Abstract
Genetic transformation is one of the most important technologies for revealing or modulating gene function. It is used widely in both functional genomics and molecular breeding of rice. Demands on its use in wild Oryza species is increasing because of their high genetic diversity. Given the difficulties in genetic crosses between distantly related species, genetic transformation offers a way to alter or transfer genetic traits in wild rice accessions. However, transformation of wild Oryza accessions by conventional methods using calli induced from scutellum tissue of embryos in mature seeds often fails. Here, we report methods using immature embryos for the genetic transformation of a broad range of Oryza species. First, we investigated the ability of callus induction and regeneration from immature embryos of 192 accessions in 20 species under several culture conditions. We regenerated plants from immature embryos of 90 accessions in 16 species. Next, we optimized the conditions of Agrobacterium infection using a vector carrying the GFP gene driven by the maize ubiquitin promoter. GFP signals were observed in 51 accessions in 11 species. We analyzed the growth and seed set of transgenic plants of O. barthii, O. glumaepatula, O. rufipogon, and O. brachyantha. The plants grew to maturity and set seeds normally. Southern blot analyses using DNA from T0 plants showed that all GFP plants were derived from independent transformation events. We confirmed that the T-DNAs were transmitted to the next generation through the segregation of GFP signals in the T1 generation. These results show that many Oryza species can be transformed by using modified immature-embryo methods. This will accelerate the use of wild Oryza accessions in molecular genetic analyses and molecular breeding.
Keywords: Agrobacterium-mediated transformation, Immature embryo, Oryza, Wild accession
Background
Rice (Oryza sativa L.) is one of the most important crops in the world. Breeding new rice cultivars with strong tolerance to environmental changes caused by global warming is an urgent goal. Because wild Oryza species have high genetic diversity and thus have better chances of being adapted to fluctuating environments, their use in basic science and breeding is increasing (Morishima and Oka 1970; Vaughan 1994). Wild Oryza accessions have been studied to analyze their drought tolerance (Zhang et al. 2006), shattering (Ishii et al. 2013), awn formation (Ikemoto et al. 2017), zinc accumulation (Ishikawa et al. 2017), salt tolerance (Yichie et al. 2018), yield (Fan et al. 2019), and disease resistance (Kariya et al. 2019). These studies identified genes regulating potential agriculturally important traits. The accumulation of whole genome sequences of wild Oryza species (Oryzabase, https://shigen.nig.ac.jp/rice/oryzabase/; Gramene, http://www.gramene.org) provides opportunities in both basic and applied studies (Stein et al. 2018; Zhao et al. 2018; Shenton et al. 2020).
Genetic transformation is one of the most important technologies for revealing or modulating gene function, and is often used in functional genomics and molecular breeding. Several methods for transferring genes into rice (Shimamoto et al. 1989) have been developed, including particle bombardment (Toriyama et al. 1988), polyethylene glycol-mediated (Zhang and Wu 1988), and Agrobacterium-mediated methods (Potrykus 1991; Hiei et al. 1994). Through the improvement of Agrobacterium-mediated methods, transformation efficiency has become high, and transgenes with single or low copy numbers or large DNA segments with defined ends can be integrated into host genomes (Hiei et al. 1994). Agrobacterium-mediated transformation in rice uses two main types of starting materials: calli induced from scutellum tissue of embryos in mature seeds, and immature embryos at approximately 10 days after fertilization. The former method is more convenient because we can collect and stock enough mature seeds to run experiments at any time. The latter method can be applied to a much wider range of cultivated rice (Hiei and Komari 2008) and is applicable to various crop species, including maize (Ishida et al. 1996), wheat (Ishida et al. 2005), and sorghum (Sato-Izawa et al. 2018). Thus, the immature embryo method may be applicable to wild Oryza species, for which the mature embryo method is often unsuccessful.
Oryza comprises two cultivated species (O. sativa and O. glaberrima) and 21 wild species (Vaughan 1989, 1994) with 9 genome types—AA, BB, CC, BBCC, CCDD, EE, FF, GG, and HHJJ—according to cytogenetic observations of interspecific F1 hybrids and total genomic DNA hybridization (Khush et al. 2001). The two cultivated species have the AA genome. The introduction of agronomically useful genes from wild accessions into cultivated species is highly valuable. However, the introduction of those genes from wild accessions with the AA genome into cultivated species by genetic crosses followed by repeated backcrosses is time consuming and labor intensive. In addition, genes linked to genes for reproductive barriers or for undesirable characters are difficult to introduce alone by crossing. The introduction of genes from wild accessions with other genomes by genetic crosses is even harder. Usually, crosses between different genome types do not produce stable offspring. In some cases, aneuploid plants can be obtained but their chromosome composition is unstable and not healthy. Genetic transformation offers promise in the transfer of genes from wild accessions, especially with genomes other than AA, and allows the functional analysis of genes in wild Oryza accessions. The Japanese National Institute of Genetics (NIG) holds in Oryzabase and distributes more than 1700 accessions of wild Oryza species. Here, we used the immature embryo method for the genetic transformation of wild rice accessions from a broad range of species with various media and Agrobacterium infection conditions and show that many Oryza species can be transformed by the immature embryo method.
Materials and Methods
Plant Materials
Seeds of wild Oryza species were supplied by NIG. Plants were grown under natural light in a greenhouse and transplanted into a paddy field in June in Mishima, Japan. At 1 month after transplanting, daylength was controlled 12.5 h light / 11.5 h dark to induce flowering. Oryza sativa ‘Nipponbare’ was grown as a control in similar conditions but under natural daylength. At 8 to 10 days after flowering, immature embryos were isolated.
Plant Regeneration from Immature Embryos
Regeneration tests using immature embryos were conducted according to Hiei and Komari (2008). All culture media are detailed in Supplemental Table 1. Ovaries were sterilized in 70% ethanol (v/v) for 1 min, followed by 15 min in 50% (v/v) sodium hypochlorite solution with shaking, and washed 5 times in sterile distilled water. Hiei and Komari (2008) used two types of callus-inducing media, nN6C and CCMC, to induce calli from japonica and indica cultivars, respectively. The immature embryos were isolated under a stereomicroscope on a clean bench, placed on nN6C or CCMC medium containing 25 mg/L meropenem, and cultured at 32 °C for 7 days. Then they were transferred to the same fresh media and cultured for another 7 days under the same conditions. Next, we tested regeneration of shoots and roots in four combinations of media, designated A to D (Supplemental Table 2). Calli formed on the immature embryos were transferred to either regeneration medium (N6R) or pre-regeneration medium (NBPRC). On N6R, calli were cultured at 32 °C for 2 weeks, and regenerated plantlets were transferred to rooting medium (N6F). On NBPRC, calli were cultured at 32 °C for 1 week, and were then transferred to regeneration medium (RNM). After 2 weeks on RNM, regenerated plantlets were transferred to rooting medium (MSI). Plants which formed roots on N6F or MSI were eventually transferred to soil in pots and grown to maturity in the greenhouse.
Binary Vectors and Transformation of Agrobacterium tumefaciens
The binary plasmid vector (pPUG1–1) used in this study contains a hygromycin phosphotransferase (hpt) gene under the control of the cauliflower mosaic virus 35S promoter, and a green fluorescent protein (GFP) gene under the control of the maize ubiquitin promoter. The vector was transferred into Agrobacterium tumefaciens strains LAB4404 and EHA105 by electroporation in an E. coli pulser (Bio-Rad).
Agrobacterium-Mediated Transformation
Agrobacterium-mediated transformation was conducted according to Hiei and Komari (2008). All media included 25 mg/L meropenem instead of cefotaxime and carbenicillin. Strain LBA4404 or EHA105 harboring pPUG1–1 was grown on LB medium supplemented with 50 mg/L hygromycin at 28 °C for 3 days. The immature embryos were held in a water bath at 43 °C for 30 min, and then cooled in water. They were then centrifuged in an angle rotor at either 1100× or 20,000×g for 10 min at 22 °C. They were placed on co-cultivation medium containing 100 μM acetosyringone with the scutellum side up, and 5 μL of Agrobacterium suspension was dropped on them. After incubation for 7 days at 25 °C in the dark, plants were regenerated as described in the previous section, with the addition of hygromycin into all media for selection.
Observation of GFP Fluorescence
GFP fluorescence was observed under a fluorescence microscope with a GFP filter (SZX16, Olympus).
Extraction of Rice Genomic DNA and Southern Blot Analysis
Total genomic DNA was extracted from ca. 1 g of fresh leaf of each transgenic line with a DNeasy Plant Maxi Kit (Qiagen) according to the manufacturer’s protocol. PCR analysis to detect the full-length hpt gene was performed using GoTaq DNA polymerase (Promega) with the primer set 5′-ATGAAAAAGCCTGAACTCACCGCG-3′ / 5′-CTATTCCTTTGCCCTCGGACGAGT-3′.
Southern blot analysis was performed with an ECL Direct Nucleic Acid Labelling and Detection System (GE Healthcare) according to the manufacturer’s protocol. Genomic DNA (5 μg) was digested by HindIII (Takara) and separated by electrophoresis. For the preparation of hpt probe, the full-length hpt gene was amplified with the primers used for PCR analysis above. Southern blot hybridization signals were detected on an Image Quant LAS 4000 Mini imager (GE Healthcare).
Results and Discussion
Optimization of Culture Media for Regeneration of Plantlets from Immature Embryos from Wild Oryza Accessions
First we tested the response of immature embryos to callus induction. From 192 accessions in 20 wild Oryza species, 138 accessions in 19 species formed callus on either or both media (Table 1). Using these 138 accessions, we tested the regeneration of shoots and roots in conditions A and B, and then using accessions which did not respond to either, in conditions C and D. Twenty-eight accessions regenerated from immature embryo calli in conditions A and B, 18 only in condition A, and 38 only in condition B (Table 2). Of 54 accessions which did not respond to A or B, 4 regenerated under C and 2 under D (Table 2).
Table 1.
Callus induction from immature embryos of wild Oryza accessions on different callus-inducing media
| Type of medium | nN6C, CCMC | nN6C | CCMC | No callus induced on either |
|---|---|---|---|---|
| O. barthii, AA | W1473, W1583 | W1467, W1605, W1643 | W0747, W1588, W1702 | W1063, W1416 |
| O. glumaepatula, AA | W1189, W1191, W2165, W2184, W2203 | W1167, W1169, W1477, W2145, W2160, W2201 | W1183, W1185, W2140, W2149, W2173, W2192 | W1187 |
| O. longistaminata, AA | W1573 | |||
| O. meridionalis, AA | W1635, W2100, W2112 | W1297, W1300, W1631, W1638 | W1625, W2071, W2077, W2080, W2081, W2105 | W2079, W2103 |
| O. rufipogon, AA | W0107, W0593, W0625, W1230, W1239, W1294, W1551, W1685, W1852, W1865, W1965, W2014, W2265 | W0629, W0630, W1235, W1739, W1807, W1866, W1944, W1962, W2003, W2051 | W0102, W0106, W0137, W0610, W0621, W1114, W1666, W1669, W1681, W1690, W1718, W1921, W1945, W2078, W2109, W2263 | W0120, W0128, W1802, W1825, W2050 |
| O. punctata (2x), BB | W1515, W1582, W1586, W1593 | W1514, W1577, W1590, W1592 | ||
| O. eichingeri, CC | W1525 | W1526 | ||
| O. officinalis, CC | W1315 | W1291 | W0002, W0614, W1301, W1830, W1930 | W0065, W0566, W1131, W1200, W1252, W1302, W1308, W1361, W1814 |
| O. rhizomatis, CC | W1805, W1812 | W1808 | ||
| O. minuta, BBCC | W1323 | W0016, W1213 | W0051, W1318, W1319, W1329, W1331, W1336, W1342 | W0045, W1328 |
| O. punctata (4x), BBCC | W1474(B) | W1024, W1408, W1564 | W0043, W1023, W1145, W1409 | |
| O. alta, CCDD | W0017, W0018 | W1147 | ||
| O. grandiglumis, CCDD | W1483 | W1480(B), W2220 | W0613, W1195, W1476 | W1247 |
| O. latifolia, CCDD | W1177, W1197 | W0047, W1539 | W0048, W0542, W1166, W1168, W1181, W1184 | |
| O. australiensis, EE | W0008, W1639 | W1632, W2082, W2084 | W1296, W1628, W1630, W2086, W2104 | |
| O. brachyantha, FF | W1401 | W0654 | W0656, W1407(B), W1706, W1711 | W1403 |
| O. granulata, GG | W0003 | W0067(B) | W0004, W0615 | |
| O. meyeriana, GG | W2068 | W1348 | ||
| O. longiglumis, HHJJ | W1227 | W1218, W1219, W1220, W1222, W1223, W1224, W1228, W1229 | ||
| O. ridleyi, HHJJ | W0001, W2033, W2035 | |||
| Sub-subtotal | 40 | 43 | 55 | 54 |
| Subtotal | 138 | |||
| Total | 192 | |||
Table 2.
Regeneration from immature embryos of wild Oryza accessions on 4 medium combinations (A–D)
| Species, genome type | Accession | A and B | A alone | B alone | C | D |
|---|---|---|---|---|---|---|
| O. barthii, AA | W0747 | 0/8 | 4/16 | |||
| W1467 | 5/8 | 0/8 | ||||
| W1473 | 4/16 | |||||
| W1583 | 6/16 | |||||
| W1588 | 0/8 | 3/8 | ||||
| W1643 | 5/8 | 0/8 | ||||
| W1702 | 0/8 | 6/8 | ||||
| O. glumaepatula, AA | W1183 | 0/8 | 6/8 | |||
| W1189 | 5/16 | |||||
| W1191 | 10/16 | |||||
| W1477 | 0/8 | 0/8 | 4/8 | 0/8 | ||
| W2140 | 0/8 | 6/8 | ||||
| W2145 | 0/8 | 0/8 | 2/8 | 0/7 | ||
| W2149 | 0/8 | 8/16 | ||||
| W2160 | 0/8 | 0/8 | 3/8 | 0/8 | ||
| W2165 | 13/16 | |||||
| W2173 | 0/8 | 10/16 | ||||
| W2184 | 12/16 | |||||
| W2192 | 0/8 | 5/8 | ||||
| W2201 | 0/7 | 0/8 | 2/8 | 0/7 | ||
| W2203 | 9/16 | |||||
| O. longistaminata, AA | W1573 | 2/40 | ||||
| O. meridionalis, AA | W1300 | 2/16 | 0/8 | |||
| W1625 | 0/8 | 3/24 | ||||
| W1631 | 2/12 | 0/8 | ||||
| W1635 | 3/16 | |||||
| W2071 | 0/8 | 1/16 | ||||
| W2080 | 0/8 | 1/16 | ||||
| W2105 | 0/8 | 2/16 | ||||
| O. rufipogon, AA | W0102 | 0/8 | 4/8 | |||
| W0106 | 0/8 | 3/8 | ||||
| W0107 | 12/16 | |||||
| W0137 | 0/8 | 7/16 | ||||
| W0593 | 10/16 | |||||
| W0610 | 0/8 | 6/16 | ||||
| W0621 | 0/7 | 2/8 | ||||
| W0625 | 7/16 | |||||
| W0629 | 3/8 | 0/8 | ||||
| W1230 | 2/16 | |||||
| W1239 | 11/16 | |||||
| W1551 | 13/16 | |||||
| W1666 | 0/8 | 5/9 | ||||
| W1669 | 0/8 | 6/8 | ||||
| W1681 | 0/8 | 4/8 | ||||
| W1685 | 12/16 | |||||
| W1690 | 0/8 | 4/8 | ||||
| W1718 | 0/8 | 4/8 | ||||
| W1739 | 3/8 | 0/7 | ||||
| W1852 | 5/16 | |||||
| W1865 | 12/16 | |||||
| W1944 | 3/8 | 0/8 | ||||
| W1945 | 0/8 | 4/8 | ||||
| W1962 | 3/8 | 0/8 | ||||
| W2014 | 11/16 | |||||
| W2051 | 4/8 | 0/8 | ||||
| W2263 | 0/8 | 5/9 | ||||
| W2265 | 13/16 | |||||
| O. punctata (2x), BB | W1582 | 5/16 | ||||
| W1586 | 4/16 | |||||
| W1590 | 3/8 | 0/8 | ||||
| W1592 | 2/8 | 0/8 | ||||
| W1593 | 4/16 | |||||
| O. eichingeri, CC | W1525 | 2/24 | 0/16 | |||
| O. officinalis, CC | W0614 | 0/8 | 2/15 | |||
| W1301 | 0/8 | 3/16 | ||||
| W1830 | 0/7 | 1/16 | ||||
| W1930 | 0/8 | 2/15 | ||||
| O. rhizomatis, CC | W1808 | 0/8 | 3/16 | |||
| W1812 | 4/16 | 0/16 | ||||
| O. minuta, BBCC | W0051 | 0/8 | 3/16 | |||
| W1213 | 2/8 | 0/8 | ||||
| W1318 | 0/8 | 0/8 | 0/8 | 1/8 | ||
| W1323 | 3/15 | |||||
| W1331 | 0/8 | 0/8 | 0/8 | 1/8 | ||
| O. punctata (4x), BBCC | W1474(B) | 2/16 | ||||
| W1564 | 0/16 | 2/16 | ||||
| O. alta, CCDD | W0017 | 2/16 | 0/12 | |||
| O. grandiglumis, CCDD | W0613 | 0/8 | 4/8 | |||
| W1195 | 0/8 | 3/16 | ||||
| W1476 | 0/8 | 4/16 | ||||
| W1480(B) | 3/16 | 0/8 | ||||
| W1483 | 6/16 | |||||
| W2220 | 4/16 | 0/8 | ||||
| O. latifolia, CCDD | W1177 | 2/16 | ||||
| W1539 | 1/24 | 0/16 | ||||
| O. brachyantha, FF | W0656 | 0/8 | 5/8 | |||
| W1407(B) | 0/8 | 4/8 | ||||
| W1706 | 0/8 | 3/8 | ||||
| W1711 | 0/8 | 5/8 | ||||
| O. longiglumis, HHJJ | W1227 | 2/32 | ||||
| Subtotal | 28 | 18 | 38 | 4 | 2 | |
| Total | 90 | |||||
number of regenerated plants / total number of tested immature embryos
Overall, among the 192 accessions tested, 90 formed calli from immature embryos and regenerated in tissue culture under one or more of the conditions tested. These accessions include AA, BB, CC, BBCC, CCDD, FF, and HHJJ genome species. Thus, we have identified conditions of tissue culture and the recovery of plantlets suitable for a wide range of Oryza species except those with EE and GG genomes. Condition B covered the widest range of wild Oryza accessions, followed by condition A. Very few of the accessions that did not respond to condition A or B responded to condition C or D. In particular, 4 accessions of O. glumaepatula (W1477, W2145, W2160, and W2201) responded to condition C, while other accessions of the same species responded to A or B. Thus, each species may have a genotype-dependent optimal condition for the regeneration of plantlets from immature embryos.
In general, species with the AA genome had a high regeneration success rate, notably O. barthii, O. glumaepatula, O. meridionalis, and O. rufipogon (Table 3). Although the success rate in O. longistaminata was 100%, we tested only one accession. As O. longistaminata generally propagates vegetatively through rhizomes rather than sexually, the immature embryo method is not particularly applicable for its transformation. We could not regenerate plantlets from immature embryos of O. australiensis, O. granulata, O. meyeriana, or O. ridleyi. Of 10 O. australiensis (EE) accessions tested, 5 formed calli and green spots but did not form shoots. Of 4 accessions of O. granulata and 2 of O. meyeriana (both GG) tested, 2 and 1 accessions, respectively, produced calli but did not form green spots or regenerate shoots and roots. It is well known that the plant hormone cytokinin plays an important role in shoot regeneration from calli. Further consideration of medium conditions, such as the concentration of cytokinin, may be necessary to induce regeneration from immature embryos of wild Oryza species with EE and GG genomes.
Table 3.
Summary of regeneration tests from immature embryos of wild Oryza species
| Species, genome type | No. accessions tested | No. accessions with callus inductiona | No. accessions with regenerationb | Percentage of regenerated accessionsc |
|---|---|---|---|---|
| O. barthii, AA | 10 | 8 (80%) | 7 (88%) | 70% |
| O. glumaepatula, AA | 18 | 17 (94%) | 14 (82%) | 78% |
| O. longistaminata, AA | 1 | 1 (100%) | 1 (100%) | 100% |
| O. meridionalis, AA | 15 | 13 (87%) | 7 (54%) | 47% |
| O. rufipogon, AA | 44 | 39 (89%) | 28 (72%) | 64% |
| O. punctata (2x), BB | 8 | 8 (100%) | 5 (63%) | 63% |
| O. eichingeri, CC | 2 | 1 (50%) | 1 (100%) | 50% |
| O. officinalis, CC | 16 | 7 (44%) | 4 (57%) | 25% |
| O. rhizomatis, CC | 3 | 3 (100%) | 2 (67%) | 67% |
| O. minuta, BBCC | 12 | 10 (83%) | 5 (50%) | 42% |
| O. punctata (4x), BBCC | 8 | 4 (50%) | 2 (50%) | 25% |
| O. alta, CCDD | 3 | 2 (67%) | 1 (50%) | 33% |
| O. grandiglumis, CCDD | 7 | 6 (86%) | 6 (100%) | 86% |
| O. latifolia, CCDD | 10 | 4 (40%) | 2 (50%) | 20% |
| O. australiensis, EE | 10 | 5 (50%) | 0 (0%) | 0% |
| O. brachyantha, FF | 7 | 6 (86%) | 4 (67%) | 57% |
| O. granulata, GG | 4 | 2 (50%) | 0 (0%) | 0% |
| O. meyeriana, GG | 2 | 1 (50%) | 0 (0%) | 0% |
| O. longiglumis, HHJJ | 9 | 1 (11%) | 1 (100%) | 11% |
| O. ridleyi, HHJJ | 3 | 0 (0%) | 0 (0%) | 0% |
| Total | 192 | 138 (72%) | 90 (65%) | 47% |
a100 × number of accessions with induced calli / total number of tested accessions
b100 × number of regenerated accessions / number of accessions with induced calli
c100 × number of regenerated accessions / total number of tested accessions
Optimization of Agrobacterium Infection of Immature Embryo Calli from Wild Oryza Accessions
Next, we compared the conditions of Agrobacterium infection of calli. Heat treatment and centrifugation of calli before Agrobacterium infection can increase the transformation efficiency in cultivated rice, maize, wheat, and sorghum (Hiei et al. 2006; Ishida et al. 2014; Sato-Izawa et al. 2018). Transformation efficiency is also affected by Agrobacterium strains (Hiei and Komari 2008). Therefore, we tested heat treatment, two conditions of centrifugation, and two Agrobacterium strains (Table 4). To monitor infection, we used a vector carrying the GFP driven by the ubiquitin promoter. Following infection, we counted the number of immature embryos with GFP-positive spots at 10 days (Fig. 1a). Using calli induced on immature embryos of Nipponbare, we showed that neither the speed of centrifugation nor the Agrobacterium strain affected the efficiency of infection (Table 4), possibly owing to very high infection efficiency in all conditions tested. Next, we infected calli of 16 wild Oryza accessions, one per species in which regeneration was achieved, and measured infection (Table 4). As wild accessions tend to flower gradually in a panicle, it is difficult to collect many ovaries at 8 to 10 day after pollination (DAP) at the same time, so we used 8 to 25 immature embryos of one accession of each species. GFP fluorescence showed that 11 out of 16 accessions were infected in one or more conditions tested (Table 4). This rate is considerably lower than that of Nipponbare. Nipponbare panicles flower in a brief period, so one can collect many immature embryos at once at the ideal stage for Agrobacterium infection. This difference could be a reason for the high infection ratio in Nipponbare. Another possibility is that the developmental window of immature embryos which can accept Agrobacterium infection may be broader in Nipponbare than in wild accessions.
Table 4.
Efficiency of Agrobacterium infection of calli induced from immature embryos of 16 wild Oryza species
| Strain Centrifugation | LBA4404 | EHA105 | |||
|---|---|---|---|---|---|
| 1100×g | 20,000×g | 1100×g | 20,000×g | ||
| 1 | O. barthii, W1467, AA | 1/9 | 2/8 | 0/8 | 1/8 |
| 2 | O. glumaepatula, W2184, AA | 3/8 | 3/9 | 1/8 | 2/8 |
| 3 | O. longistaminata, W1573, AA | 0/8 | 0/8 | 0/8 | 1/8 |
| 4 | O. meridionalis, W2080, AA | 0/9 | 1/8 | 0/8 | 0/8 |
| 5 | O. rufipogon, W1551, AA | 3/8 | 2/8 | 0/8 | 1/8 |
| 6 | O. punctata 2x, W1582, BB | 2/8 | 0/8 | 0/8 | 0/8 |
| 7 | O. eichingeri, W1525, CC | 0/8 | 0/8 | 1/8 | 0/8 |
| 8 | O. officinalis, W1301, CC | 0/16 | 0/16 | 0/17 | 0/16 |
| 9 | O. rhizomatis, W1808, CC | 0/16 | 0/15 | 0/16 | 0/17 |
| 10 | O. minuta, W0051, BBCC | 0/8 | 0/8 | 1/8 | 0/8 |
| 11 | O. punctata 4x, W1564, BBCC | 0/8 | 0/8 | 1/8 | 0/8 |
| 12 | O. alta, W0017, CCDD | 1/8 | 1/8 | 0/8 | 0/8 |
| 13 | O. grandiglumis, W2220, CCDD | 0/16 | 0/18 | 0/16 | 0/18 |
| 14 | O. latifolia, W1177, CCDD | 0/24 | 0/25 | 0/24 | 0/24 |
| 15 | O. brachyantha, W1711, FF | 2/9 | 1/8 | 0/8 | 1/8 |
| 16 | O. longiglumis, W1227, HHJJ | 0/24 | 0/24 | 0/25 | 0/24 |
| 17 | O. sativa, Nipponbare, AA | 88/90 | 76/79 | 77/83 | 81/85 |
number of GFP positive calli / total number of tested immature embryos
Fig. 1.
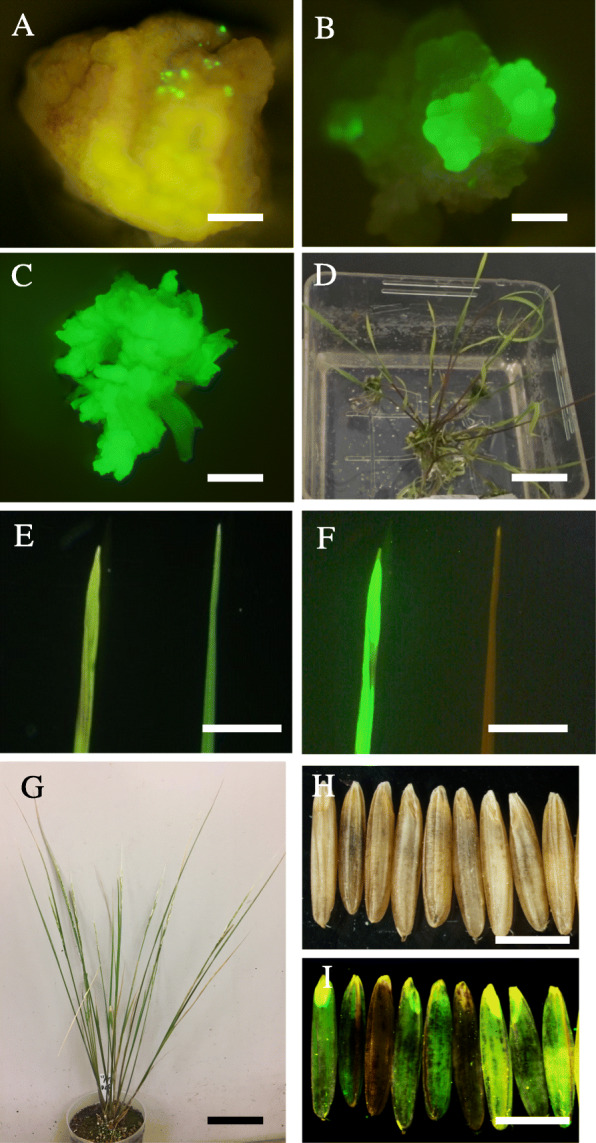
Agrobacterium-mediated transformation of Oryza brachyantha W1711. a At 10 days after infection, GFP signals were observed in immature embryos. Bar = 1 mm. b Callus was cultured for 2 weeks on selection medium. Part of the callus showed a GFP signal. Bar = 2 mm. c After 2 weeks’ culture on regeneration media, shoots with strong GFP signals were formed. Bar = 3 mm. d Regenerated plants on rooting medium. Bar = 2 cm. e, f Leaves through e bright field and f GFP filters: left, transgenic; right, non-transgenic. Bar = 1 cm. g Transgenic plants growing to maturity without morphological abnormalities. Bar = 5 cm. h, i T1 seeds of a transgenic plant through h bright field and i GFP filters. T1 seeds with and without GFP signal were segregated. Bar = 4 mm
Next, we tested Agrobacterium infection in 60 more wild accessions (Table 5), mainly under conditions in which an accession of the same species was infected (Table 4). Of the 76 accessions in 16 species, infection was successful in 51 accessions in 11 species. In most species (O. barthii, O. glumaepatula, O. longistaminata, O. rufipogon, O. punctata (2x), O. eichingeri, O. minuta, O. punctata (4x), O. alta, and O. brachyantha), infection and GFP fluorescence had a relatively high success rate (≥50%). However, in O. meridionalis, only 1 among 7 accessions showed a GFP signal. This result suggests that this species may have a mechanism that suppresses Agrobacterium infection in these conditions. Infection was not successful in O. officinalis, O. rhizomatis, O. grandiglumis, O. latifolia, or O. longiglumis. However, only 1 or 2 accessions were used for these species, so it is not clear whether this is a general tendency in each species. Further consideration of the conditions of infection, such as cold shock treatment, may be necessary (Sato-Izawa et al. 2018).
Table 5.
Efficiency of Agrobacterium infection of calli induced from immature embryos of 76 accessions of wild Oryza
| Species, genome type | Strain | LBA4404 | EHA105 | Infection efficiencya | ||
|---|---|---|---|---|---|---|
| Centrifugation | 1100×g | 20,000×g | 1100×g | 20,000×g | ||
| O. barthii, AA | W1467b | 1/9 | 2/8 | 0/8 | 1/8 | 3/6 (50%) |
| W1473 | 0/8 | 0/8 | ||||
| W1583 | 1/8 | |||||
| W1588 | 0/8 | 0/8 | ||||
| W1643 | 0/8 | 0/8 | ||||
| W1702 | 2/9 | |||||
| O. glumaepatula, AA | W1183 | 1/8 | 13/14 (93%) | |||
| W1189 | 0/8 | 0/8 | ||||
| W1191 | 2/8 | |||||
| W1477 | 1/8 | 3/16 | ||||
| W2140 | 1/8 | 1/8 | ||||
| W2145 | 1/8 | |||||
| W2149 | 2/8 | |||||
| W2160 | 2/8 | 3/16 | ||||
| W2165 | 1/8 | |||||
| W2173 | 3/16 | 2/16 | ||||
| W2184b | 3/8 | 3/9 | 1/8 | 2/8 | ||
| W2192 | 2/8 | |||||
| W2201 | 1/8 | 1/8 | ||||
| W2203 | 1/8 | 2/8 | ||||
| O. longistaminata, AA | W1573b | 0/8 | 0/8 | 0/8 | 1/8 | 1/1 (100%) |
| O. meridionalis, AA | W1300 | 0/9 | 0/8 | 1/7 (14%) | ||
| W1625 | 0/8 | 0/8 | ||||
| W1631 | 0/8 | 0/8 | ||||
| W1635 | 0/8 | 0/7 | ||||
| W2071 | 0/8 | 0/8 | ||||
| W2080b | 0/9 | 1/8 | 0/8 | 0/8 | ||
| W2105 | 0/8 | 0/8 | ||||
| O. rufipogon, AA | W0102 | 1/8 | 21/26 (81%) | |||
| W0106 | 1/8 | |||||
| W0107 | 2/8 | 2/16 | ||||
| W0137 | 0/8 | 0/8 | ||||
| W0593 | 1/8 | |||||
| W0610 | 1/8 | 1/8 | ||||
| W0621 | 1/8 | 1/8 | ||||
| W0625 | 1/9 | 1/8 | ||||
| W0629 | 1/8 | 1/8 | ||||
| W1230 | 0/8 | 0/8 | ||||
| W1239 | 2/8 | |||||
| W1551b | 3/8 | 2/8 | 0/8 | 1/8 | ||
| W1666 | 1/8 | |||||
| W1669 | 2/8 | |||||
| W1681 | 1/8 | |||||
| W1685 | 2/8 | 2/8 | ||||
| W1718 | 0/8 | 0/8 | ||||
| W1739 | 0/8 | 0/8 | ||||
| W1852 | 1/8 | |||||
| W1865 | 2/8 | 1/8 | ||||
| W1945 | 1/8 | |||||
| W1962 | 0/8 | 0/8 | ||||
| W2014 | 2/8 | 1/8 | ||||
| W2051 | 1/8 | |||||
| W2263 | 1/8 | |||||
| W2265 | 1/8 | 2/8 | ||||
| O. punctata (2x), BB | W1582b | 2/8 | 0/8 | 0/8 | 0/8 | 3/4 (75%) |
| W1586 | 1/16 | |||||
| W1592 | 0/8 | 0/8 | ||||
| W1593 | 1/16 | |||||
| O. eichingeri, CC | W1525b | 0/8 | 0/8 | 1/8 | 0/8 | 1/1 (100%) |
| O. officinalis, CC | W1301b | 0/16 | 0/16 | 0/17 | 0/16 | 0/2 (0%) |
| W1830 | 0/8 | 0/8 | ||||
| O. rhizomatis, CC | W1808b | 0/16 | 0/15 | 0/16 | 0/17 | 0/2 (0%) |
| W1812 | 0/8 | 0/8 | ||||
| O. minuta, BBCC | W0051b | 0/8 | 0/8 | 1/8 | 0/8 | 2/3 (67%) |
| W1323 | 1/8 | |||||
| W1331 | 0/8 | 0/8 | ||||
| O. punctata (4x), BBCC | W1474(B) | 1/8 | 2/2 (100%) | |||
| W1564b | 0/8 | 0/8 | 1/8 | 0/8 | ||
| O. alta, CCDD | W0017b | 1/8 | 1/8 | 0/8 | 0/8 | 1/1 (100%) |
| O. grandiglumis, CCDD | W2220b | 0/16 | 0/18 | 0/16 | 0/18 | 0/1 (0%) |
| O. latifolia, CCDD | W1177b | 0/24 | 0/25 | 0/24 | 0/24 | 0/1 (0%) |
| O. brachyantha, FF | W0656 | 2/8 | 2/8 | 2/8 | 3/4 (75%) | |
| W1407(B) | 1/8 | |||||
| W1706 | 0/8 | 0/8 | ||||
| W1711b | 2/9 | 1/8 | 0/8 | 1/8 | ||
| O. longiglumis, HHJJ | W1227b | 0/24 | 0/24 | 0/25 | 0/24 | 0/1 (0%) |
| Total | 51/76 (67%) | |||||
number of GFP positive calli / total number of tested immature embryo
a100 × number of GFP positive accessions / total number of tested accessions
bsame data as Table 4
Generation of Transgenic Plants and Inheritance of Transgene in Wild Oryza Accessions
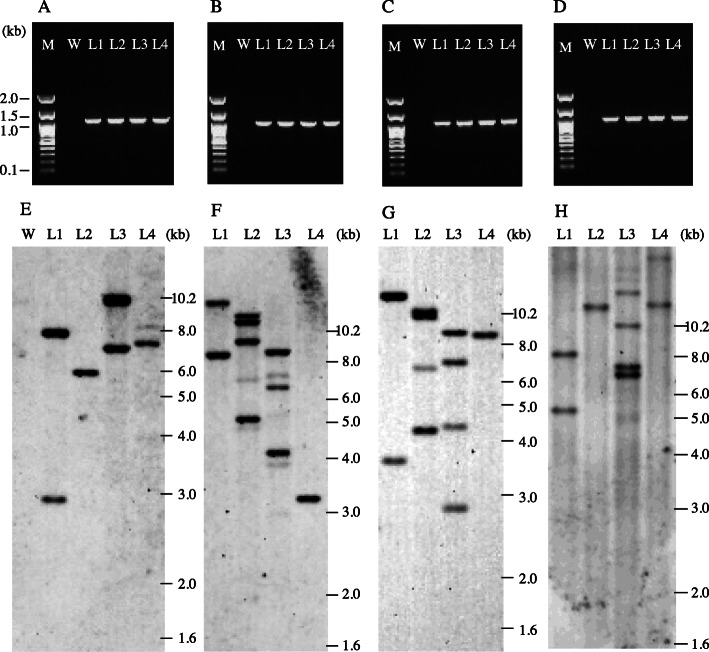
Next, we tried to generate transgenic plants expressing GFP by introducing the same vector used in the previous analysis into O. barthii (2 accessions), O. glumaepatula (8), O. rufipogon (15), and O. brachyantha (1) (Table 6). The transformation of O. brachyantha W1711 is shown as an example (Fig. 1a–i). A GFP signal was observed at 10 days after infection (Fig. 1a). Plants were selected on callus-inducing medium with antibiotics (Fig. 1b) and regenerated (Fig. 1c). Regenerated shoots were transferred to rooting medium (Fig. 1d) and then to pots with soil (Fig. 1g) and were grown to maturity. The regenerated T0 plants expressed GFP in leaves (Fig. 1e, f) and roots (Fig. 2). We confirmed the introduction of the transgene in regenerated T0 plants of O. barthii W1467, O. glumaepatula W2184, O. rufipogon W1551, and O. brachyantha W1711 by PCR (Fig. 3a–d) and found that the transformation events were all independent, with 1 to 4 T-DNA insertion loci, on the basis of the number of bands in Southern blot analysis (Fig. 3e–h).
Table 6.
Accessions used to generate transgenic plants expressing GFP
| Species, genome type | Accessions |
|---|---|
| O. barthii, AA | W1467, W1702 |
| O. glumaepatula, AA | W1477, W2160, W2165, W2173, W2184, W2192, W2201, W2203 |
| O. rufipogon, AA | W0106, W0107, W0610, W0621, W0625, W0629, W1239, W1551, W1666, W1669, W1681, W1685, W1865, W2014, W2265 |
| O. brachyantha, FF | W1711 |
Fig. 2.
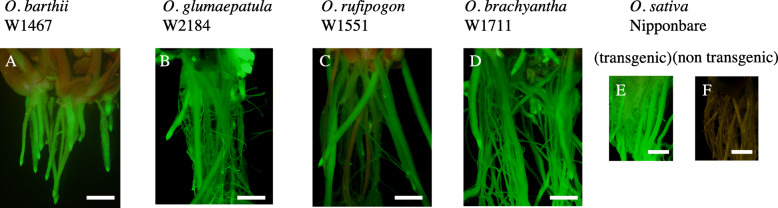
Agrobacterium-mediated transformation of wild Oryza. a–e) GFP fluorescence in roots of transgenic wild rice: aO. barthii W1467, bO. glumaepatula W2184, cO. rufipogon W1551, dO. brachyantha W1711, and eO. sativa Nipponbare. f Roots in non-transgenic Nipponbare as a negative control. Bar = 2 mm
Fig. 3.
Detection of transgenes in four lines of T0 plants from 4 accessions. a, eO. barthii W1467, b, fO. glumaepatula W2184, c, gO. rufipogon W1551, and d, hO. brachyantha W1711. a–d PCR amplification of full-length hpt gene from genomic DNA prepared from each independent line of transgenic plants (T0). Lanes: M, molecular weight marker; W, non-transgenic control; L1–L4: 4 independent lines of each transgenic wild Oryza accession. e–h Southern blot analysis of transgenic T0 plants with hpt probe. Lanes: W, non-transgenic control; L1–L4, T0 plants of the same lines in (a–d)
We obtained five or more regenerated plants from all accessions tested (Table 6). All grew to maturity without any discernable morphological abnormalities. All plants were fertile and set T1 seeds by self-pollination. We confirmed the transmission and segregation of the transgene in T1 seeds as GFP fluorescence in grains (Fig. 1h, i). From the results of Southern blot analysis, we picked two T1 lines with the transgene at either one or two loci from O. barthii W1467, O. glumaepatula W2184, O. rufipogon W1551, and O. brachyantha W1711 and tested its inheritance by observing GFP fluorescence of T1 grains (Table 7). The ratios of GFP-positive and -negative grains all fitted the expected 3:1 or 15:1 ratio, confirming the transmission of the transgene in a Mendelian manner.
Table 7.
Inheritance and segregation of the transgenes
| Species | Line numbera | GFP gene in T1 seeds | Segregation ratio (χ2, P) | |
|---|---|---|---|---|
| Positive | Negative | |||
| O. barthii, W1467 | L1 | 56 | 4 | 15:1 (0.02, 0.89) |
| L2 | 23 | 7 | 3:1 (0.04, 0.83) | |
| O. glumaepatula, W2184 | L1 | 57 | 3 | 15:1 (0.16, 0.69) |
| L4 | 22 | 8 | 3:1 (0.04, 0.83) | |
| O. rufipogon, W1551 | L1 | 56 | 4 | 15:1 (0.02, 0.89) |
| L4 | 22 | 8 | 3:1 (0.04, 0.83) | |
| O. brachyantha, W1711 | L1 | 57 | 3 | 15:1 (0.16, 0.69) |
| L2 | 23 | 7 | 3:1 (0.04, 0.83) | |
aLine number is the same as in Fig. 3
Conclusion
Our results show that a wide range of wild Oryza accessions, including those distantly related to cultivated species, can be genetically transformed by Agrobacterium by using the immature embryo method (Hiei and Komari 2008). Modification of the method increased the number of accessions that regenerated. Further attention to medium composition and conditions of infection will broaden the range of wild Oryza accessions that can be transformed.
We generated transgenic plants from O. rufipogon accessions W0106 and W1681 by the immature embryo method but not by the conventional scutellum callus method (data not shown). This difference supports the superiority of the immature embryo method for the transformation of wild Oryza accessions. This method opens the door to genome editing, accelerating the study of wild Oryza genetic resources for molecular genetic analysis and future use in molecular breeding.
Supplementary information
Additional file 1: Supplemental Table 1. Composition of media (1 L) used in this study. Supplemental Table 2. Work flow of test of regeneration from callus derived from immature embryos of wild Oryza species.
Acknowledgements
We thank Dr. Hiei Y and members of the Plant Innovation Center of Japan Tobacco for their technical assistance and tips for immature embryo culture. The wild Oryza accessions used in this study were distributed from NIG, which is supported by the National Bioresearch Project, AMED, Japan.
Abbreviations
- hpt
hygromycin phosphotransferase
- GFP
green fluorescent protein
- DAP
day after pollination
Authors’ Contributions
SSS, KT, MNT, TS, SO, KNT, and YY performed the experiments. KN and YS conceived and designed the study. All authors read and approved the final manuscript.
Funding
This work was supported by funding from the National Bioresearch Project’s Fundamental Technologies Upgrade Program to YS from AMED, Japan, and supported by a grant from RDA, Korea (PJ01321803).
Availability of Data and Materials
All datasets are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12284-020-00394-4.
References
- Fan F, Long W, Liu M, Yuan H, Pan G, Li N, Li S. Quantitative trait locus mapping of the combining ability for yield-related traits in wild rice Oryza longistaminata. J Agric Food Chem. 2019;14:8766–8772. doi: 10.1021/acs.jafc.9b02224. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ishida Y, Kasaoka K, Komari T. Improved frequency of transformation in rice and maize by treatment of immature embryos with centrifugation and heat prior to infection with Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult. 2006;87:233–243. doi: 10.1007/s11240-006-9157-4. [DOI] [Google Scholar]
- Hiei Y, Komari T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nature Prot. 2008;3:824–834. doi: 10.1038/nprot.2008.46. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Ikemoto M, Otsuka M, Thanh PT, Phan PDT, Ishikawa R, Ishii T. Gene interaction at seed-awning loci in the genetic background of wild rice. Genes Gen Sys. 2017;1:21–26. doi: 10.1266/ggs.16-00058. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Hiei Y, Komari T. Advances in wheat genetics: from genome to field. Tokyo: Springer; 2005. [Google Scholar]
- Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat Biotechnol. 1996;14:745–750. doi: 10.1038/nbt0696-745. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Tsunashima M, Hiei Y, Komari T. Wheat (Triticum aestivum L.) transformation using immature embryos. In: Wang K, editor. Agrobacterium protocols. New York: Springer; 2014. pp. 189–198. [DOI] [PubMed] [Google Scholar]
- Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R, Ishikawa R, Ashikari M. OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet. 2013;45:462–465. doi: 10.1038/ng.2567. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Iwata M, Taniko K, Monden G, Miyazaki N, Orn C, Tsujimura Y, Yoshida S, Ma JF, Ishii T. Detection of quantitative trait loci controlling grain zinc concentration using Australian wild rice, Oryza meridionalis, a potential genetic resource for biofortification of rice. PLoS One. 2017;12(10):e0187224. doi: 10.1371/journal.pone.0187224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya K, Murata K, Kokubo Y, Ube N, Ueno K, Yabuta Y, Teraishi M, Okumoto Y, Mori N, Ishihara A. Variation of diterpenoid phytoalexin oryzalexin a production in cultivated and wild rice. Phytochemistry. 2019;166:112057. doi: 10.1016/j.phytochem.2019.112057. [DOI] [PubMed] [Google Scholar]
- Khush GS, Brar DS, Hardy B (2001) Rice genetics IV. IRRI, Science Pub Inc, NH.
- Morishima H, Oka H. A survey of genetic variations in the populations of wild Oryza species and their cultivated relatives. Japan J Genet. 1970;45:371–385. doi: 10.1266/jjg.45.371. [DOI] [Google Scholar]
- Potrykus I. Gene transfer to plants: assessment of published approaches and results. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:205–225. doi: 10.1146/annurev.pp.42.060191.001225. [DOI] [Google Scholar]
- Sato-Izawa K, Tokue K, Ezura H. Development of a stable Agrobacterium-mediated transformation protocol for Sorghum bicolor Tx430. Plant Biotechnol (Tokyo) 2018;25:181–185. doi: 10.5511/plantbiotechnology.18.0424a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton M, Kobayashi M, Terashima S, Ohyanagi H, Copetti D, Hernández-Hernández T, Zhang J, Ohmido N, Fujita M, Toyada A, Ikawa H, Fujiyama A, Furuumi H, Miyabayashi T, Kubo T, Kudrna D, Wing R, Yano K, Nonomura K, Sato Y, Kurata N (2020) Evolution and diversity of the rice Oryza officinalis complex, across continents genome types, and ploidy levels. Genome Biol Evol. 10.1093/gbe/evaa037 [DOI] [PMC free article] [PubMed]
- Shimamoto K, Terada R, Izawa T, Fujimoto H. Fertile transgenic rice plants regenerated from transformed protoplasts. Nature. 1989;338:274–276. doi: 10.1038/338274a0. [DOI] [Google Scholar]
- Stein JC, Yu Y, Wing RA. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet. 2018;50:285–296. doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- Toriyama K, Arimoto Y, Uchimiya H, Hinata K. Transgenic rice plants after direct gene transfer into protoplasts. Bioresour Technol. 1988;6:1072–1074. [Google Scholar]
- Vaughan DA (1989) The genus Oryza L. current status of taxonomy. IRRI res pap Ser
- Vaughan DA. Wild relatives of rice: genetic resources hand book. Philippines: IRRI; 1994. [Google Scholar]
- Yichie Y, Brien C, Berger B, Roberts TH, Atwell BJ. Salinity tolerance in Australian wild Oryza species varies widely and matches that observed in O. sativa. Rice. 2018;11:66. doi: 10.1186/s12284-018-0257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu R. Efficient regeneration of transgenic plants from rice protoplasts and correctly regulated expression of the foreign gene in the plants. Theor App Genet. 1988;76:835–840. doi: 10.1007/BF00273668. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou S, Fu Y, Su Z, Wang X, Sun C. Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.) Plant Mol Biol. 2006;62:247–259. doi: 10.1007/s11103-006-9018-x. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, Zhan Q, Lu Y, Zhang L, Huang T, Wang Y, Fan D, Zhao Y, Qang Z, Zhou C, Chen J, Zhu C, Li W, Weng Q, Xu Q, Wang ZX, Wei X, Han B, Huang X. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet. 2018;50:278–284. doi: 10.1038/s41588-018-0041-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental Table 1. Composition of media (1 L) used in this study. Supplemental Table 2. Work flow of test of regeneration from callus derived from immature embryos of wild Oryza species.
Data Availability Statement
All datasets are available from the corresponding author on reasonable request.