Abstract
Accumulating studies have demonstrated serum exosomal microRNAs (miRNAs) represent novel biomarkers for various diseases. In this study, we aimed to explore the feasibility of using serum exosomal miRNAs as novel serological biomarkers for steroid-induced osteonecrosis of femoral head (SONFH). We identified the characters of exosomes which were obtained from fresh serum of 5 systemic lupus erythematosus (SLE) patients without SONFH, 5 SLE patients with SONFH (SLE-SONFH) and 5 healthy ones. Comprehensive exosomal miRNA sequencing was performed to profile the differentially expressed miRNAs in the three groups. We then validated the expression levels of selected miRNAs by qRT-PCR. Furthermore, KEGG pathway, GO annotation, protein-protein interaction (PPI) network, module analysis and miRNAs-mRNAs interaction network were built to analyze the potential targets and mechanism. Sequencing data conveyed that hsa-miR-135b-5p, hsa-miR-150-5p, hsa-miR-509-3-5p, hsa-miR-514a-3p and hsa-miR-708-5p were significantly differentially expressed in the three groups. The results of qRT-PCR for the first time confirmed that the expression of hsa-miR-135b-5p was strikingly up-regulated in SLE-SONFH group which were consistent with miRNA sequencing results. In addition, bioinformatics analysis indicated that the enriched functions and pathways of the most differentially expressed miRNAs including Wnt, MAPK as well as Hippo signaling pathway. The top five hub genes (FGF2, PTEN, HACE1, VAMP2, and CBL) were part of module of the PPI network, which consisted of 713 nodes and 2191 edges. In conclusion, this study provides a novel and fundamental serum exosomal miRNAs profile of SONFH and hsa-miR-135b-5p may be identified as a unique diagnostic biomarker for SONFH.
Keywords: Steroid-induced osteonecrosis of the femoral head (SONFH), serum exosomal miRNAs, hsa-miR-135b-5p, biomarkers
Introduction
Steroid-induced osteonecrosis of the femoral head (SONFH) is characterized by collapse of the femoral head, severe osteoarthritis and unavoidable artificial joint arthroplasty [1,2]. Diagnosis and intervention at the early stages (Association Research Circulation Osseous [ARCO] stages I and II) not only play an imperative roles in protecting femoral head collapse, but are also essentials for successful treatment of SONFH [3,4]. Although early-stage SONFH can be conclusively diagnosed by magnetic resonance imaging (MRI), the cost and inconvenience of MRI pose a heavy burden on the patients and healthcare system, especially in China [5,6]. Therefore, it is urgent to develop an economical and convenient laboratory test for initial screening and monitoring of SONFH development among high-risk SONFH patients who are receiving steroid therapy.
Exosomes are described as nanosized (30-150 nm) membrane vesicles released from multiple cells into the extracellular environment, and they remain stable in biological fluids such as blood, urine, and saliva [7]. Recent reports have shown presence of several nucleic acids in the exosomal lumen, including mRNAs, microRNAs (miRNAs), and long-non-coding RNAs [8,9]. Furthermore, exosomal miRNAs have been proven to remain stable in blood due to the ability of exosomes to protect the structure and function of miRNAs against degradation by an RNase [10]. Therefore, specific exosomal miRNAs in serum may represent diagnostic or prognostic biomarkers in numerous diseases [11-13]; however, the role of serum exosomal miRNAs in SONFH has not yet been reported.
The aim of our study was to investigate the feasibility of using serum exosomal miRNAs as novel serological biomarkers for the diagnosis of SONFH. We identified differentially expressed miRNAs and predicted their functions and pathways by conducting miRNA sequencing (miRNA-seq) of samples from patients with systemic lupus erythematosus (SLE), SLE-SONFH, and healthy participants. We then verified the findings of miRNA-seq by qRT-PCR and identified hsa-miR-135b-5p as a potential serological biomarker for the diagnosis of SONFH.
Materials and methods
Patients
In this study, a total of 66 individuals (22 SLE patients who underwent similar steroid treatment but without secondary SONFH, 27 SLE-SONFH patients, and 17 healthy controls) were recruited from the First Affiliated Hospital of Guangzhou University of Chinese Medicine between August 2017 and October 2017. Serum exosomes from 5 SLE patients, 5 SLE-SONFH patients, and 5 healthy individuals were subjected to miRNA sequencing. Another cohort of 17 SLE patients, 22 SLE-SONFH patients, and 12 healthy individuals were subjected to further qRT-PCR verification. Diagnosis standards for SLE and SONFH were established as described previously [14,15]. This study was reviewed and approved by the Ethics Committee of Guangzhou University of Chinese Medicine (No. ZYYECK [2017]033). The study was performed in accordance with the guidelines of the Declaration of Helsinki. Written informed consent was obtained from each participant prior to recruitment.
Isolation of exosomes from serum
Peripheral blood samples were collected and an initial spin was performed at 3,000 g for 15 min at 4°C to remove possible cellular debris. The supernatants were transferred to labeled tubes and stored at -80°C for further analysis. Serum exosomes were prepared using ExoQuick® Exosome Isolation and RNA Purification Kit (for Serum & Plasma, Cat. EQ806A-1, SBI, CA, USA) according to the manufacturer’s instructions. Briefly, the frozen serum samples were thawed rapidly and centrifuged at 12,000 g for 10 min at 4°C. Then 500 μL of supernatant was mixed thoroughly with 120 μL ExoQuick solution and kept on ice for 30 min. The mixture was centrifuged at 1,500 g for 30 min at 4°C. The exosome pellet remained at the bottom of the tube after the supernatant was discarded. Finally, the exosome pellets were resuspended in 1× phosphate-buffered saline (PBS) or 350 μL phenol-free lysis buffer for further experiments.
Transmission electron microscopy (TEM)
Morphological features of the isolated exosomes were visualized using TEM, to confirm the vesicular shape of exosomes and to obtain an estimate of diameter [16]. Briefly, the freshly isolated exosomes were resuspended in 1× PBS, and 10 μL of the suspension was pipetted onto a 400 mesh copper grid with carbon-coated formvar film and incubated for 2 min. The excess liquid was drained from the edge of the grid by blotting. Subsequently, the grid was carefully stained with a drop of 2% aqueous solution of phosphotungstic acid for approximately 2 min and followed by blotting to drain excessive liquid. Exosome images were acquired using a TEM with an accelerating voltage of 80 kV.
Nanoparticle-tracking analysis (NTA)
Particle size distribution and relative concentration of purified exosomes were determined by NTA using a NanoSight Technology NS300 Instrument equipped with a 405 nm laser and a syringe pump [17]. Briefly, exosomes were diluted in particle-free PBS and evenly mixed to achieve an acceptable concentration (ranging from 106 to 109 particles/mL). Next, an approximately 0.3 mL mixture was loaded into the NanoSight sample chamber through the syringe. The Brownian motion of the particles was recorded and analyzed using NTA 2.1 software.
Flow cytometry analysis of the exosomes
Flow cytometry was used to detect molecules on the surface of exosomes [18]. Exosomal proteins were extracted and dissolved in a lysis buffer, and the protein concentration was determined in triplicate using the Bradford assay. The samples were suspended in PBS and stained with antibodies. The expression of two principal exosomal markers, CD63 and CD81, were identified according to the instrument’s operation procedures (BD AccuriTM C6 flow cytometer). Anti-CD63 (BD 557288) and anti-CD81 (BD 551108) antibodies were used for exosome detection in this study.
RNA extraction and miRNA sequencing
Based on the manufacturer’s instructions (ExoQuick® Exosome Isolation and RNA Purification Kit for Serum & Plasma, Cat. EQ806A-1, SBI, CA, USA), 350 μL phenol-free lysis buffer was added to resuspend the exosome pellets. For further purification, 200 μL of 100% ethanol was added to the resuspended exosomes; the solution was then transferred to a spin column and centrifuged at 13,000 rpm for 1 min. Next, the flow-through was diacard, 200 μL of wash buffer was added, and the preparations were centrifuged at 13,000 rpm for 1 min. The wash step was repeated one more time. To elute the exosomal RNA, 30 μL of elution buffer was added directly onto the membrane of the spin column. Next, the samples were centrifuged first at 2,000 rpm for 2 min, and then at 13,000 rpm for 1 min. The quantity and purity of all samples were assessed using NanoDrop spectrophotometer and Agilent Bioanalyzer. Sequencing analysis was performed using Illumina HiSeq 2500 platform. Sequencing data showing a fold change > 1.5 and P < 0.05 were regarded as differentially expressed. Raw sequence reads are available if required.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
The database for comprehensive functional annotations of the identified target genes was applied to investigate GO (http://www.geneontology.org/) enrichment and KEGG (http://www.genome.jp/) signaling pathways.
Protein-protein interaction network construction and app analysis
The target genes regulated by candidate miRNAs were mapped to the STRING (http://string.embl.de/) database to construct the protein-protein interaction (PPI) network. A combined score > 0.4 was set as the threshold for significant protein pairs. Subsequently, PPI networks were visualized and analyzed using Cytoscape3.6.0 software. Next, molecular complex detection (MCODE) was used to identify significant modules of the PPI network with degree cutoff ≥ 2, node score cutoff ≥ 0.2, κ-core ≥ 2, and max depth =100. Finally, hub protein nodes of the PPI network with the degree algorithm of cytoHubba were identified.
Analysis of miRNAs-mRNAs interaction
The target genes of candidate miRNAs were predicted using TargetScan (http://www.targetscan.org), miRDB (http://mirdb.org), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw), and miRWalk (http://zmf.umm.uni-heidelberg.de). Specifically, only target genes that were recorded in all four databases were selected as the ultimate target genes. The regulatory networks of candidate miRNAs and their target genes were constructed by using the open-source software Cytoscape3.6.0.
Validation of exosomal miRNA expression by qRT-PCR
To validate the miRNA sequencing results, the selected candidate exosomal miRNAs were further examined individually by qRT-PCR in all three groups. Fifty one exosomal RNA samples (12 from healthy controls, 22 from SLE-SONFH patients, and 17 from SLE patients) were reverse transcribed and quantified using the Bulge-Loop™ miRNA qRT-PCR Starter Kit (RIBOBIO, Guangzhou, China) following the manufacturer’s instructions. U6 was used as an internal normalization control. The thermocycling conditions for PCR were started with initial denaturation at 95°C for 20 s, followed by 40 cycles of denaturation at 95°C denaturation for 10 s, annealing at 60°C for 20 s, extension at 70°C for 10 s, and melt curve stage to confirm primer specificity. The data were analyzed using the ΔCt method to calculate the relative expression levels of miRNAs from three independent experiments. The primer sequences are listed in Table 1.
Table 1.
The top five miRNAs were selected to qRT-PCR validation
| miRNA ID | Forward 5’-3’ | RT Primer |
|---|---|---|
| hsa-miR-135b-5p | CGCGTATGGCTTTTCATTCCT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACAT |
| hsa-miR-150-5p | GCGTCTCCCAACCCTTGTA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACTGG |
| hsa-miR-509-3-5p | GCGTACTGCAGACGTGGCA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCATGAT |
| hsa-miR-514a-3p | GCGCGATTGACACTTCTGTG | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCTACT |
| hsa-miR-708-5p | GCGCGAAGGAGCTTACAATCTA | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCAGC |
| U6 | AGAGAAGATTAGCATGGCCCCTG | ATCCAGTGCAGGGTCCGAGG |
Statistical analysis
All data were averaged and presented as the mean ± SD from three independent experiments and analyzed by using the SPSS 18.0 software. The differences of miRNAs expression levels among the healthy controls, SLE and SLE-SONFH groups were detected by one-way analysis of variance. P-values less than 0.05 was considered statistically significant.
Results
Characteristics of exosomes in serum
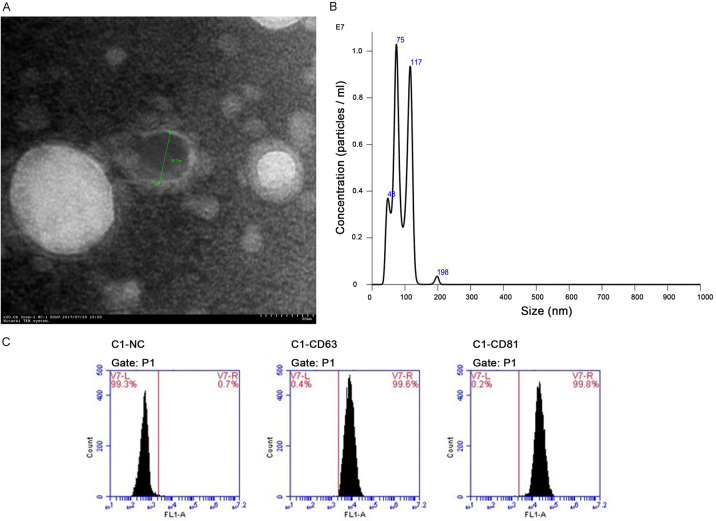
To confirm that the particles isolated from serum were indeed exosomes, the vesicles were characterized by TEM, NanoSight and flow cytometry analysis. TEM images revealed that the diameter of the serum exosomes ranged from 50 to 100 nm, corresponding to the conventional size range of exosomes (Figure 1A). The NTA measurements showed that the isolated serum exosomes had a predominant size range of 75-117 nm (Figure 1B). As expected, flow cytometry analysis demonstrated that CD63 and CD81, two commonly used exosomal markers, were abundantly expressed in the isolated particles (Figure 1C). This evidence clearly showed the main characteristics of exosomes, confirming the successful isolation of exosomes from serum samples.
Figure 1.
Characteristics of isolated exosomes from serum samples. A. Morphology of serum-derived exosome was visualized by TEM, indicating the diameter of isolated exosome in 50-100 nm. B. Size distribution of serum-derived exosome was analyzed using NTA, which were most abundant in 75-117 nm. C. The exosome-specific proteins CD63 and CD81 were detected in the serum exosomes by flow cytometry analysis.
Differential expression profiles of miRNAs in serum exosomes
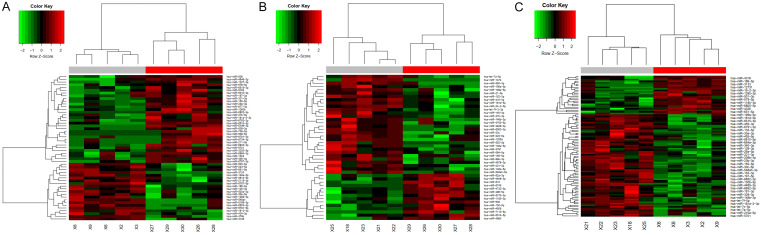
A total of 2,589 miRNAs were identified in human serum exosomes after normalization of the expression profiling data. With a threshold of absolute value of fold change > 4 and P < 0.01, differentially expressed miRNAs could be distinguished between the three groups. In our study, the results showed that there were 35 differentially abundant miRNAs (6 upregulated and 29 downregulated) between the healthy group and SLE group, 25 differentially abundant miRNAs (15 upregulated and 10 downregulated) between the healthy group and SLE-SONFH group, and 39 differentially abundant miRNAs (27 upregulated and 12 downregulated) between the SLE group and SLE-SONFH group. Detailed information about differentially abundant miRNAs is summarized in Tables 2, 3 and 4. Hierarchical clustering was carried out to show statistically significant miRNAs, with up-regulated miRNAs represented in red and down-regulated miRNAs shown in green (Figure 2A-C).
Table 2.
Differentially abundant miRNAs between the healthy controls and SLE group
| MiRNA | Gene ID | P-value | Log2 (fold change) | Deregulation |
|---|---|---|---|---|
| hsa-miR-514a-3p | MIMAT0002883 | 8.61E-06 | -8.5516 | down |
| hsa-miR-508-3p | MIMAT0002880 | 9.21E-06 | -8.9729 | down |
| hsa-miR-506-3p | MIMAT0002878 | 1.37E-05 | -11.3567 | down |
| hsa-miR-509-3-5p | MIMAT0004975 | 1.58E-05 | -7.7828 | down |
| hsa-miR-150-5p | MIMAT0000451 | 1.61E-05 | -2.4691 | down |
| hsa-miR-509-5p | MIMAT0004779 | 4.51E-05 | -9.2071 | down |
| hsa-miR-6514-3p | MIMAT0025485 | 5.71E-05 | -3.7636 | down |
| hsa-miR-202-5p | MIMAT0002810 | 0.00022039 | -5.3795 | down |
| hsa-miR-151a-5p | MIMAT0004697 | 0.000434535 | -3.8707 | down |
| hsa-miR-151b | MIMAT0010214 | 0.000492571 | -3.4268 | down |
| hsa-miR-877-3p | MIMAT0004950 | 0.000514125 | -2.2556 | down |
| hsa-miR-5189-3p | MIMAT0027088 | 0.000562383 | 3.2721 | up |
| hsa-miR-206 | MIMAT0000462 | 0.000925783 | -3.5403 | down |
| hsa-miR-514a-5p | MIMAT0022702 | 0.001790034 | -7.7937 | down |
| hsa-miR-1229-3p | MIMAT0005584 | 0.002640975 | -2.4973 | down |
| hsa-miR-509-3p | MIMAT0002881 | 0.002841417 | -3.8138 | down |
| hsa-miR-514b-3p | MIMAT0015088 | 0.002847948 | -7.467 | down |
| hsa-miR-3200-5p | MIMAT0017392 | 0.00297655 | -2.4266 | down |
| hsa-miR-4646-3p | MIMAT0019708 | 0.002981539 | -2.591 | down |
| hsa-miR-490-3p | MIMAT0002806 | 0.00301346 | 3.3147 | up |
| hsa-miR-23b-3p | MIMAT0000418 | 0.003520948 | -1.667 | down |
| hsa-miR-211-5p | MIMAT0000268 | 0.003738115 | -4.7785 | down |
| hsa-miR-6802-3p | MIMAT0027505 | 0.004947436 | -6.3743 | down |
| hsa-miR-223-5p | MIMAT0004570 | 0.005369297 | 1.2621 | up |
| hsa-miR-483-3p | MIMAT0002173 | 0.005698786 | -2.8201 | down |
| hsa-miR-2115-3p | MIMAT0011159 | 0.005869434 | 1.6358 | up |
| hsa-miR-202-3p | MIMAT0002811 | 0.005942291 | -3.4083 | down |
| hsa-miR-5196-3p | MIMAT0021129 | 0.006206503 | -6.3102 | down |
| hsa-miR-6772-3p | MIMAT0027445 | 0.006646644 | -2.2905 | down |
| hsa-miR-550a-3p | MIMAT0003257 | 0.006963959 | -2.1157 | down |
| hsa-miR-365b-3p | MIMAT0022834 | 0.007487356 | -1.9136 | down |
| hsa-miR-365a-3p | MIMAT0000710 | 0.007514356 | -1.9136 | down |
| hsa-miR-4433b-5p | MIMAT0030413 | 0.008119829 | -1.7364 | down |
| hsa-miR-4482-3p | MIMAT0020958 | 0.008706458 | 5.686 | up |
| hsa-miR-4535 | MIMAT0019075 | 0.009086014 | 3.3337 | Up |
Abbreviations: SLE, systemic lupus erythematosus.
Table 3.
Differentially abundant miRNAs between the healthy controls and SLE-SONFH group
| MiRNA | Gene location | P-value | Log2 (fold change) | Deregulation |
|---|---|---|---|---|
| hsa-miR-200b-3p | MIMAT0000318 | 0.000142774 | 4.7488 | up |
| hsa-miR-1255a | MIMAT0005906 | 0.000725732 | 3.8131 | up |
| hsa-miR-508-3p | MIMAT0002880 | 0.000745767 | -5.1503 | down |
| hsa-miR-429 | MIMAT0001536 | 0.000756843 | 4.0216 | up |
| hsa-miR-200c-3p | MIMAT0000617 | 0.000983032 | 3.4799 | up |
| hsa-miR-202-5p | MIMAT0002810 | 0.00126593 | -4.4991 | down |
| hsa-miR-151b | MIMAT0010214 | 0.001293991 | -3.3583 | down |
| hsa-miR-30c-2-3p | MIMAT0004550 | 0.001510032 | 2.9678 | up |
| hsa-miR-4483 | MIMAT0019017 | 0.001756596 | 6.9071 | up |
| hsa-miR-200a-5p | MIMAT0001620 | 0.001778037 | 3.4564 | up |
| hsa-miR-147b | MIMAT0004928 | 0.002218591 | 6.9167 | up |
| hsa-miR-4508 | MIMAT0019045 | 0.00327728 | -1.6615 | down |
| hsa-miR-151a-5p | MIMAT0004697 | 0.003683284 | -2.9793 | down |
| hsa-miR-506-3p | MIMAT0002878 | 0.003739407 | -5.5287 | down |
| hsa-miR-4521 | MIMAT0019058 | 0.004652257 | 3.7125 | up |
| hsa-miR-514a-5p | MIMAT0022702 | 0.004665005 | -4.3382 | down |
| hsa-miR-6777-3p | MIMAT0027455 | 0.005366505 | -2.0024 | down |
| hsa-miR-514a-3p | MIMAT0002883 | 0.007350538 | -5.0629 | down |
| hsa-miR-1250-5p | MIMAT0005902 | 0.008026723 | 1.9886 | up |
| hsa-miR-7113-5p | MIMAT0028123 | 0.008277439 | -6.1471 | down |
| hsa-miR-3065-5p | MIMAT0015066 | 0.008752663 | 2.7963 | up |
| hsa-miR-455-5p | MIMAT0003150 | 0.009208877 | 2.0743 | up |
| hsa-miR-199b-5p | MIMAT0000263 | 0.00935067 | 2.185 | up |
| hsa-miR-548ah-5p | MIMAT0018972 | 0.009661312 | 5.7675 | up |
| hsa-miR-708-5p | MIMAT0004926 | 0.009857857 | 4.164 | Up |
Abbreviations: SLE-SONFH, systemic lupus erythematosus with osteonecrosis of the femoral head.
Table 4.
Differentially abundant miRNAs between the SLE group and SLE-SONFH group
| MiRNA | Gene location | P-value | Log2 (fold change) | Deregulation |
|---|---|---|---|---|
| hsa-miR-455-3p | MIMAT0004784 | 4.70E-05 | 4.8643 | up |
| hsa-miR-4485-3p | MIMAT0019019 | 5.57E-05 | 3.9998 | up |
| hsa-miR-135b-5p | MIMAT0000758 | 6.79E-05 | 10.4639 | up |
| hsa-miR-30c-2-3p | MIMAT0004550 | 8.77E-05 | 3.796 | up |
| hsa-miR-4483 | MIMAT0019017 | 0.000173848 | 10.0754 | up |
| hsa-miR-708-5p | MIMAT0004926 | 0.00017683 | 10.0678 | up |
| hsa-miR-6501-5p | MIMAT0025458 | 0.00033296 | -2.6116 | down |
| hsa-miR-23b-3p | MIMAT0000418 | 0.000857341 | 2.142 | up |
| hsa-miR-193b-3p | MIMAT0002819 | 0.001526017 | 3.1634 | up |
| hsa-miR-455-5p | MIMAT0003150 | 0.001702779 | 2.505 | up |
| hsa-miR-4467 | MIMAT0018994 | 0.001794009 | -2.3038 | down |
| hsa-miR-4535 | MIMAT0019075 | 0.002149908 | -4.4279 | down |
| hsa-miR-4521 | MIMAT0019058 | 0.002362243 | 4.1367 | up |
| hsa-miR-3176 | MIMAT0015053 | 0.002693648 | -1.6141 | down |
| hsa-miR-31-5p | MIMAT0000089 | 0.002932769 | 4.1642 | up |
| hsa-miR-509-3-5p | MIMAT0004975 | 0.003067103 | 3.6547 | up |
| hsa-miR-4669 | MIMAT0019749 | 0.003094169 | -3.9558 | down |
| hsa-miR-122-3p | MIMAT0004590 | 0.003171144 | 4.0524 | up |
| hsa-miR-651-5p | MIMAT0003321 | 0.003457186 | -1.3648 | down |
| hsa-miR-200b-3p | MIMAT0000318 | 0.003587793 | 3.615 | up |
| hsa-miR-6772-3p | MIMAT0027445 | 0.003860249 | 2.7274 | up |
| hsa-miR-200c-3p | MIMAT0000617 | 0.003871634 | 3.0423 | up |
| hsa-miR-885-5p | MIMAT0004947 | 0.003878096 | 3.9627 | up |
| hsa-miR-514a-3p | MIMAT0002883 | 0.003902225 | 3.4887 | up |
| hsa-miR-4508 | MIMAT0019045 | 0.004279357 | -1.5857 | down |
| hsa-miR-483-3p | MIMAT0002173 | 0.004878581 | 3.1705 | up |
| hsa-miR-365a-3p | MIMAT0000710 | 0.005356032 | 2.2475 | up |
| hsa-miR-365b-3p | MIMAT0022834 | 0.005359588 | 2.2475 | up |
| hsa-miR-615-3p | MIMAT0003283 | 0.005401801 | 2.6966 | up |
| hsa-miR-214-3p | MIMAT0000271 | 0.00570578 | 2.6142 | up |
| hsa-miR-6805-5p | MIMAT0027510 | 0.006028297 | -1.8917 | down |
| hsa-miR-34a-5p | MIMAT0000255 | 0.006209209 | 3.4157 | up |
| hsa-miR-4326 | MIMAT0016888 | 0.006807003 | -1.3048 | down |
| hsa-miR-3188 | MIMAT0015070 | 0.008157864 | 3.2567 | up |
| hsa-miR-5096 | MIMAT0020603 | 0.008577331 | -2.4489 | down |
| hsa-miR-6715a-3p | MIMAT0025841 | 0.008599554 | 3.9059 | up |
| hsa-miR-145-5p | MIMAT0000437 | 0.00865197 | 2.4039 | up |
| hsa-miR-4492 | MIMAT0019027 | 0.00906535 | -1.6476 | down |
| hsa-miR-1285-3p | MIMAT0005876 | 0.009112779 | -1.0781 | Down |
Abbreviations: SLE, systemic lupus erythematosus; SLE-SONFH, systemic lupus erythematosus with osteonecrosis of the femoral head.
Figure 2.
Heatmap showing differentially abundant miRNAs. A. Hierarchical cluster analysis of the differentially expressed miRNAs between the healthy controls and SLE group. B. Hierarchical cluster analysis of the differentially expressed miRNAs between the healthy controls and SLE-SONFH group. C. Hierarchical cluster analysis of the differentially expressed miRNAs between the SLE and SLE-SONFH group. Red strip indicates relatively high expression, and green strip indicates relatively low expression.
GO and KEGG pathway enrichment
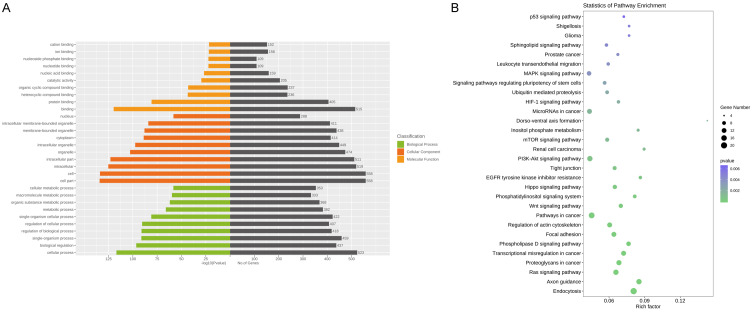
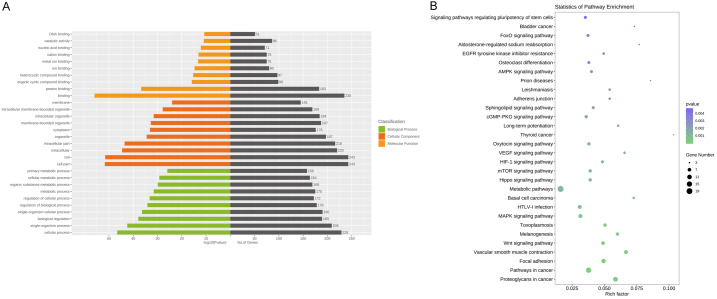
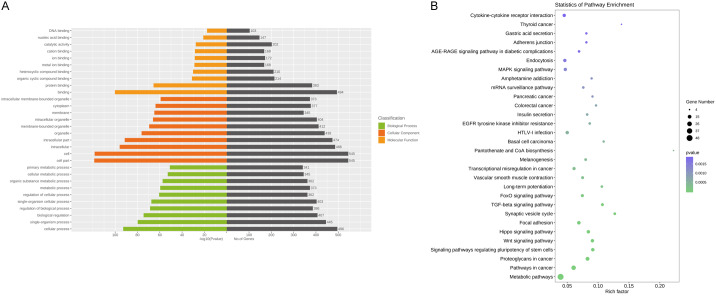
Following the miRNA-seq analysis, five most significantly expressed miRNAs (hsa-miR-135b-5p, hsa-miR-150-5p, hsa-miR-509-3-5p, hsa-miR-514a-3p, and hsa-miR-708-5p) were selected as candidate miRNAs based on the fold change ranking among the three groups. Target genes were further predicted by TargetScan, miRDB, miRTarBase and miRWalk. Furthermore, GO annotation and KEGG pathway enrichment analyses were performed using Gene Ontology and KEGG pathway databases to explore the potential targets of these five selected exosomal miRNAs. As shown in Figure 3A, 3B, candidate target genes in the normal and SLE group mainly involved in GO terms about cell (P=7.05×10-135), cell part (P=1.59×10-134) and KEGG pathway about Ras signaling pathway (P=7.34×10-6). In the normal and SLE-SONFH groups, the GO terms primarily focused on binding (P=1.16×10-56). Meanwhile, the Wnt signaling pathway (P=1.23×10-4) and MAPK signaling pathway (P=7.47×10-4) were found to be the most significantly involved pathways in SONFH pathogenesis (Figure 4A, 4B). Biomolecular information and pathways in SLE and SLE-SONFH groups showed that GO terms were closely related to cell part (P=1.74×10-119) and cell (P=3.76×10-119) (Figure 5A, 5B). The SONFH molecular pathogenic pathway in KEGG primarily involved the Wnt signaling pathway (P=1.53×10-6) and Hippo signaling pathway (p=3.27×10-6).
Figure 3.
Functional and pathway enrichment analysis of the different miRNA co-expression genes in healthy controls and SLE group. A. Based on enrichment score, GO terms were divided into biological process, cellular component and molecular function. Bar graph displayed GO enrichment results with candidate gene numbers. B. Scatterplot of enriched KEGG pathway showing statistics of pathway enrichment.
Figure 4.
Functional and pathway enrichment analysis of the different miRNA co-expression genes in healthy controls and SLE-SONFH group. A. Based on enrichment score, GO terms were divided into biological process, cellular component and molecular function. Bar graph displayed GO enrichment results with candidate gene numbers. B. Scatterplot of enriched KEGG pathway showing statistics of pathway enrichment.
Figure 5.
Functional and pathway enrichment analysis of the different miRNA co-expression genes in SLE and SLE-SONFH group. A. Based on enrichment score, GO terms were divided into biological process, cellular component and molecular function. Bar graph displayed GO enrichment results with candidate gene numbers. B. Scatterplot of enriched KEGG pathway showing statistics of pathway enrichment.
PPI network and modules
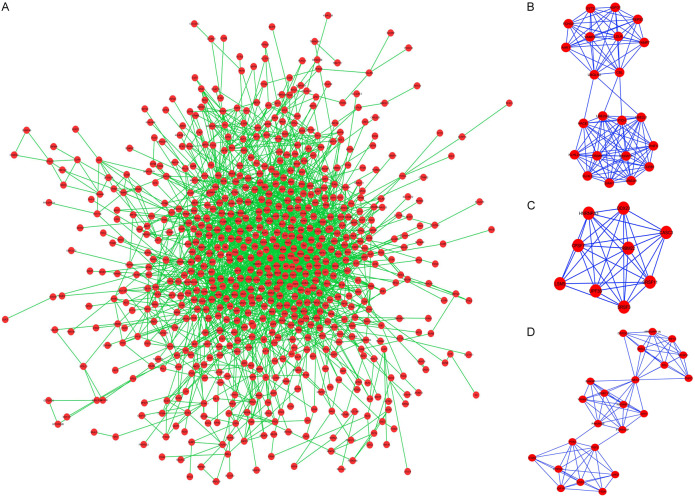
In order to explore the internal relationships and interactions among the target mRNAs, the PPI network was constructed using the STRING database. A PPI network with statistical significance consisted of 713 nodes, and 2191 edges were generated with the set of 1,082 mRNAs (Figure 6A). Hub genes of the PPI network through the degree algorithm of cytoHubba were identified as fibroblast growth factor 2 (FGF2), phosphatase and tensin homologue (PTEN), HECT domain and Ankyrin repeat containing E3 ubiquitin protein ligase 1 (HACE1), vesicle-associated membrane protein 2 (VAMP2), and calcineurin B-like protein (CBL). The top 20 hub genes of the overall PPI network are shown in Table 5.
Figure 6.
Protein-protein interaction network analysis for the proteins coded by the predicted target genes. A. PPI network of target genes. B-D. The top three significant modules selceted from the PPI network.
Table 5.
Top 20 hub genes of overall PPI network
| Rank | Gene | Score | Rank | Gene | Score |
|---|---|---|---|---|---|
| 1 | FGF2 | 46 | 10 | RAB11A | 28 |
| 2 | PTEN | 44 | 12 | ITCH | 26 |
| 3 | HACE1 | 40 | 12 | SP1 | 26 |
| 4 | VAMP2 | 34 | 12 | IGF1R | 26 |
| 5 | CBL | 33 | 12 | PTPN11 | 26 |
| 6 | PTK2 | 32 | 16 | PRKCA | 25 |
| 7 | PRKAR2B | 30 | 16 | CAMK2G | 25 |
| 8 | PPP1CC | 29 | 18 | SMAD2 | 24 |
| 8 | JAK2 | 29 | 19 | MYB | 23 |
| 10 | RAP1B | 28 | 20 | SOS1 | 22 |
Abbreviations: PPI, protein-protein interaction.
Three modules were formed in the PPI network with MCODE score ≥ 7: module 1 with MCODE score of 10.952 (nodes =22), module 2 with MCODE score of 8.5 (nodes =9) and module 3 with MCODE score of 7.545 (nodes =23) (Figure 6B-D). Hub genes, namely, VAMP2, CBL, and HACE1, were present in module 1, and PTEN and FGF2 were present in module 3.
miRNAs-mRNAs interaction network
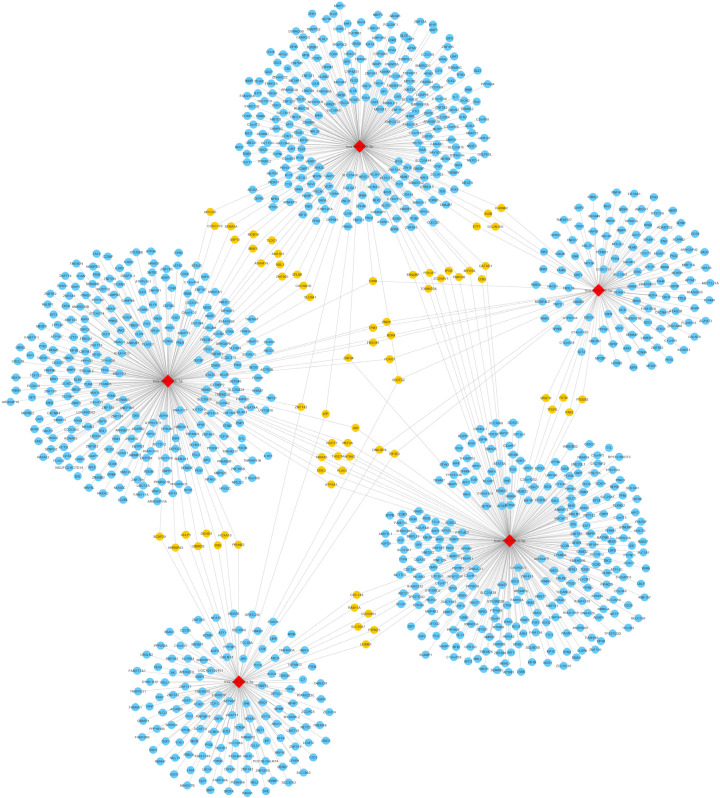
To investigate the pathogenesis of different miRNAs in SONFH, an interaction network between five selected exosomal miRNAs and their predicted target genes was established (Figure 7). In the network, five exosomal miRNAs were associated with 1081 target genes. From these results we observed that a single miRNA targeted on a couple of genes and one single gene may also bind to several related miRNAs, indicating that miRNAs and their target genes were mutually cross-linked. Moreover, hsa-miR-135b-5p was associated with 297 target genes, making it the most prominent miRNA that could play an important role in the pathogenesis of SONFH.
Figure 7.
A network of the interactions of five candidate miRNAs and their target genes. The circular blue nodes represent predicted mRNAs, the diamond red nodes represent the miRNAs, and the circular yellow nodes on behalf of two and more than two miRNAs interact with genes.
Confirmation of candidate miRNAs by qRT-PCR
Taken together, the top five most highly differentially expressed exosomal miRNAs (hsa-miR-135b-5p, hsa-miR-150-5p, hsa-miR-509-3-5p, hsa-miR-514a-3p, and hsa-miR-708-5p) in the three groups were initially selected for qRT-PCR validation using an independent cohort of serum-derived exosomes obtained from 12 healthy subjects, 22 SLE-SONFH patients and 17 SLE patients. When compared with the normal control and SLE groups, samples derived from SLE-SONFH patients showed higher expression levels of hsa-miR-135b-5p (Figure 8A). Meanwhile, we found that the expression level of hsa-miR-135b-5p was not significantly different between the normal control and SLE groups (Figure 8A). In addition, none of the remaining four exosomal miRNAs showed any significant differences in expression among the three groups (Figure 8B-E).
Figure 8.
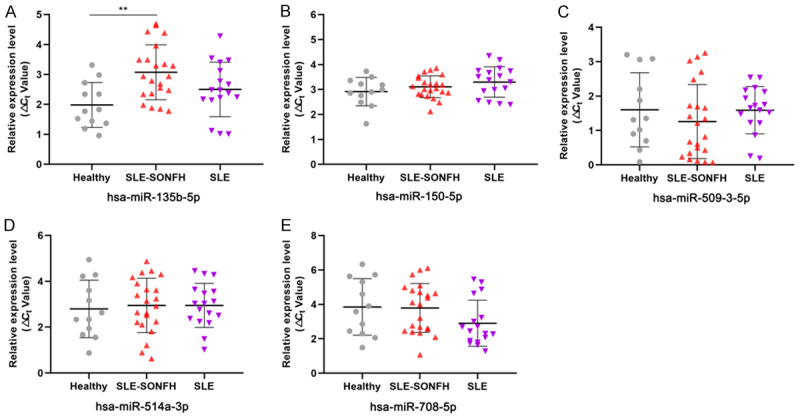
qRT-PCR validation of the expression levels of five differentially expressed miRNAs in the healthy, SLE-SONFH and SLE groups. A. Hsa-miR-135b-5p was significantly up-regulated in SLE-SONFH patients compared to healthy controls (P < 0.05). B. Expression of hsa-miR-150-5p in the healthy, SLE group and SLE-SONFH groups. C. Expression of hsa-miR-509-3-5p in the healthy, SLE grouo and SLE-SONFH groups. D. Expression of hsa-miR-514a-3p in the healthy, SLE group and SLE-SONFH groups. E. Expression of hsa-miR-708-5p in the healthy, SLE group and SLE-SONFH groups.
Discussion
SONFH is a progressive debilitating disease strongly associated with corticosteroid treatment; it frequently occurs in patients between 20 and 50 years of age. Among these patients, SLE and renal transplantation complicated with SONFH make up 3-41% [19] and 4-40% [20], respectively. The risk of SONFH in patients with SLE is 10 times higher compared to the general population [21]. Recently, a retrospective study found that 19% out of 190 SLE patients were more susceptible to SONFH when treated with steroid doses ≥ 40 mg/day during the first month [22]. Similarly, another study reported that the risk of SONFH in patients with SLE would remarkably increase in response to treatment with prednisolone > 30 mg/day within the first month [23]. Therefore, greater attention and vigilance is needed to prevent the occurrence of SONFH in the next 12 months when using high doses of steroids to treat diseases over a short time [24].
In recent years, liquid biopsy has been featured as a promising technique and is now widely used to improve the diagnosis and prognosis of multiple diseases. Exosomes are small secreted extracellular vesicles that are found in almost all biological fluids. Accumulating evidence indicates that exosomes contain a specific composition of lipids, proteins and nucleic acids [25]. Importantly, exosomes are considered critical mediators of intercellular communication because they can carry and deliver multiple messages to distal and surrounding cells. In addition, exosomes can be obtained in a non-invasive way to make it possible to use them in various clinical settings. Therefore, exosome-based liquid biopsy has emerged as a potent non-invasive tool for detecting and tracking the occurrence and development of diseases [26]. MiRNAs are small, noncoding RNAs that are abundant in exosomes and bind to the 3’ or 5’ non-coding region of mRNA to inhibit mRNA translation or promote its degradation. As a result, miRNAs play a significant role in various physiological and pathological processes including cell proliferation, differentiation, and apoptosis [27-29]. Exosomal miRNAs are protected from degradation due to the protection of the double lipid membrane, which allows them to maintain their biological activity and to remain stable in body fluids (blood, saliva, and urine) [30-32]. Therefore, exosomal miRNAs could be used as potential biomarkers for the early detection of diseases. Recent studies have provided novel insights into the potential contribution of miRNAs in serum [33,34], bone tissues [35,36] and cell lines [37,38] to the pathogenesis of SONFH.
This is the first study to reveal the potential use of the exosomal miRNAs as diagnostic markers of SONFH. In our study, we systematically purified and identified the characteristics of serum exosomes from SLE-SONFH, SLE patients, and healthy controls. miRNA-seq profiling indicated that five common human miRNAs were differentially expressed among the three groups, suggesting that they may have a role in the pathogenesis of SONFH. qRT-PCR validation identified hsa-miR-135b-5p as significantly and differentially expressed miRNA in SLE-SONFH patients compared to the healthy controls. Our results suggest that altered expression of miRNAs may be considered as potential candidate biomarkers for differentiating SONFH patients from healthy subjects. Previous studies indicated that miR-135b-5p could be a prognostic biomarker in breast cancer [39] and lupus nephritis [40]. In addition, by directly repressing MEF2C expression, miR-135b-5p could promote the proliferation and migration of vascular smooth muscle cells [41], which might play an important role in atherosclerosis pathogenesis. It was reported that the expression of miR-708-5p was significantly decreased in osteosarcoma samples compared to non-neoplastic bone samples. Moreover, overall survival analysis showed that decreased expression levels of miR-708-5p were related to a poor prognosis and lower patient survival rate [42]. Hsa-miR-135b-5p targets osteocalcin (OCN), bone sialoprotein (BSP), runt-related transcription factor 2 (RUNX2), and osterix (OSX), affecting bone marrow mesenchymal stem cells (BMSCs) differentiation into osteoblasts [43]. Hsa-miR-150-5p expression is decreased in ankylosing spondylitis and serves as a potential biomarker of the disease activity and the presence of syndesmophytes [44]. Thus, these recent data suggest that these miRNAs are closely linked to bone metabolism and provide new insights for the pathogenesis of SONFH. As detecting SONFH at an early stage is important to improve treatment and clinical outcomes, these findings may be helpful in detecting early subtle abnormalities in high-risk individuals who have received corticosteroid therapy.
The pathogenesis of SONFH is the outcome of the combined action of multiple mechanisms. Our results of KEGG pathway analysis revealed that Wnt, MAPK, and Hippo signaling pathways were significantly enriched in SONFH samples. The Wnt signaling pathway is involved in the transdifferentiation between osteogenesis and adipogenesis of BMSCs. The canonical Wnt signaling pathway has been implicated in regulating osteoblastogenesis, bone formation and remodeling [45]. A noncanonical Wnt ligand, Wnt5a, potently represses adipogenesis through transcriptional suppression of peroxisome proliferator-activated receptor gamma (PPARγ) and subsequent activation of a histone methyltransferase SETDB1 [46]. The MAPK signaling pathway plays an important regulatory role in osteoblast, osteoclast, and BMSCs proliferation, growth, and differentiation [47,48]. One of the main issues in SONFH pathogenesis is insufficient blood supply. The Hippo/YAP pathway alters the growth, death and migration of vascular smooth muscle cells and endothelial cells, which may contribute to vascular remodeling in SONFH [49].
To our knowledge, this is the first report of a role for serum exosomal miRNAs in the diagnosis of SONFH. Due to the limited size of samples in this study, we only analyzed candidate miRNAs expression levels in 12 healthy subjects, 22 SLE-SONFH, and 17 SLE patients. Large-scale study should be conducted in the future. Furthermore, we did not explore the possible mechanism of hsa-miR-135b-5p in the pathogenesis of SONFH. It would be of great interest to elucidate the biological functions of hsa-miR-135b-5p in SONFH in our future work.
In conclusion, we provide the first experimental evidence that specific exosome-associated hsa-miR-135b-5p is differentially expressed between healthy individuals and SLE patients. Our results suggest that certain serum exosomal miRNAs, such as hsa-miR-135b-5p, may represent a promising noninvasive biomarker for the diagnosis of SONFH.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (NSFC) (No. 81603641), Guangdong Provincial Science and Technology Project (2017A020213030) to Peng Chen; NSFC (No. 81774339) and 2019 Key Research of Guangzhou University of Chinese Medicine (XK2019012) to Haibin Wang.
Disclosure of conflict of interest
None.
References
- 1.Kubo T, Ueshima K, Saito M, Ishida M, Arai Y, Fujiwara H. Clinical and basic research on steroid-induced osteonecrosis of the femoral head in Japan. J Orthop Sci. 2016;21:407–413. doi: 10.1016/j.jos.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Chan KL, Mok CC. Glucocorticoid-induced avascular bone necrosis: diagnosis and management. Open Orthop J. 2012;6:449–457. doi: 10.2174/1874325001206010449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu LH, Zhang QY, Sun W, Li ZR, Gao FQ. Corticosteroid-induced osteonecrosis of the femoral head: detection, diagnosis, and treatment in earlier stages. Chin Med J (Engl) 2017;130:2601–2607. doi: 10.4103/0366-6999.217094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sen RK. Management of avascular necrosis of femoral head at pre-collapse stage. Indian J Orthop. 2009;43:6–16. doi: 10.4103/0019-5413.45318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Issa K, Jauregui JJ, McElroy M, Banerjee S, Kapadia BH, Mont MA. Unnecessary magnetic resonance imaging of hips: an economic burden to patients and the healthcare system. J Arthroplasty. 2014;29:1911–1914. doi: 10.1016/j.arth.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 6.Zhu HY, Gao YC, Wang Y, Zhang CQ. Circulating exosome levels in the diagnosis of steroid-induced osteonecrosis of the femoral head. Bone Joint Res. 2016;5:276–279. doi: 10.1302/2046-3758.56.BJR-2015-0014.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor DD, Gercel-Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front Genet. 2013;4:142. doi: 10.3389/fgene.2013.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato-Kuwabara Y, Melo SA, Soares FA, Calin GA. The fusion of two worlds: non-coding RNAs and extracellular vesicles--diagnostic and therapeutic implications (Review) Int J Oncol. 2015;46:17–27. doi: 10.3892/ijo.2014.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva M, Melo SA. Non-coding RNAs in exosomes: new players in cancer biology. Curr Genomics. 2015;16:295–303. doi: 10.2174/1389202916666150707154719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Hoshino I, Matsubara H. MicroRNAs in cancer diagnosis and therapy: from bench to bedside. Surg Today. 2013;43:467–478. doi: 10.1007/s00595-012-0392-5. [DOI] [PubMed] [Google Scholar]
- 12.Qin Y, Peng Y, Zhao W, Pan J, Ksiezak-Reding H, Cardozo C, Wu Y, Divieti Pajevic P, Bonewald LF, Bauman WA, Qin W. Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: a novel mechanism in muscle-bone communication. J Biol Chem. 2017;292:11021–11033. doi: 10.1074/jbc.M116.770941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ong SG, Lee WH, Huang M, Dey D, Kodo K, Sanchez-Freire V, Gold JD, Wu JC. Cross talk of combined gene and cell therapy in ischemic heart disease: role of exosomal microRNA transfer. Circulation. 2014;130(Suppl 1):S60–69. doi: 10.1161/CIRCULATIONAHA.113.007917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu C, Gershwin ME, Chang C. Diagnostic criteria for systemic lupus erythematosus: a critical review. J Autoimmun. 2014;48-49:10–13. doi: 10.1016/j.jaut.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Arbab D, Konig DP. Atraumatic femoral head necrosis in adults. Dtsch Arztebl Int. 2016;113:31–38. doi: 10.3238/arztebl.2016.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rikkert LG, Nieuwland R, Terstappen L, Coumans FAW. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J Extracell Vesicles. 2019;8:1555419. doi: 10.1080/20013078.2018.1555419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soo CY, Song Y, Zheng Y, Campbell EC, Riches AC, Gunn-Moore F, Powis SJ. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, Ilkovics L, Hampl A, Crha I, Jandakova E, Minar L, Weinberger V, Bryja V. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles. 2015;4:25530. doi: 10.3402/jev.v4.25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasan SS, Romeo AA. Nontraumatic osteonecrosis of the humeral head. J Shoulder Elbow Surg. 2002;11:281–298. doi: 10.1067/mse.2002.124347. [DOI] [PubMed] [Google Scholar]
- 20.Orban H, Cirstoiu C, Dragusanu M, Cristescu V. Resection--reconstruction in malignant tumors of the locomotory apparatus. Chirurgia (Bucur) 2007;102:443–446. [PubMed] [Google Scholar]
- 21.Dima A, Pedersen AB, Pedersen L, Baicus C, Thomsen RW. Risk of osteonecrosis in patients with systemic lupus erythematosus: a nationwide population-based study. Eur J Intern Med. 2016;35:e23–e24. doi: 10.1016/j.ejim.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Massardo L, Jacobelli S, Leissner M, Gonzalez M, Villarroel L, Rivero S. High-dose intravenous methylprednisolone therapy associated with osteonecrosis in patients with systemic lupus erythematosus. Lupus. 1992;1:401–405. doi: 10.1177/096120339200100610. [DOI] [PubMed] [Google Scholar]
- 23.Ono K, Tohjima T, Komazawa T. Risk factors of avascular necrosis of the femoral head in patients with systemic lupus erythematosus under high-dose corticosteroid therapy. Clin Orthop Relat Res. 1992:89–97. [PubMed] [Google Scholar]
- 24.Wang F, Wang Y, Hu N, Miao X. Risk-factors, pathogenesis, and pharmaceutical approaches for treatment of steroid-induced bone infarction of femoral head. Acta Pol Pharm. 2016;73:557–563. [PubMed] [Google Scholar]
- 25.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13:328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santiago-Dieppa DR, Steinberg J, Gonda D, Cheung VJ, Carter BS, Chen CC. Extracellular vesicles as a platform for ‘liquid biopsy’ in glioblastoma patients. Expert Rev Mol Diagn. 2014;14:819–825. doi: 10.1586/14737159.2014.943193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu SC, Mato JM, Espinosa-Diez C, Lamas S. MicroRNA-mediated regulation of glutathione and methionine metabolism and its relevance for liver disease. Free Radic Biol Med. 2016;100:66–72. doi: 10.1016/j.freeradbiomed.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guanen Q, Junjie S, Baolin W, Chaoyang W, Yajuan Y, Jing L, Junpeng L, Gaili N, Zhongping W, Jun W. MiR-214 promotes cell meastasis and inhibites apoptosis of esophageal squamous cell carcinoma via PI3K/AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;105:350–361. doi: 10.1016/j.biopha.2018.05.149. [DOI] [PubMed] [Google Scholar]
- 29.Shen L, Gan M, Li Q, Wang J, Li X, Zhang S, Zhu L. MicroRNA-200b regulates preadipocyte proliferation and differentiation by targeting KLF4. Biomed Pharmacother. 2018;103:1538–1544. doi: 10.1016/j.biopha.2018.04.170. [DOI] [PubMed] [Google Scholar]
- 30.Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, Shim SG, Paik YH. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machida T, Tomofuji T, Maruyama T, Yoneda T, Ekuni D, Azuma T, Miyai H, Mizuno H, Kato H, Tsutsumi K, Uchida D, Takaki A, Okada H, Morita M. miR1246 and miR4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep. 2016;36:2375–2381. doi: 10.3892/or.2016.5021. [DOI] [PubMed] [Google Scholar]
- 32.Lin SY, Chang CH, Wu HC, Lin CC, Chang KP, Yang CR, Huang CP, Hsu WH, Chang CT, Chen CJ. Proteome profiling of urinary exosomes identifies alpha 1-antitrypsin and H2B1K as diagnostic and prognostic biomarkers for urothelial carcinoma. Sci Rep. 2016;6:34446. doi: 10.1038/srep34446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Jiang C, Li X, Wu WKK, Chen X, Zhu S, Ye C, Chan MTV, Qian W. Circulating microRNA signature of steroid-induced osteonecrosis of the femoral head. Cell Prolif. 2018;51 doi: 10.1111/cpr.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei B, Wei W. Identification of aberrantly expressed of serum microRNAs in patients with hormone-induced non-traumatic osteonecrosis of the femoral head. Biomed Pharmacother. 2015;75:191–195. doi: 10.1016/j.biopha.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan HF, Christina VR, Guo CA, Chu YW, Liu RH, Yan ZQ. Involvement of MicroRNA-210 Demethylation in Steroid-associated Osteonecrosis of the Femoral Head. Sci Rep. 2016;6:20046. doi: 10.1038/srep20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu X, Zhang Y, Guo X, Xu H, Xu Z, Duan D, Wang K. Identification of differentially expressed microRNAs involved in non-traumatic osteonecrosis through microRNA expression profiling. Gene. 2015;565:22–29. doi: 10.1016/j.gene.2015.03.072. [DOI] [PubMed] [Google Scholar]
- 37.Hao C, Yang S, Xu W, Shen JK, Ye S, Liu X, Dong Z, Xiao B, Feng Y. MiR-708 promotes steroid-induced osteonecrosis of femoral head, suppresses osteogenic differentiation by targeting SMAD3. Sci Rep. 2016;6:22599. doi: 10.1038/srep22599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang A, Ren M, Song Y, Wang X, Wang Q, Yang Q, Liu H, Du Z, Zhang G, Wang J. MicroRNA expression profiling of bone marrow mesenchymal stem cells in steroid-induced osteonecrosis of the femoral head associated with osteogenesis. Med Sci Monit. 2018;24:1813–1825. doi: 10.12659/MSM.909655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bao C, Lu Y, Chen J, Chen D, Lou W, Ding B, Xu L, Fan W. Exploring specific prognostic biomarkers in triple-negative breast cancer. Cell Death Dis. 2019;10:807. doi: 10.1038/s41419-019-2043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Vives E, Sole C, Moline T, Vidal M, Agraz I, Ordi-Ros J, Cortes-Hernandez J. The urinary exosomal miRNA expression profile is predictive of clinical response in lupus nephritis. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21041372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z, Han Y, Liu J, Jiang F, Hu H, Wang Y, Liu Q, Gong Y, Li X. MiR-135b-5p and MiR-499a-3p promote cell proliferation and migration in atherosclerosis by directly targeting MEF2C. Sci Rep. 2015;5:12276. doi: 10.1038/srep12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delsin LEA, Roberto GM, Fedatto PF, Engel EE, Scrideli CA, Tone LG, Brassesco MS. Downregulated adhesion-associated microRNAs as prognostic predictors in childhood osteosarcoma. Pathol Oncol Res. 2019;25:11–20. doi: 10.1007/s12253-017-0316-1. [DOI] [PubMed] [Google Scholar]
- 43.Sartori EM, Magro-Filho O, Silveira MDB, Li X, Fu J, Mendonca G. Modulation of micro RNA expression and osteoblast differentiation by nanotopography. Int J Oral Maxillofac Implants. 2018;33:269–280. doi: 10.11607/jomi.5372. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Sanchez C, Font-Ugalde P, Ruiz-Limon P, Lopez-Pedrera C, Castro-Villegas MC, Abalos-Aguilera MC, Barbarroja N, Arias-de LRI, Lopez-Montilla MD, Escudero-Contreras A, Lopez-Medina C, Collantes-Estevez E, Jimenez-Gomez Y. Circulating microRNAs as potential biomarkers of disease activity and structural damage in ankylosing spondylitis patients. Hum Mol Genet. 2018;27:875–890. doi: 10.1093/hmg/ddy008. [DOI] [PubMed] [Google Scholar]
- 45.Macsai CE, Foster BK, Xian CJ. Roles of Wnt signalling in bone growth, remodelling, skeletal disorders and fracture repair. J Cell Physiol. 2008;215:578–587. doi: 10.1002/jcp.21342. [DOI] [PubMed] [Google Scholar]
- 46.Takada I, Kouzmenko AP, Kato S. Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat Rev Rheumatol. 2009;5:442–447. doi: 10.1038/nrrheum.2009.137. [DOI] [PubMed] [Google Scholar]
- 47.Yang H, Guo Y, Wang D, Yang X, Ha C. Effect of TAK1 on osteogenic differentiation of mesenchymal stem cells by regulating BMP-2 via Wnt/beta-catenin and MAPK pathway. Organogenesis. 2018;14:36–45. doi: 10.1080/15476278.2018.1455010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Funakubo N, Xu X, Kukita T, Nakamura S, Miyamoto H, Kukita A. Pmepa1 induced by RANKL-p38 MAPK pathway has a novel role in osteoclastogenesis. J Cell Physiol. 2018;233:3105–3118. doi: 10.1002/jcp.26147. [DOI] [PubMed] [Google Scholar]
- 49.He J, Bao Q, Yan M, Liang J, Zhu Y, Wang C, Ai D. The role of Hippo/yes-associated protein signalling in vascular remodelling associated with cardiovascular disease. Br J Pharmacol. 2018;175:1354–1361. doi: 10.1111/bph.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]