Abstract
Purpose
The intestine has substantial role in cholesterol homeostasis due to the presence of various cholesterol transporters and gut microbiota. Bacteroides spp. are important members of gut microbiota that employ outer membrane vesicles (OMVs) to interact with host. In this regard, we evaluated the effect of Bacteroides fragilis, Bacteroides thetaiotaomicron and related OMVs on the gene expression of important cholesterol transporters, Niemann-Pick C1-Like 1 (NPC1L1), ATP-binding cassette (ABCA1), and liver X receptors (LXRs) in Caco-2 cells.
Methods
OMVs were isolated from overnight brain heart infusion (BHI) broth of bacterial standard strains using deoxycholate and assessed by Scanning electron microscopy (SEM). The relative change in genes expression was assessed by Quantitative reverse transcription PCR (RT-qPCR) based on SYBR Green and 2-∆∆ct method in Caco-2 cells that were treated with bacteria and OMVs. Data were statistically analyzed with GraphPad Prism software. Finally, pathway enrichment based on the studied genes was performed using Cytoscape plugin ClueGO.
Results
B. fragilis (P value = 0.002) and B. thetaiotaomicron (P value = 0.001) significantly reduced NPC1L1 gene expression in Caco-2 cells. Interestingly, NPC1L1 transcripts were significantly increased by both OMVs(P value = 0.04) (P value = 0.01). Also, LXRβ was significantly down regulated by B. thetaiotaomicron (P value = 0.02). ClueGO analysis on the studied genes demonstrated several functional groups which involve in lipid and cholesterol metabolism.
Conclusion
The opposite effect of B. fragilis, B. thetaiotaomicron and related OMVs on the NPC1L1 gene expression was observed in Caco-2 cells. Interestingly, these effects partially were in line with the alternation of LXRs expression. However, based on pathway enrichment analysis, further molecular investigations are required to elaborate in details the specific association between Bacteroides spp. and OMVs with regulation of cholesterol signaling pathways including cholesterol transport, lipid storage, lipid homeostasis and cholesterol homeostasis.
Keywords: Gut microbiota, Cholesterol homeostasis, Outer membrane vesicles (OMVs), Bacteroides spp., Niemann-pick C1-like 1
Introduction
Cholesterol and related metabolites have functional roles in human health varying from vital component of cell membrane, precursor of steroidal hormones, Vitamin D, primary bile acids (BA) and metabolism of the host [1]. Cholesterol homeostasis is regulated by dietary and genetic factors that affect cholesterol intestinal absorption and hepatic synthesis. It has been known that disruption of cholesterol metabolism is related to cardiovascular disease and type 2 diabetes [2]. One of the important tissues in cholesterol homeostasis is intestine where cholesterol absorption, fecal excretion and de novo synthesis take place [3, 4]. Several important proteins have been identified as cholesterol transporter in the intestine. For example, intestinal cholesterol uptake is mediated by Niemann-Pick C1-Like 1 (NPC1L1) which is located in apical surface of enterocytes [5]. Once free cholesterol that is incorporated into bile salt micelles is taken up by NPC1L1, there are various destinies for intracellular cholesterol including: (i) without any process in enterocytes, cholesterol comes back into the lumen through apical heterodimer ATP-binding cassette (ABC) transporters G5 and G8 (ABCG5/G8) [6]. (ii) in another way, cholesterol could be absorbed through chylomicron and HDL pathways which are mediated by Apo lipoprotein B and Apo-AI secretion, respectively. HDL pathway is mediated by basolateral transporter ABCA1 in enterocytes. Intracellular free cholesterol could be incorporated into Apo-AI lipoprotein and secreted as HDL component by ABCA1 [7]. These putative genes expression are under the control of liver X receptors (LXRs). Therefore, LXRs act as regulators of cholesterol homeostasis beside involvement in other metabolic processes including lipid and carbohydrate metabolism. LXRs consists of two isoforms LXRα and LXRβ which are encoded by NR1H3 and NR1H2 genes, respectively [3, 8]. Unlike to LXRβ which is ubiquitously expressed, the high expression of LXRα is identified in the liver, intestine, adipose tissue, macrophage, kidney and spleen. Two LXRs isoform have 78% identity at the amino acid level in DNA and ligand binding domain. Oxysterols (cholesterol derivatives) as LXRs endogenous ligands activate LXRs to form heterodimer with retinoid X receptor (RXR) for binding to LXR response elements (LXREs) of the target genes promoters [9, 10].
On the other hand, the impact of gut microbiota on the cholesterol metabolism has been investigated. Gut microbiota which is colonized gastrointestinal tract (GIT) with dominancy of Firmicutes and Bacteroidets bacterial phyla play significant roles in determination of human health and disease. A significant portion of cholesterol which is evaded from absorption process reach to colon where intestinal commensal bacterial metabolization and/or excretion with feces are occurred [11]. Gut microbiota metabolizes cholesterol by enzymatic reduction to corpostanol which is contributed in reduction of serum cholesterol concentration due to increase cholesterol excretion with feces. Bacteroides sp. strain D8 which belongs to B. fragilis cluster was reported as a first human cholesterol reducing isolate [12]. Also, gut microbiota influence on the BA pool which is mediated emulsification of lipids and fat digestion. Therefore, dysbiosis which is defined as imbalance of the gut microbiota composition and interaction with host, resulted in disrupted cholesterol homeostasis [13].
Bacteroides fragilis and Bacteroides thetaiotaomicron that belong to Bacteroidets phylum have important roles in host immunity and metabolism due to have high enzymatic and regulation of immune potentials [14]. One of the important ways that is employed by B. fragilis and B. thetaiotaomicron to interact with host is the production of outer membrane vesicles (OMVs). OMVs are nanosized particles which are secreted from gram negative bacteria [15]. These spherical shaped bilayer membrane vesicles originate from outer membrane and consist of bacterial compounds including lipopolysaccharide (LPS), outer membrane proteins (OMPs), phospholipids, periplasmic components, DNA, RNA, hydrolytic enzymes and signaling molecules [16]. OMVs which are released from pathogenic and non-pathogenic bacteria have been demonstrated to involvement in many processes including biofilm formation, bacterial survival, transferring of enzymes, toxins, immunological components and signaling factors, cell to cell communication and pathogenicity [16].
As mentioned above, cholesterol homeostasis is a multifactorial process that is regulated by various organs, transcriptional factors and transporters [3]. It has been demonstrated that gut microbiota has potential roles in host metabolism which could be mediated through OMVs [17]. In this regard, we aimed to evaluate the effects of two important members of gut microbiota, B. fragilis and B. thetaiotaomicron, and related OMVs on the gene expression, including the nuclear receptors (LXRα and LXRβ) and important intestinal cholesterol transporters (NPC1L1 and ABCA1) in Caco-2 cell line as a human intestinal epithelium model. Finally, pathway enrichment analysis was performed based on the studied genes to predict the possible signaling pathways which could be affected by these bacteria and their OMVs.
Materials and methods
Bacterial culture
B. fragilis ATCC 23745 and B. thetaiotaomicron CCUG 10774 were cultured either on trypticase soy agar with 5% defibrinated sheep blood or brain heart infusion (BHI) broth supplemented with hemin (5 μg/ml) (Sigma-Aldrich, USA) and menadione (1 μg/ml) (Sigma-Aldrich, USA) at 37 °C under anaerobic conditions provided 80% N2, 10% Co2 and 10% H2 using Anoxomat™ MARK II system [18].
OMVs preparation
Isolation of OMVs was performed as described previously [19]. Briefly, B. fragilis and B. thetaiotaomicron were cultured on BHI broth under anaerobic conditions for overnight. After harvesting of bacterial cell by centrifugation, OMVs were extracted through sequential centrifugation at 20000 g (90 min, 4 °C) using Tris-ethylene diamine tetraacetic acid (EDTA) - Sodium deoxycholate (Sigma-Aldrich, USA) buffers. Finally, OMVs were filtered by a 0.22-μm polyvinylidene difluoride filter (Millipore, Billerica, MA, USA) and stored in 3% sucrose solution at −20 °C. to confirm OMVs isolation, Scanning Electron Microscopy (SEM) was performed on the gold coated samples which were fixed in PBS containing 2.5% glutaraldehyde and 2% paraformaldehyde using SEM (EM3200 KYKY Technology, China) [20].
Co-cultures
Colon adenocarcinoma cells (Caco-2) IBRC C10094 were obtained from Iranian Biological Resource Center. Cells were cultivated in high glucose Dulbecco’s modified eagle medium (DMEM) (Gibco™, USA) containing 10% fetal bovine serum (FBS, Gibco™, USA), 1% non-essential amino acids (Gibco™, USA) and 1% penicillin/streptomycin (Gibco™, USA) and incubated at 37 °C in a humidified atmosphere with 5% CO2. The medium was changed every 2–3 days. After reach to confluency, the cells were seeded at a density of 2 × 105 cells in 6-well tissue culture plates. Before treatment, the culture medium was replaced with DMEM containing 1% FBS and incubated for 2 h. Caco-2 cells were stimulated with B. fragilis, B. thetaiotaomicron at multiplicity of infection (MOI = 10) and related OMVs (at protein concentration of 50 μg/ml) for overnight [20].
Reverse transcription quantitative PCR (RT-qPCR) analysis
Total RNA extraction was carried out using RNX-Pluse (CinnaGen, Iran). The extracted RNA was quantified and qualified by NanoDrop 2000 (Thermo Fisher Scientific, USA) and agarose gel electrophoresis, respectively. RNA was reverse transcribed using RevertAid first strand cDNA synthesis kit (Thermo Scientific, USA) according to manufacturers’ instructions.
RT-qPCR was performed using SYBR Green method and LightCycler® 96 SW 1.1 (Roche, Germany). Each qPCR reactions were optimized at 20 μl final volume and performed in triplicate LightCycler® 8-Tube Strips (white) (Roche, Germany). qPCR reaction contained SYBR Premix Ex Taq II (RR820L – Takara, China), 0.5 μl of each specific primer (Table 1) and 1 μl of template cDNA. The amplification condition consisted in one step of 95 °C for 60 s, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s. The relative fold change in expression normalized to Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression, as a housekeeping gene, by the 2-∆∆CT method [20].
Table 1.
The primers sequence of studied genes
| Target gene | Forward (5′ to 3′) | Reverse (5′ to 3′) | Reference |
|---|---|---|---|
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG | [39] |
| NPC1L1 | CTGGTATCACTGGAAGCGAGT | CACGCGGGTCACATTGATGA | [40] |
| ABCA1 | ACCCACCCTATGAACAACATGA | GAGTCGGGTAACGGAAACAGG | [41] |
| NR1H3 | ACACCTACATGCGTCGCAAG | GACGAGCTTCTCGATCATGCC | [42] |
| NR1H2 | AGAAGATTCGGAAACAACAGCA | GCTGGATCATTAGTTCTTGAGCC | [43] |
Statistical analyses
Data were analyzed with two-tailed t-test using GraphPad Prism software (GraphPad Software, Inc., San Diego, CA). All results demonstrate as mean ± Standard deviation (SD). In all experiments, P < 0.05 was considered statistically significant.
Functional enrichment analysis
We constructed protein-protein interaction (PPI) network of the studied genes using STRING 9.0 database (Search Tool for the Retrieval of Interacting Genes) [21]. This network was visualized and analyzed by Cytoscape (3.4.0 software) plugin network analysis based on topological parameters including betweenness centrality (BC) and node degree parameters. To determine the biological and functional correlation of the studied genes and related signaling, pathway enrichment was performed using Cytoscape plugin ClueGO (binda G) [22].
Results
The effects of B. fragilis and B. thetaiotaomicron on the mRNA levels of nuclear transcriptional factors and intestinal cholesterol transporters
The Caco-2 cell line was employed as human intestinal epithelium cells to study the effects of B. fragilis and B. thetaiotaomicron on the important genes expression which are involved in intestinal cholesterol homeostasis. Caco-2 cells were stimulated with these bacteria for overnight. Our results demonstrated that B. fragilis significantly decreased the mRNA levels of NPC1L1 in Caco-2 cells (Fig. 1a). Also, the expression of LXRβ and NPC1L1 mRNAs were significantly down regulated by B. thetaiotaomicron (Fig. 1b). We observed that B. fragilis and B. thetaiotaomicron are significantly able to downregulate NPC1L1 in Caco-2 cells.
Fig. 1.
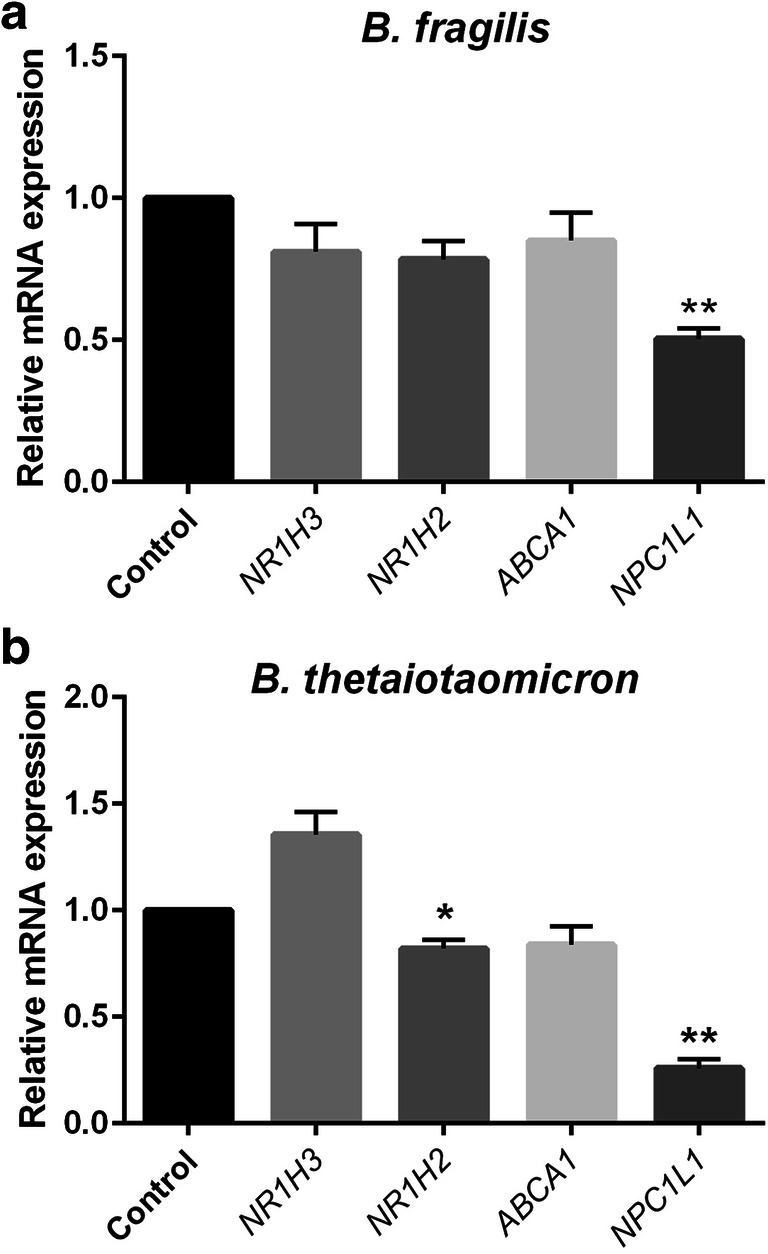
The relative gene expression of NR1H3, NR1H2, ABCA1 and NPC1L1 based on RT-qPCR experiments in Caco-2 cells treated with B. fragilis (a), B. thetaiotaomicron (b) at multiplicity of infection, MOI = 10 and PBS as control group for overnight. Values of triplicate experiments are demonstrated as mean ± SD. Statistical Significant are presented as * (P < 0.05) and,** (P < 0.01)
The effects of B. fragilis and B. thetaiotaomicron related OMVs on the mRNA levels of nuclear transcriptional factors and intestinal cholesterol transporters
B. fragilis and B. thetaiotaomicron produced OMVs with spherical shape and 30–110 nm of diameter which is determined by SEM. Mean dimension of OMVs was 85.7 ± 15.3 nm as previously reported [in press]. Caco-2 cells were challenged with B. fragilis and B. thetaiotaomicron derived OMVs to investigate the alteration of nuclear transcriptional factors, LXRs and important intestinal cholesterol transporters, ABCA1 and NPC1L1 genes expression by RT-qPCR method. Interestingly, we perceived that OMVs from B. fragilis and B. thetaiotaomicron inversely affected NPC1L1 gene expression in comparison with parental bacteria. Our experiment showed that both OMVs significantly increased NPC1L1 mRNA levels in Caco-2 cells at protein concentration of 50 μg/ml (Fig. 2a, b).
Fig. 2.
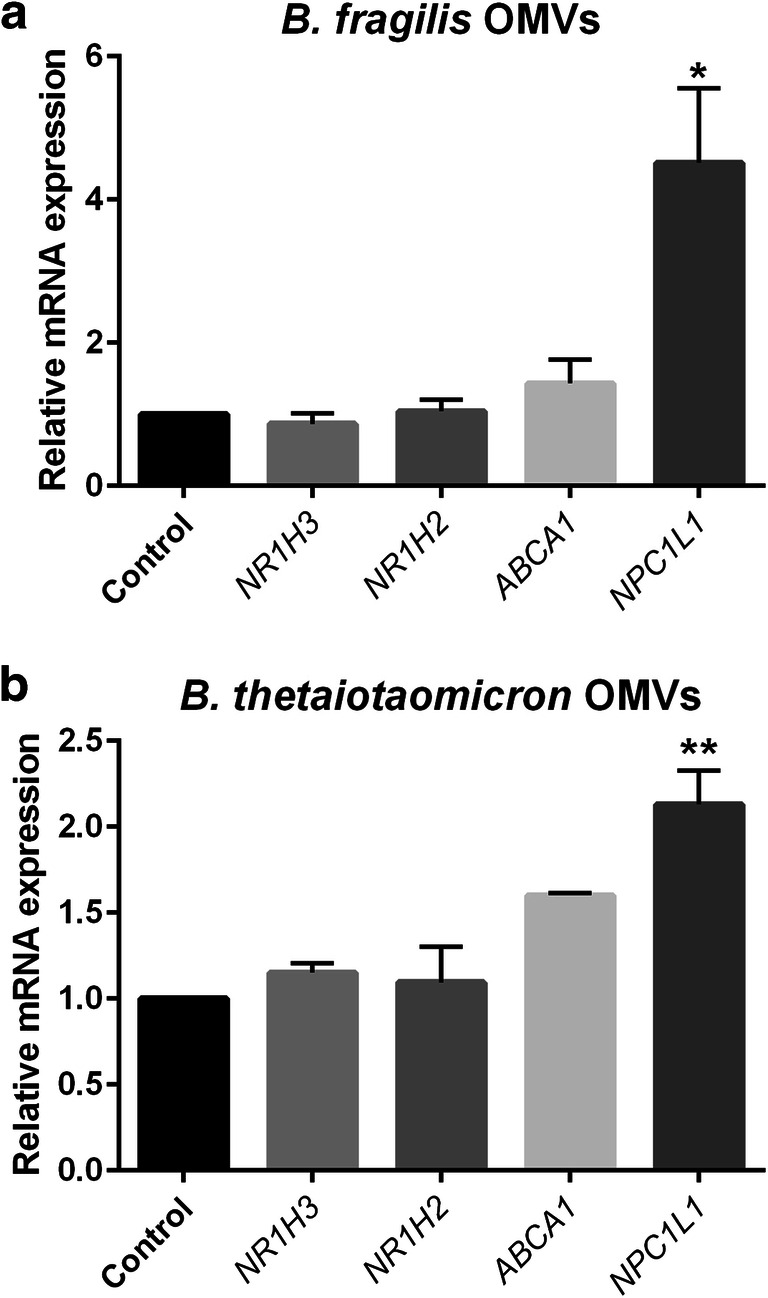
The relative mRNA expression of NR1H3, NR1H2, ABCA1 and NPC1L1 based on RT-qPCR experiments in Caco-2 cells treated with B. fragilis OMVs (a), B. thetaiotaomicron OMVs (b) at 50 μg/ml and sucrose as control group overnight. Values of triplicate experiments are demonstrated as mean ± SD. Statistical Significant are presented as * (P < 0.05) and,** (P < 0.01)
Characteristics of the studied genes in PPI network
Pathway analysis
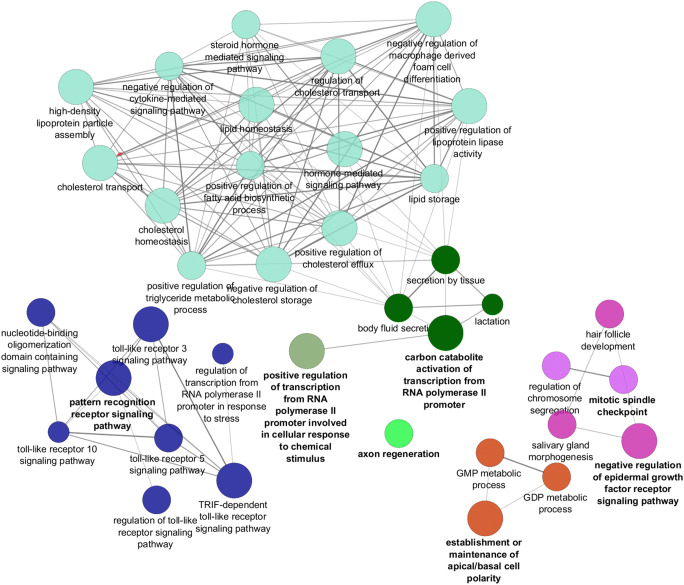
After the construction of The PPI network based on studied genes (data was not shown), the pathways enrichment was performed using Cytoscape plugin ClueGO [22]. ClueGO analysis presents pathways as a clustered network which the studied genes may be involved. Generally, 37 GO terms that were divided to 7 GO groups and 20 Pathways were identified and categorized into 5 groups based on the Kappa score (≥0.4). The main GO categories includedregulation of cholesterol transport, lipid storage, lipid homeostasis and cholesterol homeostasis (Fig. 3).
Fig. 3.
The GO terms were classified into several functional groups (different node colors) according to the kappa value. Each node represented a GO term, and the node size was proportional to the significance of the term. The most significant GO terms were labeled with a highlighted color. The edge in the nodes indicated that they shared common genes, and the width of the edge was proportional to the number of common genes
Discussion
The intestine has regulatory potentials on the cholesterol homeostasis due to the presence of substantial cholesterol transporters and a dynamic microbial community called “gut microbiota” [7, 11]. In this regard, we have investigated the effects of B. fragilis, B. thetaiotaomicron and their OMVs as interactive bacterial components on LXRs and the important intestinal cholesterol transporters in Caco-2 cell line as a human intestinal epithelium model.
It is known that gut microbiota has important regulatory role in the host functions including immune system, homeostasis and metabolism [23, 24]. Cholesterol homeostasis could be affected by gut microbiota through regulation of intestinal absorption and cholesterol metabolization. Several studies have pointed out the impact of beneficial intestinal bacteria on the intestinal cholesterol absorption in Caco-2 cell line [25, 26]. Intestinal cholesterol absorption is mainly achieved by NPC1LI apical transporter in enterocytes. NPC1LI is a crucial target for reduction of diet-induced hypercholesterolemia [5]. Ying Huang et al. showed that Lactobacillus acidophilus, Lactobacillus rhamnosus GG and Bifidobacterium lactis inhibited the gene expression of NPC1L1, while B. thetaiotaomicron did not have inhibitory effect on this gene in Caco-2 cells which were treated for 6 h at MOI 1 [25]. On the contrary, we reported the significant effects of B. fragilis and B. thetaiotaomicron in downregulating NPC1L1 transcripts. This discrepancy could be attributed to the time/MOI of co-culture and experiment condition. Based on our results B. fragilis and B. thetaiotaomicron could be considered as gut microbiota members which can reduce intestinal cholesterol uptake through reduction of NPC1LI gene expression, the main absorptive intestinal cholesterol transporter. Our finding is concordant with the role of Bacteroides sp. strain D8 which is able to reduce cholesterol and increase its excretion [12]. Therefore, it is possible that B. fragilis group participate to decrease serum cholesterol concentration by two probabilistic mechanisms including decreasing intestinal cholesterol absorption and increasing cholesterol excretion, resulted from its reduction. This hypothesis is required further investigation on the role B. fragilis group in cholesterol homeostasis.
It has been shown that other intestinal cholesterol transporters such as ABCA1 control intestinal cholesterol absorption [27]. The studies in Caco-2 cell line and animal models demonstrated that ABCA1 has important role in intestinal cholesterol absorption by HDL pathway [28, 29]. Here, we identified that B. fragilis and B. thetaiotaomicron had a modest decreasing effect on the gene expression of ABCA1 in Caco-2 cells.
The gene expression of NPC1L1 and ABCA1 are regulated by LXRs that have important regulatory role in cholesterol homeostasis [30]. Duval et al. reported NPC1L1 as new target of LXRs. Another study demonstrated that LXRs activators inhibit the expression of NPC1L1 in the intestine [31]. Ying Huang et al. observed the increase of LXR in Caco-2 cells treated with L. acidophilus and attributed the downregulation of NPC1L1 to it [25]. In the present study, we showed that B. thetaiotaomicron decreases NPC1L1 expression in line with the reduction of LXRβ transcript. Based on obtained results, we suggest that LXRβ could be a main regulator of NPC1L1 expression which is affected by B. thetaiotaomicron.
The gut microbiota-host cross talk relies on the secreted factors that are able to access to the epithelial cells and under tissues [32]. Among bacterial components, OMVs are considered as important secreted factors with regulatory potentials of immune responses and metabolism [17, 33–35]. Recently, the role of OMVs derived from B. fragilis and B. thetaiotaomicron is highlighted in gut microbiota-host interactions [20, 36, 37]. No data are available yet on the effect of OMVs on cholesterol homeostasis. Accordingly, we studied the effect of OMVs on putative genes transcripts in Caco-2 cells. Interestingly, the B. fragilis and B. thetaiotaomicron OMVs had opposite effect on NPC1L1 gene expression compared with related bacteria. Both OMVs significantly elevated NPC1L1 expression without any significant change in LXRs transcripts in Caco-2 cells. This paradox can be explained that B. fragilis and B. thetaiotaomicron could be interact with the cell surface by various components and microbe-associated molecular patterns (MAMPs) while OMVs which are able to penetrate in to the cells interact with other possible intracellular receptor and transcription factors. In this regard, the opposite effect on the gene expression between Akkermansia muciniphila, a gut microbiota member, and its OMVs was reported [38]. However, more investigation of these particles is necessary to recognize involved molecular signaling.
Also, pathway enrichment analysis based on the studied genes showed that these bacteria and related OMVs can interfere in other pathways including regulation of cholesterol transport, lipid storage, lipid homeostasis and cholesterol homeostasis. Since B. fragilis, B. thetaiotaomicron and their OMVs affected studied genes, to evaluate their role in the putative pathways will contribute to reach a comprehensive recognition of these bacteria and the effect of their OMVs on lipid (cholesterol) metabolism regulation.
Conclusion
In conclusion, B. fragilis and B. thetaiotaomicron, the two important gut microbiota members, induce significant downregulation of NPC1L1 intestinal gene expression. Interestingly, we observed the opposite effect of OMVs from B. fragilis and B. thetaiotaomicron on the expression of NPC1L1 gene. These findings suggest different regulatory effect of these bacteria and their OMVs on the gene expression of NPC1L1, the main intestinal cholesterol transporter which is responsible for intestinal cholesterol uptake. Obtained results could be considered to targeted intervention of gut microbiota composition in order to reduce intestinal cholesterol absorption in hypercholesterolemia state. Also, based on pathway enrichment analysis, further molecular studies are required to elucidate the effect of B. fragilis, B. thetaiotaomicron and related OMVs on the regulation of lipid (cholesterol) signaling pathways.
Acknowledgments
This research was funded by Iran Biotech Fund Grant 94/10243. The authors would like to thank all colleagues at Microbiology Research Center of Pasteur Institute of Iran.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sara Ahmadi Badi and Atiyyeh Motahhary contributed equally to this work.
References
- 1.van der Wulp MY, Verkade HJ, Groen AK. Regulation of cholesterol homeostasis. Mol Cell Endocrinol. 2013;368(1–2):1–16. doi: 10.1016/j.mce.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Ding J, Reynolds LM, Zeller T, Müller C, Mstat KL, Nicklas BJ et al. Alterations of a cellular cholesterol metabolism network is a molecular feature of obesity-related type 2 diabetes and cardiovascular disease. Diabetes. 2015:db141314. [DOI] [PMC free article] [PubMed]
- 3.Iqbal J, Al Qarni A, Hawwari A. Regulation of intestinal cholesterol absorption: a disease perspective. Advances in Biological Chemistry. 2017;7(01):60–75. doi: 10.4236/abc.2017.71004. [DOI] [Google Scholar]
- 4.Kruit JK, Groen AK, van Berkel TJ, Kuipers F. Emerging roles of the intestine in control of cholesterol metabolism. World J Gastroenterol: WJG. 2006;12(40):6429–6439. doi: 10.3748/wjg.v12.i40.6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betters JL, Yu L. NPC1L1 and cholesterol transport. FEBS Lett. 2010;584(13):2740–2747. doi: 10.1016/j.febslet.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abumrad NA, Davidson NO. Role of the gut in lipid homeostasis. Physiol Rev. 2012;92(3):1061–1085. doi: 10.1152/physrev.00019.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain MM. Intestinal lipid absorption and lipoprotein formation. Curr Opin Lipidol. 2014;25(3):200–206. doi: 10.1097/MOL.0000000000000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calkin AC, Tontonoz P. Liver x receptor signaling pathways and atherosclerosis. Arterioscler Thromb Vasc Biol. 2010;30(8):1513–1518. doi: 10.1161/ATVBAHA.109.191197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurencikiene J, Rydén M. Liver X receptors and fat cell metabolism. Int J Obes. 2012;36(12):1494–1502. doi: 10.1038/ijo.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baranowski M. Biological role of liver X receptors. J Physiol Pharmacol. 2008;59(Suppl 7):31–55. [PubMed] [Google Scholar]
- 11.Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2013;3(1):14–24. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gérard P, Lepercq P, Leclerc M, Gavini F, Raibaud P, Juste C. Bacteroides sp. strain D8, the first cholesterol-reducing bacterium isolated from human feces. Appl Environ Microbiol. 2007;73(18):5742–5749. doi: 10.1128/AEM.02806-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molinero N, Ruiz L, Sanchez B, Margolles A, Delgado S. Intestinal bacteria interplay with bile and cholesterol metabolism: implications on host physiology. Front Physiol. 2019;10:185. doi: 10.3389/fphys.2019.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16(5):559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Jan AT. Outer membrane vesicles (OMVs) of gram-negative bacteria: a perspective update. Front Microbiol. 2017;8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkarni HM, Jagannadham MV. Biogenesis and multifaceted roles of outer membrane vesicles from gram-negative bacteria. Microbiology. 2014;160(10):2109–2121. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- 17.Ahmadi Badi S, Moshiri A, Fateh A, Rahimi Jamnani F, Sarshar M, Vaziri F, Siadat SD. Microbiota-derived extracellular vesicles as new systemic regulators. Front Microbiol. 2017;8:1610. doi: 10.3389/fmicb.2017.01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y, Torchia MLG, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12(4):509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tafti ZSM, Moshiri A, Marvasti FE, Tarashi S, Khalili SFS, Motahhary A, et al. The effect of saturated and unsaturated fatty acids on the production of outer membrane vesicles from Bacteroides fragilis and Bacteroides thetaiotaomicron. Gastroenterology and hepatology from bed to bench. 2019;12(2):155. [PMC free article] [PubMed] [Google Scholar]
- 20.Badi SA, Siadat SD, Khatami S, Irani S. Induction Effect of Bacteroides fragilis Derived outer membrane vesicles on toll like receptors gene expression and cytokine concentrations in human intestinal epithelial cell. Cell J (Yakhteh). 2019;21(1). [DOI] [PMC free article] [PubMed]
- 21.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Assenov Y, Ramírez F, Schelhorn S-E, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2007;24(2):282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 23.Young VB. The intestinal microbiota in health and disease. Curr Opin Gastroenterol. 2012;28(1):63–69. doi: 10.1097/MOG.0b013e32834d61e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Zheng Y. The probiotic lactobacillus acidophilus reduces cholesterol absorption through the down-regulation of Niemann-Pick C1-like 1 in Caco-2 cells. Br J Nutr. 2010;103(4):473–478. doi: 10.1017/S0007114509991991. [DOI] [PubMed] [Google Scholar]
- 26.Yoon H-S, Ju J-H, Kim H-N, Park H-J, Ji Y, Lee J-E, Shin HK, Do MS, Holzapfel W. Reduction in cholesterol absorption in Caco-2 cells through the down-regulation of Niemann-Pick C1-like 1 by the putative probiotic strains lactobacillus rhamnosus BFE5264 and lactobacillus plantarum NR74 from fermented foods. Int J Food Sci Nutr. 2013;64(1):44–52. doi: 10.3109/09637486.2012.706598. [DOI] [PubMed] [Google Scholar]
- 27.Plösch T, Kosters A, Groen A, Kuipers F. The ABC of hepatic and intestinal cholesterol transport. Atherosclerosis: Diet and Drugs. Springer; 2005. pp. 465–482. [DOI] [PubMed] [Google Scholar]
- 28.Iqbal J, Anwar K, Hussain MM. Multiple, independently regulated pathways of cholesterol transport across the intestinal epithelial cells. J Biol Chem. 2003;278(34):31610–31620. doi: 10.1074/jbc.M301177200. [DOI] [PubMed] [Google Scholar]
- 29.Iqbal J, Hussain MM. Evidence for multiple complementary pathways for efficient cholesterol absorption in mice. J Lipid Res. 2005;46(7):1491–1501. doi: 10.1194/jlr.M500023-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.van der Veen JN, van Dijk TH, Vrins CL, van Meer H, Havinga R, Bijsterveld K et al. Activation of the liver X receptor stimulates trans-intestinal excretion of plasma cholesterol. Journal of Biological Chemistry. 2009:jbc. M109. 014860. [DOI] [PMC free article] [PubMed]
- 31.Duval C, Touche V, Tailleux A, Fruchart J-C, Fievet C, Clavey V, Staels B, Lestavel S. Niemann–Pick C1 like 1 gene expression is down-regulated by LXR activators in the intestine. Biochem Biophys Res Commun. 2006;340(4):1259–1263. doi: 10.1016/j.bbrc.2005.12.137. [DOI] [PubMed] [Google Scholar]
- 32.Cañas M-A, Fábrega M-J, Giménez R, Badia J, Baldomà L. Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front Microbiol. 2018;9:498. doi: 10.3389/fmicb.2018.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valguarnera E, Scott NE, Feldman M. A dual motif mediates outer-membrane translocation and packing of glycosidases into Bacteroides Outer Membrane Vesicles. bioRxiv. 2018:377861.
- 34.Behrouzi A, Vaziri F, Rad FR, Amanzadeh A, Fateh A, Moshiri A, et al. Comparative study of pathogenic and non-pathogenic Escherichia coli outer membrane vesicles and prediction of host-interactions with TLR signaling pathways. BMC research notes. 2018;11(1):539. doi: 10.1186/s13104-018-3648-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reza Aghasadeghi M, Sharifat Salmani A, Mehdi Sadat S, Javadi F, Memarnejadian A, Vahabpour R, Zabihollahi R, Moshiri A, Davar Siadat S. Application of outer membrane vesicle of Neisseria meningitidis serogroup B as a new adjuvant to induce strongly Th1-oriented responses against HIV-1. Curr HIV Res. 2011;9(8):630–635. doi: 10.2174/157016211798998772. [DOI] [PubMed] [Google Scholar]
- 36.Zakharzhevskaya NB, Vanyushkina AA, Altukhov IA, Shavarda AL, Butenko IO, Rakitina DV, Nikitina AS, Manolov AI, Egorova AN, Kulikov EE, Vishnyakov IE, Fisunov GY, Govorun VM. Outer membrane vesicles secreted by pathogenic and nonpathogenic Bacteroides fragilis represent different metabolic activities. Sci Rep. 2017;7(1):5008. doi: 10.1038/s41598-017-05264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stentz R, Horn N, Cross K, Salt L, Brearley C, Livermore DM, Carding SR. Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against β-lactam antibiotics. J Antimicrob Chemother. 2014;70(3):701–709. doi: 10.1093/jac/dku466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang C-s, Ban M, Choi E-J, Moon H-G, Jeon J-S, Kim D-K et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PloS one. 2013;8(10):e76520. [DOI] [PMC free article] [PubMed]
- 39.Zeng H, Zhang Y, Yi Q, Wu Y, Wan R, Tang L. CRIM1, a newfound cancer-related player, regulates the adhesion and migration of lung cancer cells. Growth Factors. 2015;33(5–6):384–392. doi: 10.3109/08977194.2015.1119132. [DOI] [PubMed] [Google Scholar]
- 40.Leboucher M. L'effet de l'obésité et du diabète gestationnel sur l'expression des protéines Niemann Pick C dans le placenta humain à terme. 2017. [Google Scholar]
- 41.Zhou R, Wang L, Xu X, Chen J, Hu L-h, Chen L-l et al. Danthron activates AMP-activated protein kinase and regulates lipid and glucose metabolism in vitro. Acta Pharmacologica Sinica. 2013;34(8):1061. [DOI] [PMC free article] [PubMed]
- 42.Jiang T, Ren K, Chen Q, Li H, Yao R, Hu H, Lv YC, Zhao GJ. Leonurine prevents atherosclerosis via promoting the expression of ABCA1 and ABCG1 in a Pparγ/Lxrα signaling pathway-dependent manner. Cell Physiol Biochem. 2017;43(4):1703–1717. doi: 10.1159/000484031. [DOI] [PubMed] [Google Scholar]
- 43.Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, Cooper GM, Reddy TE, Crawford GE, Myers RM. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52(1):25–36. doi: 10.1016/j.molcel.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]