Abstract
Purpose
The tumor microenvironment (TME) consists of a heterogenous cellular milieu that can influence cancer cell behavior. Its characteristics have an impact on treatments such as immunotherapy. These features can be revealed with single-cell RNA sequencing (scRNA-seq). We hypothesized that scRNA-seq analysis of gastric cancer (GC) together with paired normal tissue and peripheral blood mononuclear cells (PBMCs) would identify critical elements of cellular deregulation not apparent with other approaches.
Experimental Design
scRNA-seq was conducted on seven patients with GC and one patient with intestinal metaplasia. We sequenced 56,167 cells comprising GC (32,407 cells), paired normal tissue (18,657 cells) and PBMCs (5,103 cells). Protein expression was validated by multiplex immunofluorescence.
Results
Tumor epithelium had copy number alterations, a distinct gene expression program from normal, with intra-tumor heterogeneity. GC TME was significantly enriched for stromal cells, macrophages, dendritic cells (DCs) and Tregs. TME-exclusive stromal cells expressed distinct extracellular matrix components than normal. Macrophages were transcriptionally heterogenous and did not conform to a binary M1/M2 paradigm. Tumor-DCs had a unique gene expression program compared to PBMC DCs. TME-specific cytotoxic T cells were exhausted with two heterogenous subsets. Helper, cytotoxic T, Treg and NK cells expressed multiple immune checkpoint or costimulatory molecules. Receptor-ligand analysis revealed TME-exclusive inter-cellular communication.
Conclusions
Single-cell gene expression studies revealed widespread reprogramming across multiple cellular elements in the GC TME. Cellular remodeling was delineated by changes in cell numbers, transcriptional states and inter-cellular interactions. This characterization facilitates understanding of tumor biology and enables identification of novel targets including for immunotherapy.
INTRODUCTION
Gastric cancer (GC) is the fifth most common cancer and the third leading cause of cancer deaths worldwide (1). The current histopathologic classification scheme designates GCs as either intestinal or diffuse according to the morphology, differentiation and cohesiveness of glandular cells. Intestinal GC is preceded by changes in the gastric mucosa called the Correa cascade that progresses through inflammation, metaplasia, dysplasia and adenocarcinoma (2). Diffuse GCs lack intercellular adhesion and exhibit a diffuse invasive growth pattern. Recent integrated genomic and proteomic analyses including by the Cancer Genome Atlas (TCGA) and the Asian Cancer Research Group (ACRG) have refined the classification of GC into distinct molecular subtypes that include the intestinal and diffuse classification (3,4). Regardless of the histopathologic or molecular subtype, GCs are not isolated masses of cancer epithelial cells. Rather, these tumors have a complex morphology where cancer cells are surrounded by the tumor microenvironment (TME), a cellular milieu containing diverse cell types such as fibroblasts, endothelial and immune cells.
Increasingly, it is recognized that the cellular features of the TME play an important role in enabling tumors to proliferate and metastasize. A major component of the TME that influences tumor cell survival as well as response to treatments such as immune checkpoint blockade is the diverse and deregulated cellular states of the immune cells (5). Thus, the cellular characterization of the TME provides a more sophisticated picture of the context of tumor cell growth within its tissue of origin, characteristics of immune infiltrate and inter-cellular interactions.
The major objective of this study was to determine the specific cellular and transcriptional features that distinguish the GC TME from normal gastric tissue. We sought to define these differences at the resolution of single cells with single-cell RNA-seq (scRNA-seq). We delineated cell-specific features that are otherwise lost when using “bulk” methods in which molecular analytes cannot be attributed to their cell-of-origin. We accomplished this by using an extensive analytical framework (Figure 1A) (6–9) that revealed changes in transcriptional states, regulatory networks and intercellular communication between matched gastric tumor and normal tissue from the same patients, together with peripheral blood mononuclear cells (PBMCs) from a subset of patients. Our study identified cellular and biological features that are specific to the TME and thus offer insights which may help infer new therapeutic targets.
Figure 1:
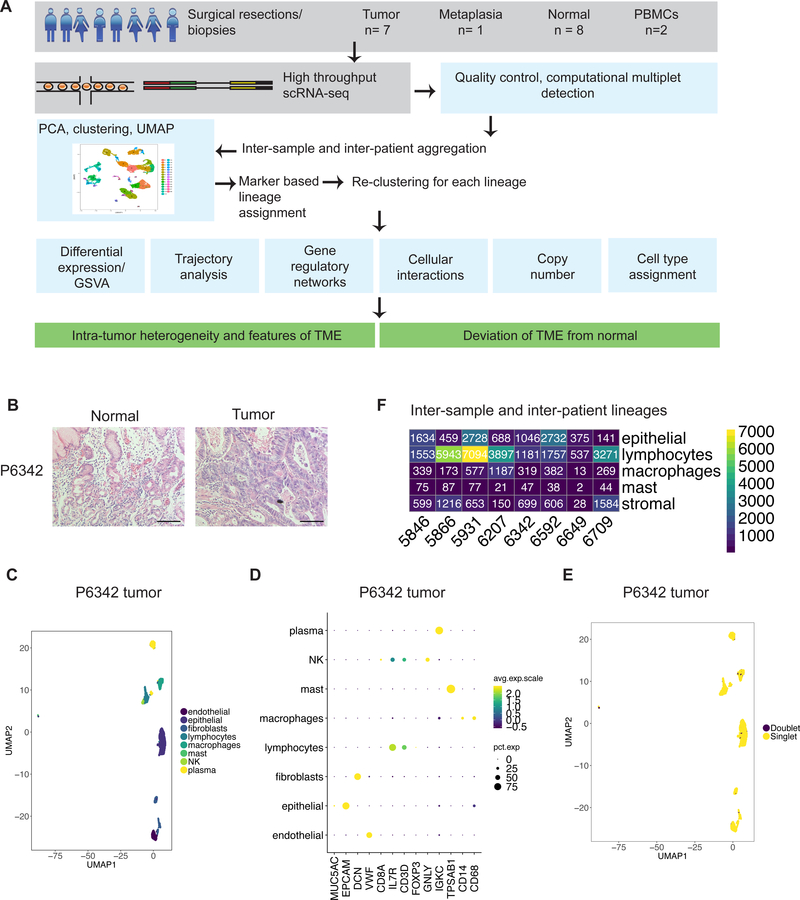
(A) Schematic representation of experimental design and analytical methods used in this study. (B) Representative images of hematoxylin and eosin staining of FFPE tissue from P6342. Scale bar indicates 50 μm. (C-F) Example of clustering analysis in tumor sample of P6342. (C) UMAP representation of dimensionally reduced data following graph-based clustering with marker-based cell type assignments. (D) Dot plot depicting expression levels of specific lineage-based marker genes together with the percentage of cells expressing the marker. (E) UMAP representation of dimensionally reduced data following graph-based clustering with computational doublet identification. (F) Heatmap depicting number of cells identified in aggregated analysis for each lineage per patient.
METHODS
Sample acquisition
All samples were acquired with informed consent under an approved institutional review board protocol from Stanford University as surgical resections or endoscopic biopsies. Matched normal tissue was obtained from sites displaced at least several centimeters from the tumor and was confirmed to lack tumor cells on histopathology review.
Tissue processing and single-cell sequencing
Tissues were collected immediately after resection and dissociated into a single-cell suspension (Supplementary Methods). PBMCs were isolated using density gradient centrifugation. Single-cell libraries were generated from cell suspensions using Chromium Single Cell 3′ Library & Gel Bead Kit v2 (10x Genomics, Pleasanton, CA, USA) as per manufacturer’s protocol and sequenced on Illumina sequencers (Illumina, San Diego, CA) (Supplementary Table S1). Cell Ranger v 3.0 (10x Genomics) ‘mkfastq’ and ‘count’ commands were used with default parameters and alignment to GRCh38 to generate matrix of unique molecular identifier (UMI) counts per gene and associated cell barcode. Datasets are available under dbGAP identifier phs001818.v1.p1.
Clustering analysis
We used Seurat (v2.3.4) (9) to create data objects from the matrix outputs. We removed cells that expressed fewer than 200 genes, had greater than 20% mitochondrial genes or had number of UMI in an outlier range indicative of potential doublets (Supplementary Table S1). We excluded genes detected in fewer than three cells. Data was normalized to log scale using the ‘NormalizeData’ function with a default scale parameter of 10000. Highly variable genes were identified using the ‘FindVariableGenes’ function with parameters for x.low.cutoff=0.0125, x.high.cutoff=6 and y.cutoff=0.5. The effects of variation in sequencing depth were regressed out by including ‘nUMI’ as a parameter in the ‘ScaleData’ function. These variable genes were used as input for PCA using the ‘RunPCA’ function. The first 20 principal components (PCs) and a resolution of 0.8 were used for clustering using ‘FindClusters’. UMAP was used for two-dimensional representation of first 20 PCs with ‘RunUMAP’.
Differential gene expression for identifying markers of a cluster relative to all other clusters or compared to a specific cluster was determined using the ‘FindAllMarkers’ or ‘FindMarkers’ functions respectively. Parameters provided for these functions were: genes detected in at least 25% cells; differential expression threshold of 0.25 log fold change using Wilcoxon rank sum test with p < 0.05 following Bonferroni correction. We compared the marker genes for each cluster to literature-based markers of cell lineages to assign a cell lineage per cluster (Supplementary Table S2).
Individual Seurat data objects were merged iteratively using the ‘MergeSeurat’ function after filtering doublets identified by DoubletFinder, an R package that enables computational identification of doublets (Supplementary methods) (10). The merged object was processed as described above with library preparation batch and number of UMIs (Supplementary Table S1) included as parameters for regression in the ‘ScaleData’ function to regress batch effects and variation in sequencing depth respectively. The ‘DoHeatmap’, ‘FeaturePlot’, ‘DimPlot’, DotPlot’, ‘VlnPlot’ were used for visualization.
For a secondary cluster analysis of each cell lineage from this aggregated dataset, clusters of interest were identified and subset using ‘SubsetData’ with parameter ‘do.clean’ set to true. Detection of variable genes, scaling with UMI regression, PCA, clustering and UMAP were repeated as described above. Following this step, we removed clusters with co-expression of cell lineage markers as multiplets (Supplementary Table S3). Proportions of each cell type relative to the total number of cells in the sample were compared for tumor and normal sites using two proportions z-test and represented as box plots after re-clustering analysis for each lineage.
A Complete description of methods is available under Supplementary Methods.
RESULTS
Cohort of gastric cancer and intestinal metaplasia
We obtained tissues from surgical resections or endoscopic biopsies from seven patients with GC and one patient with gastrointestinal metaplasia (GIM). These tissue samples represented paired gastric tumor and normal tissue from the same patient derived from the same anatomical region. Four tumors were located at the gastroesophageal junction (GEJ) and four in the body and antrum. We analyzed matched PBMCs from two patients. Based on histopathology review, the GC tumors had intestinal, diffuse or mixed features (Supplementary Table S4, Figure 1B, Supplementary Figure 1A). We classified tumors into microsatellite stability (MSS) or microsatellite instability (MSI) molecular subtypes based on expression of the DNA mismatch repair proteins MLH1, MSH2, MSH6 and PMS2 according to immunohistochemistry (IHC) (Supplementary Table S4, Supplementary Methods). Histopathology showed that three patients (P5931, P6207, P6342) had active gastritis or intestinal metaplasia in paired non-malignant tissue.
Single-cell transcriptomic profiles from gastric cancer
With scRNA-seq, we obtained transcriptional profiles of 32,407 single-cells from tumors or metaplasia, 18,657 single-cells from paired normal tissue and 5,103 PBMCs (Supplementary Table S1). To determine gene expression changes, we employed a series of analytical steps for each individual dataset including quality filtering, principal component analysis (PCA) and graph-based clustering (9,11) (Supplementary Methods). We employed uniform manifold approximation and projection (UMAP) to reduce the dimensionality of this data and allow the visualization of cell-type clusters defined by their transcriptional profiles.
Differentially expressed (DE) genes were identified as genes expressed in greater than 25% of cells in a cluster and having a log fold change greater than 0.25, using a cut-off of p < 0.05 following Bonferroni correction. We compared the DE genes from each cluster to known marker genes of various cell types (Supplementary Table S2). This information enabled us to link clusters to specific cell types including epithelial cells (expressing PGC, TFF1, MUC5AC, EPCAM, GIF, CHGA), fibroblasts (THY1, DCN, COL4A1, FAP), endothelial cells (PECAM, ENG, VWF, SELE), immune cells (PTPRC) such as CD4 T (CD3D, CD4, IL7R), cytotoxic T (CD3D, CD8A, CD8B), regulatory T (FOXP3, IL2RA), NK (NKG7, GNLY), B (MS4A1), plasma cells (immunoglobulin genes), mast cells (TPSAB1) and macrophages (CD68, CD14, FCGR3A). Examination of cells expressing markers of disparate cell types facilitated the computational detection of doublets (Figure 1C, 1D, Supplementary Methods).
Data integration for joint cell analysis across all samples
To determine the similarities and differences among all of the samples and patients, we aggregated tumor, metaplastic, normal sites and PBMCs scRNA-Seq data into a single data matrix. To reduce experimental variance, we regressed out batch effects from library preparation and variation in sequencing depth. Also, we removed computationally detected doublets (Supplementary Methods).
Clustering analysis identified 40 distinct clusters that were specific for a variety of cell types including epithelial, stromal (fibroblasts, endothelial cells), lymphocytes, macrophages and mast cells – they were found among all of the patient samples (Supplementary Figures 1B and 1C, Figure 1E). On closer examination, each cell type had multiple distinct transcriptional states. For additional characterization, we aggregated data for each cell type across samples and conducted a clustering analysis (Supplementary Methods).
Classifying tumor, normal and metaplastic epithelial cell populations
We detected differences between tumor, metaplastic and normal epithelial cells, differences in tumor epithelium derived from different patients as well as sub-clonal heterogeneity within an individual tumor. Re-clustering analysis of epithelial cells from our integrated dataset revealed three subclasses. The first subclass consisted of normal gastric epithelial cells – over 80% were derived from normal gastric samples (Figure 2A, B, C). Normal epithelial cells were detected in all samples regardless of their origin from tumor, normal or metaplastic tissue (Supplementary Figure 2C, Supplementary Table S5).
Figure 2:
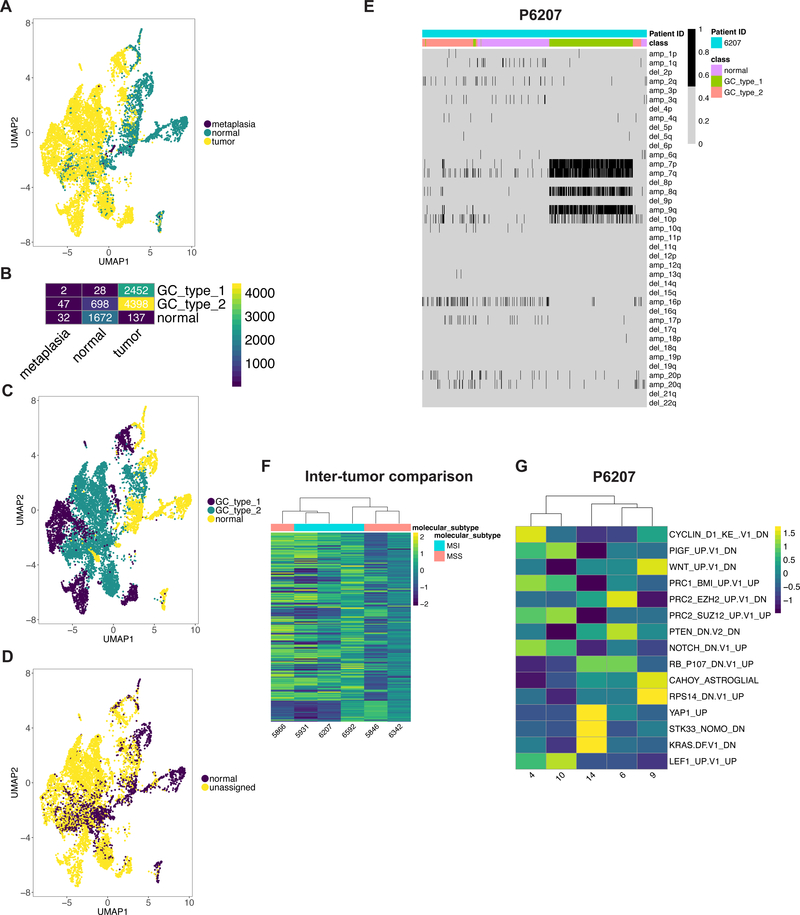
(A) UMAP representation of epithelial cells following graph-based clustering colored by sample origin. (B) Heatmap depicting number of cells per defined epithelial class according the sample origin. (C-D) UMAP representation of epithelial cells following graph-based clustering colored by (C) class (D) predicted class according to scPred. (E) Heatmap representation of statistically significant copy number changes for depicted chromosomes for epithelial cells from P6207 as a representative example. ‘amp’ denotes amplification, ‘del’ denotes deletion. (F-G) Heatmaps depicting average gene set activity of top MSigDB oncogenic c6 gene signatures following GSVA (ANOVA FDR p value < 0.05) across tumor epithelial clusters for (F) all patients and (G) all clusters for P6207.
The second subclass consisted of tumor-specific epithelial cells, which we label as the ‘GC type 1’. Approximately 98% of these cells originated from tumor samples. Interestingly, each cluster in this subclass was dominated by a single patient (Supplementary Figure 2C, Supplementary Table S5) – this indicated the extent of inter-tumor heterogeneity among all of the GCs, meaning that each individual tumor had distinct transcriptional properties.
The third subclass involved epithelial cells derived from GC as well as normal tissue, which we label as ‘GC type 2’. It also contained around 58% of cells from patient P6649 who had gastric metaplasia but did not have GC (Supplementary Figure 2C). We detected a range covering 11 to 68% of cells from normal tissues of patients with background metaplasia or gastritis on histology (P5931, P6207, P6342) (Supplementary Table 4, Supplementary figure 2C, Supplementary Table 5) in this subclass. Given their origin, these cells are likely to represent metaplastic or dysplastic epithelium, which had transcriptional features that overlapped with tumor epithelium.
As an independent determination of gastric tumor versus normal epithelial cells, we employed a different method called scPred that uses a supervised machine learning algorithm and thus provides highly accurate cell type assignment (12). We randomly subset tumor and normal site derived cells into two. One subset from each of these classes was included in the training dataset to build the scPred prediction model. We tested this model on all of the remaining cells for all samples. When we compared the scPred results to our Seurat analysis, the results were concordant. Eighty-nine percent and seventy-seven percent of cells in the ‘GC type 1’ and ‘GC type 2’ subclasses were not classified as normal gastric epithelial cells indicating that machine-learning confirms the distinction between normal, GC type 1 and GC type 2 cells (Figure 2D, Supplementary Figure 2D). Thus, scPred confirmed the transcriptional differences among the epithelial subclasses identified by our clustering analysis.
Gene expression differences among the epithelial cell subclasses
Differential expression analysis distinguished the three subclasses of epithelial cells. In normal gastric epithelium, we identified distinct mucosal populations such as pit, mucous neck, zymogen secreting chief, intrinsic factor producing parietal and neuroendocrine cells (Supplementary Figure 2B). In contrast, GC type 1 and GC type 2 subclasses downregulated some of these gastric mucosa marker genes such as MUC6, TFF2, TFF1, MUC5AC (Supplementary Table S6). The GC type 1 subclass had increased expression of intestinal mucosa markers TFF3, FABP1, SPINK4, MUC13 and REG4. The GC type 2 subclass had significantly increased expression of previously identified gastric cancer marker genes KRT7 and KRT17 (3), SOX4 and HES1 that have been implicated in metaplasia pathogenesis (13). Compared to normal cells, both GC type 1 and type 2 had upregulation of gene sets that included pathways for Myc, DNA repair and Notch signaling (Supplementary Figure 2E). However, only GC type 1 cells had higher upregulation of EMT and KRAS signaling.
Copy number alterations distinguish tumor and normal epithelium
Using scRNA-seq, one can detect amplifications or deletions at chromosome arm level by analyzing concomitant increase or decrease in gene expression respectively (14) (Supplementary Methods). To identify them, we analyzed each patient’s normal epithelial subclass cells against those from matched GC type 1 or GC type 2 subclass (Figure 2E, Supplementary Figure 3A). Copy number changes were inferred according to the posterior probability for each cell to belong to one of the two components with lower or higher gene expression indicative of deletion or amplification respectively.
A diverse spectrum of chromosome arm imbalances was present in these tumors. For patient P6207, we detected amplifications in 7p, 7q, 8q and 9q with a deletion in 10p. Patient P6342’s tumor had amplification of 20q with deletion of 4q. Patient P5846’s tumor had an amplification of 19q. Patient P5866’s tumor had deletion of 16q. Patient P5931’s tumor also had amplifications of 7p and 7q, which we have successfully identified using single-cell DNA sequencing (15). For Patient P6649 with metaplasia and P6592, samples contained only a small number of cells with significant copy number changes. We excluded the sample from patient P6709 from this analysis since we only detected 21 epithelial cells from the tumor site, possibly indicative of response to neoadjuvant chemotherapy (Supplementary Figures 2B, 1A, Supplementary Table 4). These CNA results provide additional orthogonal confirmation of the distinction between normal and tumor epithelial cells in our GC data set.
Cancer cell differences across samples and tumor clonal heterogeneity
We analyzed GC type 1 and GC type 2 cells across patients for the transcriptional activation of various oncogenic pathways. We observed significant differences in activation levels reaffirming the transcriptional heterogeneity between patients (Figure 2F). Interestingly, these activation profiles did not cluster according to differences between the molecular subtypes of MSI and MSS.
The individual patient tumors contained multiple clusters of GC type 1 and GC type 2 cells, an indication of intra-tumor sub-clonal heterogeneity. Pathway activation analysis grouped these clusters into three to five sub-populations for each patient’s tumor confirming a sub-clonal structure (Figure 2G, Supplementary Figure 4). For example, heterogeneity in P6207 was characterized by differences in cell cycle, KRAS pathway, Wnt activation (Figure 2G). We postulate that these sub-populations may provide a distinct growth advantage to the tumor.
TME reprogramming leads to macrophage states other than M1 and M2 classification
TME macrophages had distinct cellular and gene expression changes compared to normal gastric tissue consistent with the observation that macrophages acquire heterogenous phenotypes depending on their activating stimulus (16). Macrophage phenotypes are called M1 or M2 with anti and pro-tumorigenic functions respectively. However, the gene expression signatures we observed did not fall in line with either the canonical M1 or M2 classifications.
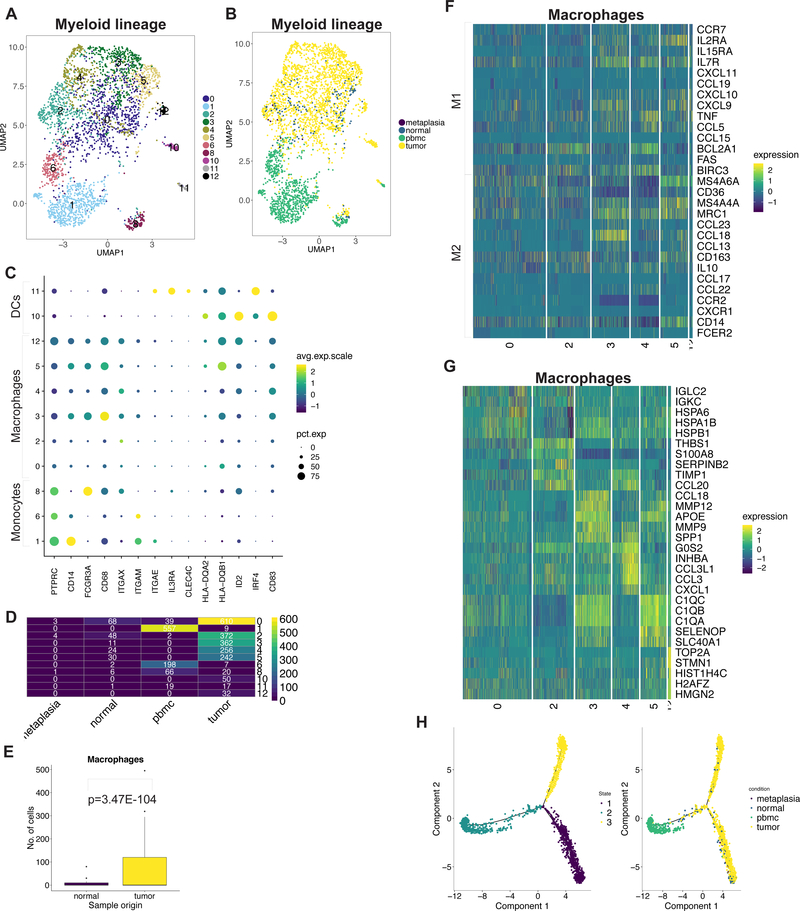
Specifically, we detected various subclasses of myeloid lineage cells as seen with 11 distinct clusters across all patients (Figure 3A-D, Supplementary Figure 5A, Supplementary Table 7). The nine monocyte-macrophage clusters were defined by marker gene expression of CD14, FCGR3A, CD68. The two dendritic cell (DC) clusters were defined by marker gene expression of CLEC4C, ID2, IRF4, CD83 (Figure 3C). Notably, macrophages were significantly enriched in tumor compared to normal gastric tissue (Figure 3E).
Figure 3:
(A) UMAP representation of macrophage cells following graph-based clustering with arbitrary cluster numbers. (B) UMAP representation colored according to the sample origin. (C) Dot plot depicting expression levels of specific genes across clusters with marker-based lineage assignments. (D) Heatmap depicting number of cells identified for each cluster according the sample origin. (E) Box plots depicting proportion of macrophages from total cells derived from tumor or normal site with p value derived from two proportions z-test. (F) Heatmap depicting expression of M1/M2 genes from each macrophage cluster. (G) Heatmap depicting top 5 highest significantly expressed genes detected from each macrophage cluster. (H) Trajectory plots of macrophages in normal and tumor tissue with monocytes from PBMCs with cells colored by identified trajectories (left) and sample origin (right).
We examined the expression of marker genes for M1 (e.g. CCL19, TNF, CCL5) and M2 (e.g. MRC1, CCL18, CCL13, CD163) states across the macrophage clusters (16) (Figure 3F). Expression of M1/M2 genes did not distinguish the clusters. Moreover, these genes were co-expressed in the same cluster. This suggests that the transcriptional heterogeneity was independent of the M1/M2 classification. We identified the significantly DE genes across all clusters to assess heterogeneous phenotypes (Supplementary Table 8). This revealed that heterogeneity was related to differences in the expression of HSP family genes, THBS1, chemokines including CCL20, CCL18, CCL3, matrix metalloproteinase genes, complement family and cell cycle regulation genes (Figure 3G). Clusters also showed differential enrichment of hallmark gene set activity confirming their distinct transcriptional programs (Supplementary Figure 5B).
PBMC monocytes clustered distinctly from tumor or normal macrophages indicating transcriptional differences (Figure 3B, D). As it stands, conventional single-cell clustering methods do not delineate the dynamic process of cell differentiation. A new type of method called trajectory analysis, uses single-cell gene expression to determine the transition among specific cell type lineages and states. Our trajectory analysis of tissue macrophages and PBMC monocytes yielded three different trajectory states (Figure 3H). Monocytes were present in a single trajectory state with few tissue macrophages. The majority of tissue macrophages differentiated along two distinct states. This suggests that tumor-infiltrating macrophages differentiate from monocytes but retain some fundamental similarities to macrophages within normal tissue.
DCs had two subclasses. One had genes that define plasmacytoid DCs including IL3RA (CD123) and CLEC4C (CD303) – this subclass was detected predominantly in PBMCs (Figure 3C, Figure 3D). A second was enriched in the TME and showed significant DE of activated DC gene markers CD83, CCR7, IL7R and ID2 (17) (Supplementary Table S9, Supplementary Figure 5C) (18). This subclass expressed the chemokines CCL22, CCL17, CCL19 and IL32 which are associated with recruitment of naïve T cells. Moreover, it highly expressed IDO1, characteristic of an immunosuppressive phenotype (19,20). This result represented a novel gene expression program in TME infiltrating DCs not previously described.
We compared activity levels of 1,558 experimentally derived immunologic gene signatures containing the term ‘macrophage’, ‘DC’ or ‘monocyte’ (Supplementary Methods, Supplementary Table S10) to these gene expression profiles. The identity of monocyte and DCs were confirmed using this approach. Also, these results provided gene set information indicating the activation phenotype of tumor-specific DCs. Each macrophage cluster was enriched for gene sets derived from a variety of experimental conditions. Hence, macrophage heterogeneity likely reflects stimulus-based context within the TME.
Regulatory genes controlling expression of a group of genes are referred to as regulons. We identified regulons for these different transcriptional cell states (6). This analysis identified transcriptional regulators such as IRF4 in DCs and also revealed a distinct set of regulatory genes defining the various macrophage populations including NFKB1, ETS2, CREM, REL, STAT1, FOXO3, etc. (Supplementary Table S11). Our data provides direct in vivo evidence that tumor-specific macrophages exist in a continuum of stimulus-dependent functional states regulated by a specific set of genes rather than the M1/M2 paradigm.
TME exhausted T cells have high CXCL13 expression and proliferation
Exhausted T cells (TEx) were a prominent feature of the gastric TME compared to normal tissue. The TME was also significantly enriched for Treg cells compared to normal. Initially, we identified CD4 helper T, CD8 T, NK, Treg, plasma and B cells using classic immunophenotyping markers. We filtered clusters with high expression of HSP family genes and lacking lineage markers (Supplementary Figure 6 A,B,C,D, Supplementary methods).
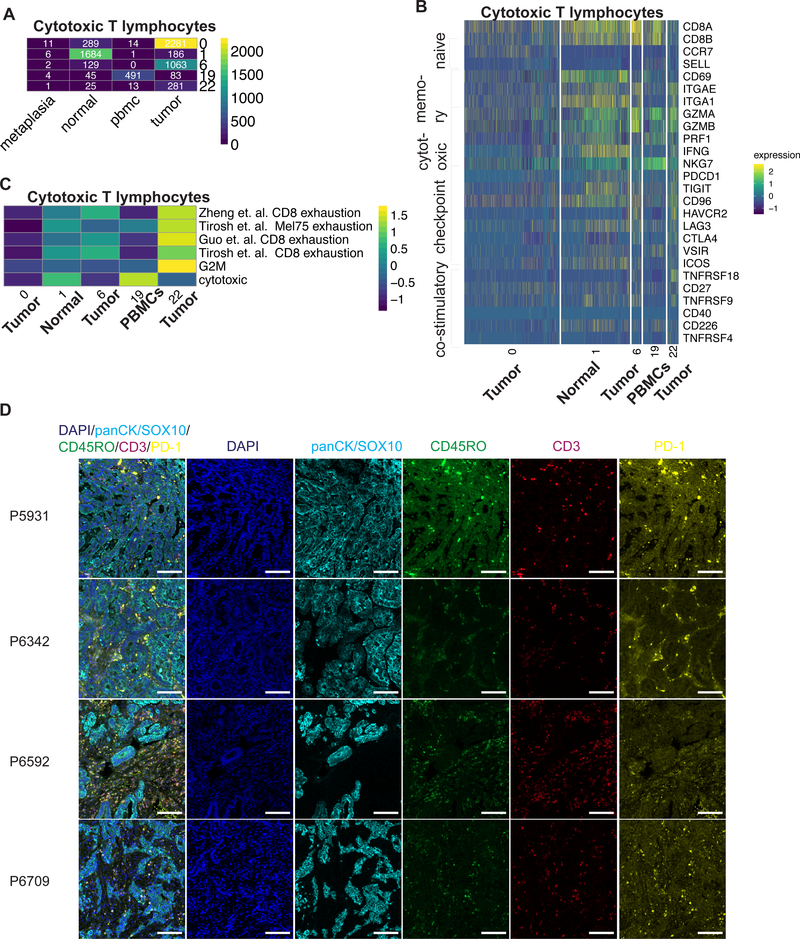
Cytotoxic CD8 T lymphocytes (CTLs) were distributed across five different clusters indicative of transcriptionally distinct cell states (Supplementary Figure 7A). They were observed among all patients with the majority detected in P5866, P5931 and P6207 (Supplementary Table 12). We examined each cluster for the dominant sample origin (normal, tumor or PBMC), expression of naïve markers (CCR7, SELL), tissue effector memory markers (CD69, ITGAE, ITGA1) and cytotoxic genes (GZMB, GZMA, PRF1, IFNG, NKG7) (Figure 4A, B). CTL subclasses included naïve tumor CTLs (cluster 0), effector normal CTLs (cluster 1), effector PBMC CTLs (cluster 19) and two subclasses of tumor effector CTLs (clusters 6, 22). Differential expression analysis identified distinct signatures among these subclasses (Supplementary Table S13).
Figure 4:
(A) Heatmap depicting number of cytotoxic T cells identified for each cluster according the sample origin. (B) Heatmap depicting expression of respective genes from each cytotoxic T cell cluster. (C) Heatmap representing average GSVA enrichment score for respective exhaustion signature for each cluster. (D) Representative images of fluorescence staining for respective markers and merged image for respective patients. Scale bar indicates 100 μm
Within cancer one observes TEx cells with reduced cytotoxic activity and expression of inhibitory receptors (21). In the TME, both CTL subclasses had low expression of PRF1 and IFNG while expressing granzymes (Figure 4B), indicative of a TEx phenotype (22). Furthermore, they expressed multiple immune checkpoints (PDCD1, TIGIT, CD96, HAVCR2 (TIM3), LAG3, CTLA4, VSIR); the expressions levels of these checkpoints is an indicator of the degree of immunosuppression. These cells also expressed multiple co-stimulatory molecules (ICOS, TNFRSF18 (GITR), CD27, TNFRSF9, CD40, CD226, TNFRSF4) (Figure 4B).
We compared the transcriptional profiles of tumor, normal and PBMC CTLs to previously published gene signatures to further understand the tumor TEx phenotype. First, we used gene signatures from T cell exposure to viruses. This experimental setup distinguishes the transcriptional program of naïve cells unexposed to virus, effector cells responding to the lymphocytic choriomeningitis virus (LMCV) and TEx cells responding to a specific clone 13 of LCMV (22). This result confirmed our assignment of naïve, effector and TEx classes across the five clusters. The two tumor-specific TEx clusters had greater enrichment for the exhaustion signatures generated with LCMV clone 13 (Supplementary Figure 7B). Also, we compared the CTL transcriptional profiles to three independently derived TEx profiles from single-cell analysis of human tumors, one from mouse tumors, together with signatures for cytotoxicity and proliferation (Figure 4C) (23–25). This result confirmed the low-cytotoxicity, high-exhaustion phenotype in both subclasses of tumor-TEx cells. Interestingly, these TEx subclasses were derived mainly from tumor samples from P5931, P6207, P6592 and P6709 that received neoadjuvant therapy (Supplementary Figure 7A).
The two subclasses of tumor-TEx cells differed in the extent of their exhaustion and proliferation gene expression program (Figure 4C). The subclass with higher proliferation (cluster 22) was associated with greater exhaustion indicative of an active immune response, which has previously been associated with terminal exhaustion (26). Immune checkpoint or costimulatory molecule gene expression was not significantly different between the two. The second subclass (cluster 6) had lower exhaustion and proliferation plus significantly higher expression of CXCL13 as previously identified in tumor but not in viral TEx (Supplementary Figure 7C) (27). These cells expressed high RBPJ, NR3C1 and BATF that are regulators of CD8 T cell fate (28). Our results thus demonstrated that effector CTLs in the TME are exhausted unlike normal tissue or PBMCs with two distinct subclasses characterized by high CXCL13 expression or proliferation.
To verify our findings, we conducted multiplex immunofluorescence staining for pan-cytokeratin/SOX-10 (epithelial cells), CD45RO (memory T cells), CD3 (T cells) and PD-1 (TEx) in tumors from four patients where adequate tissue samples were available (Figure 4D, Supplementary Figure 8A). We detected these cell lineages in all samples including cellular sub-populations of effector T cells (CD45RO, CD3 positive) and exhausted effector T cells (PD-1, CD45RO, CD3 positive) based on co-expression analysis (Supplementary Figure 8B). The stromal cells and macrophages were also apparent as they lacked expression of these markers.
Increased Tregs in the gastric TME contribute to immunosuppression
Tregs were significantly enriched in the gastric TME compared to normal gastric tissue, thus indicating an important mode of immunosuppression (Supplementary Figure 9A, B). Majority of cells were detected in P5866, P5931, P6207 and P6592 (Supplementary Table 12). The Treg cells had two distinct subclasses distinguished by higher expression of markers of proliferation (eg. MKI67, TYMS) in one sub-class (cluster 22). In addition, Tregs expressed several immune checkpoint and costimulatory molecules representing potential targets to modulate their function (Supplementary Figure 9C).
CD4 T cells were represented by four subclasses, mainly represented in P5866, P5931 and P6207 (Supplementary Table 12). Three of these were characterized by expression of naïve markers (CCR7, SELL) originating from the PBMCs or normal gastric tissue (Supplementary Figure 9D, E). Effector CD4 T cells (cluster 6) lacking naïve marker gene expression were found in both normal and tumor tissue (Supplementary Figure 9D, E). They had significantly higher expression of GZMA, GZMB, CXCL13, BATF, HLA genes (Supplementary Figure 9E). These cells are likely to represent follicular helper-like CD4 cells that express CXCL13 and are associated with tertiary lymphoid structures (29,30).
Three subclasses of NK cells were detected in PBMCs (cluster 2) and both tumor and normal tissue (clusters 13, 14) (Supplementary Figure 10A). They were derived prominently from P5866, P5931 and P6207 (Supplementary Table 12) and also contained a mix of rare populations of invariant NK and innate lymphoid cells (Supplementary Figure 10B) (31). Cells in tumor and normal sites clustered together indicating transcriptional similarity. Tumor-site cells expressed cytotoxic molecules such as GZMA, XCL2, CCL5, PRF1, CCL3, CCL4 indicating potential for an anti-tumor response (Supplementary Figure 10C). Cells also expressed several inhibitory and co-stimulatory molecules including TNFRSF18 (GITR), CD96 and KIR2DL4 expression representing targets for modulating their function.
B Cells from gastric TME and normal sites clustered together indicating transcriptional similarity (Supplementary Figure 10D). However, plasma cell clusters showed significant differences in the expression of genes encoding immunoglobulin isotypes with increased IgA encoding genes in normal tissue and IgG in TME (Supplementary Figure 10E, F).
Identification of jointly regulated genes of lymphocyte cell states
Next, we analyzed the regulatory genes or regulons that control these lymphocyte sub-populations (6) (Supplementary Table S14). The CXCL13-high tumor-TEx cells had significant enrichment of FOXO1 activity that is required for post-antigen expansion of CD8 T cells (32). In CXCL13-high CD4 T cells, the BATF gene network activity was prominently high validating their similarity to recently described CXCL13-producing helper T cells (33). We successfully identified FOXP3 and BATF gene network enrichment in Tregs confirming the accuracy of this approach. We discovered additional transcriptional regulators of TME Treg fate including KDM5B, MAF, IKZF2, SOX4, BCL3, etc. These regulons are of potential translational value given the interest in targeting epigenetics for immunotherapy (34).
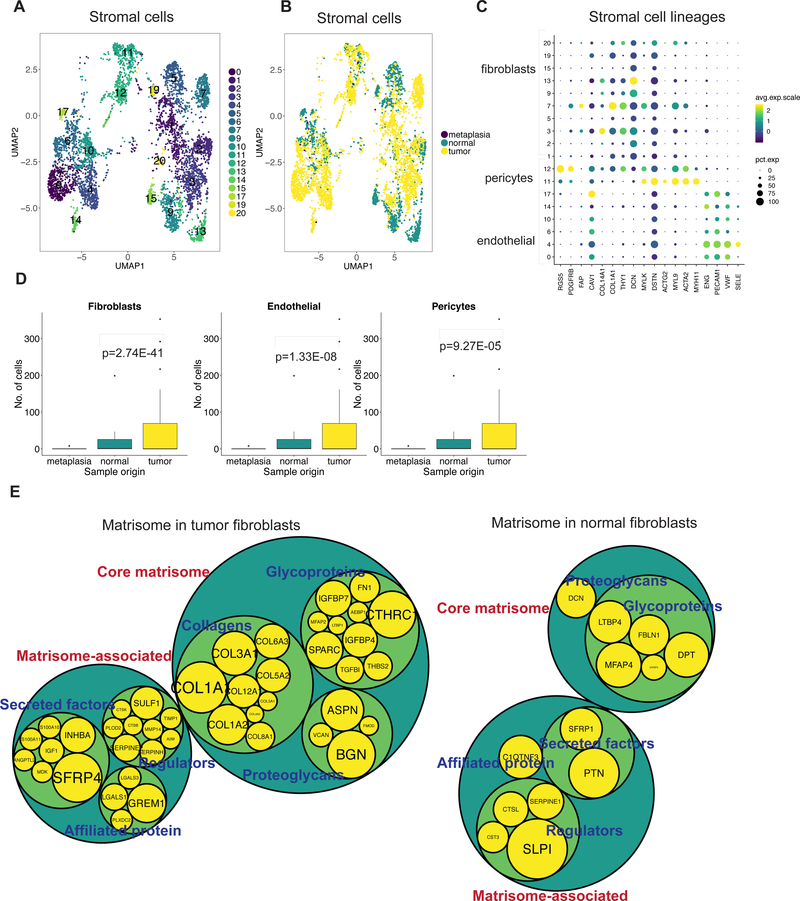
TME reprogramming of the fibroblasts, pericytes and endothelial stroma
We discovered transcriptional reprogramming of stromal cells in the tumor compared to normal that allows the generation of a tumor-specific extracellular matrix (ECM). Our analysis of the stromal cells across all samples identified fibroblasts (THY1, DCN, COL4A1, FAP), endothelial cells (PECAM, ENG, VWF, SELE), and pericytes (RSG5, PDGFRB) (Figure 5A, 5C). The observed heterogeneity among fibroblasts is likely to be driven by patient-specific factors (Supplementary Figure 11A, Supplementary Table 15). Endothelial clusters additionally differed in the expression of genes encoding secretory factors (ESM1, ANGPT2), tip cell markers (COL4A1, COL4A2, DLL4, MARCKSL1) and stalk cell markers (ACKR1, CD36, SELP, VWF) (35) and were mainly represented in P5866, P6342, P6592 and P6709 (Supplementary Figure 11B, 11C, Supplementary Table 15).
Figure 5:
(A) UMAP representation of stromal cells following graph-based clustering with arbitrary cluster numbers and (B) colored according to the sample origin. (C) Dot plot depicting expression levels of specific genes across clusters with marker-based lineage assignments. (D) Box plots depicting proportion of fibroblasts, pericytes or endothelial cells from total cells derived from tumor, normal or metaplastic site with p value derived from two proportions z-test. (E) Comparison of differentially expressed genes in tumor or normal fibroblasts to the genes of the matrisome program. Size of gene level circles is proportional to the logFC.
All three cell types were enriched in tumor tissue compared to normal (Figure 5B,5D). Stromal cells are responsible for the production and maintenance of ECM that provides mechanical support to cells and also influences their growth. Genes encoding for components or regulators of the ECM have previously been identified as the ‘matrisome’ (36,37). This gene group consists of core factors (collagens, proteoglycans and ECM glycoproteins) that make up the ECM and an associated program (ECM regulators, secretory factors and ECM-affiliated proteins). To determine the phenotypical differences of stromal cells in normal or tumor tissue, we compared their DE genes to the matrisome gene expression program. Tumor-specific fibroblasts, pericytes and endothelial cells expressed diverse ECM core and associated components (Figure 5E, Supplementary Table S16). Additionally, fibroblasts in tumors had significant overexpression of ACTA2 compared to normal tissue, indicative of their contractile ability (Supplementary Figure 11D).
Stromal cells at tumor or normal sites had significantly different regulatory genes or regulons (Supplementary Table S17). For example, tumor-specific endothelial cells had greater activity of SOX18 and SOX7 which are known regulators of a variety of endothelial cell processes (38). Tumor-specific fibroblasts had high activity of the EGR2 gene that can influence fibrosis (39). Tumor-specific pericytes were enriched for FOXF2 activity that is known to regulate pericyte differentiation (40). Hence, our approach distinguished differences in both the gene expression program and its regulators between tumor and normal stromal cells.
TME specific cellular communication has the potential to influence cell states
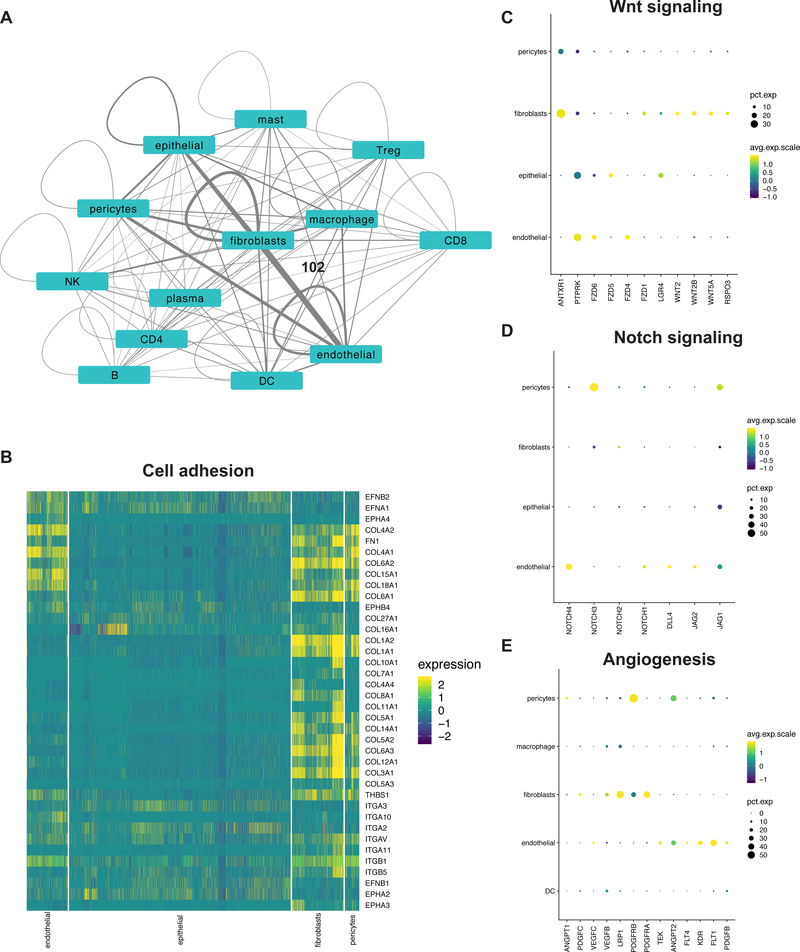
We discovered a TME-specific intercellular communications network that can potentially affect cellular behavior. First, we identified significant receptor-ligand interactions between different cell types using CellPhoneDB (8). Then, we compared these networks between tumor and normal tissue to generate a TME-specific interactome (Figure 6A, Supplementary Table 18).
Figure 6:
(A) Network depicting interactions between various cell types in tumors. Each node is a cell type and scaled edges represent the number of statistically significant detected interactions. Scale: fibroblast and endothelial edge = 102 interactions (B) Heatmap and (C-E) dot plots depicting the expression of respective genes in specific cell types.
Stromal cells were among the most prolific interactors. Prominent communication between them and epithelial cells occurred through various integrin receptor interactions with collagen, fibronectin and THBS1 ligands (Figure 6B). Bidirectional interactions between ephrin receptor family and ligands were detected in epithelial and endothelial cells. Among growth factor signaling that can promote cancer cell proliferation, we detected EGFR and MET receptors on epithelial cells together with respective ligands on stromal cells. We also detected significant EGFR interactions in fibroblasts.
Fibroblasts were a prominent source of Wnt ligands with expression of corresponding receptors on tumor epithelium, endothelial cells, fibroblasts and pericytes. This included a LGR4 – RSPO3 interaction that has the potential to regulate stemness (Figure 6C), validating our previous discovery of fibroblast-derived RSPO3 in an organoid model of gastric cancer (41).
Autocrine and paracrine Notch signaling, a known regulator of angiogenesis (42), was evident in endothelial cells. Interactions promoting Notch signaling were also significant in fibroblasts (Figure 6D). Angiogenic receptors KDR, FLT1, FLT4, PDGFB, TEK on endothelial cells and pericytes had significant autocrine and paracrine interactions with their respective ligands (Figure 6E). Among the interactome were 19 cytokines including chemokines, interleukins, tumor necrosis factors (TNFs) and their corresponding receptors that can influence immune cell fates.
DISCUSSION
For this study, we leveraged paired distal normal tissue and PBMCs to analyze the cellular dysregulation and biological changes in the GC TME. With single-cell gene expression analysis, we demonstrated that GC TME leads to a series of dramatic cellular changes compared to matched normal stomach mucosa. Specifically, we noted increases in cell numbers of stromal cells and Tregs in the TME. We also identified transcriptional cell states unique to the TME including in DCs and two subclasses of exhausted CTLs. Among our novel findings, we discovered that gene expression profiles for GC TME macrophages are heterogenous and not confined to a binary M1/M2 designation. We demonstrated that TME stromal cells encode for a specific ECM composition not found in normal tissue. We identified novel gene regulatory networks and TME-specific intercellular communication.
We validated previously described changes in normal, metaplastic and tumor epithelial cells (13,43) and were additionally able to elucidate intra-tumor heterogeneity by examining the activity of various cancer-promoting mechanisms within the tumor cells. The diversity in activation profiles suggests that targeting multiple clones with different combination strategies may be necessary to eradicate them completely.
Immune cell lineages were detected across all patients. However, a limitation of our analysis is that the majority of cells were derived from two patients (P5866 and P5931) with two sequencing replicates (Figure 1F, Supplementary table 1). Immunosuppression in GC TME was evident by the increased proportion of Tregs compared to normal tissue. We identified several checkpoint and costimulatory molecules on these cells, and their transcriptional regulators. These regulators can be investigated to understand Treg biology and to derive therapeutic targets. Indeed, recent evidence indicates that anti-CTLA4 activity is a consequence of Treg depletion in the TME (44). We detected expression of multiple immune checkpoints on cytotoxic T cells similar to other studies (23,45). These checkpoints were also detected on helper T and Treg subsets. Thus, it is important to understand the effects of immune checkpoint blockade on distinct T cell subpopulations that express the same target. Our analysis revealed transcriptional regulators responsible for these states. Plasma cells in tumor tissue expressed IgGs rather than IgAs that were detected in paired normal tissue that have been associated with a pro-cancer role by influencing myeloid cell Fc-receptors (46).
Increased immune cell signaling has previously been associated with EBV molecular subtype of GC (3). Meanwhile, MSI subtype cancers have been demonstrated to have higher responses to immune checkpoint blockade owing to the favorable immune milieu generated by higher neoantigen burden (47). In our study, immune cell subtypes did not cluster clearly according to the MSI/MSS status of samples. This resembles findings from recent studies in GC where molecular subtypes could be differentiated by gene signatures based on a combination of immune suppression, immune activation and stromal activation processes rather than the leucocyte fractions within the TME (48,49). This raises the possibility that a combination of transcriptional states in stromal and immune cells, genomic alterations in tumor cells, and their resultant interactions determines the final composition and transcriptional cell states of the TME in each molecular subtype. Utilizing scRNA-seq on a larger cohort of GC has the potential to address the mechanisms by which tumor genomics influences the TME composition and possibly responses to immunotherapy.
It has been demonstrated that neoadjuvant chemotherapy leads to increased expression of CD4, CD8, PD1, PD-L1 and TIM-3 proteins in GC TME (50). We observed presence of TEx cells in all patients that received neoadjuvant chemotherapy. Increasing the cohort size will also help to address the influence of neoadjuvant treatment on the TME transcriptional cell states that we have observed in this study.
Interactome analysis demonstrated pro-tumor effects of TME components and also the influence of cancer cells on the TME. Tumor-specific interactome represents potential treatment targets to inhibit cancer proliferation, overcome the immunosuppressive microenvironment and restore the cancer immunity cycle. Additionally, while some targets such as Wnt inhibition have previously been regarded only in the context of tumor epithelial cells, our analysis demonstrates that this might have implications for the TME.
Our study did not consider spatial context and might be affected by the dissociation process. The use of dual single-cell proteomics and transcriptomics is likely to provide a more refined analysis of immune cell sub-types (51).
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
We leveraged the power of single-cell genomics to characterize the heterogenous cell types and states in the tumor microenvironment (TME). By profiling thousands of single cells from surgical resections of gastric cancer together with paired normal mucosa and peripheral blood mononuclear cells (PBMCs), we determined the deviations in the TME from physiological conditions. Our analysis revealed a cellular reprogramming of the TME compared to normal mucosa in immune and stromal lineages. We detected transcriptional heterogeneity within macrophages and a TME-specific gene expression program in dendritic cells. Cytotoxic T cells in the TME had heterogenous profiles of exhaustion and expression of multiple immune checkpoint and costimulatory molecules. We constructed a receptor-ligand based inter-cellular communications network that was exclusive to tumor tissue. These discoveries provide information at a highly granular cellular resolution enabling advances in cancer biology, biomarker discovery and identification of treatment targets such as for immunotherapy.
ACKNOWLEDGEMENTS
We are grateful to all patients who participated in the study. We thank Christine Handy, Alison Almeda, Christina Wood-Bouwens for assistance in sample collection and documentation and Ann Renschler for assistance in sequencing. We thank Dr. Katir Patel from Ultivue, Inc. for assistance in multiplex immunofluorescence staining. This work was supported by US National Institutes of Health grants P01HG000205 (HPJ, CWB and SMG), R01HG006137 (HPJ), U01CA217875 (HPJ and AS). HPJ also received support from the American Cancer Society (124571-RSG-13–297-01), the Clayville Foundation and the Gastric Cancer Foundation.
Footnotes
Conflict of interest statement: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394–424 doi 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Correa P, Piazuelo MB. The gastric precancerous cascade. J Dig Dis 2012;13(1):2–9 doi 10.1111/j.1751-2980.2011.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513(7517):202–9 doi 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21(5):449–56 doi 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 5.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2018;175(4):998–1013 e20 doi 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14(11):1083–6 doi 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017;14(10):979–82 doi 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018;563(7731):347–53 doi 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36(5):411–20 doi 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst 2019;8(4):329–37 e4 doi 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McInnes L, Healy J. UMAP: uniform manifold approximation and projection for dimension reduction. ArXiv 2018. [Google Scholar]
- 12.Alquicira-Hernandez J, Sathe A, Ji HP, Nguyen Q, Powell JE. scPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol 2019;20(1):264 doi 10.1186/s13059-019-1862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P, Yang M, Zhang Y, Xiao S, Lai X, Tan A, et al. Dissecting the Single-Cell Transcriptome Network Underlying Gastric Premalignant Lesions and Early Gastric Cancer. Cell Rep 2019;27(6):1934–47 e5 doi 10.1016/j.celrep.2019.04.052. [DOI] [PubMed] [Google Scholar]
- 14.Muller S, Cho A, Liu SJ, Lim DA, Diaz A. CONICS integrates scRNA-seq with DNA sequencing to map gene expression to tumor sub-clones. Bioinformatics 2018;34(18):3217–9 doi 10.1093/bioinformatics/bty316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andor N, Lau BT, Catalanotti C, Kumar V, Sathe A, Belhocine K, et al. Joint single cell DNA-Seq and RNA-Seq of gastric cancer reveals subclonal signatures of genomic instability and gene expression. bioRxiv 2018. [Google Scholar]
- 16.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 2006;177(10):7303–11 doi 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 17.Collin M, McGovern N, Haniffa M. Human dendritic cell subsets. Immunology 2013;140(1):22–30 doi 10.1111/imm.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017;356(6335) doi 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietila TE, Veckman V, Lehtonen A, Lin R, Hiscott J, Julkunen I. Multiple NF-kappaB and IFN regulatory factor family transcription factors regulate CCL19 gene expression in human monocyte-derived dendritic cells. J Immunol 2007;178(1):253–61 doi 10.4049/jimmunol.178.1.253. [DOI] [PubMed] [Google Scholar]
- 20.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol 2004;4(10):762–74 doi 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 21.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15(8):486–99 doi 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 2007;27(4):670–84 doi 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352(6282):189–96 doi 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 2018;24(7):978–85 doi 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 25.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017;169(7):1342–56 e16 doi 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 26.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20(3):326–36 doi 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thommen DS, Schumacher TN. T Cell Dysfunction in Cancer. Cancer Cell 2018;33(4):547–62 doi 10.1016/j.ccell.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu B, Zhang K, Milner JJ, Toma C, Chen R, Scott-Browne JP, et al. Epigenetic landscapes reveal transcription factors that regulate CD8(+) T cell differentiation. Nat Immunol 2017;18(5):573–82 doi 10.1038/ni.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu-Trantien C, Loi S, Garaud S, Equeter C, Libin M, de Wind A, et al. CD4(+) follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest 2013;123(7):2873–92 doi 10.1172/JCI67428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi S, Watanabe T, Suzuki R, Furu M, Ito H, Ito J, et al. TGF-beta induces the differentiation of human CXCL13-producing CD4(+) T cells. Eur J Immunol 2016;46(2):360–71 doi 10.1002/eji.201546043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, et al. Molecular definition of the identity and activation of natural killer cells. Nat Immunol 2012;13(10):1000–9 doi 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedrick SM, Hess Michelini R, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nat Rev Immunol 2012;12(9):649–61 doi 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshitomi H, Kobayashi S, Miyagawa-Hayashino A, Okahata A, Doi K, Nishitani K, et al. Human Sox4 facilitates the development of CXCL13-producing helper T cells in inflammatory environments. Nat Commun 2018;9(1):3762 doi 10.1038/s41467-018-06187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn J, Rao S. Epigenetics and immunotherapy: The current state of play. Mol Immunol 2017;87:227–39 doi 10.1016/j.molimm.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Q, Eichten A, Parveen A, Adler C, Huang Y, Wang W, et al. Single-Cell Transcriptome Analyses Reveal Endothelial Cell Heterogeneity in Tumors and Changes following Antiangiogenic Treatment. Cancer Res 2018;78(9):2370–82 doi 10.1158/0008-5472.CAN-17-2728. [DOI] [PubMed] [Google Scholar]
- 36.Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics 2012;11(4):M111 014647 doi 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hynes RO, Naba A. Overview of the matrisome--an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol 2012;4(1):a004903 doi 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell 2009;16(2):180–95 doi 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharyya S, Wu M, Fang F, Tourtellotte W, Feghali-Bostwick C, Varga J. Early growth response transcription factors: key mediators of fibrosis and novel targets for anti-fibrotic therapy. Matrix Biol 2011;30(4):235–42 doi 10.1016/j.matbio.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyahi A, Nik AM, Ghiami M, Gritli-Linde A, Ponten F, Johansson BR, et al. Foxf2 Is Required for Brain Pericyte Differentiation and Development and Maintenance of the Blood-Brain Barrier. Dev Cell 2015;34(1):19–32 doi 10.1016/j.devcel.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Lau BT, Andor N, Grimes SM, Handy C, Wood-Bouwens C, et al. Single-cell transcriptome analysis identifies distinct cell types and niche signaling in a primary gastric organoid model. Sci Rep 2019;9(1):4536 doi 10.1038/s41598-019-40809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siekmann AF, Lawson ND. Notch signalling and the regulation of angiogenesis. Cell Adh Migr 2007;1(2):104–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Companioni O, Sanz-Anquela JM, Pardo ML, Puigdecanet E, Nonell L, Garcia N, et al. Gene expression study and pathway analysis of histological subtypes of intestinal metaplasia that progress to gastric cancer. PLoS One 2017;12(4):e0176043 doi 10.1371/journal.pone.0176043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang F, Du X, Liu M, Zheng P, Liu Y. Anti-CTLA-4 antibodies in cancer immunotherapy: selective depletion of intratumoral regulatory T cells or checkpoint blockade? Cell Biosci 2018;8:30 doi 10.1186/s13578-018-0229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andor N, Simonds EF, Czerwinski DK, Chen J, Grimes SM, Wood-Bouwens C, et al. Single-cell RNA-Seq of lymphoma cancers reveals malignant B cell types and co-expression of T cell immune checkpoints. Blood 2018. doi 10.1182/blood-2018-08-862292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garaud S, Zayakin P, Buisseret L, Rulle U, Silina K, de Wind A, et al. Antigen Specificity and Clinical Significance of IgG and IgA Autoantibodies Produced in situ by Tumor-Infiltrating B Cells in Breast Cancer. Front Immunol 2018;9:2660 doi 10.3389/fimmu.2018.02660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357(6349):409–13 doi 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The Immune Landscape of Cancer. Immunity 2018;48(4):812–30 e14 doi 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng D, Li M, Zhou R, Zhang J, Sun H, Shi M, et al. Tumor Microenvironment Characterization in Gastric Cancer Identifies Prognostic and Immunotherapeutically Relevant Gene Signatures. Cancer Immunol Res 2019;7(5):737–50 doi 10.1158/2326-6066.CIR-18-0436. [DOI] [PubMed] [Google Scholar]
- 50.Yu Y, Ma X, Zhang Y, Zhang Y, Ying J, Zhang W, et al. Changes in Expression of Multiple Checkpoint Molecules and Infiltration of Tumor Immune Cells after Neoadjuvant Chemotherapy in Gastric Cancer. J Cancer 2019;10(12):2754–63 doi 10.7150/jca.31755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14(9):865–8 doi 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.