis results together, it can be identified that the major differential genes play cellular functions in inflammatory response, extracellular organization, metabolism and cell adhesion.
It is believed that DNA damage response and repair capacity are closely related to radiation resistance. Ataxia-telangiectasia-mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) proteins are critical regulators of DDR [30]. ATR is activated in response to a variety of DNA damage such as single-strand break. ATM can be phosphorylated at residue S1981 after double strand damage (DSB) formation. Our study showed that the expression level of p-ATR had no significant difference between radioresistant and radiosensitive groups. Although the expressions of p-ATM and Rad51 were enhanced in NPC-R cells after γ-ray irradiation, the numbers of γ-H2AX foci were similar across the NPC and NPC-R cells at 24 h post-irradiation. Therefore, the biomarker of radioresistance might not be on the DNA damage repair pathway, in consistent with other reports [7,31].
In addition to the DNA damage repair capacity, the redox system including ROS and NOS participated in radiation-induced damage through different regulatory mechanisms [32]. Nevertheless, the endogenous NO at low-to-moderate steady can increase drug resistance and also act as a pro-growth or pro-migration signaling molecule [33]. In our study, the up-regulation of endogenous NO and down-regulation of ROS by CLIC4 increased radioresistance synergistically in NPC-R cells.
Moreover, we wonder whether CLIC4 contributes to other tumors’ radioresistance. Comparing the tumor samples and paired normal tissues, CLIC4 is highly expressed in glioblastoma (GBM), HNSC, esophageal carcinoma (ESCA), and pancreatic adenocarcinoma (PAAD), but it is lower in breast invasive carcinoma (BLCA), colon adenocarcinoma (COAD), kidney chromophobe (KICH), lung adenocarcinoma (LUAD), rectum adenocarcinoma (READ), and uterine carcinosarcoma (UCS) (Figure S6). CLIC4 has been declared as a novel target for anti-cancer therapy by enhancing cell cycle arrest and accelerating apoptosis [34]. Increasing evidence is accumulating that the expression and activity of CLIC4 have clinical relevance, while the exact mechanisms by which CLIC4 functions as a tumor suppressor or stromal activator are still open questions. It has been reported that loss of CLIC4 in tumor cells marks malignant progression in renal, ovarian, and breast cancers [35]. The results from some researches strongly implicated CLIC4 in the suppression of tumor cell growth and suggested that understanding the mechanism of repression of CLIC4 in squamous tumors is a priority for potential therapy [36]. But in some cancers with high expression of CLIC4, such as HNSC, its function and mechanisms are almost blank in tumor development and therapy.
In this study, we investigated the influence of CLIC4 on radiosensitivity through redox system. The overexpression of CLIC4 increased the radioresistance of NPC cells, whereas inhibiting CLIC4 expression enhanced radiosensitivity in NPC and NPC-R cells. We also demonstrated that CLIC4 contributes to radioresistance by activating iNOS signaling pathway. After irradiation, CLIC4 could regulate p-ATM but not Rad51. Both of them respond to DNA DSB repair. Therefore, it hard to say that CLIC4 has a direct influence on DNA damage repair.
Recent data suggest that, under a variety of cellular stress stimuli, nuclear translocation of CLIC4 is mediated by NO-induced S-nitrosylation at critical cysteine residue so that the redox-sensitive tertiary structure of CLIC4 is altered to increase its association with nuclear proteins [37,38]. Our study found that, after irradiation, CLIC4 was transported from membrane to cytoplasm and nucleus, in concurrent with the induction and nuclear translocation of iNOS protein. More importantly, when CLIC4 was knocked-down or over-expressed in NPC and NPC-R cells, ROS was increased or reduced in the irradiated cells, respectively; but opposite to ROS, NO was reduced or increased in those irradiated cells, respectively. Meanwhile, inhibition iNOS with AG relieved the radioresistance of NPC-R cells. In addition, as an upstream signaling of iNOS [39,40], NF-κB pathway positively regulates DDR signaling kinases [41]. It was also reported that CLIC4 was a transcriptional target of NF-κB [37], which is consistent with our enrichment analysis of KEGG pathway in Figure S5 that the differential expressed genes between RR and RS group are associated with NF-κB pathway.
While IMRT is the current preferred method in the radiotherapy of NPC, IMCT has dosimetric advantages with less radiation damage to normal structures, more overall survival, and local and regional recurrence-free survival for patients with NPC [4,5]. Our results showed the radioresistance to photon irradiation was invalid to carbon ions or α-particles irradiation, which provides additional positive information to the application of high-LET particle therapy of NPC.
Taken together, we applied genomics and proteomic approaches and identified differential genes or proteins in the radioresistant NPC patients and cell lines (CNE1-R and CNE2-R). High expression of CLIC4 correlated with a high relapse risk and poor prognosis in NPC patients, which implicates that CLIC4 is applicable as a potential predictive biomarker for NPC prognosis and local recurrence. Moreover, the oxidative stress response was an important factor in the generation of NPC radioresistance which could be overcome by high-LET particle ionizing radiation. These novel findings should have clinical value in distinguishing radioresistant individual from primary-care NPC patients for personalized radiotherapy.
Acknowledgements
The authors thank the operating crew of the Heavy Ion Research Facility in Lanzhou (HIRFL) for generating the carbon-ion beam used during our experiment. This study was mainly supported by the National Key R & D Program of China (No. 2017YFC0108604), the National Natural Science Foundation of China (Nos. 31770910, 31570850 and 11775052), the Research Project with Heavy Ions at NIRS-HIMAC of Japan (Proposal Nos. 17J121), and Health & Medical Collaborative Innovation Project of Guangzhou City, China (201803040003).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Mao YP, Tang LL, Chen L, Sun Y, Qi ZY, Zhou GQ, Liu LZ, Li L, Lin AH, Ma J. Prognostic factors and failure patterns in non-metastatic nasopharyngeal carcinoma after intensity-modulated radiotherapy. Chin J Cancer. 2016;35:103. doi: 10.1186/s40880-016-0167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang MX, Li J, Shen GP, Zou X, Xu JJ, Jiang R, You R, Hua YJ, Sun Y, Ma J, Hong MH, Chen MY. Intensity-modulated radiotherapy prolongs the survival of patients with nasopharyngeal carcinoma compared with conventional two-dimensional radiotherapy: a 10-year experience with a large cohort and long follow-up. Eur J Cancer. 2015;51:2587–2595. doi: 10.1016/j.ejca.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Hu J, Bao C, Gao J, Guan X, Hu W, Yang J, Hu C, Kong L, Lu JJ. Salvage treatment using carbon ion radiation in patients with locoregionally recurrent nasopharyngeal carcinoma: initial results. Cancer. 2018;124:2427–2437. doi: 10.1002/cncr.31318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis GD, Holliday EB, Kocak-Uzel E, Hernandez M, Garden AS, Rosenthal DI, Frank SJ. Intensity-modulated proton therapy for nasopharyngeal carcinoma: decreased radiation dose to normal structures and encouraging clinical outcomes. Head Neck. 2016;38(Suppl 1):E1886–1895. doi: 10.1002/hed.24341. [DOI] [PubMed] [Google Scholar]
- 6.Liu G, Zeng X, Wu B, Zhao J, Pan Y. RNA-Seq analysis of peripheral blood mononuclear cells reveals unique transcriptional signatures associated with radiotherapy response of nasopharyngeal carcinoma and prognosis of head and neck cancer. Cancer Biol Ther. 2020;21:139–146. doi: 10.1080/15384047.2019.1670521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng XP, Yi H, Li MY, Li XH, Yi B, Zhang PF, Li C, Peng F, Tang CE, Li JL, Chen ZC, Xiao ZQ. Identification of biomarkers for predicting nasopharyngeal carcinoma response to radiotherapy by proteomics. Cancer Res. 2010;70:3450–3462. doi: 10.1158/0008-5472.CAN-09-4099. [DOI] [PubMed] [Google Scholar]
- 8.Cai XZ, Zeng WQ, Xiang Y, Liu Y, Zhang HM, Li H, She S, Yang M, Xia K, Peng SF. iTRAQ-based quantitative proteomic analysis of nasopharyngeal carcinoma. J Cell Biochem. 2015;116:1431–1441. doi: 10.1002/jcb.25105. [DOI] [PubMed] [Google Scholar]
- 9.Chang JT, Chan SH, Lin CY, Lin TY, Wang HM, Liao CT, Wang TH, Lee LY, Cheng AJ. Differentially expressed genes in radioresistant nasopharyngeal cancer cells: gp96 and GDF15. Mol Cancer Ther. 2007;6:2271–2279. doi: 10.1158/1535-7163.MCT-06-0801. [DOI] [PubMed] [Google Scholar]
- 10.Chen ZT, Liang ZG, Zhu XD. A review: proteomics in nasopharyngeal carcinoma. Int J Mol Sci. 2015;16:15497–15530. doi: 10.3390/ijms160715497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jentsch TJ. Chloride channels are different. Nature. 2002;415:276–277. doi: 10.1038/415276a. [DOI] [PubMed] [Google Scholar]
- 12.Shukla A, Malik M, Cataisson C, Ho Y, Friesen T, Suh KS, Yuspa SH. TGF-beta signalling is regulated by Schnurri-2-dependent nuclear translocation of CLIC4 and consequent stabilization of phospho-Smad2 and 3. Nat Cell Biol. 2009;11:777–784. doi: 10.1038/ncb1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suh KS, Crutchley JM, Koochek A, Ryscavage A, Bhat K, Tanaka T, Oshima A, Fitzgerald P, Yuspa SH. Reciprocal modifications of CLIC4 in tumor epithelium and stroma mark malignant progression of multiple human cancers. Clin Cancer Res. 2007;13:121–131. doi: 10.1158/1078-0432.CCR-06-1562. [DOI] [PubMed] [Google Scholar]
- 14.Ponnalagu D, Rao SG, Farber J, Xin W, Hussain AT, Shah K, Tanda S, Berryman MA, Edwards JC, Singh H. Data supporting characterization of CLIC1, CLIC4, CLIC5 and DmCLIC antibodies and localization of CLICs in endoplasmic reticulum of cardiomyocytes. Data Brief. 2016;7:1038–1044. doi: 10.1016/j.dib.2016.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh H. Two decades with dimorphic chloride intracellular channels (CLICs) FEBS Lett. 2010;584:2112–2121. doi: 10.1016/j.febslet.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Suh KS, Mutoh M, Nagashima K, Fernandez-Salas E, Edwards LE, Hayes DD, Crutchley JM, Marin KG, Dumont RA, Levy JM, Cheng C, Garfield S, Yuspa SH. The organellular chloride channel protein CLIC4/mtCLIC translocates to the nucleus in response to cellular stress and accelerates apoptosis. J Biol Chem. 2004;279:4632–4641. doi: 10.1074/jbc.M311632200. [DOI] [PubMed] [Google Scholar]
- 17.Shiio Y, Suh KS, Lee H, Yuspa SH, Eisenman RN, Aebersold R. Quantitative proteomic analysis of myc-induced apoptosis: a direct role for Myc induction of the mitochondrial chloride ion channel, mtCLIC/CLIC4. J Biol Chem. 2006;281:2750–2756. doi: 10.1074/jbc.M509349200. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Salas E, Suh KS, Speransky VV, Bowers WL, Levy JM, Adams T, Pathak KR, Edwards LE, Hayes DD, Cheng C, Steven AC, Weinberg WC, Yuspa SH. mtCLIC/CLIC4, an organellular chloride channel protein, is increased by DNA damage and participates in the apoptotic response to p53. Mol Cell Biol. 2002;22:3610–3620. doi: 10.1128/MCB.22.11.3610-3620.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang TT, Lang XT, Xu CF, Wang XQ, Gong T, Yang YQ, Cui J, Bai L, Wang J, Jiang W, Zhou RB. CLICs-dependent chloride efflux is an essential and proximal upstream event for NLRP3 inflammasome activation. Nat Commun. 2017;8:202. doi: 10.1038/s41467-017-00227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dou Y, Liu Y, Zhao F, Guo Y, Li X, Wu M, Chang J, Yu C. Radiation-responsive scintillating nanotheranostics for reduced hypoxic radioresistance under ROS/NO-mediated tumor microenvironment regulation. Theranostics. 2018;8:5870–5889. doi: 10.7150/thno.27351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XD, Tu WZ, Chen D, Fu JM, Wang J, Shao CL, Zhang JH. Autophagy suppresses radiation damage by activating PARP-1 and attenuating reactive oxygen species in hepatoma cells. Int J Radiat Biol. 2019;95:1051–1057. doi: 10.1080/09553002.2019.1605461. [DOI] [PubMed] [Google Scholar]
- 22.Ren R, He M, Dong C, Xie Y, Ye S, Yuan D, Shao C. Dose response of micronuclei induced by combination radiation of alpha-particles and gamma-rays in human lymphoblast cells. Mutat Res. 2013;741-742:51–56. doi: 10.1016/j.mrfmmm.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Torikoshi M, Minohara S, Kanematsu N, Komori M, Kanazawa M, Noda K, Miyahara N, Itoh H, Endo M, Kanai T. Irradiation system for HIMAC. J Radiat Res. 2007;48(Suppl A):A15–25. doi: 10.1269/jrr.48.a15. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto H, Hayashi S, Hatashita M, Shioura H, Ohtsubo T, Kitai R, Ohnishi T, Yukawa O, Furusawa Y, Kano E. Induction of radioresistance to accelerated carbon-ion beams in recipient cells by nitric oxide excreted from irradiated donor cells of human glioblastoma. Int J Radiat Biol. 2000;76:1649–1657. doi: 10.1080/09553000050201145. [DOI] [PubMed] [Google Scholar]
- 25.Ko SH, Ryu GR, Kim S, Ahn YB, Yoon KH, Kaneto H, Ha H, Kim YS, Song KH. Inducible nitric oxide synthase-nitric oxide plays an important role in acute and severe hypoxic injury to pancreatic beta cells. Transplantation. 2008;85:323–330. doi: 10.1097/TP.0b013e31816168f9. [DOI] [PubMed] [Google Scholar]
- 26.Rajpurohit YS, Bihani SC, Waldor MK, Misra HS. Phosphorylation of deinococcus radiodurans RecA regulates its activity and may contribute to radioresistance. J Biol Chem. 2016;291:16672–16685. doi: 10.1074/jbc.M116.736389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suman S, Khan Z, Zarin M, Chandna S, Seth RK. Radioresistant Sf9 insect cells display efficient antioxidant defence against high dose gamma-radiation. Int J Radiat Biol. 2015;91:732–741. doi: 10.3109/09553002.2015.1054958. [DOI] [PubMed] [Google Scholar]
- 28.Kong L, Hu J, Guan X, Gao J, Lu R, Lu JJ. Phase I/II trial evaluating carbon ion radiotherapy for salvaging treatment of locally recurrent nasopharyngeal carcinoma. J Cancer. 2016;7:774–783. doi: 10.7150/jca.14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong L, Gao J, Hu J, Hu W, Guan X, Lu R, Lu JJ. Phase I/II trial evaluating concurrent carbon-ion radiotherapy plus chemotherapy for salvage treatment of locally recurrent nasopharyngeal carcinoma. Chin J Cancer. 2016;35:101. doi: 10.1186/s40880-016-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Awasthi P, Foiani M, Kumar A. ATM and ATR signaling at a glance. J Cell Sci. 2015;128:4255–4262. doi: 10.1242/jcs.169730. [DOI] [PubMed] [Google Scholar]
- 31.Qu C, Zhao Y, Feng G, Chen C, Tao Y, Zhou S, Liu S, Chang H, Zeng M, Xia Y. RPA3 is a potential marker of prognosis and radioresistance for nasopharyngeal carcinoma. J Cell Mol Med. 2017;21:2872–2883. doi: 10.1111/jcmm.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei JL, Wang B, Wang HH, Meng LB, Zhao Q, Li XY, Xin Y, Jiang X. Radiation-induced normal tissue damage: oxidative stress and epigenetic mechanisms. Oxid Med Cell Longev. 2019;2019:3010342. doi: 10.1155/2019/3010342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girotti AW. Nitric oxide-mediated resistance to antitumor photodynamic therapy. Photochem Photobiol. 2019 doi: 10.1111/php.13163. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suh KS, Mutoh M, Gerdes M, Yuspa SH. CLIC4, an intracellular chloride channel protein, is a novel molecular target for cancer therapy. J Investig Dermatol Symp Proc. 2005;10:105–109. doi: 10.1111/j.1087-0024.2005.200402.x. [DOI] [PubMed] [Google Scholar]
- 35.Suh KS, Crutchley JM, Koochek A, Ryscavage A, Bhat K, Tanaka T, Oshima A, Fitzgerald P, Yuspa SH. Reciprocal modifications of CLIC4 in tumor epithelium and stroma mark malignant progression of multiple human cancers. Clin Cancer Res. 2007;13:121–131. doi: 10.1158/1078-0432.CCR-06-1562. [DOI] [PubMed] [Google Scholar]
- 36.Peretti M, Angelini M, Savalli N, Florio T, Yuspa SH, Mazzanti M. Chloride channels in cancer: focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochim Biophys Acta. 2015;1848:2523–2531. doi: 10.1016/j.bbamem.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Malik M, Jividen K, Padmakumar VC, Cataisson C, Li L, Lee J, Howard OM, Yuspa SH. Inducible NOS-induced chloride intracellular channel 4 (CLIC4) nuclear translocation regulates macrophage deactivation. Proc Natl Acad Sci U S A. 2012;109:6130–6135. doi: 10.1073/pnas.1201351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik M, Shukla A, Amin P, Niedelman W, Lee J, Jividen K, Phang JM, Ding J, Suh KS, Curmi PM, Yuspa SH. S-nitrosylation regulates nuclear translocation of chloride intracellular channel protein CLIC4. J Biol Chem. 2010;285:23818–23828. doi: 10.1074/jbc.M109.091611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gul A, Kunwar B, Mazhar M, Faizi S, Ahmed D, Shah MR, Simjee SU. Rutin and rutin-conjugated gold nanoparticles ameliorate collagen-induced arthritis in rats through inhibition of NF-kappaB and iNOS activation. Int Immunopharmacol. 2018;59:310–317. doi: 10.1016/j.intimp.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 40.Shao C, Stewart V, Folkard M, Michael BD, Prise KM. Nitric oxide-mediated signaling in the bystander response of individually targeted glioma cells. Cancer Res. 2003;63:8437–8442. [PubMed] [Google Scholar]
- 41.Ozes AR, Miller DF, Ozes ON, Fang F, Liu Y, Matei D, Huang T, Nephew KP. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350–5361. doi: 10.1038/onc.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


