Abstract
Cerebral atherosclerosis contributes to dementia via unclear processes. We performed proteomic sequencing of dorsolateral prefrontal cortex in 438 older individuals and found associations between cerebral atherosclerosis and reduced synaptic signaling and RNA splicing and increased oligodendrocyte development and myelination. Consistently, single-cell RNA sequencing showed cerebral atherosclerosis associated with higher oligodendrocyte abundance. A subset of proteins and modules associated with cerebral atherosclerosis was also associated with Alzheimer’s disease, suggesting shared mechanisms.
Cerebral atherosclerosis (CA) is the accumulation of cholesterol-laden plaques in the walls of large arteries in the brain and ranges in severity from minor arterial wall thickening to significant luminal stenosis causing reduced blood flow and metabolism1. Its prevalence increases with age and the presence of vascular risk factors such as hyperlipidemia, hypertension, diabetes, and smoking1. It is a common condition and present in 7% of asymptomatic middle-aged individuals and 82% of individuals over 80 years old1,2. CA and its consequences are among the earliest changes associated with development of Alzheimer’s disease dementia (henceforth referred to as AD)3–6 and with approximately three times higher risk for AD and nine times higher risk for vascular dementia2,7–9. However, we have limited insights into molecular effects of CA on the human brain, and how these contribute to dementia.
To identify brain proteomic changes associated with CA, we performed proteomic sequencing of the dorsolateral prefrontal cortex of 438 brains with detailed neuropathological assessment of nine age-associated brain pathologies – β-amyloid, neurofibrillary tangles, CA, gross infarcts, microinfarcts, cerebral amyloid angiopathy (CAA), TDP-43, Lewy body, and hippocampal sclerosis – to identify the proteomic signature of CA independently of the eight other measured pathologies. The discovery dataset were 391 subjects from the Religious Order Study and Memory and Aging Project (ROS/MAP; Supplementary Table 1), and the replication dataset were 47 subjects from the Baltimore Longitudinal Study of Aging (BLSA; Supplementary Table 2). CA burden ranged from none to severe, with a median of mild in both datasets. CA was correlated with high plasma low-density lipoprotein and triglyceride levels, lower high-density lipoprotein level, more gross infarcts, and lower β-amyloid burden (Supplementary Figure 1). CA was associated with lower white matter integrity (β = −3.0; adjusted p=0.01; N=236) in the frontal inferior periventricular and subcortical white matter based on voxel-wise R2, an index of brain tissue integrity10, in 236 subjects with imaging data who were representative of the 391 subjects with brain proteomes (Supplementary Table 3).
Brain proteomic signature of cerebral atherosclerosis
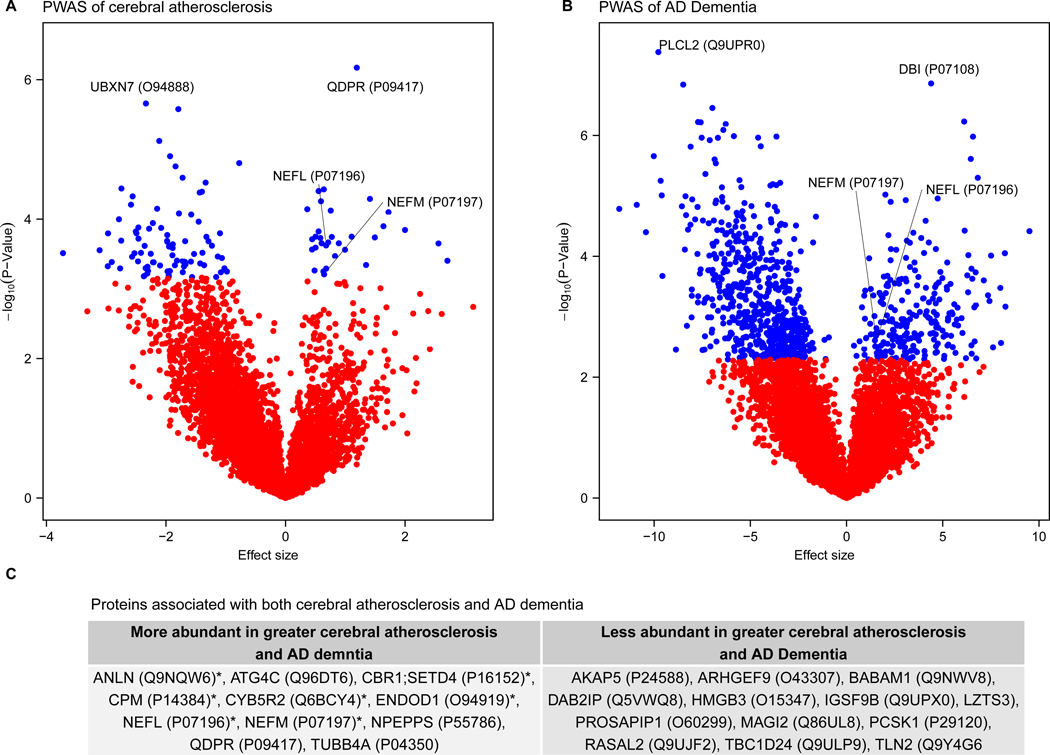
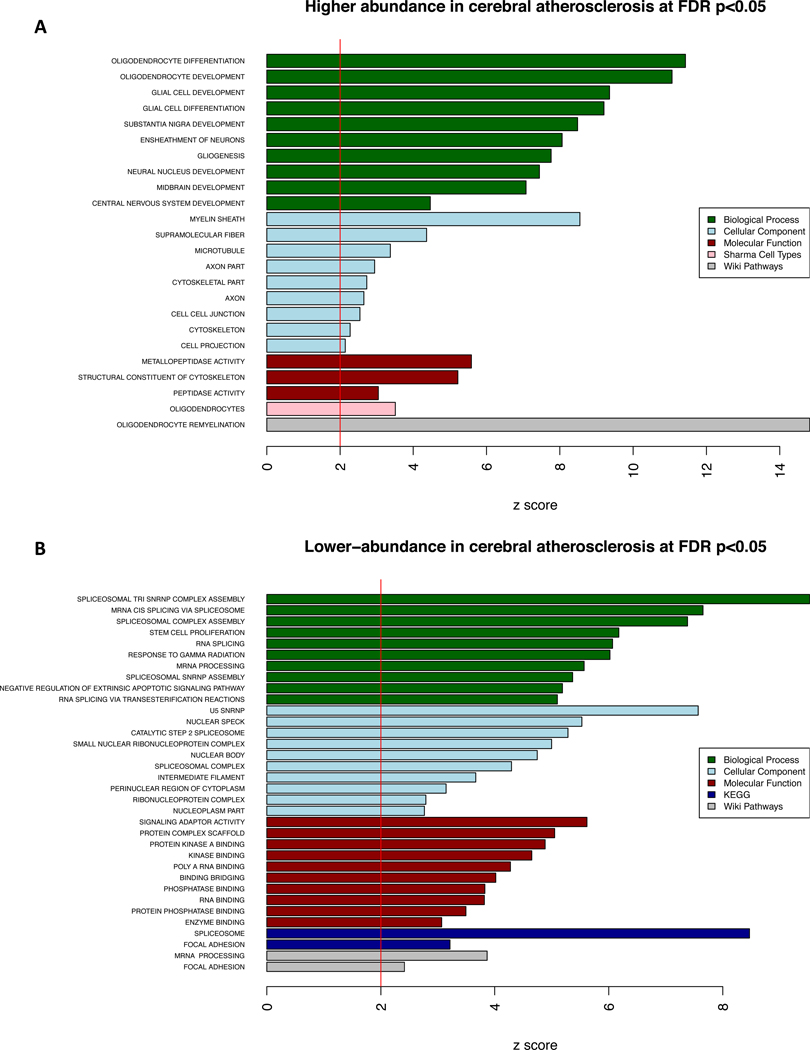
We performed a proteome-wide association study (PWAS) of CA adjusting for the presence of the other 8 measured pathologies in the discovery dataset and found 114 proteins differentially expressed at proteome-wide adjusted p<0.05 (Figure 1A, Supplementary table 4). Proteins with positive effect sizes (e.g., QDPR) were more abundant, and conversely, those with negative effect sizes (e.g., UBXN7) were less abundant with greater CA. The 32 higher-abundance proteins in CA were enriched for oligodendrocyte cell-type specific markers and oligodendrocyte differentiation, development, and re-myelination (Figure 2). The 82 lower-abundance proteins in CA were enriched for RNA spliceosome and mRNA processing (Figure 2). Next, we examined whether these findings were sensitive to the presence of vascular risk factors (i.e., predisposing factors for CA including diabetes, hypertension, and smoking) or infarction (i.e., the potential consequence of CA). These sensitivity analyses indicate that the CA-associated proteomic changes above were independent of infarcts and vascular risk factors (Supplementary Tables 5–7 and Supplementary Figures 2–3).
Figure 1:
Differential protein expression in cerebral atherosclerosis (CA) and Alzheimer’s Disease (AD). This figure summarizes the proteome-wide association study (PWAS) of CA and AD in the discovery dataset (N=375) and gives the genes associated with both CA and AD. a) Volcano plot of the PWAS of CA plotting the effect sizes and p-values estimated from linear regression of each protein and CA adjusted for clinical covariates and 8 other measured pathologies. For each protein, points are either blue or red to reflect whether the protein was or was not significantly associated with CA at proteome-wide significant level, respectively. Labels are given as gene symbols and Uniprot IDs in parentheses for the top two associated proteins and for NEFL and NEFM. b) Volcano plot of the PWAS of AD plotting the effect sizes and p-values estimated from logistic regression of each protein and AD adjusted for clinical covariates. For each protein, points are either blue or red to reflect whether the protein was or was not significantly associated with AD at proteome-wide significant level, respectively. Labels are given as gene symbols and Uniprot IDs tested, in parentheses, for the top two associated proteins and for NEFL and NEFM. c) Gene symbols and the Uniprot ID of the proteins associated with both AD and CA at proteome-wide adjusted p <0.05.
Figure 2:
Enrichment analysis of proteins associated with cerebral atherosclerosis (CA). This figure shows the results of the gene set enrichment analysis (GSEA) for the 114 proteins associated with CA independently of the 8 other measured pathologies at proteome-wide adjusted p <0.05 in the discovery dataset (N=375). a) Proteins with significantly higher-abundance in cerebral atherosclerosis (n=32) were used for GSEA, and one-sided Z-scores above 2 were plotted for results of each of the five annotation databases used. b) Proteins with significantly lower-abundance in cerebral atherosclerosis (n=82) were used for GSEA and a one-sided Z-scores above 2 were plotted for results of each of the five annotation databases used.
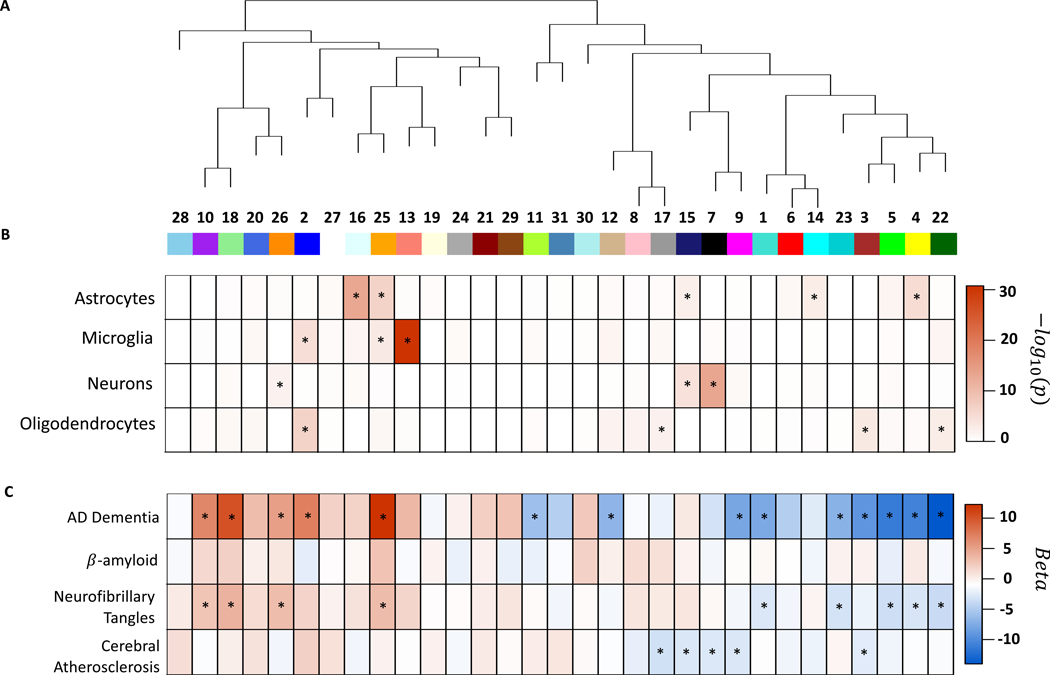
Using weighted gene co-expression network analysis, we identified 31 protein co-expression modules in the discovery dataset (Figure 3A). Among these, five modules were associated with CA independently of the other 8 measured pathologies at adjusted p<0.05, and all five modules had lower abundance in greater CA (modules 3, 7, 9, 15, and 17; Figure 3C, Supplementary Table 8). Three modules were enriched for synaptic signaling, regulation, and plasticity (modules 3, 9, and 17), and two for mRNA splicing and mRNA processing (modules 7 and 15; Supplementary Table 8). Additionally, two modules were also enriched for oligodendrocyte cell-type specific markers (modules 3 and 17), two for neuron-specific markers (modules 7 and 15), and one for astrocyte-specific markers (module 15; Figure 3B, Supplementary Table 8).
Figure 3:
Protein co-expression network analysis for brain cell types and clinical and neuropathologic outcomes. This figure summarizes the protein co-expression modules and their associations with AD, β-amyloid, neurofibrillary tangles, and cerebral atherosclerosis in the discovery dataset (N=375). a) Dendrogram for 31 protein co-expression modules derived using WGCNA. b) Brain cell-type enrichment for each module. The heatmap shows the one-sided Z-test for each brain cell-type and module and significant enrichment is indicated by an asterisk. c) Results of association testing between modules and AD dementia, β-amyloid, neurofibrillary tangles, and cerebral atherosclerosis, respectively. The heatmap gives the beta values of the regression models for each outcome using the module as the predictor adjusted for relevant covariates as described in the text. For the three neuropathologic outcomes (i.e., β-amyloid, neurofibrillary tangles, and cerebral atherosclerosis), linear regression adjusted for 8 other neuropathologies and relevant covariates was used. Benjamin-Hochberg false discovery rate were used to adjust p-values and associations with adjusted p<0.05 are indicated with asterisk. No module was associated with β-amyloid after adjusting for all the other 8 measured pathologies. Nine modules were associated with tangles after adjusting for the 8 other measured pathologies at adjusted p<0.05. Five modules were associated with cerebral atherosclerosis after adjusting for the 8 other measured pathologies.
Together, the PWAS and protein co-expression network analysis highlight that CA was associated with i) more oligodendrocyte differentiation, development, and remyelination, ii) less RNA splicing and mRNA processing in neurons and astrocytes, and iii) decreased synaptic signaling, regulation, and plasticity, independently of the 8 other measured pathologies.
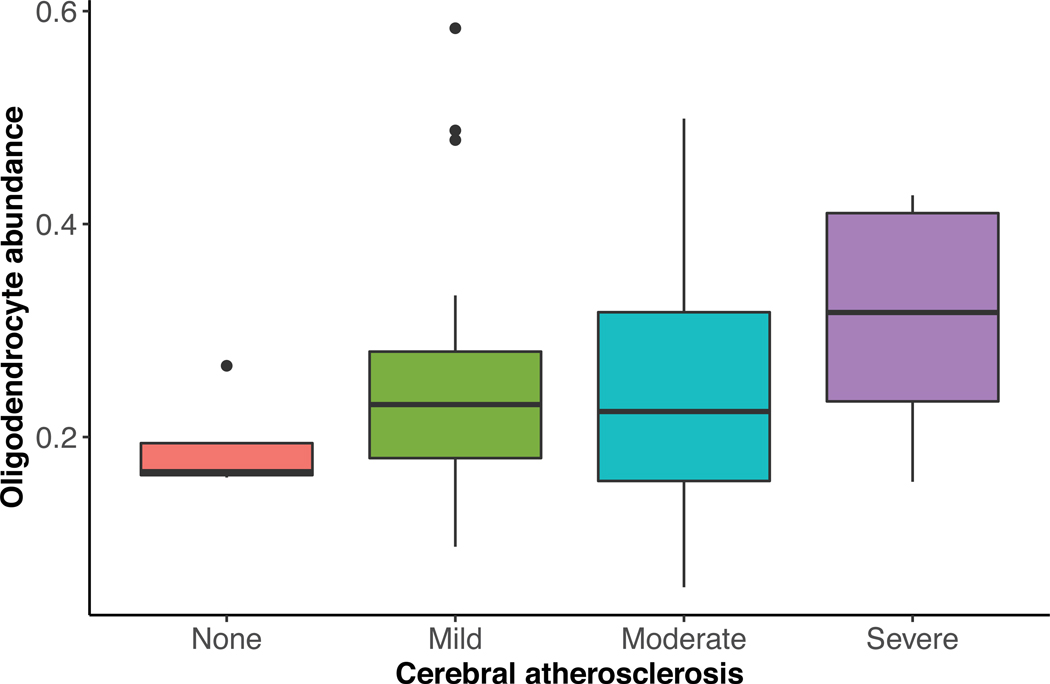
To validate our findings that CA was associated with increased oligodendrocyte differentiation, development, and re-myelination, we used human brain single-cell RNA sequencing data from non-overlapping ROS/MAP subjects11 to test whether CA was associated with higher oligodendrocyte abundance. Consistent with our findings, persons with greater CA had greater proportions of oligodendrocytes in the dorsolateral prefrontal cortex after adjusting for sex, age, and the 8 other measured pathologies (p=0.003, N=48, Extended Data Figure 1).
To replicate the proteomic findings for CA, we examined the 114 CA-associated proteins in a replication dataset (BLSA cohort). We performed a targeted replication due to sample size difference between the datasets (391 versus 47) and technical differences in proteomic profiling (i.e., trypsin versus LysC) leading to 51% of the discovery dataset proteins being quantified in both datasets (4254 of 8356 proteins). All analyses in the replication dataset were adjusted for β-amyloid, tangles, gross infarcts, microinfarcts, CAA, sex, age at death, and cell type composition in the same fashion as was done for the discovery dataset. Of the 114 CA-associated proteins, 79 proteins were measured in the replication dataset. Of these 79 proteins, 42% (33 / 79) were replicated at adjusted p<0.05 (Supplementary Table 9).
Shared molecular processes of cerebral atherosclerosis and AD
We performed PWAS and protein co-expression network analysis of AD and its hallmark pathologies – β-amyloid and neurofibrillary tangles – respectively (Figure 1B, 3C; Supplementary Table 10–12; Supplementary Figure 4–6). Subsequently, we identified 23 proteins associated with both AD and CA independently of the other 8 measured pathologies at proteome-wide adjusted p<0.05 (Figure 1C; Supplementary table 13). All 23 proteins consistently had either higher abundance in both AD and CA, or lower abundance in both, supporting the notion that CA has a detrimental effect on cognition. Of these 23 proteins, 48% were hub proteins of protein co-expression modules (Supplementary table 13). Notably, seven were hub proteins of module 2, which had higher abundance in CA and was enriched for myelination and oligodendrocytes cell-type specific markers (Supplementary Table 8). Additionally, modules 3 and 9 were associated with both AD and CA independently of the other 8 measured pathologies, and both modules had reduced abundance in CA and AD. Module 3 was enriched for synaptic regulation and oligodendrocyte cell-type specific markers, and module 9 for synaptic signaling and plasticity (Supplementary Table 8). Together, these findings suggest CA likely contributes to AD through its effects linked to reduction in synaptic signaling, regulation, and plasticity, and increase in myelination within the grey matter, which was the brain region examined.
We examined evidence for physical interaction and common cellular localization for the proteins in the modules associated with both CA and AD. We found substantial physical interactions between and among the proteins in modules 3 and 9 and the 23 differentially expressed proteins (Supplementary Figure 7, Supplementary Table 14). Additionally, many CA-associated proteins were localized in oligodendrocytes and pre- and post-synaptic regions of neurons (Supplementary Figure 7). Finally, we identified 13 non-redundant protein domains among these proteins suggesting shared physiological roles (Supplementary Table 15).
We did not find a statistical interaction between CA and β-amyloid or tangles, suggesting that CA contributes to AD independently of the two AD hallmark pathologies, consistent with prior studies showing that vascular and β-amyloid and tau pathologies are independent and additive risk factors of cognitive decline12–15.
NEFL and NEFM proteins are associated with cerebral atherosclerosis and AD
Among the 23 proteins associated with both AD and CA independently of the other 8 measured pathologies, NEFL and NEFM were noteworthy (Figure 1C). They are essential structural scaffolding proteins of neurons and released into CSF in many pathological processes that cause axonal damage16. Several studies found an association between CSF or blood NEFL level and AD diagnosis and progression17–19. Thus, we asked whether altered protein levels of NEFL and NEFM were specific to CA by performing a PWAS of each of the remaining pathologies adjusting for the other 8 measured pathologies in the discovery dataset. Strikingly, NEFL and NEFM were only associated with CA and not with β-amyloid, tangles, microinfarcts, gross infarcts, CAA, Lewy body, hippocampal sclerosis, or TDP-43 (Extended Data Figure 2A-E). We replicated the association between NEFL and NEFM proteins and CA in the replication dataset (Supplementary Table 16). Likewise, we replicated the associations between NEFL and NEFM protein abundances and AD in the replication dataset (Supplementary Table 16). NEFL and NEFM proteins showed a step-wise increase in abundance with worsening cognition from controls to MCI to AD. Notably, the associations between NEFL and NEFM proteins and AD were no longer significant when we adjusted for CA in the regression models, suggesting that CA mediates the association between NEFL, NEFM and AD. Together, these findings suggest NEFL and NEFM likely contribute to AD through CA and further studies are needed to elucidate the underlying biology of this association.
In summary, CA has proteomic effects on the human brain and contributes to AD. Furthermore, since CA has important and early contribution to AD development3,4, surrogate brain biomarkers for CA might be promising and sensitive for early AD detection.
METHODS
Study design and participants in the discovery dataset (ROS/MAP cohorts)
A total of 391 participants were from two longitudinal clinical-pathologic cohort studies of aging and Alzheimer’s disease - Religious Orders Study and Rush Memory and Aging Project (ROS/MAP) cohorts (Supplementary Table 1)20. These are participants with proteomic data that passed quality control and were included in our proteomic analyses. ROS is comprised of older Catholic priests, nuns, and monks throughout the USA. MAP recruited older lay persons from the greater Chicago area. Both studies involve detailed annual cognitive and clinical evaluations and brain autopsy. Participants provided informed consent, signed an Anatomic Gift Act and a repository consent to allow their data and biospecimens to be repurposed. The studies were approved by an Institutional Review Board of Rush University Medical Center.
Cognitive diagnosis and brain pathologies in the discovery dataset (ROS/MAP cohorts)
Final clinical diagnosis of cognitive status, including no cognitive impairment, mild cognitive impairment (MCI), Alzheimer’s disease (AD) dementia, or dementia due to other causes, was determined at the time of death by a neurologist with expertise in dementia using all available clinical data but blinded to postmortem data. The diagnosis of dementia was based on the recommendation of the National Institute of Neurological and Communicative Disorders and Stroke and the AD and Related Disorders Association21. MCI was based on accepted criteria. Case conferences including one or more neurologists and a neuropsychologist were used for consensus as necessary.
Cerebral atherosclerosis was pathologically assessed by visual inspection of the vessels in the Circle of Willis including vertebral, basilar, posterior cerebral, middle cerebral, and anterior cerebral arteries and their proximal branches9. Severity of atherosclerosis was graded with a semiquantitative scale on the basis of the extent of involvement of each artery and the number of arteries involved. Scores ranged from 0 to 3 and were treated as a semiquantitative variable in our analyses. A score of zero means no significant atherosclerosis observed. A score of 1 (mild) indicates that small amounts of luminal narrowing were observed in up to several arteries without significant occlusion. A score of 2 (moderate) means that luminal narrowing occurred in up to half of all visualized major arteries with less than 50% occlusion of any single vessel. Lastly, a score of 3 (severe) indicates that luminal narrowing occurred in more than half of all visualized arteries, and/or more than 75% occlusions of one or more vessels9.
Presence of chronic gross infarcts was determined by neuropathologic evaluation blinded to clinical data and reviewed by a board-certified neuropathologist in nine regions (midfrontal, middle temporal, entorhinal, hippocampal, inferior parietal, anterior cingulate cortices, anterior basal ganglia, thalamus, and midbrain) and treated as a dichotomous variable of present or absent 22. Presence of microinfarcts in nine brain regions was determined by neuropathological evaluation blinded to clinical data and reviewed by board-certified neuropathologist and treated as a dichotomous variable 23.
Neurofibrillary tangles and β-amyloid were identified by molecularly specific immunohistochemistry and quantified by stereology and image analysis, respectively, in eight brain regions – hippocampus, entorhinal cortex, midfrontal cortex, inferior temporal, angular gyrus, calcarine cortex, anterior cingulate cortex, and superior frontal cortex 24. Tangle density was determined using systematic sampling and was the average of the tangle densities in eight regions. β-amyloid score represents the percent area of the cortex occupied by β-amyloid and is calculated by taking the mean of β-amyloid scores in 8 brain regions. To create approximately normal distribution of tangles and β-amyloid, we took the square root of tangle density and β-amyloid score, respectively, in our analyses.
Lewy body pathology was assessed using antibodies to α-synuclein in nine regions and were scored based on four stages of distribution of α-synuclein in the brain including 0 (not present), 1 (nigral-predominant), 2 (limbic-type) and 3 (neocortical-type), and treated as present or absent in our analyses 25. Hippocampal sclerosis was identified as severe neuronal loss and gliosis in the hippocampus or subiculum using haematoxylin and eosin stain and treated as present or absent in our analyses 26. TDP-43 cytoplasmic inclusions were assessed in six regions using antibodies to phosphorylated TDP-43 27. Inclusions in each region were rated on a six-point scale and the mean of the regional scores was created. TDP-43 scores were dichotomized into absent (i.e. mean score of 0) or present (mean score >0) in our analyses. Cerebral amyloid angiopathy (CAA) was assessed in the midfrontal, midtemporal, angular, and calcarine cortices using immunostain for β-amyloid 28. Scores were averaged across these 4 regions and treated as a continuous measure.
Magnetic resonance imaging acquisition and voxelwise analysis in ROS/MAP cohorts
At autopsy brains were hemisected in accordance with standard protocol and the cerebral hemisphere with more visible pathology was immersed in 4% paraformaldehyde solution and refrigerated at 4°C29. At approximately one-month postmortem (mean=30 days; SD=19.2 days), the hemisphere from each decedent was imaged using a 3-Tesla MRI scanner after allowing the tissue to warm to room temperature. Four different scanners were used to acquire fast spin-echo T2-weighted MRI data with at least two different echo times (TE). Details of MRI acquisition have been previously reported10,30–32. The transverse relaxation time constant, T2, describes the decay rate of the transverse component of magnetization, which is sensitive to brain tissue’s free water content and paramagnetic materials, such as iron. As previously described10,30,31, we quantified the reciprocal of T2, called R2, to reduce skewness and achieve a more normally distributed signal. Individual R2 volumes were spatially registered to a cerebral hemisphere template, using first linear and then nonlinear transformations10. In order to account for inter-scanner differences in R2, we carried out a voxel-by-voxel normalization of R2 values within each group of hemispheres imaged on a given scanner by subtracting the median and dividing by the interquartile range of R2 for that scanner. In addition, we carried out spatial smoothing with full width at half maximum 2mm. Details of postmortem MRI preprocessing procedure have been described previously10,30,31. Voxelwise linear regression analysis was applied to identify brain regions where postmortem R2 was associated with cerebral atherosclerosis after controlling for age at death, sex, and years of education. Thresholds applied were first on individual voxels (p<0.01) and then on individual clusters (p <0.05 corrected for multiple comparisons) based on Random Field theory33,34.
Proteomic profiling for the discovery dataset (ROS/MAP cohorts)
The dorsolateral prefrontal cortex was chosen for examination because neuropathologic changes associated with AD are relatively late features of the disease in that region of the brain. We posit that this will enable detection of earlier changes in the brain proteome with disease. Post-mortem tissues from the dorsolateral prefrontal cortex (Brodmann area 9) was cortically dissected. Technicians were blinded to clinical outcomes of samples. Procedures for tissue homogenization were performed essentially as described previously 35. Approximately 100 mg (wet tissue weight) of brain tissue was homogenize in 8 M urea lysis buffer (8 M urea, 10 mM Tris, 100 mM NaHPO4, pH 8.5) with HALT protease and phosphatase inhibitor cocktail (ThermoFisher) using a Bullet Blender (NextAdvance). Each Rino sample tube (NextAdvance) was supplemented with ~100 μL of stainless-steel beads (0.9 to 2.0 mm blend, NextAdvance) and 500 μL of lysis buffer. Tissues were added immediately after excision and samples were then placed into the bullet blender (in 4 °C cold room). The samples were homogenized for 2 full 5 min cycles and the lysates were transferred to new Eppendorf Lobind tubes. Each sample was then sonicated for 3 cycles consisting of 5 s of active sonication at 30% amplitude followed by 15 s on ice. Samples were then centrifuged for 5 min at 15,000 g and the supernatant was transferred to a new tube. Protein concentration was determined by bicinchoninic acid (BCA) assay (Pierce). For protein digestion, 100 μg of each sample was aliquoted and volumes normalized with additional lysis buffer. An equal amount of protein from each sample was aliquoted and digested in parallel to serve as the global pooled internal standard (GIS) in each TMT batch described below. Samples were reduced with 1 mM dithiothreitol (DTT) at room temperature for 30 min, followed by 5 mM iodoacetamide (IAA) alkylation in the dark for another 30 min. Lysyl endopeptidase (Wako) at 1:100 (w/w) was added and digestion continued overnight. Samples were then 7-fold diluted with 50 mM ammonium bicarbonate (AmBic). Trypsin (Promega) was then added at 1:50 (w/w) and digestion was carried out for another 16 h. The peptide solutions were acidified to a final concentration of 1% (vol/vol) formic acid (FA) and 0.1% (vol/vol) triflouroacetic acid (TFA) and desalted with a 30 mg HLB column (Oasis). Each HLB column was rinsed with 1 mL of methanol, washed with 1 mL 50% (vol/vol) acetonitrile, and equilibrated with 2×1 mL 0.1% (vol/vol) TFA. The samples were then loaded and each column was washed with 2×1 mL 0.1% (vol/vol) TFA. Elution was performed with 2 rounds of 0.5 mL of 50% (vol/vol) acetonitrile.
Isobaric tandem mass tag (TMT) peptide labeling in the discovery dataset (ROS/MAP cohorts)
Prior to TMT labeling, samples were randomized by co-variates (age, sex, post-mortem interval, cognitive diagnosis, and pathologies) into 50 total batches (8 cases per batch). Peptides from each individual case (N=400) and the GIS pooled standard (N=100) were labeled using the TMT 10-plex kit (ThermoFisher). In each batch, TMT channels 126 and 131 were used to label GIS standards, while the 8 middle TMT channels were reserved for individual samples following randomization. Labeling was performed as described previously 35,36. Briefly, each sample (100 μg of peptides each) was re-suspended in 100 μL of 100 mM TEAB buffer. The TMT labeling reagents were equilibrated to room temperature and 256 μL anhydrous acetonitrile was added to each reagent channel and softly vortexed for 5 min and 41 μL of the corresponding TMT channels were transferred to peptide suspensions. The samples were then incubated for 1 h at room temperature. The reaction was quenched with 8 μl of 5% (vol/vol) hydroxylamine. All 10 channels were then combined and dried by SpeedVac to approximately 150 μL and diluted with 1 mL of 0.1% (vol/vol) TFA and then acidified to a final concentration of 1% (vol/vol) FA and 0.1% (vol/vol) TFA. Sep-Pak desalting was performed with a 200 mg tC18 Sep-Pak column (Waters). Each Sep-Pak column was activated with 3 mL of methanol, washed with 3 mL of 50% (vol/vol) acetonitrile, and equilibrated with 2×3 mL of 0.1% TFA. The samples were then loaded and each column was washed with 2×3 mL 0.1% (vol/vol) TFA, followed by 2 mL of 1% (vol/vol) FA. Elution was performed with 2 rounds of 1.5 mL of 50% (vol/vol) acetonitrile. The elution was dried to completeness.
High-pH off-line fractionation in the discovery dataset (ROS/MAP cohorts)
High pH fractionation was performed as previously described37 with slight modification. Dried samples were re-suspended in high pH loading buffer (0.07% vol/vol NH4OH; 0.045% vol/vol formic acid, 2% vol/vol acetonitrile) and loaded onto an Agilent ZORBAX 300Extend-C18 column (2.1mm x 150 mm with 3.5 μm beads). An Agilent 1100 HPLC system was used to carry out the fractionation. Solvent A consisted of 0.0175% (vol/vol) NH4OH; 0.01125% (vol/vol) formic acid; 2% (vol/vol) acetonitrile and solvent B consisted of 0.0175% (vol/vol) NH4OH; 0.01125% (vol/vol) formic acid; 90% (vol/vol) acetonitrile. The sample elution was performed by a 58.6 min gradient with a flow rate of 0.4 mL/min. The gradient goes 100% solvent A for 2 min, from 0% to 12% solvent B in 6 min, from 12% to 40 % over 28 min, from 40% to 44% in 4 min, from 44% to 60% in 5 min, and then kept 60% solvent B for 13.6 min. A total of 96 individual fractions were collected across the gradient and subsequently pooled into 24 fractions and dried to completeness by speedvac.
For proteomic quantification, 90% was performed using MS2 and only 10% using MS3. MS3 quantification greatly reduces the contribution of any interfering signals but has reduced sensitivity due to decreased dynamic range of reporter ion quantitation38. The use of MS2 acquisition can suffer from the presence of co-isolated and co-fragmented interfering ions that can obscure quantification; however, our use of high-pH offline fractionation with MS2 largely mitigated this issue37,38.
Mass spectrometry analysis in the discovery dataset (ROS/MAP cohorts)
All fractions were resuspended in equal volume of loading buffer (0.1% formic acid, 0.03% trifluoroacetic acid, 1% acetonitrile) and analyzed by liquid chromatography coupled to mass spectrometry essentially as described39 with slight modifications. Peptide eluents were separated on a self-packed C18 (1.9 um Dr. Maisch, Germany) fused silica column (25 cm × 75 μM internal diameter; New Objective, Woburn, MA) by a Dionex UltiMate 3000 RSLCnano liquid chromatography system (ThermoFisher Scientific) and monitored on an Orbitrap Fusion mass spectrometer (ThermoFisher Scientific). Sample elution was performed over a 180-min gradient with flow rate at 225 nL/min. The gradient goes from 3% to 7% buffer B in 5 mins, from 7% to 30% over 140 mins, from 30% to 60% in 5 mins, 60% to 99% in 2 mins, kept at 99% for 8 min and back to 1% for additional 20 min to equilibrate the column. The mass spectrometer was set to acquire in data dependent mode using the top speed workflow with a cycle time of 3 seconds. Each cycle consisted of 1 full scan followed by as many MS/MS (MS2) scans that could fit within the time window. The full scan (MS1) was performed with an m/z range of 350–1500 at 120,000 resolution (at 200 m/z) with AGC (automatic gain control) set at 4×105 and maximum injection time 50 ms. The most intense ions were selected for higher energy collision-induced dissociation (HCD) at 38% collision energy with an isolation of 0.7 m/z, a resolution of 30,000 and AGC setting of 5×104 and a maximum injection time of 100 ms. Five of the 50 TMT batches were run on the Orbitrap Fusion mass spectrometer using the SPS-MS3 method as previously described35.
Database Searches and protein quantification in the discovery dataset (ROS/MAP cohorts)
All raw files were analyzed using the Proteome Discoverer suite (version 2.3 ThermoFisher Scientific). MS2 spectra were searched against the canonical UniProtKB Human proteome database (Downloaded February 2019 with 20,338 total sequences). The Sequest HT search engine was used and parameters were specified as: fully tryptic specificity, maximum of two missed cleavages, minimum peptide length of 6, fixed modifications for TMT tags on lysine residues and peptide N-termini (+229.162932 Da) and carbamidomethylation of cysteine residues (+57.02146 Da), variable modifications for oxidation of methionine residues (+15.99492 Da) and deamidation of asparagine and glutamine (+0.984 Da), precursor mass tolerance of 20 ppm, and a fragment mass tolerance of 0.05 Da for MS2 spectra collected in the Orbitrap (0.5 Da for the MS2 from the SPS-MS3 batches). Percolator was used to filter peptide spectral matches (PSM) and peptides to a false discovery rate (FDR) of less than 1%. Following spectral assignment, peptides were assembled into proteins and were further filtered based on the combined probabilities of their constituent peptides to a final FDR of 1%. In cases of redundancy, shared peptides were assigned to the protein sequence in adherence with the principles of parsimony. Reporter ions were quantified from MS2 or MS3 scans using an integration tolerance of 20 ppm with the most confident centroid setting.
Quality control of proteomic profiles in the discovery dataset (ROS/MAP cohorts)
For each batch, the global internal standards were used to check for proteins outside of the 95% confidence interval and set to missing. Proteins with missing values in more than 50% of the 400 subjects were excluded. Each protein abundance was then scaled by a sample-specific total protein abundance to remove effects of protein loading differences, and then log2 transformed. Outlier samples were identified and removed through iterative principal component analysis (PCA). In each iteration, samples more than four standard deviations from the mean of either the first or second principal component were removed, and principal components were recalculated for the next iteration. A total of 9 subjects were removed after three rounds of PCA. After quality control, 391 subjects having 8356 proteins were included in our analyses. The normalized abundance of these 8356 proteins was the residual of the linear regression to remove effects of protein batch, MS2 versus MS3 reporter quantitation mode, sex, age at death, postmortem interval, and study (ROS vs. MAP). The normalized abundance of these proteins was used in our analyses.
Estimate of brain cell type composition in the discovery dataset (ROS/MAP cohorts)
To account for the heterogeneity of cell types in brain tissues in our analyses, we estimated the proportions of neuron, astrocyte, oligodendrocyte, and microglia using CIBERSORT pipeline and Sharma’s cell type-specific proteins as the reference 40,41. We modified the CIBERSORT pipeline by taking the absolute values of the negative regression coefficients instead of setting them to zero. This minor modification produced estimates of cell types similar to those obtained using the original CIBERSORT pipeline and RNA sequencing data. We included the proportion of each of these four cell types as covariates in our proteomic analyses.
Weighted protein co-expression network analysis in the discovery dataset (ROS/MAP cohorts)
Weighted protein co-expression network analysis was performed using Weighted Gene Co-expression Network Analysis (WGCNA) R package on normalized protein abundance in which effects of batch, sex, age at death, PMI, and study were removed 42. We applied the following parameters: soft-thresholding power of 12, “bicor” correlation type, “signed” network type, “signed” Topological Overlap Matrix, a minimum module size of 30, and p-value ratio threshold for reassigning proteins between modules of 0.05, a low propensity to merge modules with “mergeCutHeight” of 0.07, and a high sensitivity to cluster splitting (deepSplit=4). Among the 8356 proteins, 62% (5199 proteins) were assigned to 31 modules, with the largest module having 518 proteins and the smallest module having 46 proteins.
We defined hub proteins as those in the top 20% with regard to high connectivity. To determine the hub proteins for each module, the signed eigenprotein based connectivity, which is the correlation of the protein with its corresponding module eigenprotein, was calculated using the signedKME function from WGCNA package with the parameter corFnc “bicor” 43.
Protein-protein interaction and protein domain composition analyses
Lists of pairwise protein interactions were downloaded from the BioGRID database (v3.5.179, October 29, 2019) then culled for interactions containing only human gene symbols, and culled further to only include physical protein-protein interactions among the 770 gene products representing module 3, module 9, and 23 proteins associated with both cerebral atherosclerosis and AD dementia from the proteome-wide association study at FDR < 0.05. Modules 3 and 9 were chosen because they were associated with both cerebral atherosclerosis and AD dementia in our analysis. Edges and nodes among proteins with pairwise interactions among the two modules and the 23 proteins were visualized in Cytoscape44.
Enrichment analysis for pathways and gene ontology in ROS/MAP cohorts
Gene set enrichment analysis of protein co-expression modules was performed using GO-Elite with the background set to all 8356 proteins quantified in the ROS/MAP cohorts 45. Protein list per module was subjected to Fisher exact test and Z-test in the Python command line version of GO-Elite (v1.2.5) against the current annotation database for Gene Ontology Biological Process, Molecular Function, Cellular Component, Wiki Pathways, KEGG terms, and Sharma cell types downloaded in January 2019 41. Similarly, gene set enrichment analysis of the cerebral atherosclerosis-associated proteins was performed using the GO-Elite software as described above.
Clinical and pathologic characterization of the Baltimore Longitudinal Study of Aging (BLSA) cohort (replication dataset)
The replication cohort was 47 subjects from the BLSA with brain proteomic data (Supplementary Table 2). BLSA is a prospective cohort study of aging in community dwelling individuals 46. It continuously recruits healthy volunteers aged 20 or older and follows them for life regardless of changes in health or functional status. The BLSA study was approved by the Institutional Review Board and the National Institute on Aging. Cerebral atherosclerosis was assessed by visual examination of the vessels in the Circle of Willis as described previously 47. Areas of atherosclerosis were identified and then sectioned to identify the degree of stenosis by visual inspection and rated on a scale of minimal (<10%), mild (10% <mild<50%), moderate (50%≤moderate<90%), or severe (≥90%). Cerebral atherosclerosis was treated as a semi-quantitative variable in our analyses. Estimates of β-amyloid positive plaques and neurofibrillary tangles were described in detail before by O’Brien and colleagues 48. Briefly, silver staining was performed to assess severity of neuritic plaques using CERAD (score ranges from 0 to 3) and of neurofibrillary tangles using Braak (score ranges from 1 to 6), with higher scores indicating higher level of severity. Additionally, cerebral amyloid angiopathy was assessed and rated as mild (rare staining), moderate (scattered, incomplete staining), and severe (circumferential, diffuse staining) and treated as a semi-quantitative variable. Gross infarcts were assessed and quantified as absent versus present anywhere in the brain. Likewise, microinfarcts were assessed and determined as absent versus present in any locations of the brain. All BLSA participants provided written informed consent and the study was approved by the Institutional Review Board of the National Institute on Aging.
Proteomic profiling, quality control, and normalization in the BLSA replication cohort
Proteins were profiled from the dorsolateral prefrontal cortex using an analysis pipeline consisting of isobaric tandem mass tag (TMT) mass spectrometry and offline prefractionation as described in detail previously36. TMT labeling was performed following manufacturer’s protocol. Technicians were blinded to clinical outcomes of samples. MS/MS spectra were searched against a Uniprot human database and Proteome Discoverer 2.1 (ThermoFisher Scientific) as described in detail previously36. Global internal standard (GIS) mixture was checked for extreme outliers, and values outside of the range of log2(0.01) to log2(100) were excluded from analysis36. Proteins with more than four unquantifiable batches (out of a total of 8 batches) due to 0 or NA value for the GIS channel were excluded from consideration. Proteins with missing value in more than 50% of the samples were excluded. After quality control, 6533 proteins were detected36. Effects of sex, age at death, batch, and postmortem interval were regressed out using non-parametric bootstrap regression36. The normalized proteomic abundance was used for our analyses.
Statistical analysis
A proteome-wide association study (PWAS) of clinical AD dementia (no cognitive impairment versus MCI/AD dementia), neurofibrillary tangles, β-amyloid, and cerebral atherosclerosis was performed, individually, using regression and the normalized protein abundance, adjusting for sex, age at death, proportions of neuron, astrocyte, oligodendrocyte, microglia, and other covariates as specified. Continuous outcomes were visually inspected for normality of distribution and neurofibrillary tangles and β-amyloid were transformed using square root to enhance normal distribution. Breusch-Pagan test found that equal variance assumption was met for linear regression. Of note, we adjusted for sex, age at death, and cell type composition in all the PWAS. Meta-analysis of the findings from the discovery (ROS/MAP) and replication (BLSA) datasets was performed with METAL using effect size estimates and standard errors49.
Associations between protein co-expression modules and clinical AD dementia, neurofibrillary tangles, β-amyloid, and cerebral atherosclerosis, individually, were examined using linear regression, adjusting for sex, age at death, estimated proportions of neuron, astrocyte, oligodendrocyte, microglia, and other covariates as specified. Again, we adjusted for sex, age at death, and cell type composition in all of the protein co-expression module analyses. For all analyses, multiple testing adjustment was addressed with Benjamini-Hochberg (BH) false discovery rate (FDR) 50.
Extended Data
Extended Data Fig. 1. Association between cerebral atherosclerosis and oligodendrocyte abundance.
This figure summarizes the association between CA and oligodendrocyte abundance estimated from single-cell nuclear RNA sequencing of human dorsolateral prefrontal cortex in an independent dataset. Higher cerebral atherosclerosis was associated with higher oligodendrocyte abundance in the dorsolateral prefrontal cortex after adjusting for sex, age, and 8 other measured pathologies using linear regression (p=0.003, β=0.077, N=48). For each box plot, the box reflects the first and third quartile, the dark horizontal line reflects the median, the vertical lines extending from the boxes show the 1.5x the interquartile range, and points beyond the lines are outliers.
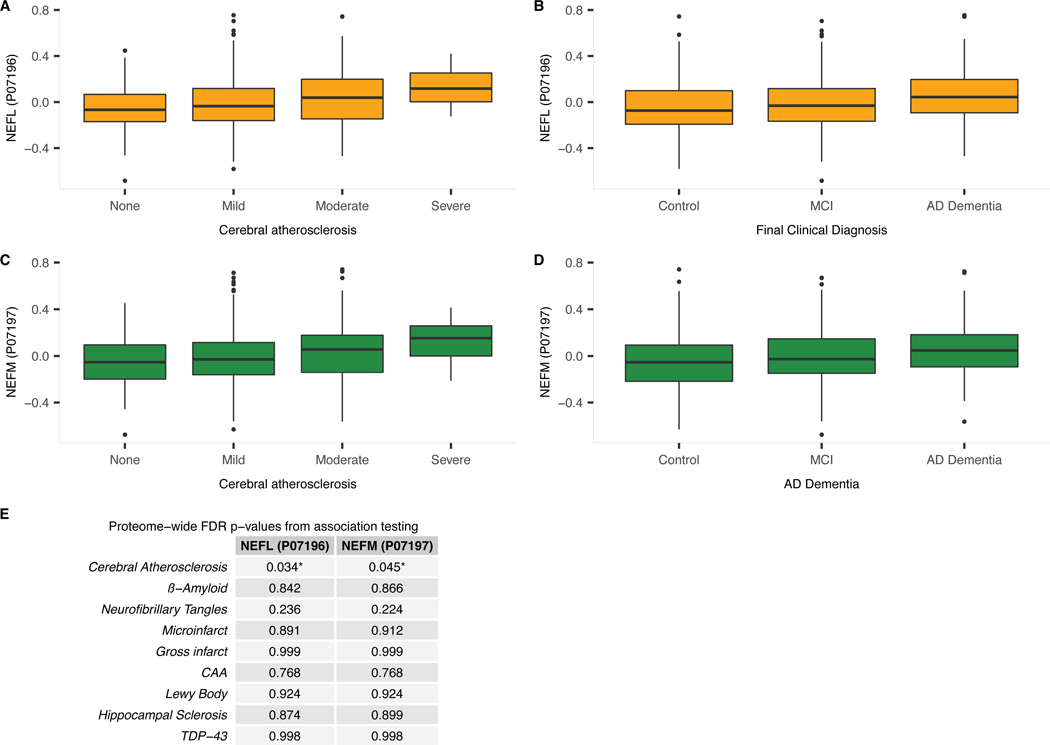
Extended Data Fig. 2. Association between NEFL or NEFM brain protein and cerebral atherosclerosis, AD dementia.
a) Boxplot of dorsolateral prefrontal cortex NEFL protein level for each level of CA (i.e., none to severe). NEFL protein levels were identified as associated with CA using linear regression adjusted for relevant covariates and 8 other pathologies (N=375; p=0.0002; adjusted p=0.034; Figure 1). b) Boxplot of dorsolateral prefrontal cortex NEFL protein level by clinical diagnosis (i.e., cognitive normal control [control], mild cognitive impairment [MCI], and AD dementia). MCI is generally regarded as an intermediate stage between cognitively normal and AD. NEFL protein levels were identified as associated with AD dementia by logistic regression adjusted for relevant covariates (N=383; p=0.0025; adjusted p=0.0322; Figure 1). c) Boxplot of dorsolateral prefrontal cortex NEFM protein level for each level of CA (i.e., none to severe). NEFM protein levels were identified as associated with CA using linear regression adjusted for relevant covariates and 8 other pathologies (N=375; p=0.0006; adjusted p=0.045). d) Boxplot of dorsolateral prefrontal cortex NEFM protein level by clinical diagnosis (i.e., cognitive normal control [control], mild cognitive impairment [MCI], and AD dementia). NEFM protein levels were identified as associated with AD dementia by logistic regression adjusted for relevant covariates (N=383; p=0.0019; adjusted p=0.028). For each boxplot, the box reflects the first and third quartile, the dark horizontal line reflects the median, the vertical lines extending from the boxes show the 1.5x the interquartile range, and points beyond the lines are outliers. e) Proteome-wide adjusted p-values for associations between NEFL and NEFM brain protein levels and each of the 9 measured pathologies adjusting for the remaining 8 measured pathologies. Associations were tested with regression and the provided p-values were adjusted for multiple testing using Benjamin-Hochberg false discovery rate.
Supplementary Material
Acknowledgements
We thank the participants of the ROS, MAP, and BLSA studies for their time and participation. Support was provided by R01 AG056533, R01 AG053960, the Accelerating Medicine Partnership for AD (U01 AG046152; U01 AG046161; U01 AG061356; U01 AG061357), the Emory Alzheimer’s Disease Research Center (P50 AG025688), and the NINDS Emory Neuroscience Core (P30 NS055077), R01 AG017917 (DAB), R01 AG015819 (DAB), RC2 AG036547 (DAB), P30 AG10161 (DAB), R01 AG042210 (JAS), and in part by the intramural program of the National Institute on Aging. APW is also supported by U01 MH115484 and I01 BX003853. TSW is also supported by RF1 AG057470, R56 AG062256, R56 AG060757, and R56 AG062633. NTS is also supported by an Alzheimer’s Association, Alzheimer’s Research UK, The Michael J. Fox Foundation for Parkinson’s Research, the Weston Brain Institute Biomarkers Across Neurodegenerative Diseases Grant (11060), R01 AG061800, and R01 AG057911.
Footnotes
Competing interests
The authors declare no competing interests.
Accession codes
Proteomic data from the discovery dataset (ROS/MAP cohorts) are available at: https://www.synapse.org/#!Synapse:syn2580853/wiki/409853.
Proteomic data from the replication dataset (BLSA cohort) are available at: https://www.synapse.org/#!Synapse:syn9884368
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
Phenotype data are available at: https://www.radc.rush.edu.
Proteomic data from the discovery dataset (ROS/MAP cohorts) are available at: https://www.synapse.org/#!Synapse:syn2580853/wiki/409853.
Proteomic data from the replication dataset (BLSA cohort) are available at: https://www.synapse.org/#!Synapse:syn9884368
References
- 1.Qureshi AI & Caplan LR Intracranial atherosclerosis. Lancet (London, England) 383, 984–998 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Roher AE, et al. Intracranial atherosclerosis as a contributing factor to Alzheimer’s disease dementia. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 7, 436–444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zlokovic BV Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nature reviews. Neuroscience 12, 723–738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iturria-Medina Y, Sotero RC, Toussaint PJ, Mateos-Perez JM & Evans AC Early role of vascular dysregulation on late-onset Alzheimer’s disease based on multifactorial data-driven analysis. Nat Commun 7, 11934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweeney MD, et al. Vascular dysfunction-The disregarded partner of Alzheimer’s disease. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 15, 158–167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snyder HM, et al. Vascular contributions to cognitive impairment and dementia including Alzheimer’s disease. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 11, 710–717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofman A, et al. Atherosclerosis, apolipoprotein E, and prevalence of dementia and Alzheimer’s disease in the Rotterdam Study. Lancet (London, England) 349, 151–154 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Beach TG, et al. Circle of Willis atherosclerosis: association with Alzheimer’s disease, neuritic plaques and neurofibrillary tangles. Acta neuropathologica 113, 13–21 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Arvanitakis Z, Capuano AW, Leurgans SE, Bennett DA & Schneider JA Relation of cerebral vessel disease to Alzheimer’s disease dementia and cognitive function in elderly people: a cross-sectional study. The Lancet. Neurology 15, 934–943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawe RJ, et al. Postmortem MRI: a novel window into the neurobiology of late life cognitive decline. Neurobiology of aging 45, 169–177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathys H, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vemuri P, et al. Vascular and amyloid pathologies are independent predictors of cognitive decline in normal elderly. Brain : a journal of neurology 138, 761–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustavsson AM, et al. Midlife Atherosclerosis and Development of Alzheimer or Vascular Dementia. Ann Neurol 87, 52–62 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesman RF, et al. Association of Intracranial Atherosclerotic Disease With Brain beta-Amyloid Deposition: Secondary Analysis of the ARIC Study. JAMA neurology (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soldan A, et al. White matter hyperintensities and CSF Alzheimer disease biomarkers in preclinical Alzheimer disease. Neurology 94, e950–e960 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalil M, et al. Neurofilaments as biomarkers in neurological disorders. Nature reviews. Neurology 14, 577–589 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Zetterberg H, et al. Association of Cerebrospinal Fluid Neurofilament Light Concentration With Alzheimer Disease Progression. JAMA neurology 73, 60–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preische O, et al. Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer’s disease. Nature medicine 25, 277–283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsson B, et al. CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis. The Lancet. Neurology 15, 673–684 (2016). [DOI] [PubMed] [Google Scholar]
REFERENCES (for Methods)
- 20.Bennett DA, et al. Religious Orders Study and Rush Memory and Aging Project. Journal of Alzheimer’s disease : JAD 64, S161–s189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKhann G, et al. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34, 939–944 (1984). [DOI] [PubMed] [Google Scholar]
- 22.Schneider JA, et al. Relation of cerebral infarctions to dementia and cognitive function in older persons. Neurology 60, 1082–1088 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Arvanitakis Z, Leurgans SE, Barnes LL, Bennett DA & Schneider JA Microinfarct pathology, dementia, and cognitive systems. Stroke 42, 722–727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett DA, Wilson RS, Boyle PA, Buchman AS & Schneider JA Relation of neuropathology to cognition in persons without cognitive impairment. Ann Neurol 72, 599–609 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider JA, et al. Cognitive impairment, decline and fluctuations in older community-dwelling subjects with Lewy bodies. Brain : a journal of neurology 135, 3005–3014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nag S, et al. Hippocampal sclerosis and TDP-43 pathology in aging and Alzheimer disease. Ann Neurol 77, 942–952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson RS, et al. TDP-43 pathology, cognitive decline, and dementia in old age. JAMA neurology 70, 1418–1424 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle PA, et al. Cerebral amyloid angiopathy and cognitive outcomes in community-based older persons. Neurology 85, 1930–1936 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider JA, Arvanitakis Z, Leurgans SE & Bennett DA The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann Neurol 66, 200–208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dawe RJ, et al. Ex vivo T2 relaxation: associations with age-related neuropathology and cognition. Neurobiology of aging 35, 1549–1561 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawe RJ, et al. Postmortem brain MRI is related to cognitive decline, independent of cerebral vessel disease in older adults. Neurobiology of aging 69, 177–184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawe RJ, Bennett DA, Schneider JA, Vasireddi SK & Arfanakis K. Postmortem MRI of human brain hemispheres: T2 relaxation times during formaldehyde fixation. Magnetic resonance in medicine 61, 810–818 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friston KJ, Worsley KJ, Frackowiak RS, Mazziotta JC & Evans AC Assessing the significance of focal activations using their spatial extent. Human brain mapping 1, 210–220 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Worsley KJ, et al. A unified statistical approach for determining significant signals in images of cerebral activation. Human brain mapping 4, 58–73 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Ping L, et al. Global quantitative analysis of the human brain proteome in Alzheimer’s and Parkinson’s Disease. Scientific data 5, 180036 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson ECB, et al. Deep proteomic network analysis of Alzheimer’s disease brain reveals alterations in RNA binding proteins and RNA splicing associated with disease. Molecular neurodegeneration 13, 52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mertins P, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nature protocols 13, 1632–1661 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McAlister GC, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Analytical chemistry 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wingo TS, et al. Integrating Next-Generation Genomic Sequencing and Mass Spectrometry To Estimate Allele-Specific Protein Abundance in Human Brain. Journal of proteome research 16, 3336–3347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nature methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma K, et al. Cell type- and brain region-resolved mouse brain proteome. Nature neuroscience 18, 1819–1831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langfelder P. & Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zambon AC, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics (Oxford, England) 28, 2209–2210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seddighi S, et al. SPARCL1 Accelerates Symptom Onset in Alzheimer’s Disease and Influences Brain Structure and Function During Aging. Journal of Alzheimer’s disease : JAD 61, 401–414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dolan H, et al. Atherosclerosis, dementia, and Alzheimer disease in the Baltimore Longitudinal Study of Aging cohort. Ann Neurol 68, 231–240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Brien RJ, et al. Neuropathologic studies of the Baltimore Longitudinal Study of Aging (BLSA). Journal of Alzheimer’s disease : JAD 18, 665–675 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willer CJ, Li Y. & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England) 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y. & Yekutieli D. The Control of the False Discovery Rate in Multiple Testing under Dependency. The Annals of Statistics 29 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.