Abstract
Objectives
To introduce bibliometric features of Iranian documents on microbiota and to provide descriptive information about retrieved documents related to the medical sciences and documents utilizing molecular techniques for microbiota detection.
Methods
This is a descriptive bibliometric study of all Iranian documents on microbiota in any language that were indexed in Scopus before 7 September 2019. We assessed the research performance through statistical analysis of the bibliometric indicators, including number of publications, citations, institutions and journals activities, co-citations and bibliographic couplings, and network analysis of co-authorships, countries’ collaborations, terms and keywords.
Results
We extracted 425 relevant documents, 260 of which pertain to the medical sciences. The most focused microbiota modulating interventions and diseases in 33 clinical trials are ‘synbiotics’ (n = 8) and ‘probiotics’ (n = 8), and ‘Obesity’ (n = 3) and ‘non-alcoholic fatty liver disease’ (n = 3), respectively. During the last decade, Iranian microbiota publications have increasingly grown with a constant upward slope, particularly in the area of medical sciences after 2016. Citation counting reveals that originals and reviews have been cited 4221 times, with an average 10.76 citations and H-index of 34. The most significant performance in publishing Iranian microbiota documents belongs to ‘Tehran University of Medical Sciences’ as the active institution (n = 89 publications) and the supporting sponsor (n = 19), ‘Microbial Pathogenesis’ as the productive journal (n = 12), ‘Seidavi A’ as the most authorships (n = 19), and ‘the United States’ as the collaborative country (n = 46).
Conclusions
The qualitative and quantitative information of this study will be a practical guidance for future study planning and policy-decision making.
Electronic supplementary material
The online version of this article (10.1007/s40200-020-00488-2) contains supplementary material, which is available to authorized users.
Keywords: Microbiota, Microbiome, Scientometrics, Bibliometrics, Iran
Introduction
Scientific production of each country has a very large impact on its economic growth which reveals the importance of the investment in research and development [1]. Even, this issue gets more significance in developing countries that want to shine in the realm of science [2]. Iran is one of the developing countries in the Middle East that has the fastest publication growth in the world even faster than China and Turkey; therefore, providing a good landscape of research trend in any specific field conducting in Iran seems to be necessary because of its financial, political and scientific purposes [3, 4]. This landscape will be achieved by a bibliometric and scientometric analysis through the literature, which provides a scientific map specifying the main subject area and document type; determining the publication development over time, the number of citations, and the most cited articles; highlighting the performance of the most active institutions, journals, and authors; finding the most repetitive terms and keywords; finally, directing other researchers and policymakers to the scientific development of literature [5–7].
Microbiota is an ecosystem of living microorganism that interacts functionally with the host and resides in and on host body sites including the skin, the respiratory, urogenital, and digestive tracts [8, 9]. The first use of the term “microbiota” in the literature returns to 1927 that discussed the soil microbiota including protozoa, fungi, bacteria, and algae [10]. Then, in the 1960s, this term attracts a great deal of attention in dental and oral medicine [11–13]. Simultaneously, in the mid-twentieth century, the studies on rumen microbiota were performed on a large scale to boost the livestock industry [14–16]. Afterwards, in the early twenty-first century, the National Institutes of Health (NIH) group [17] conducted human microbiome project which provided a sequence database of bacterial, viral and eukaryotic microbial genomes from five human body sites (the oral cavity, the nasal cavity, the gastrointestinal tract, the skin and the vagina); discovered the effect of microbiome changes residing in the human body sites on their health/diseases; and established a standard setting for handling the next-generation technologies using for metagenomic studies. So, by emerging the sequencing technology and discovering new beneficial effect of microbiota, more attention paid to this area of medical sciences, particularly to the interaction between microbiota and human [17, 18]. Recently, progressive researches in the field of microbiota have been conducted, particularly about their key role on human health through consumption of nutrients, modification of pathogens, regulation of normal physiological pathways and promoting immune system that these perspectives of microbiota have dramatically influenced on the medical science area of literature [19–22]. To our knowledge, no bibliometric and scientometric study on Iranian microbiota research was conducted until now; so, in the current study, we aim to introduce the scientific roadmap of Iranian research on microbiota from the first document exist in the Scopus to the last related publication on 7 September 2019 and provide quantitative information for study planning and policy-decision making.
Method
Data retrieval
In the current descriptive scientometric and bibliometric study, the Scopus database was chosen to retrieve all Iranian documents on microbiota because of its wide coverage of literature and also high citations record [23]. All documents indexed in Scopus from the beginning to the last published on 7 September 2019 were selected with no language limitation if their title, abstract and keywords covered the following search queries: ((TITLE-ABS-KEY (microbiota) OR TITLE-ABS-KEY (microbiome))) AND (AFFILCOUNTRY (Iran)). This query was selected to obtain all Iranian documents on microbiota. The extraction of all documents and their information from Scopus was performed in one day (7 September 2019) to avoid the disturbance in citation analysis. After completing data extraction, we split all retrieved documents into two groups through screening the titles and abstracts; the medical science related documents and the non-medical science related documents. Then, we conducted a comprehensive bibliometric analysis of all Iranian microbiota documents as our primary purpose and a descriptive analysis of medical science-related documents as our secondary purpose. To achieve our secondary purpose, we picked out all original articles related to the medical sciences and analyzed them through their study designs. Afterward, we selected clinical trials out of these articles and evaluated the interventions and diseases that were investigated in these trials and finally, provide the relation network between them. This study is the bibliometric analysis of the literature through the published articles, so ethics approval is not required.
Data analysis
All extracted data were exported into Microsoft Excel for statistical analysis of the bibliometric information and ranking the prolific data including top-cited documents, top journals, institutions, sponsors, authors, and countries which subsequently were demonstrated by Graph Pad Prism, version 8.0.1.244. We constructed the global distribution map of the countries’ collaborations in publishing Iranian microbiota documents using GunnMap 2 (http://lert.co.nz/map/). Also, we applied VOSviewer software (version 1.6.13) [24] (van Eck & Waltman, 2010; www.vosviewer.com) to visualize the density and network analysis of the co-authorships, countries’ collaborations, terms, and author keywords; and also to analyze the bibliographic coupling/co-citation of journals and documents. Besides, Science of Science Tool [25] (Sci2Team, 2009) was utilized to illustrate the interconnections network between the interventions and the diseases that were extracted from the medical science clinical trials.
Results
General features of Iranian microbiota publications
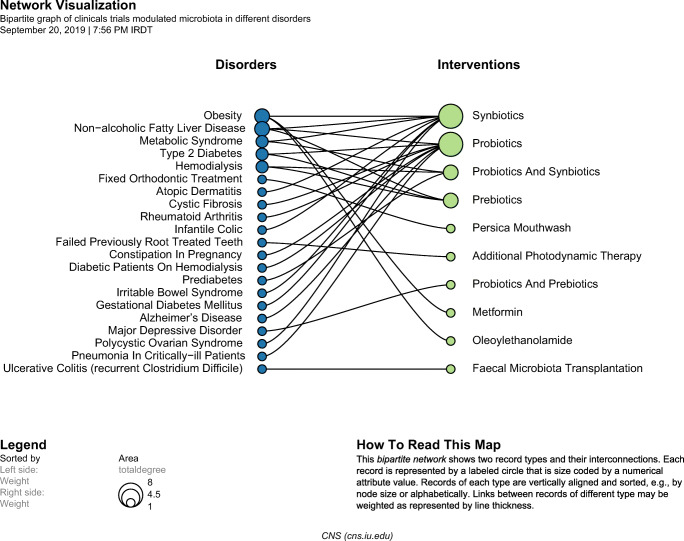
In all, 425 Iranian articles were published in the field of microbiota and microbiome, with the majority of original articles (n = 291, 68.47%). The next document type with the 23.76% share of all retrieved articles was review article (n = 101). The other document types which constitute less than 8% of the retrieved articles were demonstrated in Supplementary Fig. S1 (Online Resource 1). According to the subject area categorization of Scopus, most of these documents were in the area of Medicine (n = 204), Agricultural and Biological Sciences (n = 144) and Immunology and Microbiology (n = 91). The specific portion of other subject areas of the retrieved articles was exhibited in Supplementary Fig. S2 (Online Resource 2). The majority of retrieved documents were in English (n = 420 articles) and the remaining five articles were written in Persian. Besides, the main source of the extracted documents was journals, containing 417 Iranian microbiota documents, followed by books (n = 7 documents), and book series (n = 1 document). We also perused all 425 retrieved articles and discovered that the primary concept of 260 articles (61.17% of all Iranian microbiota articles) was about the medical sciences that interestingly, 13 articles of them were in the profession of dental and oral medicine. Next, we picked out all original articles from 260 medical sciences-related articles, which constitute 121 articles, and categorized them into the following four groups: the human studies (69 articles; 57.02%), the in-vitro studies (36 articles; 29.75%), the animal studies (13 articles; 10.74%), and others (3 articles; 2.47%). In the next step, we distinguished the study design of human studies which includes 33 clinical trials, 21 case-control studies, eight cross-sectional studies, two cohorts, one case report, and one case series; and three articles didn’t match to any of the mentioned study designs. In the next step, we extracted the following details from 33 aforementioned clinical trials and reported the results with the interconnection network visualization: the diseases from which the participants suffered and the types of microbiota modulating intervention used in these studies (Fig. 1). As is shown, 21 various diseases were assessed in these trials and 10 interventions were administered that in order of repetition include: synbiotics (8 trials); probiotics (8 trials); synbiotics in combination with probiotics (3 trials); prebiotic (3 trials); probiotics in combination with prebiotics (1 trial); Persica mouthwash (1 trial); photodynamic therapy (1 trial); metformin (1 trial); oleoylethanolamide (1 trial); and fecal microbiota transplantation (1 trial). In addition, the most-investigated diseases in these trials were as follows: Obesity (three times), nonalcoholic fatty liver disease (three times), metabolic syndrome (twice), type 2 diabetes (twice) and hemodialysis (twice).
Fig. 1.
The interconnection network between diseases and microbiota modulating interventions that are considered in clinical trials modulated microbiota in different disorders. In the first step, we picked out all original articles from 260 medical sciences-related documents, which constitute 121original articles, and categorized them into the following four groups: the human studies (69 articles; 57.02%), the in-vitro studies (36 articles; 29.75%), the animal studies (13 articles; 10.74%), and others (3 articles; 2.47%). In the next step, we distinguished the study design of all human studies and included all clinical trials dealing with microbiota modulating interventions in different disorders that finally, 33 clinical trials were carefully reviewed for data extraction
We furthermore reviewed all 260 medical science-related articles from the perspective of the molecular techniques used to detect microbiota in human studies carried out in Iran, like Real-time polymerase chain reaction (PCR) and 16S rRNA sequencing methods and disclosed that there are 14 case-control studies, 3 clinical trials, 2 cross-sectional studies and one pilot study on this subject; the distribution of various human diseases that were evaluated in these observational studies is as follows: inflammatory bowel disease (IBD) (n = 5 studies), diabetes mellitus (n = 3 studies), colorectal cancer (n = 2 studies), obesity (n = 1 study), esophageal cancer (n = 1 study), chronic urticaria (n = 1 study), recurrent kidney stone formation (n = 1 study), hospitalized acquired diarrhea (n = 1 study), and vaginal microbiome in fertile women (n = 1 study).
The first study on this subject was conducted by Nazemi M et al. [26] in 2013; they had utilized quantitative Real-time PCR (qPCR) to compare the abundance of Bifidobacteria in the intestine of diabetic patients with healthy population in Iran. In addition, the first human study in Iran using 16S rRNA sequencing technique was a case-control by Nasrollahzadeh D et al. [27] in 2015; They compared the gastric microbiota pattern in early esophageal squamous cell carcinoma with normal individuals. After that a case-control study by Heidarian F et al. [28] in 2017 compared the number and diversity of Streptococcus spp. in the stool of patients with IBD with healthy population by qPCR method. Nabizadeh E et al. [29] similarly conducted a case-control assay in 2017 using real-time PCR to detect the relative abundance of Akkermansia muciniphila, Faecalibacterium prausnitzii, Clostridium leptum, and Enterobacteriaceae in the intestine of patients with chronic urticarial compared with healthy volunteer. Navab-Moghadam F et al. [30] in 2017 evaluated the relative amount of Bacteroides fragilis, Bifidobacterium longum and Faecalibacterium prausnitzii in the stool sample of type 2 diabetic patients compared with non-diabetic control population through qPCR. Similarly, Sedighi M et al. [31] in 2017 conducted a case-control study to determine the variation in gut microbiota composition of type 2 diabetic patients and healthy population with qPCR and reported that Lactobacillus spp. was more frequent in diabetic patients while there are higher level of Bifidobacterium spp. in healthy participants. Zamani S et al. [32] in 2017 employed quantitative Real-Time PCR to detect Bacteroides fragilis residents in the intestinal of patients with ulcerative colitis (UC). In addition, Ghavami SB et al. [33] in 2018 assessed the relative amount of Methanobrevibacter smithii in the gasterointestinal tract of Iranians with IBD and healthy Iranians with qPCR and they reported lower load of Mbb. smithii in IBD. Rezasoltani S et al. [34] in 2018 evaluated the quantity of various gut bacteria in different colorectal polyps and normal colon; this case-control study targeted many types of intestinal bacteria through real-time PCR, including Streptococcus bovis/gallolyticus, Enterotoxigenic Bacteroides fragilis (ETBF), Enterococcus faecalis, Fusobacterium nucleatum, Lactobacillus spp., Porphyromonas spp., Roseburia spp., and Bifidobacterium spp. Another case-control study by Rezasoltani S et al. [35] in 2018 introduced the accuracy of various gut bacteria for early detection of colorectal cancer and adenomatous polyp using real-time PCR for quantification. After that Al-Bayati L et al. [36] in 2018 demonstrated the relative quantity of anaerobic bacterial species residing in the colon of patients with UC in compared to normal population utilizing real-time PCR. Another case-control study by Tavasoli S et al. [37] in 2019 investigated the abundance of Oxalobacter formigenes, Lactobacillus genus and Bifidobacterium genus in the gastrointestinal tract of the patients suffering from recurrent calcium kidney stone formation and normal population through quantitative real-time PCR. In addition, Mohammadzadeh N et al. [38] in 2019 employed a quantitative real-time PCR technique to compare the gut microbiome profile of hospitalized patients with diarrhea to healthy individuals. Also, Heidarian F et al. [39] in 2019 carried out a case-control study on the relative quantity of thirteen different bacterial families that were detected in the stool sample of patients with IBD and healthy participants through real-time PCR. Moossavi S et al. [40] conducted a pilot study in 2019 to demonstrate the influence of three fecal storage methods on the quantity and diversity of microbiota composition using 16S rRNA sequencing and Illumina MiSeq platform.
Besides, two cross-sectional studies utilizing molecular techniques to identify human microbiome profile were accomplished in Iran. In 2018, Nami Y et al. [41] carried out a cross-sectional study and identified various lactic acid bacteria residing in the vaginal tract of fertile women through 16S-rDNA sequencing technique. The next cross-sectional study was conducted by Mousavi SH et al. [42] in 2018 and reported the association of gut microbiota pattern e.g. bacteroidetes/firmicutes ratio with obesity in Iranian school aged population using quantitative real-time PCR to assess intestinal microbiota composition.
Additionally, We discovered that the first randomized clinical trial (RCT) utilizing molecular techniques for microbiota identification among Iranian population was conducted by Ejtahed HS et al. [43] in 2019; they reported the effect of metformin on gut microbiota composition and weight loss in obese women utilizing 16S rRNA sequencing method. The next RCT on human subjects was conducted by Payahoo L et al. [44] in 2019; they utilized quantitative real-time PCR to exhibit the effect of oleoylethanolamide consumption on the amount of Akkermansia muciniphila bacterium inhabiting in the gastrointestinal tract of obese Iranian. Also, Laffin MR et al. [45] conducted a RCT on the effect of high-amylose maize resistant starch type 2 (HAM-RS2) consumption on the abundance of Faecalibacterium genus in the end-stage chronic kidney disease by using 16 s DNA sequencing method for gut microbiota quantification. These molecular technique were also used widely in the animal intervention studies like the study by Ahmadi S et al. [46] in 2018 that they showed the effect of prebiotics on the intestinal microbiota dysbiosis induced by high fat diet in mice through 16S rRNA sequencing technique. Another experimental study with the same technique by Hosseinifard ES et al. [47] worked on the influence of psychobiotics on the gut microbiota dysbiosis induced by type 2 diabetes in rats .
Iranian microbiota articles publications over time
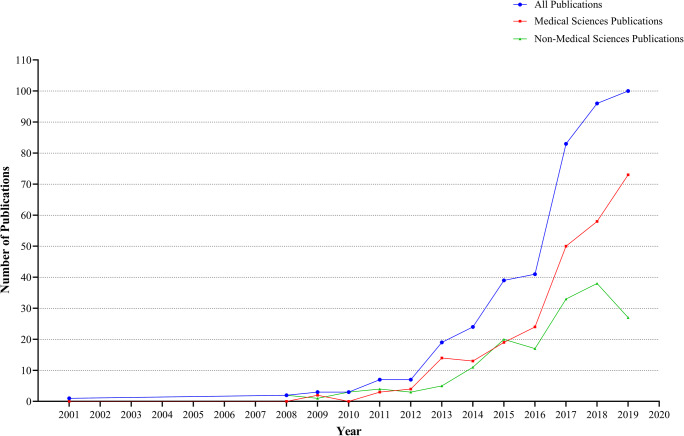
Through our search strategy, we discovered that the first Iranian attempt in the field of microbiota and microbiome research was made in 2001 by Ghazifard A. et al. [48] in the Faculty of Sciences of Esfahan University on determining bacterial and fungal components of the urban solid waste. The next attempt in this field was conducted in 2008 by Seidavi A. et al. [49] on the detection of Bifidobacterium spp. and Lactobacillus spp. by a multiplex polymerase chain reaction in the intestinal tract of broilers, followed by Ghiyasi M. et al. [50] in 2008 on the prebiotic effects on microbiota composition of broilers’ intestine that these two articles shed new light on the development of the poultry industry. Additionally, the first article in the area of medical sciences was produced in 2009 by Jajarm HH. et al. [51] on the influences of Persica mouthwash on the oral microbial composition in the cleft lip and palate individuals. Then, at least one Iranian microbiota article has been published in both medical and non-medical sciences annually, with the exception of 2010 in which no article was published in the medical sciences. As is shown in Fig. 2, at the inception of Iranian research on the microbiome, the most attention was paid to the area of non-medical sciences, but as time goes on, the role of microbiota in medical sciences has been noticed. Although the overall slope of the publications in each field has been upward during the last decade (2009–2019), the medical sciences’ papers have grown steeper, noticeably after 2016. In total, the most publishing years were 2018 and 2019 with 100 and 96 publications, respectively and we expect the gap between these two years will increase, as this field of literature gets more interest especially in the area of medical sciences and also more articles will be published by the end of 2019. The growth trend in the number of Iranian microbiota documents and their subsets, including medical science-related documents and non-medical science-related documents are depicted in Fig. 2.
Fig. 2.
The growth trend in the number of Iranian microbiota documents and their subsets, including medical science-related documents and non-medical science-related documents
Citations counting and the most cited Iranian microbiota articles
We considered the original articles and reviews for citation counting. Totally, 392 articles were accumulated, of which 296 articles cited at least once and the total citations of all included articles were counted as 4221 times. The average citations per each included articles during the study period was determined as 10.76 times that reflects the impact of Iranian microbiota articles in the literature. We also calculated H-index of the retrieved articles as 34 that indicates 34 Iranian microbiota articles have been cited 34 times or more. The growth trend in the number of citations to Iranian microbiota documents is shown in Supplementary Fig. S3 (Online Resource 3). The first citation was made in 2003, and as time goes by, the more articles were published and accordingly, the more citations were produced. The most annual citations at the date of analysis were established in 2019 and 2018 (1228 and 1166 citations, respectively); however, the statistic for 2019 will boost by the end of this year. In addition, the 10 most-cited Iranian microbiota documents are listed in Table 1. The most-cited article with 263 citations was an international collaborative original article with 140 collaborators; conducted by Henderson G et al. [52] in 2015 on the worldwide foregut microbial compositions of the ruminants and camelids and their influential factors like diets, hosts, and geography. The next most-cited article with 170 citations was an international collaborative review article between the United Kington, Canada, and Iran; conducted by Llewellyn MS et al. [53] in 2014 on the teleost microbiome researches. Although the two most cited articles in this field had a significant impact on the livestock and the aquaculture industries, half of the top 10 cited articles are in the area of medical sciences, taking the third, fourth, fifth, seventh and tenth place in this ranking, as explained below. So, the prominent article in medical sciences with 99 citations, which ranks third in the overall ranking, was an in-vivo study on diabetic rats that was carried out by Davari S et al. [54] in 2013. This experimental study assessed the influence of probiotics on impaired learning and cognitive disorders induced by diabetes and suggested that probiotics promote the hippocampal synaptic performance that this finding results in improve deteriorated memory function in diabetic rats [54]. The second prominent medical sciences-related article with 94 citations was an epidemiological transition study; conducted by Bishehsari F et al. [55] in 2014. This study reported the epidemiological information of the developing countries about colorectal cancer, including the burden of the disease, environment-related risk factors, the role of gut microbiota and nuclear receptors in tumorigenesis of cancer, the molecular pathways in pathogenesis of cancer, and the preventive strategies [55]. The third prominent article in medical sciences with a total of 89 citations was an 8-week double-blind randomized clinical trial, conducted by Vaghef-Mehrabany E et al. [56] in 2014. This trial revealed that daily administration of probiotic capsule containing Lactobacillus casei 01 reduces inflammatory activity of rheumatoid arthritis [56]. The fourth prominent medical sciences-related article with 78 citations, which ranks seventh in the overall ranking, was a review article on the role of polyphenol and probiotics in weight losing, conducted by Rastmanesh R [57] in 2011. This review article reported that Bacteroidetes/ Firmicutes ratio of the intestinal microbiota has been reduced in obese people and probiotic-rich diet cause obesity through the overgrowth of Lactobacillus species (belong to the Firmicutes phylum) in the intestine and lowering the intestinal Bacteroidetes/ Firmicutes ratio; on the other hands, polyphenol-rich diets e.g., apple, grapefruit, pear, and green tea provoke weight losing in obese people through increasing Bacteroidetes/ Firmicutes ratio of the intestinal microbiota community [57]. Finally, the fifth prominent medical sciences-related article with a total of 63 citations was a systematic review of randomized clinical trials assessing the efficacy of probiotics for bacterial vaginosis (BV) recurrences, conducted by Homayouni A et al. [58] in 2014. This systematic review disclosed that 2-month administration of probiotics containing Lactobacillus acidophilus, Lactobacillus rhamnosus GR-1, and Lactobacillus fermentum RC-14 had significantly promoted the women health through the preventive and therapeutic effects on BV [58].
Table 1.
The bibliometric characteristics of 10 Iranian microbiota documents with the most citations
| Rank | Authors | Title | Year | Source title | Cited by | Article type | CiteScore (2018) |
|---|---|---|---|---|---|---|---|
| 1 | Henderson G et al.* | Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range | 2015 | Scientific Reports | 263 | Article | 4.29 |
| 2 | Llewellyn M.S. et al.* | Teleost microbiomes: The state of the art in their characterization, manipulation and importance in aquaculture and fisheries | 2014 | Frontiers in Microbiology | 170 | Review | 4.30 |
| 3 | Davari S et al | Probiotics treatment improves diabetes-induced impairment of synaptic activity and cognitive function: Behavioral and electrophysiological proofs for microbiome-gut-brain axis | 2013 | Neuroscience | 99 | Article | 3.46 |
| 4 | Bishehsari F et al | Epidemiological transition of colorectal cancer in developing countries: Environmental factors, molecular pathways, and opportunities for prevention | 2014 | World Journal of Gastroenterology | 94 | Article | 3.43 |
| 5 | Vaghef-Mehrabany E et al | Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis | 2014 | Nutrition | 89 | Article | 3.42 |
| 6 | Ghanbari M et al | A new view of the fish gut microbiome: Advances from next-generation sequencing | 2015 | Aquaculture | 81 | Review | 3.42 |
| 7 | Rastmanesh R | High polyphenol, low probiotic diet for weight loss because of intestinal microbiota interaction | 2011 | Chemico-Biological Interactions | 78 | Review | 3.43 |
| 8 | Chavshin A.R et al | Identification of bacterial microflora in the midgut of the larvae and adult of wild caught Anopheles stephensi: A step toward finding suitable paratransgenesis candidates | 2012 | Acta Tropica | 70 | Article | 2.68 |
| 9 | Hoseinifar S.H et al | Probiotic, prebiotic and synbiotic supplements in sturgeon aquaculture: A review | 2016 | Reviews in Aquaculture | 66 | Article | 5.24 |
| 10 | Homayouni A et al | Effects of probiotics on the recurrence of bacterial vaginosis: A review | 2014 | Journal of Lower Genital Tract Disease | 63 | Review | 1.24 |
*This is an international collaborative original article
Besides, it reveals that out of 10 most cited articles, five articles with a total of 423 citations relates to the human health (medical sciences) that are explained above; three articles with a total of 317 citations relate to the aquaculture and fisheries; one article with 263 citations relates to the ruminants and camelids; and one article with 70 citations relates to the Anopheles stephensi mosquito. Among these 10 dominant articles, four were carried out in 2014, two in 2015 and one article in each of 2011, 2012, 2013, and 2016; six papers were original articles and the remaining four were reviews.
Top institutions performance in publishing Iranian microbiota articles
The productivity analysis of institutions contributed in Iranian microbiota research shows the following five universities has the most share of activity in this field: Tehran University of Medical Sciences (n = 89 publications), Tabriz University of Medical Sciences (n = 57 publications), Shahid Beheshti University of Medical Sciences (n = 56 publications), University of Tehran (n = 30 publications) and Islamic Azad University (n = 27 publications). The annual productivity of the top 5 universities is illustrated in Supplementary Fig. S4 (Online Resource 4). In comparison, Tehran University of Medical Sciences had the steepest slope in microbiota publications over time, particularly after 2015 that the years with the most publications were 2019 and 2018 (with 25 and 19 publications, respectively).
Top journals performance in publishing Iranian microbiota articles
The list of top journals publishing Iranian microbiota articles is ordered in Table 2. The journal of Microbial Pathogenesis, with the impact factor of 2.58 in 2018, produced the most Iranian microbiota publications (n = 12 papers) that covers the articles in the area of medical sciences, followed by the journal of Fish And Shellfish Immunology with the impact factor of 3.29 in 2018 (n = 11) that covers the articles in the area of medical and non-medical sciences including agricultural and biological sciences, aquatic science, and environmental science; and the journal of Poultry Science with the impact factor of 2.02 in 2018 (n = 10) that covers the articles in the area of medical sciences and non-medical sciences including agricultural and biological sciences, animal science and zoology. As ordered in Table 2, the journals with the same publications are ranked in the same place according to the dense ranking.
Table 2.
Top journals performance in publishing Iranian microbiota articles
| Rank* | Journal | Number of Publications |
|---|---|---|
| 1 | Microbial Pathogenesis | 12 |
| 2 | Fish and Shellfish Immunology | 11 |
| 3 | Poultry Science | 10 |
| 4 | Aquaculture Nutrition | 7 |
| 4 | Gastroenterology and Hepatology from Bed to Bench | 7 |
| 4 | Probiotics and Antimicrobial Proteins | 7 |
| 5 | Aquaculture | 6 |
| 5 | Frontiers in Microbiology | 6 |
| 5 | Journal of Cellular Physiology | 6 |
| 5 | Livestock Science | 6 |
| 5 | Photodiagnosis and Photodynamic Therapy | 6 |
*The journals with the same number of publications are ranked in the same place according to the dense ranking
Authors’ performance in publishing Iranian microbiota articles and their collaborations analysis
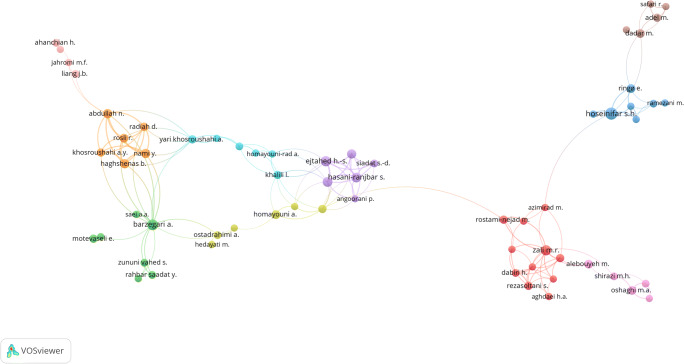
We discovered that 1828 authors had contributed in this field. The most-contributions belong to Seidavi A with 19 authorships, followed by Hoseinifar SH with 18 authorships, Barzegari A with 12 authorships, Moossavi S with 12 authorships, Siadat SD with 12 authorships, Ejtahed HS with 11 authorships, and Zali MR with 10 authorships. The clustering visualization of the largest co-authorships network between authors who contributed to at least 3 articles is mapped in Fig. 3. By considering the threshold of contributing to 3 or more articles, 123 authors were obtained that some of these authors didn’t have any co-authorship and some constructed small co-authorship networks, consisting of two to eight authors; so, we have established the largest co-authorship network (Fig. 3), comprising 63 authors that are divided into 10 clusters. In notes, there are a total of 150 collaborations between these 63 authors.
Fig. 3.
The authors’ performance clustering mapping and their co-authorships network in publishing Iranian microbiota articles. 63 authors of whom contributed to at least 3 Iranian microbiota articles constructs the largest co-authorship network and are divided into ten clusters displayed in different colors. Besides, there are a total of 150 collaborations between these 63 authors. The node size represents the number of articles published by each author and the thickness of the line between two authors represents the number of their co-authorships
Countries’ performance in publishing Iranian microbiota articles and their collaborations analysis
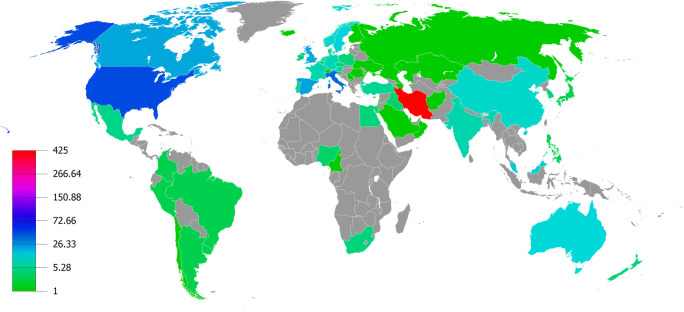
We analyzed the international collaborations of countries in publishing Iranian microbiota articles. As is illustrated in Fig. 4, the most-collaborative countries contributed in publishing these articles are located in North America and Western Europe. We also calculated all the collaborations between countries that contributed to 5 or more Iranian microbiota articles. Among 60 countries contributing in this field, 19 countries met this threshold and a total of 150 international collaborations exist between them. Then, we mapped the clustering visualization network of collaborations between these countries that as is depicted in Supplementary Fig. S5 (Online Resource 5), they were divided into three clusters that were displayed in different colors. Through this mapping analysis, we found out the most collaborative countries with Iran in publishing Iranian microbiota articles were the United States (46 collaborations), Italy (37 collaborations), Spain (23 collaborations), Canada (22 collaborations), the United Kingdom (22 collaborations), Malaysia (15 collaborations), and Sweden (14 collaborations).
Fig. 4.
The density geographical map that visualizes the volume of international collaborations of countries in publishing Iranian microbiota articles
The most repetitive terms in Iranian microbiota articles
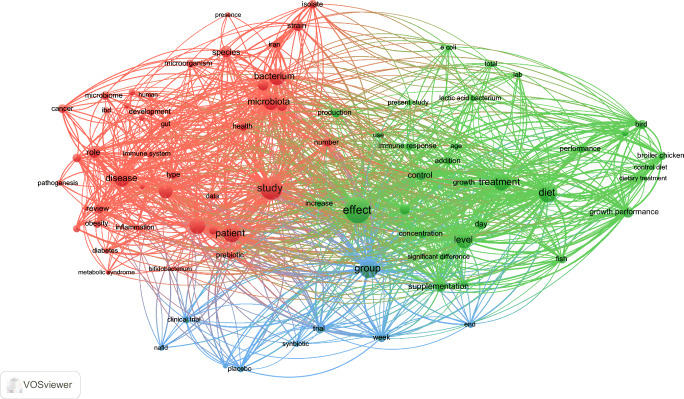
A total of 11,159 terms were utilized in the Iranian microbiota articles. To map the most repetitive terms, we consider a minimum of 30 repetitions as a threshold that 83 terms met this criterion. The clustering network mapping of the selected terms is displayed in Fig. 5. As shown, all terms were subdivided into three clusters. The most repetitive terms among all clusters are “effect” (n = 587), “study” (n = 460), “diet” (n = 352), “group” (n = 345), “patient” (n = 304), “treatment” (n = 296), “bacterium” (n = 271), “level” (n = 269), “probiotic” (n = 268), and “microbiota” (n = 266). After subdividing all terms, the most repetitive term of each cluster is as follows: “Study” in the red cluster, “effect” in the green cluster and “group” in the blue cluster. Additionally, the overlay mapping of the same terms is demonstrated in Supplementary Fig. S6 (Online Resource 6). By mapping this network, the average year of utilizing each term in the retrieved articles was displayed with different colors and therefore, the newer terms utilized in the recent microbiota and microbiome articles were identified. For example, we discovered that the most-occurred terms in the recent articles in this field are “clinical trial”, “placebo”, “prebiotic”, “inflammation”, “metabolic syndrome” and “pathogenesis” that reveals these are the most attracting field of research.
Fig. 5.
The co-occurrence clustering network mapping of the most repetitive terms with at least 30 repetitions that are retrieved from the title and abstract of Iranian microbiota articles; 83 terms meet this threshold that are subdivided into three clusters depicted with red, green and blue. The node size represents the number of repetitions of each term
The most repetitive author keywords in Iranian microbiota articles
A total of 1140 author keywords were utilized in Iranian microbiota articles. When we considered a minimum of 5 repetitions as a threshold, 43 author keywords were accumulated that two keywords didn’t have any relationship with others. The clustering network mapping of the remaining 41 keywords is illustrated in Supplementary Fig. S7 (Online Resource 7). All keywords were subdivided into six clusters. Among all clusters, the most repetitive author keywords utilized were “microbiota” (n = 50), “gut microbiota” (n = 50), “probiotics” (n = 47), “probiotic” (n = 40), “intestinal microbiota” (n = 27), “obesity” (n = 25), “microbiome” (n = 25), “synbiotic” (n = 22), “prebiotic” (n = 18), and “broiler” (n = 16). In each cluster, the most repetitive author keyword is as follows; the dark blue cluster: “microbiota”, the yellow cluster: “gut microbiota”, the purple cluster: “probiotics”, the green cluster: “probiotic”, the red cluster: “broiler” and the pale blue cluster: “diabetes” (n = 10). As described above, most of these author keywords are in the area of medical sciences that mostly emphasize the interactions between microbiota and host, particularly in the gastrointestinal tract of the host, and its effects on diseases like obesity and diabetes. Simultaneously, it seems that a lot of studies on microbiota and broilers have been carried out for aviculture purposes.
The bibliographic coupling and co-citation of journals publishing Iranian microbiota articles
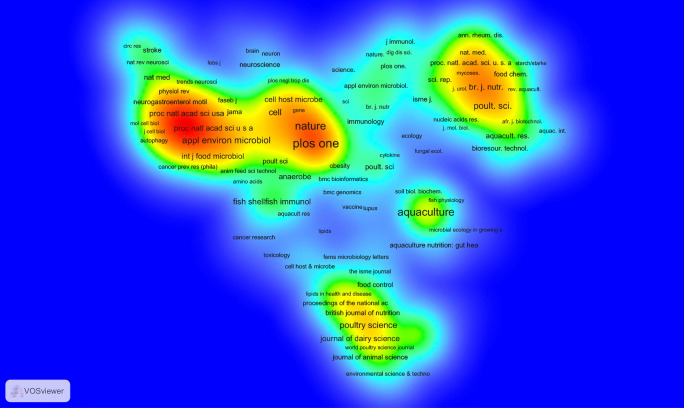
The journal bibliographic coupling analysis measures the similarities between two journals based on the frequency of the mutual references of their articles. In the current study, we considered all journals publishing at least 5 Iranian microbiota articles for the bibliographic coupling analysis. Out of a total of 262 journals, 13 journals met the threshold. The bibliographic coupling rainbow density map of the selected journals is demonstrated in Supplementary Fig. S8 (Online Resource 8). The high degree of bibliographic coupling of two journals indicates two journals probably containing the subject matter in common. As shown, the journal of ‘microbial pathogenesis’ has the most coupling strength for the Iranian microbiota documents. Instead, the journal co-citation analysis calculates the frequency of two journals’ articles that are co-cited by other articles. The co-citation rainbow density map of journals publishing at least 5 Iranian microbiota articles is depicted in Fig. 6. The high co-citation of two journals indicates that two journals have high semantic relationships; also, the high co-citation of a journal represents that the journal is the prominent source containing Iranian microbiota articles which have been co-cited by other articles e.g. ‘Plos one’ and ‘nature’ are the prominent journals in this field based on the co-citation analysis. Similarly, we conducted a bibliographic coupling analysis for the documents with 10 or more citations. This threshold was met by 115 documents (out of 425 retrieved documents) but 17 documents weren’t bibliographically coupled. The bibliographic coupling rainbow density map of the remaining 98 documents is illustrated in Supplementary Fig. S9 (Online Resource 9).
Fig. 6.
The rainbow density mapping of co-citation analysis of the journals publishing at least 5 Iranian microbiota articles. The journal co-citation analysis calculates the frequency of two journals’ articles that are co-cited by other articles. The higher co-citation of journal (from blue to red color) indicates that journal has higher semantic relationship with the field of research e.g. ‘Plos one’ and ‘nature’ are the prominent journals in this field
Discussion
The current study provides the descriptive bibliometric information about the Iranian microbiota research roadmap and as far as we know, this is the first bibliometric and scientometric analysis of the Iranian microbiota articles. This study reveals that there has been an almost permanent increase in the Iranian scientific production in the field of microbiota since 2012 and interestingly, almost two-thirds of all documents have been published during the last three years (65.64% of all documents) which indicates that this uprising area of literature has recently attracted Iranian researchers and institutions’ interest. Besides, we also found that almost half of these documents are in the area of “Medicine” that could be concluded that increasing scientific attempts were performed to identify the effects of various microbiota modulating interventions on the human health. Simultaneously, the chronological analysis of the retrieved articles demonstrated that in the late 2000s, Iranian researchers’ attention was drawn to the role of microbiota in the medical sciences; this finding can be explained by the growing global awareness of microbiota role on human life as well as understanding the metagenomics analysis of the human intestinal microbiota and also new discoveries in the efficacy of intestinal microbiota changes for diseases like inflammatory bowel disease and obesity [8, 17, 59, 60]. Also, we demonstrated that the medical sciences literature has focused on the relationships between microbiota modulating interventions (especially synbiotics and probiotics) and digestive and metabolic-related disorders like obesity, non-alcoholic fatty liver disease, metabolic syndrome, and type 2 diabetes. The country analysis shows that North America and Western Europe play an effective role in collaboration with Iranian institutions and also, the most collaboration belongs to the United State of America, whose high gross domestic product (GDP) results in high scientific co-contributions in this field [61]. It is noteworthy that the subject area evaluation of the most active institution (Tehran University of Medical Sciences), as well as the most productive journal (Microbial Pathogenesis), points out that prominently the Iranian microbiota documents have been accomplished in the area of medical sciences; that this finding is consistence with the mentioned bibliographic statistics. Besides, we discovered that the most repetitive specialized terms in these documents are ‘effect, diet, patient, treatment, bacterium, and probiotic’ that the overlay mapping analysis reveals that the terms ‘prebiotic, metabolic syndrome, inflammation, and gut microbiota’ are the hotspot of the recent studies; and the most repetitive specialized author keywords are ‘gut microbiota, intestinal microbiota, probiotics, synbiotic, prebiotic, and obesity’ that the recent research foci are on the keywords ‘immune system, dysbiosis, gut microbiota, depression, obesity, Lactobacillus, Bifidobacterium’; these findings insist that the foci of attention in the literature have been paid to the therapeutic effect of pro/pre/synbiotic products in diet on human health/diseases like obesity through the changing of intestinal microbiota composition. Probiotics are live microorganisms that if administered in an adequate quantity, promote the health of the host [62]. There are various probiotic products in the world, including dairy, cereal, fruit juice, and chocolate; while the following four probiotic dairy products are common in Iran: cheese, ice cream, yogurt, and dough. The primary microorganisms commonly used in the probiotics are lactic acid bacteria (LAB) that the two species seem to have beneficial effects on human health: BifidiBacterium lactis Bb12 and Lactobacillus acidophilus La5 [63–65]. On the other hand, prebiotic products provoke the growth of specific genus or species of intestinal microbiota that results in the host health improvement [66, 67]; and synbiotic products made up of prebiotics and probiotics combination [68]. Therefore, pro/pre/synbiotics influence on the host health/diseases through the modification of intestinal microbiota. As demonstrated in density map of terms/author keywords analysis as well as the bipartite graph of the interventions and diseases, obesity, probiotics, and synbiotics are the research focus in the Iranian microbiota literature; in addition, the worldwide scientific publications on the association between obesity and intestinal microbiome composition have been dramatically increased during the last decade [69] and the findings in this field shows obese population in comparison to the lean population have less intestinal microbiota diversity and also the proportion of various intestinal bacteria in obese people has been changed as follows: Bacteroidetes phylum has been decreased while Actinobacteria and Firmicutes phyla have been increased; also, it elucidated that during the weight loss, the Bacteroidetes proportion in the intestinal microbiota population has been increased [70–72]. Also, a cross-sectional study among Iranian school-aged children revealed that Bacteroidetes has been decreased in the gasterointestinal tract of obese children whereas the amount of Firmicutes has been increased [42]. Also, the experimental study of the obese and lean mice reported that the intestinal microbiota composition of obese mice leads to an elevated capacity to gather energy from diet and also, feacal microbiota transplantation of obese mice produces more body fat in germ-free mice than feacal microbiota transplantation of lean mice that these findings support the role of intestinal microbiota in the pathogenesis of obesity [73]. In addition, a clinical trial reported that the consumption of oleoylethanolamide supplement increases the quantity of Akkermansia muciniphila bacterium in the gastrointestinal tract of obese population and subsequently results in less dietary intake [44]. However, another clinical trial revealed that although the consumption of metformin in obese non-diabetic women causes weight loss, the gut microbiota composition after treatment remains almost unchanged [43]. Therefore, these findings shows that we need more clinical trials to discover the possible role of various gut microbiota on pathogenesis of obesity.
The NIH group has implemented the human microbiome project (HMP) program since 2007 and had dedicated $215 M to this project during ten years to provide a broad perspective of human microbiome diversity, microbiome sequences, new advanced technologies and computational tools, the dynamic interactions between microbiota and the human body and their consequences. Successfully, the HMP has provided a comprehensive practical central resource of human microbial genome sequences using 16 s rRNA sequencing and whole-genome shotgun metagenomics sequencing techniques; also, this extensive project has resulted in better understanding of the effects of human-related microbiome change on the well-being and health of the host, particularly in various health-related conditions. Also, the NIH reported that the most awards had been dedicated to understanding the microbiota role in the infectious/parasitic diseases, followed by the digestive diseases and the neoplasms and also the gastrointestinal tract was the main focuses of this project [8, 17, 18]. In the lately accomplished second phase of HMP, the NIH has focused on the differences of microbiome diversity in/on human body, genome sequences of microbiome and host, metabolomics and protein profiles of microbiome and host, as well as the changes occurred in homeostasis between microbiota and host body under three health-related conditions, including gestation and preterm labor (based on the vaginal microbiome), inflammatory bowel diseases (based on the intestinal microbiome), and prediabetes (based on the intestinal and nasal microbiome) [74]. Accordingly, the role of microbiome on well-being and health-related conditions of individuals has received more attention than ever before. With regard to the specific microbial community of each geographical region, the developing countries like Iran should move toward understanding their microbial communities and revealing the microbiome composition changes under different health-related conditions and consequently avoid wasting their resources and reduce the costs. The current study established the practical extensive outlook of Iranian microbiota research and highlighted the hotpots of bibliographic information, so other scientists could find the research foci and gaps in the Iranian microbiota literature and move in the direction of Iranian microbiota roadmap.
It is noteworthy that we retrieved all documents indexed in the Scopus database according to our search strategy that unfortunately, we may miss the related documents from other databases that are not indexed in the Scopus. However, it must keep in mind that the Scopus database has an acceptable coverage of peer-reviewed journals and publishers and also Scopus in comparison to Web of Science, another database used widely for bibliometric analysis, retrieved more citations per document [75, 76]. So the results of the current analysis will be reliable and this study can accurately demonstrate the bibliometric and scientometric features of the Iranian publications in the field of microbiota.
Conclusions
In the current study, we provide the bibliometric and scientometric characteristics of all Iranian microbiota documents that had been published before 7 September 2019 that illustrate microbiota research has received a lot of attention among Iranian scientists during the last decade; and the Iranian research focus has been moved toward the understanding of the microbiota role in the medical sciences, specifically the efficacy of the microbiota modulating products like pro/pre/synbiotics for the host health. Also, we ranked the performance of the involved institutions, journals, sponsors, authors, and countries; and visualized the hotspot of the most repetitive terms and author keywords using in the retrieved documents; therefore, we prepare practical guiding information for other researchers to move the boundaries of science and policymakers to ameliorate the human health and diseases.
Electronic supplementary material
(DOCX 1319 kb)
Acknowledgements
Not applicable.
Authors’ contributions
H.A., H.D., H.E., F.R. and P.P. conceived of the presented idea. S.D.S., B.L. and P.P. developed the theory and H.A. performed the computations. N.F., A.S., S.A.B, and S.H.R. verified the analytical methods. H.E. investigated the molecular technique aspect of retrieved articles, H.E., H.A., and H.D. contributed to the interpretation of the results and S.D.S supervised the findings of this work. H.D. and H.A. wrote the manuscript in consultation with H.E. and N.F. All authors discussed the results, provided critical feedback, helped shape the research and analysis, and contributed to the final manuscript.
Funding information
This study was funded by the Endocrinology and Metabolism Research Institute, Tehran University of Medical Sciences, Tehran, Iran.
Data availability
Not applicable.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hanieh-Sadat Ejtahed, Email: Haniejtahed@yahoo.com.
Seyed-Davar Siadat, Email: d.siadat@gmail.com.
References
- 1.Sokolov-Mladenović S, Cvetanović S, Mladenović I. R&D expenditure and economic growth: EU28 evidence for the period 2002–2012. Econ Res-Ekonomska istraživanja. 2016;29(1):1005–1020. [Google Scholar]
- 2.Sharifi V, Rahimi Movaghar A, Mohammadi M, Goodarzi R, Izadian E, Farhoudian A et al. Analysis of mental health research in the Islamic Republic of Iran over 3 decades: a scientometric study. 2008. [PubMed]
- 3.Ataie-Ashtiani B. Chinese and Iranian scientific publications: fast growth and poor ethics. Sci Eng Ethics. 2017;23(1):317–319. doi: 10.1007/s11948-016-9766-1. [DOI] [PubMed] [Google Scholar]
- 4.Akhondzadeh S. Iranian science shows world's fastest growth: ranks 17th in science production in 2012. Avicenna J Med Biotechnol. 2013;5(3):139. [PMC free article] [PubMed] [Google Scholar]
- 5.Agarwal A, Durairajanayagam D, Tatagari S, Esteves SC, Harlev A, Henkel R, Roychoudhury S, Homa S, Puchalt NG, Ramasamy R, Majzoub A, Ly KD, Tvrda E, Assidi M, Kesari K, Sharma R, Banihani S, Ko E, Abu-Elmagd M, Gosalvez J, Bashiri A. Bibliometrics: tracking research impact by selecting the appropriate metrics. Asian J Androl. 2016;18(2):296–309. doi: 10.4103/1008-682X.171582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Zou Z, Bian X, Huang Y, Wang Y, Yang C, et al. Fecal microbiota transplantation research output from 2004 to 2017: a bibliometric analysis. PeerJ. 2019;7:e6411. doi: 10.7717/peerj.6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pritchard A. Statistical bibliography or bibliometrics. J Doc. 1969;25(4):348–349. [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baudoin L, Sapinho D, Maddi A, Miotti L. Scientometric analysis of the term “microbiota” in research publications (1999-2017): A second youth of a century-old concept. FEMS Microbiol Lett. 2019;366(12). [DOI] [PubMed]
- 10.The unseen life of the soil. Science. 1927;65(1695):x–x. 10.1126/science.65.1695.0x. [DOI] [PubMed]
- 11.Crawford JJ, Shankle RJ. Application of newer methods to study the importance of root canal and oral microbiota in endodontics. Oral Surg Oral Med Oral Pathol. 1961;14(9):1109–1123. doi: 10.1016/0030-4220(61)90505-9. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons R, Socransky S, Sawyer S, Kapsimalis B, MacDonald J. The microbiota of the gingival crevice area of man—II: the predominant cultivable organisms. Arch Oral Biol. 1963;8(3):281–289. doi: 10.1016/0003-9969(63)90020-7. [DOI] [PubMed] [Google Scholar]
- 13.Socransky S, Gibbons R, Dale A, Bortnick L, Rosenthal E, Macdonald J. The microbiota of the gingival crevice area of man—I: Total microscopic and viable counts and counts of specific organisms. Arch Oral Biol. 1963;8(3):275–280. doi: 10.1016/0003-9969(63)90019-0. [DOI] [PubMed] [Google Scholar]
- 14.Mann S, Masson FM, Oxford A. Effect of feeding aureomycin to calves upon the establishment of their normal rumen microflora and microfauna. Br J Nutr. 1954;8(3):246–252. doi: 10.1079/bjn19540037. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin R, Wood W, Emery R. Conversion of glucose-C14 to propionate by the rumen microbiota. J Bacteriol. 1963;85(6):1346–1349. doi: 10.1128/jb.85.6.1346-1349.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mraz O, Cerny L, editors. Functional Composition+ Main Metabolites of Rumen Microflora in Cattle with Definite Food Intake. FOLIA MICROBIOLOGICA; 1964: INST MICROBIOLOGY, VIDENSKA 1083, PRAGUE 4 142 20, CZECH REPUBLIC.
- 17.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH human microbiome project. Genome Res. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regan NHMPATlpngLPJLAMPDDXRFLBRRPMK A review of 10 years of human microbiome research activities at the US National Institutes of Health, Fiscal Years 2007–2016. Microbiome. 2019;7:1–19. doi: 10.1186/s40168-019-0620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Mahony SM, Clarke G, Borre Y, Dinan T, Cryan J. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res. 2015;277:32–48. doi: 10.1016/j.bbr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 20.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4(137):137rv5-rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collado MC, Cernada M, Baüerl C, Vento M, Pérez-Martínez G. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes. 2012;3(4):352–365. doi: 10.4161/gmic.21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bar-Ilan J. Citations to the “Introduction to informetrics” indexed by WOS, Scopus and Google Scholar. Scientometrics. 2010;82(3):495–506. [Google Scholar]
- 24.Van Eck N, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2009;84(2):523–538. doi: 10.1007/s11192-009-0146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Team S. Science of science (Sci2) tool. Indiana University and SciTech Strategies. 2009.
- 26.Nazemi M, Amini M, Salehi R. Difference in quantities of bifidobacteria from the intestinal microflora of individuals with type 2 diabetes and healthy individuals from Iran. Journal of Isfahan Medical School. 2013;31(247):1216–1225.
- 27.Nasrollahzadeh D, Malekzadeh R, Ploner A, Shakeri R, Sotoudeh M, Fahimi S, et al. Variations of gastric corpus microbiota are associated with early esophageal squamous cell carcinoma and squamous dysplasia. Sci Rep. 2015;5:8820. doi: 10.1038/srep08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heidarian F, Noormohammadi Z, Aghdaei HA, Alebouyeh M. Relative abundance of streptococcus spp. and its association with disease activity in inflammatory bowel disease patients compared with controls. Arch Clin Infect Dis. 2017;12(2) e57291.
- 29.Nabizadeh E, Jazani NH, Bagheri M, Shahabi S. Association of altered gut microbiota composition with chronic urticaria. Ann Allergy Asthma Immunol. 2017;119(1):48–53. doi: 10.1016/j.anai.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Navab-Moghadam F, Sedighi M, Khamseh ME, Alaei-Shahmiri F, Talebi M, Razavi S, Amirmozafari N. The association of type II diabetes with gut microbiota composition. Microb Pathog. 2017;110:630–636. doi: 10.1016/j.micpath.2017.07.034. [DOI] [PubMed] [Google Scholar]
- 31.Sedighi M, Razavi S, Navab-Moghadam F, Khamseh ME, Alaei-Shahmiri F, Mehrtash A, Amirmozafari N. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb Pathog. 2017;111:362–369. doi: 10.1016/j.micpath.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 32.Zamani S, Shariati SH, Zali MR, Aghdaei HA, Asiabar AS, Bokaie S, et al. Detection of enterotoxigenic Bacteroides fragilis in patients with ulcerative colitis. Gut Pathog. 2017;9(1):53. doi: 10.1186/s13099-017-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghavami SB, Rostami E, Sephay AA, Shahrokh S, Balaii H, Aghdaei HA, et al. Alterations of the human gut Methanobrevibacter smithii as a biomarker for inflammatory bowel diseases. Microb Pathog. 2018;117:285–289. doi: 10.1016/j.micpath.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 34.Rezasoltani S, Aghdaei HA, Dabiri H, Sepahi AA, Modarressi MH, Mojarad EN. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb Pathog. 2018;124:244–249. doi: 10.1016/j.micpath.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 35.Rezasoltani S, Sharafkhah M, Aghdaei HA, Mojarad EN, Dabiri H, Sepahi AA, et al. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J Microbiol Methods. 2018;155:82–88. doi: 10.1016/j.mimet.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Al-Bayati L, Fasaei BN, Merat S, Bahonar A. Longitudinal analyses of gut-associated bacterial microbiota in ulcerative colitis patients. Arch Iran Med (AIM). 2018;21(12). [PubMed]
- 37.Tavasoli S, Alebouyeh M, Naji M, Shakiba majd G, Shabani Nashtaei M, Broumandnia N, et al. The association of the intestinal oxalate degrading bacteria with recurrent calcium kidney stone formation and hyperoxaluria: a case-control study. BJU Int. 2019;125(1):133–143. [DOI] [PubMed]
- 38.Mohammadzadeh N, Kalani BS, Bolori S, Azadegan A, Gholami A, Mohammadzadeh R, et al. Identification of an intestinal microbiota signature associated with hospitalized patients with diarrhea. Acta Microbiol Immunol Hung. 2019:1–14. [DOI] [PubMed]
- 39.Heidarian F, Alebouyeh M, Shahrokh S, Balaii H, Zali MR. Altered fecal bacterial composition correlates with disease activity in inflammatory bowel disease and the extent of IL8 induction. Curr Res Transl Med. 2019;67(2):41–50. doi: 10.1016/j.retram.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Moossavi S, Engen PA, Ghanbari R, Green SJ, Naqib A, Bishehsari F, Merat S, Poustchi H, Keshavarzian A, Malekzadeh R. Assessment of the impact of different fecal storage protocols on the microbiota diversity and composition: a pilot study. BMC Microbiol. 2019;19(1):145. doi: 10.1186/s12866-019-1519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nami Y, Haghshenas B, Khosroushahi AY. Molecular identification and probiotic potential characterization of lactic acid Bacteria isolated from human vaginal microbiota. Adv Pharm Bull. 2018;8(4):683–695. doi: 10.15171/apb.2018.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mousavi SH, Mehrara S, Barzegari A, Ostadrahimi A. Correlation of gut microbiota profile with body mass index among school age children. Iran Red Crescent Med J. 2018;20(4):e58049 (In Press).
- 43.Ejtahed H-S, Tito RY, Siadat S-D, Hasani-Ranjbar S, Hoseini-Tavassol Z, Rymenans L, et al. Metformin induces weight loss associated with gut microbiota alteration in non-diabetic obese women: a randomized double-blind clinical trial. Eur J Endocrinol. 2018;180(3):165–176(aop). [DOI] [PubMed]
- 44.Payahoo L, Khajebishak Y, Alivand MR, Soleimanzade H, Alipour S, Barzegari A, et al. Investigation the effect of oleoylethanolamide supplementation on the abundance of Akkermansia muciniphila bacterium and the dietary intakes in people with obesity: a randomized clinical trial. Appetite. 2019;141:104301. doi: 10.1016/j.appet.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 45.Laffin MR, Tayebi Khosroshahi H, Park H, Laffin LJ, Madsen K, Kafil HS, et al. Amylose resistant starch (HAM-RS2) supplementation increases the proportion of Faecalibacterium bacteria in end-stage renal disease patients: microbial analysis from a randomized placebo-controlled trial. Hemodial Int. 2019;23(3):343–347. [DOI] [PubMed]
- 46.Ahmadi S, Nagpal R, Wang S, Gagliano J, Kitzman DW, Soleimanian-Zad S, Sheikh-Zeinoddin M, Read R, Yadav H. Prebiotics from acorn and sago prevent high-fat-diet-induced insulin resistance via microbiome–gut–brain axis modulation. J Nutr Biochem. 2019;67:1–13. doi: 10.1016/j.jnutbio.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hosseinifard E-S, Morshedi M, Bavafa-Valenlia K, Saghafi-Asl M. The novel insight into anti-inflammatory and anxiolytic effects of psychobiotics in diabetic rats: possible link between gut microbiota and brain regions. Eur J Nutr. 2019;58(01):1–15. [DOI] [PubMed]
- 48.Ghazifard A, Kasra-Kermanshahi R, Far ZE. Identification of thermophilic and mesophilic bacteria and fungi in Esfahan (Iran) municipal solid waste compost. Waste Manag Res. 2001;19(3):257–261. doi: 10.1177/0734242X0101900307. [DOI] [PubMed] [Google Scholar]
- 49.Seidavi A, Mirhosseini SZ, Shivazad M, Chamani M, Sadeghi AA. The development and evaluation of a duplex polymerase chain reaction detection of Bifidobacterium spp. and Lactobacillus spp. in duodenum, jejunum, ileum and cecum of broilers. J Rapid Meth Aut Microbiol. 2008;16(1):100–112. [Google Scholar]
- 50.Ghiyasi M, Rezaei M, Sayyahzadeh H, Firouzbakhsh F, Attar A. Effects of prebiotic (Fermacto) in low protein diet on some blood parameters and intestinal microbiota of broiler chicks. Ital J Anim Sci. 2008;7(3):313–320. [Google Scholar]
- 51.Jajarm H, Jahanbin A, Mokhber N, Gooyandeh S, Mansourian A, Beitollahi J. Effects of persica mouthwash on oral microbiota of cleft lip and palate patients during fixed orthodontic treatment. J Appl Sci. 2009;9:1593–1596. [Google Scholar]
- 52.Henderson G, Cox F, Ganesh S, Jonker A, Young W, Collaborators GRC, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. 2015;5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Llewellyn MS, Boutin S, Hoseinifar SH, Derome N. Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front Microbiol. 2014;5:207. doi: 10.3389/fmicb.2014.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davari S, Talaei SA, Alaei H. Probiotics treatment improves diabetes-induced impairment of synaptic activity and cognitive function: behavioral and electrophysiological proofs for microbiome–gut–brain axis. Neuroscience. 2013;240:287–296. doi: 10.1016/j.neuroscience.2013.02.055. [DOI] [PubMed] [Google Scholar]
- 55.Bishehsari F, Mahdavinia M, Vacca M, Malekzadeh R, Mariani-Costantini R. Epidemiological transition of colorectal cancer in developing countries: environmental factors, molecular pathways, and opportunities for prevention. World J Gastroenterol: WJG. 2014;20(20):6055–6072. doi: 10.3748/wjg.v20.i20.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaghef-Mehrabany E, Alipour B, Homayouni-Rad A, Sharif S-K, Asghari-Jafarabadi M, Zavvari S. Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis. Nutrition. 2014;30(4):430–435. doi: 10.1016/j.nut.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 57.Rastmanesh R. High polyphenol, low probiotic diet for weight loss because of intestinal microbiota interaction. Chem Biol Interact. 2011;189(1–2):1–8. doi: 10.1016/j.cbi.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 58.Homayouni A, Bastani P, Ziyadi S. Mohammad-Alizadeh-Charandabi S, Ghalibaf M, Mortazavian AM et al. effects of probiotics on the recurrence of bacterial vaginosis: a review. J Low Genit Tract Dis. 2014;18(1):79–86. doi: 10.1097/LGT.0b013e31829156ec. [DOI] [PubMed] [Google Scholar]
- 59.Huang X, Fan X, Ying J, Chen S. Emerging trends and research foci in gastrointestinal microbiome. J Transl Med. 2019;17(1):67. doi: 10.1186/s12967-019-1810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao H, Wan J-Y, Wang C-Z, Li L, Wang J, Li Y, et al. Bibliometric analysis of research on the role of intestinal microbiota in obesity. PeerJ. 2018;6:e5091. doi: 10.7717/peerj.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho YA, Kim J. Effect of probiotics on blood lipid concentrations: a meta-analysis of randomized controlled trials. Medicine. 2015;94(43) e1714. [DOI] [PMC free article] [PubMed]
- 63.Sarao LK, Arora M. Probiotics, prebiotics, and microencapsulation: a review. Crit Rev Food Sci Nutr. 2017;57(2):344–371. doi: 10.1080/10408398.2014.887055. [DOI] [PubMed] [Google Scholar]
- 64.Mahmoudi R, Fakhri O, Farhoodi A, Kaboudari A, Rahimi SF. A review on probiotic dairy products as functional foods reported from Iran. Int J Food Nutr Saf. 2015;6(1):2. [Google Scholar]
- 65.Ejtahed H-S, Angoorani P, Soroush A-R, Atlasi R, Hasani-Ranjbar S, Mortazavian AM, et al. Probiotics supplementation for the obesity management; a systematic review of animal studies and clinical trials. J Funct Foods. 2019;52:228–242. [Google Scholar]
- 66.Roberfroid M, Gibson GR, Hoyles L, McCartney AL, Rastall R, Rowland I, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr. 2010;104(S2):S1–S63. doi: 10.1017/S0007114510003363. [DOI] [PubMed] [Google Scholar]
- 67.Ejtahed H-S, Soroush A-R, Siadat S-D, Hoseini-Tavassol Z, Larijani B, Hasani-Ranjbar S. Targeting obesity management through gut microbiota modulation by herbal products: a systematic review. Complement Ther Med. 2018;42:184–204. [DOI] [PubMed]
- 68.Anderson A, McNaught C, Jain P, MacFie J. Randomised clinical trial of synbiotic therapy in elective surgical patients. Gut. 2004;53(2):241–245. doi: 10.1136/gut.2003.024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ejtahed H-S, Tabatabaei-Malazy O, Soroush A-R, Hasani-Ranjbar S, Siadat S-D, Raes J, Larijani B. Worldwide trends in scientific publications on association of gut microbiota with obesity. Iran J Basic Med Sci. 2019;22(1):65–71. doi: 10.22038/ijbms.2018.30203.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 72.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 74.Integrative H. The Integrative human microbiome project. Nature. 2019;569(7758):641. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kulkarni AV, Aziz B, Shams I, Busse JW. Comparisons of citations in web of science, Scopus, and Google scholar for articles published in general medical journals. Jama. 2009;302(10):1092–1096. doi: 10.1001/jama.2009.1307. [DOI] [PubMed] [Google Scholar]
- 76.Mongeon P, Paul-Hus A. The journal coverage of web of science and Scopus: a comparative analysis. Scientometrics. 2016;106(1):213–228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1319 kb)
Data Availability Statement
Not applicable.