Abstract
F1 hybrid progenies between related subspecies often show hybrid sterility (HS) or inviability. HS is caused by failure of meiotic chromosome synapsis and sex body formation in house mouse. Previous studies identified two HS critical genomic regions named Hstx2 on Chr X and Hst1 on Chr 17 by murine forward genetic approaches. HS gene on Hst1 was reported to be Prdm9. Intersubspecific polymorphisms of Prdm9 induce HS in hybrids, and Prdm9 null mutation leads to sterility in the inbred strain. However, HS gene on Hstx2 remains unknown. Here, using knock-out studies, we showed that HS candidate genes on Hstx2 are not individually essential for spermatogenesis in B6 strain. We examined 12 genes on Hstx2: Ctag2, 4930447F04Rik, Mir743, Mir465d, Mir465c-2, Mir465b-1, Mir465c-1, Mir465, Gm1140, Gm14692, 4933436I01Rik, and Gm6812. These genes were expressed in adult testes, and showed intersubspecific polymorphisms on expressed regions. This first reverse genetic approach to identify HS gene on Hstx2 suggested that the loss of function of any one HS candidate gene does not cause complete sterility, unlike Prdm9. Thus, the mechanism(s) of HS by the HS gene on Hstx2 might be different from that of Prdm9.
Subject terms: Evolutionary genetics, Genetics
Introduction
Postzygotic reproductive isolation between related subspecies often results in hybrid incompatibility such as hybrid sterility (HS) or inviability1–4. This HS is the barrier distinguishing related species by disturbing the gametogenesis in the hybrid. HS is widely observed around intersubspecific hybrids such as yeast, plants, insects, birds, and mammals1,2,4. In 1922, Haldane’s rule was reported as the observation that if only one sex of the hybrid is inviable or sterile, that sex is the heterogametic sex (XY or ZW)5. Additionally more than 80 years ago, Dobzhansky, T. (1937) and Muller, H. (1942) proposed a two-locus model called Dobzhansky-Muller incompatibility (hereafter DMI) hypothesis that postzygotic isolation arises from a negative epistatic interaction between incompatible alleles that evolved on two independent evolutionary lineages6,7. Previous studies showed a large effect of Chr X (Chr Z) on reproductive isolation, particularly on HS or hybrid inviability8–11. In the last 30 years, several HS genes and hybrid inviability genes were identified in budding yeast, thale cress, fruit fly, and house mouse12–25. In Drosophila, HS genes: OdsH and Ovd, and hybrid inviability gene: Hmr and locate on Chr X13,14,16,21. However in house mice, these orthologous genes have not reported. Although several researchers proposed to explain this heterogametic HS or inviability as being controlled by Chr X (Chr Z) in vertebrata26–28, the X-linked (Z-linked) causative gene for Haldane’s rule and DMI has remained unclear for over a century.
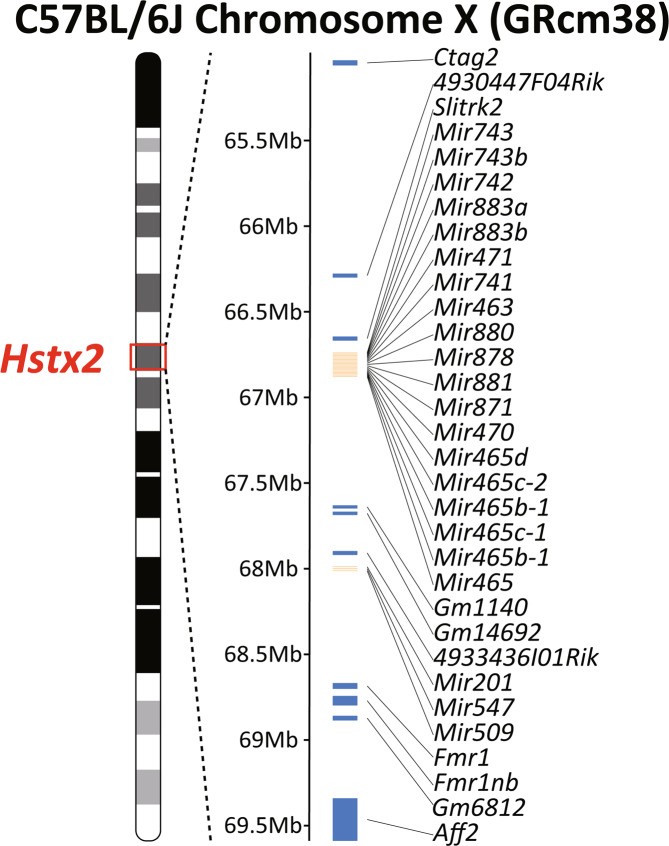
The two HS loci, Hst1 and Hstx2, were found by murine forward genetics29–32. The first HS locus in mice, Hst1 was identified as a polymorphic variant on Chr 17 between two laboratory strains, C57BL10/Sn and C6H/Di, both predominantly of Mus musculus domesticus (hereafter Mmd) origin29. To identify HS gene on Hst1, Forejt, J. et al. established HS murine recombinant inbred model using PWD/Ph (hereafter PWD) inbred strain derived from Mus musculus musculus (hereafter Mmm) subspecies and C57BL/6 (hereafter B6) inbred strain derived from Mmd subspecies33,34. PWD was established from a single pair of wild mice of the Mmm subspecies caught in 1972 in Central Bohemia, Czech Republic35. Both subspecies diverged from a common ancestor approximately 0.3 to 0.5 million years ago36. This model shows asymmetric HS, wherein (PWD × B6) F1 hybrid (mating PWD female with B6 male) shows sterility, while (B6 × PWD) F1 hybrid (mating B6 female with PWD male) shows semi-fertility35,36. To identify these HS loci, Gregorová, S. et al. also established intersubspecific chromosome substitution (consomic) strains C57BL/6-Chr#PWD/Ph/ForeJ carrying individual Mmm chromosomes or their parts on Mmd background37. The HS gene on Hst1 was identified by the forward genetic approach as PR domain containing 9 (Prdm9)24, and later was shown to control meiotic recombination hotspots38,39. Prdm9 encodes for histone H3 lysine 4 trimethyltransferase that controls the hotspots of DNA double strand breaks (DSBs) at meiotic prophase in spermatocytes. Hstx2 was found out by consomic strains C57BL/6-Chr XPWD/Ph/ForeJ carrying Mmm Chr X or their parts on Mmd background32. The 4.7 Mb (Chr X:64.9 Mb-69.6 Mb, GRCm38) HS critical region on Chr X, Hstx2, was identified by quantitative trait locus (QTL) analysis of male fertility phenotypes in these consomic strains32. They revealed that HS is also controlled by Hstx2 of PWD32.
Unlike the HS gene Prdm9 on Hst1, the HS gene on Hstx2 remains unknown. Identifying the molecular mechanism of HS controlled by Chr X can unravel the molecular mechanism of Haldane’s rule in vertebrata. Forward genetic approaches have a limit to narrow down to only 4.7 Mb Hstx2 critical region, because Hstx2 may lie on a recombination cold spot40,41. Hence, reverse genetic approach is required to identify the HS gene on Hstx2.
In a mouse model of intersubspecific hybrids between B6 and PWD, HS needs three factors, Prdm9 heterozygosity (Prdm9B6/PWD), autosomal heterozygosity and the PWD allele of Hstx2 (Hstx2PWD)42. One of HS mechanism model was explained by hybrid incompatibility between Prdm9B6/PWD and autosomal heterozygosity without DMI at least in house mouse43. Minisatellite structure of the zinc finger array coding on DNA binding sites of PRDM9 has interspecific polymorphisms between B6 and PWD43,44. The binding sites were evolutionally eroded by DNA double-strand breaks determined by PRDM9 in each strain43,44. The affinity of PRDM9B6 to B6 autosomes and the affinity of PRDM9PWD to PWD autosomes decreases with evolution, respectively43,44. This is because the evolutionary erosion of genomic DNA occurs independently in each mouse sub-strain. In hybrid, PRDM9B6-determined hotspots occur mostly on the PWD chromosome and vice versa43,44. The major meiotic consequences of asymmetric DSB hotspots result in unrepaired DNA DSBs, meiotic asynapsis and abnormal sex body formation, leading to disruption of sperm formation43. Nevertheless, only this model cannot explain the asymmetric HS because this meiotic arrest is also controlled by Hstx2. Hstx2PWD is responsible for full sterility of (PWD × B6) F1 hybrid, while the B6 allele of Hstx2 (Hstx2B6) attenuates fertility of (B6 × PWD) F1 hybrid32. However, how Hstx2 regulates HS remains elusive. Therefore, we proposed three hypotheses of the role of Hstx2. First hypothesis is that the spermatogenic function of HS gene(s) on Hstx2 is lost in (PWD × B6) F1 male but partially remains in (B6 × PWD) F1 male. Second hypothesis is that dominant-negative Hstx2PWD impedes the spermatogenesis in hybrids. Third hypothesis is that Hstx2B6 rescues fertility of hybrids. Here, to verify whether Hstx2 are essential for spermatogenesis in the first hypothesis, we aimed to delete HS gene on Hstx2 in B6 mice. We hypothesised that HS gene on Hstx2 may have similar features to Prdm9, such as expressing in adult testes45, having intersubspecific polymorphisms on expressed region between Mmm and Mmd43,44, explaining HS mechanism by single locus43 and inducing sterility by knock-out (KO) in B6 genetic background45,46. If our first hypothesis is correct, the HS gene on Hstx2 KO B6 mice would be sterile. Therefore, we aimed to identify HS gene on Hstx2 through KO study.
To identify HS candidate gene, we investigated the expression of 32 genes on Hstx2 in adult B6 testes using databases. Next, we analysed intersubspecific polymorphisms on expressed regions between B6 and PWD, also PWK/Phj (Mmm inbred strain, hereafter, PWK) by past reports and public database.
Results
Twelve HS candidate genes were identified on Hstx2
Male sterility of reciprocal hybrids between PWD and B6 is controlled by Hstx2 (64.9 Mb-69.6 Mb, GRCm38)32, which carries 10 protein-coding genes and 22 microRNA (miRNA) genes (Fig. 1). As mentioned earlier, if our hypothesis is correct, the HS gene on Hstx2 KO B6 mice must be sterile. To find out the HS gene from the 32 genes on Hstx2, we firstly screened them for expression in adult B6 testes. Testes expression was analysed using Fantom5 (http://fantom.gsc.riken.jp/5/), BioGPS (http://biogps.org/circadian/#goto=welcome) and miRbase (http://www.mirbase.org/), which revealed that of the 32 genes, 9 protein-coding genes: Ctag2, 4930447F04Rik, Gm1140, Gm14692, 4933436I01Rik, Fmr1, Fmr1nb, Gm6812, and Aff2, and all 22 miRNA genes were expressed in adult B6 testes (Table 1 and S1). The only one gene, Slitrk2 showed no expression in adult B6 testes (Table 1). Next, we analysed intersubspecific polymorphism of the genes on Hstx2. We focused on intersubspecific polymorphism on expressed regions, which are the coding region in protein-coding alleles and mature miRNA region in miRNA alleles. Bhattacharyya et al. reported intersubspecific missense single nucleotide polymorphisms (SNPs) of HS candidate genes among B6, PWD, and PWK32 (Table 1). PWK is a Mmm inbred strain and closely related to PWD47. By comparison of the B6 allele of HS candidate genes with that of the PWD allele, Ctag2, 4930447F04Rik, Slitrk2, Mir743, 4933436I01Rik, Fmr1nb, and Aff2 were found to carry missense SNPs between B6 and PWD. Fmr1 does not carry any intersubspecific missense SNPs. We excluded Slitrk2, Fmr1, Fmr1nb, and Aff2 on Hstx2 as the targets, because these genes have already been reported as not being necessary for spermatogenesis in B6 male by KO studies48–51. Additionally, we targeted Gm1140, Gm14692, Gm6812, and Mir465 cluster because Gm1140, Gm14692, Gm6812, Mir465c-2, Mir465b-1, Mir465c-1, and Mir465 also have intersubspecific missense SNPs between B6 and PWK (Table S1). The next generation sequence (NGS) data on PWK was obtained from Sanger Institute Mouse Genome Project (http://www.sanger.ac.uk/science/data/mouse-genomes-project) and we aligned PWK and B6 reference genome (GRCm38). A very recent study reported that Gm1140, Gm14692, and Gm6812 have intersubspecific missense SNPs between B6 and PWD, and Mir465 cluster shows copy number polymorphism between B6 and PWD (and PWK)40. Therefore, we added these genes to the targets.
Figure 1.
Gene map of the Hstx2 critical region on Chr X. Gene map of the Hstx2 critical region on Chr X (64.9Mb-69.6 Mb, GRCm38). Hstx2 carries 10 protein-coding genes and 22 miRNA genes.
Table 1.
Testis expression and intersubspecific missense SNPs between B6 and PWD of candidate genes on Hstx2.
| Gene | testis expression | SNP position | B6 | PWD | PWK |
|---|---|---|---|---|---|
| Ctag2 | + | 65,047,953 | T | G | G |
| 4930447F04Rik | + | 66,303,564 | A | C | C |
| Slitrk2 | − | 66,655,874 | A | G | G |
| 66,656,111 | A | G | G | ||
| Mir743 | + | 66,776,774 | T | C | C |
| 493436I01Rik | + | 67,920,137 | C | T | T |
| 67,920,143 | T | G | G | ||
| 67,920,312 | G | T | T | ||
| 67,920,431 | T | A | A | ||
| 67,920,805 | T | A | A | ||
| 67,920,818 | C | T | T | ||
| 67,920,822 | T | G | G | ||
| Fmr1 | + | — | — | — | — |
| Fmr1nb | + | 68,762,025 | C | G | G |
| 68,769,064 | T | A | A | ||
| Aff2 | + | 69,544,913 | A | G | G |
| 69,830,745 | C | T | T | ||
| 69,830,760 | C | G | G | ||
| 69,830,782 | A | T | T | ||
| 69,834,780 | G | A | A |
Taken together, we targeted 12 HS candidate genes, composed of six protein-coding genes, namely Ctag2, 4930447F04Rik, Gm1140, Gm14692, 4933436I01Rik, and Gm6812, and six miRNA genes, namely Mir743, Mir465d, Mir465c-2, Mir465b-1, Mir465c-1, and Mir465.
Generation of eight HS candidate gene/cluster KO male mice by CRISPR/Cas9 system
To investigate whether each of the 12 HS candidate genes are essential for spermatogenesis, we generated each HS candidate gene KO mice using CRISPR/Cas9 system. We respectively designed two single guide RNA (sgRNA) sites, one upstream of the target gene and the other downstream of it on B6 genetic background in order to remove all genomic region. Mir465d, Mir465c-2, Mir465b-1, Mir465c-1, and Mir465 are located on Mir465 cluster, which is composed of 6 repeats (Fig. S1); as we could not design sgRNA for each gene of the Mir465 cluster, we decided to knock out the complete Mir465 cluster. To create each of the eight HS candidate gene/cluster KO mice, we performed the microinjection of pX330, the circular plasmid carrying sgRNA and Cas9 expression units, or the electroporation of sgRNA and Cas9 nuclease into B6 mouse eggs gained by mating with B6 male or in vitro fertilisation (IVF), and then each KO F0 founder were selected for KO allele, removing all genomic region of target gene. To confirm the CRISPR/Cas9 mutations, we performed genotyping using genomic PCR from mouse tail and then confirmed the KO allele. Thus, we generated each of the eight gene/cluster KO male founders (Table 2).
Table 2.
Generation of each the HS candidate gene KO founders by CRISPR/Cas9 system.
| Gene | Number of embryos | Number of pups | Number of founders | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Injected | Transferreda | Newbornb | Weaningc | Birthrateb/a | Male | Female | Totald | Mutation efficiencyd/c | |
| Ctag2 | 83 | 80 | 31 | 31 | 38.8% | 13 | 8 | 21 | 67.7% |
| 4930447F04Rik | 84 | 82 | 27 | 27 | 32.9% | 13 | 7 | 20 | 74.1% |
| Mir743 | 155 | 146 | 41 | 38 | 28.1% | 7 | 7 | 14 | 36.8% |
| Mir465 cluster | 75 | 72 | 26 | 24 | 36.1% | 4 | 3 | 7 | 29.2% |
| Gm1140 | 266 | 249 | 38 | 36 | 15.3% | 2 | 1 | 3 | 8.33% |
| Gm14692 | 100 | 98 | 18 | 16 | 18.4% | 1 | 2 | 3 | 18.8% |
| 4933436I01Rik | 105 | 90 | 32 | 31 | 35.6% | 10 | 7 | 17 | 54.8% |
| Gm6812 | 112 | 90 | 6 | 6 | 6.67% | 1 | 2 | 3 | 50.0% |
Fertility and spermatogenesis of the eight HS candidate gene/cluster KO male mice
To investigate fertility of each candidate gene KO male founder, the sexually mature KO male F0 founder was mated with sexually mature B6 wild-type female. F0 founder have the possibility of mosaicism; therefore, these KO F0 male founder may not show full knock out of the gene in the testes. If KO allele was inherited to next generation, it would imply that the germ cell carried the KO allele and could be developed to fertilisable sperm. From mating test, each heterozygote KO female mice was born (Table 3). One of 4933496I01Rik KO male founder generated wild-type female, suggesting that the founder was mosaicism. Except for the above, all other F1 female mice that were born from mating F0 male founders with female B6, showed the KO allele. Therefore, in our experiment, the male founders would not have shown mosaicism. These results suggested that the fertilisable sperm could have developed from the germ cell carrying the KO allele. Therefore, all eight KO male genotypes, 1 HS candidate gene/cluster per each, were fertile.
Table 3.
Fertility of each the HS candidate gene KO male founders.
| Gene | Founder number | Number of F1 | |||||
|---|---|---|---|---|---|---|---|
| Male | Female | ||||||
| Hemizygote | Wild-Type | Total | Heterozygote | Wild-Type | Total | ||
| Ctag2 | #2 | 0 | 4 | 4 | 6 | 0 | 6 |
| #3 | 0 | 8 | 8 | 7 | 0 | 7 | |
| #17 | 0 | 7 | 7 | 2 | 0 | 2 | |
| 4930447F04Rik | #9 | 0 | 1 | 1 | 1 | 0 | 1 |
| #11 | 0 | 2 | 2 | 3 | 0 | 3 | |
| Mir743 | #4 | 0 | 5 | 5 | 2 | 0 | 2 |
| Mir465 cluster | #1 | 0 | 5 | 5 | 3 | 0 | 3 |
| #14 | 0 | 6 | 6 | 0 | 0 | 0 | |
| #16 | 0 | 2 | 2 | 1 | 0 | 1 | |
| Gm1140 | #8 | 0 | 3 | 3 | 3 | 0 | 6 |
| Gm14692 | #46 | 0 | 8 | 8 | 9 | 0 | 9 |
| 4933436I01Rik | #7 | 0 | 7 | 7 | 5 | 0 | 5 |
| #8 | 0 | 6 | 6 | 4 | 0 | 4 | |
| #20 | 0 | 8 | 8 | 3 | 5 | 8 | |
| Gm6812 | #6 | 0 | 4 | 4 | 9 | 0 | 9 |
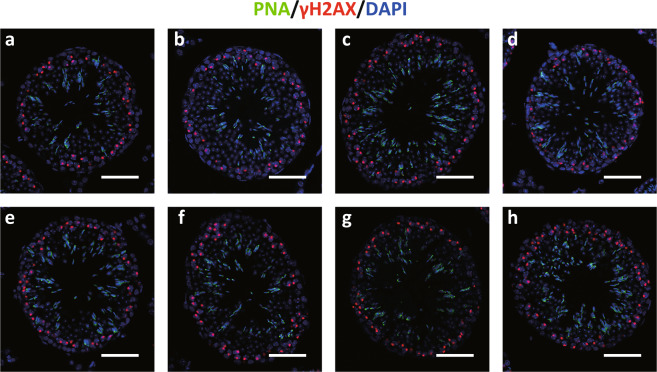
To investigate the spermatogenesis of each KO mice, histochemical analysis was performed. Peanut agglutinin (PNA) lectin histochemistry (LHC) is available as a marker of sperm. Immunohistochemistry (IHC) of phosphorylated form of the histone variant H2AX (γH2AX) is a marker of sex body on pachytene spermatocyte. IHC and PNA-LHC of testis section of each KO F0 founder showed sperm and sex body formation on these spermatocytes (Fig. 2). These results suggested that the eight candidate genes/cluster were not individually necessary for sperm formation at least in B6 genetic background.
Figure 2.
Histochemical analysis of the testis from each of the eight KO founders. PNA-LHC (green), γH2AX (red), and DAPI (blue) staining of the testis section of Ctag2 (a), 4930447F04Rik (b), Mir743 (c), Mir465 cluster (d), Gm1140 (e), Gm14692 (f), 4933436I01Rik (g), and Gm6812 (h) KO founders. All show sperm in lumen of seminiferous tubule and sex body formation. Scale bar: 50 µm.
Discussion
Our reverse genetic approach revealed that the eight HS candidates, comprising six protein-coding genes, one miRNA gene and one miRNA cluster, were not individually necessary for spermatogenesis in B6 mice. These results exclude the possibility that the loss of function of one HS gene/cluster on Hstx2 causes sterility, at least in B6 genetic background. In hybrid context between B6 and PWD, Hstx2B6 has semi-fertility but Hstx2PWD fully lacks fertility32. Therefore, we hypothesised that Hstx2B6 and Hstx2PWD are essential for spermatogenesis in both inbred strains and hybrids, and then the function of Hstx2B6 remains while that of Hstx2PWD is disrupted in the hybrids. While we confirmed whether each HS candidate KO caused severe meiotic defects and meiotic failure in B6 mice, we did not assess more subtle phenotypes, such as a mere reduction in sperm count or motility. Therefore, these HS candidate KO males might show semi-fertility.
Previous studies identified two HS loci by forward genetic approaches using consomic strains29,30, and the HS gene on Hst1 was identified as Prdm924. However, these forward genetic approaches have a limit to narrow down 4.7 Mb (Chr X:64.9 Mb-69.6 Mb, GRCm38) of Hstx2 critical region on PWD Chr X, because there may be recombination cold spot on Hstx240,41. Recently, Lustyk, D. et al. reduced 4.7 Mb region of Hstx2 to 2.7 Mb (Chr X:66.51 Mb-69.21 Mb) by backcross where SPO11-driven Cas9 nuclease was targeted by CRISPR to Hstx2 interval in female meiotic prophase40. Nevertheless, this approach could not identify HS gene on Hstx2. Therefore, a reverse genetic approach is required to identify HS gene on Hstx2.
Prdm9 is the only identified HS gene and is responsible for sterility phenotypes in male hybrids of house mouse38,39,52. Intersubspecific polymorphisms on the minisatellite cording for zinc-finger array of Prdm9, such as repeat variants and missense SNPs, and autosomal heterozygosity induce asymmetric DNA DSBs that cause sterility in hybrids43,44. Additionally, Prdm9 is expressed in adult testes and its null mutation leads to sterility in the inbred strain45,46. Therefore, we hypothesised that HS gene on Hstx2 may have similar features to Prdm9: intersubspecific polymorphisms of HS gene on Hstx2 might cause HS, and the HS gene null mutation may lead to sterility on B6 background, as in the case of Prdm9. However, some hybrid with a sufficiently high recombination rate did not show sterility by Prdm9 null mutation46, so that Prdm9 is not essential for meiosis in male mice of every species. At least in inbred male, Prdm9 is essential for spermatogenesis. In this study, we disregarded intersubspecific polymorphisms on intergenic and intron region. Therefore, another gene on Hstx2 which has other polymorphisms such as SNPs on its regulatory region might be HS gene. Additionally, we added 4 genes/cluster: Gm1140, Gm14692, Gm6812, and Mir465 cluster to the targets because these genes have intersubspecific missense SNPs between B6 and PWK. A very recent study reported that Gm1140, Gm14692 and Gm6812 have intersubspecific missense SNPs between B6 and PWD, and Mir465 cluster have copy number polymorphism between B6 and PWD40. Therefore, this report supports our selecting.
However, in our study, none of the HS candidate genes on Hstx2 was found to be essential for spermatogenesis on B6 genetic background. In our introduction, we proposed three hypotheses for the role of Hstx2. Based on our findings, we could partially exclude our first hypothesis, which was that the spermatogenic function of HS gene(s) on Hstx2 is lost in (PWD × B6) F1 male, but partially remains in (B6 × PWD) F1 male. However, in this hypothesis, the polygenic effect is retained. On Hstx2, there are a few related and adjacent genes. The coding sequence of Gm6812 is similar to that of 1700020N15Rik, which is located nearby Hstx2 (Chr X:69.9 Mb). The coding sequence of Gm6812 has only one missense SNP compared with that of 1700020N15Rik in both B6 and PWD mice. The coding sequence of Gm1140 is also same as that of Gm14692. These genes might complement each other functionally. Besides these possibilities, recent study reported that KO of several genes on Hstx2 resulted in arrested spermatogenesis in B6 mice53. The deletion of each Mir741, Mir871, and Mir880 did not affect spermatogenesis, but deletion of 6 miRNAs, Mir741, Mir463, Mir880, Mir878, and Mir871 disrupted spermatogenesis in B6 mice. Therefore, Mir463 or Mir878 might be essential for spermatogenesis, or polygene of these 6 miRNAs may have a critical role in spermatogenesis. The second hypothesis was that dominant-negative Hstx2PWD impedes the spermatogenesis in hybrids. Nineteen miRNAs on Hstx2 BAC transgenic mice carrying these autosomal genes show sterility54. Normally, these miRNAs are actively transcribed in spermatogonia and suppressed by meiotic sex chromosome inactivation in pachytene spermatocytes. In this BAC transgenic mice, these miRNAs are mis-expressed on pachytene, which induces spermatogenic defects in the BAC transgenic mice. Among the 19 miRNA genes, Mir743 and the Mir465 cluster showed intersubspecific polymorphisms between B6 and PWD. In our study, we generated Mir743 single gene KO B6 mice and Mir465 cluster KO B6 mice, which did not show male sterility. If the second hypothesis was correct, HS gene KO on Hstx2 of PWD in (PWD × B6) F1 male would not show sterility. The third hypothesis was that Hstx2B6 rescues fertility of hybrids, while Hstx2PWD cannot rescue HS. (PWD × B6) F1 show sterility caused by asymmetric DSBs43. Although the amount of asymmetric DSB in (B6 × PWD) F1 male spermatocyte is equivalent to that in (PWD × B6) F1 male, (B6 × PWD) F1 male shows semi-fertility43. This result suggests that HS in (B6 × PWD) F1 male is rescued without recovering the asymmetry of meiotic DSBs. Therefore, we propose another recovery mechanism of HS without repairing asymmetry of meiotic DSBs. We suspect that HS gene of PWD on Hstx2 cannot rescue HS in (PWD × B6) F1 male, while HS gene of B6 on Hstx2 can rescue HS in (B6 × PWD) F1 male, inducing asymmetric HS. If the third hypothesis was correct, HS gene KO on Hstx2 of B6 in (B6 × PWD) F1 male would not show sterility.
In conclusion, we used a new approach to determine HS gene on Hstx2 by reverse genetics, which revealed that Ctag2, 4930447F04Rik, Mir743, Mir465 cluster, Gm1140, Gm14692, 4933436I01Rik, and Gm6812 are not individually necessary for spermatogenesis in B6 mice. This result partially proved that the loss of function of one gene/cluster on Hstx2 does not cause complete HS. Furthermore, the HS gene on Hstx2 might cause HS by different mechanisms than that shown by Prdm9. Thus, our study contributes toward determination of the HS gene on Hstx2 by at least excluding out one possibility.
Methods
Mice
C57BL/6 J (B6) and Jcl:CD1 (ICR) inbred strain were purchased from Charles River Laboratories (Yokohama, Japan) and CLEA Japan (Tokyo, Japan). All mice were housed in plastic cages under pathogen-free conditions in a room maintained at 23.5 °C ± 2.5 °C and 52.5% ± 12.5% relative humidity under a 14-h light:10-h dark cycle. Mice had free access to commercial chow (MF; Oriental Yeast, Tokyo, Japan) and filtered water. Animal experiments were carried out in a humane manner with approval from the Institutional Animal Experiment Committee of the University of Tsukuba in accordance with the Regulations for Animal Experiments of the University of Tsukuba and Fundamental Guidelines for Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Immunohistochemistry of γH2AX and PNA lectin histochemistry
All mice were sacrificed over 10-week-old. After intraperitoneal (i.p.) injection of a fatal dose of pentobarbital, mice were transcardially perfused with phosphate-buffered saline (PBS) under anaesthesia. Testes, from which tunica albuginea were removed, were fixed overnight in 10%-Formaldehyde Neutral Buffer Solution (Nacalai Tesque, Kyoto, Japan) and then dehydrated in 70% ethanol at room temperature over 1 week. The fixed testes were embedded in paraffin and stored at room temperature until use. The testes were sectioned at a thickness of 6 µm, deparaffinised, immersed in 0.25% Triton X-100 in PBS and then the antigens were activated in Target Retrieval Solution (Dako, Santa Clara) for 10 min at 121 °C. After blocking the sections by the blocking solution [0.1% bovine serum albumin (BSA), 0.01% Tween20, 10% goat serum in PBS] for 60 min at room temperature, the sections were incubated with mouse IgG anti-γH2AX antibody (1:100, #05-636; Millipore, MA) overnight at 4 °C in a chamber with high humidity. After washing the sections twice using PBS, goat anti-mouse IgG Alexa Fluor 647 (1:200, #A28181; Invitrogen, CA) and PNA lectin (1:100, #L7381-1MG; SIGMA-ALDRICH, MO) were applied on the sections and incubated for 60 min at room temperature in a dark chamber with high humidity. The sections were washed twice with PBS and incubated with 4’,6-diamidino-2-phenylindole (DAPI) in PBS (1:500) for 10 min. After washing the sections using PBS, the sections were mounted with Prolong Gold antifade reagent with DAPI (Invitrogen). Images of seminiferous tubules were captured using a fluorescence microscope CCD camera (#BZ-X710; KEYENCE, Osaka, Japan) and processed using BZ-X analyzer software (KEYENCE).
Sequencing and genotyping
Genomic DNA was extracted from the tails of 3-week-old mice. Genotyping PCR was performed using PrimeSTAR GXL DNA Polymerase (Takara Bio, Shiga, Japan) or AmpliTaq Gold DNA Polymerase (Applied Biosystems); the primers used for this are listed in Table S2.
Generation of KO mice using CRISPR/Cas9 system
To remove all genomic region of each HS candidate gene, we respectively designed two sgRNAs, one whose target site was upstream of the target gene and the other was downstream of it on B6 genetic background. The sequences of the sgRNAs are listed in Table S3. To create KO mice, we performed microinjection or electroporation. The microinjection was performed according to previously reported method, with some modification55. Female B6 mice were i.p. injected with pregnant mare serum gonadotropin (PMSG) and human chorionic gonadotropin (hCG) with a 48-h interval, and mated with male B6 mice. The zygotes were collected from oviducts. Then, two pX330 (circular, 5 ng/µl each), targeting two CRISPR sites of each HS candidate gene, was injected into the pronuclei according to standard protocols. The injected embryos (pronuclei stage) were then transferred into pseudopregnant ICR females. The electroporation was performed as reported previously, with some modification56. The sgRNA was synthesised and purified using the GeneArt Precision gRNA Synthesis Kit (Thermo Fisher Scientific, MA) and dissolved in Opti-MEM (Thermo Fisher Scientific). PMSG and hCG were i.p. injected into female B6 mice with 48-h interval, and unfertilised oocytes were collected from their oviducts. We then performed in vitro fertilisation with these oocytes and sperm from B6 mice according to standard protocols. Five hours later, two sgRNA (25 ng/µl each) targeting two CRISPR sites of each HS candidate gene, and GeneArt Platinum Cas9 Nuclease (100 ng/µl, Thermo Fisher Scientific) were electroporated into these zygotes using the NEPA 21 electroporator (Nepa Gene, Chiba, Japan). The poring pulse was set to: 225 V, 2 ms pulse width, 50 ms pulse interval, and + 4 pulse number. The transfer pulse was set to: 20 V, 50 ms pulse width, 50 ms pulse interval, and ± 5 pulse number (attenuation rate was set to 40%). After electroporation, the developed 2-cell embryos were transferred into the oviducts of pseudopregnant ICR females.
Supplementary information
Acknowledgements
We thank the members of the Laboratory Animal Resource Center Sugiyama for helpful discussions and encouragement. This work was supported by Grants-in-Aid for Young Scientists (B) (17K14971: to S.M.), Scientific Research (B) (19H03142: to S.M.), and Scientific Research on Innovative Areas “Platform of Advanced Animal Model Support” (16H06276 to S.T., S.M., F.S.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT). We would like to thank Editage (www.editage.com) for English language editing.
Author contributions
S.M., and F.S. were responsible for all experimental design. Y.D., Y.H., Ke.K., Ka.K., H.S. and S.T. developed KO mouse models. Ke.M. and K.N. performed genotyping and confirm KO phenotype. S.A., and A.Y. provided care for PWK mouse strain. Ka.M. and F.S. administered all animal experiments.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-65986-y.
References
- 1.Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin’s corollary to Haldane’s rule. Genetics. 2007;176(2):1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2008;62(1):50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brideau NJ, Barbash DA. Functional conservation of the Drosophila hybrid incompatibility gene. Lhr. BMC Evol. Biol. 2011;11(1):57. doi: 10.1186/1471-2148-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coyne, J. A. & Orr, H. A. Speciation. (Sunderland, Massachusetts: Sinauer Associates (2004).
- 5.Haldane J. Sex ration and unisexual sterility in animal hybrids. J. Genet. 1922;12(2):101–109. doi: 10.1007/BF02983075. [DOI] [Google Scholar]
- 6.Dobzhansky, T. Genetics and the Origin of Species. (New York, Columbia University Press (1937).
- 7.Muller H. Isolating mechanisms. evolution, and temperature. Biol. Symp. 1942;6:71–125. [Google Scholar]
- 8.Presgraves DC. Sex chromosomes and speciation in. Drosophila. Trends Genet. 2008;24(7):336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slotman M, Della Torre A, Powell JR. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis. Genetics. 2004;167(1):275–287. doi: 10.1534/genetics.167.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turelli, M. & Orr, H. A. The dominance theory of Haldane’s rule. Genetics140, 389–402, https://www.genetics.org/content/140/1/389.short (1995). [DOI] [PMC free article] [PubMed]
- 11.Presgraves DC. Evaluating genomic signatures of “the large X‐effect” during complex speciation. Mol. Ecol. 2018;27(19):3822–3830. doi: 10.1111/mec.14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HY, et al. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 2008;135(6):1065–1073. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 13.Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282(5393):1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 14.Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;326(5959):1538–1541. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liénard MA, Araripe LO, Hartl DL. Neighboring genes for DNA-binding proteins rescue male sterility in Drosophila hybrids. Proc. Natl. Acad. Sci. USA. 2016;113(29):E4200–E4207. doi: 10.1073/pnas.1608337113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbash D. A., Siino D. F., Tarone A. M., Roote J. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proceedings of the National Academy of Sciences. 2003;100(9):5302–5307. doi: 10.1073/pnas.0836927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brideau NJ, et al. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314(5803):1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- 18.Phadnis N, et al. An essential cell cycle regulation gene causes hybrid inviability in Drosophila. Science. 2015;350(6267):1552–1555. doi: 10.1126/science.aac7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang S, Presgraves DC. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science. 2009;323(5915):779–782. doi: 10.1126/science.1169123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Presgraves DC, Balagopalan L, Abmayr SM, Orr HA. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature. 2003;423(6941):715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- 21.Phadnis N, Orr HA. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science. 2009;323(5912):376–379. doi: 10.1126/science.1163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cattani MV, Presgraves DC. Incompatibility between X chromosome factor and pericentric heterochromatic region causes lethality in hybrids between Drosophila melanogaster and its sibling species. Genetics. 2012;191(2):549–559. doi: 10.1534/genetics.112.139683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawamura, K., Yamamoto, M. T., & Watanabe, T. K. Hybrid lethal systems in the Drosophila melanogaster species complex. II. The Zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics133(2), 307–313, https://www.genetics.org/content/133/2/307.short (1993). [DOI] [PMC free article] [PubMed]
- 24.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323(5912):373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 25.Bomblies K, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5(9):e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larson EL, Keeble S, Vanderpool D, Dean MD, Good JM. The composite regulatory basis of the large X-effect in mouse speciation. Mol. Biol. Evol. 2016;34(2):282–295. doi: 10.1093/molbev/msw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellegren H. Genomic evidence for a large-Z effect. Proc. R. Soc. Lond. 2009;276(1655):361–366. doi: 10.1098/rspb.2008.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dufresnes C, et al. Empirical evidence for large X-effects in animals with undifferentiated sex chromosomes. Sci. Rep. 2016;6(1):1–7. doi: 10.1038/srep21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forejt J, Iványi P. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.) Genet. Res. 1974;24(2):189–206. doi: 10.1017/S0016672300015214. [DOI] [PubMed] [Google Scholar]
- 30.Gregorová S, et al. Sub-milliMorgan map of the proximal part of mouse Chromosome 17 including the hybrid sterility 1 gene. Mamm. Genome. 1996;7(2):107. doi: 10.1007/s003359900029. [DOI] [PubMed] [Google Scholar]
- 31.Dzur-Gejdosova M, Simecek P, Gregorová S, Bhattacharyya T, Forejt J. Dissecting the genetic architecture of F1 hybrid sterility in house mice. Evolution. 2012;66(11):3321–3335. doi: 10.1111/j.1558-5646.2012.01684.x. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya Tanmoy, Reifova Radka, Gregorova Sona, Simecek Petr, Gergelits Vaclav, Mistrik Martin, Martincova Iva, Pialek Jaroslav, Forejt Jiri. X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids. PLoS Genetics. 2014;10(2):e1004088. doi: 10.1371/journal.pgen.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forejt J. Hybrid sterility in the mouse. Trends Genet. 1996;12(10):412–417. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- 34.Forejt, J., Pialek, J. & Trachtulec, Z. Hybrid male sterility genes in the mouse subspecific crosses. Evolution of the house mouse, (3), 482-503, (2012).
- 35.Gregorová, S. & Forejt, J. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies–a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol (Praha), 46(1), 31-41 https://fb.cuni.cz/volume-46-2000-no-1#mus (2000). [PubMed]
- 36.Din W, et al. Origin and radiation of the house mouse: clues from nuclear genes. J. Evol. Biol. 1996;9(5):519–539. doi: 10.1046/j.1420-9101.1996.9050519.x. [DOI] [Google Scholar]
- 37.Gregorová S, et al. Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res. 2008;18(3):509–515. doi: 10.1101/gr.7160508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327(5967):836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327(5967):835–835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lustyk Diana, Kinský Slavomír, Ullrich Kristian Karsten, Yancoskie Michelle, Kašíková Lenka, Gergelits Vaclav, Sedlacek Radislav, Chan Yingguang Frank, Odenthal-Hesse Linda, Forejt Jiri, Jansa Petr. Genomic Structure of Hstx2 Modifier of Prdm9-Dependent Hybrid Male Sterility in Mice. Genetics. 2019;213(3):1047–1063. doi: 10.1534/genetics.119.302554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balcova M, et al. Hybrid sterility locus on chromosome X controls meiotic recombination rate in mouse. PLoS Genet. 2016;12(4):e1005906. doi: 10.1371/journal.pgen.1005906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharyya T, et al. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc. Natl. Acad. Sci. USA. 2013;110(6):E468–E477. doi: 10.1073/pnas.1219126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies B, et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature. 2016;530(7589):171. doi: 10.1038/nature16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smagulova F, Brick K, Pu Y, Camerini-Otero RD, Petukhova GV. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 2016;30(3):266–280. doi: 10.1101/gad.270009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438(7066):374. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 46.Mihola Ondrej, Pratto Florencia, Brick Kevin, Linhartova Eliska, Kobets Tatyana, Flachs Petr, Baker Christopher L., Sedlacek Radislav, Paigen Kenneth, Petkov Petko M., Camerini-Otero R. Daniel, Trachtulec Zdenek. Histone methyltransferase PRDM9 is not essential for meiosis in male mice. Genome Research. 2019;29(7):1078–1086. doi: 10.1101/gr.244426.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Von Deimlingi, O. H., Forejt, J. & Wienker, T. F. Allelic profile at 37 biochemical loci of two inbred strains of the house mouse derived from wild Mus musculus musculus. Lab. Anim. 22(1), 61–66, doi:10.1258%2F002367788780746610 (1988). [DOI] [PubMed]
- 48.Dickinson ME, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bakker CE, et al. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78(1):23–33. doi: 10.1016/0092-8674(94)90569-X. [DOI] [PubMed] [Google Scholar]
- 50.Miyata H, et al. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc. Natl. Acad. Sci. USA. 2016;113(28):7704–7710. doi: 10.1073/pnas.1608458113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu Y, et al. Impaired conditioned fear and enhanced long-term potentiation in Fmr2 knock-out mice. J. Neurosci. 2002;22(7):2753–2763. doi: 10.1523/JNEUROSCI.22-07-02753.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliver PL, et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5(12):e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ota H, et al. Identification of the X-linked germ cell specific miRNAs (XmiRs) and their functions. PLoS one. 2019;14(2):e0211739. doi: 10.1371/journal.pone.0211739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Royo H, et al. Silencing of X-linked microRNAs by meiotic sex chromosome inactivation. PLoS Genet. 2015;11(10):e1005461. doi: 10.1371/journal.pgen.1005461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoshino Y, et al. Simple generation of hairless mice for in vivo imaging. Exp. Anim. 2017;66(4):437–445. doi: 10.1538/expanim.17-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato Y, et al. A mutation in transcription factor MAFB causes Focal Segmental Glomerulosclerosis with Duane Retraction Syndrome. Kidney Int. 2018;94(2):396–407. doi: 10.1016/j.kint.2018.02.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.