Abstract
Background
The coronavirus disease 2019 (COVID-19) epidemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), began in Wuhan city, Hubei province, in December, 2019, and has spread throughout China. Understanding the evolving epidemiology and transmission dynamics of the outbreak beyond Hubei would provide timely information to guide intervention policy.
Methods
We collected individual information from official public sources on laboratory-confirmed cases reported outside Hubei in mainland China for the period of Jan 19 to Feb 17, 2020. We used the date of the fourth revision of the case definition (Jan 27) to divide the epidemic into two time periods (Dec 24 to Jan 27, and Jan 28 to Feb 17) as the date of symptom onset. We estimated trends in the demographic characteristics of cases and key time-to-event intervals. We used a Bayesian approach to estimate the dynamics of the net reproduction number (Rt) at the provincial level.
Findings
We collected data on 8579 cases from 30 provinces. The median age of cases was 44 years (33–56), with an increasing proportion of cases in younger age groups and in elderly people (ie, aged >64 years) as the epidemic progressed. The mean time from symptom onset to hospital admission decreased from 4·4 days (95% CI 0·0–14·0) for the period of Dec 24 to Jan 27, to 2·6 days (0·0–9·0) for the period of Jan 28 to Feb 17. The mean incubation period for the entire period was estimated at 5·2 days (1·8–12·4) and the mean serial interval at 5·1 days (1·3–11·6). The epidemic dynamics in provinces outside Hubei were highly variable but consistently included a mixture of case importations and local transmission. We estimated that the epidemic was self-sustained for less than 3 weeks, with mean Rt reaching peaks between 1·08 (95% CI 0·74–1·54) in Shenzhen city of Guangdong province and 1·71 (1·32–2·17) in Shandong province. In all the locations for which we had sufficient data coverage of Rt, Rt was estimated to be below the epidemic threshold (ie, <1) after Jan 30.
Interpretation
Our estimates of the incubation period and serial interval were similar, suggesting an early peak of infectiousness, with possible transmission before the onset of symptoms. Our results also indicate that, as the epidemic progressed, infectious individuals were isolated more quickly, thus shortening the window of transmission in the community. Overall, our findings indicate that strict containment measures, movement restrictions, and increased awareness of the population might have contributed to interrupt local transmission of SARS-CoV-2 outside Hubei province.
Funding
National Science Fund for Distinguished Young Scholars, National Institute of General Medical Sciences, and European Commission Horizon 2020.
Introduction
Since December, 2019, an increasing number of atypical pneumonia cases caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been reported in Wuhan, a city in the Chinese province of Hubei.1 As of Feb 17, 2020, 72 436 cases of coronavirus disease 2019 (COVID-19), including 1868 deaths, had been reported in mainland China.2 The outbreak has now spread to 198 countries, areas, or territories beyond China.3 On Jan 30, 2020, WHO declared the COVID-19 outbreak a public health emergency of international concern.4
An early report5 on the epidemiology of the COVID-19 outbreak included analysis of the first 425 confirmed cases detected in Wuhan up to Jan 22, 2020. Since then, the temporal dynamics and spatial dissemination of COVID-19 has changed, with 17% of cases reported outside of Hubei in mainland China as of Feb 17. In provinces outside Hubei, the COVID-19 epidemic is characterised by a mixture of local transmission and importation of cases from Hubei.6 A report6 based on the 44 672 confirmed cases detected in mainland China up to Feb 11, gave a description of the characteristics of COVID-19 cases for mainland China and in Hubei. Until now, there has been little information on the epidemiological features and transmission dynamics of the COVID-19 outbreak beyond Hubei. This information will be crucial to inform intervention policy in real-time, not only for China, but also for other countries with COVID-19 transmission.
Research in context.
Evidence before this study
On March 12, 2020, we searched PubMed and Web of Science for papers published after Jan 1, 2020, using the search terms (“nCoV” OR “COVID” OR “novel coronavirus” OR “severe acute respiratory syndrome coronavirus 2”) AND (“evolving epidemiology” OR “epidemiological characteristics” OR “transmission dynamics” OR “net reproduction number”). No language restrictions were applied. We identified 24 papers published in peer-reviewed journals. We also included two relevant papers that had yet to be indexed by PubMed or Web of Science. The study of the first confirmed 425 cases in Wuhan reported a mean incubation period of 5·2 days and a mean serial interval of 7·5 days. One study, based on 291 patients in China, estimated a median incubation period of 4 days, and another four studies estimated the incubation period of cases outside Wuhan or Hubei ranging from 5 to 6·7 days. One mathematical modelling study estimated that the median daily net reproduction number in Wuhan declined from 2·35 (95% CI 1·15–4·77) 1 week before travel restrictions were introduced on Jan 23 to 1·05 (0·41–2·39) 1 week after. A report from the Chinese Center for Disease Control and Prevention that was based on the analysis of 44 672 confirmed cases up to Feb 11, 2020, provided an overall description of the characteristics of coronavirus disease 2019 (COVID-19) cases in China as a whole and in Hubei province. However, that study did not provide estimates of key time-to-event intervals nor of the reproduction number. Until now, there has been scarce information to understand the changing epidemiological features and transmission dynamics of the COVID-19 outbreak beyond Hubei province.
Added value of this study
We collected individual information from official, publicly available sources on 8579 laboratory-confirmed cases detected outside Hubei province up to Feb 17, accounting for 69% of all 12 447 reported cases at that time. We analysed changes in demographic characteristics, key time-to-event intervals (eg, incubation period, serial interval, and time from symptom onset to hospital admission), and the dynamics of the net reproduction number (R t) at the provincial level. We found significant differences in the epidemiology of COVID-19 as the epidemic continued to spread across China. The time from symptom onset to hospital admission decreased from 4·4 days (95% CI 0·0–14·0) in the period of Dec 24 to Jan 27, to 2·6 days (0·0–9·0) in the period from Jan 28 to Feb 17. The mean incubation period was estimated at 5·2 days (95% CI 1·8–12·4) and the mean serial interval at 5·1 days (1·3–11·6). Most importantly, as of Feb 8, 2020, we estimated R t to be below the epidemic threshold in provinces outside Hubei that have reported the largest number of cases (Hunan, Shandong, Shenzhen city [Guangdong province], Anhui, Henan, Jiangsu, Jiangxi, Shaanxi, and Zhejiang).
Implications of all the available evidence
We show that the epidemiology and transmission dynamics of the COVID-19 epidemic are rapidly changing and should be closely monitored. Our findings suggest that strict containment measures, movement restrictions, and increased awareness of the population might have contributed to controlling the COVID-19 outbreak outside Hubei province in China. Given that the reproduction number is positively related to the growth rate of the epidemic and the length of the serial interval, the shorter serial interval estimated here (5·1 days vs 7·5 days) implies that transmissibility of severe acute respiratory syndrome coronavirus 2 might not be as high as previous estimates suggest. More broadly, our findings are crucial to inform intervention policy in real-time for other countries, given the international expansion of the COVID-19 epidemic.
We aimed to describe the epidemiological characteristics of the COVID-19 outbreak 50 days after it was recognised in Chinese provinces outside Hubei. We also estimated changes in key time-to-event intervals and reproduction numbers to assess whether the strict control measures put in place in China have been successful in slowing transmission.
Methods
Case definitions and surveillance
Since the outbreak of COVID-19 was first detected in Wuhan, the Chinese Center for Disease Control and Prevention (China CDC) has launched a new surveillance system, first in Wuhan, then extended to the entire country, to record information about COVID-19 cases (appendix pp 2–3).1, 5 Guidelines on the diagnosis and treatment of patients with novel coronavirus-infected pneumonia (NCIP) were published, with the first issued on Jan 15, 2020, defining a suspected NCIP case as pneumonia that fulfilled specific clinical criteria and had an epidemiological link to the Huanan seafood wholesale market in Wuhan or had travelled to Wuhan in the 14 days before symptom onset (appendix pp 2–3). The subsequent two versions of the guidelines (the first issued on Jan 18 and the second on Jan 22) removed the clinical criterion of no symptomatic improvement or deterioration after 3 days of antibiotic treatment to accelerate identification of cases. Additionally, the epidemiological link was revised to include travel history to Wuhan, direct contact with patients from Wuhan who had fever or respiratory symptoms within 14 days before symptom onset, or part of a COVID-19 cluster. In the fourth version of the guidelines (issued on Jan 27), the clinical criteria were broadened to meet any two of the three remaining clinical criteria (ie, fever, radiographic findings of pneumonia, and normal or reduced white blood cell count or reduced lymphocyte count at early stage of illness), and an epidemiological criterion was added (link with a confirmed COVID-19 case). In the fifth version issued on Feb 4, clinically diagnosed cases were defined as suspected cases with radiographic findings of pneumonia, to be used exclusively in Hubei.7
This study was approved by the institutional review board of the School of Public Health, Fudan University (Shanghai, China). All data were collected from publicly available sources. Data were deidentified, and informed consent was waived.
Data sources and collection
All of the provinces outside Hubei were included in the analysis of demographic characteristics, key time-to-event intervals, incubation period, and serial interval. Nine locations for which we had sufficient data coverage were used to estimate the net reproduction number.
Daily aggregated data on the number of cumulative cases in mainland China were extracted from the official websites of national, provincial, and municipal health commissions (appendix pp 3–10). Individual records on laboratory-confirmed COVID-19 cases were collected from two publicly available official sources: the websites of national, provincial, and municipal health commissions, and the websites of national and local government news media. Individual information was extracted by the data collection team and entered into a structured database comprising three sections: demographic characteristics; exposure and travel history; and key timelines, including exposure timeline, date of symptom onset, date of hospital admission, and date of official reporting. A well trained data collection team of about 40 members from Fudan University collected individual information (appendix pp 10, 11). Each individual record was cross-checked by three coauthors (WW, MLi, and WZ), and consensus was attained before finalising an entry in the database. Conflicting information was resolved based on the original data source; for example, if individual records on local governmental websites and governmental news media were inconsistent, we extracted the data from the governmental website. We used information on age, sex, location of detection, exposure history, dates of symptom onset, hospital admission, and official reporting to describe demographic characteristics of cases and estimate key time-to-event intervals, incubation period, serial interval, and net reproduction number. Details on the collection of individual data, the definitions of key variables, and the assessment of completeness of variables are provided in the appendix (pp 3–11).
We validated our individual records against the official line lists obtained from the websites of Shandong Provincial Health Commission, Shenzhen Municipal Health Commission, and Hunan Provincial Health Commission (appendix p 12).
Statistical analysis
We restricted analyses to provinces other than Hubei where the majority of individual records were available—ie, 8579 (98%) of 8738. We used the date of the fourth revision of the guidelines to divide the epidemic into two time periods. The first period ran from the emergence of COVID-19 in provinces outside Hubei, Dec 24 to Jan 27, when the definition of suspected cases was broadened to capture milder cases. The second period ran from Jan 28 to Feb 17. We did statistical analyses of demographic and epidemiological characteristics of confirmed cases stratified by the two epidemic periods. χ2 test was used to compare the age and sex structure. Two-sample t test was used to compare the key time-to-event intervals.
We estimated key time-to-event distributions for COVID-19 cases, including symptom onset to first health-care consultation, hospital admission, and official reporting. We estimated the time from infection to symptom onset (ie, the incubation period) by analysing COVID-19 cases with confirmed epidemiological links (clusters) identified by prospective contact tracing. The date of presumed infection was estimated from history of exposure, after excluding cases with exposure to Wuhan. When multiple exposures were reported, we considered the interval between the first and last recorded dates of exposure. We fit three parametric distributions (Weibull, gamma, and lognormal) to time-to-event data and selected the best fit based on the minimum Akaike information criterion. Confidence intervals for the estimated distributions were obtained with 2000 bootstrap simulations for either censored or non-censored data.
We analysed clusters of COVID-19 cases with epidemiological links confirmed by prospective contact tracing (all cases with travel history to Wuhan or Hubei—ie, declared visiting or residing, where excluded) to estimate the interval between onset of symptoms in a primary (index) case and the onset of symptoms in secondary cases generated by that primary case (ie, the serial interval). The serial interval was estimated by fitting a gamma distribution to the lag between the dates of symptom onset in the index and secondary cases.
Using the estimated distribution of the serial interval, we calculated the net reproduction number (R t), which is the mean number of secondary cases generated by a typical primary case at time t. Consecutive generations of cases arise after a period measured by the serial interval or by the generation time.8 We used a Bayesian approach9, 10 to estimate R t from the time series of symptom onset dates and the distribution of the serial interval, considering importations and local transmission.11 For this analysis, the last 9 days of the dataset were excluded to account for the possible incompleteness of the dataset due to reporting delays. The methods used to estimate R t are reported in detail in the appendix (p 23). Throughout, 95% CI refers to the 2·5th and 97·5th percentiles of the estimated distribution. Statistical analyses were done using R, version 3.6.0. R t was estimated from a code written by the authors, which is available online.
Role of the funding source
The funder of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
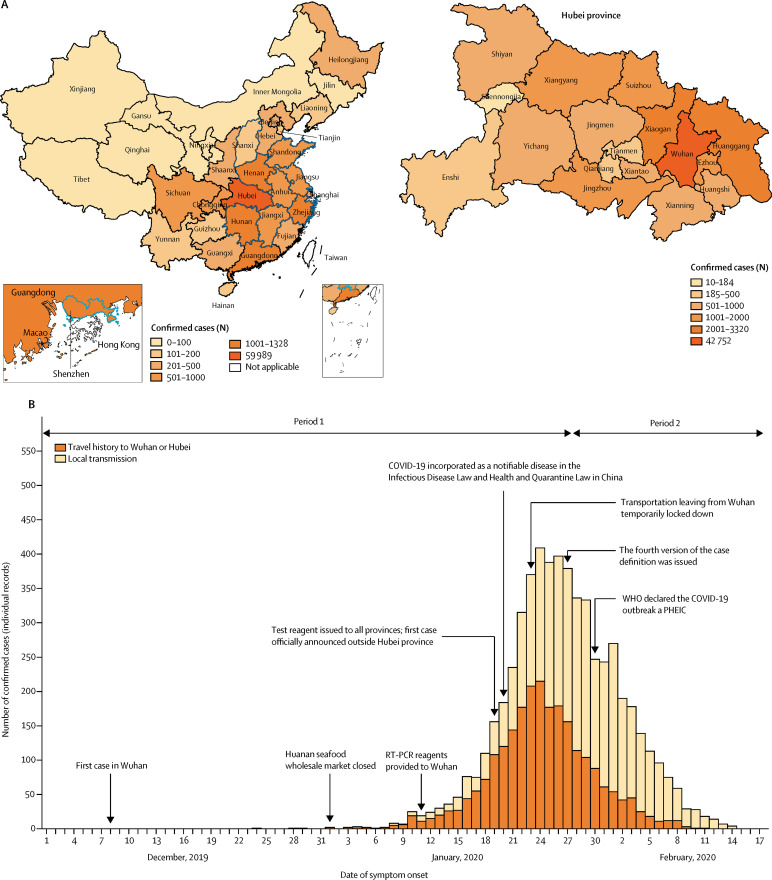
As of Feb 17, 2020, 72 436 COVID-19 cases had been reported in all 31 provinces of mainland China. 42 752 (59%) of 72 436 cases were detected in Wuhan city of Hubei province, 17 237 (24%) were detected in cities other than Wuhan in Hubei province, and 12 447 (17%) were reported in other provinces (figure 1A ). We collected individual information on 8579 laboratory-confirmed cases detected outside Hubei up to Feb 17, accounting for 69% of the 12 447 cases reported at that time. For most of the locations, there was consistency between the time series derived from our individual records and the official case reports (appendix pp 12–14).
Figure 1.
Geographical distribution and temporal dynamics of confirmed COVID-19 cases
(A) Geographical distribution of confirmed COVID-19 cases (including clinically diagnosed cases for Hubei province), officially reported as of Feb 17, 2020 (n=72 436), in each province of mainland China (left) and in each city of Hubei province (right). Provinces or cities with a blue contour indicate those selected for analysis of the net reproduction number. (B) Time series of COVID-19 cases for which we had individual records in provinces outside Hubei province, divided into cases with travel history to Wuhan or Hubei and cases of local transmission. If no information about the travel history of a patient was reported in the individual records, we assumed that the patient acquired the infection locally. This graph refers to 5683 cases with available symptom onset date (out of 8579 individual cases available in our individual records). The decline in the number of cases in the last few days of February is in part due to the delay between the date of reporting of cases and the date of symptom onset. The first period ran from Dec 24 to Jan 27. Dec 24 is the earliest symptom onset date of the cases reported outside Hubei. COVID-19=coronavirus disease 2019. PHEIC=public health emergency of international concern.
Starting at the end of December, 2019, the COVID-19 epidemic grew rapidly outside Hubei and was characterised by a mixture of local transmission and case importations from Hubei (figure 1B). The median age of cases was 44 years (33–56), with the proportions of cases that were in age groups younger than 18 years (p<0·0001) and older than 64 years (p<0·0001) increasing from the earlier to the more recent time periods (appendix p 15). Overall, the proportion of cases among individuals younger than 18 years was low (5%; table 1 ). The proportion of male cases decreased between the two epidemic periods (p=0·00012), but remained around 50% (table 1; appendix p 15).
Table 1.
Characteristics of laboratory-confirmed COVID-19 cases in provinces outside Hubei in mainland China by epidemic period, as of Feb 17, 2020
| All (n=8579) | Period 1 (Dec 24 to Jan 27; n=4210) | Period 2 (Jan 28 to Feb 17; n=2379) | Unclassified*(n=1990) | ||||
|---|---|---|---|---|---|---|---|
| Age, years | 44 (33–56) | 43 (33–55) | 46 (33–58) | 44 (31–56) | |||
| Age group, years | |||||||
| 0–6 | 124/8579 (1%) | 28/4210 (1%) | 44/2379 (2%) | 52/1990 (3%) | |||
| 7–17 | 233/8579 (3%) | 65/4210 (2%) | 81/2379 (3%) | 87/1990 (4%) | |||
| 18–24 | 477/8579 (6%) | 248/4210 (6%) | 106/2379 (4%) | 123/1990 (6%) | |||
| 25–49 | 4127/8579 (48%) | 2147/4210 (51%) | 1082/2379 (45%) | 898/1990 (45%) | |||
| 50–64 | 2113/8579 (25%) | 990/4210 (24%) | 631/2379 (27%) | 492/1990 (25%) | |||
| ≥65 | 1002/8579 (12%) | 394/4210 (9%) | 373/2379 (16%) | 235/1990 (12%) | |||
| Sex | |||||||
| Female | 3943/8579 (46%) | 1841/4210 (44%) | 1163/2379 (49%) | 939/1990 (47%) | |||
| Male | 4401/8579 (51%) | 2258/4210 (54%) | 1166/2379 (49%) | 977/1990 (49%) | |||
| Exposure history | |||||||
| Presence of at least one exposure | 6611/8579 (77%)† | 3342/4210 (79%) | 1708/2379 (72%) | 1561/1990 (78%) | |||
| Exposure to animals and seafood markets, or wild animals | 34/6611 (1%) | 23/3342 (1%) | 7/1708 (0%) | 4/1561 (0%) | |||
| Exposure to COVID-19 cases or patients with acute respiratory infections | 2978/6611 (45%) | 855/3342 (26%) | 1125/1708 (66%) | 998/1561 (64%) | |||
| Exposure to Wuhan or Hubei | 3672/6611 (56%) | 2428/3342 (73%) | 591/1708 (35%) | 653/1561 (42%) | |||
| Residence in Wuhan‡ | 1602/3672 (44%) | 1135/2428 (47%) | 216/591 (37%) | 251/653 (38%) | |||
| Visited Wuhan | 577/3672 (16%) | 413/2428 (17%) | 96/591 (16%) | 68/653 (10%) | |||
| Residence in Hubei or visit Hubei§ | 519/3672 (14%) | 222/2428 (9%) | 153/591 (26%) | 144/653 (22%) | |||
Data are median (IQR), or n/n (%). Lower denominators indicate missing data, excluded from the analysis. Percentages might not total 100% because of rounding. COVID-19=coronavirus disease 2019.
We defined two time periods as the symptom onset date; if the date of symptom onset was missing and the date of official reporting, hospital visit, or outcome was earlier than or equal to Jan 27, 2020, cases were classified into period 1, otherwise, they were unclassified.
77% of records contained specific information on the exposure.
Wuhan residents or individuals who have lived in Wuhan for at least 14 days before symptom onset.
Hubei residents or individuals who visited Hubei, without specific city where they lived or visited.
The presence of at least one known exposure was reported by 6611 (77%) of 8579 cases (table 1). In the early epidemic period, most cases reported an exposure in Wuhan or Hubei. Subsequently, an increasing number of cases reported exposure to COVID-19 cases or patients with acute respiratory infections (p<0·0001; table 1).
The time from symptom onset to hospital admission shortened as the epidemic progressed, decreasing from 4·4 days (95% CI 0·0–14·0) during the first period of the epidemic to 2·6 days (0·0–9·0) in the second period (table 2 ). A similar decreasing time trend was observed for the interval from symptom onset to first health-care consultation (table 2).
Table 2.
Key time-to-event intervals for laboratory-confirmed COVID-19 cases by epidemic period, as of Feb 17, 2020
| All | Period 1 (Dec 24 to Jan 27) | Period 2 (Jan 28 to Feb 8)* | Difference†(95% CI), p value | |
|---|---|---|---|---|
| Time from symptom onset to first health-care consultation, days‡ | 2·5 (0·0–10·0), n=2888 | 3·0 (0·0–11·1), n=1836 | 1·6 (0·0–7·0), n=1052 | 1·4 (1·2–1·6), p<0·0001 |
| Time from symptom onset to hospital admission, days‡ | 3·8 (0·0–12·0), n=2001 | 4·4 (0·0–14·0), n=1310 | 2·6 (0·0–9·0), n=691 | 1·8 (1·5–2·1), p<0·0001 |
| Time from first health-care consultation to hospital admission, days‡ | 1·5 (0·0–9·0), n=1725§ | 1·4 (0·0–9·0); n=850 | 1·4 (0·0–9·0), n=353 | 0·0 (0·2–0·4), p=0·6551 |
| Time from symptom onset to official reporting, days‡ | 7·4 (1·0–18·0), n=5024§ | 8·9 (2·0–19·8), n=2727 | 5·4 (1·0–12·0), n=2079 | 3·5 (3·3–3·7), p<0·0001 |
Data are mean (95% CI), n, unless otherwise specified.
To account for reporting delays, we excluded the last 9 days of data (ie, data after Feb 8, 2020).
Difference between periods 1 and 2; Welch two-sample t test was used to calculate the p value.
Estimated from empirical data through complete-case analysis; the estimates obtained by fitting gamma, Weibull, and lognormal distributions are reported in the appendix (pp 15–17).
Sample size may be different from the sum of the two periods because it also includes cases without recorded date of symptom onset, which was used for classification of cases into temporal periods.
We analysed the time from exposure to illness onset for 49 cases with no travel history who were identified by prospective contact tracing. These cases represented 37 clusters. We estimated a mean incubation period of 5·2 days (95% CI 1·8–12·4), with the 95th percentile of the distribution at 10·5 days. The incubation period was well approximated by a lognormal distribution (appendix pp 17–19).
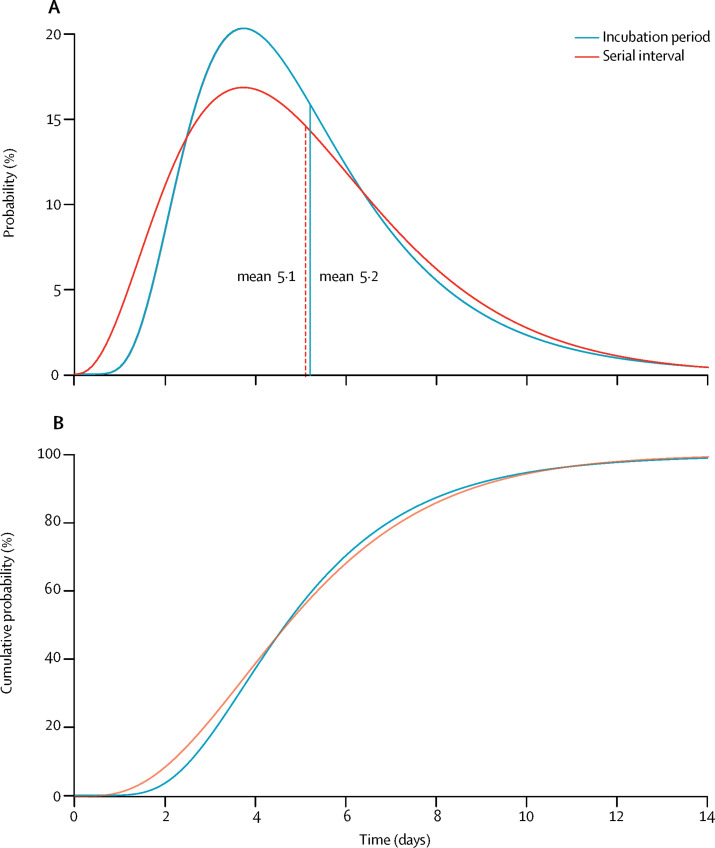
We analysed the time between symptom onset in 35 secondary cases and 28 corresponding primary cases (appendix pp 19–22). One case who reported the onset of symptoms on the same day as the index case was removed from the analysis. The serial interval followed a gamma distribution with an estimated mean of 5·1 days (95% CI 1·3–11·6). A comparison of the distribution of the incubation period and the serial interval is reported in figure 2 , and shows overlap between the two distributions. As we cannot exclude that a fraction of these secondary cases had a previous exposure to an unidentified infection source, we did a sensitivity analysis using different levels of data censoring; the resulting estimates were between 5·0 days (0·8–13·0) and 6·3 days (3·2–10·5; appendix p 22).
Figure 2.
Distributions of the incubation period and serial interval
(A) Comparison between the best-fitting distributions of the incubation period and of the serial interval; the vertical dashed lines represent the means of the two distributions. (B) Cumulative density function of the best-fitting distributions of the incubation period and the serial interval.
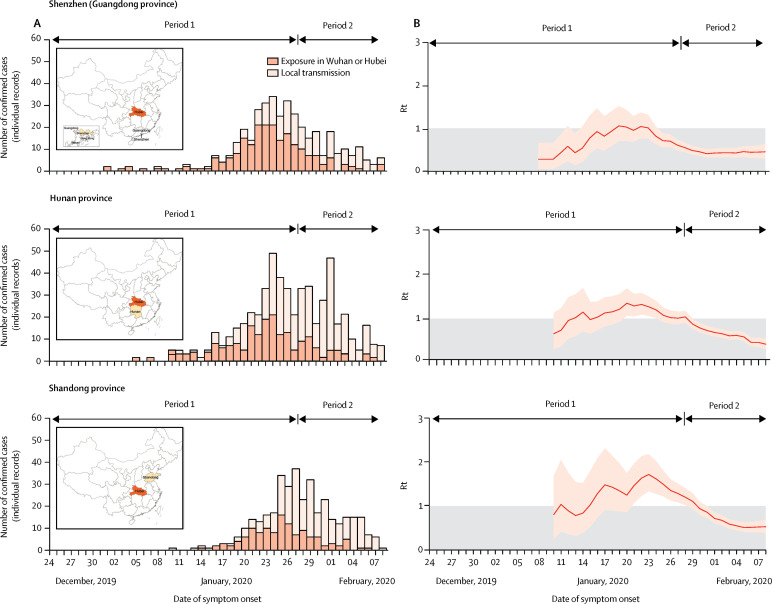
The transmission dynamics of COVID-19 differed between provinces outside Hubei. Here we report the results for a city (Shenzhen—a major city with more than 12 million inhabitants in Guangdong province) and two provinces (Hunan and Shandong) for which we have validated our individual records against the full official line lists (appendix pp 3–8). Results for the other six locations are included in the appendix (p 24). Although these locations were among those that reported the largest number of COVID-19 cases as of Feb 17, 2020,3 they showed highly different transmission patterns. Specifically, in Shenzhen, we estimated that the mean R t was above the epidemic threshold for about 1 week (approximately from Jan 18 to Jan 23), with a maximum value of 1·08 (95% CI 0·74–1·54) on Jan 22, and the outbreak was mostly sustained by cases with travel history to or from Wuhan or Hubei (figure 3 ). In Hunan province, we estimated the mean R t to be above the epidemic threshold for about 2 weeks from Jan 13 to Jan 28, with a peak value of 1·34 on Jan 20 (1·06–1·68; figure 3). Shandong province showed a slightly longer epidemic period than Hunan characterised by sustained local transmission and a larger peak mean R t of 1·71 on Jan 23 (1·32–2·17; figure 3). In these three locations, the mean R t steadily decreased and has remained below the epidemic threshold since the end of January, 2020. In general, we found that in all the analysed areas (among the most affected provinces outside Hubei), R t had decreased below 1·0 from Feb 8.
Figure 3.
Temporal dynamics of Rt in three Chinese locations outside Hubei
Period 1 ran from Dec 24 to Jan 27, and period 2 from Jan 28 to Feb 8. (A) Daily number of new cases in Shenzhen city (Guangdong province), Hunan province, or Shandong province, divided into cases with travel history to Wuhan or Hubei and cases resulting from local transmission. The insets show maps of China highlighting Hubei province (dark colour) and the analysed location (lighter colour). (B) Estimated Rt over a 4-day moving average. We excluded the last 9 days of data (ie, data after Feb 8) to account for reporting delays. We estimated the 90th percentile of the distribution of the time from onset to reporting in mainland China outside Hubei province during period 2 of the epidemic to be 9·0 days. Rt=net reproduction number.
Discussion
We have provided an assessment of the rapidly changing epidemiology and transmission dynamics of the COVID-19 outbreak beyond Hubei province in mainland China. We found significant differences in the epidemiology of COVID-19 as the epidemic continued to spread across China. We estimated that, as of Feb 8, 2020, R t has been below the epidemic threshold in the nine locations for which we had sufficient data coverage, which were among those with the largest number of reported cases (outside Hubei). This finding suggests that China has made key steps towards the interruption of COVID-19 transmission, owing to stringent containment measures, including isolation of cases, quarantine of contacts, and strict restrictions on personal movement (appendix pp 26–30), as well as to increased awareness and behavioural change of the population.
At the beginning of the epidemic, COVID-19 cases were mostly observed among elderly people.12 As the epidemic progressed, we observed a further increase in number of cases among people aged 65 years and older, as well as an increase among younger individuals (<18 years). Since Jan 28, 2020, however, the proportion of confirmed cases that are in people younger than 18 years is still only about 5%, although this age group represents approximately 20% of the Chinese population.13 From the data available here, it is not possible to ascertain whether younger individuals have a reduced risk of infection or an increased propensity for a milder clinical outcome of infection (thus resulting in a lower rate of detection). Schools in China were closed for most of the epidemic because of the 2020 Chinese New Year holidays;14 given studies15 showing that children record the largest number of contacts among all age groups on a regular weekday due to contacts at schools, it is possible that children were less exposed than normal during this time. It is unclear whether nationwide school holidays contributed to the low proportion of confirmed COVID-19 cases among school-age individuals and whether reopening of schools will lead to a change in the transmission patterns of COVID-19.16
At the beginning of the epidemic, a disproportionate fraction of COVID-19 cases were male.12 As of Feb 17, 2020, however, we observed about the same number of cases among men and women (51% of cases are male). This finding suggests either differential exposure by sex occurring at the beginning of the epidemic (most cases reported possible exposure to live markets because they were shop owners, or worked at or visited the live markets17, 18) or possible bias in the detection of the first few cases.
In provinces outside Hubei, we estimated the mean incubation period to be 5·2 days, in agreement with previous studies,5, 19, 20, 21 although estimates between 4·0 and 6·7 days have been reported.22, 23 The 95th percentile of the distribution (10·5 days) suggests contact tracing and medical observations of contacts of people with COVID-19 are important to detect individuals with a long incubation period. In these provinces, the mean serial interval was 5·1 days (95% CI 1·3–11·6). This estimate is considerably shorter than a previous estimate (7·5 days) derived from an analysis of six observations of the serial interval in Wuhan.5 Given that the basic reproduction number is positively associated with the growth rate of the epidemic and the length of the serial interval,8 the shorter mean serial interval estimated here implies that transmissibility might not be as high as initial estimates suggest. We cannot rule out, however, that the serial interval might differ between Wuhan and other Chinese provinces. The short estimate of the serial interval that we obtained might be linked to the short estimate for the interval from symptom onset to hospital admission (with a mean of about 2·6 days since Jan 28) for mainland China outside Hubei. A longer admission delay was reported in Wuhan in the early phase of the outbreak,5 which could drive longer serial intervals. It should be stressed also that serial interval estimates based on household clusters might be up to 20% shorter than the true value, as suggested by a theoretical study.10
We estimated that the serial interval was about as long as the incubation period, which is in overall agreement with an independent previous estimate.5 This finding suggests the possibility of an early peak of infectiousness, with possible transmission of SARS-CoV-2 before the onset of symptoms. If confirmed, the presence of relevant presymptomatic transmission might hamper control efforts, including contact tracing and timely isolation of index cases, as well as passenger screening at airports. By contrast, strategies based on social distancing, such as limiting mass gatherings and contact in the workplace and in schools, might still be effective.15, 24, 25 Evidence is accumulating to indicate the possibility of transmission during the incubation phase.25, 26, 27, 28, 29, 30, 31 It is worth noting also that our estimates of the incubation period and serial interval were based on analysis of cluster data, for which we assumed that the risk of infection from the community was negligible compared with that from infected individuals in the cluster. Although this assumption appears to be reasonable given the intensity of contact tracing, community transmission cannot be fully ruled out.
The results presented thus far support a change in epidemiological characteristics of the COVID-19 outbreak over time and as the epidemic expanded to multiple locations. Many of the key epidemiological time-delay distributions are different from those reported in studies focusing on the early transmission dynamics of COVID-19 in Wuhan.5 This difference is probably the consequence of the increased awareness of the public and physicians, behavioural changes of the population with respect to respiratory disease symptoms, increased health-care readiness, and elevated alert and response across mainland China.
We estimated that R t followed different patterns across China. We found that the epidemic was self-sustained for short periods of time only (no more than 3 weeks) in provinces outside Hubei that reported a large number of cases,32 and estimated that, since the end of January, 2020, R t has been below the epidemic threshold in all studied provinces. This finding is consistent with the gradual decrease in the number of detected COVID-19 cases reported across China and suggests a beneficial effect of the strict public health intervention policies implemented in China. In particular, strict social-distancing measures were implemented in all the analysed Chinese provinces and included close community management (eg, case isolation and household quarantine of close contacts), suspension of public activities, traffic restrictions, and school closure. A full list and timeline of the implemented interventions in each of the analysed locations is reported in the appendix (26–30). It should be stressed that the effectiveness of containment measures only applies while they are in place, and a relaxation of public health interventions or a substantial change in human behaviour might lead to a subsequent increase in transmission. Furthermore, our findings are based on analysis of nine of the most affected provinces in mainland China outside Hubei. Therefore, although unlikely, it is possible that other provinces reporting only a few cases could still have self-sustained outbreaks. Finally, the contribution of asymptomatic and presymptomatic infections to COVID-19 transmission remains unclear, as does the amount of asymptomatic or presymptomatic transmission captured in existing datasets.28, 29, 30, 31, 33, 34
This study is prone to the usual limitations pertaining to analysis of rapidly evolving infectious disease outbreaks, including biases due to case ascertainment, non-homogeneous sampling over time and by location, as well as hidden and unmodelled correlations. Thus, the uncertainty might be larger than estimated. Our estimates of R t are most robust, when assuming that under-reporting and the frequency of asymptomatic or presymptomatic transmission remain constant. Our estimates, however, are sensitive to marked changes in reporting rates and in the proportion of SARS-CoV-2 infections that lead to clinically identifiable symptoms. Individual records were retrieved from different data sources and thus might be affected by geographical differences in sampling of cases with specific exposures (eg, imported or locally acquired infections) and available dates of symptom onset. However, we compared our data for Shenzhen city, Hunan province, and Shandong province with official line lists and found the datasets to be of similar quality. It should also be stressed that the case definition has been changed several times since the start of the epidemic, and in particular, it was broadened on Jan 27. Therefore, because potentially more cases were identified since this change in case definition, our estimate of R t should be considered an upper bound (appendix pp 25, 26). This finding lends further support to our conclusion that R t has been decreasing since the end of January.
Despite these limitations, a real-time updated patient line list such as the one we have compiled is crucial to assess the epidemiology and transmission dynamics of an emerging pathogen, inform situational awareness, and optimise responses to the outbreak. Since Jan 20, 2020, the National Health Commission of China has incorporated COVID-19 as a notifiable disease.35 The Chinese Government has committed to timely disclosure of COVID-19 information, a decision highly praised by WHO.36 Accordingly, the local health commissions and the official data sources, from which our individual data originate, were authorised to release real-time information about epidemiological investigations on COVID-19 cases. The data collected for this analysis represent a valuable source of information and highlight the importance of publicly available records.
In conclusion, our study provides a detailed overview of the changing epidemiology and transmission dynamics of COVID-19 in mainland China outside Hubei province. Our findings suggest a slowing down of the COVID-19 outbreak in mainland China (outside Hubei province), indicating that the initial steps taken towards interruption of COVID-19 transmission might have been effective. However, the epidemic is not yet under control, and a large proportion of the population is still susceptible. The trajectory of the outbreak in China and beyond will depend on the effectiveness of control policies and human behaviour in the coming months.
Data sharing
The individual line list analysed in this study is available online.
Acknowledgments
Acknowledgments
We thank Wenkai Yang, Jingyuan Feng, Jialu Cheng, Qiuyi Xu, Haixin Ju, Xufang Bai, Zi Yu, Yumin Zhang, Wei Guo, Zeyao Zhao, Xin Chen, Sihong Zhao, Rong Du, Jiaxian Chen, Jiangnan Li, Geshu Zhang, Hong Peng, Xin Shen, Zeyu Li, and Yuheng Feng from Fudan University (Shanghai, China) for assisting with data collection. We thank Nicole Samay for her assistance in preparing the figures. HY acknowledges financial support from the National Science Fund for Distinguished Young Scholars (number 81525023), Key Emergency Project of Shanghai Science and Technology Committee (number 20411950100), and the National Science and Technology Major Project of China (numbers 2018ZX10201001–010, 2018ZX10713001–007, and 2017ZX10103009–005). MEH acknowledges financial support from the National Institute of General Medical Sciences (U54-GM111274). SM and MA acknowledge financial support from the European Commission Horizon 2020 MOOD project.
Editorial note: the Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Contributors
MA and HY designed the experiments. JZ, WW, YW, XD, XingC, MeL, WZ, XW, LY, XinhC, QW, and YL collected data. JZ, MaL, WW, YW, XD, and MA analysed data. JZ, MaL, WW, JY, KS, IML, MEH, PW, BJC, SM, CV, AV, MA, and HY interpreted the results. MA and HY wrote the Article. JZ, MaL, XW, JY, MEH, PW, BJC, CV, and AV edited and revised the Article.
Declaration of interests
BJC has received honoraria from Roche and Sanofi Pasteur. AV has received funding from Metabiota. HY has received research funding from Sanofi Pasteur, GlaxoSmithKline, Yichang HEC Changjiang Pharmaceutical Company, and Shanghai Roche Pharmaceutical Company. All other authors declare no competing interests.
Supplementary Material
References
- 1.Chinese Center for Disease Control and Prevention Epidemic update and risk assessment of 2019 novel coronavirus 2020. 2020. http://www.chinacdc.cn/yyrdgz/202001/P020200128523354919292.pdf
- 2.National Health Commission of the People's Republic of China Update on COVID-19 as of 24:00 on March 10, 2020. 2020. http://www.nhc.gov.cn/xcs/yqfkdt/202003/b4abcf83e53d4284b2981c75917385eb.shtml [DOI] [PMC free article] [PubMed]
- 3.World Health Organization Coronavirus disease 2019 (COVID-19) situation report–47. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200307-sitrep-47-covid-19.pdf?sfvrsn=27c364a4_2
- 4.World Health Organization Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV) 2020. https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov)
- 5.Li Q, Guan X, Wu P. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. published online Jan 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19)—China, 2020. China CDC Weekly. 2020;2:145–151. [PMC free article] [PubMed] [Google Scholar]
- 7.National Health Commission of the People's Republic of China Diagnosis and treatment guideline on pneumonia infection with 2019 novel coronavirus (5th trial edn) 2020. http://www.nhc.gov.cn/xcs/zhengcwj/202002/d4b895337e19445f8d728fcaf1e3e13a.shtml
- 8.Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc Biol Sci. 2007;274:599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization Ebola Response Team. Aylward B, Barboza P. Ebola virus disease in West Africa—the first 9 months of the epidemic and forward projections. N Engl J Med. 2014;371:1481–1495. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu QH, Ajelli M, Aleta A, Merler S, Moreno Y, Vespignani A. Measurability of the epidemic reproduction number in data-driven contact networks. Proc Natl Acad Sci USA. 2018;115:12680–12685. doi: 10.1073/pnas.1811115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson RN, Stockwin JE, van Gaalen RD. Improved inference of time-varying reproduction numbers during infectious disease outbreaks. Epidemics. 2019;29 doi: 10.1016/j.epidem.2019.100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen N, Zhou M, Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Bureau of Statistics China census in 2018. 2020. http://www.stats.gov.cn/tjsj/ndsj/2019/indexch.htm
- 14.Ministry of Education of the People's Republic of China Notice on the postponement of the spring semester of 2020. 2020. http://www.moe.gov.cn/jyb_xwfb/gzdt_gzdt/s5987/202001/t20200127_416672.html
- 15.Litvinova M, Liu QH, Kulikov ES, Ajelli M. Reactive school closure weakens the network of social interactions and reduces the spread of influenza. Proc Natl Acad Sci USA. 2019;116:13174–13181. doi: 10.1073/pnas.1821298116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Litvinova M, Liang Y. Age profile of susceptibility, mixing, and social distancing shape the dynamics of the novel coronavirus disease 2019 outbreak in China. medRχiv. 2020 doi: 10.1101/2020.03.19.20039107. published online March 20. (preprint). [DOI] [Google Scholar]
- 17.Huang C, Wang Y, Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu P, Hao X, Lau EHY. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Euro Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.5.2000062. published online Feb 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linton NM, Kobayashi T, Yang Y. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J Clin Med. 2020;9:e538. doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauer SA, Grantz KH, Bi Q. The Incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020 doi: 10.7326/M20-0504. published online March 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan WJ, Ni ZY, Hu Y. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. published online Feb 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian S, Hu N, Lou J. Characteristics of COVID-19 infection in Beijing. J Infect. 2020 doi: 10.1016/j.jinf.2020.02.018. published online Feb 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowling BJ, Lau EHY, Lam CLH. Effects of school closures, 2008 winter influenza season, Hong Kong. Emerg Infect Dis. 2008;14:1660–1662. doi: 10.3201/eid1410.080646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ministry of Culture and Tourism of the People's Republic of China Notice on extending the spring festival holiday of 2020. 2020. https://www.mct.gov.cn/whzx/whyw/202001/t20200127_850576.htm
- 26.Yu P, Zhu J, Zhang Z, Han Y, Huang L. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person-to-person transmission during the incubation period. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa077. published online Feb 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou L, Ruan F, Huang M. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020 doi: 10.1056/NEJMc2001737. published online Feb 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao S, Gao D, Zhuang Z. Estimating the serial interval of the novel coronavirus disease (COVID-19): a statistical analysis using the public data in Hong Kong from January 16 to February 15, 2020. medRχiv. 2020 doi: 10.1101/2020.02.21.20026559. published online Feb 21. (preprint). [DOI] [Google Scholar]
- 29.Tong ZD, Tang A, Li KF. Potential presymptomatic transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg Infect Dis. 2020;26 doi: 10.3201/eid2605.200198. published online May 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Z, Xu X, Wu Y, Wang L, Cowling BJ, Meyers LA. The serial interval of COVID-19 from publicly reported confirmed cases. medRχiv. 2020 doi: 10.1101/2020.02.19.20025452. published online March 13. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tindale L, Coombe M, Stockdale JE. Transmission interval estimates suggest pre-symptomatic spread of COVID-19. medRiv. 2020 doi: 10.1101/2020.03.03.20029983. published online March 6. (preprint). [DOI] [Google Scholar]
- 32.Kucharski AJ, Russell TW, Diamond C. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30144-4. published online March 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of 2019 novel coronavirus onboard the Princess Cruises ship, 2020. medRχiv. 2020 doi: 10.1101/2020.02.20.20025866. published online March 6: (preprint). [DOI] [Google Scholar]
- 34.Bai Y, Yao L, Wei T. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.2565. published online Feb 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Health Commission of the People's Republic of China Inclusion of 2019 novel coronavirus diseases (COVID-19) into statutory infectious disease management. 2020. http://www.nhc.gov.cn/jkj/s7916/202001/44a3b8245e8049d2837a4f27529cd386.shtml
- 36.McNeice A. WHO chief praises containment efforts. 2020. http://www.chinadaily.com.cn/a/202001/31/WS5e3335fba310128217273bff.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The individual line list analysed in this study is available online.