Abstract
Non-coding RNA has been reported to be crucial regulator for cancer progression. This work was aimed to investigate the roles and associated mechanisms of non-coding RNA activated by DNA damage (NORAD) in hepatocellular carcinoma (HCC) progression. In this work, we explored the expression of NORAD, microRNA-144-3p (miR-144-3p), and septin 2 (SEPT2) in HCC tissues and cells. Effects of knockdown of ectopic of NORAD on HCC cell proliferation, colony formation, and apoptosis were explored. Rescue experiments were conducted to explore whether NORAD regulates HCC cell behaviors via the miR-144-3p/SEPT2 axis. Moreover, the effect of NORAD on HCC tumor progression in vivo was analyzed. We showed NORAD expression was elevated in both HCC tissues and cells. NORAD knockdown inhibits HCC cell growth but promotes apoptosis, while the overexpression of NORAD has opposite effects. Besides that, we found knockdown of NPRAD inhibits tumor growth. Moreover, we showed miR-144-3p expression was inversely correlated with NORAD and SEPT2, while NORAD and SEPT2 was positively correlated in HCC tissues. Functional assays showed NORAD functions as ceRNA through binding with miR-144-3p to regulate SEPT2 expression in HCC. Collectively, we showed NORAD serves as an oncogenic lncRNA to promote HCC progression via the miR-144-3p/SEPT2 axis.
Keywords: NORAD, miR-144-3p, SEPT2, hepatocellular carcinoma
Introduction
As the fourth leading cause of cancer-related deaths worldwide, liver cancer accounts for 1,761,007 deaths each year [1,2]. Hepatocellular carcinoma (HCC) is reported to represent 85-90% of all liver cancer cases and often accompanied with cirrhosis, hepatitis B or C virus infection [1,2]. The prognosis for HCC patients is poor due to only 10-37% of them are suitable for surgery treatment [3]. Hence, it is imperative to explore the mechanisms underlying HCC progression.
As an important type of non-coding RNA (ncRNA), long non-coding RNA (lncRNA) has been recently reported to play crucial roles in tumorigenesis [4]. They have limited protein-coding capabilities but can regulate numerous gene expression mainly through competing endogenous RNA (ceRNA) mechanism via interacting with microRNA (miRNA) [5,6]. miRNAs are conserved ncRNA at the length of 18-25 nucleotides and regulated gene expression through 3’-untranslated region (3’-UTR) binding [7]. In recent years, multiplies studies revealed lncRNAs were involved in HCC tumorigenesis via functioning as ceRNA [8,9]. Numbers of lncRNAs that aberrantly expressed in HCC are large, but pathways are diverse. Bioinformatic analysis methods were revealed to be powerful tools to identify abnormally expressed lncRNAs in cancers [10,11].
Non-coding RNA activated by DNA damage (NORAD), located on exon of Chr20q11.23, was found upregulated expression in cancers [12-14]. In ovarian cancer, it was found NORAD knockdown could inhibit ovarian cancer proliferation, migration, and invasion but promotes apoptosis, while the overexpression of NORAD had opposite effects [12]. In epithelial ovarian cancer, NORAD was also found upregulated in tumor tissues and cells, and regulated proliferation, drug resistance, and tumor progression via serving as sponge for miR-155-5p [13]. In non-small cell lung cancer, NORAD was found significantly elevated expression in patients with metastasis and advance tumor stages [14]. Moreover, miR-656-3p was identified as a target for NORAD to regulate cancer cell proliferation and migration [14].
miR-144-3p was found decreased expression in castration-resistant prostate cancer tissues and cells compared with normal tissues and cells [15]. In addition, we showed that miR-144-3p overexpression inhibits the cell proliferation, and colony formation but promotes apoptosis in castration-resistant prostate cancer cells via targeting centrosomal protein 55 [15]. Moreover, miR-144-3p was shown to be overexpressed in papillary thyroid carcinoma and promotes cancer progression via targeting paired box gene 8 [16]. These results indicated that miR-144-3p has diverse roles in cancers.
In this work, we explored the expression pattern and functions of NORAD in HCC. Then, ceRNA theory was applied to understand NORAD and miRNA-mRNA crosstalk in HCC. The purpose of this work was to investigate the regulatory mechanisms of NORAD in HCC.
Materials and methods
Expression dataset
Levels of NORAD, microRNA-144-3p (miR-144-3p), and septin 2 (SEPT2) in HCC tissues and normal tissues were analyzed at StarBase. Correlation of NORAD or SEPT2 and miR-144-3p was also analyzed at StarBase.
Cell culture
Normal human hepatocyte LO2 cell and HCC cell lines (Hep3B, Huh7, BEL-7402 and HCCLM3) were purchased from Cell Bank of Chinese Academy of Sciences (Shanghai, China). These cells were incubated in DMEM (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS (Invitrogen) at 37°C moist atmosphere contains 5% CO2.
Cell transfection
NORAD expression vector (pcNORAD) and pcDNA3.1 was purchased from GeneChem (Shanghai, China). miR-144-3p mimic (5’-UACAGUAUAGAUGAUGUACU-3’) and suitable corresponding control (miR-NC, 5’-UUCUCCGAACGUGUCACGUTT-3’) was also bought from GeneChem. Small interfering RNA targeting NORAD (si-NORAD, 5’-GCGGTTGGTCTTCATTCTA-3’) or SEPT2 (si-SEPT2, 5’-GCTGCTCACAATCGTTGAT-3’) and matched controls (siR-NCs, 5’-TTCTCCGAACGTGTCACGT-3’) were obtained from GeneChem. Transfection was conducted using Lipofectamine 2000 when cell confluence reached to about 60%.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Trizol (Invitrogen) was used to isolated RNA from the cultured cells. RNA concentration was quantified with NanoDrop-1000, then equal amount of RNA sample (1 μg) was reverse transcribed into complementary DNA with PrimerScript kit (Takara, Dalian, Liaoning, China). qRT-PCR was performed at QuanStudio 6 (Thermo Fisher Scientific, Inc.) using SYBR regent (Takara). Primer sequences were: NORAD: F: 5’-TGATAGGATACATCTTGGACATGGA-3’, R: 5’-AACCTAATGAACAAGTCCTGACATACA-3’; SEPT2: F: 5’-GGTGACGCTATCAACTGCAGAG-3’, R: 5’-ATGATGTGCCGCCTGTTCAAGC-3’; GAPDH: F: 5’-GAAGAGAGAGACCCTCACGCTG-3’, R: 5’-ACTGTGAGGAGGGGAGATTCAGT-3’; miR-144-3p: F: 5-GCGCGCTACAGTATAGATGATG-3’, R: 5’-GCTGTCAACGATACGCTACG-3’; U6 snRNA: F: 5’-CTCGCTTCGGCAGCACA-3’, R: 5-AACGCTTCACGAATTTGCGT-3’. The 2-ΔΔCt method was used to calculate the relative expression levels. PCR amplification conditions were: 95°C for 5 sec, 60°C for 30 s, 72°C for 30 s for 40 cycles.
Western blot
Cells were washed with PBS and treated with RIPA lysis buffer in supplement with protease inhibitors. Protein concentration was determined by BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific), and 50 μg samples were then isolated with 10% SDS-PAGE and transferred into PVDF membrane. Membrane was incubated with skim-free milk and then primary antibodies (anti-Bax: ab182733; anti-Bcl2: ab196495; anti-GAPDH: ab181602; Abcam, Cambridge, MA, USA) at 4°C for overnight. Then, membrane was treated with HRP-conjugated secondary antibody (ab6721, Abcam) at 37°C for 40 min. Protein signals were treated with ECL reagent (Beyotime, Haimen, Jiangsu, China) and analyzed with Image J software.
Cell proliferation assay
Cell proliferation capacity was measured with cell counting kit-8 (CCK-8) kit purchased from Beyotime. Cells transfected were seeded into 96-well plates at the density of 2 × 103 cells/well and cultured for indicated time until the adding of 10 μl CCK-8 reagent. Optical density of each well was measured at 450 nm using microplate reader (Bio-Rad, Hercules, CA, USA).
Colony formation assay
To detect the colonies numbers, 1,000 cells were seeded in 6-well plates and maintained at 37°C incubator for 14 days. Polyoxymethylene was used to fix colonies for 30 min, which were further stained with 0.1% crystal violet for 10 min and counted with microscope (Nikon, Tokyo, Japan).
Flow cytometry assay
2 × 105 cells were collected, suspended with binding buffer, and stained with Annexin V-FITC and propidium iodide (PI) at dark for 20 min. Subsequently, cell apoptosis rate was measured using flow cytometry (Beckman Coulter, Miami, FL, USA) by counting 2 × 104 cells.
Luciferase activity reporter assay
The StarBase (http://starbase.sysu.edu.cn/) was applied to analyze the miRNA target that can bind with NORAD and the mRNA target that can bind with miRNA. Through bioinformatic analysis, miR-144-3p and SEPT2 were selected for followingly analyses. The wild-type (wt) and mutant (mt) sequence of NORAD and SEPT2 were synthesized by Sangon (Shanghai, China) and inserted into pGL3 (Promega, Madison, WI, USA) to generate wt/mt-NORAD/SEPT2. Cells were transfected with luciferase activity vectors and miRNAs with lipofectamine 2000 for 48 h. Then, relative luciferase activity was measured with Dual-Luciferase Reporter Assay System (Promega) after normalization to Renilla luciferase activity.
RIP immunoprecipitation (RIP) assay
RIP assay was performed using EZ-Magna RiP Kit (Millipore, Billerica, MA, USA) followingly the manufacturer’s protocols. Cells were lysed in RIP lysis buffer and then incubated with magnetic beads conjugated with anti-Ago2 or anti-IgG. qRT-PCR was performed to measure the enrichment of NORAD, miR-144-3p, or SEPT2 after RNA isolation using Trizol reagent.
Animal study
Animal study protocol was approved by Tianjin First Central Hospital. HCCLM3 transfected with lenti-shRNA NORAD (sh-NORAD) or control (sh-NC) were injected into buttocks of mice. Tumor length and width were measured every 1 week. Mice were killed after 4 weeks to measure tumor weight and then photo the tumor tissues.
Immunohistochemical (IHC) analysis
For hematoxylin-eosin stain (HE) staining, tissues obtained from mice were embedded and sectioned, and stained with haematoxylin and eosin. For IHC studies, tissues samples were incubated with anti-Ki67 (ab16667) and secondary antibody. Signal was visualized using 3’-diaminobenzidine chromogen (DAB), and then counterstained with haematoxylin.
Statistical analysis
Data were presented as means ± SD after analyzing at SPSS 17.0 (SPSS Inc, Chicago, IL, USA). Student’s t-test and one-way ANOVA with Tukey’s post hoc test were applied for difference analysis in groups. P values less than 0.05 was regarded as statistically significant.
Results
NORAD was highly expressed in HCC tissues and cells
To analyze the roles of NORAD in HCC, we explored its expression by analyzing data from StarBase. We showed that NORAD expression was significantly upregulated in HCC tissues compared with normal tissues (Figure 1A). Moreover, qRT-PCR analysis result showed that NORAD levels were also elevated expression in HCC cells compared with normal cell line (Figure 1B).
Figure 1.
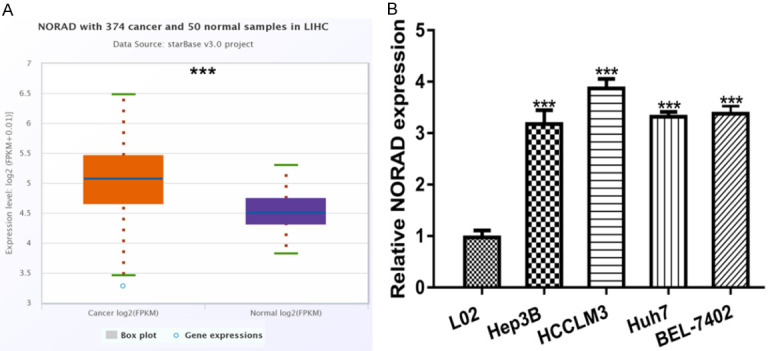
NORAD exhibits remarkedly high expression in HCC tissues and cells. A. NORAD expression in HCC tissues and normal tissues. B. NORAD expression in HCC cells and normal cell. NORAD: non-coding RNA activated by DNA damage; HCC: hepatocellular carcinoma.
Knockdown of NORAD inhibits HCC cell proliferation, colony formation but promotes apoptosis
To detect the function of NORAD in HCC, the HCC cell with highest NORAD expression was selected for loss-of-function experiments. The introduction of si-NORAD decreased NORAD expression in HCC cell (Figure 2A). Through CCK-8 assay and colony formation assay, we showed the knockdown of NORAD inhibits HCC cell proliferation and colony formation (Figure 2B and 2C). In addition, the flow cytometry assay revealed that NORAD knockdown promotes HCC cell apoptosis (Figure 2D). Western blot showed Bax expression was increased, whereas Bcl-2 expression was decreased by si-NORAD (Figure 2E).
Figure 2.
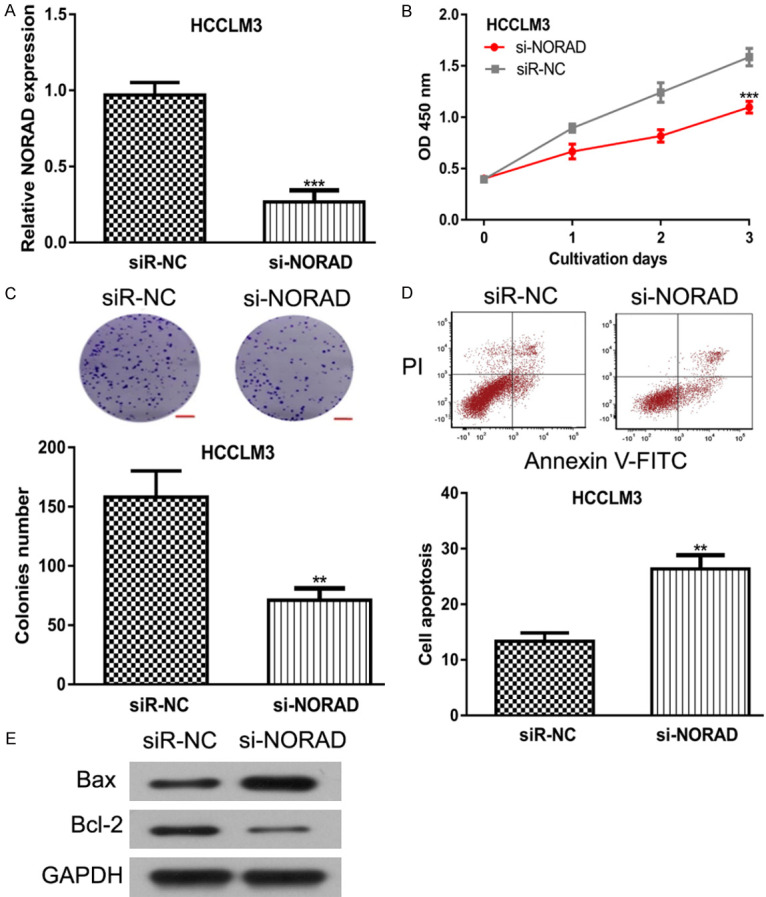
NORAD knockdown inhibits HCC cell proliferation, colony formation but promotes apoptosis. After si-NORAD transfection, (A) NORAD expression was suppressed, (B) Cell proliferation rate was inhibited, (C) Colony formation ability was inhibited, while (D) Cell apoptosis rate was stimulated, and (E) Bax expression was increased and Bcl-2 expression was decreased in HCC cells. NORAD: non-coding RNA activated by DNA damage; HCC: hepatocellular carcinoma; si-NORAD: small interfering RNA against NORAD; siR-NC: negative control siRNA.
Overexpression of NORAD promotes HCC cell proliferation, colony formation but inhibits apoptosis
In addition, gain-of-function experiments were applied to further understand the biological roles of NORAD in HCC. NORAD expression level was significantly increased by pcNORAD in HCC cell compared with pcDNA3.1 (Figure 3A). NORAD overexpression enhanced HCC cell proliferation rate as indicated by CCK-8 assay (Figure 3B). Colony formation assay further confirmed that NORAD overexpression could increase the colonies number formed in pcNORAD transfected groups (Figure 3C). Furthermore, we showed the overexpression of NORAD inhibits cell apoptosis in HCC (Figure 3D). Western blot showed Bax expression was inhibited, whereas Bcl-2 expression was elevated by pcNORAD (Figure 3E).
Figure 3.
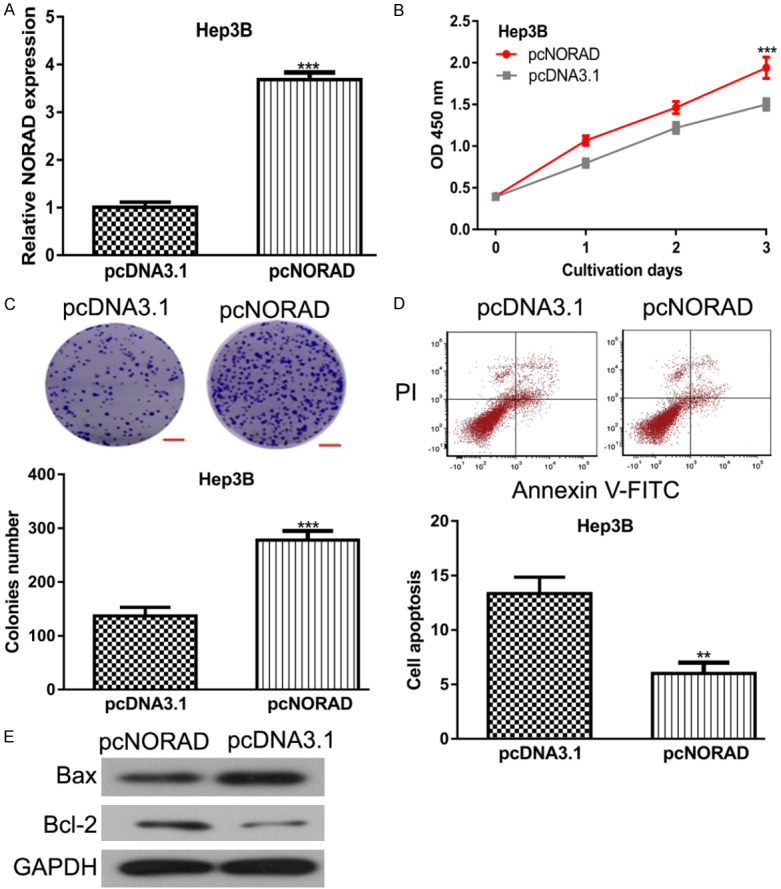
NORAD overexpression promotes HCC cell proliferation, colony formation but inhibits apoptosis. After pcNORAD transfection, (A) NORAD expression was increased, (B) Cell proliferation rate was elevated, (C) Colony formation ability was enhanced, while (D) Cell apoptosis rate was inhibited, and (E) Bax expression was decreased and Bcl-2 expression was increased in HCC cells. NORAD: non-coding RNA activated by DNA damage; HCC: hepatocellular carcinoma.
NORAD binds with miR-144-3p to regulate SEPT2 expression
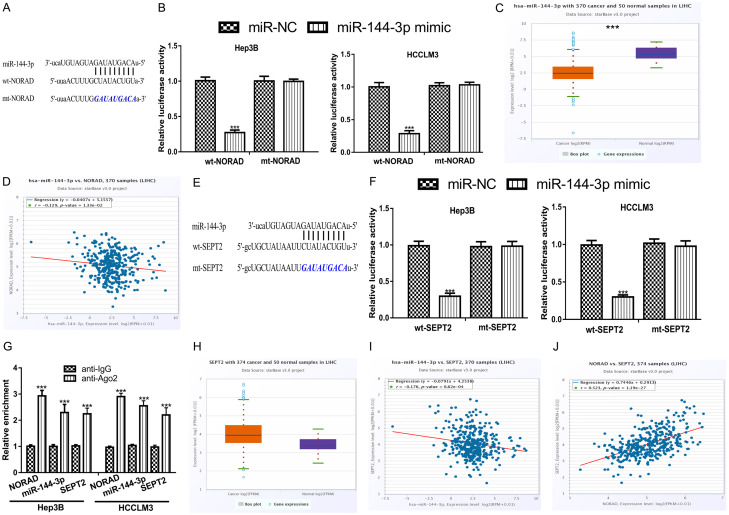
To determine the effects of NORAD in HCC, we explored the downstream targets of NORAD. We found miR-144-3p was a putative target of NORAD (Figure 4A). To validate the interaction of NORAD and miR-144-3p, luciferase activity reporter assay was conducted. We showed luciferase activity in HCC cells harboring wt-NORAD was decreased by miR-144-3p mimic (Figure 4B). Furthermore, we showed miR-144-3p levels were decreased in HCC tissues compared with normal tissues (Figure 4C). Expression correlation assay showed NORAD and miR-144-3p expression was negatively correlated in HCC tissues (Figure 4D). In addition, we analyzed the targets for miR-144-3p and revealed SEPT2 was a possible target (Figure 4E). Luciferase activity reporter assay demonstrated that miR-144-3p reduced the luciferase activity in HCC cells transfected with wt-SEPT2, but not those with mt-SEPT2 (Figure 4F). In addition, we found SEPT2 expression was elevated in HCC tissues compared with normal tissues (Figure 4G). RIP assay showed NORAD, miR-144-3p, and SEPT1 was co-enriched in anti-Ago2 groups compared with anti-IgG groups (Figure 4H). Meanwhile, miR-144-3p and SEPT2 was revealed to be inversely correlated in HCC tissues (Figure 4I). More importantly, we revealed NORAD and SEPT2 was positively correlated in HCC tissues (Figure 4J). These results indicated that NORAD could bind with miR-144-3p to regulate SEPT2 expression in HCC.
Figure 4.
NORAD and SEPT2 shared binding site in miR-144-3p. A. Binding region between NORAD and miR-144-3p. B. Luciferase activity in HCC cells harboring wt/mt-NORAD and miR-144-3p mimic or miR-NC showed luciferase activity can be decreased by miR-144-30 mimic in cells with wt-NORAD. C. miR-144-3p expression was decreased in HCC tissues compared with normal tissues. D. Negatively correction of NORAD and miR-144-3p in HCC tissues. E. Binding region between SEPT2 and miR-144-3p. F. Luciferase activity in HCC cells harboring wt/mt-SEPT2 and miR-144-3p mimic or miR-NC showed luciferase activity can be decreased by miR-144-30 mimic in cells with wt-SEPT2. G. SEPT2 expression was increased in HCC tissues compared with normal tissues. H. RIP assay indicated that NORAD, miR-144-3p, and SEPT2 were co-enriched in anti-Ago2 group compared with anti-IgG group. I. Negatively correction of SEPT2 and miR-144-3p in HCC tissues. J. Positively correction of SEPT2 and NORAD in HCC tissues. NORAD: non-coding RNA activated by DNA damage; HCC: hepatocellular carcinoma; miR-144-3p: microRNA-144-3p; SEPT2: septin 2; wt: wild-type; mt: mutant; miR-NC: negative control microRNA.
Effects of NORAD/miR-144-3p/SEPT2 axis on HCC cells
To further investigate the roles of NORAD/miR-144-3p/SEPT2 axis in HCC, we performed rescue experiments. The results of qRT-PCR indicated that SEPT2 expression was increased by pcNORAD and miR-144-3p mimic but decreased by si-SEPT2 (Figure 5A). Function assays showed cell proliferation and colony formation were decreased, while cell apoptosis was stimulated by miR-144-3p mimic and si-SEPT2 (Figure 5B-D). In addition, we showed the effects of NORAD overexpression on HCC cell behaviors could be partially reversed by either miR-144-3p overexpression or SEPT2 knockdown (Figure 5B-D).
Figure 5.
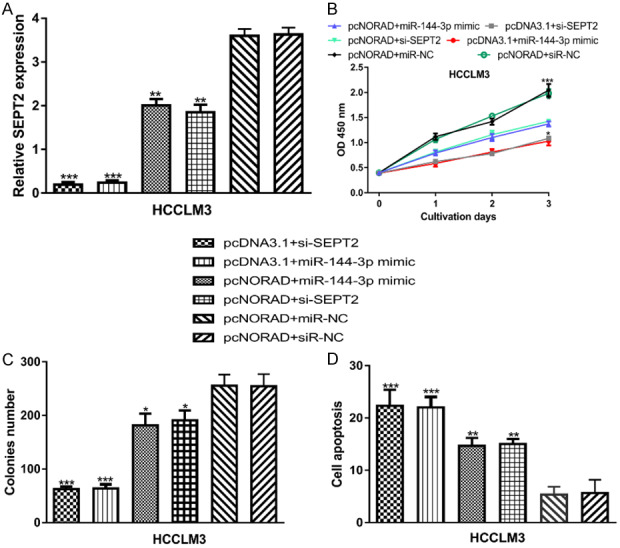
NORAD regulates HCC cell behaviors via regulating miR-144-3p/SEPT2 axis. (A) NORAD expression, (B) Cell proliferation, (C) Colony formation, and (D) Cell apoptosis in HCC cell with pcDNA3.1+miR-144-3p mimic, pcDNA3.1+si-SEPT2, pcNORAD+miR-144-3p mimic, pcNORAD+si-SEPT2, pcNORAD+miR-NC, or pcNORAD+siR-NC transfection. NORAD: non-coding RNA activated by DNA damage; HCC: hepatocellular carcinoma; miR-144-3p: microRNA-144-3p; SEPT2: septin 2; miR-NC: negative control microRNA; siR-NC: negative control siRNA.
NORAD knockdown inhibits HCC tumor growth in vivo
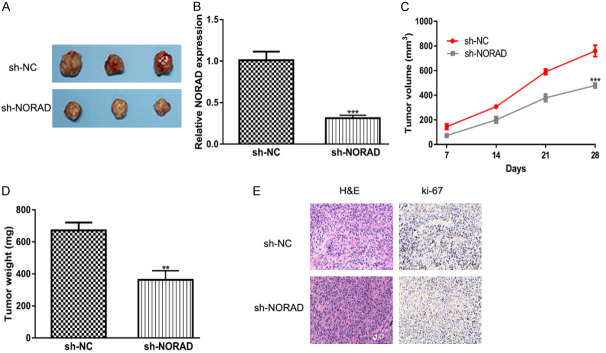
Furthermore, animal experiments were conducted to detect the roles of NORAD on HCC tumor growth. As shown in Figure 6A, tumors formed in the sh-NORAD groups were smaller than those formed in the sh-NC groups. In addition, the knockdown of NORAD inhibits NORAD expression, tumor volume and weight compared with normal groups (Figure 6B-D). Moreover, IHC assays showed that NORAD knockdown group had lower ki-67 levels (Figure 6E).
Figure 6.
NORAD knockdown inhibits HCC tumor growth in vivo. A. The images of dissected tumors from sh-NORAD groups and sh-NC groups. B. NORAD expression level was decreased in sh-NORAD groups compared with sh-NC groups. C. Tumor volume was lower in sh-NORAD groups than in sh-NC groups. D. Tumor weight was lower in sh-NORAD groups than in sh-NC groups. E. Tumur tissue samples were immunostained for H&E and ki-67. NORAD: non-coding RNA activated by DNA damage; HCC: hepatocellular carcinoma; sh-NORAD: small hairpin RNA against NORAD; sh-NC: negative control shRNA.
Discussion
Increasing evidence indicated lncRNAs play crucial roles in regulating carcinogenesis, and may be developed as novel therapeutic targets for cancer [4-6]. In this work, we provided novel evidence that lncRNA NORAD was upregulated in HCC tumor tissues and cells, and knockdown of NORAD could exert anti-tumor effects in HCC through regulating the miR-144-3p/SEPT2 axis.
At first, we showed that NORAD was elevated expression in HCC tissues compared with normal tissues by analyzing the data obtained from StarBase. In addition, qRT-PCR method showed NORAD was also upregulated in the HCC cells investigated compared with normal cell line. Then, investigations to explore the biological roles of NORAD in HCC were performed. We showed overexpression of NORAD promotes HCC cell growth by inhibiting apoptosis, while the knockdown of NORAD caused opposite effects on HCC cell behaviors. In addition, the knockdown of NORAD inhibits HCC tumor growth in vivo.
Next, we tried to explore the mechanisms by which NORAD exerts the oncogenic role in HCC through analyzing its downstream targets. We showed miR-144-3p was a target for NORAD, while SEPT2 was a putative target for miR-144-3p. miR-144-3p was selected for analysis due to miR-144-3p ranks top among all the prediction miRNA target results of NORAD using StarBase. Moreover, miR-144-3p was found can be directly regulated by lncRNAs including NEAT1, TUG1, and so on [17,18]. The luciferase activity reporter assay confirmed the interaction of NORAD or SEPT2 with miR-144-3p. Moreover, we showed miR-144-3p expression was negatively correlated with NORAD or SEPT2 in HCC tissues. More importantly, we found NORAD and SEPT2 was positively correlated in HCC tissues. Our work here showed that miR-144-3p overexpression could inhibit cancer cell proliferation, colony formation but promote apoptosis, suggesting the tumor suppressive role of miR-144-3p. SEPT2 was found overexpressed in biliary tract cancer and promoted cancer progression via negatively regulated by miR-140-5p, indicating the oncogenic role of SEPT2 [19]. Our work presented here also revealed the oncogenic role of SEPT2 in HCC. In future work, the expression level of NORAD and its associations with overall survival, disease-free survival, and clinicopathological features of HCC patients should be validated to strength the importance of NORAD in HCC.
Conclusions
In summary, we showed that NORAD could stimulate HCC cell proliferation and colony formation but inhibit apoptosis with the mechanisms of absorbing miR-144-3p to promote SEPT2 expression. Our work provided novel evidence in understanding the mechanisms underlying HCC progression.
Acknowledgements
This work was supported by Tianjin Science and Technology Plan Project (19ZXDBSY00010).
Disclosure of conflict of interest
None.
References
- 1.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.O’Suilleabhain CB, Poon RT, Yong JL, Ooi GC, Tso WK, Fan ST. Factors predictive of 5-year survival after transarterial chemoembolization for inoperable hepatocellular carcinoma. Br J Surg. 2003;90:325–331. doi: 10.1002/bjs.4045. [DOI] [PubMed] [Google Scholar]
- 4.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Zhu S, Meng N, He Y, Lu R, Yan GR. ncRNA-encoded peptides or proteins and cancer. Mol Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lara JC, Arzate-Mejía RG, Recillas-Targa F. Enhancer RNAs: insights into their biological role. Epigenet Insights. 2019;12:2516865719846093. doi: 10.1177/2516865719846093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaheer U, Faheem M, Qadri I, Begum N, Yassine HM, Al Thani AA, Mathew S. Expression profile of microRNA: an emerging hallmark of cancer. Curr Pharm Des. 2019;25:642–653. doi: 10.2174/1386207322666190325122821. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, Shi X, Ye G, Xu Y, Xu J, Lu J, Lu W. Up-regulated long non-coding RNA DUXAP8 promotes cell growth through repressing Krüppel-like factor 2 expression in human hepatocellular carcinoma. Onco Targets Ther. 2019;12:7429–7436. doi: 10.2147/OTT.S214336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Zhou Y, Yang L, Ma Y, Peng X, Yang S, Li H, Liu J. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J Cell Physiol. 2020;235:3402–3413. doi: 10.1002/jcp.29230. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Tang D, Zhang Z, Wang Z. Identification of aberrantly expressed lncRNA and the associated TF-mRNA network in hepatocellular carcinoma. J Cell Biochem. 2020;121:1491–1503. doi: 10.1002/jcb.29384. [DOI] [PubMed] [Google Scholar]
- 11.Zhao X, Tang DY, Zuo X, Zhang TD, Wang C. Identification of lncRNA-miRNA-mRNA regulatory network associated with epithelial ovarian cancer cisplatin-resistant. J Cell Physiol. 2019;234:19886–19894. doi: 10.1002/jcp.28587. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Yan Y, Chen Y, Li J, Yang J. Involvement of NORAD/miR-608/STAT3 axis in carcinostasis effects of physcion 8-O-β-glucopyranoside on ovarian cancer cells. Artif Cells Nanomed Biotechnol. 2019;47:2855–2865. doi: 10.1080/21691401.2019.1637884. [DOI] [PubMed] [Google Scholar]
- 13.Tong L, Ao Y, Zhang H, Wang K, Wang Y, Ma Q. Long noncoding RNA NORAD is upregulated in epithelial ovarian cancer and its downregulation suppressed cancer cell functions by competing with miR-155-5p. Cancer Med. 2019;8:4782–4791. doi: 10.1002/cam4.2350. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Chen T, Qin S, Gu Y, Pan H, Bian D. Long non-coding RNA NORAD promotes the occurrence and development of non-small cell lung cancer by adsorbing MiR-656-3p. Mol Genet Genomic Med. 2019;7:e757. doi: 10.1002/mgg3.757. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.You B, Zhang KC. MicroRNA-144-3p inhibits cell proliferation and promotes apoptosis in castration-resistant prostate cancer by targeting CEP55. Eur Rev Med Pharmacol Sci. 2018;22:7660–7670. doi: 10.26355/eurrev_201811_16383. [DOI] [PubMed] [Google Scholar]
- 16.Liu C, Su C, Chen Y, Li G. MiR-144-3p promotes the tumor growth and metastasis of papillary thyroid carcinoma by targeting paired box gene 8. Cancer Cell Int. 2018;18:54. doi: 10.1186/s12935-018-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei JL, Wu CJ, Chen JJ, Shang FT, Guo SG, Zhang XC, Liu H. LncRNA NEAT1 promotes the progression of sepsis-induced myocardial cell injury by sponging miR-144-3p. Eur Rev Med Pharmacol Sci. 2020;24:851–61. doi: 10.26355/eurrev_202001_20069. [DOI] [PubMed] [Google Scholar]
- 18.Wang P, Yang Z, Ye T, Shao F, Li J, Sun N, He J. lncTUG1/miR-144-3p affect the radiosensitivity of esophageal squamous cell carcinoma by competitively regulating c-MET. J Exp Clin Cancer Res. 2020;39:7. doi: 10.1186/s13046-019-1519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Zhang W, Tang H, Qian H, Yang J, Zhu Z, Ren P, Lu B. Septin 2 accelerates the progression of biliary tract cancer and is negatively regulated by mir-140-5p. Gene. 2016;589:20–26. doi: 10.1016/j.gene.2016.05.005. [DOI] [PubMed] [Google Scholar]