Abstract
There is a lack of a well-established approach for assessment of early treatment outcomes for modern therapies for pancreatic ductal adenocarcinoma (PDAC) e.g. dinaciclib or dendritic cell (DC) vaccination. Here, we developed multivariate models using MRI texture features to detect treatment effects following dinaciclib drug or DC vaccine therapy in a transgenic mouse model of PDAC including 21 LSL-KrasG12D; LSL-Trp53R172H; Pdx-1-Cre (KPC) mice used as untreated control subjects (n=8) or treated with dinaciclib (n=7) or DC vaccine (n=6). Support vector machines (SVM) technique was performed to build a linear classifier with three variables for detection of tumor tissue changes following drug or vaccine treatments. Besides, multivariate regression models were generated with five variables to predict survival behavior and histopathological tumor markers (Fibrosis, CK19, and Ki67). The diagnostic performance was evaluated using accuracy, area under the receiver operating characteristic curve (AUC) and decision curve analyses. The regression models were evaluated with adjusted r-squared (Radj 2). SVM classifier successfully distinguished changes in tumor tissue with an accuracy of 95.24% and AUC of 0.93. The multivariate models generated with five variables were strongly associated with histopathological tumor markers, fibrosis (Radj 2=0.82, P<0.001), CK19 (Radj 2=0.92, P<0.001) and Ki67 (Radj 2=0.97, P<0.001). Furthermore, the multivariate regression model successfully predicted survival of KPC mice by interpreting tumor characteristics from MRI data (Radj 2=0.91, P<0.001). The results demonstrated that MRI texture features had great potential to generate diagnosis and prognosis models for monitoring early treatment response following dinaciclib drug or DC vaccine treatment and also predicting histopathological tumor markers and long-term clinical outcomes.
Keywords: Dendritic cell vaccine, dinaciclib, machine learning, magnetic resonance imaging, pancreatic ductal adenocarcinoma, texture analysis
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive and lethal types of cancer with an expected incidence rate of 3.22% among new cancer patients yet makes up to 7.54% of all cancer-related deaths in the United States [1]. Despite advances in oncology over the last decade, the survival rate of PDAC has not significantly improved (<9%) [2]. As most patients are diagnosed at advanced stages, only 20% of the PDAC patients are suitable for surgical resection [2]. Therefore, novel treatment approaches and imaging biomarkers that would expedite the assessment of treatment outcomes remain to be an important focus of cancer research.
Recent developments in technology and advancements in cancer biology have offered a promising therapeutic option with the use of immunotherapy [3,4]. The unique ability of dendritic cells (DCs) for cross-presenting tumor-associated antigens to CD8+ T cells in the draining lymph nodes makes these cells an ideal candidate for cancer vaccine studies including pancreatic cancer [5,6]. Recently, several drugs have received FDA approval for cancer treatment. Among them, dinaciclib, a potent small-molecule inhibitor of cyclin-dependent kinases 1, 2, 5, and 9 have a good safety profile [7]. Besides, dinaciclib has been well-tolerated in initial clinical trials and showed clinical efficacy against several malignancies [8,9], and also demonstrated inhibition of pancreatic cancer growth and progression in murine xenograft models [10,11].
Tumor size is a widely used metric for assessment of cancer biology [12], characterization of tumor types [13], evaluation of therapeutic responses and survival [14,15]. However, it is not sufficiently sensitive for modern therapies including immunotherapy due to the infiltration of immune cells, intratumoral hemorrhage, intracellular and vasogenic oedema [16-18]. Therefore, novel approaches that reflect biological or metabolic tissue profiles are required for dynamic monitoring of therapeutic outcomes.
Texture analysis is an approach to determine underlying tissue characteristics by analyzing radiological data which offered a new procedure for diagnosis, evaluation of disease severity and prediction of survival [19-23]. Besides, quantitative analysis of medical images has enabled revealing complex structures of tumor tissues and demonstrated potential benefits in the field of oncology [18,24-26]. Despite wide usage in clinical cancer studies, the technical feasibility of MRI texture analysis has not been well investigated in animal studies which can facilitate translation of gold standard histology and empower integration of precision medicine into clinical research.
The purpose of our study was to evaluate the technical feasibility of quantitative MRI texture features for detection of early treatment response by interpreting underlying structural characteristics of tumor tissue and prediction of overall survival (OS) following dinaciclib or DC vaccine therapy with multivariate classification and regression models in a KPC mouse model of PDAC.
Materials and methods
In our pre-clinical studies, we followed regulations determined by the Institutional Animal Care and Use Committee of Northwestern University and treated animals humanely while regularly monitoring the quality of life of the subjects.
Animal model and treatments
In this study, LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre (KPC) mouse model was preferred due to high similarity in pathophysiological and biological characteristics to human PDAC [27-29]. The mouse strains p53LSL−R270H (strain number 01XM3), KrasLSL−G12D (strain number 01XJ6), and Pdx-1-Cre (strain number 01XL5) were purchased from Jackson Laboratory (Bar Harbor, ME). KPC mice were generated and characterized by performing the procedure described in [27]. Twenty-one KPC mice were randomly selected for untreated control (n=8) and treatment groups e.g. DC vaccine (n=6) and dinaciclib (n=7). After tumors reached a detectable stage by MRI (tumor diameter > 2 mm, ~3 months of age), 3×106 DCs pulsed with irradiated KPC cell lysates were injected intraperitoneally to mice in DC vaccine treatment group once a week for three weeks. Besides, the mice in the dinaciclib treatment group received dinaciclib (40 mg/kg) intraperitoneally twice a week for three weeks after the detection of a pancreatic tumor on MRI.
MRI acquisition and processing
Starting one week of the completion of the treatment (~4 weeks after tumor detection), KPC mice were scanned once in every two weeks with a 7T Bruker small-bore MRI scanner equipped with a commercial mouse coil (Clinscan, Ettlingen, Germany) using MRI sequences listed in Table 1 until KPC mice reach to one of the humane endpoints [30]. During the imaging procedure, mice were restrained in the supine position and kept under anesthesia administered by an automated delivery system (Isoflurane Vaporizer, Rockmart, GA). A waterbed heating system (SA Instruments, Stony Brook, NY) was also utilized to adjust body temperature while MRI sequences were triggered according to the respiratory rate.
Table 1.
MRI acquisition parameters used on 7T Bruker MRI scanner system with commercial mouse coil
| MRI Sequence | TR (ms) | TE (ms) | FA (°) | Thickness/Gap (mm) | Averages | Resolution (µm) |
|---|---|---|---|---|---|---|
| T1w (axial) | 630 | 20 | 90 | 0.7/0.7 | 2 | 156.25 |
| T2w (coronal) | 1600 | 37 | - | 1.0/2.0 | 1 | 96.35 |
| T2w (axial) | 2066 | 40 | - | 0.8/0.8 | 2 | 117.19 |
MRI data acquired one week after completion of the treatments, (~4 weeks after tumor detection) were examined to detect treatment-related changes in tumor tissue and predict the survival time of the KPC mice. Besides, we analyzed MRI data acquired before euthanasia to predict histopathological tumor markers (fibrosis, CK19, and Ki67). On the basis of T1w and T2w MRI images, pancreatic tumors were outlined on T2w MRI slice including maximal tumor diameter with a consensus of two experienced radiologists under the instruction of a senior radiologist. MRI signal intensity within the region of interest (ROI) was discretized using a fixed bin size approach before computing the quantitative MRI texture features.
Histology
After KPC mice were euthanized, pancreatic tumor sections were dissected by performing surgery in our laboratory. A tumor block was fixed in 10% formalin and embedded in paraffin. A single 5 μm thick pancreatic tissue slide from each KPC mouse was then analyzed with histology staining (Masson’s trichrome for fibrosis) and immunostaining (CK19 and Ki67) in which CK19 is an adenocarcinoma marker associated with PDAC prognosis and Ki67 is used as a marker of tumor cell proliferation. The generated histology slides were then evaluated by an experienced pathologist with more than 10 years of experience in gastrointestinal oncology. Afterward, histology images were quantitatively analyzed using ImageJ at 20× magnification level to measure fibrosis percentage, CK19 positive (CK19*) area, and the number of Ki67 positive (Ki67*) cells [31].
Feature extraction and selection
The quantitative features of tumor tissues were computed from ROIs using seven feature extraction methods, e.g. first-order statistics (FoS), co-occurrence matrix (CM), run-length matrix (RM), shape features (SP), local binary patterns (LBP), fractal analysis (FA), the histogram of oriented gradients (HoG) and two filters, gradient (GD) and wavelet transform (WT) using Matlab® (v9.1, MathWorks, MA) [20,21,32]. Moreover, power, variance, FoS, CM, and RM features were computed from GD and WT images. The extracted 152 texture features from T2w MRI data (FoS, 6 features; CM, 6 features; RM, 7 features; FA, 1 feature; SP, 9 features, LBP, 10 features; HoG, 6 features; GD, 25 features; WT, 88 features) were standardized using z-score normalization method.
To remove associated features, we performed a feature selection approach that evaluates cross-correlation coefficients. The features demonstrating a strong correlation (|r| > 0.8) were removed from the set. Afterward, key variables were selected from this set based on the performance of the support vector machines (SVM) classification and multivariate regression models by increasing the complexity of the models by adopting an exhaustive search procedure.
Multivariate modeling and statistical analysis
A linear SVM classification model was generated with leave-one-out cross-validation by evaluating the performance of the generated models with accuracy and area under the receiver operating curve (AUC). The performance of the SVM classifier was visualized using receiver operating characteristics (ROC) and decision curve plots.
To predict survival time of the KPC mice and histopathological tumor markers, separate multivariate regression models were generated with identified key variables following an exhaustive search procedure. The performance of the generated model was evaluated by computing adjusted r squared (Radj 2) values. The two-tailed student T-test was performed to evaluate the statistical difference as accepting P<0.05 as statistically significant. Besides, Kaplan Meier analysis was used to evaluate the survival behavior of the KPC mice used as untreated control and treated with dinaciclib or DC vaccine therapy.
Results
A total of 152 texture features was computed from T2w MRI data using seven feature extraction methods and two filters to detect early treatment effects on tumor tissue (Figure 1A). After discarding strongly associated texture features (|r| > 0.8), 45 variables have remained in the feature set. The correlation of the remaining features was visualized using a heatmap representation in Figure 1B. The z scores and diagnostic performances of the features were presented in Figure 1C. Afterward, these features were further examined by adopting an exhaustive search procedure to generate linear SVM classifiers. The key variables were determined with the assessment of cross-validation accuracy and AUC. The final SVM classification model included three texture features (Table 2) demonstrated a diagnostic accuracy of 95.24%, an AUC of 0.92, a sensitivity of 100%, and specificity of 87.5% for detecting treatment-related changes on tumor tissue. The posterior probability of each subject is visualized in Figure 2A and ROC curves are utilized to demonstrate the predictive performance of the SVM classifier in Figure 2B. The linear separation plane of the generated SVM classification model is visualized in Figure 2C. The decision curve plot is presented in Figure 2D to demonstrate the diagnostic advantages of the generated model in clinically acceptable terms.
Figure 1.
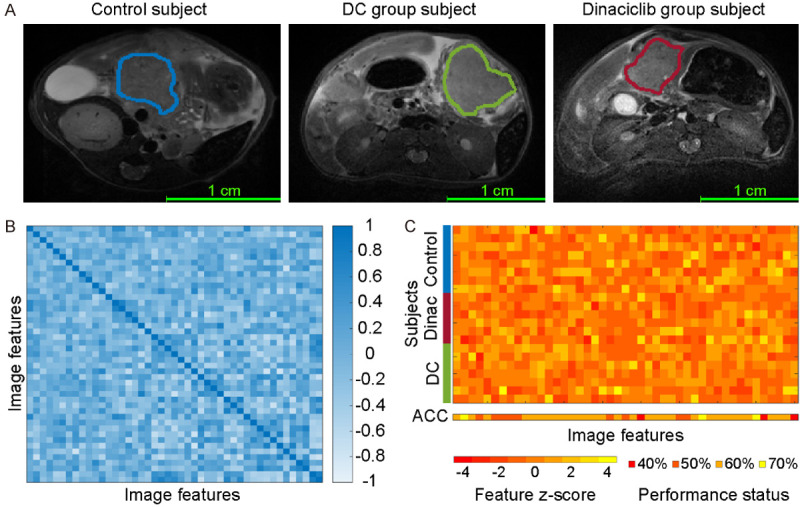
The quantitative MRI texture features to characterize underlying tumor structure. Representative MRI images for PDAC tumors from untreated control and treatment (DC vaccine and dinaciclib) groups were presented in (A). The heatmap representation demonstrates the association between computed texture features identified following correlation analysis (B). The z-scores of candidate features and diagnostic accuracies (ACC) of each variable in a univariate model were presented in (C). Multivariable models are performed to improve early detection of treatment effects on tumor tissues due to observed lower accuracy with univariate models.
Table 2.
The quantitative MRI texture features used to build classification and regression models
| Treatment Effects | Overall Survival | Trichrome | CK19 marker | Ki67 marker | ||
|---|---|---|---|---|---|---|
| Features | 1 | FoS mean (h) | FoS mean | FA fractal dimension | RM LRE | RM LGLE |
| 2 | FoS mean (a) | RM SRE | LBP | FoS kurtosis (h) | RM LGLE of GD | |
| 3 | RM nonuniformity (v) | FoS 3rd moment (d) | RM SRE (a) | FoS contrast (a) | FA fractal dimension | |
| 4 | FoS contrast (a) | RM LGLRE | RM SRE (a) | FoS entropy (h) | ||
| 5 | RM LGLRE (v) | FoS kurtosis of HoG | RM RLE (a) | RM SRE (d) |
Abbreviations: CM: co-occurrence matrix; FA: fractal anisotropy; FoS: first-order statistics; GD: gradient image; HoG: histogram of oriented gradients; LBP: local binary patterns; LRE: long-run emphasis; LGLE: low gray-level emphasis; LGLRE: low gray level run emphasis; RLE: run-length emphasis; RM: run-length matrix; SRE: short-run emphasis. (a): approximate wavelet coefficients; (v): vertical wavelet coefficients; (h): horizontal wavelet coefficients; (d): diagonal wavelet coefficients.
Figure 2.
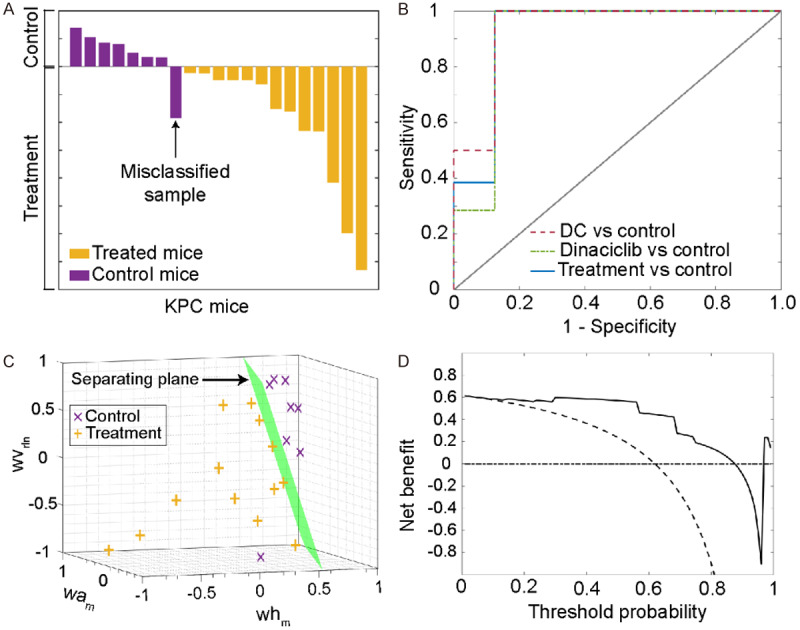
The diagnostic performance of the generated classification model to detect changes in the tumor microenvironment. (A) represents posterior probabilities of each sample in control and treatment (dinaciclib and DC vaccine) groups that were computed with support vector machines classification model. Only one KPC mice used as untreated control subject was misclassified. The receiver operating curves (ROC) for the developed model are given in (B). The model was utilized to generate a ROC curve with different groups (DC vs. control, dinaciclib vs. control, Treatment (DC and dinaciclib vs. control). (C) visualizes the generated separation surface (green) to differentiate pancreatic tumors according to the status of treatment. (D) shows the benefit curve of the generated classification model according to decision curve plot analysis (solid line: prediction model, dashed line: assume all subjects are treated, dotted-dashed line: assume no subjects were treated). Abbreviations: wvrln: run-length nonuniformity of vertical wavelet coefficients; wam: mean of approximate wavelet coefficients; whm: mean of horizontal wavelet coefficients.
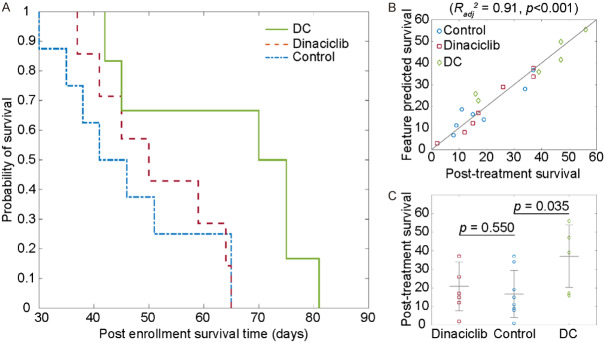
The survival behavior of the KPC mice was demonstrated using Kaplan-Meier plot (Figure 3A). The survival time of the KPC mice after detection of the tumors was 46±13 days for KPC mice in the untreated control group and 58±15 days for the KPC mice in dinaciclib or DC vaccine treatment groups (P=0.091). Moreover, the mice treated with DC vaccine had an average survival time of 65±17 days while the survival time of KPC mice treated with dinaciclib was 52±11 days. Despite no significant difference between untreated or treated groups, treated mice had longer survival time than untreated mice (treatment vs. control; 27±17 days vs. 16±13 days). Besides, the mice treated with DC vaccines demonstrated significantly improved survival time (P=0.035) yet not mice treated with dinaciclib (P=0.550). Afterward, the association between quantitative MRI texture features and survival behavior of the KPC mice was evaluated by performing multivariate analysis while sequentially increasing the complexity of the regression model. The final regression model generated with five texture features (Table 2) has obtained Radj 2 of 0.91 (P<0.001). The response of the regression model is visualized in Figure 3B and post-treatment survival data is presented in Figure 3C.
Figure 3.
Survival behavior and prediction analysis of the samples. Post enrolment survival behavior of the KPC mice was demonstrated with Kaplan-Meier plot (A). The prediction performance of the generated multivariable model for survival of the PDAC mice (B). The model had a strong correlation with observed survival behavior of KPC mice including untreated control and treated with DC vaccine or dinaciclib (Radj 2=0.91). The statistical assessment of post-treatment survival (C). The survival of KPC mice treated with DC vaccine had significantly longer (P=0.035) while there was no significant improvement for KPC mice treated with dinaciclib (P=0.55).
Throughout the pathological analysis of pancreatic tumor tissue, we observed a lower level of fibrosis (Figure 4A) and Ki67* area (Figure 4B) and also a higher number of CK19* cells (Figure 4C) for KPC mice treated with DC vaccine or dinaciclib compared to untreated control mice. However, the fibrosis percentage of mice treated with dinaciclib and Ki67* cells of KPC mice treated with DC vaccine was not significantly different than untreated control mice. Afterward, we investigated the association between MRI texture features and histopathological tumor markers by performing multivariate analysis while increasing the complexity of the models incrementally. The final regression models, constructed with five MRI texture features to predict fibrosis percentage, demonstrated a strong correlation (r=0.93) with measured fibrosis percentage in KPC mice including treatment and control groups. The generated model obtained Radj 2 of 0.82 (P<0.001). Besides, the regression model constructed with five variables had a strong correlation (r=0.98) with measured CK19* area which corresponds to Radj 2 of 0.92 (P<0.001). Moreover, a strong correlation was observed between predicted and measured Ki67* cells. The model integrating five variables obtained Radj 2 of 0.97 (P<0.001). The quantitative features used to generate multivariable regression models were listed in Table 2 and the behavior of the regression models with an increasing number of independent variables were presented in Figure 5A-C. Moreover, residual plots of generated regression models for histopathological tumor markers are shown in Figure 5D-F.
Figure 4.
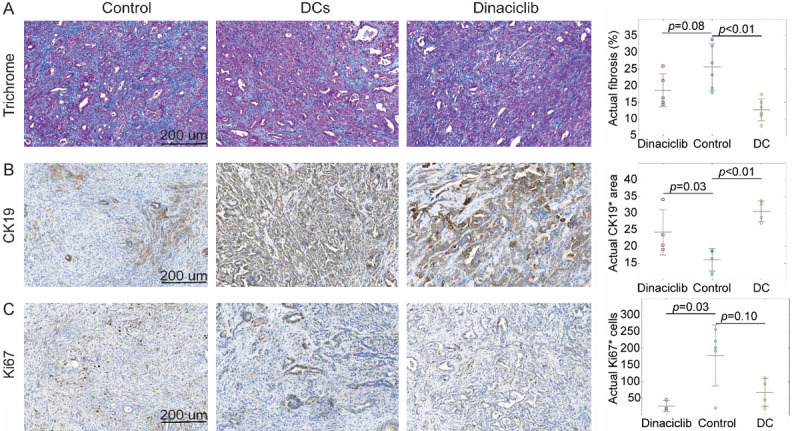
Histopathological analysis of the tumor tissues with trichrome staining (A), CK19 (B) and Ki67 (C) immunostainings for assessment of treatment effects. The subjects treated with DC vaccine had significantly lower fibrosis (P<0.01) and higher CK19* (P<0.01) area while there were no significant differences for Ki67* (P > 0.05) cells. Besides, the subjects treated with dinaciclib had significantly higher CK19* area (P=0.05) and lower number Ki67* (P=0.03) cells while no significant difference was observed in fibrosis pepercentage (P > 0.05).
Figure 5.
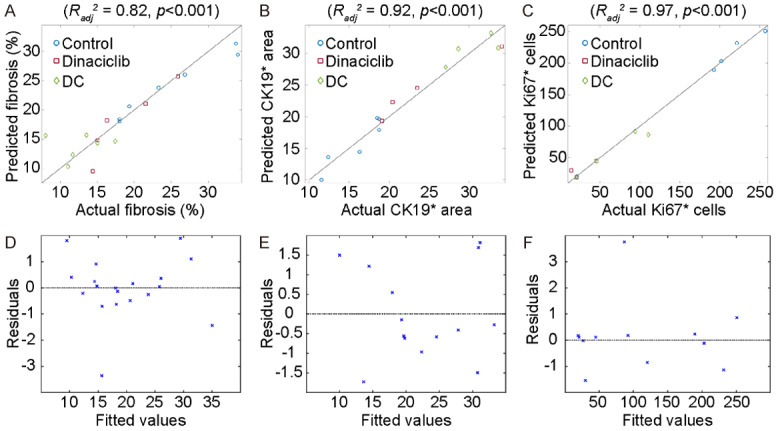
Performance of the regression models that predict histopathological tumor markers (fibrosis, CK19*, and Ki67*). The generated models had a strong correlation with the measured values of the histopathological tumor markers, (A) fibrosis performance (Radj 2=0.82), (B) CK19* area (Radj 2=0.92), and (C) number of Ki67* cells (Radj 2=0.97). Moreover, the residuals of the models were given in (D-F) for fibrosis percentage, CK19* area, and the number of Ki67* cells, respectively.
Discussion
In this study, we investigated the technical feasibility of quantitative MRI texture features for detection of early treatment response describing the treatment-related changes on tumor tissue and prediction of OS following dinaciclib or DC vaccine therapy by performing multivariate analysis. The generated multivariable classification and regression models demonstrated that T2w MRI texture features may serve as noninvasive imaging biomarkers to monitor tumor microenvironment changes and predict therapeutic outcomes for dinaciclib or DC vaccine treatment in KPC mouse model of PDAC.
A classification model, integrated with three texture features, detected treatment-related changes in tumor tissue with an accuracy of 95.24%, sensitivity of 100%, and specificity of 87.5%. Besides, the multivariable regression model constructed with five variables demonstrated a positive correlation with the survival time of KPC mice (Radj 2=0.91). Moreover, three separate multivariable models were developed to predict histopathological tumor markers using five texture features. These regression models obtained a favorable prediction performance for collagen and tumor cell proliferation, and also long-term clinical outcomes.
In recent years, the potential of quantitative image analysis approaches for PDAC was investigated in several studies [33-35]. Choi investigated the potential value of T2w MRI texture to identify long-term clinical outcomes of PDAC patients after 18.5 months of pancreatectomy [33]. Their results demonstrated that entropy and tumor size were associated with OS of PDAC patients while only tumor size was statistically significant. Another study examined the texture of 18F-FDG-PET/CT images of twenty-six PDAC patients prior and posterior radiation therapy to link tumor heterogeneity and OS [34]. The authors stated that two clinical features (age and node stage) and three texture features (homogeneity, variance and cluster tendency) were important prognostic factors for OS of PDAC patients. Furthermore, Attiyeh examined quantitative features to describe the association of CT data and OS in PDAC patients by generating two models that combine image features with CA19-9 (AUC of 0.69) and CA19-9 plus Brennan score (AUC of 0.74) [35].
However, quantitative image analysis approaches performed in these studies were not correlated with gold standard histological data. Despite the promising results of clinical research studies analyzing MRI texture, pre-clinical and clinical studies that benefit animal models engineered to mimic genetic behavior of the human disease remain to be an important focus of cancer research for quantitative analysis. Moreover, animal models are utilized to demonstrate technical feasibility of MRI texture analysis and findings of these studies are validated with gold standard histology which ultimately can be applied for precision medicine in clinical practice [36].
In this study, our goal was to investigate the potential of quantitative MRI features for detection of therapeutic changes in tumor microenvironment and prediction of longterm clinical outcomes following dinaciclib or DC vaccine treatment. The multivariable classification model including three features detected structural changes in tumor tissue caused by drug or vaccine therapies (accuracy of 95.24%). Despite significantly improved OS of KPC mice treated with DC vaccine (P=0.035), there was no statistical difference between control and dinaciclib treatment groups (P=0.55). The multivariable regression model was developed with five texture features to predict KPC mice OS which demonstrated a strong association with survival function (Radj 2=0.91). Besides, we analyzed the characteristics of MRI data to develop regression models to predict histopathological tumor markers measured by histology staining or immunostaining. These models demonstrated a positive correlation with fibrosis percentages (Radj 2=0.82), CK19* area (Radj 2=0.92), and the number of Ki67* cells (Radj 2=0.97).
Our study had several limitations. First, pancreatic tumors were outlined with a manual segmentation approach that requires user interaction and increases pre-processing time. Further studies will benefit adopting an automated approach for tumor segmentation to reduce processing time and potential user bias. However, manual segmentation of the tumor tissue with two radiologists under the supervision of a senior radiologist is a standard approach for preclinical studies. Second, our study included a lower number of subjects due to the nature of preclinical studies; however, it was comparable to other published preclinical studies. Besides, our study was aiming to evaluate the technical feasibility of the texture analysis which had a statistical power of more than 0.8 at a significance level of 0.05. Therefore, future studies that overcome these limitations will be needed for advancing the results of this study.
In conclusion, our study demonstrated that texture-based MRI imaging features had remarkable potential to interpret tissue characteristics and may be used to identify early treatment response to dinaciclib or DC vaccine therapy by interpreting pancreatic tumor tissue characteristics obtained from histopathological tumor markers and prediction of long-term clinical outcomes.
Acknowledgements
This study was supported by the National Cancer Institute (grants R01CA209886, R01CA196967), by 2019 Harold E. Eisenberg Foundation Scholar Award and by the Fishel Fellowship Award at the Robert H. Lurie Comprehensive Cancer Center.
Disclosure of conflict of interest
None.
Abbreviations
- CM
co-occurrence matrix
- FA
fractal anisotropy
- FoS
first-order statistics
- GD
gradient image
- HoG
histogram of oriented gradients
- LBP
local binary patterns
- LRE
long-run emphasis
- LGLE
low gray-level emphasis
- LGLRE
low gray level run emphasis
- RLE
run-length emphasis
- RM
run-length matrix
- SRE
short-run emphasis
- (a)
approximate wavelet coefficients
- (v)
vertical wavelet coefficients
- (h)
horizontal wavelet coefficients
- (d)
diagonal wavelet coefficients
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 3.Aleksandra A, Alice D, Marco F. Pancreatic ductal adenocarcinoma: current and evolving therapies. Int J Mol Sci. 2017;18:1338. doi: 10.3390/ijms18071338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butterfield LH. The society for immunotherapy of cancer biomarkers task force recommendations review. Semin Cancer Biol. 2018;52:12–15. doi: 10.1016/j.semcancer.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desch AN, Randolph GJ, Murphy K, Gautier EL, Kedl RM, Lahoud MH, Caminschi I, Shortman K, Henson PM, Jakubzick CV. CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell-associated antigen. J Exp Med. 2011;208:1789–1797. doi: 10.1084/jem.20110538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shikhar M, Carolyn DB, Steve C, Elizabeth GM, Colleen AC, Mingli L, Gina S, Mohamed LS, Michelle HN, Melanie BT, Chrystal MP, Andres MS, Michael IN, Mark PR, Zihai L, David JC. Vaccination with poly (IC:LC) and peptide-pulsed autologous dendritic cells in patients with pancreatic cancer. J Hematol Oncol. 2017;10:1–13. doi: 10.1186/s13045-017-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, Seghezzi W, Paruch K, Dwyer MP, Doll R, Nomeir A, Windsor W, Fischmann T, Wang Y, Oft M, Chen T, Kirschmeier P, Lees EM. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther. 2010;9:2344–2353. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 8.Zhenghu C, Zhenyu W, Jonathan CP, Yang Y, Shayahati B, Jiaxiong L, Ting H, Yanling Z, Xin X, Hong Z, Joanna SY, Shangfeng L, Jianhua Y. Multiple CDK inhibitor dinaciclib suppresses neuroblastoma growth via inhibiting CDK2 and CDK9 activity. Sci Rep. 2016;6:29090. doi: 10.1038/srep29090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker A, Gregory GP, Verbrugge I, Kats L, Hilton JJ, Vidacs E, Lee EM, Lock RB, Zuber J, Shortt J, Johnstone RW. The CDK9 inhibitor dinaciclib exerts potent apoptotic and antitumor effects in preclinical models of MLL-rearranged acute myeloid leukemia. Cancer Res. 2016;76:1158–1169. doi: 10.1158/0008-5472.CAN-15-1070. [DOI] [PubMed] [Google Scholar]
- 10.Hu C, Dadon T, Chenna V, Yabuuchi S, Bannerji R, Booher R, Strack P, Azad N, Nelkin BD, Maitra A. Combined inhibition of cyclin-dependent kinases (dinaciclib) and AKT (MK-2206) blocks pancreatic tumor growth and metastases in patient-derived xenograft models. Mol Cancer Ther. 2015;14:1532–9. doi: 10.1158/1535-7163.MCT-15-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldmann G, Mishra A, Bisht S, Karikari C, Garrido-Laguna I, Rasheed Z, Ottenhof NA, Dadon T, Alvarez H, Fendrich V, Rajeshkumar NV, Matsui W, Brossart P, Hidalgo M, Bannerji R, Maitra A, Nelkin BD. Cyclin-dependent kinase inhibitor dinaciclib (SCH727965) inhibits pancreatic cancer growth and progression in murine xenograft models. Cancer Biol Ther. 2011;12:598–609. doi: 10.4161/cbt.12.7.16475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pigula M, Huang HC, Mallidi S, Anbil S, Liu J, Mai Z, Hasan T. Size-dependent tumor response to photodynamic therapy and irinotecan monotherapies revealed by longitudinal ultrasound monitoring in an orthotopic pancreatic cancer model. Photochem Photobiol. 2019;95:378–386. doi: 10.1111/php.13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens WR, Johnson CD, Stephens DH, Batts KP. CT findings in hepatocellular carcinoma: correlation of tumor characteristics with causative factors, tumor size, and histologic tumor grade. Radiology. 1994;191:531–537. doi: 10.1148/radiology.191.2.8153335. [DOI] [PubMed] [Google Scholar]
- 14.Yao FY, Ferrell L, Bass NM, Watson JJ, Bacchetti P, Venook A, Ascher NL, Roberts JP. Liver transplantation for hepatocellular carcinoma: expansion of the tumor size limits does not adversely impact survival. Hepatology. 2001;33:1394–1403. doi: 10.1053/jhep.2001.24563. [DOI] [PubMed] [Google Scholar]
- 15.Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade, and necrosis: the SSIGN score. J Urol. 2002;168:2395–2400. doi: 10.1016/S0022-5347(05)64153-5. [DOI] [PubMed] [Google Scholar]
- 16.Gerwing M, Herrmann K, Helfen A, Schliemann C, Berdel W, Eisenblätter M, Wildgruber M. The beginning of the end for conventional RECIST - novel therapies require novel imaging approaches. Nat Rev Clin Oncol. 2019;16:442–458. doi: 10.1038/s41571-019-0169-5. [DOI] [PubMed] [Google Scholar]
- 17.Kuhl CK, Alparslan Y, Schmoee J, Sequeira B, Keulers A, Brümmendorf TH, Keil S. Validity of RECIST version 1.1 for response assessment in metastatic cancer: a prospective, multireader study. Radiology. 2019;290:349–356. doi: 10.1148/radiol.2018180648. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, Liang C, Tian J, Liang C. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281:947–957. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 19.Eresen A, Hafsa NE, Alic L, Birch SM, Griffin JF, Kornegay JN, Ji JX. Muscle percentage index as a marker of disease severity in golden retriever muscular dystrophy. Muscle Nerve. 2019;60:621–628. doi: 10.1002/mus.26657. [DOI] [PubMed] [Google Scholar]
- 20.Li K, Xiao J, Yang J, Li M, Xiong X, Nian Y, Qiao L, Wang H, Eresen A, Zhang Z, Hu X, Wang J, Chen W. Association of radiomic imaging features and gene expression profile as prognostic factors in pancreatic ductal adenocarcinoma. Am J Transl Res. 2019;11:4491–4499. [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Eresen A, Lu Y, Yang J, Shangguan J, Velichko Y, Yaghmai V, Zhang Z. Radiomics signature for the preoperative assessment of stage in advanced colon cancer. Am J Cancer Res. 2019;9:1429–1438. [PMC free article] [PubMed] [Google Scholar]
- 22.Eilaghi A, Baig S, Zhang Y, Zhang J, Karanicolas P, Gallinger S, Khalvati F, Haider MA. CT texture features are associated with overall survival in pancreatic ductal adenocarcinoma - a quantitative analysis. BMC Med Imaging. 2017;17:38. doi: 10.1186/s12880-017-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalvati F, Zhang Y, Baig S, Lobo-Mueller EM, Karanicolas P, Gallinger S, Haider MA. Prognostic value of CT radiomic features in resectable pancreatic ductal adenocarcinoma. Sci Rep. 2019;9:5449. doi: 10.1038/s41598-019-41728-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanduleanu S, Woodruff HC, de Jong EEC, van Timmeren JE, Jochems A, Dubois L, Lambin P. Tracking tumor biology with radiomics: a systematic review utilizing a radiomics quality score. Radiother Oncol. 2018;127:349–360. doi: 10.1016/j.radonc.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Horvat N, Veeraraghavan H, Khan M, Blazic I, Zheng J, Capanu M, Sala E, Garcia-Aguilar J, Gollub MJ, Petkovska I. MR imaging of rectal cancer: radiomics analysis to assess treatment response after neoadjuvant therapy. Radiology. 2018;287:833–843. doi: 10.1148/radiol.2018172300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji GW, Zhang YD, Zhang H, Zhu FP, Wang K, Xia YX, Zhang YD, Jiang WJ, Li XC, Wang XH. Biliary tract cancer at CT: a radiomics-based model to predict lymph node metastasis and survival outcomes. Radiology. 2019;290:90–98. doi: 10.1148/radiol.2018181408. [DOI] [PubMed] [Google Scholar]
- 27.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53 R172H and Kras G12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 28.Courtin A, Richards FM, Bapiro TE, Bramhall JL, Neesse A, Cook N, Krippendorff BF, Tuveson DA, Jodrell DI. Anti-tumour efficacy of capecitabine in a genetically engineered mouse model of pancreatic cancer. PLoS One. 2013;8:e67330. doi: 10.1371/journal.pone.0067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai Z, Shi Y, Wang J, Qiu L, Teng G, Zhang F, Yang X. Multi-modality imaging-monitored creation of rat orthotopic pancreatic head cancer with obstructive jaundice. Oncotarget. 2017;8:54277–54284. doi: 10.18632/oncotarget.17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stokes WS. Humane endpoints for laboratory animals used in regulatory testing. ILAR J. 2002;43(Suppl):S31–S38. [PubMed] [Google Scholar]
- 31.Caroline AS, Wayne SR, Kevin WE. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. ArXiv e-prints; 2016. [Google Scholar]
- 33.Choi M, Lee Y, Yoon S, Choi JI, Jung S, Rha S. MRI of pancreatic ductal adenocarcinoma: texture analysis of T2-weighted images for predicting long-term outcome. Abdom Radiol (NY) 2019;44:122–130. doi: 10.1007/s00261-018-1681-2. [DOI] [PubMed] [Google Scholar]
- 34.Yue Y, Osipov A, Fraass B, Sandler H, Zhang X, Nissen N, Hendifar A, Tuli R. Identifying prognostic intratumor heterogeneity using pre- and post-radiotherapy 18F-FDG PET images for pancreatic cancer patients. J Gastrointest Oncol. 2017;8:127–138. doi: 10.21037/jgo.2016.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Attiyeh MA, Chakraborty J, Doussot A, Langdon-Embry L, Mainarich S, Gönen M, Balachandran VP, D’Angelica MI, DeMatteo RP, Jarnagin WR, Kingham TP, Allen PJ, Simpson AL, Do RK. Survival prediction in pancreatic ductal adenocarcinoma by quantitative computed tomography image analysis. Ann Surg Oncol. 2018;25:1034–1042. doi: 10.1245/s10434-017-6323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lloyd KC, Robinson PN, MacRae CA. Animal-based studies will be essential for precision medicine. Sci Transl Med. 2016;8:352ed12. doi: 10.1126/scitranslmed.aaf5474. [DOI] [PubMed] [Google Scholar]