Abstract
Carbapenem-resistant Enterobacterales (CRE) is an increasing problem worldwide. Here, we examined the clonal relatedness of 71 non-repetitive CRE isolates collected in a university hospital in Tehran, Iran, between February 2015 and March 2016. Pulsed-field gel electrophoresis (PFGE) and MLST were used for epidemiological analysis. Screening for antibiotic resistance genes, PCR-based replicon typing, conjugation experiments, and optical DNA mapping were also performed. Among all 71 isolates, 47 isolates of Klebsiella pneumoniae (66.2%), eight Escherichia coli (11.2%), five Serratia marcescens (7%), and two Enterobacter cloacae (2.8%) harbored blaNDM–1 and blaOXA–48 genes together or alone. PFGE analysis revealed that most of the OXA-48- and NDM-1-producing K. pneumoniae and all of OXA-48-producing S. marcescens were clonally related, while all eight E. coli and two E. cloacae isolates were clonally unrelated. The predominant clones of carbapenemase-producing K. pneumoniae associated with outbreaks within the hospital were ST147 (n = 13) and ST893 (n = 10). Plasmids carrying blaNDM–1 and blaOXA–48 were successfully transferred to an E. coli K12-recipient strain. The blaOXA–48 gene was located on an IncL/M conjugative plasmid, while the blaNDM–1 gene was located on both IncFII ∼86-kb to ∼140-kb and IncA/C conjugative plasmids. Our findings provide novel epidemiologic data on carbapenemase-producing Enterobacterales (CPE) in Iran and highlight the importance of horizontal gene transfer in the dissemination of blaNDM–1 and blaOXA–48 genes. The occurrence and transmission of distinct K. pneumoniae clones call for improved infection control to prevent further spread of these pathogens in Iran.
Keywords: carbapenemase-producing Enterobacterales, PFGE, MLST, ST147, optical DNA mapping
Introduction
Carbapenems are broad-spectrum beta-lactam agents that are frequently used as a last resort to treat serious infections caused by multidrug-resistant Enterobacterales. Resistance to carbapenems mainly depends on the production of carbapenemase enzymes. Carbapenemase-producing Enterobacterales (CPE) are increasingly reported and represent a major public health threat (Tängdén and Giske, 2015). The most clinically significant carbapenemases in Enterobacterales include the class A (KPC type), class B (metallo-β-lactamases [MBLs] [i.e., VIM, IMP, and NDM types]), and class D carbapenem-hydrolyzing β-lactamases (OXA-48-like enzymes) (Nordmann et al., 2011; Tängdén and Giske, 2015). NDM-1 and OXA-48 β-lactamases were initially identified in India and Turkey, respectively, and then spread to various countries worldwide including India, the Middle East, and Mediterranean countries (Yong et al., 2009; Johnson and Woodford, 2013; Sartor et al., 2014; Jamal et al., 2016; Solgi et al., 2017b). There is a lot of pilgrimage tourism and business travel between Iran and neighboring countries such as Iraq, Afghanistan, Pakistan, Turkey, and the Persian Gulf, so travelers with CPE colonization may be the vectors for spread of resistant strains. In the scope of outbreaks in Iran, diverse sequence types (STs) of dominant OXA-48- and NDM-producing Klebsiella pneumoniae have been identified in outbreaks or solitary case reports (STs 11, 893, 147, and 915) (Solgi et al., 2017a, 2018). VIM-2-producing K. pneumoniae ST23 has been reported in Iran more recently (Mohammad Ali Tabrizi et al., 2018).
The dissemination of OXA-48 and NDM-1 among Enterobacterales is mediated by the rapid spread of broad host-range conjugative plasmids. The blaNDM–1 gene has been detected on plasmids of various incompatibility groups: IncF, IncA/C, IncL/M, IncH, IncN, and IncX3 or untypeable (Voulgari et al., 2014). The blaOXA–48 gene has also been carried by various plasmids types including IncL/M, IncN, and IncA/C (Guo et al., 2016). Up until today, only one study has reported the finding of the prevalence and distribution of carbapenem resistance among Enterobacterales isolates in Iran (Shahcheraghi et al., 2017). However, limited data about the sequence type of CRE isolates that has spread in Iran were available.
Here, we investigated the prevalence of ESBL and carbapenemase genes, to explore the distribution of plasmid replicons, and molecular epidemiology of CPE isolated in an Iranian hospital.
Materials and Methods
Bacterial Strains
In this cross-sectional study, a total of 71 non-repetitive carbapenem-resistant Enterobacterales (CRE) clinical isolates resistant to at least one of the carbapenems (imipenem, meropenem, or ertapenem) were collected at the Loghman Hakim Educational Hospital, a 496-bed university hospital in Tehran (Iran) between February 2015 and March 2016. All isolates were identified by standard biochemical tests and API 20E (bioMérieux, Marcy-l’Etoile, France).
Antimicrobial Susceptibility Testing and Phenotypic Assay
Antimicrobial susceptibility testing of 10 antibiotics (imipenem, meropenem, ertapenem, cefepime, cefotaxime, ceftazidime, aztreonam, amikacin, gentamicin, and ciprofloxacin) was done by a standard disk diffusion method according to the Clinical and Laboratory Standards Institute [CLSI] (2017) guidelines. The minimal inhibitory concentration (MIC) determinations for carbapenems (imipenem, meropenem, and ertapenem) were performed by gradient test strips (Liofilchem, Italy) based on Clinical and Laboratory Standards Institute [CLSI] (2017) guidelines. MICs of colistin were determined by broth macrodilution method using colistin sulfate (Sigma-Aldrich), and EUCAST breakpoints were used for interpretation (EUCAST, 2017). Escherichia coli ATCC 25922 was used as quality control. Initial screening for the presence of carbapenemases was done by the modified Hodge test (MHT) test by following the Clinical and Laboratory Standards Institute [CLSI] (2017) guideline.
Molecular Detection of Genes Encoding Carbapenemases and ESBLs
Plasmid DNA was extracted using the Gene JET Plasmid Maxi-Prep Kit (Thermo Scientific). The presence of genes encoding carbapenemases (blaKPC, blaGES, blaVIM, blaIMP, blaNDM, and blaOXA–48) and extended-spectrum β-lactamases (ESBL) (blaCTX–M) and further beta-lactamases (blaTEM, blaSHV) were detected by PCR amplification using specific primers as described previously (Poirel et al., 2011; Shahcheraghi et al., 2013), followed by sequencing (Macrogen Research, Seoul, South Korea).
Molecular Typing
The genetic relatedness of CPE isolates was investigated by pulsed-field gel electrophoresis (PFGE). The genomic DNA of the CPE isolates and reference marker Salmonella serotype Braenderup strain H9812 were digested by XbaI endonuclease, which was performed with a CHEF-DRIII system (Bio-Rad Laboratories) as previously described (Tenover et al., 1995). A similarity coefficient was obtained using Dice coefficients. Cluster analysis was done with the unweighted pair group method with arithmetic averages (UPGMA). Isolates that exhibited similarity cut-off ≥80% of their banding patterns were considered to belong to the same clonal lineage (pulsotypes). Multilocus sequence typing (MLST) was performed according to the protocol described on the Pasteur Institute MLST website1 for K. pneumoniae, MLST website for E. coli2, and MLST website for Enterobacter cloacae3.
Conjugation Experiments and PCR-Based Replicon Typing
Conjugation experiments were done using the blaNDM–1 and blaOXA–48 producers as the donors and E. coli K12 [F– lac + Nal (r)] as the recipient strain (Filter mating). Isolates LO35, LO89, LO112, LO179, LO271, and LO273 which harbored only the blaNDM–1 or blaOXA–48 gene, and isolates LO149, LO155, and LO204, which harbored the blaOXA–48 and blaNDM–1 genes, were selected and used. Transconjugants were selected on a MacConkey agar plate containing 32 mg/L nalidixic acid (Sigma-Aldrich) and 1 mg/L MEM (MAST, Merseyside, United Kingdom) (Lyimo et al., 2016) and were confirmed to have blaNDM–1 and blaOXA–48 by PCR analysis. PCR-based replicon typing analysis (PBRT) was performed to determine the plasmid incompatibility (Inc) groups for all CPE strains and the obtained transconjugants (Carattoli et al., 2005).
Plasmid Extraction
Plasmid DNA was prepared from an overnight culture with NucleoBond® Xtra Midi kit for isolates according to the manufacturer’s description for high-copy plasmid purification (Müller et al., 2016a). Eluted plasmid DNA is then precipitated with isopropanol and washed with 70%; the dried pellet was reconstituted TE buffer, pH 8.0. The DNA concentration and purity were determined using the Qubit 3.0 Fluorometer.
Optical DNA Mapping in Nanochannels for Plasmid Analysis
The presence of the blaNDM–1 gene on plasmids from isolates LO94, LO204, LO271, LO247, LO64, LO63, LO89, and LO149 was investigated using optical DNA mapping (Müller and Westerlund, 2017). For this, Cas9 enzyme (PNA Bio Inc., Newbury Park, CA, United States) was used to make a site-specific cut at the blaNDM–1 gene (target gene sequence was 5′-CGGTATGGACGCGCTGCATG-3′, RNA was synthesized by Dharmacon Inc., Lafayette, CO, United States) on the plasmids (Müller et al., 2016b). Cas9 will cut all the blaNDM–1 gene-carrying plasmids in each isolate at the same location which would show as a consensus cut site in the ODM data. For the plasmids not carrying the blaNDM–1 gene, we expected randomly distributed cuts.
After the Cas9 reaction, the plasmids were stained using YOYO-1 and Netropsin which created an emission intensity pattern along the DNA, with dark AT-rich regions and bright GC-rich regions (Nyberg et al., 2012; Nilsson et al., 2014). Netropsin prevents the binding of the fluorescent YOYO-1 to AT-rich regions which results in the formation of a variation in intensity, a DNA barcode. Plasmids were stretched to their full contour lengths by confining them in 100 × 150-nm2 nanofluidic channels and imaged using an EMCCD camera. For each of the eight isolates, hundreds of plasmids were imaged and analyzed. The barcodes were aligned, clustered based on similarity, and compared among the isolates using custom-built MATLAB routines (Müller et al., 2016a). Lambda phage DNA was used as an internal control to correlate the length in pixels with the length in base pairs, and this correlation factor was then used to estimate plasmid sizes.
Results
Bacterial Isolates
During the study period, 71 clinical CRE isolates were collected from 44 male and 27 female patients. These isolates mainly belonged to the species K. pneumoniae (56/71, 78.8%), E. coli (8/71, 11.2%), Serratia marcescens (5/71, 7%), and E. cloacae (2/71, 2.8%). Twenty-two isolates (31%) were isolated from an ICU poisoning ward, whereas the remaining of isolates were recovered from other ward. The majority of the isolates were from urine (30/71, 42.2%) and tracheal (24/71, 33.8%) specimens. Other sample types included blood (6/71; 8.4%), wound secretions (5/71; 7%), sputum (3/71; 4.2%), catheter (2/71; 2.8%), and cerebrospinal fluid (1/71; 1.4%).
Antimicrobial Susceptibility
Susceptibility profiles against ten antimicrobials agents are listed in Table 1. As expected, the majority of the CRE isolates exhibited resistance to most β-lactams. Most of the isolates were also resistant to ciprofloxacin (70/71 98.6%) and gentamicin (42/71 59.1%). On the other hand, most of them were susceptible to amikacin (46/71 64.8%), and all isolates were susceptible to colistin, with MICs ≤ 1 mg/L. Based on phenotypic detection, 40 out of the 62 isolates (64.5%) were positive for MHT.
TABLE 1.
Clinical information and molecular characteristics of 71 carbapenem-resistant Enterobacterales isolated from a university hospital in Tehran, Iran.
| Patient/Strain | Species | Carbapenemase genes | Associated β-lactamases | Inc groupa | ST | Specimen | Hospitalization unit | Resistance phenotype | MIC (μ g/ml) | ||
| ERT | MEM | IPM | |||||||||
| LO-1 | K. pneumoniae | NDM-1, OXA-48 | CTX-M-15, TEM-1, SHV-106 | IncFII, IncL/M | ST15 | Tracheal | Emergency ICU | CAZ, CTX, FEP, CIP | 8 | 8 | 8 |
| LO-7 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-199 | IncFII | ST893 | Tracheal | Poisoning ICU | CAZ, CTX, FEP, GEN, CIP | 8 | 8 | 8 |
| LO-8 | K. pneumoniae | – | CTX-M-15, TEM-1 | ND | ND | Wound | Poisoning ICU | CAZ, CTX, FEP, CIP | 4 | 1 | >4 |
| LO-17 | K. pneumoniae | NDM-1 | CTX-M-15, SHV-1 | IncFII | ND | Urine | Nerves of men | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 64 |
| LO-20 | K. pneumoniae | – | CTX-M-15, TEM-1, SHV-1 | ND | ND | Tracheal | Poisoning ICU | CAZ, CTX, FEP, CIP | 4 | 0/5 | >4 |
| LO-21 | K. pneumoniae | – | CTX-M-15, TEM-1, SHV-1 | ND | ND | Tracheal | Poisoning ICU | CAZ, CTX, FEP, CIP | 2 | 1 | >4 |
| LO-30 | K. pneumoniae | – | CTX-M-15, TEM-1, SHV-1 | ND | ND | Urine | Poisoning ICU | CAZ, CTX, FEP, CIP | 4 | 0/5 | >4 |
| LO-36 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-1 | IncFII | ND | Urine | Poisoning ICU | CAZ, CTX, FEP, GEN, CIP | 8 | 4 | >4 |
| LO-56 | K. pneumoniae | – | CTX-M-15, TEM-1 | ND | ND | Wound | Surgery | CAZ, CTX, FEP, CIP | 2 | 1 | >4 |
| LO-63 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-11 | UT | ST147 | Wound | Surgery | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 32 |
| LO-64 | K. pneumoniae | NDM-1, OXA-48 | CTX-M-15, TEM-1, SHV-199 | IncL/M | ST893 | Tracheal | Poisoning ICU | CAZ, CTX, FEP, CIP | 8 | 32 | 64 |
| LO-68 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-11 | IncFII | ST147 | Sputum | Infectious | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 256 |
| LO-70 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-1 | IncFII | ND | Tracheal | General ICU | CAZ, CTX, FEP, CIP | 8 | 8 | 8 |
| LO-77 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-11 | IncFII | ST147 | Tracheal | General ICU | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 8 | 32 |
| LO-78 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-12 | UT | ST147 | Tracheal | Emergency ICU | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 32 |
| LO-80 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-11 | IncFII | ST147 | Urine | Infectious | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 64 |
| LO-82 | K. pneumoniae | NDM-1, OXA-48 | CTX-M-15, TEM-1, SHV-199 | IncL/M | ST893 | Urine | Internal emergency | CAZ, CTX, FEP, AM, GEN, CIP | 8 | 32 | 256 |
| LO-88 | K. pneumoniae | OXA-48 | CTX-M-15, TEM-1, SHV-1 | IncL/M | ND | Urine | Internal emergency | CAZ, CTX, FEP, GEN, CIP | 8 | 4 | 4 |
| LO-89 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-11 | IncFII | ST147 | Urine | Outpatient | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 16 | 32 |
| LO-91 | K. pneumoniae | OXA-48 | CTX-M-15, TEM-1, SHV-1 | IncL/M | ST377 | Tracheal | Emergency ICU | CAZ, CTX, FEP, AM, GEN, CIP | 8 | 8 | 8 |
| LO-94 | K. pneumoniae | NDM-1 | CTX-M-15, SHV-199 | UT | ST16 | Tracheal | Infectious | CAZ, CTX, FEP, GEN, CIP | 8 | 4 | 4 |
| LO-95 | K. pneumoniae | OXA-48 | CTX-M-15, TEM-1, SHV-199 | IncL/M | ST893 | Tracheal | Internal emergency | CAZ, CTX, FEP, GEN, CIP | 8 | 2 | >4 |
| LO-97 | K. pneumoniae | OXA-48 | CTX-M-15, TEM-1, SHV | IncL/M | ST16 | Urine | Emergency ICU | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 2 | >4 |
| LO-106 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-1 | IncFII | ND | Blood | Poisoning ICU | CAZ, CTX, FEP, AMK, GEN, CIP | 4 | 4 | 2 |
| LO-110 | K. pneumoniae | OXA-48 | CTX-M-15, TEM-1, SHV-199 | IncL/M | ST893 | Tracheal | Poisoning ICU | CAZ, CTX, FEP, CIP | 4 | 2 | >4 |
| LO-114 | K. pneumoniae | NDM-1 | CTX-M-15, SHV-1 | IncFII | ST657 | Tracheal | Internal emergency | CAZ, CTX, FEP, CIP | 8 | 8 | 8 |
| LO-119 | K. pneumoniae | – | CTX-M-15, TEM-1, SHV-1 | ND | ND | Urine | Emergency ICU | CAZ, CTX, FEP, GEN, CIP | 2 | 1 | >4 |
| LO-121 | K. pneumoniae | OXA-48 | TEM-1, SHV-199 | IncL/M | ST893 | Sputum | Internal emergency | CAZ, CTX, FEP, CIP | 8 | 8 | 8 |
| LO-123 | K. pneumoniae | NDM-1 | CTX-M-15, SHV-1 | IncFII | ST35 | Tracheal | Poisoning ICU | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 32 |
| LO-125 | K. pneumoniae | OXA-48 | CTX-M-15, TEM-1, SHV-182 | IncL/M | ST11 | Urine | Internal emergency | CAZ, CTX, FEP, CIP | 4 | 2 | >4 |
| LO-126 | K. pneumoniae | OXA-48 | CTX-M-15, TEM-1, SHV-199 | IncL/M | ST893 | Urine | Poisoning ICU | CAZ, CTX, FEP, CIP | 4 | 1 | >4 |
| LO-147 | K. pneumoniae | NDM-1, OXA-48 | CTX-M-15, TEM-1, SHV-106 | IncFII, IncL/M | ST15 | Tracheal | General ICU | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 32 |
| LO-149 | K. pneumoniae | NDM-1, OXA-48 | CTX-M-15, TEM-1, SHV-106 | IncFII, IncL/M | ST15 | Tracheal | Poisoning ICU | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 32 |
| LO-154 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-199 | UT | ST16 | Urine | Internal emergency | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 32 |
| LO-155 | K. pneumoniae | NDM-1, OXA-48 | CTX-M-15, TEM-1, SHV-199 | IncL/M | ST893 | Tracheal | Poisoning ICU | CAZ, CTX, FEP, CIP | 8 | 32 | 32 |
| LO-179 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-199 | IncFII | ST16 | Urine | Infectious | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 32 |
| LO-181 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-1 | IncFII | ST147 | Urine | Surgery | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 16 | 8 |
| LO-191 | K. pneumoniae | NDM-1 | – | IncFII | ST1308 | Wound | Surgery | CAZ, CTX, FEP | 8 | 4 | 4 |
| LO-204 | K. pneumoniae | NDM-1, OXA-48 | CTX-M-15, TEM-1, SHV-106 | IncFII, IncL/M | ST15 | Catheter | Surgery | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 256 |
| LO-216 | K. pneumoniae | – | CTX-M-15, SHV-199 | ND | ND | Urine | Internal emergency | CAZ, CTX, FEP, CIP | 2 | 0/5 | >4 |
| LO-217 | K. pneumoniae | NDM-1 | CTX-M-15, SHV-199 | IncFII | ST16 | Urine | General ICU | CAZ, CTX, FEP, GEN, CIP | 8 | 16 | 24 |
| LO-246 | K. pneumoniae | – | CTX-M-15, TEM-1, SHV-1 | ND | ND | Tracheal | Poisoning ICU | CAZ, CTX, FEP, AMK, GEN, CIP | 4 | 2 | >4 |
| LO-247 | K. pneumoniae | NDM-1, OXA-48 | CTX-M-15, TEM-1, SHV-199 | IncFII | ND | Catheter | Neurosurgery | CAZ, CTX, FEP, CIP | 8 | 8 | 4 |
| LO-251 | K. pneumoniae | NDM-1, OXA-48 | CTX-M-15, TEM-1, SHV-199 | IncL/M | ST893 | Tracheal | Poisoning ICU | CAZ, CTX, FEP, CIP | 8 | 4 | 2 |
| LO-261 | K. pneumoniae | OXA-48 | TEM-1, SHV-199 | IncL/M | ST893 | Tracheal | Infectious | CAZ, CTX, FEP, CIP | 8 | 4 | 4 |
| LO-262 | K. pneumoniae | NDM-1 | TEM-1, SHV-172 | IncFII | ST147 | Urine | Neurosurgery | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 256 |
| LO-263 | K. pneumoniae | NDM-1, OXA-48 | TEM-1, SHV-12 | IncL/M | ST147 | Urine | Neurosurgery | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 128 |
| LO-264 | K. pneumoniae | – | CTX-M-15, TEM-1, SHV-1 | ND | ND | Tracheal | Poisoning ICU | CAZ, CTX, FEP, GEN, CIP | 4 | ND | ND |
| LO-268 | K. pneumoniae | OXA-48 | TEM-1 | IncL/M | ST23 | Sputum | Neurosurgery | CAZ, CTX, FEP, GEN, CIP | 8 | 4 | 4 |
| LO-269 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-11 | IncFII | ST147 | Urine | Internal emergency | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 16 | 4 |
| LO-270 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-11 | IncFII | ST147 | Urine | Infectious | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 16 | 4 |
| LO-271 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-12 | IncFII | ST147 | Blood | Infectious | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 16 | 32 |
| LO-272 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1, SHV-1 | IncFII | ST377 | Urine | Infectious | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 128 |
| LO-277 | K. pneumoniae | NDM-1 | CTX-M-15, TEM-1 | UT | ST2012 | Cerebrospinal fluid | Infectious | CAZ, CTX, FEP, CIP | 8 | 8 | 8 |
| LO-278 | K. pneumoniae | OXA-48 | CTX-M-15, TEM-1, SHV-1 | IncL/M | ST377 | Blood | Infectious | CAZ, CTX, FEP, GEN, CIP | 8 | 16 | 8 |
| LO-279 | K. pneumoniae | NDM-1 | CTX-M-15, SHV-11 | IncFII | ST147 | Blood | Infectious | CAZ, CTX, FEP, GEN, CIP | 8 | 32 | 32 |
| LO-4 | E. coli | OXA-48 | CTX-M-15 | IncL/M | ND | Urine | Internal of women | CAZ, CTX, FEP, CIP | 1 | 0/125 | >4 |
| LO-35 | E. coli | OXA-48 | CTX-M-15 | IncL/M | ST410 | Urine | Poisoning ICU | CAZ, CTX, FEP, GEN, CIP | 0/5 | 0/125 | >4 |
| LO-96 | E. coli | OXA-48 | CTX-M-15, TEM-1 | IncL/M | ND | Wound | Infectious | CAZ, CTX, FEP, GEN, CIP | 1 | 0/5 | >4 |
| LO-175 | E. coli | OXA-48 | CTX-M-15, TEM-1 | IncL/M | ST1431 | Urine | Emergency ICU | CAZ, CTX, FEP, CIP | 2 | 0/5 | >4 |
| LO-180 | E. coli | OXA-48 | CTX-M-15, TEM-1 | IncL/M | ST3134 | Urine | Outpatient | CAZ, CTX, FEP, AMK, GEN, CIP | 1 | 0/125 | >4 |
| LO-183 | E. coli | OXA-48 | – | IncL/M | ST5114 | Urine | Outpatient | CAZ, CTX, FEP, CIP | 2 | 0/125 | >4 |
| LO-231 | E. coli | NDM-1 | CTX-M-15, TEM-1 | IncA/C | ST131 | Urine | Internal emergency | CAZ, CTX, FEP, GEN, CIP | 2 | 1 | >4 |
| LO-233 | E. coli | OXA-48 | CTX-M-15 | IncL/M | ST5114 | Urine | Emergency ICU | CAZ, CTX, FEP, CIP | 1 | 0/125 | >4 |
| LO-112 | S. marcescens | OXA-48 | CTX-M-15, TEM-1, SHV-12 | IncL/M | – | Blood | Poisoning ICU | CAZ, CTX, FEP, CIP | 8 | 16 | 4 |
| LO-113 | S. marcescens | OXA-48 | CTX-M-15, TEM-1, SHV-12 | IncL/M | – | Blood | Poisoning ICU | CAZ, CTX, FEP, CIP | 8 | 32 | 32 |
| LO-133 | S. marcescens | OXA-48 | CTX-M-15, TEM-1, SHV-12 | IncL/M | – | Tracheal | Poisoning ICU | CAZ, CTX, FEP, CIP | 8 | 16 | 8 |
| LO-166 | S. marcescens | OXA-48 | CTX-M-15, TEM-1, SHV-12 | IncL/M | – | Tracheal | Poisoning ICU | CAZ, CTX, FEP, CIP | 8 | 16 | 16 |
| LO-207 | S. marcescens | OXA-48 | CTX-M-15, TEM-1, SHV-12 | IncL/M | – | Tracheal | Poisoning ICU | CAZ, CTX, FEP, CIP | 8 | 16 | 16 |
| LO-273 | E. cloacae | NDM-1 | CTX-M-15, TEM-1 | IncFII | ST78 | Urine | Outpatient | CAZ, CTX, FEP, AMK, GEN, CIP | 8 | 32 | 32 |
| N-20-LO | E. cloacae | NDM-1 | CTX-M-15, TEM-1 | IncFII | ST175 | Urine | General ICU | CAZ, CTX, FEP, GEN, CIP | 8 | 4 | 4 |
aIncompatibility (Inc) group. ND, not determined; UT, untypeable.; F, female; M, male; MIC, minimal inhibitory concentrations; S, susceptible; IPM, imipenem; MEM, meropenem; ETP, ertapenem; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; ATM, aztreonam; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; CST, colistin. Only carbapenemase producing isolates were MLST analyzed.
Carbapenemase and ESBL Genes
The genotyping results of carbapenemase and ESBL genes among CRE isolates are shown in Table 1. Of the 71 CRE isolates, 62 were carbapenemase producers. Among the 62 carbapenemase-producing isolates, 29 were found positive for the blaNDM–1 gene, 23 were positive for the blaOXA–48 gene, and ten of the blaNDM–1-positive isolates co-harbored blaOXA–48 genes. Among the blaNDM–1-positive Enterobacterales species, 26 K. pneumoniae isolates, two E. cloacae isolates, and a single E. coli isolate were identified. The twenty-three blaOXA–48 producers were K. pneumoniae (n = 11), E. coli (n = 7), and S. marcescens (n = 5). All the ten isolates co-producing blaNDM–1 and blaOXA–48 were K. pneumoniae. Other carbapenemase genes (blaGES, blaKPC blaVIM, and blaIMP) were not detected. Among the 71 CRE isolates, 91.5% (65/71) ESBL producers were observed. Out of 65 ESBL producers, 64 (98.4%) harbored blaCTX–M–15 and eight (12.3%) harbored blaSHV–12; furthermore, a lot of isolates harbored additional blaTEM/SHV genes.
Clonal Relationship of CRE Isolates
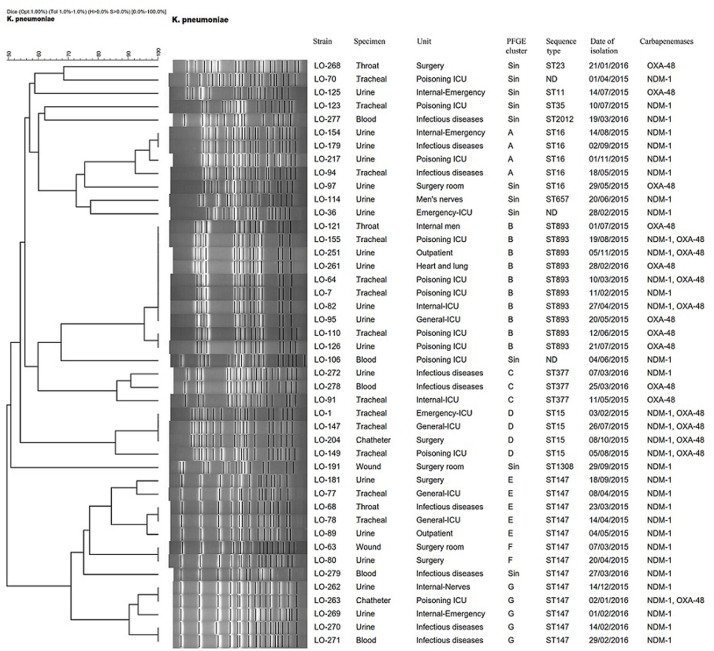
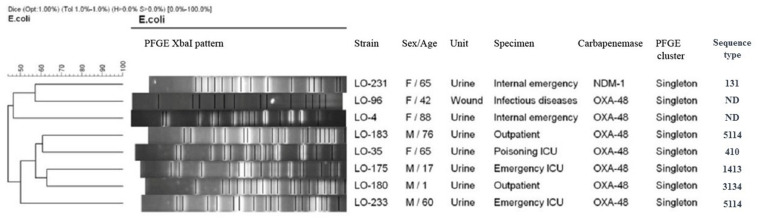
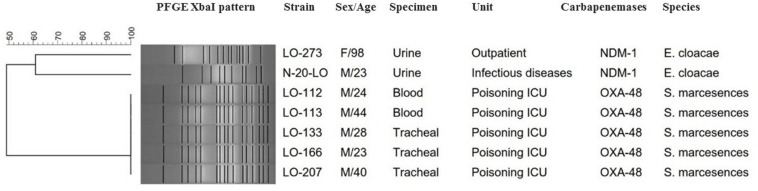
Based on a cutoff of 80% genetic similarity, PFGE revealed that 44 carbapenemase-positive K. pneumoniae isolates could be categorized in seven clusters A (4 isolates), B (10 isolates), C (3 isolates), D (4 isolates), E (5 isolates), F (2 isolates), and G (5 isolates), while 11 isolates appeared to be singletons (Figure 1). Clusters E, F, and G belonged to ST147, while clusters A, B, C, and D were categorized as ST16, ST893, ST377, and ST15, respectively. The eight NDM-1- and OXA-48-producing E. coli isolates were clonally unrelated by PFGE (Figure 2), including two belonging to the same sequence type (ST5114). The PFGE patterns of five OXA-48-positive S. marcescens isolates showed 100% similarity, but the two NDM-1-positive E. cloacae had distinct PFGE patterns (Figure 3).
FIGURE 1.
Dendrogram based on PFGE of 44 isolates of CPKP and their ST determined via MLST. ND, not determined.
FIGURE 2.
Dendrogram based on PFGE of 8 isolates of carbapenemase-producing Escherichia coli and their ST determined via MLST. ND, not determined.
FIGURE 3.
Serratia marcescens and E nterobacter cloacae are grouped together in the same dendrogram for comparison. Dendrogram based on PFGE of 5 isolates of OXA-48-producing S. marcescens and 2 NDM-1-producing E. cloacae.
Plasmid Replicon Typing and Conjugation Assay
The blaNDM–1 gene was identified on an IncFII-type plasmid for twenty-six K. pneumoniae and two E. cloacae isolates and on an IncA/C-type plasmid for a single E. coli isolate, while the blaOXA–48 gene was identified on an IncL/M-type plasmid for nineteen K. pneumoniae, seven E. coli, and five S. marcescens isolates. In the six K. pneumoniae isolates, we could not identify the incompatibility group.
Conjugation experiments revealed that all of the NDM-1 and OXA-48 plasmids were successfully transferred to E. coli K12, conferring resistance to carbapenems and cephalosporins in transconjugants. In addition, co-transfer of blaNDM–1, blaOXA–48, and other resistance determinants (blaCTX–M, blaTEM, and blaSHV) was observed in several isolates (Table 2). Plasmid gel extraction followed by PCR amplification of the transconjugants revealed that the blaOXA–48 gene was harbored on transferable plasmids belonging to the IncL/M incompatibility group, while the blaNDM–1 gene was located on conjugative plasmids. Transconjugant Tc-Lo204 had two different plasmids, and the size of one plasmid was ∼140 kb with blaNDM–1 and the other one was ∼135 kb with blaOXA–48. Notably, all blaOXA–48-positive conjugative plasmids co-harbored beta-lactamase gene blaCTX–M–15.
TABLE 2.
Microbiological characteristics of nine clinical CPE isolates and their transconjugants.
| Isolate | Species | ST | MIC (mg/L) | Antimicrobial resistance phenotype | β-lactamase(s) | Size of plasmids | Inc group | ||
| ERT | MEM | IPM | |||||||
| LO-35 | E. coli | 410 | 0.5 | 0/125 | >4 | CAZ, CTX, FEP, CIP | OXA-48, CTX-M-15 | ∼39 kb | IncL/M |
| Tc-LO-35a | – | 0/125 | 0/125 | >4 | CAZ, CTX, FEP | OXA-48, CTX-M-15 | ∼39 kb | IncL/M | |
| LO-89 | K. pneumoniae | ST147 | 8 | 16 | 32 | CAZ, CTX, FEP, AMK, GEN, CIP | NDM-1, CTX-M-15, TEM, SHV | 104.8 ± 3.6 | IncFII |
| Tc-LO-89a | – | 4 | 8 | ND | CAZ, CTX, FEP | NDM-1, CTX-M-15, TEM | – | IncFII | |
| LO-112 | S. marcescens | – | 8 | 16 | 4 | CAZ, CTX, FEP, CIP | OXA-48, CTX-M-15, TEM, SHV | ∼39 kb | IncL/M |
| Tc-LO-112a | – | 4 | 4 | 2 | CAZ, CTX, FEP | OXA-48, CTX-M-15, TEM | ∼39 kb | IncL/M | |
| LO-149 | K. pneumoniae | ST15 | 8 | 32 | 32 | CAZ, CTX, FEP, AMK, GEN, CIP | NDM-1, OXA-48, CTX-M-15, TEM, SHV | 130.6 ± 3.2 | IncFII, IncL/M |
| Tc-LO-149a | – | 8 | 16 | 8 | CAZ, CTX, FEP, AMK, GEN | NDM-1, SHV | 130.6 ± 3.2 | IncFII | |
| LO-155 | K. pneumoniae | ST893 | 8 | 32 | 32 | CAZ, CTX, FEP, CIP | NDM-1, OXA-48, CTX-M-15, TEM, SHV | – | IncL/M |
| Tc-LO-155a | – | 8 | 8 | >4 | CAZ, CTX, FEP | OXA-48, CTX-M-15, TEM | – | IncL/M | |
| LO-179 | K. pneumoniae | ST16 | 8 | 32 | 24 | CAZ, CTX, FEP, AMK, GEN, CIP | NDM-1, CTX-M-15, TEM, SHV | – | IncFII |
| Tc-LO-179a | – | 4 | 4 | ND | CAZ, CTX, FEP, AMK, GEN | NDM-1, TEM | – | IncFII | |
| LO-204 | K. pneumoniae | ST15 | 8 | 32 | 256 | CAZ, CTX, FEP, AMK, GEN, CIP | NDM-1, OXA-48, CTX-M-15, TEM, SHV | 140.2 ± 3.2 135.1 ± 3.0 | IncFII, IncL/M |
| Tc-LO-204a | – | 8 | ND | ND | CAZ, CTX, FEP, AMK, GEN | NDM-1, OXA-48, TEM, SHV | 140.2 ± 3.2 135.1 ± 3.0 | IncFII, IncL/M | |
| LO-271 | K. pneumoniae | ST147 | 8 | 16 | 32 | CAZ, CTX, FEP, AMK, GEN, CIP | NDM-1, CTX-M-15, TEM, SHV | 107.4 ± 4.6 86.3 ± 4.8 | IncFII |
| Tc-LO-271a | – | 4 | 8 | ND | CAZ, CTX, FEP | NDM-1, CTX-M-15, TEM | – | IncFII | |
| LO-273 | E. cloacae | ST78 | 8 | 32 | 24 | CAZ, CTX, FEP, AMK, GEN, CIP | NDM-1, CTX-M-15 | – | IncFII |
| Tc-LO-273a | – | 8 | ND | ND | CAZ, CTX, FEP, AMK, GEN | NDM-1 | ∼50 kb | IncFII | |
MIC, minimal inhibitory concentrations; ERT, ertapenem, MEM, meropenem; IPM, imipenem; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; AMK, amikacin; GEN, gentamicin, CIP, ciprofloxacin, CST, colistin. aTc, E. coli K12 transconjugants selected in media containing 1 μg/ml MEM. Plasmid size for LO-89, LO-149, LO-204, and LO-271 isolates was estimated by ODM; that for other isolates was estimated by plasmid preparation.
Optical DNA Mapping
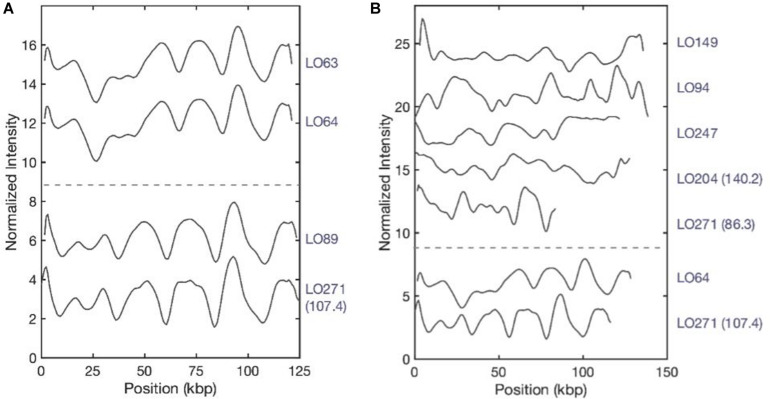
The presence of the blaNDM–1 gene on plasmids of isolates LO94, LO204, LO271, LO247, LO64, LO63, LO89, and LO149 was characterized using optical DNA mapping (ODM). Table 3 presents a summary of the ODM data for the blaNDM–1-carrying plasmids in these eight K. pneumoniae strains. DNA barcodes for each isolate were clustered based on similarity, and clusters with consensus cut sites (with at least nine barcodes) were used to infer the Cas9 cutting, suggesting the presence of the blaNDM–1 gene on the plasmids (Müller and Westerlund, 2017). For isolate LO271, two plasmids (∼86 kb and ∼107 kb) carrying the blaNDM–1 gene were identified. For isolate LO204, two plasmids of length ∼140 kb and ∼135 kb were found; however, only the ∼140-kb plasmid carried the blaNDM–1 gene. The remaining six isolates carried only one plasmid in the size range ∼110 kb to ∼130 kb carrying the blaNDM–1 gene.
TABLE 3.
Clinical and ODM information about blaNDM–1-carrying plasmids in eight K. pneumoniae strains isolated from Loghman hospital in Tehran.
| Strain no. | MIC (μ g/ml) | Species | ST | blaNDM–1 carrying plasmids (kbp) | |||
| ERT | MEM | IPM | |||||
| LO-94 | 8 | 4 | 4 | K. pneumoniae | 16 | 126.3 + 5.1 | |
| LO-204 | 8 | 32 | 256 | K. pneumoniae | 15 | 140.2 ± 3.2 | 135.1 ± 3.0* |
| LO-271 | 8 | 16 | 32 | K. pneumoniae | 147 | 86.3 ± 4.8 | 107.4 ± 4.6 |
| LO-247* | 8 | 8 | 4 | K. pneumoniae | ND* | 110.7 ± 2.5 | |
| LO-64 | 8 | 32 | 48 | K. pneumoniae | 893 | 125.4 ± 3.0 | |
| LO-63 | 8 | 32 | 32 | K. pneumoniae | 147 | 122.7 ± 2.9 | |
| LO-89 | 8 | 16 | 32 | K. pneumoniae | 147 | 104.8 ± 3.6 | |
| LO-149 | 8 | 32 | 32 | K. pneumoniae | 15 | 130.6 ± 3.2 | |
*Did not carry blaNDM–1. ND, not determined.
After plasmid size estimation and blaNDM–1 gene detection, we compared the consensus barcodes among the eight isolates (Figure 4). The ODM assay showed that identical plasmids with the same size (∼125 kb) and the same location of the blaNDM–1 were found in LO63 and LO64 (Figure 4A). These isolates belong to sequence types ST147 and ST893, respectively (Figure 1), suggesting a possible transmission of plasmid from one strain to the other. Similarly structured plasmids were found in LO89 and LO271 (∼107 kb) (Figure 4A); they both belong to the same sequence type, ST147. By visual inspection, it appears that large regions of the plasmids of isolates LO63, LO64, LO89, and LO271 (Figure 4A) are similar, further accentuated by the fact that the blaNDM–1 gene is located at the same position. There are however, other regions that are not the same, and the size differs (plasmids from LO63, LO64, and LO89 were ∼125 kb while the plasmid from LO271 was ∼107 kb). The plasmids from the other isolates do not match among each other (Figure 4B) or with the plasmids in Figure 4A. In total, we therefore found seven different plasmids carrying the blaNDM–1 gene.
FIGURE 4.
Plasmid barcodes of blaNDM–1-carrying plasmids in eight Klebsiella pneumoniae strains. Since the plasmids are linearized by Cas9-targeting blaNDM–1, all barcode ends are where we locate the blaNDM–1 gene. For the samples containing two blaNDM–1 plasmids, sizes are written in brackets to differentiate the plasmids. (A) Identical plasmids with the same sizes (∼125 kb) and the same location of blaNDM–1. (B) Plasmids encoding blaNDM–1 that do not match among each other. LO-271 (107.4) and LO-64 were plotted here for reference.
Discussion
Herein, we found 71 CRE in a period of 1 year with a lot of CPE species from patients in the same hospital in Tehran, Iran, and major dissemination of the blaNDM–1 and blaOXA–48 genes, which might be considered endemic in the geographical area, through the spread of conjugative plasmids.
The co-occurrence of NDM-1- and OXA-48-producing Enterobacterales species is also considerable since the identification of NDM-1 and OXA-48 producers in Iran (Solgi et al., 2017b), Lebanon (Dandachi et al., 2016), and Kuwait (Jamal et al., 2015) shows that these carbapenemases, known to be widespread in the Indian subcontinent, may also be widespread in the Middle East. In our study, the majority of the NDM-1- and OXA-48-producing Enterobacterales isolates co-harbored at least one ESBL gene which is concordant with previous reports (Torres-González et al., 2015; Solgi et al., 2017a). In this study, nine carbapenem-resistant K. pneumoniae were identified; this may be due to other resistance mechanisms (e.g., more rare carbapenemases, porin loss, AmpC enzymes) that were not investigated in detail in this study.
The plasmid incompatibility types IncFII and IncA/C were identified among the NDM-1-producing isolates, while only IncL/M was detected among OXA-48 producers (Table 1). These replicon types have been reported in Enterobacterales species in many regions of the world (Brañas et al., 2015; Guo et al., 2016; Kieffer et al., 2016; Solgi et al., 2017b). Also, Weber et al. (2019) demonstrated that the potential transmission of mobilized Tn125-like transposons with blaNDM–1 into different plasmids among Enterobacterales species (Weber et al., 2019). Conjugation assays were successful for all CPE isolates and allowed the identification of blaOXA–48-carrying plasmids belonging to the IncL/M incompatibility group in all transconjugants, with the exception of Tc-LO-149 (Table 2). Also, analysis of transconjugants showed that the blaNDM–1 carried on transferable plasmids belonging to the IncFII and IncA/C incompatibility group, respectively.
The identification of conjugative plasmids harboring blaNDM–1 and blaOXA–48 genes in CRE isolates shows that these plasmids contribute to the dissemination of carbapenemase genes among Enterobacterales species. Therefore, resistance to carbapenems in CRE isolates is likely to be associated with the spread of these genes in this hospital, which is consistent with previous studies (Jamal et al., 2016; Kieffer et al., 2016; Solgi et al., 2017a).
Pulsed-field gel electrophoresis revealed that different clones of carbapenemase-producing K. pneumoniae (CPKP) were present, and there were two predominated clones that were identified as ST147 and ST893, comprising 13 and 10 isolates, respectively. ST147 and ST893 have been circulating in this hospital setting during the period of investigation, indicating two separate outbreaks, with the ICU poisoning acting as the epicenter. Indeed, hospital outbreaks of ST147 NDM-1-producing K. pneumoniae are common in Europe (Bogaerts et al., 2011; Giske et al., 2012), whereas the outbreak of OXA-48-producing ST893 K. pneumoniae was only reported from Isfahan, Iran (Solgi et al., 2018).
The dominant endemic sequence type K. pneumoniae in our study was ST147 which co-harbored NDM-1 and blaCTX–M–15, blaTEM–1, and blaSHV–11,12,172 genes. As an internationally successful sequence type, ST147 has previously been linked to the spread of ESBLs (especially CTX-M-15), OXA-48, VIM, and KPC and recently also to NDM-1 in various countries (Bogaerts et al., 2011; Messaoudi et al., 2017). In addition, ST893, the second most common sequence type in this study that co-harbored blaCTX–M–15, blaTEM–1, and blaSHV–199, has also only been reported in Iran among CPKP isolates which has been associated with ESBL and carbapenemase genes (Solgi et al., 2018). Several other STs were found among CPKP isolates, including ST16 (cluster A), ST377 (cluster C), ST15 (cluster D), ST11, ST23, ST35, ST2012, ST657, and ST1308.
The four isolates of ST15 (cluster D) were isolated from patients in four ward. All isolates carried blaNDM–1 in combination of blaOXA–48 and blaCTX–M–15, blaTEM–1, and blaSHV–106 genes. K. pneumoniae ST15 represents a single locus variant of ST14 and is currently widely disseminated among CTX-M-15- and OXA-48- or NDM-1-producing K. pneumoniae isolates in different geographical regions (Poirel et al., 2014; Kieffer et al., 2016; Ben et al., 2017; Jelić et al., 2017).
The four NDM-1- and one OXA-48-producing K. pneumoniae in our study belonged to ST16 and were positive for blaCTX–M–15 and blaSHV–199 genes. It is noteworthy that OXA-48-producing ST16 have also been described in K. pneumoniae that caused outbreaks in two hospitals in different regions of Spain (Oteo et al., 2013). Furthermore, two OXA-48- and one NDM-1-producing K. pneumoniae were isolated from three patients in two different ward. They belonged to ST377, which has previously not been reported as a carbapenemase producer. Finally, one OXA-48-producing K. pneumoniae isolate that co-carried blaCTX–M–15, blaTEM–1, and blaSHV–182 was identified as ST11. The blaOXA–48-harboring IncL/M plasmids have been mainly described in K. pneumoniae ST11 in different countries including, Spain (Brañas et al., 2015), Taiwan (Ma et al., 2015), and Greece (Voulgari et al., 2013).
Considering this study and our previous study in Isfahan province (Solgi et al., 2018), the main K. pneumoniae STs that were identified in Iran were ST893, ST11, and ST147. This scenario suggests that these STs have likely been circulating in Iran in recent years. Our results show that, in general, the population structure of CP E. coli is more diverse than that of CPKP, which is essentially similar to the findings of other studies (Sartor et al., 2014; Kieffer et al., 2016; Solgi et al., 2017b). We detected E. coli ST410, ST1431, ST3134, and ST5114 which have been reported as harboring blaOXA–48 and ESBL genes. Moreover, we identified only one ST131 of E. coli which harbored blaNDM–1, blaCTX–M–15, and blaTEM–1 genes. The association of NDM-1 and ESBL genes with the pandemic clone ST131 has been previously reported from several countries (Peirano et al., 2011, 2014).
The two NDM-1-positive E. cloacae isolates were genetically not related and belonged to two STs, ST78 and ST175, both also carried blaCTX–M–15 and blaTEM–1 genes, while the five S. marcescens isolates were considered identical (>99% similarity). Interestingly, looking at the hospitalization ward from which the patients originated, several infections were detected at the ICU poisoning, with a total of five patients harboring this OXA-48-producing S. marcescens strain which co-carried further beta-lactamase genes (blaCTX–M–15, blaSHV–12, and blaTEM–1). Our results showed that this OXA-48-producing S. marcescens strain was isolated among inpatients who shared a room. Therefore, it is possible that the spread of this strain from patient to patient occurred. To the best of our knowledge, this is the first report of an outbreak of OXA-48-producing S. marcescens that co-harbored ESBL genes in Iran. A small hospital outbreak linked to OXA-48-producing S. marcescens has been previously reported in Lebanon (Hammoudi et al., 2014). The exact mechanism of CPE spread in Iran is not well understood. Our previous study in July to November 2015 in two university hospitals in Iran showed that the rate of fecal carriage of CRE among inpatients is high (37.9%) and predominant species were K. pneumoniae, E. coli, E. cloacae, and Proteus mirabilis, which harbored the blaNDM–1 and blaOXA–48 genes (Solgi et al., 2017a). The circulation of blaNDM–1 and blaOXA–48 carbapenemase genes in the general population may result in a further spread by traveling and continuous introduction into the hospitals.
In conclusion, findings of extensive analysis of plasmids in the present study showed the enormous potential of spread of carbapenemase genes by horizontal gene transfer via plasmids and we identified the conjugative plasmids carrying the blaNDM–1 and blaOXA–48 genes in different Enterobacterales species that co-produce ESBLs. Here, in one Tehran hospital, we report two separate outbreaks of NDM-1-producing ST147 and OXA-48-producing ST893 K. pneumoniae STs. Furthermore, an outbreak with OXA-48-producing S. marcescens was observed. It is necessary to continue epidemiological and active surveillance to improve the control and prevention of infections associated with CPE isolates in healthcare facilities.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The study was approved by the research and the Ethics Committee of the Pasteur Institute of Iran (No. 1395.51). No ethical approval was obtained for using the clinical isolates since they were collected during the routine diagnostic laboratory at our hospital.
Author Contributions
HS and FS designed the study. HS, SN, Y-LL, GG, VN, and AN carried out the experiments. HS, CG, FB, FW, and FS analyzed the data. HS, CG, FW, and FS wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of the Laboratory of Microbiology of Loghman Hakim Educational Hospital, Tehran, for providing isolates included in this study. We are grateful to the group of Tobias Ambjörnsson, Lund University, for providing the software for analyzing the optical DNA mapping data. We also like to thank Dr. Sriram Kesarimangalam for fabricating the nanofluidic devices.
Funding. This work was supported by the Pasteur Institute of Iran (project No: IR.PII.REC.1395.51).
References
- Ben T. F., Alonso C. A., Achour W., Ruiz-Ripa L., Torres C., Ben H. A. (2017). First Description of KPC-2-Producing Escherichia coli and ST15 OXA-48-Positive Klebsiella pneumoniae in Tunisia. Microb. Drug. Resist. 23 365–375. 10.1089/mdr.2016.0090 [DOI] [PubMed] [Google Scholar]
- Bogaerts P., Bouchahrouf W., De-Castro R. R., Deplano A., Berhin C., Piérard D., et al. (2011). Emergence of NDM-1-producing Enterobacteriaceae in Belgium. Antimicrob. Agents. Chemother. 55 3036–3038. 10.1128/AAC.00049-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brañas P., Villa J., Viedma E., Mingorance J., Orellana M. A., Chaves F. (2015). Molecular epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital in Madrid: successful establishment of an OXA-48 ST11 clone. Int. J. Antimicrob. Agents. 46 111–116. 10.1016/j.ijantimicag.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2017). Performance Standards For Antimicrobial Susceptibility Testing, 27th Edn, Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Dandachi I., Salem Sokhn E., Najem E., Azar E., Daoud Z. (2016). Carriage of beta-lactamase-producing Enterobacteriaceae among nursing home residents in north Lebanon. Int. J. Infect. Dis. 45 24–31. 10.1016/j.ijid.2016.02.007 [DOI] [PubMed] [Google Scholar]
- EUCAST (2017). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 7.0, Valid from 2017-01-01. Available online at: http://www.eucast.org/clinical_breakpoints/ (accessed February 24, 2017). [Google Scholar]
- Giske C. G., Fröding I., Hasan C. M., Turlej-Rogacka A., Toleman M., Livermore D., et al. (2012). Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob. Agents. Chemother. 56 2735–2738. 10.1128/AAC.06142-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., An J., Ma Y., Ye L., Luo Y., Tao C., et al. (2016). Nosocomial outbreak of OXA-48-producing Klebsiella pneumoniae in a Chinese hospital: clonal transmission of ST147 and ST383. PLoS One 11:160754 10.1371/journal.pone.0160754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoudi D., Ayoub M., Moubareck C., Aires J., Adaime A., Barakat A. (2014). Countrywide spread of OXA-48 carbapenemase in lebanone surveillance and genetic characterization of carbapenem-non- susceptible Enterobacteriaceae in 10 hospitals over a one-year period. Int. J. Infect. Dis. 29 139–144. 10.1016/j.ijid.2014.07.017 [DOI] [PubMed] [Google Scholar]
- Jamal W. Y., Albert M. J., Khodakhast F., Poirel L., Rotimi V. O. (2015). Emergence of new sequence type OXA-48 carbapenemase-producing Enterobacteriaceae in Kuwait. Microb. Drug Resist. 21 329–334. 10.1089/mdr.2014.0123 [DOI] [PubMed] [Google Scholar]
- Jamal W. Y., Albert M. J., Rotimi V. O. (2016). High prevalence of New Delhi Metallo-β-lactamase-1 (NDM-1) producers among carbapenem-resistant Enterobacteriaceae in Kuwait. PLoS One 11:e0152638 10.1371/journal.pone.0152638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelić M., Škrlin J., Bejuk D., Košćak I., Butić I., Gužvinec M., et al. (2017). Characterization of isolates associated with emergence of OXA-48-producing Klebsiella pneumoniae in Croatia. Microb. Drug Resist. 24 973–979. 10.1089/mdr.2017.0168 [DOI] [PubMed] [Google Scholar]
- Johnson A. P., Woodford N. (2013). Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J. Med. Microbiol. 62 499–513. 10.1099/jmm.0.052555-0 [DOI] [PubMed] [Google Scholar]
- Kieffer N., Nordmann P., Aires-de-Sousa M., Poirel L. (2016). High prevalence of carbapenemase-producing Enterobacteriaceae among hospitalized children in Luanda, Angola. Antimicrob. Agents. Chemother. 60 6189–6192. 10.1128/AAC.01201-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyimo B., Buza J., Subbiah M., Temba S., Kipasika H., Smith W., et al. (2016). IncF Plasmids are commonly carried by antibiotic resistant Escherichia coli Isolated from drinking water sources in northern tanzania. Int. J. Microbiol. 2016 1–7. 10.1155/2016/3103672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Wang J. T., Wu T. L., Siu L. K., Chuang Y. C., Lin J. C., et al. (2015). Emergence of OXA-48-Producing Klebsiella pneumoniae in Taiwan. PLoS One 10:e0139152 10.1371/journal.pone.0139152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi A., Haenni M., Mansour W., Saras E., Bel Haj, Khalifa A., et al. (2017). ST147 NDM-1-producing Klebsiella pneumoniae spread in two Tunisian hospitals. J. Antimicrob. Chemother. 72 315–316. 10.1093/jac/dkw401 [DOI] [PubMed] [Google Scholar]
- Mohammad Ali Tabrizi A., Badmasti F., Shahcheraghi F., Azizi O. (2018). Outbreak of hypervirulent Klebsiella pneumoniae harbouring blaVIM-2 among mechanically-ventilated drug-poisoning patients with high mortality rate in Iran. J. Glob. Antimicrob. Resist. 15 93–98. 10.1016/j.jgar.2018.06.020 [DOI] [PubMed] [Google Scholar]
- Müller V., Karami N., Nyberg L. K., Pichler C., Torche Pedreschi P. C., Quaderi S., et al. (2016a). Rapid tracing of resistance plasmids in a nosocomial outbreak using optical DNA mapping. ACS Infect. Dis. 2 322–328. 10.1021/acsinfecdis.6b00017 [DOI] [PubMed] [Google Scholar]
- Müller V., Rajer F., Frykholm K., Nyberg L. K., Quaderi S., Fritzsche J., et al. (2016b). Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci. Rep. 6:37938 10.1038/srep37938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller V., Westerlund F. (2017). Optical DNA mapping in nanofluidic devices: principles and applications. Lab. Chip. 17 579–590. 10.1039/C6LC01439A [DOI] [PubMed] [Google Scholar]
- Nilsson A. N., Emilsson G., Nyberg L. K., Noble C., Stadler L. S., Fritzsche J., et al. (2014). Competitive binding-based optical DNA mapping for fast identification of bacteria-multi-ligand transfer matrix theory and experimental applications on Escherichia coli. Nucleic Acids. Res. 42:e118 10.1007/978-1-62703-553-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Naas T., Poirel L. (2011). Global spread of carbapenemase producing Enterobacteriaceae. Emerg. Infect. Dis. 17 1791–1798. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg L. K., Persson F., Berg J., Bergström J., Fransson E., Olsson L., et al. (2012). A single-step competitive binding assay for mapping of single DNA molecules. Biochem. Biophys. Res. Commun. 417 404–408. 10.1016/j.bbrc.2011.11.128 [DOI] [PubMed] [Google Scholar]
- Oteo J., Hernández J., Espasa M., Fleites A., Sáez D., Bautista V., et al. (2013). Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J. Antimicrob. Chemother. 68 317–321. 10.1093/jac/dks383 [DOI] [PubMed] [Google Scholar]
- Peirano G., Bradford P. A., Kazmierczak K. M., Badal R. E., Hackel M., Hoban D. J., et al. (2014). Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg. Infect. Dis. 20 1928–1931. 10.3201/eid2011.141388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano G., Schreckenberger P. C., Pitout J. D. D. (2011). Characteristics of NDM-1-producing Escherichia coli isolates that belong to the successful and virulent clone ST131. Antimicrob. Agents. Chemother. 55 2986–2988. 10.1128/AAC.01763-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Walsh T. R., Cuvillier V., Nordmann P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70 119–123. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Poirel L., Yilmaz M., Istanbullu A., Arslan F., Mert A., Bernabeu S., et al. (2014). Spread of NDM-1-Producing Enterobacteriaceae in a neonatal intensive care unit in istanbul, Turkey. Antimicrob. Agents. Chemother. 58 2929–2933. 10.1128/AAC.02047-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor A., Raza M. W., Abbasi S., Day K. M., Perry J. D., Paterson D. L., et al. (2014). Molecular epidemiology of NDM-1-producing Enterobacteriaceae and Acinetobacter baumannii isolates from Pakistan. Antimicrob. Agents. Chemother. 58 5589–5593. 10.1128/AAC.02425-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahcheraghi F., Aslani M. M., Mahmoudi H., Karimitabar Z., Solgi H., Bahador A., et al. (2017). Molecular study of carbapenemase genes in clinical isolates of Enterobacteriaceae resistant to carbapenems and determining their clonal relationship using pulsed-field gel electrophoresis. J. Med. Microbiol. 66 570–576. 10.1099/jmm.0.000467 [DOI] [PubMed] [Google Scholar]
- Shahcheraghi F., Nobari S., Rahmati Ghezelgeh F., Nasiri S., Owlia P., Nikbin V. S., et al. (2013). First report of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Iran. Microb. Drug Resist. 19 30–36. 10.1089/mdr.2012.0078 [DOI] [PubMed] [Google Scholar]
- Solgi H., Badmasti F., Aminzadeh Z., Giske C. G., Pourahmad M., Vaziri F., et al. (2017a). Molecular characterization of intestinal carriage of carbapenem-resistant Enterobacteriaceae among inpatients at two Iranian university hospitals: first report of co-production of blaNDM-7 and blaOXA-48. Eur. J. Clin. Microbiol. Infect. Dis. 36 2127–2135. 10.1007/s10096-017-3035-3 [DOI] [PubMed] [Google Scholar]
- Solgi H., Badmasti F., Giske C. G., Aghamohammad S., Shahcheraghi F. (2018). Molecular epidemiology of NDM-1- and OXA-48-producing Klebsiella pneumoniae in an Iranian hospital: clonal dissemination of ST11 and ST893. J. Antimicrob. Chemother. 73 1517–1524. 10.1093/jac/dky081 [DOI] [PubMed] [Google Scholar]
- Solgi H., Giske C. G., Badmasti F., Aghamohammad S., Havaei S. A., Sabeti S., et al. (2017b). Emergence of carbapenem resistant Escherichia coli isolates producing blaNDM and blaOXA-48-like carried on IncA/C and IncL/M plasmids at two Iranian university hospitals. Infect. Genet. Evol. 55 318–323. 10.1016/j.meegid.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Tängdén T., Giske C. G. (2015). Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J. Intern. Med. 277 501–512. 10.1111/joim.12342 [DOI] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33 2233–2239. 10.1128/JCM.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-González P., Bobadilla-Del Valle M., Tovar-Calderón E., Leal-Vega F., Hernández-Cruz A., Martínez-Gamboa A. (2015). Outbreak caused by Enterobacteriaceae harboring NDM-1 metallo-β-lactamase carried in an IncFII plasmid in a tertiary care hospital in Mexico City. Antimicrob. Agents. Chemother. 59 7080–7083. 10.1128/AAC.00055-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulgari E., Gartzonika C., Vrioni G., Politi L., Priavali E., Levidiotou-Stefanou S., et al. (2014). The Balkan region: NDM-1-producing Klebsiella pneumoniae ST11 clonal strain causing outbreaks in Greece. J. Antimicrob. Chemother. 69 2091–2097. 10.1093/jac/dku105 [DOI] [PubMed] [Google Scholar]
- Voulgari E., Zarkotou O., Ranellou K., Karageorgopoulos D. E., Vrioni G., Mamali V., et al. (2013). Outbreak of OXA-48 carbapenemase-producing Klebsiella pneumoniae in Greece involving an ST11 clone. J. Antimicrob. Chemother. 68 84–88. 10.1093/jac/dks356 [DOI] [PubMed] [Google Scholar]
- Weber R. E., Pietsch M., Frühauf A., Pfeifer Y., Martin M., Luft D., et al. (2019). IS26-mediated transfer of blaNDM-1 as the main route of resistance transmission during a polyclonal, multispecies outbreak in a german hospital. Front. Microbiol. 17:2817 10.3389/fmicb.2019.02817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new Metallo-β-Lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents. Chemother. 53 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation, to any qualified researcher.