Abstract
The genus Polyalthiopsis Chaowasku (Annonaceae) was a poorly known monotypic genus from Vietnam that was recently segregated from the highly polyphyletic genus Polyalthia s.l. The sister clade relationship between Polyalthiopsis and Miliusa was not well established in previous study. The phylogenetic position of two Polyalthia spp. from China, P. chinensis S.K.Wu ex P.T.Li and P. verrucipes C.Y.Wu ex P.T.Li, remains unresolved and is shown here to be phylogenetically affiliated with Polyalthiopsis. Phylogenetic analyses of six chloroplast regions (matK, ndhF, psbA-trnH, rbcL, trnL-F and ycf1; ca.7.3 kb, 60 accessions) unambiguously placed Polyalthia chinensis and P. verrucipes in the same clade with Polyalthiopsis floribunda (PP = 1, MPBS = 97%); the entire clade is sister to Miliusa with weak to strong support (PP = 1, MPBS = 54%). Polyalthia chinensis and P. verrucipes share several diagnostic characters with Polyalthiopsis floribunda, including the raised midrib on the upper surface of the leaf in vivo, conspicuous foliar glands when dried, petiole with transverse striations when dried and axillary inflorescences. The two species differ from Polyalthiopsis floribunda in having fewer flowers per inflorescence, longer linear petals and two ovules per carpel. On the basis of the combined molecular phylogenetic and morphological data, we propose two new combinations, Polyalthiopsis chinensis (S.K.Wu ex P.T.Li) B.Xue & Y.H.Tan and Polyalthiopsis verrucipes (C.Y.Wu ex P.T.Li) B.Xue & Y.H.Tan. The protologue of Polyalthia verrucipes did not include a description of the flowers, which we provide here. An updated description for the genus Polyalthiopsis and a key to species in the genus Polyalthiopsis is also provided.
Keywords: Annonaceae , molecular phylogeny, morphology, Polyalthia chinensis, Polyalthia verrucipes, Polyalthiopsis
Introduction
The genus Polyalthia Blume has historically been the source of considerable taxonomic confusion in Annonaceae due to its highly polyphyletic status (van Setten and Koek-Noorman 1992; Doyle and Le Thomas 1994; Doyle et al. 2000). Recent molecular phylogenetic studies have clarified generic circumscriptions and resulted in the segregation of disparate elements from the previously highly polyphyletic genus Polyalthia s.l., including removal of species now placed in several new genera–Maasia Mols & al. (Mols et al. 2008), Huberantha Chaowasku (Chaowasku et al. 2012 [as “Hubera”], Chaowasku et al. 2015); Wuodendron B.Xue, Y.H.Tan & T.Chaowasku (Xue et al. 2018) and Polyalthiopsis Chaowasku (Chaowasku et al. 2018); the transfer of species to Fenerivia Diels (Saunders et al. 2011), Marsypopetalum Scheff. (Xue et al. 2011), Monoon Miq. (Xue et al. 2012), Goniothalamus (Blume) Hook. f. & Thomson (Tang et al. 2013), Meiogyne Miq. (Xue et al. 2014) and Wangia X.Guo & R.M.K.Saunders (Xue et al. 2016). The circumscription of Polyalthia s.str. was consequently redefined (Xue et al. 2012).
Amongst the newly segregated genera, Polyalthiopsis Chaowasku is a poorly known monotypic genus from Vietnam (Chaowasku et al. 2018). The type species Polyalthiopsis floribunda is known from only two field collections (Poilane 10052, P, A, BO, CMUB, HN, K, L, P; and Chaowasku 128, CMUB). The species was first collected in 1924 and described under the name Polyalthia floribunda Jovet-Ast (Jovet-Ast 1940). I.M. Turner (2016) subsequently transferred the name to Huberantha. Ninety years after the first collection, Chaowasku collected this species again in 2014 and was able to sequence it for a phylogenetic study. It was shown not to be congeneric with Huberantha and was instead retrieved (without statistical support) as sister to Miliusa, leading Chaowasku et al. (2018) to erect a monotypic genus, Polyalthiopsis Chaowasku, to accommodate it. With only one Polyalthiopsis species and limited DNA regions used in the phylogenetic study, the sister relationship between Polyalthiopsis and Miliusa was not well established. It is also difficult to identify important diagnostic characters for Polyalthiopsis with only one flowering collection and a single monocarp available for taxonomic comparison.
Several species names remain unresolved in Polyalthia and await assignment to specific genera (Xue 2013; Xue et al. 2012), including the Chinese endemics Polyalthia chinensis S.K.Wu ex P.T.Li and P. verrucipes C.Y.Wu ex P.T.Li. As with P. floribunda, these two species are represented by very few collections and lack adequate flowering and fruiting descriptions.
The name Polyalthia chinensis was published in 1976, based on a flowering collection (Qinghai-Xizang Exped. 74-4451, KUN & PE) from Mêdog, Xizang, China, in 1974 (Li 1976; Li and Gilbert 2011). A second sterile specimen was subsequently collected in 1983 (B. S. Li & S. Z. Cheng 2668, PE). Until now, the species was only represented by these two collections.
The name Polyalthia verrucipes was published in 1976, based on a fruiting collection (C.W. Wang 76321, IBSC, NAS, A, PE) from Menghai, Yunnan, China, in 1936 (Li 1976; Li and Gilbert 2011). A second collection with immature fruits was collected in 1957 (Sino-Russia Exped. 9527, KUN & PE). Although Hou and Li (2007) cited further collections (S. K. Wu 1345, 1375, KUN; X. L. Hou 112, CANT, IBSC; and T. X. Sun 200037, CANT), we failed to locate those specimens in the cited herbaria.
The relationship between these two species has been controversial. Both species are represented by very few collections, with P. chinensis lacking fruiting specimens and P. verrucipes lacking flowering specimens, rendering morphological comparison problematic. Based on the foliar glands and leaf venation, Hou and Li (2007) regarded the name P. chinensis as synonymous with P. verrucipes, although this treatment was rejected by Li and Gilbert (2011) in the Flora of China without explanation. Li and Gilbert (2011) included identical floral descriptions in their treatment of P. verrucipes and P. chinensis, but with no clear indication of the source of this information, casting some doubt over the floral description of P. verrucipes.
With limited morphological characters, especially the lack of flowers in P. verrucipes and the limited material available, the relationship between P. chinensis and P. verrucipes and their taxonomic placement has never been resolved. We therefore, carried out several field explorations to search for these two species. This resulted in new collections of Polyalthia verrucipes, including flowers, enabling clarification of the relationship between P. chinensis and P. verrucipes, as well as their phylogenetic position. As a consequence, we were able to enlarge the poorly known genus Polyalthiopsis, supplementing available descriptions and providing better support for its sister relationship.
Phylogenetic analysis
Taxon and DNA region sampling
Two accessions of Polyalthia chinensis (B. S. Li & S. Z. Cheng 2668, PE; and Qinghai-Xizang Exped. 74-4451, KUN), as well as two accessions of Polyalthia verrucipes (Sino-Russia Exped. 9527, PE; and Y.H. Tan MH1603, IBSC) were sampled and integrated with data of 56 Annonaceae accessions from previous datasets (Chaowasku et al. 2018; Guo et al. 2014; Xue et al. 2016, 2018). The final dataset comprised a total of 60 accessions of Annonaceae representing all major clades in the family, including 44 accessions representing 29 genera from subfam. Malmeoideae, 12 accessions representing 11 genera from subfam. Annonoideae, three species from subfam. Ambavioideae and one species of Anaxagorea A. Saint.-Hilaire. (subfam. Anaxagoreoideae). For Miliuseae, representatives of all currently accepted genera were included.
Six chloroplast DNA regions (matK, ndhF, rbcL, psbA-trnH and trnL-F and ycf1) were sequenced for the above-mentioned four collections of Polyalthia chinensis and P. verrucipes. The samples, localities and GenBank accession numbers are listed in Appendix I.
DNA extraction, amplification and sequencing
Genomic DNA was extracted from herbarium materials using a modified cetyl trimethyl ammonium bromide (CTAB) method (Doyle and Doyle 1987). A single amplification protocol was used for amplification of the chloroplast regions: template denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 sec; primer annealing at 50 °C for 1 min; and primer extension at 72 °C for 1 min, followed by a final extension step at 72 °C for 10 min. The primers used to amplify the psbA-trnH intergenic spacer were psbAF (Sang et al. 1997) and trnH2 (Tate and Simpson 2003); other primers are the same as those used by Thomas et al. (2012). PCR products were visualised using agarose gel electrophoresis. Successful amplifications were purified and sequenced on an Applied Biosystems 3730xl DNA Analyzer at Sangon Biotech (Shanghai) Co. Ltd., Guangzhou, China.
Alignment and phylogenetic analyses
Sequences were assembled and edited using Geneious ver. 5.4.3 (Drummond et al. 2010) and pre-aligned with the MAFFT (Katoh et al. 2002) plugin in Geneious using the automatic algorithm selection and default settings and, subsequently, manually checked and optimised. An inversion of 15 positions in psbA-trnH was identified and reverse complemented in the alignment, following a strategy previously applied by Pirie et al. (2006), to retain substitution information in the fragments.
Maximum parsimony (MP) analyses of the seven combined regions were conducted using PAUP ver. 4.0b10 (Swofford 2003). All characters were weighted equally and gaps treated as missing data. The most parsimonious trees were obtained with heuristic searches of 1,000 replicates of random stepwise sequence addition, tree bisection-reconnection (TBR) branch swapping with no limit to the number of trees saved. Bootstrap support (BS) was calculated following Müller (2005), with 10,000 simple stepwise addition replicates with TBR branch swapping and no more than 10 trees saved per replicate.
Bayesian analysis was performed using NSF Extreme Science & Engineering Discovery Environment (XSEDE) application of MrBayes ver. 3.2.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) provided by the CIPRES Science Gateway (Miller et al. 2010). PartitionFinder2 was used to test the dataset for partitions (model of evolution: mrbayes; model of selection: AICc; scheme search: greedy) (Guindon et al. 2010; Lanfear et al. 2012, 2016). The best partition scheme suggested six partitions, based on DNA region identity with GTR+G chosen for matK, psbA-trnH, trnL-F and ycf1 regions and GTR+I+G selected for the ndhF and rbcL regions. Two independent Metropolis-coupled Markov Chain Monte Carlo (MCMC) analyses were run. Each search used three incrementally heated and one cold Markov chain and was run for 10 million generations and sampled every 1,000th generation. The temperature parameter was set to 0.08. The mean branch length prior was set from the default mean (0.1) to 0.01 (brlenspr = unconstrained: exponential (100.0)) to reduce the likelihood of stochastic entrapment in local tree length optima (Brown et al. 2010). Convergence was assessed using the standard deviation of split frequencies, with values < 0.01 interpreted as indicating good convergence. Tracer ver. 1.6 (Rambaut et al. 2014) was used to determine whether the parameter samples were drawn from a stationary, unimodal distribution and whether adequate effective sample sizes (ESS) for each parameter (ESS > 200) were reached. The first 25% of samples (2,500 trees) were discarded as burn-in and the post-burn-in samples summarised as a 50% majority-rule consensus tree.
Morphological studies
Comparative morphological data were obtained from specimens deposited in KUN, IBSC and PE herbaria and from published literature. Field surveys were carried out in Menghai County, Yunnan Province, with voucher specimens deposited in HITBC and IBSC.
Results
The concatenated alignment of the 60-terminal dataset consisted of 7,334 characters. The MP heuristic search retrieved four equally most parsimonious trees of 3,519 steps (consistency index, CI = 0.664; retention index, RI = 0.709).
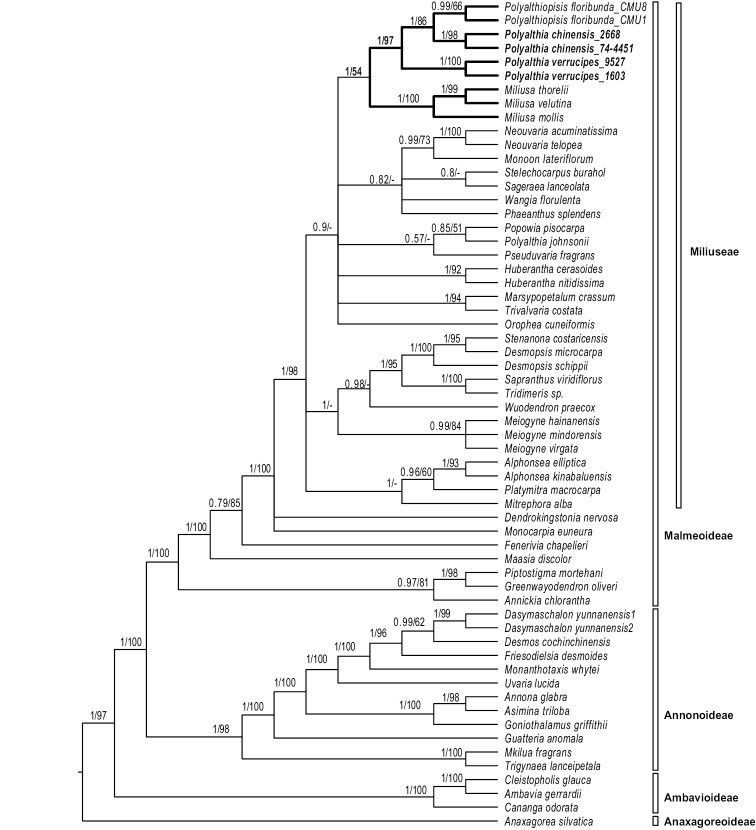
The MP and Bayesian analyses resulted in similar topologies. The 50% majority-rule consensus tree resulting from the Bayesian analyses under the six partitioned model is shown as Fig. 1. The results are consistent with previous phylogenetic analyses of the family, with the backbone of the tribe Miliuseae unresolved as in previous studies.
Figure 1.
Bayesian 50% majority-rule consensus tree under partitioned models (cpDNA data: matK, ndhF, psbA-trnH, rbcL and trnL-F; 60 taxa) showing Annonaceae subfams. Anaxagoreoideae, Ambavioideae and Annonoideae. Numbers at the nodes indicate Bayesian posterior probabilities and maximum parsimony bootstrap values (> 50%) in that order.
Polyalthia chinensis and P. verrucipes are not retrieved in the same clade as Polyalthia johnsonii, but were strongly supported as members of the same clade as Polyalthiopsis floribunda (PP [posterior probability] = 1, MPBS = 97%), with the entire clade sister to Miliusa with weak to strong support (PP = 1, MPBS = 54%).
Discussion
Polyalthia chinensis was regarded as a synonym of P. verrucipes by Hou and Li (2007). The phylogenetic positions of these two species are quite distinct, with the following relationship: (Polyalthia verrucipes, (Polyalthia chinensis, Polyalthiopsis floribunda)) (Fig. 1). Our field collection of the flowers of Polyalthia verrucipes provides further evidence for the distinction between these two species.
Although these two species resemble each other vegetatively (Fig. 2A, C), they differ in the number of flowers per inflorescence, the length and thickness of the pedicel and the colour of the petals. The inflorescences of Polyalthia chinensis have 1–2 flowers (Fig. 2A, C), whereas those of Polyalthia verrucipes comprise a solitary flower (Figs 2D, 3F, G). The pedicel of P. chinensis is slender and ca. 7 mm long, whereas that of P. verrucipes is stout and shorter than 2 mm. The petals of P. chinensis are green (Li 1976), whereas those of P. verrucipes are white (Fig. 3F–H). The leaf also differs slightly, with the leaf lamina of P. chinensis (2.5–3.8 cm) narrower than that of P. verrucipes (2.5–5 cm) and slightly thinner. The morphological data are therefore congruent with the phylogenetic topology and our phylogenetic and morphological analyses support the hypothesis that both species are not conspecific, as suggested by Li and Gilbert (2011).
Figure 2.
Morphological comparison between Polyalthiopsis chinensis and P. verrucipesA type specimen of P. chinensis (Qinghai-Xizang Exped. 74-4451, PE) B infloresence of P. chinensis (Qinghai-Xizang Exped. 74-4451, PE) C type specimen of P. verrucipes (C. W. Wang 76321, A) D inflorescence of P. verrucipes (B. Xue & H.B. Ding 311, IBSC) E adaxial leaf surface of P. chinensis (Qinghai-Xizang Exped. 74-4451, KUN) F abaxial leaf surface of P. chinensis (Qinghai-Xizang Exped. 74-4451, KUN) G adaxial leaf surface of P. verrucipes (Yunnan Exped. 9527, KUN) H abaxial leaf surface of P. verrucipes (Yunnan Exped. 9527, KUN).
Figure 3.
Morphology of Polyalthiopsis verrucipes comb. nov. A trunk, showing greyish bark B a branch, showing the leaf lamina C adaxial leaf surface, showing the raised midrib D close-up of the abaxial surface of dried leaf, showing glands E petiole with transverse striations when dried F lateral view of the flower G top view of the flower H bottom view of the flower I adaxial and abaxial view of the stamen J carpel K longitudinal section of a developing carpel, showing two developing ovules L developing young fruits M single dried monocarp, showing the two seeds 01187409 (C. W. Wang 76321, PE) N cylindrical seed, showing longitudinal groove around circumference (C. W. Wang 76321, PE). – Photos: A, C–E, I–N, by Bine Xue; B, by Yun-Hong Tan; G–H by Hong-Bo Ding.
The monotypic genus Polyalthiopsis Chaowasku was published in 2018, based on Polyalthia floribunda collected in Vietnam (Chaowasku et al. 2018). It was reconstructed as the sister group of Miliusa, but without statistical support. Polyalthiopsis, Huberantha and Miliusa have previously been retrieved as an unsupported to weakly supported clade in Chaowasku et al. (2018). Although Chaowasku et al. (2018) mentioned that a more comprehensive phylogenetic study, using the whole plastome sequence data, demonstrates the same topology with strong support, the result has yet to be published. Polyalthiopsis is also retrieved as sister to Miliusa in this study, with weak support in the maximum parsimony analysis (MPBS = 54%), but strong support in the Bayesian analysis (PP = 1). This sister relationship was also well supported in Xue et al. (2020) (ML BS = 86%, suppl. material 1: fig. S1). The relationship between Huberantha and the Polyalthiopsis-Miliusa collective clade is, however, not retrieved in this study. The long-recognised sister relationship between Miliusa and Huberantha in previous studies (Mols et al. 2008; Saunders et al. 2011; Xue et al. 2011, 2012; Chaowasku et al. 2012, 2014; Chatrou et al. 2012; Guo et al. 2017) can be redefined here following the inclusion of Polyalthiopsis.
Polyalthiopsis Chaowasku is easily distinguished from most of the other genera in the tribe Miliuseae by its raised midrib on the adaxial leaf surface. When dry, such an adaxial leaf midrib appears slightly sunken. The raised midrib on the adaxial leaf surface is rarely observed in the Annonaceae, but is known from Artabotrys (Sinclair 1955; Turner 2012), Cremastosperma (Pirie 2005), Cyathocalyx (Surveswaran et al. 2010), Isolona (Couvreur 2009), Monodora (Couvreur 2009), Mezzettia (van der Heijden and Kessler 1990), Pseudephedranthus (Erkens et al. 2017) and Stelechocarpus (Chaowasku et al. 2013; van Heusden 1995). Another distinct feature of Polyalthiopsis floribunda is the dried petiole with multiple transverse striations (Chaowasku et al. 2018). Not many Annonaceae species have this pronounced drying artifact. One more distinct feature is the obvious foliar glands on the leaf surface when dried (obvious in fig. 2C in Chaowasku et al. 2018). Foliar glands are also observed in Wuodendron B.Xue, Y.H.Tan & Chaowasku in Miliuseae (Xue et al. 2018).
Based on one species with only two collections, the genus is not well described and compared and, hence, it is difficult to identify important diagnostic characters.
Polyalthia chinensis and P. verrucipes are retrieved in the same clade as Polyalthiopsis floribunda in the molecular phylogeny (Fig. 1). Sterile material of these three species is very similar. The leaves are elliptic with a cuneate base and acute to acuminate apex, with brochidodromous venation and reticulate tertiary veins. The leaf midrib in all three species is furthermore raised adaxially in vivo (Fig. 3C; raised midrib still visible in the specimen of P. chinensis), with multiple transverse striations on the dried petiole (Figs 3E, 4C) and obvious foliar glands on dried leaf surface (Figs 2E–H, 3D, 4B). Although all three species have axillary inflorescences, the number of flowers per inflorescence differs: P. chinensis has one to two flower(s) per inflorescence (Fig. 2A, B), Polyalthia verrucipes has only one flower per inflorescence (Figs 2D, 3F–H), while Polyalthiopsis floribunda has 1–5 flower(s) per inflorescence. The shape of the petal also differs: the petals of Polyalthia chinensis and P. verrucipes are linear (Figs 2B, D, 3F–H, 4D, E), while those of Polyalthiopsis floribunda are elliptic-ovate (Chaowasku et al. 2018). The carpel characters of Polyalthia chinensis and P. verrucipes also differ greatly from those of Polyalthiopsis floribunda: the former two species have two ovules per ovary and hence two seeds in each monocarp (Fig. 3K, M), whereas Polyalthiopsis floribunda has only one ovule per ovary (Jovet-Ast 1940; Chaowasku et al. 2018).
Figure 4.
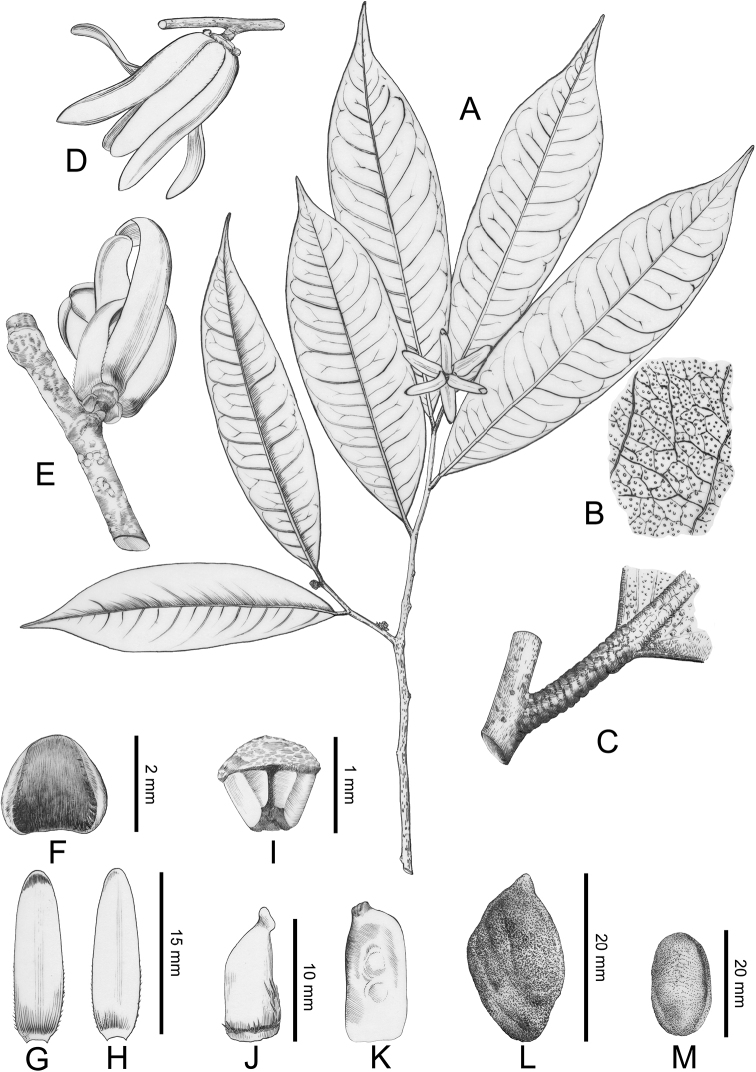
Polyalthiopsis verrucipes comb. nov. A flowering branch B close-up of adaxial surface of leaf, showing glands C close-up of leaf petiole, showing the transverse striations on dried petiole D lateral view of the flower E adaxial view of the flower F sepal G outer petal H inner petal I stamen J carpel K longitudinal section of the developing carpel, showing two lateral ovules L a dried monocarp M a seed. Drawn by Ding-Han Cui. (A–K from B. Xue & H. B. Ding XB311, IBSC; L, M from C. W. Wang 76321, PE)
In conclusion, Polyalthia chinensis, P. verrucipes and Polyalthiopsis floribunda share axillary inflorescences, a raised midrib on the adaxial leaf surface (Fig. 3C), petioles with transverse striations when dry (Fig. 3E) and foliar glands on dried leaf surface (Figs 2E–H, 3D, 4B). These characters render the three species distinctive from other species in the tribe and are thus diagnostic for the enlarged genus Polyalthiopsis.
The present phylogenetic study shows that Polyalthia chinensis is strongly supported as sister to Polyalthiopsis floribunda (PP = 1, MPBS = 86%). The collective clade is strongly supported as sister to Polyalthia verrucipes (PP = 1, MPBS = 97%). The whole clade (comprising the three species) is weakly to strongly supported (PP = 1, MPBS = 54%) as sister to Miliusa. The morphological and molecular phylogenetic data therefore support the transfer of Polyalthia verrucipes and P. chinensis to Polyalthiopsis and the new nomenclatural combinations are proposed here.
As Polyalthia verrucipes was published based on fruiting material only (Li 1976), with the newly collected flowers, an updated description is presented. It is noteworthy that the floral description of P. verrucipes, published by Li and Gilbert (2011), does not correspond with the material we collected in the field, but is instead similar to that of P. chinensis.
As more species were included in the genus Polyalthiopsis, an updated description and a key to the three species is also provided.
Taxonomy
Polyalthiopsis
Chaowasku, Ann. Bot. Fennici 55: 130. 2018.
959A71D2-149F-5E9B-BA95-45393380B607
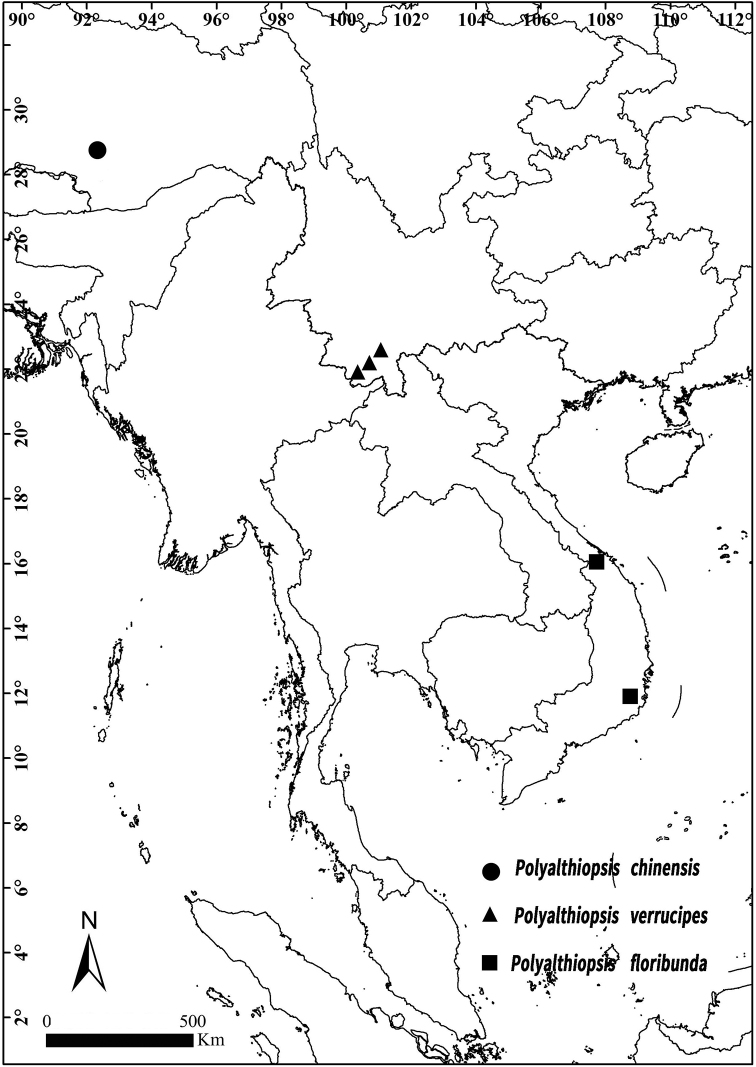
Figure 5.
Distribution of Polyalthopsis chinensis, P. floribunda and P. verrucipes.
Type species.
Polyalthiopsis floribunda (Jovet-Ast) Chaowasku – Polyalthia floribunda Jovet-Ast, Notul. Syst. 9: 75. 1940. – Huberantha floribunda (Jovet-Ast) I.M.Turner, Webbia 71: 229. 2016. – Type: Vietnam. Phanrang Prov., Tra Ca, 10 March 1924, Poilane 10052 (holotype P [barcode no. P00411080]; isotypes A[barcode no. A00351290], BO?, CMUB, HN, K[barcode no. K000608178], L[barcode no. L3728819], P [barcode no. P00411081; P00411082]), in flower.
Description.
Medium-sized to large trees. Young twigs glabrous. Leaves petiolate, blade elliptic, with glandular dots observable when dry, base cuneate, apex acute to bluntly (caudate-)acuminate; petiole with transverse striations when dry; upper surface of midrib raised in living plants, becoming slightly sunken when dry, lower surface of midrib raised; secondary veins rather faint in living plants, becoming slightly raised on both sides when dry, leaf venation brochidodromous; tertiary veins reticulate. Flower(s) in 1- to 5-flowered inflorescences, bisexual, pedicellate; inflorescences axillary, peduncle inconspicuous, bracts present. Sepals broadly ovate-triangular. Petals membranous-papyraceous to leathery. Outer petals elliptic-ovate or linear-lanceolate. Inner petals (narrowly) elliptic-ovate or linear-lanceolate. Stamens numerous per flower, connective truncate, covering thecae. Carpels numerous per flower; ovaries with 1 or more line(s) of hairs; stigma terete; ovule(s) 1 or 2 per ovary, sub-basal or lateral. Monocarps oblong to rhomboidal or cylindrical, stipitate, glabrous. Seed(s) 1or 2 per monocarp, cylindrical, surface smooth, raphe broadly sunken and partially slightly raised in middle, endosperm ruminations lamelliform.
Distribution.
Three species, known from Xizang, Yunnan Provinces of China and Thừa Thiên-Hu, Ninh Thuận Provinces of Vietnam (Fig. 5).
Polyalthiopsis chinensis
(S.K.Wu ex P.T.Li) B.Xue & Y.H.Tan comb. nov.
B6164F55-50B2-53DF-9D9D-E7AC47339DDF
urn:lsid:ipni.org:names:77209705-1
Polyalthia chinensis S.K.Wu & P.T.Li in Acta Phytotax. Sin. 14 (1): 108, t. 4. 1976. Basionym
Type.
China. Xizang: Mêdog, 20 Auguest 1974, Qinghai-Xizang Exped. 74-4451 (holotype, PE! [barcode no., PE01187290]; isotypes, PE! [[barcode no., PE01187291, PE01187292, PE01187293], KUN! [barcode no., KUN0677650]).
Distribution and habitat.
Known from Mêdog in Xingzang Province (Fig. 5), growing in rain forests, at low to medium elevations (800–1000 m a.s.l.).
Phenology.
Flowering in August.
Additional specimens examined.
B. S. Li & S. Z. Cheng 2668 (PE).
Preliminary IUCN conservation status.
DD (IUCN 2012). This species is only represented by two collections in Mêdog in Xizang Province. As Mêdog is not well explored, we tentatively recommend the conservation status as Data Deficient.
Polyalthiopsis verrucipes
(C.Y.Wu ex P.T.Li) B.Xue & Y.H.Tan comb. nov.
682DD7C5-A379-546E-909A-FCE806978D83
urn:lsid:ipni.org:names:77209706-1
Polyalthia verrucipes C.Y.Wu ex P.T.Li in Acta Phytotax. Sin. 14 (1): 110. 1976. Basionym
Type.
Lectotype (designated here). China. Yunnan: Menghai, July 1936, C. W. Wang 76321 (IBSC! [barcode no. IBSC0003386]; isolectotypes, A [barcode no. A00039580, photo!], IBSC! [barcode no. IBSC0003386], PE! [barcode no. PE01187287, PE01187470], NAS[barcode no. NAS00321991, photo!]).
Description.
Trees to 15 m tall (Fig. 3A). Branches greyish-black, glabrous. Petiole 3–7 mm long, 1–2 mm in diameter, glabrous, with transverse striations when dry (Figs 3E, 4C); leaf laminas oblong to oblong-lanceolate, 10–17 × 2.5–5 cm, base broadly cuneate or obtuse, apex acuminate (Figs 3B, 4A), both surfaces glabrous, thinly leathery, densely verrucate with foliar glands when dry (Figs 2G, H, 3D, 4B); upper surface of midrib raised when fresh (Fig. 3C), becoming flat or slightly sunken when dry (Fig. 2G), lower surface of midrib raised; secondary veins 15–18 on each side of midrib, delicate and prominent on both surfaces; tertiary veins reticulate. Inflorescences axillary, with solitary flower (Figs 2D, 3F, H, 4A, D, E). Pedicel 1–2 mm long, hispid, with one ovate bracteole at top, 2–3 ovate bracteoles at base (Fig. 3F, L). Sepals ovate, 2 × 2 mm, slightly reflexed, ciliate (Fig. 4F). Petals 6, valvate, free, in 2 whorls; white, linear, both whorls subequal, ca. 16 × 3–5 mm, thinly leathery, glabrous, slightly ciliate (Figs 3F–H, 4G, H). Stamens 40–50 per flower, ca. 1 mm long (Figs 3I, 4I); connective truncate. Carpels 12–16 per flower, oblong, glabrous; stigma ovoid, puberulent; ovary with 1 or 2 line(s) of hairs (Figs 3J, 4J); ovules 2 per ovary, lateral (Figs 3K, 4K). Fruiting pedicel becoming longer and thicker, 5–7 mm long, ca. 3 mm in diameter; monocarp stipes 7–9 mm long; monocarps oblong to rhomboid, ca. 2 cm long, 1 cm in diameter (Figs 3M, 4M). Seeds 2 per monocarp, yellowish, semi-ellipsoid to ellipsoid, ca. 18 cm long, 8 mm in diameter, endosperm rumination lamelliform (Figs 3M, N, 4M).
Distribution and habitat.
Known from several localities in Yunnan Province (Fig. 5): Hei-long-tan and Manxi in Meng-hai County and Kun-man in Meng-yang Town, Jinghong, growing in rain forests, at medium elevations (1300–1800 m a.s.l.).
Phenology.
Flowering in February to March; fruiting from April to July.
Additional specimens examined.
China. Yunnan: Kun-man, Xiao-meng-yang, 27 April 1957, Yunnan Exped. 9527 (IBSC, KUN, PE); Man-xi, Menghai, 16 March 2016, Y.H. Tan MH1603 (HITBC, IBSC); 5 March 2019, B. Xue & H.B. Ding 311, 312, 313 (HITBC, IBSC, KUN).
Preliminary IUCN conservation status.
CR A2ac, C2(a)(i) (IUCN 2012). Polyalthiopsis verrucipes was assessed as EN A2c by the China Plant Specialist Group (2004). Prior to this study, P. verrucipes was only represented in herbaria by two collections from Yunnan, China (two localities, both of which have subsequently been severely deforested). Our field survey in 2016 identified one population with dozens of individuals of dbh ca. 10 cm and dozens of young treelets in Manxi village, Menghai County. We made a second visit to the location in 2019 and found only a few individuals with dbh larger than 10 cm and few treelets. Herbicide had been used in that location. The bark at the bottoms of the tree trunks was damaged. The local farmers appear to clear the forests in this way for tea plantation and it is anticipated that the trees with damaged bark could not survive. We hope additional undocumented sub-populations will be found and protected, although further field investigation is needed to better understand the current status of populations. At present, we recommend that this species be regarded as critically endangered (CR) based on current IUCN Red List Categories and Criteria (IUCN 2012).
Key to Polyalthiopsis
| 1a | Inflorescences 1–5-flowered; petals elliptic-ovate; ovule 1 per carpel; distributed in Vietnam | P. floribunda |
| 1b | Inflorescences 1–2-flowered; petals linear; ovules 2 per carpel; distributed in China | 2 |
| 2a | Inflorescences 1–2-flowered; pedicel to 5–7 mm long; flowers green; distributed in Xizang, China | P. chinensis |
| 2b | Inflorescences with a single flower; pedicel 1–2 mm long; flowers white; distributed in Yunnan, China | P. verrucipes |
Supplementary Material
Acknowledgements
This research was supported by the National Natural Science Foundation of China [Grant no. 31872646, awarded to Bine Xue and Grant no. 31970223, awarded to Yunhong Tan]; Forestry Scientific Technology Innovation Project of Guangdong Province (No.2020KJCX010); a project of the Lancang-Mekong Cooperation (LMC) Special Fund (Biodiversity Monitoring and Network Construction along Lancang-Mekong River Basin project); the CAS 135 programme [No. 2017XTBG-F03]; and a project supported by the biodiversity investigation, observation and assessment programme (2019–2023) of the Ministry of Ecology and Environment of China. We are grateful to Jing-hua Wang and En-de Liu in KUN and Qin Ban and Xin-tang Ma in PE for permission to access their collections and for permission to use the leaf materials; Ji Mo and Kai-rong Pu for helping in the field in Menghai; Richard Saunders, Daniel Thomas, Thomas Couvreur and an anonymous reviewer for improving the manuscript.
Appendix 1
Voucher information and GenBank accession numbers for samples used in this study (–, missing data; *, newly generated sequences). Voucher data are given for accessions for which DNA sequences were newly obtained, using the following format: species, origin, voucher and Genbank accession numbers for matK, ndhF, rbcL, psbA-trnH and trnL-F and ycf1. For DNA sequences published in previous studies, voucher information is available from GenBank.
Alphonsea elliptica Hook. f. & Thomson, AY518807, JQ690401, JQ690402, –, AY319078, JQ690403; Alphonsea kinabaluensis J. Sinclair, AY518811, –, –, AY318968, AY319080, –; Ambavia gerrardii (Baill.) Le Thomas, AY220435, AY218168, –, –, AY220411(intron)AY220358(spacer), –; Anaxagorea silvatica R. E. Fr., AY743477, EF179280, –, AY743439, AY743458, –; Annickia chlorantha (Oliv.) Setten & Maas, AY841393, AY841401, –, AY841594, AY841671, –; Annona glabra L., DQ125050, EF179281, –, AY841596, AY841673, –; Asimina triloba (L.) Dunal, AY743479, EF179287, –, AY743441, AY743460, –; Cananga odorata (Lam.) Hook. f. & Thomson, AY841394, AY841403, –, AY841602, AY841680, –; Cleistopholis glauca Pierre ex Engl. & Diels, AY841395, AY841404, –, AY841603, AY841681, –; Dasymaschalon yunnanense (Hu) Bân 1, JQ768560, JQ768598, JQ768639, JQ768680, JQ768720, –; Dendrokingstonia nervosa (Hook. f. & Thomson) Rauschert, KJ418392, KJ418386, KJ418400, KJ418382, KJ418407, –; Desmopsis microcarpa R. E. Fr., AY518804, JX544771, AY841461, AY319059, AY319173, JX544758; Desmopsis schippii Standl., AY518805, JQ723786, –, AY319060, AY319174, –; Desmos cochinchinensis Lour., JQ768568, JQ768604, –, JQ768688, JQ768728, –; Dasymaschalon yunnanense (Hu) Bân 2, KF680919, KF680919, –, –, –, –; Fenerivia chapelieri (Baill.) R. M. K. Saunders, JF810375, JQ723788, –, JF810387, JF810399, –; Friesodielsia desmoides (Craib) Steenis, JQ768577, JQ768612, –, JQ768696, JQ768738, –; Goniothalamus griffithii Hook. f. & Thomson, AY743484, EF179296, –, AY743446, AY743465, –; Greenwayodendron oliveri (Engl.) Verdc., AY743489, AY841408, AY841465, AY743451, AY743470, –; Guatteria anomala R. E. Fr., AY740913, EF179298, –, AY740962, AY741011, –; Hubera cerasoides (Roxb.) Chaowasku, AY518854, JQ723810, KF709055, AY319017, AY319131, JQ723950; Hubera nitidissima (Dunal) Chaowasku, KF682110, KF682116, KF709056, KF682103, KF682105, –; Maasia discolor (Diels) Mols P. J. A. Kessler & Rogstad, AY518872, AY841416, AY841500, AY319021, AY841584, –; Marsypopetalum crassum (R. Parker) B. Xue & R. M. K. Saunders, HQ286571, JQ723792, KF709057, HQ286577, HQ286583, JQ723929; Meiogyne hainanensis (Merr.) Bân, JQ723773, –, –, JQ723860, JQ723913, JQ723936; Meiogyne mindorensis (Merr.) Heusden, JQ723776, JQ723800, –, JQ723863, JQ723916, JQ723939; Meiogyne virgata (Blume) Miq., AY518798, JQ723805, JX544784, AY318982, AY319094, JQ723945; Miliusa mollis Pierre, AY518851, JQ690503, JQ690504, –, AY319102, JQ690505;Miliusa thorelii Finet & Gagnep.,AY518846,JQ690519,JQ690520,-,AY319104,JQ690521; Miliusa velutina (Dunal) Hook. f. & Thomson, AY518847, JQ690536, JQ690537, AY318993, AY319105, JQ690538; Mitrephora alba Ridl., AY518855, JQ723807, KF709058, AY318994, AY319106, JQ723947; Mkilua fragrans Verdc., DQ125060, EF179303, –, AY841634, AY841712, –; Monanthotaxis whytei (Stapf) Verdc., EF179278, EF179304, –, AY841635, AY841713, –; Monocarpia euneura Miq., AY518865, AY841412, AY841477, AY318998, AY319111, –; Monoon lateriflorum (Blume) B. Xue & R. M. K. Saunders, JQ723783, JQ723811, KF709060, JQ723870, JQ723923, JQ723951; Neo-uvaria parallelivenia (Boerl.) H. Okada & K. Ueda, AY518794, –, –, AY319000, AY319113, –; Neo-uvaria telopea Chaowasku, JX544751, JX544778, JX544791, JX544755, JX544783, JX544766; Orophea cuneiformis King, KF682112, KF682119, –, –, KF682107, –; Phaeanthus splendens Miq., AY518864, JX544777, JX544790, JX544754, AY319126, JX544765; Piptostigma mortehani De Wild., AY743492, AY841415, –, AY743454, AY743473, –; Platymitra macrocarpa Boerl., AY518812, JQ723809, KF709062, AY319013, AY319127, JQ723949; Polyalthia johnsonii (F. Muell.) B. Xue & R. M. K. Saunders, JQ723767, JQ723791, KF709063, JQ723854, JQ723907, JQ723928; Polyalthiopsis chinensis (S.K.Wu ex P.T.Li) B.Xue [= Polyalthia chinensis S.K.Wu & P.T.Li, Polyalthia chinensis_74-4451], China, Xizang Province, Qinghai-Xizang Exped. 74-4451 (KUN), MT239203*, –, –, –,MT239213*, –; Polyalthiopsis chinensis (S.K.Wu ex P.T.Li) B.Xue [= Polyalthia chinensis S.K.Wu & P.T.Li, Polyalthia chinensis_2668], China, Xizang Province, B. S. Li & S. Z. Cheng 2668 (PE), MT239201*, –, –, –, MT239211*, –; Polyalthiopsis verrucipes (C.Y.Wu ex P.T.Li) B.Xue [= Polyalthia verrucipes C.Y.Wu ex P.T.Li, Polyalthia verrucipes_1603], China, Yunnan Province, Y.H. Tan MH1603 (IBSC), MT239202*, MT239205*, MT239207*, MT239209*, MT239212*, MT239215*; Polyalthiopsis verrucipes (C.Y.Wu ex P.T.Li) B.Xue [= Polyalthia verrucipes C.Y.Wu ex P.T.Li, Polyalthia verrucipes_9527], China, Yunnan Province, Sino-Russia Exped. 9527 (PE), MT239200*, MT239204*, MT239206*, MT239208*, MT239210*, MT239214*; Polyalthiopsis floribunda (Jovet-Ast) Chaowasku, Chaowasku 168 (CMUB), MG264583, MG264588, MG264570, MG264580, MG264575, –; Polyalthiopsis floribunda (Jovet-Ast) Chaowasku, Chaowasku 128 (CMUB), MG264585, MG264590, MG264572, –, MG264577, –; Popowia pisocarpa (Blume) Endl., AY518862, JQ723812, KF709065, AY319044, AY319158, JQ723953; Pseuduvaria fragrans Y. C. F. Su, JQ723784, JQ723813, XXXXXXXX, JQ723871, JQ723924, JQ723954; Sageraea lanceolata Miq., AY518799, JX544774, JX544787, AY319050, AY319164, JX544762; Sapranthus viridiflorus G. E. Schatz, AY743493, AY841422, AY841515, AY319051, AY319165, JQ723955; Stelechocarpus burahol (Blume) Hook. f. & Thomson, AY518803, JQ723814, KF709067, AY319053, AY319167, JQ723956; Stenanona costaricensis R. E. Fr., AY518801, JX544772, AY841516, AY319069, AY319183, JX544759; Tridimeris sp., JX544750, JX544773, JX544786, JX544753, JX544782, JX544761; Trigynaea lanceipetala D. M. Johnson & N. A. Murray, AY743487, EF179309, –, AY743449, AY743468, –; Trivalvaria costata (Hook. f. & Thomson) I. M. Turner, HQ286574, JQ723815, XXXXXXXX, HQ286580, HQ286586, JQ723957; Uvaria lucida Benth. subsp. virens (N. E. Br.) Verdc., AY238966, EF179310, –, AY238957, EF179319, –; Wangia florulenta (C. Y. Wu ex P. T. Li) Bine Xue, KX495154, KX495158, KX495156, KX495162, KX495160; Wuodendron praecox (Hook. f. & Thomson) B.Xue, Y.H.Tan & X.L.Hou, MF687367, MF687369, MF687371, MF687373, MF687375, MF687377.
Citation
Xue B, Ding H-B, Yao G, Shao Y-Y, Fan X-J, Tan Y-H (2020) From Polyalthia to Polyalthiopsis (Annonaceae): transfer of species enlarges a previously monotypic genus. PhytoKeys 148: 71–91. https://doi.org/10.3897/phytokeys.148.50929
References
- Brown JM, Hedtke SM, Lemmon AR, Lemmon EM. (2010) When trees grow too long: Investigating the causes of highly inaccurate Bayesian branch-length estimates. Systematic Biology 59(2): 145–161. 10.1093/sysbio/syp081 [DOI] [PubMed] [Google Scholar]
- Chaowasku T, Johnson DM, Van der Ham RWJM, Chatrou LW. (2012) Characterization of Hubera (Annonaceae), a new genus segregated from Polyalthia and allied to Miliusa Phytotaxa 69(1): 33–56. 10.11646/phytotaxa.69.1.6 [DOI]
- Chaowasku T, Ham RWJMVD, Chatrou LW. (2013) Integrative systematics supports the establishment of Winitia, a new genus of Annonaceae (Malmeoideae, Miliuseae) allied to Stelechocarpus and Sageraea Systematics and Biodiversity 11(2): 195–207. 10.1080/14772000.2013.806370 [DOI]
- Chaowasku T, Thomas DC, van der Ham RWJM, Smets EF, Mols JB, Chatrou LW. (2014) A plastid DNA phylogeny of tribe Miliuseae: Insights into relationships and character evolution in one of the most recalcitrant major clades of Annonaceae. American Journal of Botany 101(4): 691–709. 10.3732/ajb.1300403 [DOI] [PubMed] [Google Scholar]
- Chaowasku T, Johnson DM, van der Ham RWJM, Chatrou LW. (2015) Huberantha, a replacement name for Hubera (Annonaceae: Malmeoideae: Miliuseae). Kew Bulletin 70(2): 1–23. 10.1007/s12225-015-9571-z [DOI] [Google Scholar]
- Chaowasku T, Damthongdee A, Jongsook H, Nuraliev MS, Ngo DT, Le HT, Lithanatudom P, Osathanunkul M, Deroin T, Xue B, Wipasa J. (2018) Genus Huberantha (Annonaceae) revisited: Erection of Polyalthiopsis, a new genus for H. floribunda, with a new combination H. luensis. Annales Botanici Fennici 55(1–3): 121–136. 10.5735/085.055.0114 [DOI] [Google Scholar]
- Chatrou LW, Pirie MD, Erkens RHJ, Couvreur TLP, Neubig KM, Abbott JR, Mols JB, Maas JW, Saunders RMK, Chase MW. (2012) A new subfamilial and tribal classification of the pantropical flowering plant family Annonaceae informed by molecular phylogenetics. Botanical Journal of the Linnean Society 169(1): 5–40. 10.1111/j.1095-8339.2012.01235.x [DOI] [Google Scholar]
- China Plant Specialist Group (2004) Polyalthia verrucipes The IUCN Red List of Threatened Species 2004: e.T46415A11053994. 10.2305/IUCN.UK.2004.RLTS.T46415A11053994.en [DOI]
- Couvreur TLP. (2009) Monograph of the syncarpous African genera Isolona and Monodora (Annonaceae). Systematic Botany Monographs 87: 1–150. [Google Scholar]
- Doyle JJ, Doyle JL. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19(1): 11–15. [Google Scholar]
- Doyle JA, Le Thomas A. (1994) Cladistic analysis and pollen evolution in Annonaceae. Acta Botanica Gallica 141(2): 149–170. 10.1080/12538078.1994.10515148 [DOI] [Google Scholar]
- Doyle JA, Bygrave P, Le Thomas A. (2000) Implications of molecular data for pollen evolution in Annonaceae. In: Harley MM, Morton CM, Blackmore S. (Eds) Pollen and Spores: Morphology and Biology.Royal Botanic Gardens, Kew, 259–284.
- Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S. (2010) Geneious version 5.1. http://www.geneious.com/
- Erkens RHJ, Oosterhof J, Wester LYT, Maas PJM. (2017) Revisions of Ruizodendron and Pseudephedranthus (Annonaceae) including a new species and an overview of most up-to-date revisions of Neotropical Annonaceae genera. PhytoKeys 86: 75–96. 10.3897/phytokeys.86.13773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology 59(3): 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Guo X, Wang J, Xue B, Thomas DC, Su YC, Tan YH, Saunders RM. (2014) Reassessing the taxonomic status of two enigmatic Desmos species (Annonaceae): Morphological and molecular phylogenetic support for a new genus, Wangia. Journal of Systematics and Evolution 52(1): 1–15. 10.1111/jse.12064 [DOI] [Google Scholar]
- Guo X, Tang CC, Thomas DC, Couvreur TLP, Saunders RMK. (2017) A mega-phylogeny of the Annonaceae: Taxonomic placement of five enigmatic genera and support for a new tribe, Phoenicantheae. Scientific Reports 7(1): 1–7323. 10.1038/s41598-017-07252-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XL, Li PT. (2007) Three synonyms of Annonaceae in China. Zhiwu Fenlei Xuebao 45(3): 369–375. 10.1360/aps06012 [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford, England) 17(8): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- IUCN (2012) IUCN Red List Categories and Criteria: version 3.1. Second edition. IUCN, Gland, Switzerland and Cambridge, UK, 32 pp. [Google Scholar]
- Jovet-Ast MS. (1940) Annonacees Nouvelles D’indochine. Notulae Systematicae. Herbier Du Museum de Paris, 73–88.
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30(14): 3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S. (2012) PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29(6): 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. (2016) PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34(3): 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Li PT. (1976) Some notes on the Annonaceae of China. Acta Phytotaxonomica Sinica 14(1): 96–113. [Google Scholar]
- Li PT, Gilbert MG. (2011) Annonaceae. In: Wu ZY, Raven PH, Hong DY. (Eds) Flora of China.Science Press, Beijing & Missouri Botanical Garden Press, St. Louis, 672–713.
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). IEEE, New Orleans, 8 pp 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Mols JB, Kessler PJA, Rogstad SH, Saunders RMK. (2008) Reassignment of six Polyalthia species to the new genus Masia (Annonaceae): Molecular and morphological congruence. Systematic Botany 33(3): 490–494. 10.1600/036364408785679752 [DOI] [Google Scholar]
- Müller K. (2005) The efficiency of different search strategies in estimating parsimony jackknife, bootstrap, and Bremer support. BMC Evolutionary Biology 5(1): 1–58. 10.1186/1471-2148-5-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirie MD. (2005) Cremastosperma (and other evolutionary digressions): Molecular phylogenetic, biogeographic, and taxonomic studies in Neotropical Annonaceae. PhD thesis, National Herbarium Netherland, Utrecht University, Utrecht.
- Pirie MD, Chatrou LW, Mols JB, Erkens RHJ, Oosterhof J. (2006) ‘Andean-centred’ genera in the short-branch clade of Annonaceae: Testing biogeographical hypotheses using phylogeny reconstruction and molecular dating. Journal of Biogeography 33(1): 31–46. 10.1111/j.1365-2699.2005.01388.x [DOI] [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. (2014) Tracer v1.6. http://beast.bio.ed.ac.uk/Tracer
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany 84(8): 1120–1136. 10.2307/2446155 [DOI] [PubMed] [Google Scholar]
- Saunders RMK, Su YCF, Xue B. (2011) Phylogenetic affinities of Polyalthia species (Annonaceae) with columellar-sulcate pollen: Enlarging the Madagascan endemic genus Fenerivia Taxon 60(5): 1407–1416. 10.1002/tax.605016 [DOI]
- Sinclair J. (1955) A revision of the Malayan Annonaceae. Gardens’ Bulletin (Singapore) 14(2): 149–516. [Google Scholar]
- Surveswaran S, Wang RJ, Su YCF, Saunders RMK. (2010) Generic delimitation and historical biogeography in the early-divergent ‘ambavioid’ lineage of Annonaceae: Cananga, Cyathocalyx and Drepananthus Taxon 59(6): 1721–1734. 10.1002/tax.596007 [DOI]
- Swofford DL. (2003) PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4.0b10. Sinauer Associates, Sunderland.
- Tang C, Xue B, Saunders RMK. (2013) A new species of Goniothalamus (Annonaceae) from Palawan, and a new nomenclatural combination in the genus from Fiji. PhytoKeys 32: 27–35. 10.3897/phytokeys.32.6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JA, Simpson BB. (2003) Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Systematic Botany 28(4): 723–737. [Google Scholar]
- Thomas DC, Surveswaran S, Xue B, Sankowsky G, Mols JB, Kessler PJA, Saunders RMK. (2012) Molecular phylogenetics and historical biogeography of the Meiogyne-Fitzalania clade (Annonaceae): Generic paraphyly and late Miocene-Pliocene diversification in Australasia and the Pacific. Taxon 61(3): 559–575. 10.1002/tax.613006 [DOI] [Google Scholar]
- Turner IM. (2012) Annonaceae of Borneo: A review of the climbing species. Gardens’ Bulletin (Singapore) 64(2): 371–479. [Google Scholar]
- Turner IM. (2016) New combinations and new synonyms in Asian Annonaceae with a bibliographical note. Webbia: Journal of Plant Taxonomy and Geography 71(2): 229–232. 10.1080/00837792.2016.1241516 [DOI] [Google Scholar]
- van der Heijden E, Kessler PJA. (1990) Studies on the tribe Saccopetaleae (Annonaceae) – III. Revision of the genus Mezzettia Beccari. Blumea 35: 217–228. [Google Scholar]
- van Heusden ECH. (1995) Revision of the southeast Asian genus Stelechocarpus (Annonaceae). Blumea 40(2): 429–438. [Google Scholar]
- van Setten AK, Koek-Noorman J. (1992) Fruits and seeds of Annonaceae. Morphology and its significance for classification. 142: 1–101. [pl. 101–150] [Google Scholar]
- Xue B. (2013) Molecular phylogenetics of Polyalthia (Annonaceae): identifying clades and morphological synapomorphies in a large polyphyletic genus. PhD thesis, the University of Hong Kong, Hong Kong.
- Xue B, Su YCF, Mols JB, Kessler PJA, Saunders RMK. (2011) Further fragmentation of the polyphyletic genus Polyalthia (Annonaceae): Molecular phylogenetic support for a broader delimitation of Marsypopetalum. Systematics and Biodiversity 9(1): 17–26. 10.1080/14772000.2010.542497 [DOI] [Google Scholar]
- Xue B, Su YCF, Thomas DC, Saunders RMK. (2012) Pruning the polyphyletic genus Polyalthia (Annonaceae) and resurrecting the genus Monoon Taxon 61(5): 1021–1039. 10.1002/tax.615009 [DOI]
- Xue B, Thomas DC, Chaowasku T, Johnson DM, Saunders RMK. (2014) Molecular phylogenetic support for the taxonomic merger of Fitzalania and Meiogyne (Annonaceae): New nomenclatural combinations under the conserved name Meiogyne. Systematic Botany 39(2): 396–404. 10.1600/036364414X680825 [DOI] [Google Scholar]
- Xue B, Tan YH, Ye XE. (2016) The identity of Polyalthia florulenta (Annonaceae): A second species of Wangia in China. Phytotaxa 283(2): 163–171. 10.11646/phytotaxa.283.2.5 [DOI] [Google Scholar]
- Xue B, Tan YH, Thomas DC, Chaowasku T, Hou XL, Saunders RMK. (2018) A new Annonaceae genus, Wuodendron, provides support for a post-boreotropical origin of the Asian-Neotropical disjunction in the tribe Miliuseae. Taxon 67(2): 250–266. 10.12705/672.2 [DOI] [Google Scholar]
- Xue B, Guo X, Landis JB, Sun M, Tang CC, Soltis PS, Soltis DE, Saunders RMK. (2020) Accelerated diversification correlated with functional traits shapes extant diversity of the early divergent angiosperm family Annonaceae. Molecular Phylogenetics and Evolution 142(106659): 1–15. 10.1016/j.ympev.2019.106659 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.