Abstract
BRCA1-associated protein 1 (BAP1) is a member of the ubiquitin C-terminal hydrolase family of deubiquitinating enzymes and is implicated in transcriptional regulation. The BAP1 gene is mutated in 5%-15% of patients with clear cell renal cell carcinoma (ccRCC), the most common form of renal cancer, which suggests that BAP1 is a tumor suppressor. However, whether BAP1 influences the progression of ccRCC tumors expressing wildtype (WT) BAP1 is unclear. Here, we identified DIDO1 as a bona fide substrate for BAP1. DIDO1 is a component of the centrosome proteins and plays an essential role in spindle assembly. BAP1 binds to DIDO1 and stabilizes DIDO1 through de-ubiquitination. BAP1 contributes to chromosome stability partially via DIDO1. A positive correlation was identified between BAP1 and DIDO1 expression in ccRCC tissues. Downregulation of both BAP1-loss and DIDO1 protein expression in ccRCC was associated with adverse clinicopathological features. This study revealed a novel mechanism involving BAP1 in the regulation of DIDO1 stability, and the results also provide insight into the relationship between BAP1 mutations and chromosome instability in ccRCC.
Keywords: BAP1, DIDO1, clear cell renal cell carcinoma, chromosome instability, de-ubiquitination
Introduction
Renal cell carcinoma (RCC) is the most lethal of the common urologic cancers and constitutes 2%-3% of all adult malignant neoplasms [1,2]. Clear cell renal cell carcinoma (ccRCC) is the most common type of RCC, accounting for 70%-80% of all kidney cancers. Despite the emergence of novel targeted therapies over the last decade, including antiangiogenetic drugs and mammalian target of rapamycin inhibitors, the prognosis of ccRCC patients with metastasis or relapse remains poor, with a 5-year survival rate of less than 10% [3]. Therefore, a better understanding of the mechanisms underlying RCC tumorigenesis is required to help in the development and improvement of treatment options for RCC patients.
Recent analyses of the ccRCC tumor genome using next-generation sequencing revealed frequent gene mutations, including BAP1, PBRM1, SETD2, and KDM5C [4-6]. Such genes encode proteins involved in chromatin regulation and function as tumor suppressors. The BAP1 gene encodes the BRCA1-associated protein 1 (BAP1), a deubiquitinating enzyme, that exerts its tumor suppressor activity through deubiquitinating activity and nuclear localization. De-ubiquitination involves the NH2-terminal ubiquitin COOH-terminal hydrolase (UCH) domain, while nuclear localization involves a nuclear localization signal (NLS). As previously reported, BAP1-deficient cancer cells are more vulnerable to γ-radiation and more sensitive to olaparib, which indicates that radiotherapy and PARP inhibitors may be more effective in cases with BAP1 mutations than in cases with wildtype BAP1 [7,8]. However, how a BAP1 mutation contributes to the initiation and progression of ccRCC remains poorly understood.
The ubiquitin ligases select substrates for ubiquitin conjugation, which is reversed by the action of deubiquitinating enzymes [9]. BAP1 is a nuclear deubiquitinating enzyme that was originally identified as a BRCA1-binding protein in a yeast two-hybrid screen [10,11]. BAP1 has been linked to the de-ubiquitination of several cellular substrates, including the transcriptional regulator host cell factor 1 (HCF1), histone H2Aub, Ino80, and γ-tubulin [12-16]. However, very few BAP1 targets have been identified and functionally explored in ccRCC. The death inducer-obliterator 1 protein (DIDO1), the shortest splicing variant encoded by the DIDO gene, regulates the maintenance of mouse embryonic stem cells [17]. The DIDO gene encodes three splicing variants (DIDO1, DIDO2, and DIDO3) and has been implicated in apoptosis and development [18-20]. A recent study demonstrated that targeted disruption of the DIDO gene gives rise to centrosome amplification, a weakened spindle-assembly checkpoint (SAC) and division defects that challenge chromosome stability [21].
In this study, DIDO1 was identified as a BAP1 interactor. BAP1-loss expression correlated with DIDO1 downregulation in ccRCC. Moreover, the de-ubiquitination of DIDO1 by BAP1 plays a significant role in the regulation of mitotic progression and the prevention of chromosome instability.
Methods
Cell culture and transfection
786-O, 769-P and 293T cells were obtained from the American Type Culture Collection. 786-O and 769-P cells were cultured in RPMI 1640 medium with 10% fetal bovine serum. 293T cells were cultured in DMEM with 10% fetal bovine serum. Cells were transiently transfected with plasmids or siRNAs using Lipofectamine 3000 or RNAiMax Transfection Reagent (Invitrogen) according to the manufacturer’s instructions.
Expression constructs
The BAP1 and DIDO1 cDNAs were purchased from Genechem, and subcloned into pCIN4-FLAG-HA and pCMV-Myc expression vectors. The cDNA for DIDO1 was also subcloned into PCIN4-mCherry vectors to create a mCherry-DIDO1 fusion protein. BAP1 and DIDO1 mutants were generated by the KOD-Plus Mutagenesis Kit (TOYOBO). All the constructs were verified by DNA sequencing.
RNA interference
The negative control and specific siRNAs for BAP1 and DIDO1 were purchased from GenePharma. Transfection of siRNAs was performed following the manufacturer’s instructions. siRNA sequence information is provided in Supplementary Table 1.
Immunoprecipitation
For immunoprecipitation of the FLAG-tagged proteins, transfected cells were lysed with BC100 buffer 24 h after transfection. Whole-cell lysates were immunoprecipitated by overnight incubation with monoclonal anti-FLAG antibody conjugated M2 agarose beads (Sigma). After three washes with FLAG lysis buffer, followed by two washes with BC100 buffer, the bound proteins were eluted from the beads with FLAG Peptide (Sigma)/BC100 and were subjected to Western blot (WB) analysis. For immunoprecipitation of the endogenous proteins, cells were lysed with cell lysis buffer (Cell Signaling), and the lysates were centrifuged. The supernatant was precleared with protein A/G beads (Sigma) and incubated overnight with the indicated antibody at 4°C. The immunocomplexes were then incubated for 2 h at 4°C with protein A/G beads. After centrifugation, the pellets were collected and washed fi a times with lysis buffer, resuspended in sample buffer, and further analyzed by SDS-PAGE.
Western blotting
Cell lysates or immune-precipitates were subjected to SDS-PAGE, and then proteins were transferred onto nitrocellulose membranes (GE Healthcare). The membranes were blocked in Tris-buffered saline (pH 7.4) containing 5% nonfat milk and 0.1% Tween-20, washed three times in Tris-buffered saline containing 0.1% Tween-20, and incubated with the primary antibody overnight at 4°C. The membranes were then incubated with the secondary antibody for 1 h at room temperature. Antibody binding was visualized using the ECL Chemiluminescence System (Santa Cruz). Information regarding the primary antibodies used in this study is provided in Supplementary Table 2.
Quantitative RT-PCR
Total RNA from transiently transfected cells was extracted using the TRIzol reagent (Invitrogen), and cDNA was reverse transcribed using the Superscript RT Kit (TOYOBO), according to the manufacturer’s instructions. Primer sequence information is provided in Supplementary Table 1. PCR amplification was performed using the SYBR Green PCR Master Mix Kit (TOYOBO). Endogenous GAPDH was used for normalization.
Protein complex purification
The epitope-tagging strategy to isolate BAP1-containing protein complexes from human cells was performed as previously described, with some modifications. Briefly, to obtain a FLAG-HA-BAP1-expressing cell line, 786-O cells were transfected with pCIN4-FLAG-HA-BAP1 constructs and selected for 2 weeks in 1 mg/ml G418. The tagged BAP1 protein levels were detected by WB analysis. The stable cell lines were chosen to expand for protein complex purification. For purification, the cells were lysed on ice for 2 h in BC100 buffer (20 mM Tris-Cl, pH 7.9, 100 mM NaCl, 0.2 mM EDTA, 20% glycerol) containing 0.2% Triton X-100 and fresh protease inhibitor. The homogenate was centrifuged for 30 min at 12000 rpm at 4°C. Cleared lysates were filtered through 0.45 μM spin filters (Millipore) and immunoprecipitated by anti-FLAG antibody-conjugated M2 agarose (Sigma). The bound polypeptides eluted with the FLAG Peptide (Sigma) were further affinity purified by anti-HA antibody-conjugated agarose (Sigma). The final elutes from the HA-beads with HA peptides were resolved by SDS-PAGE on a 4%-20% gradient gel (Bio-Rad) for Coomassie Blue staining. Gel bands were cut out from the gel and subjected to mass-spectrometric sequencing.
Immunofluorescence
Cells cultured on coverslips in 24-well plates were fixed in 4% paraformaldehyde for 10 min and permeabilized in 0.2% Triton X-100 solution for 5 min. The coverslips were blocked with 2% BSA plus 5% goat serum for 1 h, and subsequently incubated with primary antibodies against HA, α-tubulin, and γ-tubulin, which was followed by sequential incubation with fluorescent secondary antibodies (Alexa488 goat anti-mouse, Alexa 488 goat anti-rabbit, or Alexa 546 goat anti-mouse; Invitrogen). Finally, cells were counterstained with DAPI to reveal the nuclei. Fluorescence images were captured and processed using a fluorescence microscope.
Clinical specimens and immunohistochemistry
Tissue samples were obtained from patients with previously untreated, nonmetastatic ccRCC who underwent radical nephrectomy at the Department of Urology, Zhongnan Hospital of Wuhan University from August 2017 to December 2018. Two specimens were collected from each patient; one specimen was taken from a normal region of the kidney (at least 0.5 cm from the tumor) and another specimen was taken from the tumor. Two pathologists simultaneously confirmed the histological diagnosis by examining hematoxylin and eosin stained sections of the specimens. The pathological stage was determined according to the American Joint Committee on Cancer (AJCC), and the tumor grade was classified using the Fuhrman grading system [22]. All patients gave informed consent.
All specimens were fixed in formalin for up to 24 h immediately after they were collected. The specimens were then dehydrated, paraffinized, and embedded in paraffin blocks. Tissue sections were cut at a thickness of 3-4 μm and were air-dried overnight. The sections were deparaffinized, rehydrated, and subjected to heat-induced antigen retrieval with sodium citrate buffer (10 mM sodium citrate, 0.05% Tween-20 pH 6.0) before being incubated with 3% hydrogen peroxide for 5 min to block endogenous peroxidase activity. Sections were then incubated, first with the appropriate primary antibody, and subsequently with biotinylated goat anti-mouse IgG. To detect a signal, the VECTASTAIN ABC kit (Vector Laboratories) was used according to the manufacturer’s instructions. The slides were further counterstained with hematoxylin. Appropriate positive and negative controls were utilized for each immune-stain run.
Statistical analysis
Experiments were performed in at least triplicates, unless otherwise stated. All statistical tests were two-sided and were performed using GraphPad Prism (Graphpad Software). Student’s t-tests were used to analyze the data for most experiments. The relationship between BAP1 and DIDO1 expression was analyzed by the Spearman rank correlation. Differences between the expression of BAP1 or DIDO1 and clinicopathological features were assessed by Fisher’s exact test. Values with P<0.05 were considered statistically significant.
Results
BAP1 interacts with DIDO1
To investigate the cellular functions and to identify the molecular mediators of BAP1 in RCC, we isolated the BAP1 complex from 786-O RCC cells using TAP methods and determined the proteins present in the complex using mass spectrometry (Figure 1A, 1B). Notably, HCF1, RBBP7, and MCRS1 were detected in abundance in the complex, verifying the efficiency of this approach. We also found that DIDO1 was present in the purified BAP1 complex (Figure 1B). Since the function of BAP1 in DIDO1 regulation has not been previously reported, we selected DIDO1 for subsequent analyses. To verify that DIDO1 is a bona fio BAP1 interactor, we first examined whether BAP1 can interact with DIDO1 in cells. FLAG-HA (FH)-DIDO1 and Myc-BAP1 constructs were co-expressed in 293T cells. Cells were subsequently harvested for coimmunoprecipitation (Co-IP) with the anti-FLAG antibody. As shown in Figure 1C, Myc-BAP1 was immunoprecipitated by FH-DIDO1, suggesting an exogenous interaction between these two proteins. In addition, a reciprocal Co-IP assay was performed using lysates of 293T cells that were co-transfected with FH-BAP1 and Myc-DIDO1 constructs. The results indicated that FH-BAP1 was able to immune-precipitate Myc-DIDO1 (Figure 1D). Next, we investigated whether endogenous BAP1 and DIDO1 interact with each other. Immunoprecipitation using the anti-DIDO1 antibody was performed using cell lysates prepared from 786-O cells. As shown in Figure 1E, endogenous BAP1 was efficiently coimmunoprecipitated with endogenous DIDO1. Moreover, reciprocal immunoprecipitation experiments confirmed endogenous DIDO1 was co-immunoprecipitated with endogenous BAP1, confirming an endogenous interaction between these proteins (Figure 1F). To investigate whether BAP1 colocalizes with DIDO1 in vivo, 786-O cells transfected with FH-BAP1 and mCherry-BAP1 were immune-stained with the anti-HA antibody and visualized by confocal microscopy. As shown in Figure 1G, BAP1 and DIDO1 were colocalized in the nucleus. Taken together, these results indicate that BAP1 forms a complex with DIDO1 in vivo.
Figure 1.
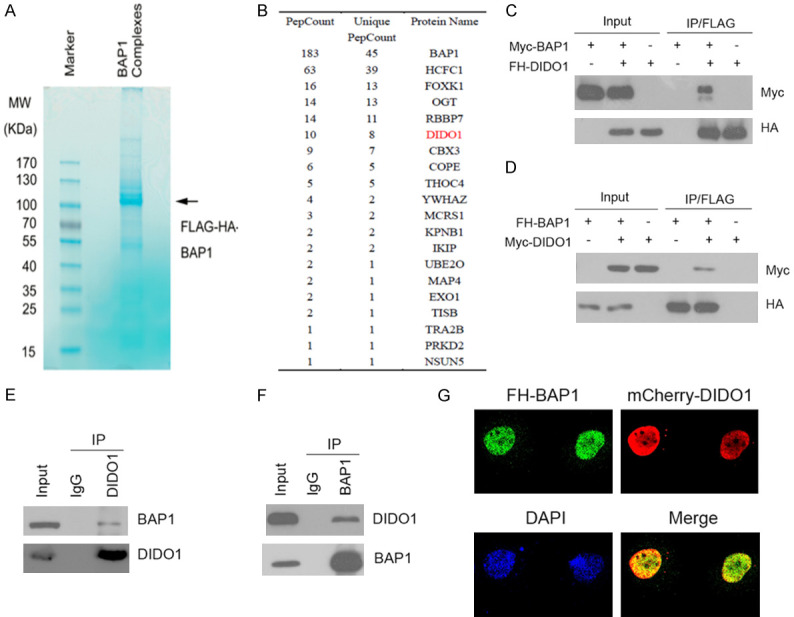
BAP1 interacts with DIDO1. A, B. Tandem affinity purification of the BAP1-containing protein complex was conducted using 786-O cells stably expressing FH-BAP1. A. Associated proteins were separated by SDS-PAGE and visualized by Coomassie Blue (CB) staining. B. The number of total/unique peptides identified by mass spectrometry analysis is shown in the table. C. 293T cells were co-transfected with FH-DIDO1 and Myc-BAP1 constructs. After 24 h, cell lysates were prepared for Co-IP with an anti-FLAG antibody and for WB analyses. D. FH-BAP1 was immunoprecipitated from 293T cells expressing Myc-DIDO1 and FH-BAP1. The immune-precipitates were subjected to WB analyses with the indicated antibodies. E, F. Immunoprecipitation using anti-DIDO1 or anti-BAP1 antibody was performed using cell lysates prepared from 786-O cells. The immune-precipitates were subjected to WB analyses with the indicated antibodies. G. 786-O cells transfected with FH-BAP1 and mCherry-DIDO1 constructs were immune-stained with the anti-HA antibody and visualized by confocal microscopy. 50 cells were counted per experiment, and 15 cells show colocalization of BAP1 and DIDO1.
Identification of the mutual-binding regions of BAP1 and DIDO1
To gain more insight into the BAP1-DIDO1 interaction, we determined which region of BAP1 mediates its interaction with DIDO1. BAP1 is composed of an N-terminal UCH domain, an HBM motif, a region that mediates the association with BRCA1 (BR), a 60 residue helical motif that shares conservation with UCH37 (ULD for UCH37 like domain), and a NLS (Figure 2A). We determined the region of BAP1 that binds to DIDO1. We generated BAP1 deletion mutants and tested their ability to interact with DIDO1 by Co-IP assay. The results showed that BAP1-FL, BAP1-1-692, and BAP1-1-597, but not others, were able to immune-precipitate Myc-DIDO1 (Figure 2C), suggesting that the UCH domain was responsible for binding to DIDO1.
Figure 2.
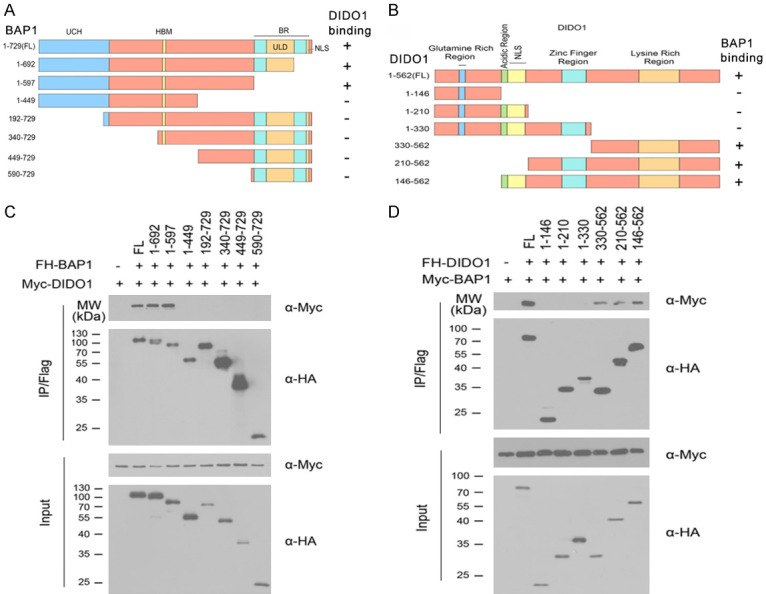
Identification of the mutual-binding regions of BAP1 and DIDO1. A. Schematic representation of BAP1 deletion mutants used in the study. The ability of each BAP1 fragment to bind to DIDO1 is indicated (+: binding, -: no binding). B. 293T cells were co-transfected with Myc-DIDO1 and the indicated FH-BAP1 or deletion mutants. Cell lysates were prepared and subjected to immunoprecipitation using the anti-FLAG antibody, followed by WB analyses with indicated antibodies. C. Schematic representation of DIDO1 deletion mutants. The ability of each DIDO1 fragment to bind to BAP1 is indicated. D. 293T cells were co-transfected with Myc-BAP1 in combination with a series of FH-DIDO1 deletion mutants. Cell lysates were immunoprecipitated with anti-FLAG antibody, followed by WB analyses with indicated antibodies.
DIDO1 protein includes glutamine rich region, acidic region, NLS, zinc finger region and lysine rich region (Figure 2B). As shown in Figure 2D, Co-IP assay showed that DIDO1-FL, DIDO1-330-562, DIDO1-210-562, and DIDO1-146-562 were able to bind to BAP1. Therefore, we concluded that the region of DIDO1 (330-562aa) is required for the interaction with BAP1.
BAP1 stabilizes DIDO1 by de-ubiquitination
Since BAP1 is a deubiquitinating enzyme, we explored whether BAP1 deubiquitinates and stabilizes DIDO1. First, we assessed whether BAP1 stabilizes DIDO1. We generated a BAP1 mutant in which the highly conserved Cys residues (C91) in its UCH domain were mutated to Ser (BAP1-C91S). The C91S mutation has been shown to abrogate BAP1’s de-ubiquitinase activity. As shown in Figure 3A, ectopic expression of BAP1-WT, but not the BAP1-C91S mutant (lacking catalytic activity), increased the co-expressed DIDO1 protein level in a dose-dependent manner, indicating that its de-ubiquitinse activity is required for promoting DIDO1 stabilization. Moreover, ectopic expression of BAP1-WT, but not the BAP1-C91S mutant in 786-O cells, resulted in a moderate increase of the protein level of endogenous DIDO1 (Figure 3B). Next, we depleted the endogenous BAP1 with two specific siRNAs in 786-O and 769-P cells and observed that DIDO1 protein was markedly downregulated (Figure 3C, 3D). To exclude the possibility that DIDO1 protein downregulation resulted from transcriptional downregulation, we performed qRT-PCR to measure the mRNA level of BAP1 and DIDO1 in BAP1-depleted 786-O and 769-P cells. In contrast to the significant decrease in BAP1 mRNA levels, DIDO1 mRNA levels in BAP1-depleted 786-O and 769-P cells stayed at a level similar to that of the control cells (Figure 3E, 3F), suggesting that the effect of BAP1 on DIDO1 is not mediated through the upregulation of DIDO1 mRNA expression. To determine whether BAP1 increases DIDO1 by extending its half-life, the protein level of DIDO1 was monitored after treatment with the protein synthesis inhibitor cycloheximide (CHX). In the absence of de novo protein synthesis, the half-life of endogenous DIDO1 protein was much shorter in the BAP1-depleted cells than that in control cells (Figure 3G, 3H), further suggesting that BAP1 regulates DIDO1 at the posttranslational level.
Figure 3.
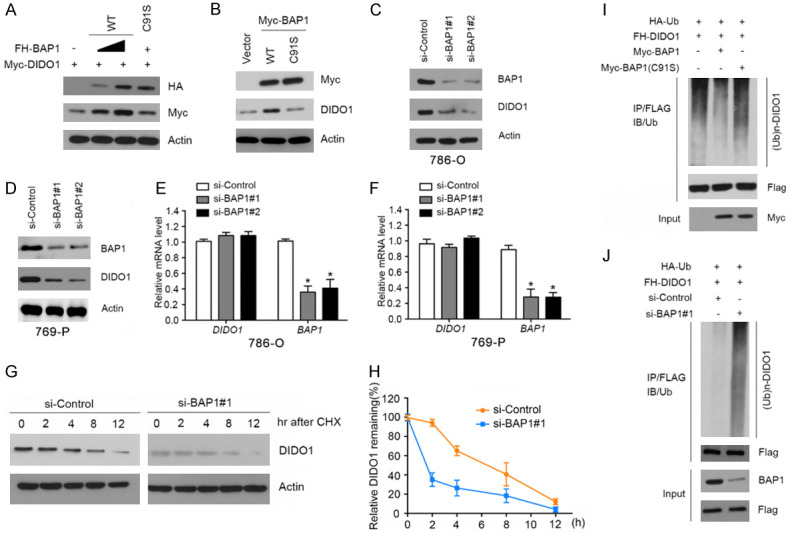
BAP1 stabilizes DIDO1 by de-ubiquitination. A. 293T cells were transfected with Myc-DIDO1 and increasing amounts of FH-BAP1-WT or C91S mutant constructs. 24 h after transfection, cells were harvested for WB analyses. B. 786-O cells were transfected with Myc-BAP1-WT or C91S mutant constructs. 24 h after transfection, cells were harvested for WB analyses. C, D. 786-O and 769-P cells were transfected with the negative control or two independent BAP1 siRNAs, respectively. 48 h after transfection, cells were harvested for WB analyses. E, F. qRT-PCR measurement of the mRNA levels of BAP1 and DIDO1 in BAP1-depleted 786-O and 769-P cells. GAPDH was used for normalization. The mean values (S.D.) of three independent experiments are shown (*, P<0.05). G, H. 786-O cells were transfected with the negative control or BAP1 siRNAs. 48 h after transfection, cells were collected at various times after cycloheximide (CHX) treatment and then were subjected to WB analyses. At each time point, the relative intensities of DIDO1 were first normalized to the intensities of actin and then to the value of the 0-hr time point. Similar results were obtained from two independent experiments. I. HA-Ub and FH-DIDO1 along with Myc-BAP1-WT or C91S constructs were co-transfected into 786-O cells. 24 h after transfection, cells were treated with 20 μM MG132 for 6 h. FH-MDIDO1 protein was immunoprecipitated with anti-FLAG antibody. The ubiquitinated forms of DIDO1 were analyzed by WB with anti-HA antibody. J. BAP1 was depleted by siRNAs in 786-O cells transiently expressing HA-Ub and FH-DIDO1. Cells were treated with 20 μM MG132 for 6 h. FH-DIDO1 protein was immunoprecipitated and subjected to WB analyses.
To further determine whether BAP1 promotes DIDO1 stabilization through the regulation of DIDO1 de-ubiquitination, HA-ubiquitin and FH-DIDO1 constructs were co-expressed with BAP1-WT or C91S mutant in 786-O cells. As shown in Figure 3I, ectopically expressing BAP1-WT, but not the C91S mutant, markedly reduced the DIDO1 poly-ubiquitination. Conversely, depletion of BAP1 using siRNAs increased DIDO1 ubiquitination (Figure 3J), indicating that BAP1 is responsible for DIDO1 de-ubiquitination. Taken together, these data demonstrate that the de-ubiquitinase BAP1 maintains DIDO1 protein steady-state level through promoting DIDO1 de-ubiquitination in ccRCC cells.
BAP1 contributes to chromosome stability partially via DIDO1
Defects in chromosome segregation result in chromosomal instability and aneuploidy, which are hallmarks of many cancers. Accumulating evidence suggests that BAP1 is critical for maintaining chromosome stability, and DIDO1 depletion in cells causes abnormal spindles and aneuploidy. These observations led us to explore whether BAP1 maintains chromosome stability via DIDO1. HK-2, an immortalized proximal tubule epithelial cell line from normal adult human kidney, was used in these experiments. We depleted BAP1 or DIDO1 in HK-2 cells using siRNAs. WB analyses confirmed that BAP1 or DIDO1 protein was efficiently depleted after siRNA transfection (Figure 4A). Then the cells were stained with α/γ-tubulin and DAPI. A significant proportion of multipolar spindle formation in BAP1 or DIDO1-depleted HK-2 cells was observed (Figure 4B, 4D). We also observed that a significant multipolar spindle formation in BAP1 or DIDO1-depleted HK-2 cells (Figure 4C, 4E). To determine whether BAP1 exerts its activity for chromosome stability by stabilizing DIDO1, we generated HK-2 cell lines stably overexpressing FH-DIDO1 (Figure 4F). Compared to control cells, BAP1 depletion-induced multipolar spindle formation was partially rescued in HK-2 cells stably overexpressing FH-DIDO1 (Figure 4G, 4H). Taken together, these results suggest that BAP1 and DIDO1 were both required for proper mitotic progression, and BAP1 contributes to chromosome stability at least in part via DIDO1.
Figure 4.
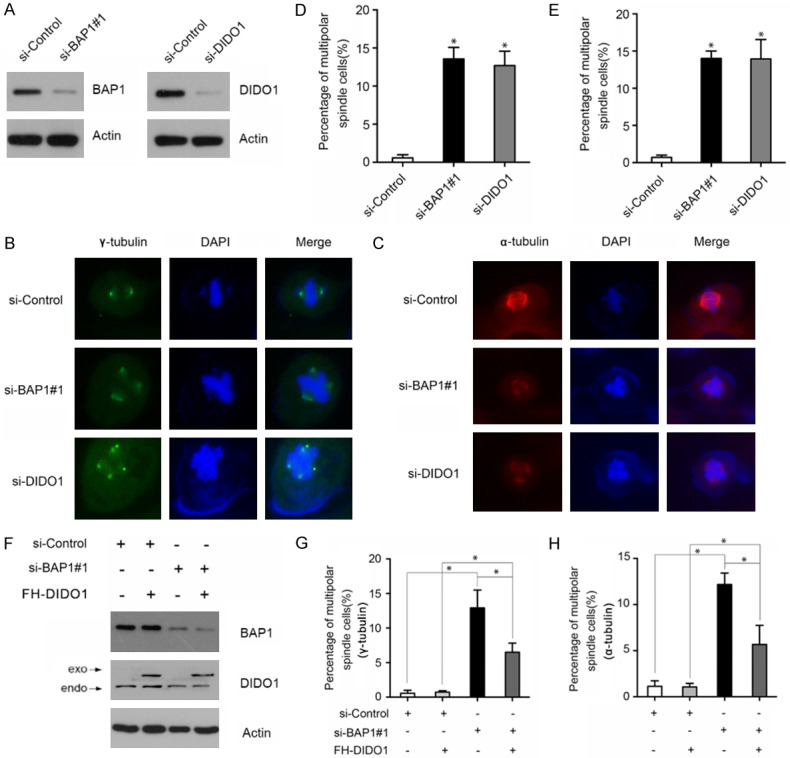
Stabilization of DIDO1 by BAP1 contributes to chromosome stability. A. BAP1 and DIDO1 protein levels were assessed by WB analyses in HK-2 cells transfected with si-BAP1 (#1) or si-DIDO1. B. Representative images of mitotic spindles observed in HK-2 cells transfected with BAP1 or DIDO1 siRNAs and stained for γ-tubulin (green) and DAPI (blue) as indicated. C. The HK-2 cells transfected with BAP1 or DIDO1 siRNAs were stained for α-tubulin (red) and DAPI (blue) as indicated. D, E. The percentage of cells with abnormal mitotic spindles is shown. The mean values (S.D.) were obtained from three independent experiments, with at least 50 mitotic cells counted per experiment (*, P<0.05). F. HK-2 cells stably expressing FH-DIDO1 were transfected with BAP1 siRNAs. Cells were harvested for WB analyses. G, H. The percentage of abnormal mitotic spindle cells is shown. The mean values (S.D.) were obtained from three independent experiments, with at least 50 mitotic cells counted per experiment (*, P<0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
BAP1 protein expression correlates with DIDO1 expression in ccRCC patients
BAP1 is mutated in 5-15% of sporadic ccRCCs, and the majority of BAP1 mutations in ccRCCs lead to BAP protein loss in tumor tissues. We investigated whether DIDO1 is downregulated in BAP1-loss tumors. First, we analyzed the expression of BAP1 and DIDO1 in 34 paired tissues from ccRCC patients by immunohistochemistry. The clinical and pathologic characteristics of the patients are shown in Supplementary Table 3. We scored the staining of both BAP1 and DIDO1 from 0 to 3 and designated scores 0-1 as negative and scores 2-3 as positive. In 34 paired ccRCC cases, BAP1 and DIDO1 were positive in adjacent normal tissues, but 4/34 were BAP1-negative in tumor tissues, and 6/34 were DIDO1-negative in tumor tissues (Figure 5A). The expression of BAP1 and DIDO1 was also strongly correlated (Figure 5B, r=0.7606, P<0.001). In the tumor from case 1, both BAP1 and DIDO1 were stained strongly in ccRCC cells. The tumor from case 2 showed ccRCC cells with BAP1 loss and weak staining of DIDO1 (Figure 5C). We also observed that two ccRCCs that were BAP1 positive still showed weak DIDO1 staining, suggesting that other mechanisms such as epigenetic silencing may downregulate DIDO1.
Figure 5.
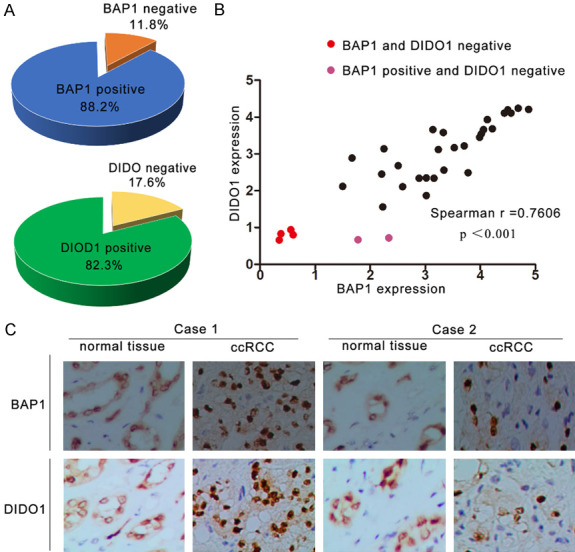
BAP1 protein was positively correlated with DIDO1 in ccRCC. A. BAP1 and DIDO1 staining patterns in 34 paired ccRCC cases. B. BAP1 and DIDO1 expression positively correlated (r=0.7606, P<0.001). Staining intensities were quantified by morphometry. The x/y axis represents the expression ratio of the indicated proteins between paired tumor and normal tissues. According to the depth of immunohistochemical staining, the expression of BAP1 and DIDO1 in renal cell carcinoma was evaluated by points of 0-3, and the deeper the staining, the higher the score. The expression of 0-1 was considered to be negative, while 2-3 was considered to be positive. C. Representative IHC of normal and tumor tissues with positive or negative staining of BAP1 and DIDO1. Scale bar =50 μm.
Discussion
Recurrent BAP1 mutations in ccRCC have been confirmed by several independent genome-wide sequencing studies [8,23,24]. The clinical implication of BAP1 loss has been explored and shows that ccRCC with BAP1 mutations is associated with worse clinical outcomes and aggressive clinicopathological features [4,5,25,26]. Although frequent mutations of BAP1 in ccRCC have been identified, the functional impact of BAP loss is still poorly understood. In this study, we demonstrated for the first time that DIDO1 is a bona fide substrate for BAP1. Our study also provides new insights into our understanding of BAP1-mutated ccRCC.
Studies of the DIDO1 gene and its variants support their important function in numerous cellular processes, such as apoptosis and chromosomal segregation, particularly in the hematopoietic system [18-21,27-29]. A previous study showed that the cellular level of DIDO1 is tightly regulated by the ubiquitin-proteasome pathway, and ubiquitin E3 ligase WPP2 targets DIDO1 for proteasomal-dependent degradation in the nucleus [29]. Thus, it is possible that DIDO1 abundance and activity is fine-tuned by a subtle balance between ubiquitination and de-ubiquitination. In our study, we demonstrated that BAP1 binds to DIDO1 and stabilizes DIDO1 by de-ubiquitination. A previous study showed DIDO1 associated with polarization, which indicates that DIDO1 is dispensable in cell polarization [29]. Our results showed that depletion of BAP1 or DIDO1 in a normal renal epithelial cell line causes abnormal mitotic spindles, suggesting these two proteins are both essential for maintaining chromosome stability in renal epithelial cells.
Our results also showed exogenous overexpressed DIDO1 only partially rescued BAP1 depletion-induced mitotic abnormality in HK-2 cells. A recent study revealed that BAP1 deubiquitinates DIDO1 and MCRS1, that BAP1 binds to MCRS1 and stabilizes MCRS1 by de-ubiquitination. And that BAP1 contributes to chromosome stability partially via MCRS1 [30]. Another study showed BAP1 deubiquitinates and stabilizes Ino80, which is involved in various chromosomal processes, including DNA repair, telomere regulation, centromere stability and chromosome segregation [16]. Another study showed de-ubiquitination of γ-tubulin by BAP1 prevents chromosome instability in breast cancer cells [15]. Therefore, it is possible that BAP1 maintains chromosome stability by modulating the ubiquitination status of various substrates.
Our tandem affinity purification test revealed that in addition to interacting with DIDO1, BAP1 also potentially interacts with Forkhead box protein K1 (FOXK1), 14-3-3 protein zeta/delta (YWHAZ), and karyopherin β1 (KPNB1). FOXK1 was overexpressed in 16 cancerous human tissue types, induced tumor cell EMT, maintained the invasive potential of colorectal cancer and appeared to have a crucial role in the metastatic progression of human carcinomas [31]. YWHAZ is included in the 14-3-3 family of proteins, which are a family of evolutionarily highly conserved acidic proteins expressed in all eukaryotic organisms. YWHAZ has been implicated in the initiation and progression of cancer and has been shown to be overexpressed in multiple cancer tissues and cell lines [32]. KPNB1 is a nuclear transport receptor belonging to the karyopherin family that is involved in transporting proteins through the nuclear pore. KPNB1 is required for proteostasis maintenance and its inhibition induces apoptosis in glioblastoma cells [33]. It is possible these proteins are molecular mediators of BAP1 activity, but the detailed molecular mechanisms and clinical relevance in ccRCC remain to be determined through further study. A better understanding of the molecular mechanism and clinical consequence of BAP1 loss in ccRCC will help identify targets for drug development.
Acknowledgements
This work was in part supported by the National Natural Science Foundation of China (81803013), Joint research fund of Health Commission of Hubei Province (WJ2019H018) and the Fundamental Research Funds for the Central Universities (2042019kf0137).
Disclosure of conflict of interest
None.
Abbreviations
- BAP1
BRCA1 associated protein1
- DIDO1
death inducer-obliterator 1 protein
- ccRCC
clear cell renal cell carcinoma
- DUB
de-ubiquitination enzymes
- Co-IP
co-immunoprecipitation
- IHC
Immunohistochemical
- WB
Western Blotting
Supporting Information
References
- 1.Flanigan RC, Campbell SC, Clark JI, Picken MM. Metastatic renal cell carcinoma. Curr Treat Options Oncol. 2003;4:385–390. doi: 10.1007/s11864-003-0039-2. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen YB, Gonen M, Liu H, Takeda S, Voss MH, Tickoo SK, Reuter VE, Russo P, Cheng EH, Sander C, Motzer RJ, Hsieh JJ. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19:3259–67. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapur P, Peña-Llopis S, Christie A, Zhrebker L, Pavía-Jiménez A, Rathmell WK, Xie XJ, Brugarolas J. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol. 2013;14:159–167. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gossage L, Murtaza M, Slatter AF, Lichtenstein CP, Warren A, Haynes B, Marass F, Roberts I, Shanahan SJ, Claas A, Dunham A, May AP, Rosenfeld N, Forshew T, Eisen T. Clinical and pathological impact of VHL, PBRM1, BAP1, SETD2, KDM6A, and JARID1c in clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2014;53:38–51. doi: 10.1002/gcc.22116. [DOI] [PubMed] [Google Scholar]
- 7.Piva F, Santoni M, Matrana MR, Satti S, Giulietti M, Occhipinti G, Massari F, Cheng L, Lopez-Beltran A, Scarpelli M, Principato G, Cascinu S, Montironi R. BAP1, PBRM1 and SETD2 in clear-cell renal cell carcinoma: molecular diagnostics and possible targets for personalized therapies. Expert Rev Mol Diagn. 2015;15:1201–1210. doi: 10.1586/14737159.2015.1068122. [DOI] [PubMed] [Google Scholar]
- 8.Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, Leng N, Pavía-Jiménez A, Wang S, Yamasaki T, Zhrebker L, Sivanand S, Spence P, Kinch L, Hambuch T, Jain S, Lotan Y, Margulis V, Sagalowsky AI, Summerour PB, Kabbani W, Wong SW, Grishin N, Laurent M, Xie XJ, Haudenschild CD, Ross MT, Bentley DR, Kapur P, Brugarolas J. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz DC, Wilkinson KD, Maul GG, Barlev N, Berger SL, Prendergast GC, Rauscher FJ 3rd. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 12.Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, Wu J, Phu L, Katavolos P, LaFave LM, Abdel-Wahab O, Modrusan Z, Seshagiri S, Dong K, Lin Z, Balazs M, Suriben R, Newton K, Hymowitz S, Garcia-Manero G, Martin F, Levine RL, Dixit VM. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarrizi R, Menard JA, Belting M, Massoumi R. Deubiquitination of gammatubulin by BAP1 prevents chromosome instability in breast cancer cells. Cancer Res. 2014;74:6499–6508. doi: 10.1158/0008-5472.CAN-14-0221. [DOI] [PubMed] [Google Scholar]
- 16.Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat Commun. 2014;5:5128. doi: 10.1038/ncomms6128. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Kim H, Liang J, Lu W, Ouyang B, Liu D, Songyang Z. The death-inducer obliterator1 (Dido1) gene regulates embryonic stem cell self-renewal. J Biol Chem. 2014;289:4778–4786. doi: 10.1074/jbc.M113.486290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Domingo D, Leonardo E, Grandien A, Martínez P, Albar JP, Izpisúa-Belmonte JC, Martínez-A C. DIO-1 is a gene involved in onset of apoptosis invitro, whose misexpression disrupts limb development. Proc Natl Acad Sci U S A. 1999;96:7992–7997. doi: 10.1073/pnas.96.14.7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Domingo D, Ramírez D, González de Buitrago G, Martínez-A C. Death inducer-obliterator 1 triggers apoptosis after nuclear translocation and caspase up-regulation. Mol Cell Biol. 2003;23:3216–3225. doi: 10.1128/MCB.23.9.3216-3225.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fütterer A, Campanero MR, Leonardo E, Criado LM, Flores JM, Hernández JM, San Miguel JF, Martínez-A C. Didogene expression alterations are implicated in the induction of hematological myeloid neoplasms. J Clin Invest. 2005;115:2351–2362. doi: 10.1172/JCI24177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trachana V, van Wely KH, Guerrero AA, Fütterer A, Martínez-A C. Dido disruption leads to centrosome amplification and mitotic checkpoint defects compromising chromosome stability. Proc Natl Acad Sci U S A. 2007;104:2691–2696. doi: 10.1073/pnas.0611132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, Ogawa S. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 25.Kapur P, Christie A, Raman JD, Then MT, Nuhn P, Buchner A, Bastian P, Seitz C, Shariat SF, Bensalah K, Rioux-Leclercq N, Xie XJ, Lotan Y, Margulis V, Brugarolas J. BAP1 immunohistochemistry predicts outcomes in a multi-institutional cohort with clear cell renal cell carcinoma. J Urol. 2014;191:603–610. doi: 10.1016/j.juro.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 26.Joseph RW, Kapur P, Serie DJ, Eckel-Passow JE, Parasramka M, Ho T, Cheville JC, Frenkel E, Rakheja D, Brugarolas J, Parker A. Loss of BAP1 protein expression is an independent marker of poor prognosis in patients with low-risk clear cell renal cell carcinoma. Cancer. 2014;120:1059–1067. doi: 10.1002/cncr.28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojas AM, Sanchez-Pulido L, Fütterer A, van Wely KH, Martinez-A C, Valencia A. Death inducer obliterator protein 1 in the context of DNA regulation. Sequence analyses of distant homologues point to a novel functional role. FEBS J. 2005;272:3505–3511. doi: 10.1111/j.1742-4658.2005.04759.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Chen Y, Chen Z, Wu Q, Ke WJ, Wu QL. Gambogic acid induces death inducer-obliterator 1-mediated apoptosis in Jurkat T cells. Acta Pharmacol Sin. 2008;29:349–354. doi: 10.1111/j.1745-7254.2008.00762.x. [DOI] [PubMed] [Google Scholar]
- 29.Fütterer A, de Celis J, Navajas R, Almonacid L, Gutiérrez J, Talavera-Gutiérrez A, Pacios-Bras C, Bernascone I, Martin-Belmonte F, Martinéz-A C. DIDO as a switchboard that regulates self-renewal and differentiation in embryonic stem cells. Stem Cell Reports. 2017;8:1062–1075. doi: 10.1016/j.stemcr.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng J, Ma J, Li W, Mo R, Zhang P, Gao K, Jin X, Xiao J, Wang C, Fan J. Stabilization of MCRS1 by BAP1 prevents chromosome instability in renal cell carcinoma. Cancer Lett. 2015;369:167–174. doi: 10.1016/j.canlet.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Peng Y, Zhang P, Huang X, Yan Q, Wu M, Xie R, Wu Y, Zhang M, Nan Q, Zhao J, Li A, Xiong J, Ren Y, Bai Y, Chen Y, Liu S, Wang J. Direct regulation of FOXK1 by C-jun promotes proliferation, invasion and metastasis in gastric cancer cells. Direct regulation of FOXK1 by C-jun promotes proliferation, invasion and metastasis in gastric cancer cells. Cell Death Dis. 2016;7:e2480. doi: 10.1038/cddis.2016.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimura Y, Komatsu S, Ichikawa D, Nagata H, Hirajima S, Takeshita H, Kawaguchi T, Arita T, Konishi H, Kashimoto K, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br J Cancer. 2013;108:1324–1331. doi: 10.1038/bjc.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu ZC, Liu JW, Li K, Zheng J, Xiong ZQ. KPNB1 inhibition disrupts proteostasis and triggers unfolded protein response-mediated apoptosis in glioblastoma cells. Oncogene. 2018;37:2936–2952. doi: 10.1038/s41388-018-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


