Abstract
Background: We previously constructed AdEasy system for rapid generation of recombinant adenovirus expressing coding genes. However, it is unclear if AdEasy system could be used for exogenously expression of long noncoding RNAs (lncRNAs). Here we investigated how to overexpress lncRNA H19 with AdEasy system and identified the effect of overexpression H19 on mesenchymal stem cells (MSCs) osteogenic differentiation. Methods: H19 fragment 1 and H19 fragment 2 were amplified from mouse genomic DNA separately and then connected for full-length H19. H19 was firstly subcloned to homemade pMOK plasmid, then H19 was cut off from pMOK-H19 and subcloned to recombinant adenovirus plasmid. After homologous recombination in AdEasier cells (BJ5183 cell), packing in mammalian packaging cell line and amplification in 293pTP cells, high titer AdH19 was obtained. Immortalized mouse adipose-derived progenitors (iMADs) were infected by AdH19 with different infection rate, the expression of H19, H19 related microRNAs (miRs) and osteogenic differentiation markers were determined by TqPCR. Alkaline phosphatase (ALP) activities and matrix mineralization were determined by ALP assays and Alizarin red S staining respectively. Results: AdEasy system was suitable for rapid generation and production of H19, AdH19 can effectively overexpress H19 and serve as functional lncRNA in mesenchymal stem cells (MSCs). Higher dosage of AdH19 inhibited osteogenic differentiation of MSCs, however, lower dosage of AdH19 promoted osteogenic differentiation of MSCs. Conclusions: We firstly reported the method for the generation of functional lncRNA with AdEasy system, and identified the biphasic effect of H19 on MSCs osteogenic differentiation.
Keywords: lncRNA H19, AdEasy system, mesenchymal stem cells (MSCs), osteogenic differentiation
Introduction
With the development of whole genome and transcriptome sequencing technologies, non-coding RNAs (ncRNAs), especially long non-coding RNAs (lncRNAs) were recognized as regulatory RNAs [1-5]. Recently, lncRNAs were reported to regulate the target gene expression in the initiation of transcription, during the transcription, post-transcription and post-transcription modification etc. [4,6]. However, be different from coding genes, lncRNA coding no protein, it is difficult for the investigators to mimic the expression or identify the function of lncRNAs [5,7,8]. Therefore, it is necessary to construct a vector which can simulate the expression of lncRNA exogenously.
Recombinant adenoviruses are replication-defective adenoviral vectors, and have been widely used as gene transfer vehicles in many areas [9-11]. We previously optimized and constructed the AdEasy system which simplifies the production and generation of recombinant adenoviruses, at the same time, AdEasy system hold the advantage of high titer, easy for preparation, can transduce a wide variety of cells or tissues [9,11,12]. What is more, recently, we optimized the procedures of adenovirus generation from the aspects of packing, amplification etc. [13,14] However, it is not clear whether AdEasy system can effectively express lncRNA as a ncRNA vehicle.
LncRNA H19 (H19) was firstly isolated and reported in 1980s by 4 different laboratories, which is one of most widely and deeply studied lncRNAs so far [15-22]. As imprinted genes, H19-Igf2 locus was define as the paradigm for the study of genomic imprinting [23,24]. Recently, H19 was reported play an essential role in the osteogenic differentiation process [25-27]. Huang [27] et al found that H19 promotes osteoblast differentiation via TGF-β1/Smad3/HDAC signaling pathway by deriving miR-675. Liang [26] et al identified that H19 acting as ceRNA to promote osteogenic differentiation by targeting Wnt signaling pathway. In our previous study [28], we found the biphasic expression of H19 in BMP9 induced osteogenic differentiation of MSCs, which indicated the biphasic effect of H19 in regulating osteogenic differentiation of MSCs. However, it is not clear whether H19 regulate osteogenic differentiation of MSCs in a dosage dependent manner.
In the present study, we successfully constructed the recombinant adenovirus expression H19 with AdEasy system, and identified AdH19 mediated exogenous expression of H19 could act as functional lncRNA. What is more, by using different dosage of AdH19 we identified the biphasic effect of H19 on osteogenic differentiation of MSCs, which is helpful for further exploring the regulatory function of H19 in osteogenic differentiation of MSCs.
Materials and methods
Cell culture and chemicals
Human embryo kidney 293 cell line (HEK 293) was purchased from American Type Culture Collection (ATCC, Manassas, VA), 293pTP cells were derived from HEK 293 cell which express high level of human Ad5 pTP gene as described previously [13]. RAPA (rapid adenovirus production and amplification) cell line was derived for 293T cell which express high level of EA1 and pTP genes as previously characterized [14]. The mouse embryonic fibroblasts (MEFs) and immortalized mouse adipose-derived (iMAD) cells are mouse mesenchymal stem cells as reported respectively [29,30]. Cells were maintained in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Gemini Bio-Products, West Sacramento, CA), 100 U/ml penicillin, and 100 mg/ml streptomycin, maintained at 37°C in a humidified 5% carbon dioxide (CO2) atmosphere [29-32]. Restriction enzymes and ligation buffers were supplied by New England Biolabs (Ipswich, MA). Unless indicated otherwise, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Thermo-Fisher Scientific (Waltham, MA).
Genomic DNA extraction
The MEF cells were cultured in 100 mm dishes, at 80% confluence, cells were spun down and collected in 1.5 ml tube, then washed with PBS twice. DNA extraction buffer (244.5 μl ddH2O, 10N NaOH 5 μl, 0.5M EDTA (PH 8.0) 0.5 μl) was used to lysis the cells. MEFs were resuspended with DNA extraction buffer, and then keep in vibrating metal bath for 20 minutes. When the mixture was cool down at room temperature, PC8 (phenol/chloroform, pH 8.0) was used to purification the DNA, after precipitation, the DNA pipet was dissolved in ddH2O. At the same time whole genomic DNA was checked in PAGE (Polyacrylamide Gel Electrophoresis) gel.
The amplification of whole H19 fragment
According to the mouse lncRNA H19 (NR_130973.1), whole H19 fragment primers were designed as: Fwd: 5’-atcatcgatGGGTGTGGGAGGGGGGTGGGGGGTGGGGGTGGGGGGTATCGGGGAAACTG-3’, Rev: 5’-acgacgcgtAATGACTGTAACTGTATTTATTGATG-3’. Two fragment primers were respectively designed as: Fragment 1: Fwd: 5’-atcatcgatGGGTGTGGGAGGGGGGTGGGGGGTGGGGGTGGGGGGTATCGGGGAAACTG-3’, Rev: 5’-gagaaatgaagacatgagttaattg-3’, Fragment 2: Fwd: 5’-gggtaagtgtctgtcccgctcgtggtcacc-3’, Rev: 5’-acgacgcgtAATGACTGTAACTGTATTTATTGATG-3’. Cleavage sites were contained in primers. An overlap was included for two fragments design. A touchdown cycling program was as follows: 94°C for 2 minutes for 1 cycle; 92°C for 20 seconds, 65°C for 30 seconds, and 72°C for 13 cycles decreasing 1°C per cycle; and then at 92°C for 20 seconds, 60°C for 30 seconds, and 72°C for 20 seconds for 20-25 cycles, depending on the abundance of a given gene. PCR products were resolved on 1.5% agarose gels.
Construction and generation of recombinant adenoviral vector
The full-length H19 was firstly subcloned to homemade pMOK plasmid (MluI and BglII), which contains diversity restriction endonuclease sites. The length of pMOK was around 3000 base pair, which was approximate to the full-length of H19. The full-length of H19 was cut off from pMOK-H19 between XbaI and SalI restriction endonuclease sites, and then subcloned into an adenoviral shuttle vector pAdtrace-Tox, resulting in pAdtrace-H19, as previously characterized [11,31-34]. Bacteria screening, plasmid screening and DNA sequencing were carried out to confirm the veracity of the clone. After linearized with restriction enzyme EcoRV, pAdtrace-H19 was transfected into AdEasier cells (BJ5183 cell), generating recombinant adenovirus pAd5-H19. After linearized with restriction enzyme PacI, recombinant adenovirus pAd5-H19 was transfected into RAPA cells and generating recombinant adenoviruses AdH19. AdH19 was amplified in 293pTP cells, and high titer AdH19 was collected as previously described [11], the packing and amplification process was shown in Supplementary Figure 1.
Semi-quantitative PCR
H19 screening primers were designed by using the Primer3 program. H19 fragment 1 screening primers were as follows: Fwd: 5’-GCAGTGCAGGGTGTCACCAGAAGG-3’, Rev: 5’-CAATTAACTCATGTCTTCATTTCTC-3’. H19 fragment 2 screening primers are as follows: Fwd: 5’-CATCTTCATGGCCAACTCTGCCTG-3’, Rev: 5’-CATCAATAAATACAGTTACAGTCATT-3’. A touchdown cycling program was as follows: 94°C for 2 minutes for 1 cycle; 92°C for 20 seconds, 65°C for 30 seconds, and 72°C for 10 cycles decreasing 1°C per cycle; and then at 92°C for 20 seconds, 57°C for 30 seconds, and 72°C for 20 seconds for 15-20 cycles, depending on the abundance of a given gene. PCR products were resolved on 1.5% agarose gels.
RNA isolation and touchdown quantitative real-time PCR (TqPCR)
The immortalized multipotent adipose-derived (iMAD) mesenchymal stem cells (MSCs) were seeded in 60 mm dishes, at 40 to 50% confluence, cells were infected with AdH19, AdRFP was used as control. Cells were harvested at desired time points for subsequent analysis. Total RNA was isolated with TRIZOL Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Reverse transcription reactions were performed using hexamer and M-MuLV Reverse Transcriptase (New England Biolabs, Ipswich, MA). PCR primers were designed by using the Primer3 Plus program [35]. The resultant cDNA products were diluted 10- to 100-fold and used as PCR templates. The quantitative PCR analysis was performed using our optimized TqPCR protocol [28,36]. Briefly, qPCR reactions were set up with SYBR Green (Bimake, Houston, TX) master mix according to the manufacturer’s instructions. The cycling program was modifed by incorporating 4 cycles of touchdown steps prior to the regular cycling program as previously described [36]. The dissolve curve did not determine any nonspecific amplification. All sample values were normalized to GAPDH expression by using the 2-ΔΔCt method. The TqPCR primer sequences are listed in Table 1.
Table 1.
List of TqPCR primers
| Gene | qPCR Primer Sequences | |
|---|---|---|
|
| ||
| Forward | Reverse | |
| mouse GAPDH | GCCTCGTCCCGTAGACAAAA | TTCCCATTCTCGGCCTTGAC |
| mouse Runx2 | CCGGTCTCCTTCCAGGAT | GGGAACTGCTGTGGCTTC |
| mouse BSP | AGGGAACTGACCAGTGTTGG | ACTCAACGGTGCTGCTTTTT |
| mouse OCN | CCAAGCAGGAGGGCAATA | TCGTCACAAGCAGGGTCA |
| mouse OPN | CCTCCCGGTGAAAGTGAC | CTGTGGCGCAAGGAGATT |
| mmu-H19-Fragment1 | CAGAGTCCGTGGCCAAGG | CGCCTTCAGTGACTGGCA |
| mmu-H19-Fragment2 | GCTGGGAAGGGTTCGACC | TGTGCCATTCTGCTGCGA |
| mmu-miR-675 | ATTCCCATGAGGCACTGCGG | ATGTTCTGTCTGTCATGCCA |
| mmu-miR-106b | CCTGCTGGGACTAAAGTGCT | TACCCACAGTGCGGTAGC |
| mmu-miR-125a | CCCTTTAACCTGTGAGGACGT | GGCTCCCAAGAACCTCACC |
| mmu-miR-449b | AGACTCGGGTAGGCAGTGT | GTGGCAGGGTAGCTGTGG |
| mmu-miR-17 | CAAAGTGCTTACAGTGCAGGT | GTGCCCTCACTGCAGTAGA |
| mmu-miR-449a | TGTGATGGCTTGGCAGTGT | TTAGCTGGTGCCGCTCAC |
| mmu-miR-34a | TGGCAGTGTCTTAGCTGGT | CAATGTGCAGCACTTCTAGGG |
| mmu-miR-107 | TCAGCTTCTTTACAGTGTTGCC | AGCCCTGTACAATGCTGCT |
| mmu-miR-27b | AGGTGCAGAGCTTAGCTGA | GCCACTGTGAACAAAGCGG |
| mmu-miR-199b | CGGGAGCGGAGAGGGCCAGA | TACAAGGATGATGAGCCGAG |
| mmu-miR-132 | ACCGACGCCTGGCCCGGGCA | GCAGCGATACCTCGAGGGCG |
| mmu-miR-93 | CCCCTCTTGGACCTTAGTCA | CACTATCAGAGGTTGTGTCC |
| mmu-miR-129-1 | CACAGCTGTCTCCTTTGGAT | AGACCAAGCCTCCCGTAGAT |
| mmu-miR-let-7d | GACCAGCAAGAATAAAATGG | AACGTATGCTGGTATAATAA |
Alkaline phosphatase (ALP) assay
The ALP activities were assessed using the modified Great Escape SEAP chemiluminescence assay (BD Clontech) and/or histochemical staining, as described previously [31,32,37,38]. For ALP histochemical staining, the cells were induced for osteogenic differentiation using osteogenic medium (containing ascorbic acid (50 mg/ml) and β-glycerophosphate (10 mM)). At the scheduled time, cells were fixed with 0.05% glutaradehyde at room temperature for 10 min. After washing with distilled water, cells were stained subjected to histochemical staining with a mixture of 0.1 mg/mL of napthol AS-MX phosphate and 0.6 mg/mL of Fast Blue BB salt. Histochemical staining was recorded using bright light microscopy.
For the chemiluminescence assay, the cells were lysed by the Cell Culture Lysis Buffer (Promega, Madison, WI). Then 5 μl Cell Lysis Buffer, 5 ul substrate (BD Clontech) and 15 μl Lupo Buffer were mixed well under a light-proof condition, and incubated at room temperature for 20 minutes before measuring chemiluminescence signals. Each assay condition was performed in triplicate, and the results were repeated in at least 3 independent experiments. The ALP activity was normalized by total cellular protein concentrations among the samples.
Matrix mineralization assay (alizarin red S staining)
The iMAD cells were seeded in 24-well plates and cultured in the presence of ascorbic acid (50 mg/ml) and β-glycerophosphate (10 mM). On day 15 after infection, cells were treated with 4% paraformaldehyde for 30 minutes. After washing with PBS, cells were incubated with 2% alizarin red S for 30 minutes, followed by extensive washing with distilled water [31,37-39]. The staining of calcium mineral deposits was recorded using bright light microscopy.
Clustering analysis
The clustering analysis was carried out with the using of Multiple Experiment Viewer (MeV) software. H19 related microRNAs (miRNAs) were detected by TqPCR. Relative normalized expression of the miRNAs were subjected to MeV software.
Statistical analysis
All quantitative experiments were performed in triplicate and/or repeated 3 times. Data were expressed as mean ± standard deviation (SD). The one-way analysis of variance or paired t-test were used to analyze statistical significance. A value of P<0.05 was considered statistically significant.
Results
Schematic depiction of the generation of AdH19
The process of construction of pAdtrace-H19 was shown in Figure 1. Briefly, whole mouse genomic DNA was extracted from MEF cells. H19 fragment 1 and fragment 2 were amplified by touchdown PCR respectively, and then H19 fragment 1 and fragment 2 were digested by restriction endonuclease and ligated by ligation buffer stepwisely, which result in full-length H19. Full-length H19 was firstly cloned into homemade plasmid pMOK which possessing a similar base pair with H19. Then, full-length H19 was cut off from pMOK-H19 and subcloned to pAdTrace-Tox plasmid, which generating pAdtrace-H19. After recombinant in AdEasier cells (BJ5183 cell), packing in RAPA cells and amplification in 293pTP cells, high titre AdH19 were generated.
Figure 1.
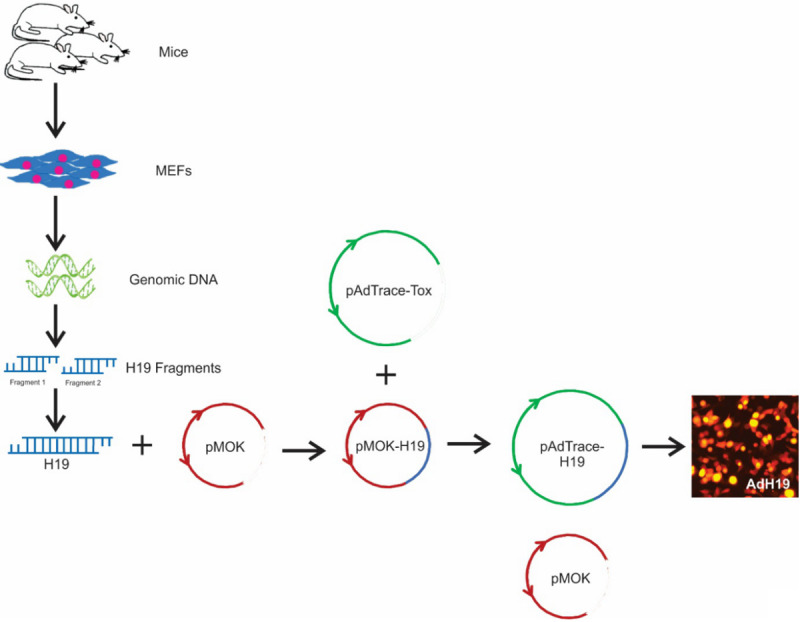
Schematic depiction of the construction of recombinant adenovirus vector. Whole mouse genomic DNA were extracted from mouse embryonic fibroblasts (MEFs), which were isolated from new born mice. Then, H19 fragment 1 and H19 fragment 2 were amplified from mouse genome separately and then connected for full-length H19. Full-length H19 was firstly subcloned to pMOK plasmid, and then full-length H19 was cut off from pMOK-H19 and subcloned to recombinant adenovirus vector. After recombinant in AdEasier cells (BJ5183 cell), packing in RAPA cells and amplification in 293pTP cells, high titre AdH19 were generated.
AdH19 effectively overexpress H19 and miR-675 in MSCs
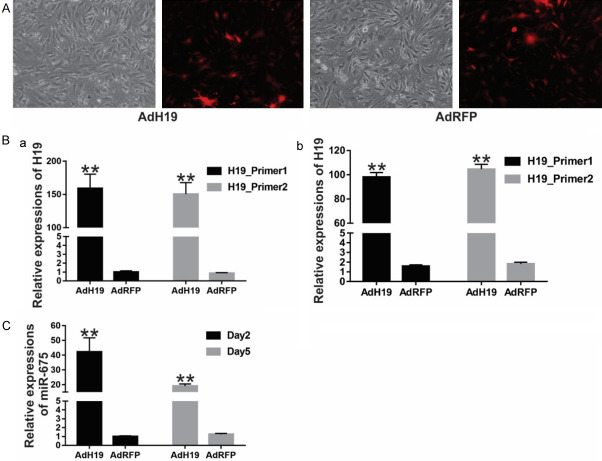
The iMAD cells were cultured and infected by recombinant adenoviruses expression H19 and RFP respectively. As shown in Figure 2A, the red fluorescence indicated the effective infection at day 2. The H19 expressions in AdH19 group were upregulated significantly at 2 days and 5 days after transduction compared with AdRFP group using Tq-PCR analysis (Figure 2B). At the same time, we determined the expressions of microRNA-675 (miR-675), which is encoded by H19, and found that H19 could be effectively upregulated by AdH19 at day 2 and day 5 respectively. These results suggest AdH19 can effectively mediate the overexpression of H19 and H19 coded miR-675.
Figure 2.
AdH19 mediated overexpression of H19 in MSCs. A: IMAD cells were infected with high titre AdH19 or AdH19. Two days later, bright light and fluorescence microscope examination showed the transduction efficiency of recombinant adenoviruses in iMAD cells, AdFRP as controls (100×). B, C: AdH19 mediated overexpression of H19 (with 2 different pair of primers) and H19 encoded microRNA miR-675 at day 2 and day 5. IMAD cells were infected with AdH19 or AdRFP, at indicated time points, Total RNA was isolated and subjected to TqPCR analysis, relative expressions of H19 or miR675 were normalized with the reference gene GAPDH, **P<0.01.
AdH19 function as competitive ceRNA
One of the most important function of lncRNA is acting as ceRNA. H19 was reported working as functional lncRNA by several authors. Here, we investigated whether AdH19 influence the expressions of miR-675, miR-125a, miR-199b, miR-132, miR-27b, miR-34a, miR-449a, miR-449b, miR93, miR-106b, miR-107, miR129-1, miR-17 and miR-let-7d. iMAD cells were infected with AdH19, AdRFP was used as control, total RNAs were isolated 2 days after infection. The expressions of miRs were detected by Tq-PCR. In accordance with the previous reports [24,40-51], miR-675, miR125a, miR-199b were upregulated by AdH19, and miR-132, miR-27b, miR-34a, miR-449a, miR-449b, miR-93, miR-106b, miR-107, miR-129-1, miR-17 and miR-let-7d were downregulated by AdH19 at different levels (Figure 3). These results indicate that except working as pre-miR of miR-675, AdH19 can also working as ceRNA to competitively combine with some miRs.
Figure 3.
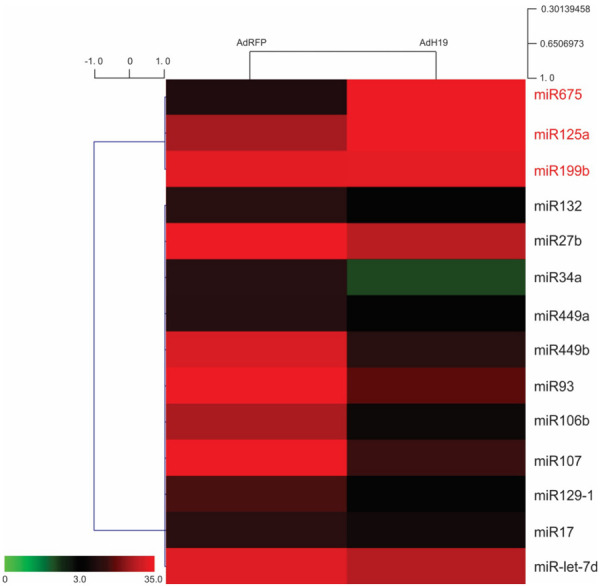
Functional identification of AdH19 mediated overexpression of H19. Subconfluent iMADs were infected with AdH19 or AdRFP for 2 days, total RNA was isolated from the infected iMADs and subjected to TqPCR analysis of the selected 14 miRNAs which may be associated with H19. The relative normalized expression of these miRNAs was subjected to clustering analysis, 3 miRs are upregulated and 11 miRs are downregulated.
H19 and osteogenic differentiation markers expression during the osteogenic differentiation of MSCs
In consideration the regulatory function of H19 in the process of osteogenic differentiation, we firstly explored the expression of H19 and osteogenic differentiation markers expression level in different time point. With the stimulation of osteogenic medium, we found that H19 expression level increased gradually from day 1 to day 3, and reach the peak level at day 3, then the expression of H19 decreased gradually and maintained in a relatively high level at day 7 and day 9 (Figure 4A). Meanwhile, the expression of early osteogenic differentiation marker Runx2 increased from day 3 to day 5 and back to the basal level at day 7 (Figure 4B), late osteogenic differentiation markers increased gradually from day 3 to day 9 (Figure 4C-E). These results suggest that a low or medium level of H19 is necessary for osteogenic differentiation of MSCs.
Figure 4.
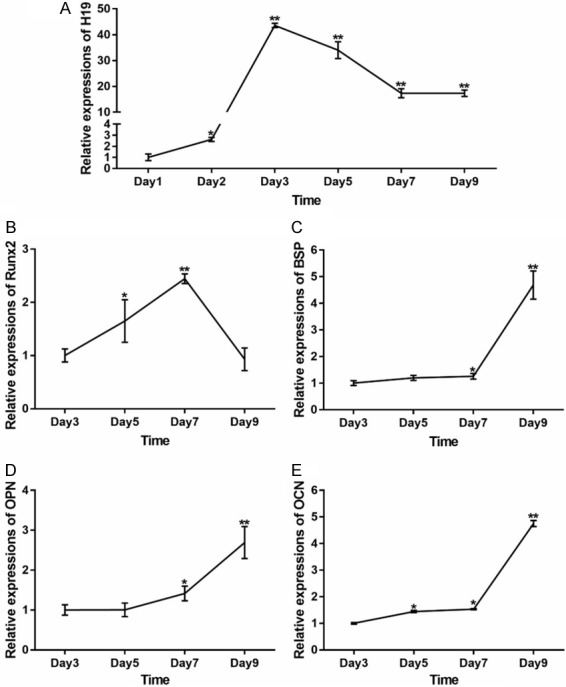
Expression levels of H19 and osteogenic differentiation markers during osteogenic differentiation of MSCs. IMAD cells were cultured in osteogenic medium, at the indicated time points, total RNA was isolated and subjected to TqPCR analysis. (A) Expression level of H19 (A) and osteogenic differentiation markers (B-E) at different time point. All samples were normalized with the reference gene GAPDH. Each assay condition was done in triplicate. Relative expression was calculated by dividing the relative expression values (i.e., gene/GAPDH). **P<0.01, *P<0.05, Runx2, runt-related transcription factor 2; BSP, bone sialoprotein; OPN, osteopontin; OCN, osteocalcin.
AdH19 mediated overexpression of H19 in MSCs is in a dosage dependent manner
IMAD cells were infected with AdH19 or AdRFP in three dosage (low, medium, high), as shown in Figure 5A, the infection rate of low dosage was about 20%, 40% for medium dosage and 80% for high dosage. Total RNA was isolated at day 2 and was subjected to reverse transcription and qPCR analysis, the expression level of H19 were positively corelated with the infection rate, as for high dosage, the expression level of H19 was around 285 times higher than control group (Figure 5B). These data suggest that adenovirus mediated overexpression of H19 in MSCs is in a dosage dependent manner.
Figure 5.
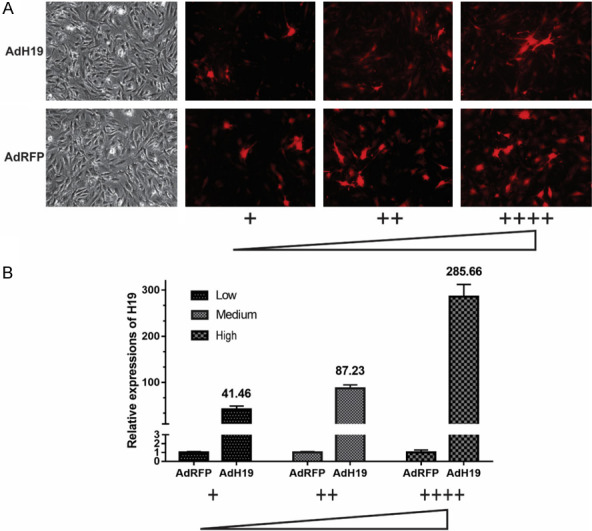
AdH19 mediated overexpression of H19 in MSCs is in a dosage dependent manner. A: Bright light and fluorescence microscope examination showed the transduction efficiency of recombinant adenoviruses in iMAD cells two days after infection, AdFRP as controls (100×). The infection rate of low dosage was about 20%, 40% for medium dosage and 80% for high dosage. B: Expression levels of H19 in different infection rate at day 2. All samples were normalized with the reference gene GAPDH. Each assay condition was done in triplicate, **P<0.01.
AdH19 biphasic regulating osteogenic differentiation of MSCs
We further explored the effect of different expression level of H19 on osteogenic differentiation of MSCs. IMAD cells were incubated in osteogenic medium and infected with AdH19 or AdRFP in low, medium or high dosage. Total RNA was isolated at the indicated time points and was subjected to reverse transcription and qPCR analysis. As shown in Figure 6, the expression of Runx2 at day 7, OPN and OCN at day 9 were upregulated in low dosage of AdH19, and downregulated in medium or high dosage of AdH19 compared with control group.
Figure 6.
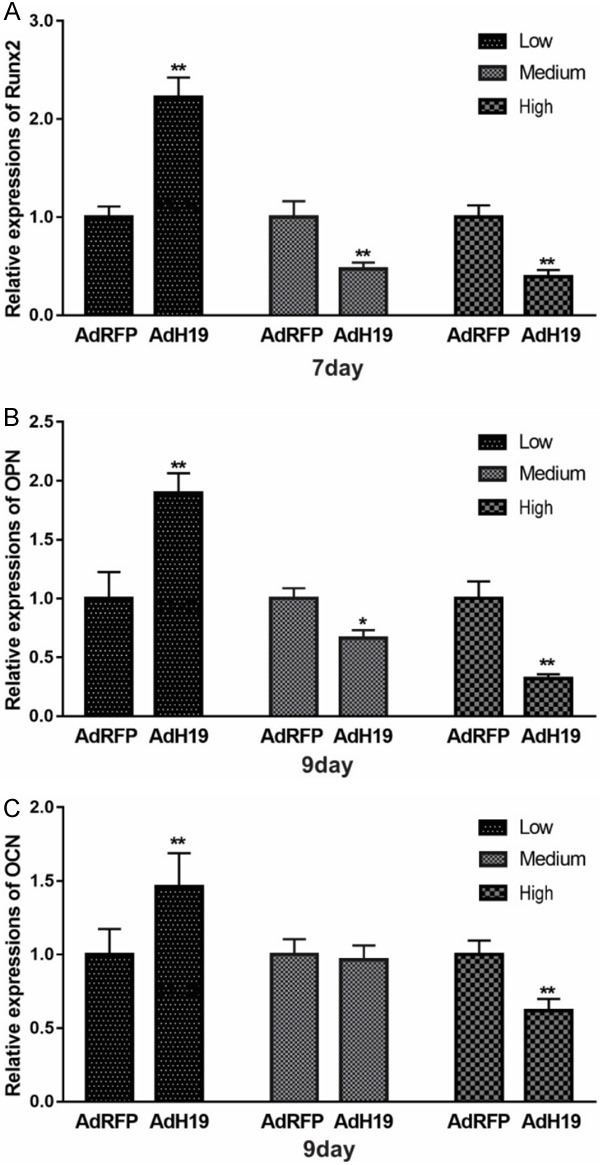
AdH19 mediated overexpression of H19 biphasic regulating osteogenic differentiation markers expression during osteogenic differentiation of MSCs. IMAD cells were cultured in osteogenic medium, at the indicated time points, total RNA was isolated and subjected to TqPCR analysis. A: Expression level of Runx2 at day 7. B: Expression level of OPN at day 9. C: Expression level of OCN at day 9. Each assay condition was done in triplicate. **P<0.01, Runx2, runt-related transcription factor 2; OPN, osteopontin; OCN, osteocalcin.
Then we analyzed the effect of different dosage of AdH19 on early osteogenic differentiation of MSCs. We found that low dosage of AdH19 dramatically upregulated ALP activities both qualitatively and quantitatively, however high dosage of AdH19 significantly downregulated ALP activities both qualitatively and quantitatively (Figure 7A, 7B). Similarly, late osteogenic differentiation marker matrix mineralization was determined at day 15. As exhibited in Figure 7C, low dosage of AdH19 promoted matrix mineralization of MSCs, however, medium and high dosage of AdH19 inhibited matrix mineralization of MSCs. These data indicated that low dosage of AdH19 promoted osteogenic differentiation of MSCs, and high dosage of AdH19 inhibited osteogenic differentiation of MSCs, namely, AdH19 biphasic regulating osteogenic differentiation of MSCs.
Figure 7.
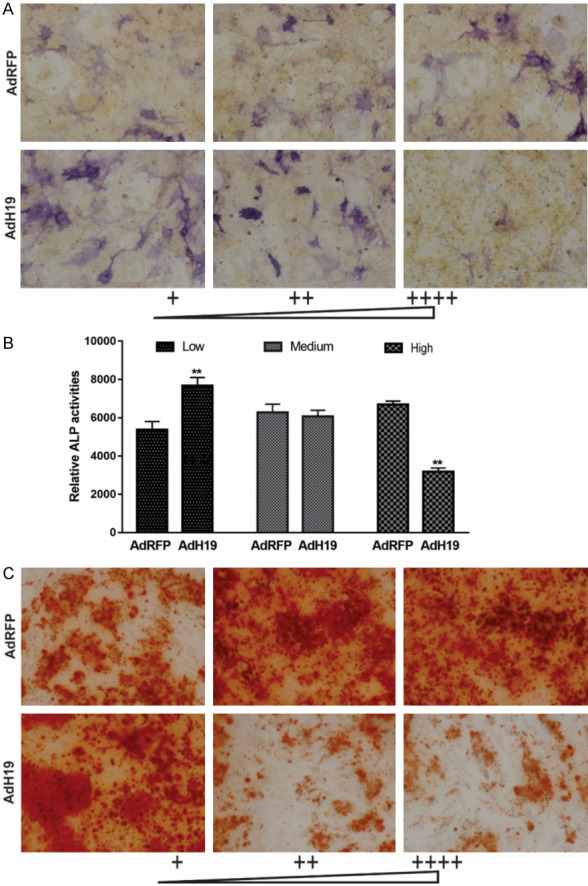
AdH19 mediated overexpression of H19 biphasic regulating early and late osteogenic differentiation of MSCs. (A, B) AdH19 mediated overexpression of H19 biphasic regulating ALP activity in MSCs. Subconfluent iMAD cells were infected with AdH19 or AdRFP with different dosage. At day 7 after infection, the infected cells were subjected to ALP activity assays by either histochemical staining (100×) (A) or quantitative bioluminescence assay (B). Each assay conditions were done in triplicate. Representative staining is shown, **P<0.01. (C) AdH19 mediated overexpression of H19 biphasic regulating matrix mineralization of MSCs. Subconfluent iMAD cells were infected with AdH19 or AdRFP with different dosage. At day 15 after infection, the infected cells were subjected to Alizarin Red S staining. Each assay condition was done in triplicate. Representative microscopic images are shown (100×).
Discussion
Current evidence shows that only about 1.2% the mammalian genome codes for amino acids in proteins, however, vast majority of the genome is transcribed, which is well beyond the boundaries of known genes [3,52]. Therefore many investigators pay attention to identify the function of ncRNA, especially lncRNA [53-55]. Owing to lncRNA coding no protein, regulating multiple stages during both transcription and translation [5,6,53], it is essential to mimic the overexpression of lncRNA exogenously. Here, in order to explore an effective method to overexpression lncRNA exogenously, we choose H19 as an example, which is one of the best known lncRNA and hold the potential to regulate several processes during cancer developing, cell proliferation etc. [16,27,56-58], and confirmed that AdEasy system could overexpression H19 in a stable, replicable, simple and convenient manner.
Full-length H19 was generated by two separate fragments containing specific enzyme sites, which were amplified from mouse genomic DNA respectively. We firstly subcloned full-length H19 into homemade plasmid pMOK, which hold a variety of restriction endonuclease sites, and then subcloned full-length H19 into recombinant adenovirus vector, that was because we could not subclone full-length H19 into recombinant adenovirus vector directly or subclone H19 fragment 1 and H19 fragment 2 into recombinant adenovirus vector stepwisely. These results indicate the importance of diversity of restriction endonuclease sites in vectors.
Recombinant adenoviruses are replication-defective adenoviral vectors which are used for gene therapy, vaccine therapy and basic biology [9,10,59]. During the past decades, we constructed AdEasy system for rapid generation and production of recombinant adenovirus vectors, at the same time, we optimized the procedures of adenovirus generation from the aspects of packing, amplification [11,13,14,34]. In the present study, we identified that H19 could be subcloned into our AdEasy system, and generating adenovirus overexpression H19 with the using of our optimized pTP293 cells and rapid adenovirus production and amplification (RAPA) cells [13,14], which indicate the optimized procedures of AdEasy system are applicable for exogenous generation lncRNA. In other words, this is the first time to report that lncRNA could be overexpressed exogenously with AdEasy system, which is helpful for next identify the function of lncRNAs.
H19 has been identified nearly four decades ago, the diverse of H19 functions, including regulating embryonal development and growth [23], acting as oncogene [60,61] or tumor suppressor [16,17], working as modular scaffold of histone modification complexes [22] etc. have been reported during the past years. However, one of H19’s most important function is acting with microRNAs. While function as nocoding RNA, H19 also encoding miR-675 [46,62,63]. Huang [27] et al reported that H19-miR-675 axis regulated osteoblast differentiation through TGF-β signaling pathway. Dudek [64] et al found that H19-miR-675 regulated type-II collagen expression. Steck [65] et al found that H19-miR-675 was associated with the anabolism and catabolism of osteoarthritis cartilage. Here, we found that adenovirus mediated overexpression of H19 up-regulated miR-675 dramatically, which indicates adenovirus mediated overexpression of H19 is functional.
Meanwhile, H19 was found to act as ceRNA or molecular sponge [7,24,51,66,67]. In the present study, we also explored whether adenovirus mediated overexpression of H19 acting as ceRNA. Among the 14 miRs which we selected may associated with H19 [24,40-51], we found that 3 miRs (miR-675, miR-125, miR-199b) were upregulated by overexpression of H19, and 11 miRs (miR-132, miR-27b, miR34a, miR-449a, miR-449b, miR93, miR-106b, miR-107, miR-129-1, miR-17 and miR-let-7d) were downregulated by overexpression of H19. Be consistent with the previous reports, H19 was found to be act as ceRNA or molecular sponge of miR-107 [21,43], miR-let-7 [24,44,45], miR-93 [68], miR-106b [69], miR-17 [51]. These results suggest that adenovirus mediated overexpression of H19 could work as functional ceRNA or molecular sponge.
Recently, H19 regulating osteogenic differentiation of MSCs were reported [25-27]. We found that the expression level of H19 in MSCs is relatively higher than other genes [28], at the same time, H19 is upregulated during the early stage, and sustained in a relatively high level during the late stage of osteogenic differentiation of MSCs. So, we asking if higher expression level of H19 could promote osteogenic differentiation. Here, with the advantage of semi-quantitative feature of adenovirus, we identified that adenovirus mediated low overexpression level of H19 could promote osteogenic differentiation of MSCs, nevertheless, high expression level of H19 inhibited osteogenic differentiation of MSCs. In summary, we found the biphasic effect of H19 in regulating osteogenic differentiation of MSCs, which suggest the flexibility of lncRNA in regulating physical or pathological processes.
Conclusion
We firstly reported AdEasy system mediated overexpression of H19 could act as functional lncRNA, and further identified the biphasic effect of H19 in regulating osteogenic differentiation of MSCs. These results are helpful for further identify the function and regulatory mechanisms of H19 in osteogenic differentiation of MSCs.
Acknowledgements
We would like to thank the staffs and scholars in Molecular Oncology Laboratory, Medical Center, The University of Chicago for the contribution to this work. The reported work was supported in part by the National Natural Science Foundation of China (NSFC) (#81371972, #81572142 and #81972069). This project was also supported by Natural Science Foundation of Chongqing Science and Technology Commission (#cstc2018jcyjAX0088 and #cstc2017shmsA0787), Major Project of Chongqing Health and Family Planning Commission (#2015-1-12), Cultivating Program of The First Affiliated Hospital of Chongqing Medical University (#2018PYJJ-11) and Pre-NSFC research program of Chongqing Medical University. JL was a recipient of the Predoctoral Fellowship from the China Scholarship Council, the Graduate Research and Innovation Project from Chongqing Education Commission and the Outstanding Predoctorate Research Fellowship from Chongqing Medical University (#CYB15098). Funding sources were not involved in the study design, in the collection, analysis and interpretation of data; in writing of the report; and in the decision to submit the paper for publication.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Strausberg RL, Levy S, Rogers YH. Emerging DNA sequencing technologies for human genomic medicine. Drug Discov Today. 2008;13:569–577. doi: 10.1016/j.drudis.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 2.Chan EY. Advances in sequencing technology. Mutat Res. 2005;573:13–40. doi: 10.1016/j.mrfmmm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, Rozowsky JS, Gerstein MB, Wahlestedt C, Hayashizaki Y, Carninci P, Gingeras TR, Mattick JS. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev. 2013;22:2240–2253. doi: 10.1089/scd.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 8.Leone S, Santoro R. Challenges in the analysis of long noncoding RNA functionality. FEBS Lett. 2016;590:2342–2353. doi: 10.1002/1873-3468.12308. [DOI] [PubMed] [Google Scholar]
- 9.Breyer B, Jiang W, Cheng H, Zhou L, Paul R, Feng T, He TC. Adenoviral vector-mediated gene transfer for human gene therapy. Curr Gene Ther. 2001;1:149–162. doi: 10.2174/1566523013348689. [DOI] [PubMed] [Google Scholar]
- 10.Graham FL, Prevec L. Adenovirus-based expression vectors and recombinant vaccines. Biotechnology. 1992;20:363–390. doi: 10.1016/b978-0-7506-9265-6.50022-1. [DOI] [PubMed] [Google Scholar]
- 11.Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, Vogelstein B, He TC. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 12.Lee CS, Bishop ES, Zhang R, Yu X, Farina EM, Yan S, Zhao C, Zheng Z, Shu Y, Wu X, Lei J, Li Y, Zhang W, Yang C, Wu K, Wu Y, Ho S, Athiviraham A, Lee MJ, Wolf JM, Reid RR, He TC. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu N, Zhang H, Deng F, Li R, Zhang W, Chen X, Wen S, Wang N, Zhang J, Yin L, Liao Z, Zhang Z, Zhang Q, Yan Z, Liu W, Wu D, Ye J, Deng Y, Yang K, Luu HH, Haydon RC, He TC. Overexpression of Ad5 precursor terminal protein accelerates recombinant adenovirus packaging and amplification in HEK-293 packaging cells. Gene Ther. 2014;21:629–637. doi: 10.1038/gt.2014.40. [DOI] [PubMed] [Google Scholar]
- 14.Wei Q, Fan J, Liao J, Zou Y, Song D, Liu J, Cui J, Liu F, Ma C, Hu X, Li L, Yu Y, Qu X, Chen L, Yu X, Zhang Z, Zhao C, Zeng Z, Zhang R, Yan S, Wu X, Shu Y, Reid RR, Lee MJ, Wolf JM, He TC. Engineering the rapid adenovirus production and amplification (RAPA) cell line to expedite the generation of recombinant adenoviruses. Cell Physiol Biochem. 2017;41:2383–2398. doi: 10.1159/000475909. [DOI] [PubMed] [Google Scholar]
- 15.Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113:188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- 16.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 17.Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, Hensle T, Weiss L, McMorrow L, Loew T, Kraus W, Gerald W, Tycko B. Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nat Genet. 1994;7:440–7. doi: 10.1038/ng0794-440. [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi T, Sullivan MJ, Ogawa O, Reeve AE. Epigenetic changes encompassing the IGF2H19 locus associated with relaxation of IGF2 imprinting and silencing of H19 in Wilms tumor. Proc Natl Acad Sci U S A. 1995;92:2159–63. doi: 10.1073/pnas.92.6.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Li G, Zhang JF. The role of long non-coding RNA H19 in musculoskeletal system: a new player in an old game. Exp Cell Res. 2017;360:61–65. doi: 10.1016/j.yexcr.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Qian B, Wang DM, Gu XS, Zhou K, Wu J, Zhang CY, He XY. LncRNA H19 serves as a ceRNA and participates in non-small cell lung cancer development by regulating microRNA-107. Eur Rev Med Pharmacol Sci. 2018;22:5946–5953. doi: 10.26355/eurrev_201809_15925. [DOI] [PubMed] [Google Scholar]
- 22.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 24.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Zheng Y, Jin C, Li X, Jia L, Li W. Long non-coding RNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci Rep. 2016;6:28897. doi: 10.1038/srep28897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang WC, Fu WM, Wang YB, Sun YX, Xu LL, Wong CW, Chan KM, Li G, Waye MM, Zhang JF. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep. 2016;6:20121. doi: 10.1038/srep20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Zheng Y, Jia L, Li W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-beta1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells. 2015;33:3481–3492. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 28.Liao J, Yu X, Hu X, Fan J, Wang J, Zhang Z, Zhao C, Zeng Z, Shu Y, Zhang R, Yan S, Li Y, Zhang W, Cui J, Ma C, Li L, Yu Y, Wu T, Wu X, Lei J, Wang J, Yang C, Wu K, Wu Y, Tang J, He BC, Deng ZL, Luu HH, Haydon RC, Reid RR, Lee MJ, Wolf JM, Huang W, He TC. lncRNA H19 mediates BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) through Notch signaling. Oncotarget. 2017;8:53581–53601. doi: 10.18632/oncotarget.18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu S, Wang J, Ye J, Zou Y, Zhu Y, Wei Q, Wang X, Tang S, Liu H, Fan J, Zhang F, Farina EM, Mohammed MM, Song D, Liao J, Huang J, Guo D, Lu M, Liu F, Liu J, Li L, Ma C, Hu X, Lee MJ, Reid RR, Ameer GA, Zhou D, He T. Bone morphogenetic protein 9 (BMP9) induces effective bone formation from reversibly immortalized multipotent adipose-derived (iMAD) mesenchymal stem cells. Am J Transl Res. 2016;8:3710–3730. [PMC free article] [PubMed] [Google Scholar]
- 30.Rameshwar P, Huang E, Bi Y, Jiang W, Luo X, Yang K, Gao JL, Gao Y, Luo Q, Shi Q, Kim SH, Liu X, Li M, Hu N, Liu H, Cui J, Zhang W, Li R, Chen X, Shen J, Kong Y, Zhang J, Wang J, Luo J, He BC, Wang H, Reid RR, Luu HH, Haydon RC, Yang L, He TC. Conditionally immortalized mouse embryonic fibroblasts retain proliferative activity without compromising multipotent differentiation potential. PLoS One. 2012;7:e32428. doi: 10.1371/journal.pone.0032428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao J, Wei Q, Zou Y, Fan J, Song D, Cui J, Zhang W, Zhu Y, Ma C, Hu X, Qu X, Chen L, Yu X, Zhang Z, Wang C, Zhao C, Zeng Z, Zhang R, Yan S, Wu T, Wu X, Shu Y, Lei J, Li Y, Luu HH, Lee MJ, Reid RR, Ameer GA, Wolf JM, He TC, Huang W. Notch signaling augments BMP9-induced bone formation by promoting the osteogenesis-angiogenesis coupling process in mesenchymal stem cells (MSCs) Cell Physiol Biochem. 2017;41:1905–1923. doi: 10.1159/000471945. [DOI] [PubMed] [Google Scholar]
- 32.Liao J, Hu N, Zhou N, Lin L, Zhao C, Yi S, Fan T, Bao W, Liang X, Chen H, Xu W, Chen C, Cheng Q, Zeng Y, Si W, Yang Z, Huang W. Sox9 potentiates BMP2-induced chondrogenic differentiation and inhibits BMP2-induced osteogenic differentiation. PLoS One. 2014;9:e89025. doi: 10.1371/journal.pone.0089025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu N, Jiang D, Huang E, Liu X, Li R, Liang X, Kim SH, Chen X, Gao JL, Zhang H, Zhang W, Kong YH, Zhang J, Wang J, Shui W, Luo X, Liu B, Cui J, Rogers MR, Shen J, Zhao C, Wang N, Wu N, Luu HH, Haydon RC, He TC, Huang W. BMP9-regulated angiogenic signaling plays an important role in the osteogenic differentiation of mesenchymal progenitor cells. J Cell Sci. 2012;126:532–541. doi: 10.1242/jcs.114231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, Wang J, Deng F, Yan Z, Xia Y, Wang Z, Ye J, Deng Y, Zhang Z, Qiao M, Li R, Denduluri SK, Wei Q, Zhao L, Lu S, Wang X, Tang S, Liu H, Luu HH, Haydon RC, He TC, Jiang L. TqPCR: a touchdown qPCR assay with significantly improved detection sensitivity and amplification efficiency of SYBR green qPCR. PLoS One. 2015;10:e0132666. doi: 10.1371/journal.pone.0132666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng H, Jiang W, Phillips FM, Haydon RC, Peng Y, Zhou L, Luu HH, An N, Breyer B, Vanichakarn P, Szatkowski JP, Park JY, He TC. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs) J Bone Joint Surg Am. 2003;85:1544–1552. doi: 10.2106/00004623-200308000-00017. [DOI] [PubMed] [Google Scholar]
- 38.Kang Q, Sun MH, Cheng H, Peng Y, Montag AG, Deyrup AT, Jiang W, Luu HH, Luo J, Szatkowski JP, Vanichakarn P, Park JY, Li Y, Haydon RC, He TC. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther. 2004;11:1312–1320. doi: 10.1038/sj.gt.3302298. [DOI] [PubMed] [Google Scholar]
- 39.Sharff KA, Song WX, Luo X, Tang N, Luo J, Chen J, Bi Y, He BC, Huang J, Li X, Jiang W, Zhu GH, Su Y, He Y, Shen J, Wang Y, Chen L, Zuo GW, Liu B, Pan X, Reid RR, Luu HH, Haydon RC, He TC. Hey1 basic helix-loop-helix protein plays an important role in mediating BMP9-induced osteogenic differentiation of mesenchymal progenitor cells. J Biol Chem. 2009;284:649–659. doi: 10.1074/jbc.M806389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercey O, Popa A, Cavard A, Paquet A, Chevalier B, Pons N, Magnone V, Zangari J, Brest P, Zaragosi LE, Ponzio G, Lebrigand K, Barbry P, Marcet B. Characterizing isomiR variants within the microRNA-34/449 family. FEBS Lett. 2017;591:693–705. doi: 10.1002/1873-3468.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandbothe M, Buurman R, Reich N, Greiwe L, Vajen B, Gurlevik E, Schaffer V, Eilers M, Kuhnel F, Vaquero A, Longerich T, Roessler S, Schirmacher P, Manns MP, Illig T, Schlegelberger B, Skawran B. The microRNA-449 family inhibits TGF-beta-mediated liver cancer cell migration by targeting SOX4. J Hepatol. 2017;66:1012–1021. doi: 10.1016/j.jhep.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Wildung M, Esser TU, Grausam KB, Wiedwald C, Volceanov-Hahn L, Riedel D, Beuermann S, Li L, Zylla J, Guenther AK, Wienken M, Ercetin E, Han Z, Bremmer F, Shomroni O, Andreas S, Zhao H, Lize M. Transcription factor TAp73 and microRNA-449 complement each other to support multiciliogenesis. Cell Death Differ. 2019;26:2740–2757. doi: 10.1038/s41418-019-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui J, Mo J, Luo M, Yu Q, Zhou S, Li T, Zhang Y, Luo W. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:12400–12409. [PMC free article] [PubMed] [Google Scholar]
- 44.Kou N, Liu S, Li X, Li W, Zhong W, Gui L, Chai S, Ren X, Na R, Zeng T, Liu H. H19 facilitates tongue squamous cell carcinoma migration and invasion via sponging miR-let-7. Oncol Res. 2019;27:173–182. doi: 10.3727/096504018X15202945197589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao L, Zhang Z, Li Y, Zhao P, Chen Y. LncRNA H19/miR-let-7 axis participates in the regulation of ox-LDL-induced endothelial cell injury via targeting periostin. Int Immunopharmacol. 2019;72:496–503. doi: 10.1016/j.intimp.2019.04.042. [DOI] [PubMed] [Google Scholar]
- 46.Muller V, Oliveira-Ferrer L, Steinbach B, Pantel K, Schwarzenbach H. Interplay of lncRNA H19/miR-675 and lncRNA NEAT1/miR-204 in breast cancer. Mol Oncol. 2019;13:1137–1149. doi: 10.1002/1878-0261.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park H, Park H, Pak HJ, Yang DY, Kim YH, Choi WJ, Park SJ, Cho JA, Lee KW. miR-34a inhibits differentiation of human adipose tissue-derived stem cells by regulating cell cycle and senescence induction. Differentiation. 2015;90:91–100. doi: 10.1016/j.diff.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 48.You L, Pan L, Chen L, Gu W, Chen J. MiR-27a is essential for the shift from osteogenic differentiation to adipogenic differentiation of mesenchymal stem cells in postmenopausal osteoporosis. Cell Physiol Biochem. 2016;39:253–265. doi: 10.1159/000445621. [DOI] [PubMed] [Google Scholar]
- 49.Dong S, Yang B, Guo H, Kang F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem Biophys Res Commun. 2012;418:587–591. doi: 10.1016/j.bbrc.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 50.Fang S, Deng Y, Gu P, Fan X. MicroRNAs regulate bone development and regeneration. Int J Mol Sci. 2015;16:8227–8253. doi: 10.3390/ijms16048227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L, Yang J, Zhu X, Li D, Lv Z, Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283:2326–2339. doi: 10.1111/febs.13741. [DOI] [PubMed] [Google Scholar]
- 52.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kashi K, Henderson L, Bonetti A, Carninci P. Discovery and functional analysis of lncRNAs: methodologies to investigate an uncharacterized transcriptome. Biochim Biophys Acta. 2016;1859:3–15. doi: 10.1016/j.bbagrm.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 54.Blythe AJ, Fox AH, Bond CS. The ins and outs of lncRNA structure: how, why and what comes next? Biochim Biophys Acta. 2016;1859:46–58. doi: 10.1016/j.bbagrm.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 55.Li YP, Wang Y. Large noncoding RNAs are promising regulators in embryonic stem cells. J Genet Genomics. 2015;42:99–105. doi: 10.1016/j.jgg.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giovarelli M, Bucci G, Ramos A, Bordo D, Wilusz CJ, Chen CY, Puppo M, Briata P, Gherzi R. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A. 2014;111:E5023–5028. doi: 10.1073/pnas.1415098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee DF, Su J, Kim HS, Chang B, Papatsenko D, Zhao R, Yuan Y, Gingold J, Xia W, Darr H, Mirzayans R, Hung MC, Schaniel C, Lemischka IR. Modeling familial cancer with induced pluripotent stem cells. Cell. 2015;161:240–254. doi: 10.1016/j.cell.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lattanzi L, Salvatori G, Coletta M, Sonnino C, Cusella De Angelis MG, Gioglio L, Murry CE, Kelly R, Ferrari G, Molinaro M, Crescenzi M, Mavilio F, Cossu G. High efficiency myogenic conversion of human fibroblasts by adenoviral vector-mediated MyoD gene transfer. An alternative strategy for ex vivo gene therapy of primary myopathies. J Clin Invest. 1998;101:2119–2128. doi: 10.1172/JCI1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li S, Hua Y, Jin J, Wang H, Du M, Zhu L, Chu H, Zhang Z, Wang M. Association of genetic variants in lncRNA H19 with risk of colorectal cancer in a Chinese population. Oncotarget. 2016;7:25470–25477. doi: 10.18632/oncotarget.8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, Zhou Y, Huang T, Cheng AS, Yu J, Kang W, To KF. The interplay of lncRNA-H19 and its binding partners in physiological process and gastric carcinogenesis. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarnowski M, Tkacz M, Czerewaty M, Poniewierska-Baran A, Grymula K, Ratajczak MZ. 5Azacytidine inhibits human rhabdomyosarcoma cell growth by downregulating insulinlike growth factor 2 expression and reactivating the H19 gene product miR675, which negatively affects insulinlike growth factors and insulin signaling. Int J Oncol. 2015;46:2241–2250. doi: 10.3892/ijo.2015.2906. [DOI] [PubMed] [Google Scholar]
- 64.Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285:24381–24387. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steck E, Boeuf S, Gabler J, Werth N, Schnatzer P, Diederichs S, Richter W. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med (Berl) 2012;90:1185–1195. doi: 10.1007/s00109-012-0895-y. [DOI] [PubMed] [Google Scholar]
- 66.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen K, Ma Y, Wu S, Zhuang Y, Liu X, Lv L, Zhang G. Construction and analysis of a lncRNAmiRNAmRNA network based on competitive endogenous RNA reveals functional lncRNAs in diabetic cardiomyopathy. Mol Med Rep. 2019;20:1393–1403. doi: 10.3892/mmr.2019.10361. [DOI] [PubMed] [Google Scholar]
- 68.Li JP, Xiang Y, Fan LJ, Yao A, Li H, Liao XH. Long noncoding RNA H19 competitively binds miR-93-5p to regulate STAT3 expression in breast cancer. J Cell Biochem. 2019;120:3137–3148. doi: 10.1002/jcb.27578. [DOI] [PubMed] [Google Scholar]
- 69.Wei J, Gan Y, Peng D, Jiang X, Kitazawa R, Xiang Y, Dai Y, Tang Y, Yang J. Long non-coding RNA H19 promotes TDRG1 expression and cisplatin resistance by sequestering miRNA-106b-5p in seminoma. Cancer Med. 2018;7:6247–6257. doi: 10.1002/cam4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.