Abstract
Pancreatic cancer (PC) is one of the top deaths causing cancers with low 5-year survival rate. Long non-coding RNAs (lncRNAs) are recognized as a crucial type of nonprotein-coding transcripts implicated in tumorigenesis. Emerging evidence has implied that LINC00152 exerts the potential oncogenic functions in various cancers. Nevertheless, the role of LINC00152 in PC remains elusive. In the present study, we found that LINC00152 was significantly up-regulated while miR-150 was down-regulated both in tissues and cell lines of PC, indicating their negative correlation in PC progression. Functionally, overexpression of LINC00152 promoted cell proliferation, migration and invasion, while LINC00152 knockdown reversed these effects. Mechanistic experiments reveal that miR-150 acted as a target of LINC00152 confirmed by luciferase reporter assay. Moreover, inhibition of miR-150 could markedly attenuate the suppression of cell proliferation, migration and invasion by knocking down LINC00152. Altogether, our findings concluded that LINC00152 facilitated PC progression through inhibiting miR-150 expression, indicating an innovative therapeutic target for PC.
Keywords: LINC00152, miR-150, pancreatic cancer
Introduction
As one of the most death causing malignancies, pancreatic cancer (PC) with a 5-year survival rate of less than 5% is predicted to have 55,440 new cases and 44,330 deaths in United States [1]. Similarly, according to the investigation by the National Office for Cancer Prevention and Control of China, about 90,100 new cases and 79,400 deaths will be occurred every year in China [2]. With the current diagnostic and therapeutic strategies, PC are usually diagnosed at an advanced stage and metastasis has already been occurred [3]. No effective treatment could be achieved to improve the outcome of PC patients [4]. The lack of the meaningful biomarkers for PC significantly hampers the early diagnosis and effective treatment of PC, which arouses the urgent needs to explore the mechanisms in PC occurrence and progression [4].
With the development of sequencing techniques and bioinformatics methods, a majority (>90%) portion of a genome is recognized to be transcribed into non-coding RNAs, which codes no proteins [5]. Among the non-coding RNAs, those with more than 200nt in length was defined as long non-coding RNAs (lncRNAs) [6-8]. Even the controversial biological properties of the reported lncRNAs, the important regulatory function of lncRNAs has been widely recognized and investigated [9]. For instance, a series of biological process, such as cell cycle, cell differentiation, epigenetic regulation, cancer initiation and metastasis, and development of drug resistance, have been proved to be modulated by lncRNAs [10-12]. Specifically, aberrant expression of lncRNAs was also found in various cancers, which play important roles in the cancer development, metastasis, recurrence and poor outcomes [13-17]. For example, Hui et al. recently found that lncRNA AGAP2-AS1 promoted PC cell growth and migration through suppressing ANKRD1 and ANGPTL4 [3]. Jiang et al. revealed that lncRNA HOTAIR was associated with the susceptibility of PC [18]. LINC00152, transcribed from chromosome region 2p11.2, was firstly found overexpressed in gastric cancer and worked as an oncogene in many other cancers as well, such as gliomas, liver, lung, and colorectal cancer [19-21]. LINC00152 promoted the cell proliferation and invasion ability of these cancer cells by regulating various targets, including EGFR, EZH2, miR-16, and miR-139-5p [22-26]. These findings implied the important role and complicated interaction of LINC00152 in different cancers. However, the function and role of LINC00152 in PC remains little known.
MicroRNA (miRNAs) is a small type of non-coding RNAs with 20-24 nucleotides in length [27]. miRNAs regulate gene expression through degrading the targeting mRNAs or suppressing the translation efficiency of the targeting mRNAs [28]. In varies types of cancers, miRNAs play important roles in cell proliferation, migration and invasion [29-31]. They might be recognized as oncogenes or tumor suppressors in different cancers. For instance, Feng et al. discovered that miR-150 suppressed the tumor development in colorectal cancer by targeting c-Myb [32]. Recently, Srivastava et al. found that miR-150 inhibited the growth and malignant behavior in PC cells by regulating MUC4, indicating the important ole of miR-150 in PC development [28]. In addition, lncRNAs have been reported to fulfill its function by negatively regulating miRNAs in tumor progression [13,17,33]. However, there is no investigation of the interaction between LINC00152 and miR-150 in PC cells.
In the present study, we demonstrated that LINC00152 was up-regulated in PC and the miR-150 was down-regulated in PC, which were negatively correlated. The interaction between LINC00152 and miR-150, and the mechanism related to the development of PC cells were investigated. Our findings will help to elucidate the important functions of lncRNAs and miRNAs in gene regulation and tumorigenesis of pancreatic cancer, which might provide potential powerful biomarkers for early diagnosis and therapeutic targets of pancreatic cancer.
Materials and methods
Tissue specimens and cell lines
Pancreatic cancer tissues and adjacent normal tissues from 28 patients were collected during tumor resection with their signed statement of informed consent. Then, the tissues specimens were stored at -80°C in liquid nitrogen before usage. The study was approved by the ethics committee of Hunan Cancer Hospital.
Four pancreatic cancer cell lines, including BxPC3, Panc1, AsPC1, and SW1990, as well as normal pancreatic duct epithelial cell line HPDE6-C7, were purchased from the American Type Culture Collection (ATCC, Manassas, VA). HEK293T cells were obtained from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). All the cell lines were cultured with RPMI 1640 medium or DMEM medium supplemented with 10% fetal bovine serum (FBS) in a humidified chamber with 5% CO2 at 37°C.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The TRIzol reagent (Invitrogen, Carlsbad, CA) was used to extract the total RNA from the clinical tissues and cell lines. Then, complementary DNA (cDNA) was synthesized with the reverse transcription kit (Invitrogen) according to the manufacturer’s protocol. Then, samples are amplified and analyzed by SYBR Green PCR Kit (Toyobo, Nipro, Osaka, Japan) according to the manufacturer’s protocol. Finally, the 7500 Fast Real-time PCR System (Applied Biosystems, Waltham, MA) for qRT-PCR analysis. All PCR assays were carried out at least three times. The expression levels of target genes were normalized to corresponding loading controls and calculated by using the 2-ΔΔCt method. The primers sequences were shown as follows: LINC00152: 5’-AGTTACGGAGGACCCAGCAA-3’ (sense) and 5’-GGGCTGAGTCGTGATTTTCG-3’ (antisense); miR-152: 5’-AGCGCAGGTTCTGTGATACACT-3’ (sense) and 5’-AACGCTTCACGAATTTGCGT-3’ (antisense); GAPDH: 5’-CCAGGTGGTCTCCTCTGA-3’ (sense) and 5’-GCTGTAGCCAAATCGTTGT-3’ (antisense); U6: 5’-CTCGCTTCGGCAGCACA-3’ (sense) and 5’-AACGCTTCACGAATTTGCGT-3’ (antisense).
Cell transfection
All the small interfering RNAs (siRNA), including siRNA LINC00152 (siLINC00152) and scrambled control siRNA (siNC) were synthesized by GenePharma (Shanghai, China). The sequences were shown as follows: siLINC00152: 5’-UCUAUGUGUCUUAAUCCCUUGUCCU-3’; siNC: 5’-GACACGCGACUUGUACCAC-3’. Besides, pcDNA3.1 empty vector (Invitrogen, Shanghai, China) inserting with the cDNA sequences of LINC00152 was subjected to construct overexpressing plasmid of LINC00152. Moreover, the mimics and inhibitors of miR-150 and the negative controls were obtained from GenePharma. Lipofectamine 2000 transfection reagent (Invitrogen) was used for transfection according to the manufacturer’s instructions.
CCK-8 assay
Cell lines were seeded in 96-well plates with density of 2000 cells per well and then cultured with different treatment. Then, cell proliferation was analyzed with CCK-8 assay (Dojindo Laboratories, Kumanoto, Japan) according to the manufacturer’s instruction. Briefly, after the different treatment, 10 µL of the CCK-8 solution was added into each well, followed by another 2 h incubation at 37°C. The absorbance at 490 nm was read to calculate the cell viability rates on a microplate reader.
Colony formation assay
In a 6-well plate, different treated cells were cultured with a density of 5000 cells/well in the medium for 15 days. Finally, the cells were fixed using methanol and then stained with 0.1% crystal violet. The visible colonies were counted, and all the experiments were repeated at three times.
EDU assay
The cell proliferation was also tested using EDU assay, with a EDU assay kit (RiboBio, Nanjing, China). Briefly, the treated cells were stained with Hoechst 33342, and then imaged by a fluorescence microscope (Olympus Corp., Tokyo., Japan). The images were analyzed using the Image J software (National Institutes of Health, Bethesda, MD).
Wound healing assay
Typically, the cells were cultured in 6-well plates to reach a 90% confluence. Then, a 200 µL pipette was used to scratch the cell layer to form a wound. After 48 h, the wound width was measured to demonstrate the wound healing ability of the tested cells.
Transwell assay
For migration ability evaluation, a two-chamber with a membrane filter inserts (8 µm pore sieze; Corning, Cambridge, MA) was used. The 1000 cells were firstly plated in the top chamber in serum-free medium. The lower chamber was filled with medium containing 10% FBS. After 48 h incubation at 37°C, the cells on the top chamber were removed and the cells on the lower chamber were stained with 1% crystal violet after the fixation by methanol. The stained cells were counted under the microscope and four visual fields were randomly chosen to count. Three independent experiments were performed for each group.
For the invasion assay, the procedure was similar to the migration transwell experiment, except that a layer of Matrigel (BD Biosciences) was coated on the chamber membrane.
Luciferase reporter assay
The wide-type and mutant of LINC00152 were cloned into the pmirGLO luciferase vector (Promega, Madison, WI). HEK293T cells were co-transfected with miR-150 mimics and pmirGLO luciferase vectors by Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. After 48 h incubation, the luciferase reporter activity was measured by Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol. Each experiment was performed in triplicate and repeated at least three times.
Generation of xenografts
The transfected SW1990 cells (2 × 106 in 0.1 ml PBS) was subcutaneously injected to six-week-old BALB/c female athymic nude mice (Vitalriver, Beijing, China) to generate xenografts. The animal experiment was approved by the Animal Ethics Committee of Hunan Cancer Hospital. The volume of xenografts was recorded every 5 days (tumor volume = (length × width2)/2). After 25 days, the mice were sacrificed, and the tumor samples were collected for routine IHC.
Histological examination
The tumor tissues from the mice were fixed in 10% formalin and embedded in paraffin. The sections were first stained with hematoxylin and eosin. Then, anti- Ki67 (diluted at 1:100, Santa Cruz Biotechnology), anti-ZEB1 (diluted at 1:100, Abcam, Cambridge, MA, USA) and anti-E-cadherin (diluted at 1:100, Abcam, Cambridge, MA, USA) were used to perform the immunohistochemical staining, according to the manufacture’s instruction.
Statistical analysis
Data are presented as the means ± standard deviation (SD) and each group were repeated at least three times. Student’s t-test or one-way ANOVA was used to compare the difference. P < 0.05 was defined as statistically significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001).
Results
LINC00152 was highly expressed and miR-150 was down-regulated in pancreatic tissues and cell lines
To investigate whether there is the association of LINC00152 and miR-150 in PC, the expression patterns of LINC00152 and miR-150 in PC tissues and normal adjacent tissues were analyzed by qRT-PCR assay. As shown in Figure 1A and 1B, LINC00152 was notably up-regulated but miR-150 was significantly down-regulated in PC tissues compared with the normal tissues. Subsequently, Pearson correlation analysis also demonstrated the negative relationship between LINC00152 and miR-150 in PC tissues (Figure 1C). Following that, the expression patterns of LINC00152 and miR-150 were also examined in five PC cell lines, including SW1990, BxPC-3, AsPC1, Panc1, and HS-766T, as well as the normal pancreatic duct epithelial cell line HPDE6-C7. The results displayed that the expression of LINC00152 was dramatically enhanced in PC cell lines compared with the HPDE6-C7 cells (Figure 1D), inversely, the expression of miR-150 was significantly decreased in pancreatic cancer cell lines compared with the HPDE6-C7 cells (Figure 1E), indicating their negative regulation on PC progression.
Figure 1.
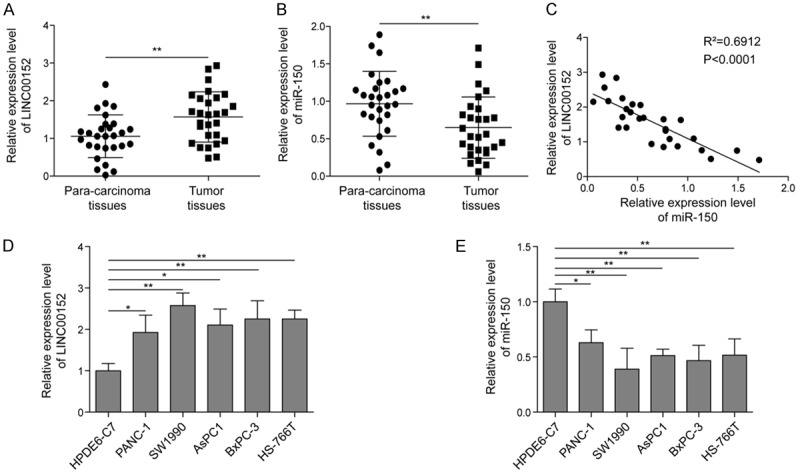
Expression of LINC00152 and miR-150 in PC tissues and cell lines. A, B. The expression levels of LINC00152 and miR-150 in PC tissues (n=28) and adjacent normal tissues (n=28) by qRT-PCR assay. C. The Pearson correlation analysis between LINC00152 and miR-150 in PC tissues. D, E. The expression patterns of LINC00152 and miR-150 in five different PC cell lines, including BxPC3, Panc1, AsPC1, Sw1990, and HS-766T, as well as the normal pancreatic cell line HPDE6-C7 by qRT-PCR analysis. The mean ± SD in the graph presents the relative levels from three replications. *P < 0.05, **P < 0.01.
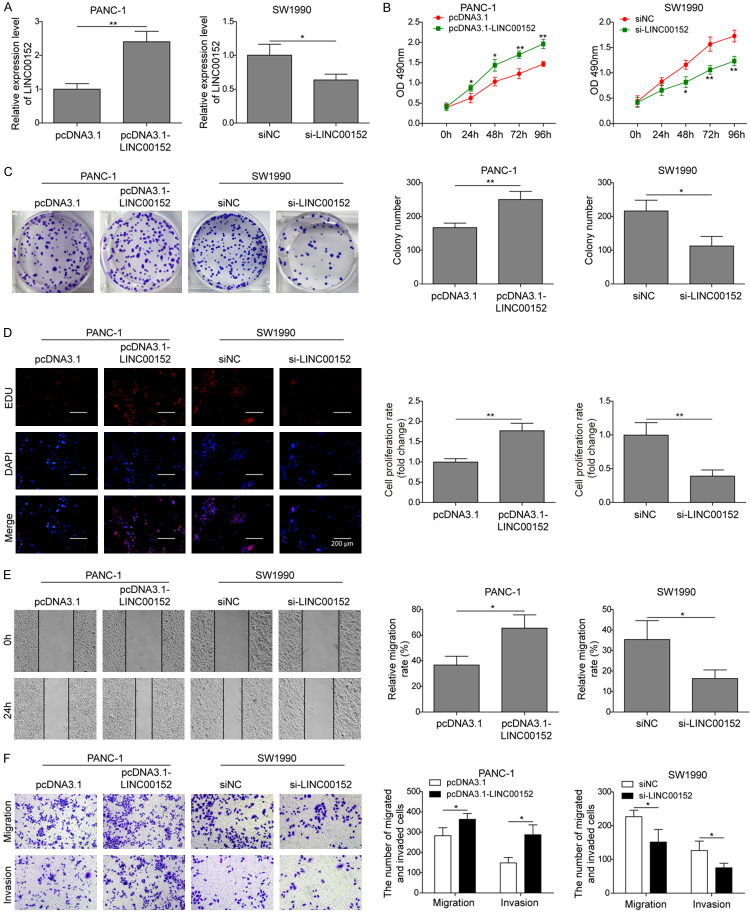
LINC00152 promotes proliferation, migration and invasion of PC cells
To explore the role of LINC00152 in PC tumorigenesis, we overexpressed or silenced LINC00152 in Panc1 and SW1990 cells by transfecting with pcDNA3.1-LINC00152 and si-LINC00152, respectively (Figure 2A), which was based on the determination of LINC00152 expression levels in five PC cell lines. The results demonstrated the successful overexpression or knockdown in LINC00152 in Panc1 and SW1990 cells, respectively. By evaluating cell viability using CCK-8 assay, we found that overexpression of LINC00152 significantly promoted cell viability of Panc1 cells, while knockdown of LINC00152 noticeably caused the suppression of cell viability in SW1990 cells (Figure 2B). We also tested the colony-forming efficiency of Panc1 and SW1990 cells by colony formation assay after transfection with pcDNA3.1-LINC00152 and si-LINC00152, respectively. The results showed that overexpression or knockdown of LINC00152 promoted or inhibited the number of colony formation, respectively (Figure 2C). Additionally, EDU assay also verified the similar outcome with overexpression or knockdown of LINC00152 in PC cell lines (Figure 2D). Furthermore, wound healing assay was used to investigate the impact of LINC00152 on cell migration ability. As shown in Figure 2E, overexpression or knockdown of LINC00152 showed narrower or broader wound width compared with the control groups, respectively. Meanwhile, the impacts of LINC00152 on migration and invasion of PC cells using transwell assay further validated that overexpression of LINC00152 significantly promoted cell migration and invasion ability. Inversely, knockdown of LINC00152 presented the opposite effects on cell migration and invasion (Figure 2F). Taken together, these results indicated that LINC00152 promoted cell proliferation, migration and invasion of PC cells.
Figure 2.
Effects of LINC00152 on cell proliferation, migration and invasion of PC cells. (A) The expression of LINC00152 in Panc1 and SW1990 cells, cells treated with pcDNA3.1 or pcDNA3.1-LINC00152, and cells treated with siNC and siLINC00152 was detected by qRT-PCR. (B-D) CCK-8, colony formation, EDU assays were used to determine the capacity of cell proliferation of the cells described for (A), respectively. (E) Cell migration by the four groups was detected by wound healing. (F) The transwell assays were used to determine cell migration and invasion ability of Panc1 cells transfected with pcDNA3.1 or pcDNA3.1-LINC00152, and SW1990 cells transfected with siNC or si-LINC00152. The mean ± SD in the graph presents the relative levels from three replications. *P < 0.05, **P < 0.01.
LINC00152 acts as a sponge of miR-150
As indicated by the negative correlations between LINC00152 and miR-150 in PC tissues and cells, we hypothesized that LINC00152 might function as a sponge of miR-150. Bioinformatic analysis (Starbase) also implied the complementary binding sites between LINC00152 and miR-150. Therefore, a dual luciferase assay was used to verify this hypothesis. The results showed that transfection of miR-150 mimics significantly decreased the luciferase activity of LINC00152-WT, whereas, no changes were observed in the luciferase activity of LINC00152-MUT (Figure 3A). Furthermore, overexpression of LINC00152 in Panc1 cells significantly decreased miR-150 expression. However, knockdown of LINC00152 notably enhanced the expression of miR-150 in SW1990 cells (Figure 3B). Subsequently, miR-150 mimics was used to transfect both Panc1 and SW1990 cells to up-regulate the expression of miR-150 (Figure 3C). Interestingly, overexpression of miR-150 dramatically reduced LINC00152 expression in both Panc1 and SW1990 cells (Figure 3D). Therefore, these results demonstrated that LINC00152 inhibited miR-150 by binding with it in PC.
Figure 3.
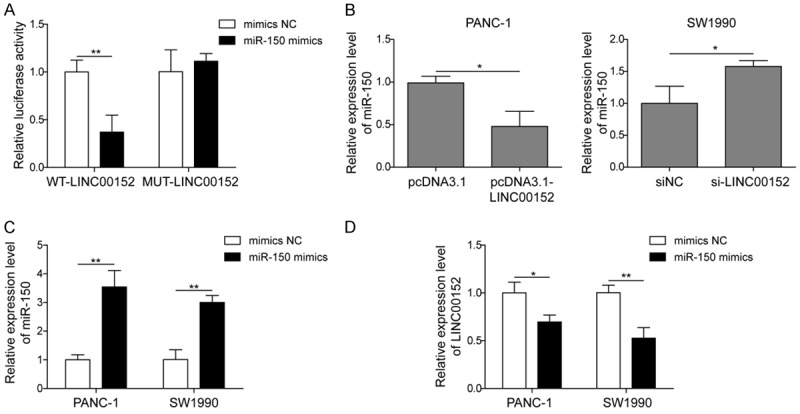
LINC00152 acts as a sponge for miR-150. A. HEK293T cells were co-transfected with luciferase reporter gene vector containing WT-LINC00152 or MUT-LINC00152 and miR-150 mimics or mimics-NC. B. The expression of miR-150 in Panc1 and SW1990 cells transfected with pcDNA3.1 or pcDNA3.1-LINC00152, siNC or si-LINC00152 was determined by qRT-PCR, respectively. C. The expression of miR-150 in Panc1 and SW1990 cells transfected with miR-150 mimics or mimics NC was measured by qRT-PCR. D. The expression of LINC00152 in Panc1 and SW1990 cells transfected with miR-150 mimics or mimics NC was measured by qRT-PCR. The mean ± SD in the graph presents the relative levels from three replications. *P < 0.05, **P < 0.01.
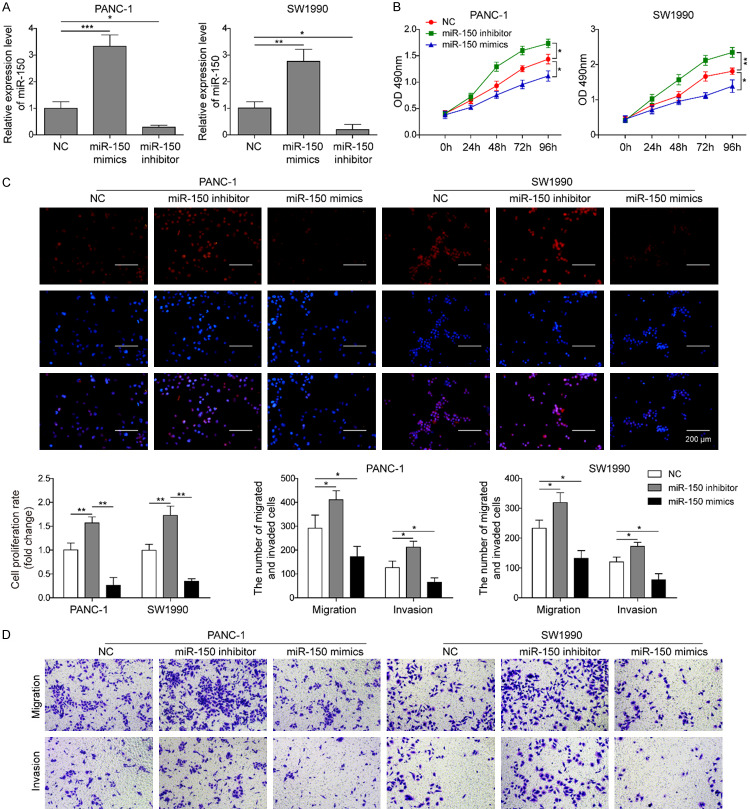
miR-150 suppresses the cell proliferation, migration and invasion of PC cells
To verify the role of miR-150 in the development of PC, miR-150 mimic, miR-150 inhibitor or the negative control were transfected into Panc1 and SW1990 cells. As presented in Figure 4A, miR-150 mimic notably up-regulated the expression of miR-150 in both cell lines, whereas miR-150 inhibitor significantly silenced the expression of miR-150 in both cell lines compared with the negative control group. Through the CCK-8 (Figure 4B) and EDU assays (Figure 4C), we found that overexpression of miR-150 significantly suppressed cell proliferation of Panc1 and SW1990 cells. On the contrary, silencing of miR-150 in Panc1 and SW1990 cells significantly promoted cell proliferation. Moreover, overexpression of miR-150 in Panc1 and SW1990 cells also significantly suppressed cell migration and invasion (Figure 4D). Reversely, knockdown of miR-150 dramatically promoted cell migration and invasion (Figure 4D). Therefore, the data suggested that miR-150 may exert tumor-suppressive effects on PC.
Figure 4.
miR-150 suppresses cell proliferation, migration and invasion. A. qRT-PCR analysis of miR-150 in Panc1 and SW1990 cells transfected with mimic NC, miR-150 mimics or miR-150 inhibitor. B. The cell viability of Panc1 and SW1990 cells transfected with mimics NC, miR-150 mimics or miR-150 inhibitor was evaluated by CCK-8 assay at 24, 48, 72, and 96 hours. C. The effects of miR-150 in Panc1 and SW1990 cells on proliferation was determined using EDU assay. D. The cell migration and invasion ability of Panc1 and SW1990 cells transfected with mimics NC, miR-150 mimic or miR-150 inhibitor was evaluated by transwell assay. The mean ± SD in the graph presents the relative levels from three replications. *P < 0.05, **P < 0.01, ***P < 0.001.
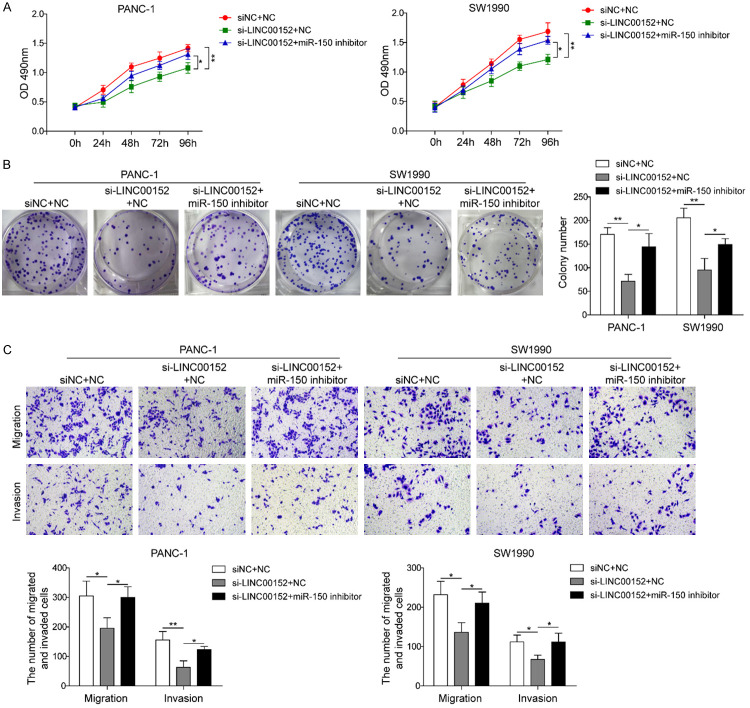
miR-150 reverses the effect of LINC00152 on PC cells
To uncover whether LINC00152 affected the disease development of pancreatic cancer cells through sponging miR-150, miR-150 inhibitor or negative control (NC) vector was co-transfected into PC cells after silencing LINC00152. As shown in Figure 5A and 5B, miR-150 inhibitor reversed the dramatic reduction by silencing LINC00152 on cell viability and colony formation. Similarly, miR-150 inhibitor also abolished the suppressive effects of LINC00152 knockdown on cell migration and invasion in PC cells (Figure 5C). These results indicated that LINC00152 might regulate PC progression via targeting miR-150.
Figure 5.
miR-150 reverses the effect of LINC00152 on PC cells. A. The CCK-8 assay for Panc1 and SW1990 cells transfected with siNC + mimics NC, si-LINC00152 + mimics NC, and si-LINC00152 + miR-150 inhibitor at 24, 48, 72, and 96 hours. B. The cell proliferation by the three groups was detected by colony formation assay. C. The transwell assays of cell migration and invasion were performed in PANC-1 and SW1990 cells transfected with siNC + mimics NC, si-LINC00152 + mimics NC, and si-LINC00152 + miR-150 inhibitor. All results were presented as the mean ± SD and experiment were performed in triplicate. *P < 0.05, **P < 0.01. NC: negative control.
ZEB1 is a direct target of miR-150 in PC cells
In order to investigate the molecular mechanism of miR-150 in PC cells, the potential target of miR-150 was identified through bioinformatics analysis. The results from the analysis using StarBase v2.0 (http://starbase.sysu.edu.cn/) demonstrated that ZEB1 was a target of miR-150 (Figure 6A). Next, the dual-luciferase reporter assay was performed to validate the relationship between ZEB1 and miR-150 in PC cells. As shown in Figure 6B, the results showed that overexpression of miR-150 in cells transfected with the wild-type plasmid significantly suppressed luciferase activity. In contrast, overexpression of miR-150 in cells transfected with the mutant plasmid had no effect on the luciferase activity. Furthermore, we performed qRT-PCR and western blot to determine the expression level of ZEB1 in PC cells after different treatments. We found that the ZEB1 protein level was increased when the PC cells were transfected with miR-150 inhibitor compared with control cells (Figure 6C and 6D). However, transfection with miR-150 mimics significantly suppressed the expression level of ZEB1 in PC cells (Figure 6C and 6D). Taken together, these results indicated that ZEB1 is the functional target of miR-150 in PC cells.
Figure 6.
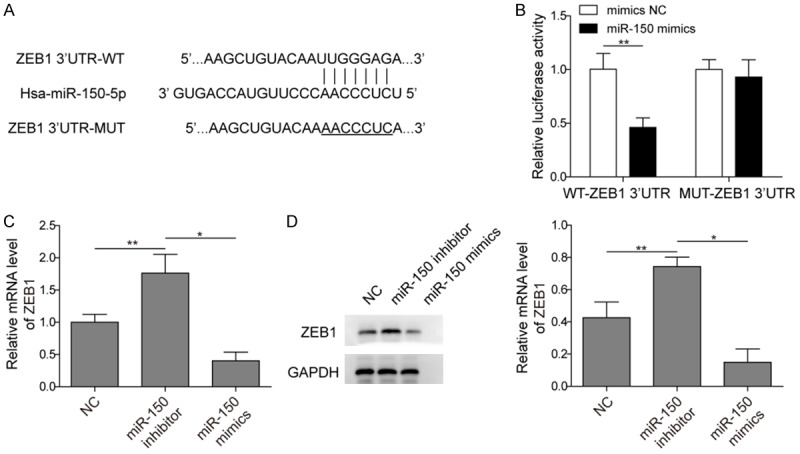
ZEB1 is a direct target of miR-150 in PC cells. A. Predicted miR-150 target sequence in the 3’-UTR of ZEB1 mRNA. B. miR-150 suppressed the luciferase activity of the wild-type ZEB1 3’-UTR expression vector but had no effect on the one with mutant ZEB1. C. The mRNA expression of ZEB1 in the PC cells after transfection with NC, miR-150 inhibitor, and miR-150 mimics, respectively. D. The protein level of ZEB1 in the PC cells after transfection with NC, miR-150 inhibitor, and miR-150 mimics, respectively (The original western blot images reference to Supplementary Figure 1). All results were presented as the mean ± SD and experiment were performed in triplicate. *P < 0.05, **P < 0.01. NC: negative control.
LINC00152 promotes tumor growth of pancreatic cancer in vivo by targeting miR-150
In order to confirm the tumor promotion effect of LINC00152, SW1990 cells transfected with si-NC or si-LINC00152 were injected subcutaneously into nude mice to establish the xenograft model in vivo. Each group contained 6 mice, and the tumor volumes were recorded every 5 days until 25 days. As shown in Figure 7A-C, silencing of LINC00152 significantly inhibited the tumor growth. HE staining also demonstrated that the damage of pancreatic tissues in the si-LINC00152 group was significantly attenuated (Figure 7D). Moreover, IHC analysis of Ki67, ZEB1 and E-cadherin presented that knockdown of LINC00152 dramatically caused the reduction of Ki-67 and ZEB1, while induced the increased of E-cadherin in PC tissues (Figure 7D). Furthermore, we analyzed the expression level of LINC00152 and miR-150 in these two groups by qRT-PCR. The results showed that the expression of LINC00152 was significantly suppressed, whereas the expression of miR-150 was markedly enhanced in the tumor tissues transfected with si-LINC00152 (Figure 7E). Overall, these results indicated that LINC00152 may promote PC progression by sponging miR-150.
Figure 7.
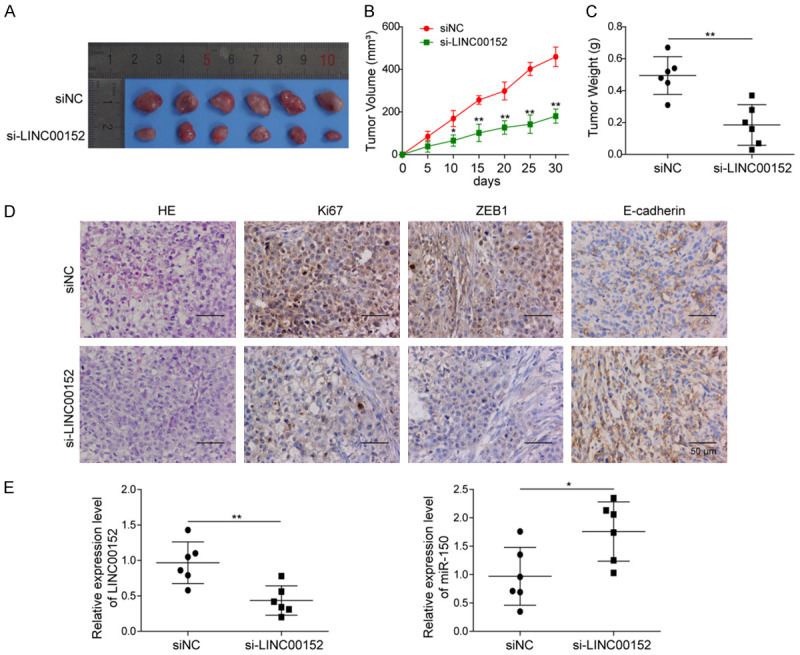
LINC00152 promoted PC tumor growth by sponging miR-150 in mice. A. Representative images of xenograft were shown after transfecting with si-NC or si-LINC00152. B, C. Tumor growth was monitored by analyzing tumor volume and weight. D. HE and immunohistochemical staining were performed to analyze the Ki-67, ZEB1, and E-cadherin expressions in xenografts. E. The expressions of LINC00152 and miR-150 in xenografts by qRT-PCR. The mean ± SD in the graph presents the relative levels from three replications. *P < 0.05, **P < 0.01.
Discussion
Pancreatic cancer leads to the extremely poor prognosis (with 5-year survival rate less than 10%) due to the limitation of biomarkers for early diagnosis and potential potent therapeutic targets [34]. The first-line chemotherapeutics for pancreatic cancer is usually effective but the resistance is generated quickly, causing the recurrence with the addition of metastasis [35]. To improve the survival rate of the PC patients, the discovery of the potential biomarkers and therapeutics targets attracts great attention for both scientists and pharmaceutical companies.
lncRNAs have been recognized as an important molecular star in several biological process, such as cell development and human diseases [11,12,17,19]. Specifically, a large number of lncRNAs have been found to be involved in different types of cancer, impacting the tumor initiation, metastasis, drug resistance, and invasion [8,13,18,33]. The mechanisms of these lncRNAs in cancer progression for gene regulation include genomic imprinting, RNA decay, sponge-like miRNAs, and chromatin modification [36,37]. Several lncRNAs, such as HOTTIP, NORAD, PVT1, and SNHG15, were all found to play important roles in PC development [38-41]. For example, Hui et al. found that TUG1 regulates the cell cycle, proliferation, and apoptosis of PC cells by suppressing RND3 and MT2A [42]. LINC00152 has been widely demonstrated aberrant expression in many cancer types, such as gastric cancer, lung cancer, renal cancer, hepatocellular carcinoma, and gallbladder cancer [19,22,24-26]. For instance, LINC00152 was found to be involved in cell cycle arrest, cell migration and invasion, and apoptosis in gastric cancer [22]. Up-regulation of LINC00152 in glioma cells significantly promoted cell proliferation and invasion by targeting miR-16 [20]. As an oncogene, LINC00152 was also found overexpressed in colorectal cancer to promote cell proliferation, metastasis, and induce 5-FU resistance by suppressing miR-139-5p [19]. In this study, we determined the expression level of the LINC00152 in human PC tissues and found that LINC00152 was significantly overexpressed in PC tissues compared with the normal health tissues. The analysis of LINC00152 in PC cell lines also confirmed the overexpression of LINC00152 in this PC cells than the normal cell lines. The up-regulation of LINC00152 in PC cell lines was positively correlated with the cell proliferation rate, invasion and migration ability, indicating that LINC00152 may have potential as a novel important prognostic indicator for PC.
Several tumor-related pathways, such as EGFP, PI3K/AKT signaling, mTOR, are one type of targets of LINC00152 to regulate the cancer development and progression [19]. However, miRNA sponge is another new mechanism that LncRNAs might be used to regulate the target genes and related cancer development [43]. LINC00152 has been reported as a miRNA sponge to bind with several miRNAs in cancer cells [19,20]. For example, in renal cell carcinoma, LINC00152 was found to repress P16 expression through interacting with miR-205 to promote the renal cancer cell progression [44]. In glioma cells, LINC00152 was recognized as a molecular sponge for miR-16 to enhance the cell proliferation and invasion of glioma [20]. In the present study, we found that the expression of miR-150 has exact opposite expression levels in PC tissues and cell lines to those of LINC00152 through qRT-PCR analysis, suggesting that LINC00152 might be a direct target of miR-150.
miR-150 was reported to play a suppressive role in many cancers, such as colorectal cancer, leukemia, melanoma, and pancreatic cancer [28,32,45]. It is usually downregulated and associated with the poor proliferation, invasion and metastatic ability of cancer cells. miR-150 has relative low expression in malignant pancreatic tissues, and overexpression of miR-150 could significantly inhibit the growth, clonogenicity, migration and invasion of PC cells [28]. We speculated that the function of LINC00152 in PC cells is fulfilled by regulating the miR-150 pathway. The results from the luciferase and qRT-PCR analysis demonstrated that the binding of LINC00152 to miR-150. The following functional and mechanistic assays confirmed that LINC00152 promotes the cell proliferation, migration and invasion by competitively sponging miR-150. The in vivo results also demonstrated that LINC00152 silencing could significantly inhibit the PC development.
In summary, our work demonstrates that LINC00152 is overexpressed in PC, which is negatively correlated with the expression of miR-150. Both of them could be used as potential biomarker for predicting the prognosis of PC. LINC00152 promotes to the PC cell proliferation, migration, and invasion by inhibiting miR-150, uncovering a novel miRNA sponge pathway of LINC00152/miR-150 in PC cells. All these results suggested that LINC00152 may be a potent potential diagnostic biomarker and therapeutic target for PC.
Acknowledgements
This work was supported by the grants from the Natural Science Foundation of Hunan Province (grant number 2018JJ2246); Foundation of Hunan Province (grant number 2019JJ80084) and the Hunan Health and Family Planning Commission Research Fund (grant number B2017-085).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Hui B, Ji H, Xu Y, Wang J, Ma Z, Zhang C, Wang K, Zhou Y. RREB1-induced upregulation of the lncRNA AGAP2-AS1 regulates the proliferation and migration of pancreatic cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death Dis. 2019;10:207. doi: 10.1038/s41419-019-1384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Leveille N, Melo CA, Rooijers K, Diaz-Lagares A, Melo SA, Korkmaz G, Lopes R, Akbari Moqadam F, Maia AR, Wijchers PJ, Geeven G, den Boer ML, Kalluri R, de Laat W, Esteller M, Agami R. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Q, Wang J, Wu X, Ma R, Zhang T, Jin S, Han Z, Tan R, Peng J, Liu G, Li Y, Wang Y. LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res. 2015;43:D193–196. doi: 10.1093/nar/gku1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng Z, Zhang C, Duan C. Functions and mechanisms of long noncoding RNAs in lung cancer. Onco Targets Ther. 2016;9:4411–4424. doi: 10.2147/OTT.S109549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khanduja JS, Calvo IA, Joh RI, Hill IT, Motamedi M. Nuclear noncoding RNAs and genome stability. Mol Cell. 2016;63:7–20. doi: 10.1016/j.molcel.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 12.Mehra M, Chauhan R. Long noncoding RNAs as a key player in hepatocellular carcinoma. Biomark Cancer. 2017;9:1179299X17737301. doi: 10.1177/1179299X17737301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S, Zhang W, Liu K, Liu Y. LncRNA SNHG16 promotes tumor growth of pancreatic cancer by targeting miR-218-5p. Biomed Pharmacother. 2019;114:108862. doi: 10.1016/j.biopha.2019.108862. [DOI] [PubMed] [Google Scholar]
- 14.Chu P, Wang Q, Wang Z, Gao C. Long non-coding RNA highly up-regulated in liver cancer protects tumor necrosis factor-alpha-induced inflammatory injury by down-regulation of microRNA-101 in ATDC5 cells. Int Immunopharmacol. 2019;72:148–158. doi: 10.1016/j.intimp.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Wang F, Zhang S. Knockdown of lncRNA MNX1-AS1 suppresses cell proliferation, migration, and invasion in prostate cancer. FEBS Open Bio. 2019;9:851–858. doi: 10.1002/2211-5463.12611. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Wang R, Feng N, Wang Y, Gao S, Zhang F, Qian Y, Gao M, Yu H, Zhou B, Qian B. SNPs in LncRNA genes are associated with non-small cell lung cancer in a Chinese population. J Clin Lab Anal. 2019;33:e22858. doi: 10.1002/jcla.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Yin L, Li H, Liu LH, Xiao T. The LncRNA LINC00963 facilitates osteosarcoma proliferation and invasion by suppressing miR-204-3p/FN1 axis. Cancer Biol Ther. 2019:1–8. doi: 10.1080/15384047.2019.1598766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang D, Xu L, Ni J, Zhang J, Cai M, Shen L. Functional polymorphisms in LncRNA HOTAIR contribute to susceptibility of pancreatic cancer. Cancer Cell Int. 2019;19:47. doi: 10.1186/s12935-019-0761-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bian Z, Zhang J, Li M, Feng Y, Yao S, Song M, Qi X, Fei B, Yin Y, Hua D, Huang Z. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis. 2017;6:395. doi: 10.1038/s41389-017-0008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Li D, Gao Y, Tang W, Iw L, Cao Y, Hao B. Long Intergenic Noncoding RNA 00152 Promotes Glioma Cell Proliferation and Invasion by Interacting with MiR-16. Cell Physiol Biochem. 2018;46:1055–1064. doi: 10.1159/000488836. [DOI] [PubMed] [Google Scholar]
- 21.Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441–5447. doi: 10.1007/s13277-014-1709-3. [DOI] [PubMed] [Google Scholar]
- 22.Chen WM, Huang MD, Sun DP, Kong R, Xu TP, Xia R, Zhang EB, Shu YQ. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7:9773–9787. doi: 10.18632/oncotarget.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007–2012. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 24.Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G, Sun B. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813–42824. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai Q, Wang ZQ, Wang SH, Li C, Zhu ZG, Quan ZW, Zhang WJ. Upregulation of long non-coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. Am J Transl Res. 2016;8:4068–4081. [PMC free article] [PubMed] [Google Scholar]
- 26.Chen QN, Chen X, Chen ZY, Nie FQ, Wei CC, Ma HW, Wan L, Yan S, Ren SN, Wang ZX. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer. 2017;16:17. doi: 10.1186/s12943-017-0581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava SK, Bhardwaj A, Singh S, Arora S, Wang B, Grizzle WE, Singh AP. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011;32:1832–1839. doi: 10.1093/carcin/bgr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nahand JS, Taghizadeh-Boroujeni S, Karimzadeh M, Borran S, Pourhanifeh MH, Moghoofei M, Bokharaei-Salim F, Karampoor S, Jafari A, Asemi Z, Tbibzadeh A, Namdar A, Mirzaei H. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol. 2019;234:17064–17099. doi: 10.1002/jcp.28457. [DOI] [PubMed] [Google Scholar]
- 30.Yang CX, Sedhom W, Song J, Lu SL. The role of microRNAs in recurrence and metastasis of head and neck squamous cell carcinoma. Cancers (Basel) 2019;11:395. doi: 10.3390/cancers11030395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cristobal I, Sanz-Alvarez M, Luque M, Carames C, Rojo F, Garcia-Foncillas J. The role of microRNAs in hepatoblastoma tumors. Cancers (Basel) 2019;11:409. doi: 10.3390/cancers11030409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng J, Yang Y, Zhang P, Wang F, Ma Y, Qin H, Wang Y. miR-150 functions as a tumour suppressor in human colorectal cancer by targeting c-Myb. J Cell Mol Med. 2014;18:2125–2134. doi: 10.1111/jcmm.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi W, Zhang C, Ning Z, Hua Y, Li Y, Chen L, Liu L, Chen Z, Meng Z. Long non-coding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression. J Exp Clin Cancer Res. 2019;38:60. doi: 10.1186/s13046-019-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang R, Nie W, Yao K, Chou J. Depletion of the lncRNA RP11-567G11.1 inhibits pancreatic cancer progression. Biomed Pharmacother. 2019;112:108685. doi: 10.1016/j.biopha.2019.108685. [DOI] [PubMed] [Google Scholar]
- 35.Avan A, Quint K, Nicolini F, Funel N, Frampton AE, Maftouh M, Pelliccioni S, Schuurhuis GJ, Peters GJ, Giovannetti E. Enhancement of the antiproliferative activity of gemcitabine by modulation of c-Met pathway in pancreatic cancer. Curr Pharm Des. 2013;19:940–950. [PubMed] [Google Scholar]
- 36.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17:756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 37.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 38.Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao X, Li W, Zheng S, Ye H, Wang L, He Z, Lin Q, Li Z, Chen R. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81. doi: 10.1016/j.canlet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, Cheng D, Chen H, Deng X, Peng C, Shen B. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. 2017;16:169. doi: 10.1186/s12943-017-0738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida K, Toden S, Ravindranathan P, Han H, Goel A. Curcumin sensitizes pancreatic cancer cells to gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1 expression. Carcinogenesis. 2017;38:1036–1046. doi: 10.1093/carcin/bgx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Z, Huang H, Wang J, Zhou Y, Pu F, Zhao Q, Peng P, Hui B, Ji H, Wang K. Long non-coding RNA SNHG15 inhibits P15 and KLF2 expression to promote pancreatic cancer proliferation through EZH2-mediated H3K27me3. Oncotarget. 2017;8:84153–84167. doi: 10.18632/oncotarget.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hui B, Xu Y, Zhao B, Ji H, Ma Z, Xu S, He Z, Wang K, Lu J. Overexpressed long noncoding RNA TUG1 affects the cell cycle, proliferation, and apoptosis of pancreatic cancer partly through suppressing RND3 and MT2A. Onco Targets Ther. 2019;12:1043–1057. doi: 10.2147/OTT.S188396. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T, Li X. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7:13479–13490. doi: 10.18632/oncotarget.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Liu J, Bai H, Dang Y, Lv P, Wu S. Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am J Cancer Res. 2017;7:312–322. [PMC free article] [PubMed] [Google Scholar]
- 45.Fang ZH, Wang SL, Zhao JT, Lin ZJ, Chen LY, Su R, Xie ST, Carter BZ, Xu B. miR-150 exerts antileukemia activity in vitro and in vivo through regulating genes in multiple pathways. Cell Death Dis. 2016;7:e2371. doi: 10.1038/cddis.2016.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.