Abstract
Objective:
To determine the mechanism by which tranilast induces miR-200c expression in leiomyoma smooth muscle cells (LSMC).
DESIGN:
Experimental study
SETTING:
Academic research laboratory
PATIENT(S):
Women undergoing hysterectomy for leiomyoma.
Intervention:
Blockade of RelA/p65.
MAIN OUTCOME MEASURE(S):
The effects of tranilast and blockade of RelA/p65 on miR-200c expression.
Results:
Tranilast, an inflammation inhibitor dose-dependently induced miR-200c in LSMC and myometrium smooth muscle cells (MSMC) with a more profound effect in LSMC as compared with MSMC. The treatment of LSMC with Bay 117082, an inhibitor of IκB phosphorylation, further enhanced miR-200c induction by tranilast. The knockdown of RelA/p65 by siRNA also induced miR-200c expression in LSMC. Although tranilast had no effect on total RelA/p65 protein levels in LSMC, it significantly induced RelA/p65 phosphorylation at S536, while reducing its activity as well as its nuclear translocation. ChIP assay indicated that tranilast reduces the binding ability of RelA/p65 to miR-200c promoter resulting in miR-200c induction. Tranilast also inhibited interleukin 8 (IL8) expression in LSMC. The induction of miR-200c by tranilast in part mediates the inhibitory effect of tranilast on the expression of IL8 and cyclin-dependent Kinase 2 (CDK2) in LSMC.
Conclusion:
Induction of miR-200c by tranilast in LSMC is mediated through a transcriptional mechanism involving inhibition of nuclear factor kappa-B (NF-kB) signaling pathway. These results highlight the significance of inflammation in the pathogenesis of leiomyoma and the potential utility of anti-inflammatory drugs for treatment of leiomyomas.
Keywords: Tranilast, Leiomyoma, miR-200, NF-kB, CDK2
Capsule
Tranilast induces miR-200c expression through blockade of the activity and nuclear translocation of RelA/p65 in leiomyoma smooth muscle cells.
Introduction
Uterine leiomyomas are the most common benign fibrotic tumors affecting 40–70% of women during their reproductive years (1, 2). In addition to surgical intervention which accounts for the most common indication for all hysterectomies performed in the United States annually (1, 2), several hormonal therapies have been used as alternative interventions for management of their growth and associated symptoms such as pelvic pressure, chronic pelvic pain, abnormal uterine bleeding as well as infertility and pregnancy complications (1, 3). Although their etiology is unknown, these tumors whose growth is dependent on ovarian steroids are characterized by increased angiogenesis, excess deposition of the extracellular matrix (ECM), inflammation as well as elevated expression of pro-fibrotic cytokines such as transforming growth factor-β3 (TGF-β3), Activin-A and platelet-derived growth factor (PDGF) (4–6). In addition, excess accumulation of mast cells along with increased secretory products from these cells has been considered as being pivotal in the pathogenesis of several fibrotic disorders, including leiomyomas (7).
Accumulated proteomic and genomic studies have indicated that altered expression of small non-coding RNAs, including microRNAs (miRNAs), play a key role in cellular events leading to leiomyoma development and growth (5, 8–12). More specifically, our laboratory and others have provided evidence that the expression of miR-200 and miR-29 family are suppressed in leiomyoma and their altered expression at least in part accounts for molecular mechanism underlying ECM accumulation, angiogenesis and inflammation (13–17).
Tranilast (N-3, 4-dimethoxycinnamoyl anthranilic acid) is an orally administered synthetic drug with anti-allergic properties has been used clinically for treatment of several respiratory disorders as well as keloids and hypertrophic scars (18, 19). Recent reports indicate that tranilast not only inhibits the release of inflammatory mediators from mast cells, but also inhibits collagen bio-synthesis, growth factor expression, cell growth and TGFβ–induced transformation of fibroblasts into myofibroblastic phenotype (19). Subsequent studies have confirmed the anti-proliferative or anti-tumor effect of tranilast in several cells including prostate, breast, glioma, gastric, pancreatic and other tumors (18, 19). We and other groups have also demonstrated that tranilast inhibited the rate of cell proliferation and the expression of several genes functionally involved in cell cycle progression [CDK2, cyclin-dependent kinase inhibitor p21(Waf1/Cip1), p53, proliferating cell nuclear antigen (PCNA) and cyclin D1 (CCND1)], collagen type I (COL1) and collagen type III alpha 1 chain (COL3A1), profibrotic cytokines [transforming growth factor-β3 (TGF-β3) and Activin-A], and enzymes catalyzing epigenetic modifications [DNA Methyltransferase 1 (DNMT1) and enhancer of zeste homolog 2 (EZH2)] in isolated LSMC (16, 20, 21). Furthermore, we reported that tranilast induced miR-29c levels through an epigenetic mechanism in LSMC (16).
Although NF-κB is comprised of multiple subunits, RelA/p65 has received the most attention especially in studies of its phosphorylation (22). Phosphorylation of RelA/p65 causes a conformational change resulting in alteration of its protein-protein interactions, ubiquitination as well as stability (23). Among the 11 identified phosphorylation sites in RelA/p65, four sites (S205, T254, S276 and S281) were found within the N-terminal REL homology domain (RHD), two (S311 and S316) in the linker region and five (T435, S468, T505, S529 and S536) located in the C-terminal transactivation domain (TAD). Significant research has focused on S536 (22), and a number of kinases have been identified which could phosphorylate RelA/p65 at S536 including IKKα, IKKβ, IKKε, ribosomal subunit S6 kinase 1 (RSK1), casein kinase 1γ1 (CK1γ1) and NF-κB activating kinase (NAK)/TANK-binding kinase 1 (TBK1) (24–27). Because RelA/p65 phosphorylation at S536 has been reported to enhance its transcriptional activity and nuclear translocation (24, 28–31), the aim of this study was to determine the effects of tranilast on RelA/p65 phosphorylation at S536.
Leiomyoma express significantly lower levels of miR-200c whose target genes include IL8, CDK2, zinc finger E-Box binding homeobox 1 (ZEB1), zinc finger E-Box binding homeobox 2 (ZEB2), vascular endothelial growth factor A (VEGFA), tissue inhibitor of metalloproteinases 2 (TIMP2), fibulin 5 (FBLN5) and inhibitor of nuclear factor kappa B kinase subunit beta (IKBKB) (17, 32, 33) all of which influence the outcome of a number of cellular events in leiomyomas. The objective of the present study was therefore to determine if tranilast which has anti-inflammatory properties alters the expression of miR-200c and secondly, to determine if the NF-kB pathway could mediate the effects of tranilast on miR-200c.
Materials and Methods
Primary Myometrium and Leiomyoma Smooth Muscle Cells Isolation
Primary MSMC and LSMC were isolated from fresh specimens as previously described (33). Fresh tissues used for cells isolation were collected from patients not taking any hormonal medications for at least three months prior to surgery at Harbor-UCLA Medical Center with prior approval from Institutional Review Board (#036247). Briefly, MSMC and LSMC were cultured in DMEM supplemented with 10% fetal bovine serum until reaching confluence with a change of media every 2–3 days. Cells at passages p1 to p3 were used for all experiments. Cell culture experiments were performed at least three times using MSMC or LSMC isolated from different patients as indicated in the figure legends. Overall, 5 primary MSMC and 22 primary LSMC were used in this study. All supplies for isolation and cell culture were purchased from Sigma-Aldrich (St. Louis, MO) and Fisher Scientific (Atlanta, GA).
siRNA Transfection
LSMC cultured as above and at sub-confluence were transfected with 50 nM of siRNA negative control (siNC) or siRNA against RelA/p65 (sip65; Santa Cruz Biotechnology, Dallas, Texas) for 72–96 hours using PureFection transfection reagent (System Biosciences, Inc., Mountain View, CA) according to the manufacturer’s protocol.
Loss-of-function of miR-200c
LSMC were seeded at a cell density of 5 × 104/well in six-well plates and at sub-confluence transfected with 50 nM of anti-miR negative control (aNC) or anti-miR-200c (a-miR-200c) (Applied Biosystems, Carlsbad, CA) for 96 hours using PureFection transfection reagent (System Biosciences, Inc.) according to the manufacturer’s protocol.
RNA Isolation and qRT-PCR Analysis
Total RNA was extracted from LSMC using Trizol (Thermo Fisher Scientific, Waltham, MA) and their quantity and quality was determined (ND-1000 Spectrophotometer, NanoDrop Technologies, Wilmington, DE) as previously described (11, 32, 34). Subsequently, 1 μg RNA was reverse-transcribed using random primers for IL8 (Fisher Scientific). The miR-200c primer design and PCR conditions has been described previously (35). Quantitative RT-PCR was carried out using SYBR gene expression master mixes (Applied Biosystems). Reactions were incubated for 10 min at 95°C followed by 40 cycles for 15 seconds at 95°C and 1 min at 60°C. The levels of mRNA and miRNA were quantified using the Invitrogen StepOne System and normalized to FBXW2 (36) and RNU6B, respectively. All reactions were run in triplicate and relative expression was determined using the comparative cycle threshold method (2−ΔΔCT), as recommended by the supplier (Applied Biosystems). Abundance values were expressed as fold changes compared to the corresponding control group. The primer sequences used were as follows: IL8 (sense, 5’-CTTGGCAGCCTTCCTGATTT-3’; antisense, 5’-TTCTTTAGCACTCCTTGGCAAAA-3’); and FBXW2 (sense, 5’-CCTCGTCTCTAAACAGTGGAATAA-3’; antisense, 5’-GCGTCCTGAACAGAATCATCTA-3’). miR-200c (sense, 5’-AGTAATACTGCCGGGTAATGA-3’; antisense, 5’-GGTCCAGTTTTTTTTTTTTTTTCCA-3’); and RNU6B (sense, 5’-ATTGGAACGATACAGAGAAGATTAG-3’; antisense, 5’-AATATGGAACGCTTCACGAAT-3’).
Immunoblotting
Total protein isolated from LSMC following treatment conditions was subjected to immunoblotting as previously described (12, 37). Briefly, samples were suspended in RIPA buffer containing 1 mM EDTA and EGTA (Boston BioProducts, Ashland, MA) supplemented with 1 mM PMSF and a complete protease inhibitor mixture (Roche Diagnostics, Indianapolis, IN), sonicated, and centrifuged at 4°C for 10 min at 14,000 rpm. The concentration of protein was determined using the BCA™ Protein Assay Kit (Thermo Scientific Pierce, Rockford, IL). Equal aliquots (Fifty micrograms) of total protein for each sample were denatured with SDS-PAGE sample buffer, and separated by electrophoresis on an SDS polyacrylamide gel. After transferring the samples to a nitrocellulose membrane, the membrane was blocked with TBS-Tween + 5% milk, and probed with the following primary antibodies: RelA/p65 (Cell Signaling Technology, Inc., Danvers, MA), p-RelA/p65 (Ser 536), Calpain, c-Jun, IL8 and CDK2 (Santa Cruz Biotechnology). The membranes were washed with TBS containing 0.1%Tween-20 wash buffer after each antibody incubation cycle. SuperSignal West Pico Chemiluminescent Substrate™ (Thermo Scientific Pierce) was used for detection, and photographic emulsion was used to identify the protein bands, which were subsequently quantified by densitometry. The membranes were also stripped and probed with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (Santa Cruz Biotechnology) serving as the loading control. The densities of the specific protein bands were quantified with a scanning densitometer (Bio-Rad GS-800, Hercules, CA), and the results were expressed as means ± SEM normalized to GAPDH.
Luciferase Reporter Assays
LSMC were seeded in six-well plates until reaching sub-confluence and transiently cotransfected with a luciferase reporter plasmid (1 μg/well) containing nuclear factor kappa-B (NF-kB) conserved binding sequences for NF-kB activity detection (Signosis, Sunnyvale, CA) and pRL-TK plasmid (Promega, Madison, WI) encoding Renilla luciferase (0.2 μg/well) as a control for differences in transfection efficiency using PureFection transfection reagent. Firefly and Renilla luciferase activities were measured after 8 hrs of tranilast (200 μM) treatment using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity and the level of induction was reported as the mean ± SEM of three experiments performed in duplicates and compared with a ratio in cells treated with vehicle control (DMSO) set at 1.
Subcellular Fractionation
LSMC were treated with tranilast (200 μM) for 36 hrs. Cytoplasmic and nuclear proteins were fractionated following the protocol of the subcellular protein fractionation kit (Thermal Scientific).
Chromatin Immunoprecipitation
Following treatment of LSMC and MSMC with tranilast (200 μM) for 36h, ChIP analysis was carried out using Simple ChIP enzymatic chromatin IP kit (Cell Signaling Technology) according to the manufacturer’s protocol. PCR-amplification was performed using primers designed to amplify RelA/p65 binding site on miR-200c promoter (forward: 5’-GGGATGAGGGTGGGTAAATC-3’, reverse: 5’-GCCTCTGAGCCACCTTC-3’), under PCR condition of 95°C for 5 min, 35 cycles of 94°C for 30s, 55°C for 30s, and 72°C for 30s. PCR a mplified products were electrophoresed on 2% agarose gel and visualized under ultraviolet illumination. Band densities were quantified through normalization to the corresponding inputs and percent input method was used to quantitate the values of the immunoprecipitated DNA.
Enzyme-linked immunosorbent assay
Collected culture conditioned media and cells lysates were centrifuged, supernatants were collected and total protein content was determined by BCA™ Protein Assay Kit (Thermo Scientific Pierce). The IL8 content in culture media was determined following the protocol of Human IL8 ELISA MAX Deluxe Set (BioLegend, San Diego, CA) with detection limit of 15.6 pg/ml and sensitivity of 8 pg/ml. The level of IL8 was reported as pg/mg of protein or as fold change compared to control experiments.
Statistical analysis
Throughout the text, all data are presented as mean ± SEM and analyzed by PRISM software (Graph-Pad, San Diego, CA). Dataset normality was determined by the Kolmogorove–Smirnoff test. Comparisons involving two groups were analyzed using unpaired Student’s t-tests as appropriate. One-way ANOVA was used for comparisons involving multiple groups. Statistical significance was established at P<0.05.
Results
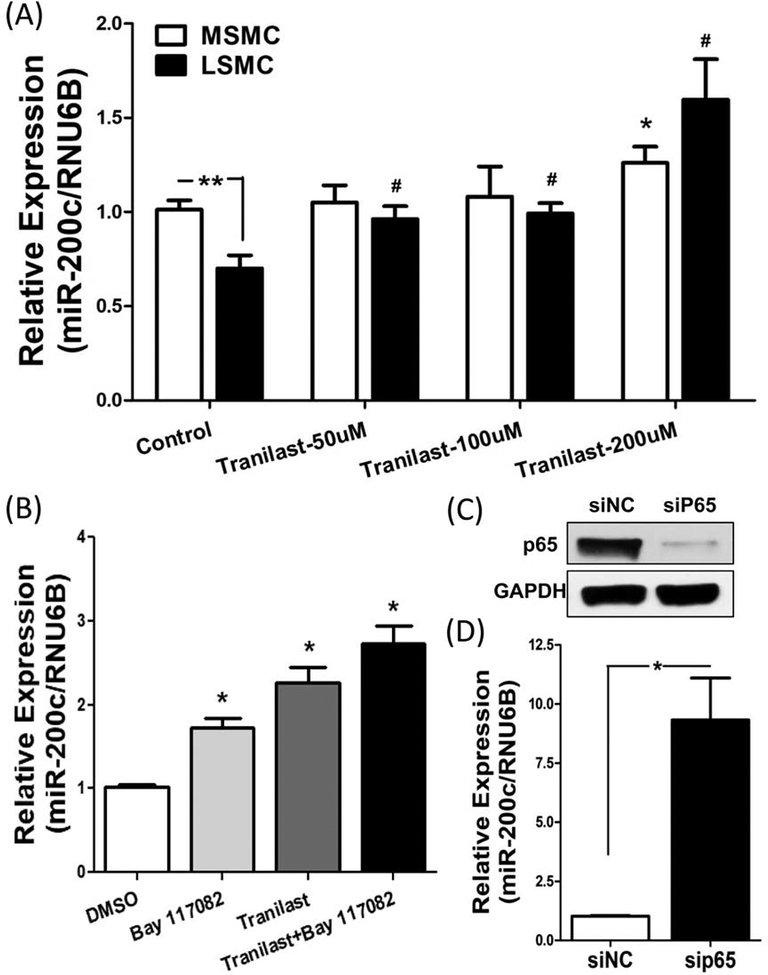
This study was initiated to determine whether tranilast has any effect on miR-200c expression in leiomyoma. As such, isolated paired MSMC and LSMC were treated with tranilast and miR-200c expression was detected by qRT-PCR. In support of our previous report (17), miR-200c expression was significantly lower in LSMC as compared to MSMC and tranilast dose dependently increased the expression of miR-200c (P < 0.05; Fig. 1A) in both LSMC and MSMC but with a more profound effect in LSMC as compared to MSMC. Because tranilast is an anti-inflammatory agent (38), we next examined the molecular mechanism by which tranilast induces miR-200c expression in LSMC. Isolated LSMC were treated with Bay 117082, an inhibitor of IκB phosphorylation, along with tranilast. The concentration of tranilast (200 μM) selected was based on other studies (39–41) and dose–response experiments carried out in our previous work (16). Similar to tranilast, Bay 117082 not only induced miR-200c expression, but also enhanced the effect of tranilast on miR-200c expression in LSMC (P < 0.05; Fig. 1B).
Figure 1.
(A) qRT-PCR analysis of miR-200c expression following treatment of paired MSMC and LSMC with different doses of tranilast for 48 hrs (N=5). (B) The relative expression of miR-200c was determined following treatment of LSMC with 200 μM tranilast for 48 h with or without Bay 117082 (10 μM) for the last 24 h (N=7). (C) Shows the effect of RelA/p65 knockdown through transfection of LSMC with siRNA against RelA/p65 for 96 hrs on RelA/p65 protein abundance (N=4). (D) The level of miR-200c as determined by qRT-PCR is shown following transfection of LSMC with siRNA against RelA/p65 for 96 hrs (N=4). The results are presented as mean ± SEM of independent experiments as mentioned above using cells isolated from different patients in each set. P values (*P<0.05) are indicated by corresponding lines.
Next, siRNA was used to knock down the protein expression of RelA/p65 in LSMC (P < 0.05; Fig. 1C). As a result of RelA/p65 knockdown the expression of miR-200c was significantly increased (P < 0.05; Fig. 1D), indicating that miR-200c is under regulation of RelA/p65.
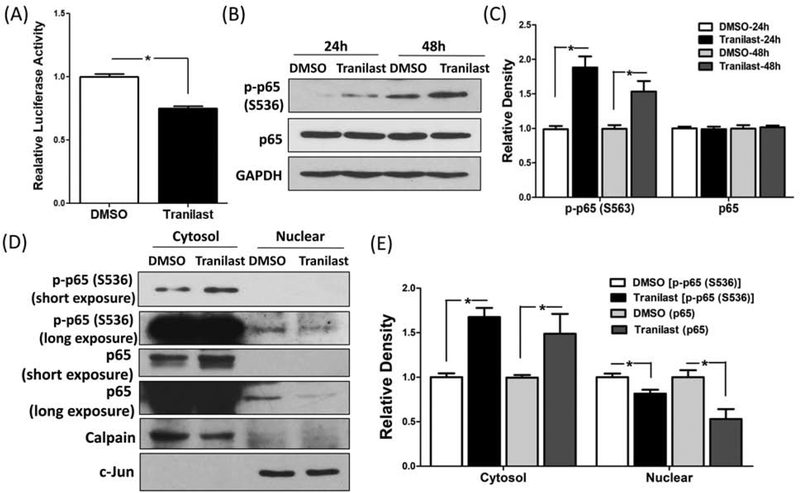
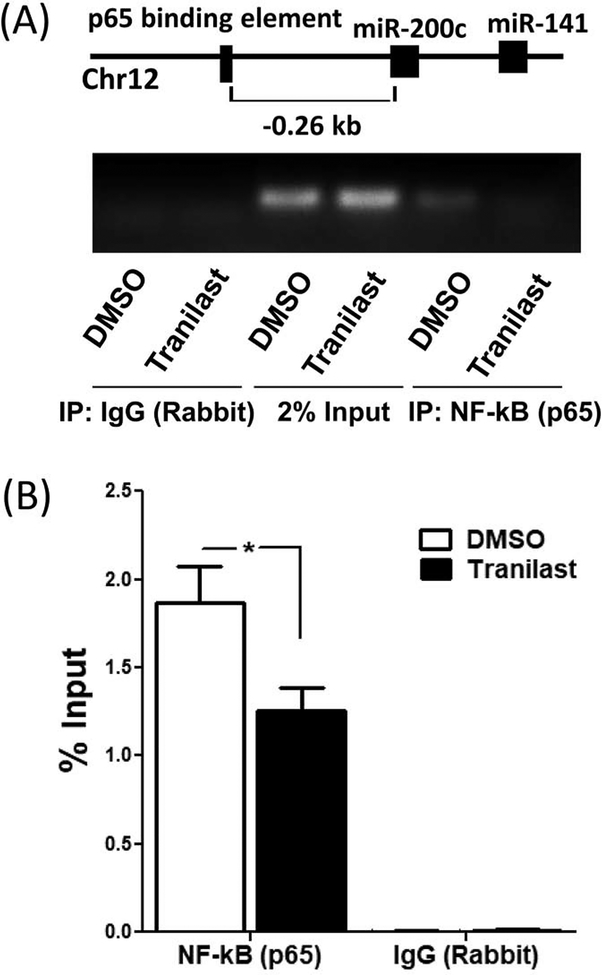
Additionally, using luciferase reporter assay we demonstrated that tranilast significantly reduced RelA/p65 activity in LSMC (P < 0.05; Fig. 2A). Previous reports indicated that the phosphorylated RelA/p65 at serine 536 is involved in RelA/p65 activity (22). As such, to further our understanding of the molecular mechanism of tranilast action in LSMC, we assessed the levels of RelA/p65 phosphorylation at serine 536. As shown in figure 2B and 2C, treatment of LSMC with tranilast for 24 and 48 hours increased the expression of phosphorylated RelA/p65 at serine 536 (P < 0.05). However, tranilast had no effect on total protein abundance of RelA/p65 (Fig. 2B and 2C). Using subfractionation of cellular and nuclear protein, tranilast significantly induced RelA/p65 protein and its phosphorylation at serine 536 in the cytosolic fraction and decreased them in the nuclear fraction (P < 0.05; Fig. 2D and 2E). Furthermore, ChIP assay demonstrated that the binding ability of RelA/p65 to miR-200c promoter was reduced by tranilast (P < 0.05; Fig. 3), thus promoting the induction of miR-200c.
Figure 2.
(A) The level of RelA/p65 activity in LSMC transfected with a luciferase reporter construct containing preserved RelA/p65 binding sites and pRL-TK plasmid as control for transfection efficiency. Cells were treated with tranilast (200 μM) for 8 hrs. The ratio of Firefly:Renilla was determined and reported as relative luciferase activity as compared to control (DMSO) which was independently set as 1 (N=3). (B–C) Immunoblotting analysis of phosphorylated RelA/p65 at serine 536 and total RelA/p65 following treatment of LSMC with 200 μM tranilast for 24 hrs and 48 hrs (N=3). The relative band densities are shown in (C). (D) Shows protein levels of phosphorylated RelA/p65 at serine 536 and RelA/p65 in LSMC following treatment with 200 μM tranilast for 36 hrs (N=3). Cells were harvested and sub-fractionated into cytosol and nuclear proteins and subjected to immunoblotting analysis with Calpain and c-Jun serving as markers for the respective subcellular fractions. (E) The band density of phosphorylated RelA/p65 at serine 536 and RelA/p65 was semi-quantified and normalized to Calpain (cytosol) or c-Jun (nuclear). The results are presented as mean ± SEM of independent experiments as mentioned above using cells isolated from different patients in each set. P values (*P < 0.05) indicated by the corresponding lines.
Figure 3.
(A) The graphical depiction of the miR-200c promoter with the location of RelA/p65 binding element. ChIP assay with RelA/p65 or control IgG was performed on chromatin derived from LSMC after 36 hrs incubation with tranilast (200 μM) or vehicle DMSO (N=4). Primers specific to the depicted RelA/p65 binding element were used for PCR amplification. 2% inputs are indicated. The result is a representative from four sets of independent experiments using LSMC isolated from four patients. (B) The quantitative data is presented by percent input method as mean ± SEM with P values (*P < 0.05) indicated by corresponding lines.
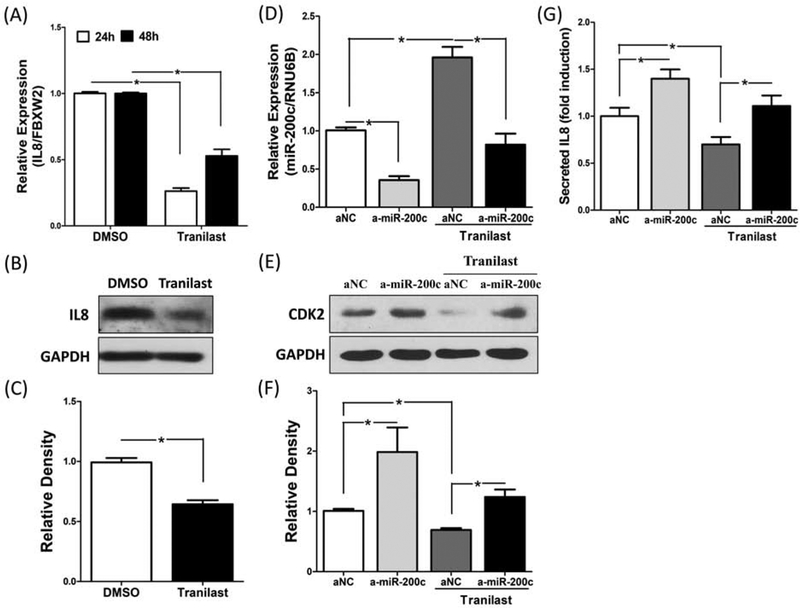
Our previous work had demonstrated that IL8 is under regulation of RelA/p65 and miR-200c through targeting IKBKB in LSMC (32), therefore we next examined the effect of tranilast on IL8 expression. In support of previous published data (42), the treatment of tranilast in LSMC significantly repressed IL8 mRNA and protein expression (P < 0.05; Fig. 4A, 4B and 4C). Because CDK2 is also down-regulated by tranilast (16, 20) and is a direct target of miR-200c (33), in order to determine the physiological significance of miR-200c in mediating the effects of tranilast in LSMC, IL8 and CDK2 were measured following transfection of LSMC with anti-miR-200c oligonucleotides or anti-miR negative control combined with tranilast or DMSO. As shown in figure 4D to 4G, knockdown of miR-200c resulted in partial attenuation of the tranilast-induced inhibitory effect on IL8 and CDK2 expression (P <0.05).
Figure 4.
(A) mRNA expression of IL8 in LSMC treated with Tranilast (200 μM) or DMSO as control for 24h to 48h (N=6). (B) A representative immunoblotting of IL8 expression in LSMC treated with Tranilast (200 μM) for 48h (N=5). (C) Demonstrates the relative band densities of IL8. (D) LSMC were transfected with anti-miR scrambled oligonucleotides (aNC) or anti-miR-200c oligonucleotides (a-miR-200c) for 96 hours with DMSO or tranilast (200 μM) for the last 48 hours and the levels of miR-200c was determined (N=4). (E) A representative of gel demonstrating CDK2 expression following transfection with anti-miR scrambled oligonucleotides (aNC) or anti-miR-200c oligonucleotides (a-miR-200c) in LSMC for 96 hours with DMSO or tranilast (200 μM) for the last 48 hours (N=4). (F) The bar plot represents the mean relative band densities of CDK2. (G) Secreted IL8 was determined from collected medium following transfection with anti-miR scrambled oligonucleotides (aNC) or anti-miR-200c oligonucleotides (amiR-200c) in LSMC for 96 hours with DMSO or tranilast (200 μM) for the last 48 hours (N=4). The results are presented as mean ± SEM of independent experiments using cells isolated from different patients in each set. P values (*P < 0.05) indicated by the corresponding lines.
Discussion
Our results demonstrate for the first time that tranilast significantly induces miR-200c expression in MSMC and LSMC, but more profoundly in LSMC. This differential effect may be due to the presence of greater degree of inflammation in leiomyoma as compared with myometrium (5, 14). The induction of miR-200c is mediated at least in part through a mechanism involving repression of RelA/p65 nuclear translocation which results in decreased RelA/p65 activity and transcriptional regulation of miR-200c promoter. ChIP analysis further indicated that tranilast reduced the binding of RelA/p65 to miR-200c promoter. Because IL8 and CDK2 are involved in tranilast-mediated suppression of LSMC inflammation and proliferation (16, 20), as a proof of principle we selected IL8 and CDK2 for further investigation and demonstrated that the tranilast mediated inhibition of IL8 and CDK2 expression is at least in part through induction of miR-200c in LSMC.
The significance of tranilast-induced miR-200c expression relates to miR-200c regulatory function on expression of many target genes including several epithelial–mesenchymal transition (EMT)-, cell cycle-, ECM-, inflammation- and apoptosis-related genes (17, 32, 43–46). We have previously reported that the lower expression of miR-200c in leiomyoma was inversely correlated with several of its target genes expression such as CDK2, ZEB1, ZEB2, VEGFA, TIMP2, FBLN5, IL8 and IKBKB (17, 32, 33). Moreover, we demonstrated that miR-200c regulates IL8 expression indirectly through downregulation of NF-kB signaling pathway by targeting IKBKB. Functionally, repression of IL8 after gain-of-function of miR-200c in LSMC mirrors the profile in fibroids characterized by elevated IL8 expression and lower miR-200c (17, 32, 34).
Expression profiling has demonstrated that there is aberrant expression of many miRNAs in leiomyomas as compared to myometrium (13, 17, 34, 47, 48); however, the regulatory mechanisms behind the altered expression of miRNAs in leiomyoma remain mostly unclear. Recent studies have reported that the expression of miR-200 family is under regulation by DNA methylation and histone modifications, leading to progression of glioblastoma, pancreatic adenocarcinoma and basal type of breast cancer (49, 50). Additionally, several transcription factors including ZEB1, ZEB2, krüppel-like factor 5 (KLF5), achaete-scute family BHLH transcription factor 2 (ASCL2), nanog homeobox (NANOG), octamer-binding protein 4 (Oct4), SRY-box containing gene 2 (Sox2), hepatocyte nuclear factor-1β (HNF-1β), specificity protein 1 (Sp1) and c-Jun have been identified as regulators of miR-200 family expression by acting directly on its promoter (51–57). To our knowledge this is the first demonstration that the transcription factor RelA/p65 also regulates miR-200c expression and the effect of tranilast on miR-200c induction is mediated through this pathway in LSMC.
Tranilast, an inhibitor of tryptase, has been demonstrated to have various anti-proliferative and anti-inflammatory effects (19), and recently has been reported to inhibit cytokine-induced NF-κB activity and transcriptional ability on inflammatory associated genes expression (42, 58, 59). Tryptase acting as serine proteases is stored in large amounts in mast cells and has been demonstrated to stimulate angiogenesis via several mechanisms including activation of c-Kit (CD117), high affinity IgE receptor (FcεRI) and protease-activated receptor-2 (PAR-2) (60–63). Accordingly, the correlation between mast cells, chronic inflammation and tumor development has been established (64–66). Previous studies demonstrated that mast cells are frequently abundant in leiomyomas (67–69). Additionally, another tryptase inhibitor, nafamostat mesylate, has been reported to inhibit pancreatic cancer through blockade of nuclear NF-kB activation (70). In this regard, activation of NF-kB signaling pathway has been identified in a variety of tumors including leiomyoma and demonstrated to play an important role in the modulation of cell proliferation, inflammatory responses and oncogenesis (5, 71, 72). Our previous work also indicated that leiomyomas express elevated levels of phosphorylated p65 at serine 536 as compared with matched myometrium (14).
Although RelA/p65 phosphorylation at S536 has been reported to enhance its transcriptional activity and nuclear translocation (24, 28–31), our results unexpectedly indicated that treatment of LSMC for 24 and 48 hours with tranilast elevated phosphorylation of RelA/p65 at serine 536. In addition, the pattern of phosphorylated RelA/p65 at serine 536 in cellular and nuclear fraction was similar to that of RelA/p65 which is accumulated in cellular fraction, but reduced in nuclear fraction in response to treatment of LSMC with tranilast. In agreement with our results, several reports suggested that S536 phosphorylation may inhibit nuclear translocation of RelA/p65, resulting in repression of NF-κB signaling pathway (73, 74). Moreover, several studies have identified another role for S536 phosphorylation, namely promotion of proteasomal degradation of RelA/p65, thereby reducing its protein stability and transcriptional activity (25, 27). Collectively, the conditions and consequences of RelA/p65 phosphorylation at S536 are cell and physiologic condition specific.
Our data indicates that tranilast reduced NF-κB activity (figure 2A), induced p65 phosphorylation at S536 (figure 2B) and repressed mRNA expression of IL8 (figure 4A) and CDK2 (16) in a short period of treatment, whereas the induction of miR-200c by tranilast in LSMC required a significantly longer treatment (48 hours) (figure 1A and 1B), implying that miR-200c might have an accessory role in mediating the action of tranilast in LSMC. In support of this interpretation, knockdown of miR-200c enhanced protein levels of CDK2 and secreted IL8, and partially relieved tranilast-mediated inhibition of IL8 and CDK2 expression (figure 4E, 4F and 4G). These results suggest that miR-200c mediates at least some of the effects of tranilast on several downstream target genes; however, the effects of tranilast on other miRNAs expression and target genes warrant further investigation.
In addition to tranilast, other agents such as green tea extracts, halofuginone and vitamin D have been evaluated as alternative leiomyoma therapeutic approaches based on their effects on alteration of mast cell activation, expression of proliferation associated genes, pro-fibrotic cytokines and ECM regulation (1, 75–77). Whether these agents also exert their effects on miR-200c or other anti-fibrotic miRNAs remains to be determined.
In summary, our data provides support for direct effects of tranilast on miR-200c induction through a molecular mechanism involving altered transcriptional ability of RelA/p65 in LSMC. However, whether tranilast through this and/or other mechanisms influences the expression of other anti-fibrotic and anti-inflammatory miRNAs remains to be determined. Because the anti-fibrotic and anti-inflammatory properties of tranilast has been extensively studied under both in vitro and in vivo conditions (19), the need for an animal model is warranted to further support the potential effectiveness of tranilast for treatment of leiomyomas.
Acknowledgements:
This study was supported by NIH (HD088868).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Segars JH, Parrott EC, Nagel JD, Guo XC, Gao X, Birnbaum LS et al. Proceedings from the Third National Institutes of Health International Congress on Advances in Uterine Leiomyoma Research: comprehensive review, conference summary and future recommendations. Hum Reprod Update 2014;20:309–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doherty L, Mutlu L, Sinclair D, Taylor H. Uterine fibroids: clinical manifestations and contemporary management. Reprod Sci 2014;21:1067–92. [DOI] [PubMed] [Google Scholar]
- 3.Islam MS, Protic O, Stortoni P, Grechi G, Lamanna P, Petraglia F et al. Complex networks of multiple factors in the pathogenesis of uterine leiomyoma. Fertil Steril 2013;100:178–93. [DOI] [PubMed] [Google Scholar]
- 4.Islam MS, Ciavattini A, Petraglia F, Castellucci M, Ciarmela P. Extracellular matrix in uterine leiomyoma pathogenesis: a potential target for future therapeutics. Hum Reprod Update 2018;24:59–85. [DOI] [PubMed] [Google Scholar]
- 5.Chegini N Proinflammatory and profibrotic mediators: principal effectors of leiomyoma development as a fibrotic disorder. Semin Reprod Med 2010;28:180–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor DK, Holthouser K, Segars JH, Leppert PC. Recent scientific advances in leiomyoma (uterine fibroids) research facilitates better understanding and management. F1000Res 2015;4:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo X, Chegini N. The expression and potential regulatory function of microRNAs in the pathogenesis of leiomyoma. Semin Reprod Med 2008;26:500–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo X, Pan Q, Liu L, Chegini N. Genomic and proteomic profiling II: comparative assessment of gene expression profiles in leiomyomas, keloids, and surgically-induced scars. Reprod Biol Endocrinol 2007;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med 2008;12:227–40. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Marsh EE, Lin Z, Yin P, Milad M, Chakravarti D, Bulun SE. Differential expression of microRNA species in human uterine leiomyoma versus normal myometrium. Fertil Steril 2008;89:1771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang TD, Khorram O. Expression Profiling of lncRNAs, miRNAs, and mRNAs and Their Differential Expression in Leiomyoma Using Next-Generation RNA Sequencing. Reprod Sci 2018;25:246–55. [DOI] [PubMed] [Google Scholar]
- 12.Chuang TD, Xie Y, Yan W, Khorram O. Next-generation sequencing reveals differentially expressed small noncoding RNAs in uterine leiomyoma. Fertil Steril 2018;109:919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiang W, Liu Z, Serna VA, Druschitz SA, Liu Y, Espona-Fiedler M et al. Down-regulation of miR-29b is essential for pathogenesis of uterine leiomyoma. Endocrinology 2014;155:663–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuang TD, Khorram O. Mechanisms underlying aberrant expression of miR-29c in uterine leiomyoma. Fertil Steril 2016;105:236–45.e1. [DOI] [PubMed] [Google Scholar]
- 15.Marsh EE, Steinberg ML, Parker JB, Wu J, Chakravarti D, Bulun SE. Decreased expression of microRNA-29 family in leiomyoma contributes to increased major fibrillar collagen production. Fertil Steril 2016;106:766–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang TD, Khorram O. Tranilast Inhibits Genes Functionally Involved in Cell Proliferation, Fibrosis, and Epigenetic Regulation and Epigenetically Induces miR-29c Expression in Leiomyoma Cells. Reprod Sci 2017;24:1253–63. [DOI] [PubMed] [Google Scholar]
- 17.Chuang TD, Panda H, Luo X, Chegini N. miR-200c is aberrantly expressed in leiomyomas in an ethnic-dependent manner and targets ZEBs, VEGFA, TIMP2, and FBLN5. Endocr Relat Cancer 2012;19:541–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogosnitzky M, Danks R, Kardash E. Therapeutic potential of tranilast, an anti-allergy drug, in proliferative disorders. Anticancer Res 2012;32:2471–8. [PubMed] [Google Scholar]
- 19.Darakhshan S, Pour AB. Tranilast: a review of its therapeutic applications. Pharmacol Res 2015;91:15–28. [DOI] [PubMed] [Google Scholar]
- 20.Shime H, Kariya M, Orii A, Momma C, Kanamori T, Fukuhara K et al. Tranilast inhibits the proliferation of uterine leiomyoma cells in vitro through G1 arrest associated with the induction of p21(waf1) and p53. J Clin Endocrinol Metab 2002;87:5610–7. [DOI] [PubMed] [Google Scholar]
- 21.Islam MS, Protic O, Ciavattini A, Giannubilo SR, Tranquilli AL, Catherino WH et al. Tranilast, an orally active antiallergic compound, inhibits extracellular matrix production in human uterine leiomyoma and myometrial cells. Fertil Steril 2014;102:597–606. [DOI] [PubMed] [Google Scholar]
- 22.Christian F, Smith EL, Carmody RJ. The Regulation of NF-kappaB Subunits by Phosphorylation. Cells 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milanovic M, Kracht M, Schmitz ML. The cytokine-induced conformational switch of nuclear factor kappaB p65 is mediated by p65 phosphorylation. Biochem J 2014;457:401–13. [DOI] [PubMed] [Google Scholar]
- 24.Buss H, Dorrie A, Schmitz ML, Hoffmann E, Resch K, Kracht M. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-{kappa}B at serine 536 is mediated by multiple protein kinases including I{kappa}B kinase (IKK)-{alpha}, IKK{beta}, IKK{epsilon}, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J Biol Chem 2004;279:55633–43. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature 2005;434:1138–43. [DOI] [PubMed] [Google Scholar]
- 26.Sizemore N, Lerner N, Dombrowski N, Sakurai H, Stark GR. Distinct roles of the Ikappa B kinase alpha and beta subunits in liberating nuclear factor kappa B (NF-kappa B) from Ikappa B and in phosphorylating the p65 subunit of NF-kappa B. J Biol Chem 2002;277:3863–9. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Hu L, Tong X, Ye X. Casein kinase 1gamma1 inhibits the RIG-I/TLR signaling pathway through phosphorylating p65 and promoting its degradation. J Immunol 2014;192:1855–61. [DOI] [PubMed] [Google Scholar]
- 28.Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L et al. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol 2005;25:7966–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Cai Y, Shao LJ, Siddiqui J, Palanisamy N, Li R et al. Activation of NF-{kappa}B by TMPRSS2/ERG Fusion Isoforms through Toll-Like Receptor-4. Cancer Res 2011;71:1325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F, Tang E, Guan K, Wang CY. IKK beta plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J Immunol 2003;170:5630–5. [DOI] [PubMed] [Google Scholar]
- 31.Delhase M, Kim SY, Lee H, Naiki-Ito A, Chen Y, Ahn ER et al. TANK-binding kinase 1 (TBK1) controls cell survival through PAI-2/serpinB2 and transglutaminase 2. Proc Natl Acad Sci U S A 2012;109:E177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chuang TD, Khorram O. miR-200c Regulates IL8 Expression by Targeting IKBKB: A Potential Mediator of Inflammation in Leiomyoma Pathogenesis. PLoS One 2014;9:e95370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chuang TD, Khorram O. Regulation of Cell Cycle Regulatory Proteins by MicroRNAs in Uterine Leiomyoma. Reprod Sci 2019;26:250–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chuang TD, Luo X, Panda H, Chegini N. miR-93/106b and their host gene, MCM7, are differentially expressed in leiomyomas and functionally target F3 and IL-8. Mol Endocrinol 2012;26:1028–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busk PK. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics 2014;15:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almeida TA, Quispe-Ricalde A, Montes de Oca F, Foronda P, Hernandez MM. A high-throughput open-array qPCR gene panel to identify housekeeping genes suitable for myometrium and leiomyoma expression analysis. Gynecol Oncol 2014;134:138–43. [DOI] [PubMed] [Google Scholar]
- 37.Chuang TD, Sakurai R, Gong M, Khorram O, Rehan VK. Role of miR-29 in mediating offspring lung phenotype in a rodent model of intrauterine growth restriction. Am J Physiol Regul Integr Comp Physiol 2018;315:R1017–r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ammendola M, Leporini C, Marech I, Gadaleta CD, Scognamillo G, Sacco R et al. Targeting Mast Cells Tryptase in Tumor Microenvironment: A Potential Antiangiogenetic Strategy. Biomed Res Int 2014;2014:154702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prud’homme GJ, Glinka Y, Toulina A, Ace O, Subramaniam V, Jothy S. Breast cancer stem-like cells are inhibited by a non-toxic aryl hydrocarbon receptor agonist. PLoS One 2010;5:e13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin UH, Kim SB, Safe S. Omeprazole Inhibits Pancreatic Cancer Cell Invasion through a Nongenomic Aryl Hydrocarbon Receptor Pathway. Chem Res Toxicol 2015;28:907–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suwa S, Kasubata A, Kato M, Iida M, Watanabe K, Miura O et al. The tryptophan derivative, tranilast, and conditioned medium with indoleamine 2,3-dioxygenase-expressing cells inhibit the proliferation of lymphoid malignancies. Int J Oncol 2015;46:1369–76. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Kan M, Li A, Hou L, Jia H, Xin Y et al. Inhibitory Effects of Tranilast on Cytokine, Chemokine, Adhesion Molecule, and Matrix Metalloproteinase Expression in Human Corneal Fibroblasts Exposed to Poly(I:C). Curr Eye Res 2016;41:1400–7. [DOI] [PubMed] [Google Scholar]
- 43.Fan YC, Mei PJ, Chen C, Miao FA, Zhang H, Li ZL. MiR-29c inhibits glioma cell proliferation, migration, invasion and angiogenesis. J Neurooncol 2013;115:179–88. [DOI] [PubMed] [Google Scholar]
- 44.Matsuo M, Nakada C, Tsukamoto Y, Noguchi T, Uchida T, Hijiya N et al. MiR-29c is downregulated in gastric carcinomas and regulates cell proliferation by targeting RCC2. Mol Cancer 2013;12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ et al. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci U S A 2008;105:5874–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG et al. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun 2011;411:586–92. [DOI] [PubMed] [Google Scholar]
- 47.Peng Y, Laser J, Shi G, Mittal K, Melamed J, Lee P et al. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res 2008;6:663–73. [DOI] [PubMed] [Google Scholar]
- 48.Fitzgerald JB, Chennathukuzhi V, Koohestani F, Nowak RA, Christenson LK. Role of microRNA-21 and programmed cell death 4 in the pathogenesis of human uterine leiomyomas. Fertil Steril 2012;98:726–34 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ning X, Shi Z, Liu X, Zhang A, Han L, Jiang K et al. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett 2015;359:198–205. [DOI] [PubMed] [Google Scholar]
- 50.Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. Embo j 2011;30:770–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian Y, Pan Q, Shang Y, Zhu R, Ye J, Liu Y et al. MicroRNA-200 (miR-200) cluster regulation by achaete scute-like 2 (Ascl2): impact on the epithelial-mesenchymal transition in colon cancer cells. J Biol Chem 2014;289:36101–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan Q, Meng L, Ye J, Wei X, Shang Y, Tian Y et al. Transcriptional repression of miR-200 family members by Nanog in colon cancer cells induces epithelial-mesenchymal transition (EMT). Cancer Lett 2017;392:26–38. [DOI] [PubMed] [Google Scholar]
- 53.Wang G, Guo X, Hong W, Liu Q, Wei T, Lu C et al. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc Natl Acad Sci U S A 2013;110:2858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B, Zhang Z, Xia S, Xing C, Ci X, Li X et al. KLF5 activates microRNA 200 transcription to maintain epithelial characteristics and prevent induced epithelial-mesenchymal transition in epithelial cells. Mol Cell Biol 2013;33:4919–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 2008;68:7846–54. [DOI] [PubMed] [Google Scholar]
- 56.Hajarnis SS, Patel V, Aboudehen K, Attanasio M, Cobo-Stark P, Pontoglio M et al. Transcription Factor Hepatocyte Nuclear Factor-1beta (HNF-1beta) Regulates MicroRNA-200 Expression through a Long Noncoding RNA. J Biol Chem 2015;290:24793–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong X, Zheng L, Shen J, Zhang D, Xiong M, Zhang Y et al. Suppression of MicroRNA 200 Family Expression by Oncogenic KRAS Activation Promotes Cell Survival and Epithelial-Mesenchymal Transition in KRAS-Driven Cancer. Mol Cell Biol 2016;36:2742–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chikaraishi A, Hirahashi J, Takase O, Marumo T, Hishikawa K, Hayashi M et al. Tranilast inhibits interleukin-1beta-induced monocyte chemoattractant protein-1 expression in rat mesangial cells. Eur J Pharmacol 2001;427:151–8. [DOI] [PubMed] [Google Scholar]
- 59.Spiecker M, Lorenz I, Marx N, Darius H. Tranilast inhibits cytokine-induced nuclear factor kappaB activation in vascular endothelial cells. Mol Pharmacol 2002;62:856–63. [DOI] [PubMed] [Google Scholar]
- 60.Blair RJ, Meng H, Marchese MJ, Ren S, Schwartz LB, Tonnesen MG et al. Human mast cells stimulate vascular tube formation. Tryptase is a novel, potent angiogenic factor. J Clin Invest 1997;99:2691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milia AF, Salis MB, Stacca T, Pinna A, Madeddu P, Trevisani M et al. Protease-activated receptor-2 stimulates angiogenesis and accelerates hemodynamic recovery in a mouse model of hindlimb ischemia. Circ Res 2002;91:346–52. [DOI] [PubMed] [Google Scholar]
- 62.Ribatti D, Ranieri G, Nico B, Benagiano V, Crivellato E. Tryptase and chymase are angiogenic in vivo in the chorioallantoic membrane assay. Int J Dev Biol 2011;55:99–102. [DOI] [PubMed] [Google Scholar]
- 63.Moon TC, Lee E, Baek SH, Murakami M, Kudo I, Kim NS et al. Degranulation and cytokine expression in human cord blood-derived mast cells cultured in serum-free medium with recombinant human stem cell factor. Mol Cells 2003;16:154–60. [PubMed] [Google Scholar]
- 64.Crivellato E, Ribatti D. The mast cell: an evolutionary perspective. Biol Rev Camb Philos Soc 2010;85:347–60. [DOI] [PubMed] [Google Scholar]
- 65.Komi DEA, Redegeld FA. Role of Mast Cells in Shaping the Tumor Microenvironment. Clin Rev Allergy Immunol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ribatti D, Tamma R, Vacca A. Mast Cells and Angiogenesis in Human Plasma Cell Malignancies. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maluf HM, Gersell DJ. Uterine leiomyomas with high content of mast cells. Arch Pathol Lab Med 1994;118:712–4. [PubMed] [Google Scholar]
- 68.Orii A, Mori A, Zhai YL, Toki T, Nikaido T, Fujii S. Mast cells in smooth muscle tumors of the uterus. Int J Gynecol Pathol 1998;17:336–42. [DOI] [PubMed] [Google Scholar]
- 69.Yavuz E, Gulluoglu MG, Akbas N, Tuzlali S, Ilhan R, Iplikci A et al. The values of intratumoral mast cell count and Ki-67 immunoreactivity index in differential diagnosis of uterine smooth muscle neoplasms. Pathol Int 2001;51:938–41. [DOI] [PubMed] [Google Scholar]
- 70.Fujiwara Y, Furukawa K, Haruki K, Shimada Y, Iida T, Shiba H et al. Nafamostat mesilate can prevent adhesion, invasion and peritoneal dissemination of pancreatic cancer thorough nuclear factor kappa-B inhibition. J Hepatobiliary Pancreat Sci 2011;18:731–9. [DOI] [PubMed] [Google Scholar]
- 71.DiDonato JA, Mercurio F, Karin M. NF-kappaB and the link between inflammation and cancer. Immunol Rev 2012;246:379–400. [DOI] [PubMed] [Google Scholar]
- 72.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer 2013;12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mattioli I, Sebald A, Bucher C, Charles RP, Nakano H, Doi T et al. Transient and selective NF-kappa B p65 serine 536 phosphorylation induced by T cell costimulation is mediated by I kappa B kinase beta and controls the kinetics of p65 nuclear import. J Immunol 2004;172:6336–44. [DOI] [PubMed] [Google Scholar]
- 74.Pradere JP, Hernandez C, Koppe C, Friedman RA, Luedde T, Schwabe RF. Negative regulation of NF-kappaB p65 activity by serine 536 phosphorylation. Sci Signal 2016;9:ra85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Leo V, Morgante G, La Marca A, Musacchio MC, Sorace M, Cavicchioli C et al. A benefit-risk assessment of medical treatment for uterine leiomyomas. Drug Saf 2002;25:759–79. [DOI] [PubMed] [Google Scholar]
- 76.Grudzien MM, Low PS, Manning PC, Arredondo M, Belton RJ, Jr., Nowak RA. The antifibrotic drug halofuginone inhibits proliferation and collagen production by human leiomyoma and myometrial smooth muscle cells. Fertil Steril 2010;93:1290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brakta S, Diamond JS, Al-Hendy A, Diamond MP, Halder SK. Role of vitamin D in uterine fibroid biology. Fertil Steril 2015;104:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]