Abstract
Aims:
To evaluate the clinical utility of first and second trimester prenatal screening biomarkers for early pregnancy prediction of gestational diabetes mellitus (GDM) risk in nulliparous women
Methods:
We conducted a population-based cohort study of nulliparous women participating in the California Prenatal Screening Program from 2009–2011 (n=105,379). GDM was ascertained from hospital discharge records or birth certificates. Models including maternal characteristics and prenatal screening biomarkers were developed and validated. Risk stratification and reclassification were performed to assess clinical utility of the biomarkers.
Results:
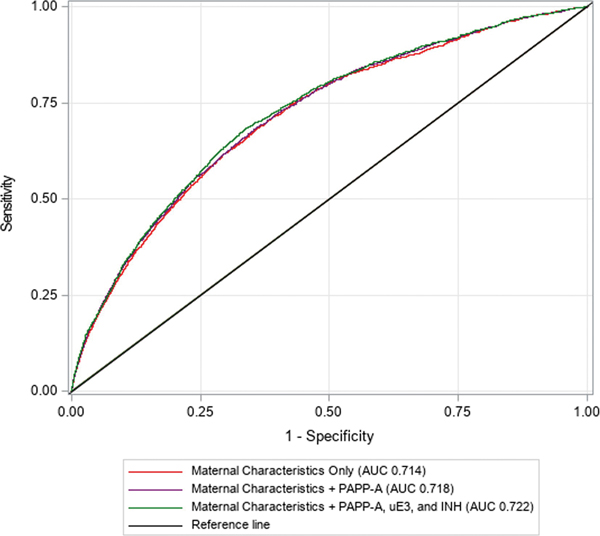
Decreased levels of first trimester pregnancy-associated plasma protein A (PAPP-A) and increased levels of second trimester unconjugated estriol (uE3) and dimeric inhibin A (INH) were associated with GDM. The addition of PAPP-A only and PAPP-A, uE3, and INH to maternal characteristics resulted in small, yet significant, increases in AUC (maternal characteristics only: AUC 0.714 (95% CI 0.703–0.724), maternal characteristics + PAPP-A: AUC 0.718 (95% CI 0.707–0.728), maternal characteristics + PAPP-A, uE3, and INH: AUC 0.722 (0.712–0.733)); however, no net improvement in classification was observed.
Conclusions:
PAPP-A, uE3, and INH have limited clinical utility for prediction of GDM risk in nulliparous women. Utility of other readily accessible clinical biomarkers in predicting GDM risk warrants further investigation.
Keywords: gestational diabetes mellitus, prenatal screening, biomarkers, risk management, prediction model, clinical utility
1. Introduction
Gestational diabetes mellitus (GDM) is a common complication of pregnancy associated with significant maternal and neonatal morbidities, including increased risk of pre-eclampsia, macrosomia, and subsequent type 2 diabetes.[1] In most developed countries, universal glucose testing is performed at 24–28 weeks’ gestation to identify women with overt glucose intolerance.[2] Ideally, women at high risk of developing GDM would be identified earlier in pregnancy to facilitate preventive intervention, enhance antenatal care, and improve clinical outcomes through reduced maternal and fetal exposure to metabolic alterations and potential epigenetic malprogramming.[3–5] Metabolic derangements other than glucose intolerance appear to play a role in the pathogenesis of GDM and, therefore, could be of use for prediction of GDM risk.[5–7] Numerous biomarkers associated with early pregnancy metabolic derangements, such as adipokines and inflammatory mediators, have been evaluated for their potential to add to the prediction of later pregnancy glucose intolerance.[5, 6] Although some biomarker-enhanced risk prediction models for GDM have shown promise, lack of external validation and translation into applications that can be incorporated into routine prenatal care has limited their clinical utility.[8]
First and early second trimester maternal serum multi-marker screening is routinely offered to women during pregnancy and has been validated as an effective way to calculate fetal risk for chromosomal aneuploidy and open neural tube defects.[9, 10] Abnormal levels of prenatal screening biomarkers, including pregnancy-associated plasma protein A (PAPP-A), human chorionic gonadotrophin (hCG), α-fetoprotein (AFP), unconjugated estriol (uE3), and dimeric inhibin A (INH), have been shown to be associated with a number of adverse obstetric outcomes in the absence of aneuploidy and structural anomalies.[10] Because of the close ties between these biomarkers and placental functioning and metabolism, they could also be of value in screening for GDM.[10–13] Many studies have found significant associations between aberrant levels of prenatal screening biomarkers and subsequent GDM development;[13] however, few have assessed their predictive ability in GDM risk prediction models.[13–15]
In the present study, we aim to build on our previous research[4] by incorporating first and second trimester prenatal screening biomarkers into clinical models for early pregnancy prediction of GDM risk in nulliparous women, a population with unique risk profiles.[4, 16] We assessed the clinical utility of these biomarkers using risk reclassification methods. Incorporating biomarkers that precede the onset of hyperglycemia into a risk prediction model for GDM may facilitate earlier risk assessment, screening, and diagnosis, thereby reducing the risk for adverse maternal and infant outcomes through targeted intervention.[6]
2. Subjects, Materials and Methods
2.1. Study Population
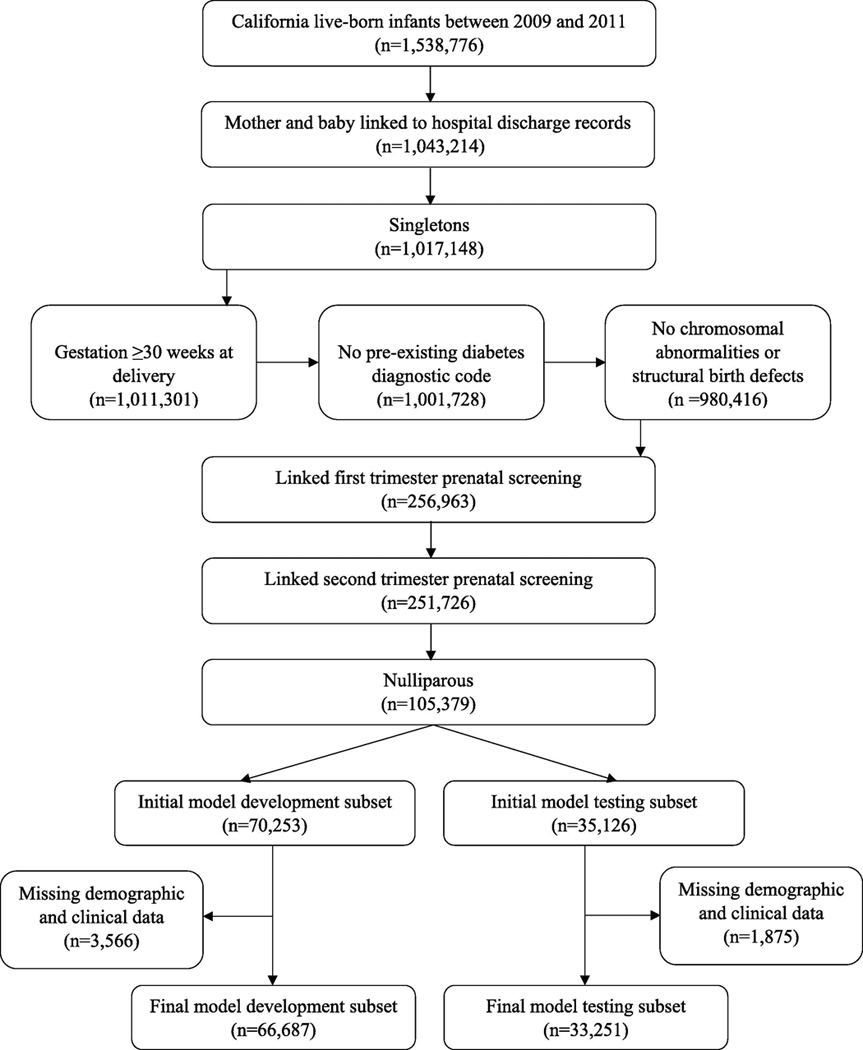
Our study population was drawn from live births in California from 2009–2011 in a birth cohort file maintained by the California Office of Statewide Health Planning and Development (n=1,538,776) (Figure 1).[17] Participants were included after linkage of infant birth and death certificates and mother and infant hospital discharge records one year prior to delivery and one year post-delivery (n=1,043,214). Women carrying multiples, those who delivered prior to routine administration of GDM glucose testing (<30 weeks’ gestation), those with pre-existing type 1 or type 2 diabetes mellitus, and women carrying fetuses with chromosomal abnormalities or major structural birth defects were excluded. Number of fetuses (i.e., singleton or multiple) and gestational age at delivery were ascertained from birth certificates. Pre-existing type 1 or type 2 diabetes mellitus was ascertained from International Classification of Diseases, 9th Revision, Clinical Modification (ICD-9-CM) diagnostic codes 648.0 (‘diabetes mellitus complicating pregnancy’) or 250 (‘diabetes mellitus’) listed on hospital discharge records or birth certificate denotation of diagnosis of diabetes prior to pregnancy. Information on chromosomal abnormalities was obtained from the California Chromosomal Defect Registry.[18] Major structural birth defects were identified from hospital discharge records (Supplementary Table 1).[19]
Figure 1.
Sample selection.
We restricted the sample to nulliparous women who participated in both first and early second trimester (<20 weeks’ gestation) prenatal screening through the California Prenatal Screening (PNS) Program administered by the Division of Genetic Disease Screening within the California Department of Public Health (n=105,379).[20] Parity was ascertained from birth certificates. PNS records were linked to birth certificates through probabilistic matching based on mother’s name, date of birth, social security number, delivery date, PNS accession date, telephone number, street address, city, and zip code (IBM Web Sphere Quality Stage Version 7.5). Linkages were verified with clerical review and post-match queries.[21] For analytic purposes, we randomly divided the final cohort into a development subset (2/3 of total: n=70,253 women) and a testing subset (1/3 of total: n=35,126 women). 5% of women were excluded in both the model development and testing subsets due to missing demographic and clinical data (Supplementary Tables 2 and 3). The characteristics of women who were excluded were similar to those who were included.
2.2. Data Collection
As part of routine prenatal screening, first trimester PAPP-A and hCG were measured in serum samples collected between 10–14 weeks’ gestation and second trimester AFP, hCG, uE3, and INH were measured in serum samples collected between 15–20 weeks’ gestation.[20] Following collection, samples were sent to one of eight regional laboratories in California under contract with the California PNS Program for testing using an automated analytical system and a uniform protocol (PAPP-A, hCG, AFP, and uE3: AutoDELFIA, Perkin Elmer Life Sciences, Waltham, MA; INH: Beckman Coulter assay system, Brea, CA).[22–24] Within each laboratory, multiple of the median (MoM) biomarker values were calculated by dividing each individual’s raw biomarker value by the median biomarker value for the patient population with the same gestational age analyzed within that laboratory. This conversion allowed for the fact that prenatal screening biomarker levels differ by gestational age and permitted consistent interpretation of testing results across laboratories.[25] Adjustments for maternal weight, race/ethnicity, smoking status, and pre-existing diabetes were also performed.[26, 27] Results were then electronically transferred and stored at the California Department of Public Health.[24]
Maternal demographic and clinical characteristics known to be associated with GDM development were captured from birth certificate and hospital discharge records obtained from the California Office of Statewide Health Planning and Development, as previously described.[4] GDM was identified using ICD-9-CM code 648.8 (‘pregnancy complicated by abnormal glucose tolerance’) or birth certificate denotation of diabetes diagnosis during pregnancy.
2.3. Statistical Analysis
2.3.1. Model Development
GDM risk prediction models were developed using the model development subset. Development and validation of a model for early pregnancy prediction of GDM risk based on demographic and clinical risk factors for nulliparous women has been described previously.[4] This model included five risk factors (race/ethnicity, age at delivery, pre-pregnancy body mass index (BMI), family history of diabetes, and pre-existing hypertension) and had an area under the receiver operating characteristic curve (AUC) of 0.732 (95% confidence interval (CI) 0.728–0.735) in an internal validation cohort and 0.710 (95% CI 0.672–0.749) in an external validation cohort. To meet regression assumptions for the new models including demographic and clinical risk factors and prenatal screening biomarkers, each biomarker was natural log-transformed. Within the model development subset, the association between GDM and each biomarker was first assessed using univariate logistic regression. Biomarkers found to be significantly associated with GDM (p<0.001) were then included in separate multivariable logistic regression models including maternal demographic and clinical characteristics and first trimester biomarkers only and maternal demographic and clinical characteristics and first and early second trimester biomarkers. Due to the large sample size, a more stringent alpha level (p <0.001) was used for assessing significant associations.[4] We assessed pair-wise correlations between biomarkers using Spearman (non-parametric) rank correlation to investigate the independent effect of each biomarker within the prediction model.
2.3.2. Model Testing
Models were tested within the model testing subset. Predictive accuracy was assessed using both discrimination and calibration statistics. We evaluated model discrimination using AUC. Change in AUC (ΔAUC) was quantified using the DeLong, DeLong, and Clark-Pearson non-parametric approach.[28] Calibration, related to goodness of fit, was assessed by plotting and comparing each woman’s predicted risk of GDM to her observed outcome.[4, 29] Model over-/under-estimation and overfitting were assessed using the calibration slope and intercept of the linear predictor.[4, 8, 30]
2.3.3. Risk Stratification
Risk stratification, a method for categorizing patients into risk groups based on their personal risk factors, is important for clinical decision making.[31] As such, we converted individual predicted risks produced from the developed models into high or low risk groups using a previously determined predicted risk threshold of 6%.[4] Sensitivity, specificity, positive predictive value, negative predictive value, and correct classification rate were computed for each model at the chosen threshold.
2.3.4. Clinical Risk Reclassification
The predictive value of a biomarker is often based on its ability to increase the AUC of an established multivariable risk prediction model. However, it is often difficult for a biomarker that is clinically relevant, yet has only a moderate independent association with the outcome, to significantly change the value of AUC. Clinical interpretation of the magnitude of this change relative to the baseline model is also challenging.[32, 33] Risk reclassification, which assesses differences in patient risk group classification between the established risk prediction model and a new model including a biomarker, can be used to more meaningfully evaluate the clinical utility of the biomarker.[32] Changes in risk reclassification were visualized using reclassification tables and quantified using categorical net reclassification improvement (NRI) indices.[32, 34] NRI ranges from −2 to 2, with zero indicating that no net improvement in classification resulted from the addition of a biomarker to an established risk prediction model.[35] Negative NRIs result from more individuals being placed in the wrong risk category (e.g., women with GDM categorized as low risk or women without GDM categorized as high risk) with the addition of a biomarker to an established risk prediction model than the established risk prediction model without the biomarker.[36] As no clinical guidelines for GDM risk categorization currently exist, the 6% predicted risk threshold used to stratify high and low risk groups may be considered arbitrary. Therefore, we also assessed risk reclassification at 3% and 12% predicted risk thresholds.[36, 37]
All statistical analyses were performed using SAS software, Version 9.4 (SAS Institute Inc., Cary, NC, USA). Risk reclassification measures were calculated using the SAS Macro written by Kevin F. Kennedy and Michael J. Pencina.[34] Methods and protocols for this study were approved by the Committee for the Protection of Human Subjects within the Health and Human Services Agency of the State of California. Data provided to the researchers by California Office of Statewide Health Planning and Development (Protocol # 12–09-0702) and California Biobank Program (Screening Information System request no. 476) were deidentified and determined not to qualify as human subjects research by the University of Iowa Institutional Review Board (IRB no.: 201706737).
3. Results
3.1. Cohort Characteristics
Demographic and clinical characteristics of the study population are shown in Supplementary Table 4. Within the total nulliparous population, 7.5% of women were diagnosed with GDM. The population was racially and ethnically diverse, with most women identifying as White, not Hispanic (34.3%), Hispanic (34.5%), or Asian (20.7%). The median age at delivery was 29 years and the median pre-pregnancy BMI was 23.2. Around 1% of the population reported having a family history of diabetes and preexisting hypertension. First and second trimester biomarker median MoM values ranged from 0.98 (uE3) to 1.07 (PAPP-A). These characteristics remained consistent after randomly dividing the total population into model development and testing subsets.
3.2. Model Development
The association between maternal demographic and clinical characteristics, first trimester prenatal screening biomarkers, and second trimester prenatal screening biomarkers and GDM in the model development subset are summarized in Table 1. Consistent with our previous findings,[4] race/ethnicity, age at delivery, pre-pregnancy BMI, family history of diabetes, and pre-existing hypertension were significant risk factors for GDM. Decreased first trimester PAPP-A MoM values and increased second trimester uE3 and INH MoM values were associated with increased odds of GDM (PAPP-A: OR 0.78 (95% CI 0.74–0.82); uE3: OR 1.58 (95% CI 1.41–1.77); INH: OR 1.35 (95% CI 1.26–1.45)). Two separate GDM risk prediction models were developed combining maternal demographic and clinical characteristics with significant first trimester (PAPP-A) and first and second trimester (PAPP-A, uE3, and INH) prenatal screening biomarkers. All biomarkers remained significant after adjustment for maternal demographic and clinical characteristics and/or other biomarkers (maternal characteristics + PAPP-A model: PAPP-A: aOR 0.75 (95% CI 0.71–0.79); maternal characteristics + PAPP-A, uE3, and INH model: PAPP-A: aOR 0.71 (95% CI 0.67–0.75), uE3: aOR 1.71 (95% CI 1.50–1.94), INH: aOR 1.24 (95% CI 1.15–1.34)). PAPP-A, uE3, and INH were not found to be highly correlated with each other in the pair-wise comparison of biomarkers (Supplementary Table 5).
Table 1.
Association between maternal demographic and clinical characteristics, first trimester prenatal screening biomarkers, and second trimester prenatal screening biomarkers and gestational diabetes mellitus (GDM) in the model development subset (n=66,687).
| No GDM n (%) | GDM n (%) | OR (95% CI) | Maternal demographic and clinical characteristics only model aOR (95% CI) | Maternal demographic and clinical characteristics + PAPP-A model aOR (95% CI) | Maternal demographic and clinical characteristics + PAPP-A, uE3, and INH model aOR (95% CI) | |
|---|---|---|---|---|---|---|
| Sample Size | 61,813 (92.7) | 4,874 (7.3) | ||||
| Demographic and Clinical Characteristics | ||||||
| Race/ethnicity | ||||||
| White, not Hispanic | 21,772 (35.2) | 1,189 (24.4) | REF | REF | REF | REF |
| Hispanic | 21,548 (34.9) | 1,468 (30.1) | 1.24 (1.15–1.34)** | 1.54 (1.41–1.68)** | 1.54 (1.42–1.68)** | 1.53 (1.40–1.66)** |
| Black | 1,932 (3.1) | 98 (2.0) | 0.94 (0.77–1.15) | 0.97 (0.78–1.21) | 0.99 (0.79–1.23) | 0.98 (0.79–1.22) |
| Asian | 12,015 (19.4) | 1,749 (35.9) | 2.58 (2.39–2.78)** | 3.34 (3.08–3.62)** | 3.34 (3.08–3.62)** | 3.25 (3.00–3.52)** |
| AI/AN | 131 (0.2) | -- | 1.82 (1.05–3.17) | 1.71 (0.93–3.16) | 1.74 (0.94–3.21) | 1.72 (0.93–3.19) |
| H/PI | 126 (0.2) | 15 (0.3) | 2.11 (1.25–3.56) | 1.76 (1.01–3.07) | 1.81 (1.03–3.16) | 1.76 (1.00–3.08) |
| Other racial group* | 4,289 (6.9) | 343 (7.0) | 1.46 (1.30–1.65)** | 1.59 (1.40–1.80)** | 1.60 (1.41–1.82)** | 1.59 (1.40–1.81)** |
| Age at delivery (years)†a | 29.0 (24.0–32.0) | 31.0 (27.0–34.0) | 1.08 (1.07–1.08)** | 1.08 (1.08–1.09)** | 1.08 (1.08–1.09)** | 1.08 (1.08–1.09)** |
| Pre-pregnancy BMI (kg/m2)†b | 23.0 (20.8–26.4) | 25.0 (21.9–29.8) | 1.08 (1.07–1.08)** | 1.10 (1.10–1.11)** | 1.10 (1.10–1.11)** | 1.10 (1.09–1.11)** |
| Family history of diabetes | 416 (0.7) | 80 (1.6) | 2.47 (1.94–3.14)** | 2.22 (1.72–2.86)** | 2.21 (1.72–2.85)** | 2.21 (1.72–2.85)** |
| Pre-existing hypertension | 559 (0.9) | 127 (2.6) | 2.93 (2.41–3.56)** | 1.66 (1.35–2.04)** | 1.64 (1.33–2.01)** | 1.64 (1.33–2.01)** |
| First Trimester Biomarkers† | ||||||
| PAPP-A MoM | 1.07 (0.74–1.55) | 0.99 (0.66–1.45) | 0.78 (0.74–0.82)** | - | 0.75 (0.71–0.79)** | 0.71 (0.67–0.75)** |
| hCG MoM | 1.02 (0.78–1.34) | 1.00 (0.76–1.31) | 0.90 (0.84–0.97) | - | - | - |
| Second Trimester Biomarkers† | ||||||
| AFP MoM | 1.01 (0.82–1.24) | 1.01 (0.82–1.26) | 1.11 (1.01–1.22) | - | - | - |
| hCG MoM | 1.00 (0.72–1.38) | 1.00 (0.72–1.40) | 1.05 (0.99–1.11) | - | - | - |
| uE3 MoM | 0.98 (0.83–1.14) | 1.00 (0.85–1.18) | 1.58 (1.41–1.77)** | - | - | 1.71 (1.50–1.94)** |
| INH MoM | 1.04 (0.81–1.35) | 1.09 (0.85–1.41) | 1.35 (1.26–1.45)** | - | - | 1.24 (1.15–1.34)** |
OR, odds ratio; aOR, adjusted odds ratio; AI/AN, American Indian/Alaska Native; H/PI, Hawaiian/Pacific Islander; BMI, body mass index; PAPP-A, pregnancy-associated plasma protein-A; MoM, multiple of the median; hCG, human chorionic gonadotropin; AFP, α-fetoprotein; uE3, unconjugated estriol; INH, inhibin.
Odds ratios and p-values were estimated using univariate logistic regression. Adjusted odds ratios and p-values were estimated using multivariable logistic regression. Each variable was adjusted for all other variables within the model.
Median (interquartile range (IQR): Q1 (25%)–Q3 (75%)) were calculated using non-transformed variables. Univariate and multivariable logistic regression was performed using log-transformed variables.
- Not retained in the model; -- Data suppressed (n<15).
Includes two or more races and race unknown.
Data are expressed as median (IQR).
Odds ratios were calculated per year of age.
Odds ratios were calculated per kg/m2.
p<0.001
3.3. Model Testing
Incremental improvements in discrimination were observed with the addition of PAPP-A only and PAPP-A, uE3, and INH to the maternal demographic and clinical characteristics model (maternal characteristics only: AUC 0.714 (95% CI 0.703–0.724), maternal characteristics + PAPP-A: AUC 0.718 (95% CI 0.707–0.728), maternal characteristics + PAPP-A, uE3, and INH: AUC 0.722 (95% CI 0.712–0.733) (Figure 2). The changes in AUC were small, yet significant (maternal characteristics + PAPP-A: ΔAUC: 0.004, p<0.001; maternal characteristics + PAPP-A, uE3, and INH: ΔAUC 0.009, p<0.001). All three models were well-calibrated (i.e., intercepts were close to 0 and slopes were close to 1) within the model testing subset (Supplementary Figure 1).
Figure 2.
Receiver operating characteristic curves for gestational diabetes risk prediction models within the model testing subset (n=33,251).
AUC, area under the receiver operating characteristic curve; PAPP-A, pregnancy-associated plasma protein A; uE3, unconjugated estriol; INH, dimeric inhibin A. Maternal characteristics only model includes the following variables: race/ethnicity, age at delivery (natural cubic spline transformed), pre-pregnancy body mass index (BMI) (natural cubic spline transformed), family history of diabetes, and pre-existing hypertension.Maternal characteristics plus first trimester biomarker model includes the following variables: race/ethnicity, age at delivery (natural cubic spline transformed), prepregnancy BMI (natural cubic spline transformed), family history of diabetes, pre-existing hypertension, and first trimester PAPP-A multiple of the median (MoM) (natural log-transformed). Maternal characteristics plus first and second trimester biomarkers model includes the following variables: a: race/ethnicity, age at delivery (natural cubic spline transformed), pre-pregnancy BMI (natural cubic spline transformed), family history of diabetes, pre-existing hypertension, first trimester PAPP-A MoM (natural log-transformed), second trimester uE3 MoM (natural log-transformed), and second trimester INH MoM (natural log-transformed).
3.4. Risk Stratification
The performance of the risk stratification strategy for each model is outlined in Supplementary Table 6. At a 6% predicted risk threshold, 47.1%, 46.8%, and 46.4% of women (maternal characteristics only, maternal characteristics + PAPP-A, and maternal characteristics + PAPP-A, uE3, and INH) were considered high risk for GDM, respectively. All three models showed similar performance, with moderate sensitivity and specificity (maternal characteristics only: 76.2% and 55.2%, maternal characteristics + PAPP-A: 75.7% and 55.5%, and maternal characteristics + PAPP-A, uE3, and INH: 76.1% and 56.0%) and correct classification of around 57% of GDM cases and non-cases.
3.5. Clinical Risk Reclassification
At a 6% predicted risk threshold, the maternal characteristics + PAPP-A and maternal characteristics + PAPP-A, uE3, and INH models placed more women without GDM into the low risk category than the maternal characteristics only model (maternal characteristics only: 55.24%, maternal characteristics + PAPP-A: 55.52%, maternal characteristics + PAPP-A, uE3, and INH: 55.99%) (Table 2). However, the models including prenatal screening biomarkers also placed fewer women with GDM into the high risk category than the maternal characteristics only model (maternal characteristics only: 76.19%, maternal characteristics + PAPP-A:
Table 2.
Comparison of risk classifications for nulliparous women with and without gestational diabetes mellitus based the maternal characteristics only, maternal characteristics + PAPP-A, and maternal characteristics + PAPP-A, uE3, and INH models within the model testing subset (n=33,251).
| Risk Category | Model | |||||
|---|---|---|---|---|---|---|
| Maternal Characteristics Only | Maternal Characteristics + PAPP-A | Maternal Characteristics + PAPP-A, uE3, and INH | ||||
| No GDM (n= 30,731) % | GDM (n= 2,520) % | No GDM (n= 30,731) % | GDM (n= 2,520) % | No GDM (n= 30,731) % | GDM (n= 2,520) % | |
| Low (<6% predicted risk) | 55.24 | 23.81 | 55.52 | 24.29 | 55.99 | 23.93 |
| High (>6% predicted risk) | 44.76 | 76.19 | 44.48 | 75.71 | 44.01 | 76.07 |
| Total | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
GDM, gestational diabetes mellitus; PAPP-A, pregnancy-associated plasma protein A; uE3, unconjugated estriol; INH, dimeric inhibin A.
Model includes the following variables: race/ethnicity, age at delivery (natural cubic spline transformed), pre-pregnancy BMI (natural cubic spline transformed), family history of diabetes, and pre-existing hypertension.
Model includes the following variables: race/ethnicity, age at delivery (natural cubic spline transformed), pre-pregnancy BMI (natural cubic spline transformed), family history of diabetes, pre-existing hypertension, and first trimester PAPP-A MoM (natural log-transformed).
Model includes the following variables: a: race/ethnicity, age at delivery (natural cubic spline transformed), pre-pregnancy BMI (natural cubic spline transformed), family history of diabetes, pre-existing hypertension, first trimester PAPP-A MoM (natural log-transformed), second trimester uE3 MoM (natural log-transformed), and second trimester INH MoM (natural log-transformed).
75.71%, maternal characteristics + PAPP-A, uE3, and INH: 76.07%). Overall, the addition of prenatal screening biomarkers to the maternal characteristics only model resulted in no net improvement in classification (maternal characteristics + PAPP-A: categorical NRI −0.002 (95% CI −0.010–0.007); maternal characteristics + PAPP-A, uE3, and INH: categorical NRI 0.006 (95% CI −0.003–0.016)) (Supplementary Table 7). The categorical NRIs were similar when the predicted risk threshold was adjusted to 3% and 12%.
4. Discussion
We rigorously evaluated the clinical utility of first and second trimester prenatal screening biomarkers for early pregnancy prediction of GDM risk within a large cohort of nulliparous women. Decreased levels of first trimester PAPP-A and increased levels of second trimester uE3 and INH were significantly associated with GDM development. The addition of PAPP-A only and PAPP-A, uE3, and INH to a previously validated model including maternal demographic and clinical characteristics offered incremental improvements in predictive accuracy, with significant increases in AUC. However, the increases in predictive accuracy with the addition of prenatal screening biomarkers were small compared to GDM risk prediction based on maternal characteristics alone and no net improvement in classification was observed, indicating that these biomarkers had limited clinical utility.
A limited number of studies to-date have examined the predictive ability of first trimester prenatal screening biomarkers in GDM risk prediction models and findings have been conflicting.[13–15] Lovati et al. found that incorporating first trimester PAPP-A in a clinical risk factor model increased the predictive ability of the model from AUC 0.60 (95% CI 0.56–0.64) (clinical risk factors only) to AUC 0.70 (95% CI 0.66–0.74) (clinical risk factors plus PAPP-A).[38] Two more recently conducted studies found similar results, with increases in model predictive performance observed when PAPP-A and other first trimester biomarkers (adiponectin, mean arterial pressure, and uterine artery pulsatility index) were added to models including traditional GDM risk factors.[39, 40] In contrast, Syngelaki et al. and Xiao et al. showed that models including first trimester prenatal screening biomarkers in conjunction with traditional risk factors did not improve GDM screening performance over models including traditional risk factors alone (Syngelaki et al.: maternal risk factors alone: AUC 0.841, maternal risk factors + PAPP-A: AUC 0.841; Xiao et al.: maternal risk factors alone: AUC 0.684, maternal risk factors + PAPP-A: AUC 0.686).[41, 42] All but one of these studies (Syngelaki et al.) used a case-control study design, which is ineffective for risk modeling as the sample is biased (i.e., the study sample has a higher proportion of cases than the population of interest).[13, 43]
Development of models predicting the risk of various obstetric complications has been increasing. However, clinical implementation is lacking, which may be due to limited evaluation of prediction model performance, impact, and usefulness in clinical practice.[44] Although our findings suggest that the integration of prenatal screening biomarkers into clinical models for early prediction of GDM risk did not have substantial net improvement in classification, other readily accessible clinical biomarkers representing etiological pathways associated with GDM development may be of value. The clinical utility of biochemical markers of long-term maternal glycemic control, fat and steroid metabolism, and systemic inflammation, as well as anthropometric measurements, such as maternal adiposity and blood pressure,[45] for early pregnancy prediction of GDM risk should be assessed in future studies with risk reclassification methods.
To our knowledge, this is the first study to assess the value of combining demographic and clinical risk factors and both first and early second trimester prenatal screening biomarkers in GDM risk prediction. The size of our study cohort enabled us to build and validate adequately powered prediction models. Utilization of a racially and ethnically diverse cohort supports the generalizability of our findings. We are also the first to use risk reclassification methods, which may be more clinically meaningful than change in AUC,[33] to evaluate the clinical utility of first and second trimester prenatal screening biomarkers in early pregnancy prediction of GDM risk. The California PNS Program is one of the largest and most comprehensive prenatal screening programs in the world.[24, 46] The centralization of its laboratories and standardization of patient follow-up allows this program to attain optimal quality control,[46] adding credence to the measurements of biomarkers used in our analysis.
Although our study has many strengths, we also acknowledge its limitations. Our study focused on nulliparous women, so the findings may not be generalizable to multiparous women. While our sample had a diverse racial/ethnic makeup, it is possible that factors such as poverty, access to medical care, failure to facilitate informed choice, acculturation and language skills, risk perception, and values and beliefs could have affected women’s ability and/or choice to undergo prenatal screening.[47] Furthermore, this bias may have been exacerbated through our selection of women who had received prenatal screening in both the first and second trimester. Because our data source was administrative and medical charts were inaccessible, inaccurate reporting of demographic and clinical variables, including GDM, is possible. However, use of both hospital discharge and birth certificate data has been shown to be an accurate source of information for both pre-existing and gestational diabetes.[48]
We did not have data on exactly how or when GDM was screened and diagnosed, which may have caused some misclassification. GDM screening is routinely performed at 24–28 weeks’ gestation in the United States,[1] and the 50-g oral glucose challenge test (two-step approach) is the most commonly used screening test.[49] The prevalence of GDM within our study population was similar to previously reported estimates within the United States.[50] To ensure that women in our study population had the opportunity to be screened for GDM, we excluded women who delivered prior to 30 weeks’ gestation. All women included in this study participated in both first and second trimester prenatal screening, indicating that they were receiving care around the time that GDM screening would have been performed.
Risk prediction algorithms for GDM are not currently used in clinical practice, and as such, clinically meaningful predicted risk cutoffs were not available. Individual predicted risks were converted into high or low risk groups using a 6% predicted risk threshold, which we have previously validated.[4] Risk classification statistics were similar when we adjusted the threshold to 3% and 12%. A complete case analysis was utilized in model development and testing, which may impart bias. However, as only a small percentage of women were missing predictor data, it is likely that any bias introduced through use of this method was minor.[4]
3.1. Conclusion
First and second trimester prenatal screening biomarkers, PAPP-A, uE3, and INH, have limited clinical utility for early pregnancy prediction of GDM risk in nulliparous women. The utility of other readily accessible clinical biomarkers in predicting GDM risk warrants investigation in future studies. Incorporating biomarkers that precede the onset of hyperglycemia into a risk prediction model for GDM may facilitate earlier risk assessment, screening, and diagnosis.
Supplementary Material
Highlights.
Identifying women at risk for gestational diabetes early in pregnancy is ideal
Prenatal screening biomarkers may be of value in screening for gestational diabetes
To assess clinical utility, risk stratification and reclassification were performed
No net classification improvement indicated limited clinical utility of biomarkers
Utility of other biomarkers in predicting gestational diabetes risk is warranted
Acknowledgments
Funding
This work was supported by the National Institutes of Health [grant number 5 T32 HL087738-14 which supports B.M.S.]. The content is solely the responsibility of the authors and does not necessarily represent the official views of this agency. This work was also supported by the California Preterm Birth Initiative within the University of California, San Francisco (UCSF7027075). Members of the California Preterm Birth Initiative participated as authors on this manuscript and are specifically noted. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Declaration of Interest
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].ACOG Practice Bulletin No. 190: Gestational Diabetes Mellitus. Obstetrics and gynecology. 2018;131:e49–e64. [DOI] [PubMed] [Google Scholar]
- [2].Rani PR, Begum J. Screening and Diagnosis of Gestational Diabetes Mellitus, Where Do We Stand. J Clin Diagn Res. 2016;10:QE01–QE4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].North RA, McCowan LME, Dekker GA, Poston L, Chan EHY, Stewart AW, et al. Clinical risk prediction for pre-eclampsia in nulliparous women: development of model in international prospective cohort. BMJ (Clinical research ed). 2011;342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Donovan BM, Breheny PJ, Robinson JG, Baer RJ, Saftlas AF, Bao W, et al. Development and validation of a clinical model for preconception and early pregnancy risk prediction of gestational diabetes mellitus in nulliparous women. PLoS One. 2019;14:e0215173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brink HS, van der Lely AJ, van der Linden J. The potential role of biomarkers in predicting gestational diabetes. Endocr Connect. 2016;5:R26–R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Correa PJ, Vargas JF, Sen S, Illanes SE. Prediction of gestational diabetes early in pregnancy: targeting the long-term complications. Gynecologic and obstetric investigation. 2014;77:145–9. [DOI] [PubMed] [Google Scholar]
- [7].Leiva A, Pardo F, Ramirez MA, Farias M, Casanello P, Sobrevia L. Fetoplacental vascular endothelial dysfunction as an early phenomenon in the programming of human adult diseases in subjects born from gestational diabetes mellitus or obesity in pregnancy. Experimental diabetes research. 2011;2011:349286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lamain-de Ruiter M, Kwee A, Naaktgeboren CA, de Groot I, Evers IM, Groenendaal F, et al. External validation of prognostic models to predict risk of gestational diabetes mellitus in one Dutch cohort: prospective multicentre cohort study. BMJ (Clinical research ed). 2016;354:i4338. [DOI] [PubMed] [Google Scholar]
- [9].Driscoll DA, Gross SJ, for the Professional Practice Guidelines C. Screening for fetal aneuploidy and neural tube defects. Genetics in Medicine. 2009;11:818–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lakhi N, Govind A, Moretti M, Jones J. Maternal serum analytes as markers of adverse obstetric outcome. The Obstetrician & Gynaecologist. 2012;14:267–73. [Google Scholar]
- [11].Sweeting A, Park F, Hyett J. The first trimester: prediction and prevention of the great obstetrical syndromes. Best practice & research Clinical obstetrics & gynaecology. 2015;29:183–93. [DOI] [PubMed] [Google Scholar]
- [12].Jelliffe-Pawlowski LL, Baer RJ, Blumenfeld YJ, Ryckman KK, O’Brodovich HM, Gould JB, et al. Maternal characteristics and mid-pregnancy serum biomarkers as risk factors for subtypes of preterm birth. BJOG : an international journal of obstetrics and gynaecology. 2015;122:1484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Donovan BM, Nidey NL, Jasper EA, Robinson JG, Bao W, Saftlas AF, et al. First trimester prenatal screening biomarkers and gestational diabetes mellitus: A systematic review and meta-analysis. PLoS One. 2018;13:e0201319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Eleftheriades M, Papastefanou I, Lambrinoudaki I, Kappou D, Lavranos D, Akalestos A, et al. Elevated placental growth factor concentrations at 11–14 weeks of gestation to predict gestational diabetes mellitus. Metabolism: clinical and experimental. 2014;63:1419–25. [DOI] [PubMed] [Google Scholar]
- [15].Talasaz ZH, Sadeghi R, Askari F, Dadgar S, Vatanchi A. First trimesters Pregnancy-Associated Plasma Protein-A levels value to Predict Gestational diabetes Mellitus: A systematic review and meta-analysis of the literature. Taiwanese journal of obstetrics & gynecology. 2018;57:181–9. [DOI] [PubMed] [Google Scholar]
- [16].Miranda ML, Edwards SE, Myers ER. Adverse birth outcomes among nulliparous vs. multiparous women. Public Health Rep. 2011;126:797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].California Office of Statewide Health Planning and Development. Available data files. 2019. [https://oshpd.ca.gov/data-andreports/research-data-request-information/]. Accessed 19 August 2019.
- [18].California Department of Public Health. About California Birth Defects Monitoring Program. 2019. [https://www.cdph.ca.gov/Programs/CFH/DGDS/Pages/cbdmp/about.aspx]. Accessed 17 October 2019.
- [19].Baer RJ, Norton ME, Shaw GM, Flessel MC, Goldman S, Currier RJ, et al. Risk of selected structural abnormalities in infants after increased nuchal translucency measurement. American journal of obstetrics and gynecology. 2014;211:675e1–19. [DOI] [PubMed] [Google Scholar]
- [20].California Department of Public Health Genetic Disease Screening Program. The California Prenatal Screening Program. 2017. [https://www.cdph.ca.gov/Programs/CFH/DGDS/CDPH%20Document%20Library/PNS%20Documents/Patient%20Booklet%20Consent_ENG-ADA.pdf]. Accessed 20 June 2019.
- [21].Barradas DT, Dietz PM, Pearl M, England LJ, Callaghan WM, Kharrazi M. Validation of Obstetric Estimate Using Early Ultrasound: 2007 California Birth Certificates. Paediatric and perinatal epidemiology. 2014;28:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jelliffe-Pawlowski LL, Shaw GM, Currier RJ, Stevenson DK, Baer RJ, O’Brodovich HM, et al. Association of early-preterm birth with abnormal levels of routinely collected first- and second-trimester biomarkers. American journal of obstetrics and gynecology. 2013;208:492e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kazerouni NN, Currier RJ, Flessel M, Goldman S, Hennigan C, Hodgkinson C, et al. Detection rate of quadruple-marker screening determined by clinical follow-up and registry data in the statewide California program, July 2007 to February 2009. Prenatal diagnosis. 2011;31:901–6. [DOI] [PubMed] [Google Scholar]
- [24].Cunningham GC, Tompkinison DG. Cost and effectiveness of the California triple marker prenatal screening program. Genetics in medicine : official journal of the American College of Medical Genetics. 1999;1:199–206. [DOI] [PubMed] [Google Scholar]
- [25].Shiefa S, Amargandhi M, Bhupendra J, Moulali S, Kristine T. First Trimester Maternal Serum Screening Using Biochemical Markers PAPP-A and Free beta-hCG for Down Syndrome, Patau Syndrome and Edward Syndrome. Indian journal of clinical biochemistry : IJCB. 2013;28:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baer RJ, Lyell DJ, Norton ME, Currier RJ, Jelliffe-Pawlowski LL. First trimester pregnancy-associated plasma protein-A and birth weight. European journal of obstetrics, gynecology, and reproductive biology. 2016;198:1–6. [DOI] [PubMed] [Google Scholar]
- [27].Currier R, Wu N, Van Meter K, Goldman S, Lorey F, Flessel M. Integrated and first trimester prenatal screening in California: program implementation and patient choice for follow-up services. Prenatal diagnosis. 2012;32:1077–83. [DOI] [PubMed] [Google Scholar]
- [28].DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- [29].Han K, Song K, Choi BW. How to Develop, Validate, and Compare Clinical Prediction Models Involving Radiological Parameters: Study Design and Statistical Methods. Korean J Radiol. 2016;17:339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology (Cambridge, Mass). 2010;21:128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dom Dera J Risk Stratification: A Two-Step Process for Identifying Your Sickest Patients. Fam Pract Manag. 2019;26:21–6. [PubMed] [Google Scholar]
- [32].Pencina MJ, D’Agostino RB Sr., D’Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Statistics in medicine. 2008;27:157–72; discussion 207–12. [DOI] [PubMed] [Google Scholar]
- [33].Leening MJ, Vedder MM, Witteman JC, Pencina MJ, Steyerberg EW. Net reclassification improvement: computation, interpretation, and controversies: a literature review and clinician’s guide. Ann Intern Med. 2014;160:122–31. [DOI] [PubMed] [Google Scholar]
- [34].Kennedy KF, Pencina MJ. A SAS® macro to compute added predictive ability of new markers predicting a dichotomous outcome. 2010. [Google Scholar]
- [35].Afshinnia F, Rajendiran TM, Wernisch S, Soni T, Jadoon A, Karnovsky A, et al. Lipidomics and Biomarker Discovery in Kidney Disease. Semin Nephrol. 2018;38:127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cook NR. Clinically relevant measures of fit? A note of caution. American journal of epidemiology. 2012;176:488–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cook NR, Paynter NP. Performance of reclassification statistics in comparing risk prediction models. Biom J. 2011;53:237–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lovati E, Beneventi F, Simonetta M, Laneri M, Quarleri L, Scudeller L, et al. Gestational diabetes mellitus: Including serum pregnancy-associated plasma protein-A testing in the clinical management of primiparous women? A case–control study. Diabetes research and clinical practice. 2013;100:340–7. [DOI] [PubMed] [Google Scholar]
- [39].Sweeting AN, Wong J, Appelblom H, Ross GP, Kouru H, Williams PF, et al. A first trimester prediction model for gestational diabetes utilizing aneuploidy and pre-eclampsia screening markers. The journal of maternal-fetal & neonatal medicine : the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. 2017:1–9. [DOI] [PubMed] [Google Scholar]
- [40].Farina A, Eklund E, Bernabini D, Paladino M, Righetti F, Monti G, et al. A First-Trimester Biomarker Panel for Predicting the Development of Gestational Diabetes. Reproductive sciences (Thousand Oaks, Calif). 2017;24:954–9. [DOI] [PubMed] [Google Scholar]
- [41].Syngelaki A, Kotecha R, Pastides A, Wright A, Nicolaides KH. First-trimester biochemical markers of placentation in screening for gestational diabetes mellitus. Metabolism: clinical and experimental. 2015;64:1485–9. [DOI] [PubMed] [Google Scholar]
- [42].Xiao D, Chenhong W, Yanbin X, Lu Z. Gestational diabetes mellitus and first trimester pregnancy-associated plasma protein A: A case-control study in a Chinese population. Journal of diabetes investigation. 2018;9:204–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rose S, van der Laan MJ. A note on risk prediction for case-control studies. U.C. Berkeley Division of Biostatistics Working Paper Series. Working Paper 241. 2008. [http://biostats.bepress.com/ucbbiostat/paper241]. Accessed 22 June 2019. [Google Scholar]
- [44].Kleinrouweler CE, Cheong-See FM, Collins GS, Kwee A, Thangaratinam S, Khan KS, et al. Prognostic models in obstetrics: available, but far from applicable. American Journal of Obstetrics & Gynecology. 2016;214:79–90.e36. [DOI] [PubMed] [Google Scholar]
- [45].Poon LC, David McIntyre H, Hyett JA, Fonseca EBD, Hod M. The first-trimester of pregnancy - a window of opportunity for prediction and prevention of pregnancy complications and future life. Diabetes research and clinical practice. 2018. [DOI] [PubMed] [Google Scholar]
- [46].Kazerouni NN, Currier B, Malm L, Riggle S, Hodgkinson C, Smith S, et al. Triple-marker prenatal screening program for chromosomal defects. Obstetrics and gynecology. 2009;114:50–8. [DOI] [PubMed] [Google Scholar]
- [47].Norton ME, Hopkins LM, Pena S, Krantz D, Caughey AB. First Trimester Combined Screening: Experience with an Instant Results Approach. American journal of obstetrics and gynecology. 2007;196:606e1–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lydon-Rochelle MT, Holt VL, Cardenas V, Nelson JC, Easterling TR, Gardella C, et al. The reporting of pre-existing maternal medical conditions and complications of pregnancy on birth certificates and in hospital discharge data. American journal of obstetrics and gynecology. 2005;193:125–34. [DOI] [PubMed] [Google Scholar]
- [49].U.S. Preventive Services Task Force. Final recommendation statement: Gestational diabetes mellitus, screening. 2014. [https://www.uspreventiveservicestaskforce.org/Page/Document/RecommendationStatementFinal/gestational-diabetes-mellitusscreening]. Accessed 26 January 2020.
- [50].Deputy NP, Kim SY, Conrey EJ, Bullard KM. Prevalence and Changes in Preexisting Diabetes and Gestational Diabetes Among Women Who Had a Live Birth - United States, 2012–2016. MMWR Morb Mortal Wkly Rep. 2018;67:1201–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.