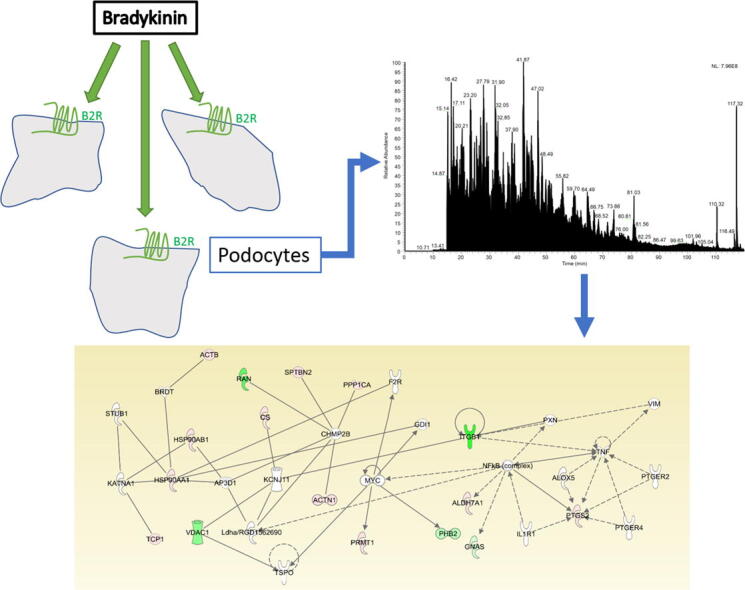
Graphical abstract
Keywords: Cyclooxygenase-2, ERK1/2, Liquid Chromatography Mass Spectrometry, Bradykinin 2 receptor, Podocyte proteome
Abstract
Podocyte damage is one of the hallmarks of diabetic nephropathy leading to proteinuria and kidney damage. The underlying mechanisms of podocyte injury are not well defined. Bradykinin (BK) was shown to contribute to diabetic kidney disease. Here, we evaluated the temporal changes in proteome profile and inflammatory signals of podocytes in response to BK (10−7M). Protein profile was evaluated by liquid chromatography mass Spectrometry (LC-MS/MS) analysis. Proteome profile analysis of podocytes treated with BK (10−7M) for 3 and 6 h, revealed 61 proteins that were differentially altered compared to unstimulated control podocytes. Pathway enrichment analysis suggested inhibition of cell death pathways, engagement of cytoskeletal elements and activation of inflammatory pathways. One of the inflammatory proteins that was identified to be induced by BK treatment is Prostaglandin (PG) H Synthase-2 (Cyclooxygenase-2, COX-2). In addition, BK significantly induced the production and release of PGE2 and this effect was inhibited by both COX-2 and MEK Kinase inhibitors, demonstrating that the production of PGE2 by BK is mediated via COX-2 and MAPK-dependent mechanisms. These findings provide a global understanding of the effector modulated proteome in response to BK and also reveal BK as an important modulator of inflammation and a potential player in podocyte injury.
Background
Diabetic kidney disease (DKD) is the leading cause of end-stage kidney disease [1], and it has long been recognized that the high mortality due to cardiovascular disease in diabetics with nephropathy suggested a strong association between microvascular and macrovascular pathologies in diabetes [2]. Podocyte loss has been identified as a hallmark for the progression of DKD, where the loss of podocytes has been shown to be correlated with an increase in albumin excretion rate (AER) [3], [4]. Hemodynamic injury within the glomerulus such as the dysregulation of glomerular filtration rate (GFR) has been such as a key feature contributing to development and progression of DKD [5].
Podocytes are epithelial cells lining the Bowman’s capsule in the glomerulus, as they form the third barrier of filtration preceded by the fenestrated endothelial glomerular cells and the glomerular basement membrane [6]. Podocytes possess a large number of foot processes forming slits, which filters fluid after passing through the first two barriers of filtration. The slit diaphragm serves as a signaling hub for the podocytes, with many structural proteins play their role in regulating the reactive elements of the glomerular filtration system, and thus are implicated in renal damage when dysfunctions occur with these proteins. Anatomically, it has been established that podocyte injury is a hallmark of diabetic nephropathy [3], while mesangial matrix expansion [7] and reduction in glomerular endothelial fenestration have been correlated with its progression [8]. The maladaptive factors that are accountable for these signals leading to DKD are yet not fully defined.
The Kallikrein-Kinin system has been implicated as a key player in the development of renal injury in DKD. Higher plasma prekallikrein levels were associated with higher AER in type 1 diabetic patients [9]. The AER and glomerular and tubular injury were attenuated in diabetic bradykinin 2 receptor (B2R−/−) null mice compared to wild-type diabetic B2R+/+ mice [10], [11]. In addition, previous findings from our renal global genome profiling study identified a number of dysregulated oxidative stress genes in diabetic B2R−/− mice known to be essential to the pathogenesis of DKD [12].
It is of note that the comparative proteomics discipline is increasingly utilized to identify new biomarkers in disease and experimental states. The advent of advanced liquid chromatography/tandem mass spectrometry (LC-MS/MS) proteomics platforms has made possible the construction of the protein profile of a cell at a given time and physiological/pathological state [13]. Hays et al. detected proteins belonging to the cytoskeletal activity functional network utilizing proteomics analysis of conditionally immortalized murine podocytes [14]. In addition, utilizing a combined proteome and transcriptome analysis of podocytes revealed a number of significantly altered proteins are linked to podocyte cytoskeleton and protein transport [15]. Furthermore, in a recent study by Rinchen et al in native mouse podocytes using a combined proteomic and transcriptomic approach provided new insights into candidate markers of podocyte biology [16]. A potential role for engagement of bradykinin 2 receptor (B2R) in podocyte biology has not been fully explored. In the present manuscript, we use advanced proteomics data analysis to determine the protein profile of cultured rat podocytes treated with bradykinin (BK). Pathway enrichment analysis was then performed to broaden our understanding of the cellular processes and networks occurring as a result of BK treatment, and to identify key players in the signaling pathway downstream of activation of B2R.
Material and methods
Materials
Bradykinin (SC090), culture and reagents were all purchased from Sigma-Aldrich (MO, St Louis, USA). DC ProteinTM assay, electrophoresis reagents and ClarityTM Western ECL were purchased from Bio-Rad (Hercules, California, USA). PD 98,059 and Ibuprofen were ordered from Cayman Co (MI, Ann Arbor, USA). Antibodies used as follow: Anti-β-Actin (Abcam, Cambridge, UK, ab8227), Phospho-p44/42 MAPK (ERK1/2) rabbit mAb (Cell Signaling Technology, Danvers, MA, USA, 4370 s), p42/44 MAPK rabbit mAb (ERK1/2) (Cell Signaling Technology, 4695 s), Peroxidase-conjugated affinipure donkey anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA, USA) 711-035-152), or affinipure donkey anti-mouse IgG (Jackson Immune, 715–035-150), mouse monoclonal antibody anti-COX-2 (clone COX214). HPLC water was purchased from Avantor (Valley Center, PA, USA), acetonitrile (ACN) from Thermo Fisher Scientific (Waltham, MA, USA), trypsin/Lys-C mixture was produced by Promega (Madison, WI, United States) and formic acid (FA) was obtained from (TCI America, Portland, OR, USA).
Mass spectrometry
Extraction and tryptic digestion of proteins
Conditionally immortalized rat podocytes grown in RPMI 1640 culture media containing insulin, PS, Glutamine, fetal bovine serum (FBS) and HEPES were used in our studies. The starvation media was deprived of FBS and Insulin. Serum starved cells were treated with BK (10−7M) for 3 and 6 h. The concentration of BK was selected based on our prior studies assessing the effects of various doses of BK on renal and vascular cells. With respect to the two time points (3 and 6 h), we were interested in determining the temporal changes in early response differentially expressed proteins in podocytes as a result of BK stimulation. Beads beating method was next employed for cell lysis and protein extraction. Briefly, zirconium beads (diameter 0.4 mm) and a 200 μL aliquot of aqueous solution containing 5% sodium deoxycholate (SDC) were added to the cell samples. The mixture was homogenized using Beadbug microtube homogenizer at 4 °C at 4000 rpm. Six times of 30-second-homogenization plus another 30-second-pause were performed to break the cells. The cell lysate was sonicated for 30 min at 0 °C to enhance the extraction of proteins. After centrifuging at 14,800 g for ten minutes, the supernatant was diluted ten X with 50 mM ammonium bicarbonate buffer. BCA protein assay kit (Thermo Scientific; Rockford, IL) was used for Protein determination. To reduce the protein sample, 1.5 μL of DTT (200 mM) was added for 45 min at 60 °C. 6 μL of Iodoacetamide (200 mM) to alkylate the sample was added in the dark for 45 min at 37.5 °C. Tryptic digestion was conducted by incubating with trypsin (0.4 µg) 37 °C for 18 h and microwave-digested for 30 min at 45 °C. To quench the digestion, 0.5 μL of formic acid (FA) was added. Meanwhile, SDC detergent was precipitated in the acidic condition. Samples were then mixed thoroughly and centrifuged 10 min at a speed of 14,800g. Digested peptides (Supernatant) were dried and mixed with 0.1% formic acid and 2% acetonitrile (ACN) for LC-MS/MS analysis.
LC-MS/MS assay
Data from LC-MS/MS was collected utilizing the Dionex Ultimate 3000 nano-LC system (Thermo Fisher Scientific) interfaced to an LTQ Orbitrap Velos mass spectrometer with a nano-ESI source (Thermo Fisher Scientific). A 1 µg aliquot of tryptic digests was analyzed by the LC-MS/MS. Generated peptides were purified utilizing the C18 PepMap 100 trap column, 3 µL/min flow rate. Peptide separation was done with a C18 Acclaim PepMap RSLC column (75 µm I.D. × 15 cm, 2 µm particle sizes, 100 Å pore sizes) with a flow rate of 350 nl/min. Mobile phase comprised of solution A (2% ACN/0.1% FA in HPLC water) and solution B (99.9% ACN/0.1% FA). The separation method was used according to our previously published protocol [13]. The MS was operated in positive ion mode. Two scan events at data dependent acquisition mode were employed in the MS/MS analysis. The detailed methodology for the scan events were used according to our previously published methods [13].
LC-MS/MS data analysis
The data from Liquid chromatography-electrospray ionization-tandem mass spectrometry LC-ESI-MS/MS was utilized to produce mascot generic format file (*.mgf) employing the Proteome Discover version 1.2 software (Thermo Scientific). Data were examined utilizing the SwissProt database (Rattus) in MASCOT version 2.4 (Matrix Science Inc., MA, USA). MASCOT identified peptides and proteins were reduced by PeptideProphet and ProteinProphet algorithms. Peptide identifications with probabilities higher than 95% were accepted, whereas those with probabilities higher than 99% containing a minimum of 2 peptides were allowed.
Pathway enrichment analysis
Pathway Studio software (version 11. Elsevier) along with Ingenuity Pathway Analysis software (IPA, Qiagen, Ingenuity® Systems) were used to assess functional correlations among the different treatment cohorts to assess associations, relationships, biological processes and molecular functions within the different treatment groups. For the statistical analysis, Fisher’s test was employed to evaluate nonrandom associations (protein–protein interaction and pathways). Gene symbols were imported for each experimental data set and its corresponding log2 fold change expression values were uploaded for functional analysis using IPA and pathway studio knowledgebase [13].
Cell treatment and analysis
Quiescent podocytes cultured in 6-well plates were treated with BK (10−7 M) in the presence and absence of MEK (MAP kinase kinase) inhibitor (PD-98059, 25x10−6 M) and/or a non-selective COX-1/2 inhibitor, Ibuprofen (10−6 M) for various time points. PGE2 levels were measured in the supernatants by Enzyme-immunoassay (EIA) as described [17]. Podocytes were lysed in 200 μL of lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholic acid, 1% NP40, plus protease and phosphatase inhibitors leupeptin 2 μg/ml, aprotinin 2 μg/ml, sodium orthovanadate 2 mM, PMSF 1 mM, Benzamidine 1 mM, sodium pyrophosphate 1 mM, and sodium fluoride 10 mM). Concentration of protein was assessed using the Lowry assay with BSA as standard. For western blotting of COX-2 levels and phosphorylation of ERK1/2, 20–30 μg of total proteins were separated by SDS-PAGE (polyacrylamide gel electrophoresis) as previously described [18], [19]. Protein bands were revealed by ECL kit on the Bio-Rad ChemiDoc MP imaging System. Protein bands were quantified using ImageJ software (NIH, USA), and a ratio to actin or total ERK1/2 was determined and expressed relative to untreated cells as control.
Statistical analysis
Simple descriptive statistics were initially conducted on the mean for each group and for each time point. The normality of the data was determined graphically using the Q-Q and numerically utilizing Shapiro-Wilk test for normality. When normality was met, independent t-test was conducted to compare the means between the groups and p-values were reported based on the test of homogeneity. When the normality of the data could not be assumed the alternative nonparametric Mann-Whitney U test was conducted as an alternative approach. Data were expressed as mean ± SE and significance was considered at p < 0.05.
Availability of Proteomic data. The proteomics data generated from LC-MS/MS was deposited in the PRIDE Archive (http://www.ebi.ac.uk/pride/archive/) via the PRIDE partner repository with the data set identifier PXD010015. Username: reviewer26015@ebi.ac.uk Password: drhw12lo.
Results
Proteomic analysis
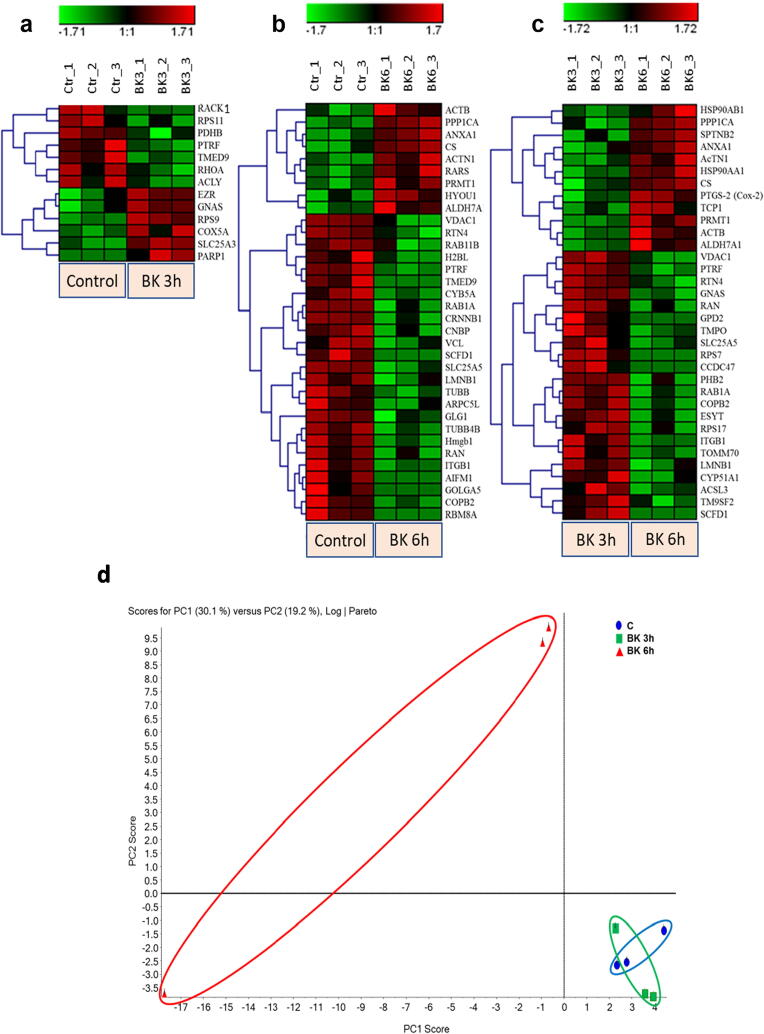
Proteome profiling of podocyte cell protein abundance was done using LC-MS/MS followed by MASCOT analysis of the resulting protein spectra from control and BK stimulated cells. Analysis showed a regulation of 280 proteins, reduced to 61 proteins when adjusting for significance (p < 0.05). These hits were split among 3 groups comparing the change in protein profile between each defined experimental condition, resulting in the analytical groups, control vs 3 h BK treatment (Fig. 1a), control vs 6 h BK treatment (Fig. 1b), and 3 h vs 6 h BK treatment (Fig. 1c). The pattern of significant protein upregulation and downregulation in the 3 analytical groups is shown in Table 1. A full list of the significantly modified proteins by BK (p < 0.05) along with their accession numbers, protein symbols, fold change, and log2 fold change are presented in Table 1, Table 2, Table 3. The hierarchal clustering of the expressed protein profile in control and BK treated cells are depicted by the heat maps shown in Fig. 1, where a row represents a protein and a column represents one of the triplicates for each experimental group. Some observed trends in the different heat maps is an increase in regulated proteins past the 3-hour BK treatment point, and the overlapping of a number of the modified proteins between the “Control vs BK 6 h” and “BK 3 h vs BK 6 h” groups. A number of proteins that are of particular interest are dysregulated by BK treatment including PTGS-2 (COX-2), RhoA, β-Catenin and Integrin-β1.
Fig. 1.
Hierarchical clustering (heat maps) and X showing the different proteins expression profile in the 3 experimental groups: (a) Control vs 3 h BK, (b) Control vs 6 h BK, and (c) 3 h vs 6 h BK. Each row represents a different protein while each column represents one triplicate from the corresponding experimental group. Green color represents downregulation of protein expression, whereas the red color represents upregulation of protein expression. Color intensity reflects the expression level of the proteins (scale). The label on the right-hand side of the heat maps represents the accession number of the proteins. (d) Principal Component Analysis (PCA) of BK-stimulated podocytes. The total normalized expression data of the proteins was used to depict the scatter plots of the first (X) and the second (Y) principal components. The numbering of the nodes is an identification of the samples. Letters used in the labels of the nodes represent the repeats. Blue oval encircles the control samples, green oval encircles the BK 3 h stimulated samples, and red oval encircles the BK 6 h stimulated samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Comparative list of proteins in the “Control vs 3 h BK treatment” experimental group.
| Accession | Protein Name | Symbol | p-value | Fold change | Log2 Fold change |
|---|---|---|---|---|---|
| P16638 | ATP Citrate Lyase | ACLY | 0.049 | 0.149 | −2.748 |
| P11240 | Cytochrome c oxidase subunit 5A | COX5A | 0.031 | 5.344 | 2.418 |
| P31977 | Ezrin | EZR | 0.033 | 1.358 | 0.442 |
| P63095 | GNAS Complex locus | GNAS | 0.034 | 1.318 | 0.399 |
| P85125 | Polymerase I and transcript release factor release factor | PTRF | 0.021 | 0.662 | −0.596 |
| P49432 | Pyruvate dehydrogenase | PDHB | 0.037 | 0.376 | −1.409 |
| P61589 | Ras homolog family member A | RHOA | 0.047 | 0.507 | −0.981 |
| P63245 | Receptor for activated C Kinase1 | RACK1 | 0.044 | 0.306 | −1.707 |
| P62282 | Ribosomal Protein S11 | RPS11 | 0.042 | 0.186 | −2.424 |
| P29314 | Ribosomal Protein S9 | RPS9 | 0.002 | 1.844 | 0.883 |
| P16036 | Solute carrier family 25 | SLC25A3 | 0.003 | 2.562 | 1.357 |
| Q5I0E7 | Transmembrane p24 trafficking protein 9 | TMED9 | 0.014 | 0.104 | −3.262 |
Table 2.
Comparative list of proteins in the “Control vs 6 h BK treatment” experimental group.
| Accession | Protein Name | Symbol | p-value | Fold change | Log2 Fold change |
|---|---|---|---|---|---|
| P60711 | Actin Beta | ACTB | 0.043 | 2.159 | 1.111 |
| Q9Z1P2 | Actinin alpha 1 | ACTN1 | 0.008 | 1.959 | 0.970 |
| Q64057 | Aldehyde Dehydrogenase 7 family member A1 | ALDH7A1 | 0.040 | 3.771 | 1.915 |
| P07150 | Annexin A1 | ANXA1 | 0.004 | 2.013 | 1.009 |
| Q9JM53 | Apoptosis inducing factor, mitochondrial | AIFM1 | 0.016 | 0.001 | −10.751 |
| P40329 | arginyl-tRNA synthetase | RARS | 0.015 | 2.237 | 1.161 |
| A1L108 | Actin-related protein 2/3 complex subunit 5-like protein | ARPC5L | 0.042 | 0.186 | −2.425 |
| Q9WU82 | Catenin Beta 1 | CTNNB1 | 0.009 | 0.171 | −2.545 |
| P62634 | CCHC-type zinc finger nucleic acid binding protein | CNBP | 0.024 | 0.198 | −2.339 |
| Q8VHF5 | Citrate Synthase | CS | 0.009 | 5.960 | 2.575 |
| O35142 | Coatomer protein complex subunit beta 2 | COPB2 | 0.017 | 0.101 | −3.308 |
| P00173 | Cytochrome b5 Type A | CYB5A | 0.008 | 0.082 | −3.609 |
| Q62638 | Golgi glycoprotein 1 | GLG1 | 0.026 | 0.346 | −1.531 |
| Q3ZU82 | Golgin 5A | GOLGA5 | 0.042 | 0.001 | −10.961 |
| P63159 | High mobility group box 1 | Hmgb1 | 0.009 | 0.112 | −3.159 |
| Q00715 | Histone Cluster 1, H2bl | – | 0.039 | 0.721 | −0.471 |
| Q63617 | Hypoxia up-regulated 1 | HYOU1 | 0.027 | 1.539 | 0.622 |
| P49134 | Integrin subunit Beta 1 | ITGB1 | 0.014 | 0.152 | −2.713 |
| P70615 | Lamin B1 | LMNB1 | 0.038 | 0.312 | −1.681 |
| P85125 | Polymerase I and transcript release factor release factor | PTRF | 0.002 | 0.297 | −1.750 |
| Q63009 | Protein arginine methyltransferase 1 | PRMT1 | 0.020 | 4.450 | 2.154 |
| P62138 | Protein Phosphatase 1 | PPP1CA | 0.001 | 3.394 | 1.763 |
| O35509 | RAB11B, member RAS oncogene family | RAB11B | 0.046 | 0.454 | −1.138 |
| Q6NYB7 | RAB1A, member RAS oncogene family | RAB1A | 0.015 | 0.314 | −1.670 |
| P62828 | RAN, member RAS oncogene family | RAN | 0.045 | 0.224 | −2.161 |
| Q9JK11 | Reticulon 4 | RTN4 | 0.018 | 0.497 | −1.007 |
| Q27W01 | RNA binding motif protein 8A | RBM8A | 0.008 | 0.001 | −10.174 |
| Q62991 | Sec 1 family domain | SCFD1 | 0.006 | 0.000 | −11.026 |
| Q09073 | Solute carrier family 25 | SLC25A5 | 0.003 | 0.608 | −0.717 |
| Q5I0E7 | Transmembrane p24 trafficking protein 9 | TMED9 | 0.006 | 0.000 | −11.362 |
| Q6P9T8 | Tubulin Beta 4B class Ivb | TUBB4B | 0.007 | 0.783 | −0.353 |
| P69897 | Tubulin Beta class I | TUBB | 0.020 | 0.895 | −0.160 |
| P85972 | Vinculin | VCL | 0.028 | 0.242 | −2.049 |
| Q9Z2L0 | Voltage dependent anion channel 1 | VDAC1 | 0.019 | 0.409 | −1.289 |
Table 3.
Comparative list of proteins in the “3hl vs 6 h BK treatment” experimental group.
| Accession | Protein Name | Symbol | p-value | Fold change | Log2 Fold change |
|---|---|---|---|---|---|
| P60711 | Actin Beta | ACTB | 0.020 | 2.608 | 1.383 |
| Q9Z1P2 | Actinin alpha 1 | ACTN1 | 0.014 | 1.698 | 0.764 |
| Q63151 | acyl-CoA synthetase long-chain family member 3 | ACSL3 | 0.044 | 0.246 | −2.025 |
| Q64057 | Aldehyde Dehydrogenase 7 family member A1 | ALDH7A1 | 0.035 | 3.099 | 1.632 |
| P07150 | Annexin A1 | ANXA1 | 0.027 | 1.572 | 0.652 |
| Q8VHF5 | Citrate Synthase | CS | 0.020 | 2.966 | 1.569 |
| O35142 | Coatomer protein complex subunit beta 2 | COPB2 | 0.008 | 0.139 | −2.851 |
| Q5U2X6 | Coiled-coil domain containing 47 | CCDC47 | 0.029 | 0.000 | −11.177 |
| Q64654 | Cytochrome P450 family 51 subfamily A member 1 | CYP51A1 | 0.045 | 0.203 | −2.297 |
| Q9Z1X1 | Extended synaptotagmin 1 | ESYT1 | 0.016 | 0.177 | −2.494 |
| P35571 | Glycerol-3-phosphate dehydrogenase 2 | GPD2 | 0.041 | 0.000 | −11.906 |
| P63095 | GNAS Complex locus | GNAS | 0.004 | 0.615 | −0.700 |
| P82995 | hsp90 alpha family class A member 1 | HSP90AA1 | 0.004 | 2.293 | 1.197 |
| P34058 | hsp90 alpha family class B member 1 | HSP90AB1 | 0.044 | 2.382 | 1.252 |
| P49134 | Integrin subunit Beta 1 | ITGB1 | 0.009 | 0.120 | −3.055 |
| P70615 | Lamin B1 | LMNB1 | 0.042 | 0.325 | −1.623 |
| P85125 | Polymerase I and transcript release factor release factor | PTRF | 0.007 | 0.449 | −1.154 |
| Q5XIH7 | Prohibitin 2 | PHB2 | 0.035 | 0.494 | −1.018 |
| P35355 | Prostaglandin-endoperoxide synthase 2 | PTGS2 | 0.016 | 1.666 | 0.737 |
| Q63009 | Protein arginine methyltransferase 1 | PRMT1 | 0.007 | 15.617 | 3.965 |
| P62138 | Protein Phosphatase 1 | PPP1CA | 0.015 | 2.926 | 1.549 |
| Q6NYB7 | RAB1A, member RAS oncogene family | RAB1A | 0.011 | 0.268 | −1.899 |
| P62828 | RAN, member RAS oncogene family | RAN | 0.038 | 0.198 | −2.334 |
| Q9JK11 | Reticulon 4 | RTN4 | 0.018 | 0.473 | −1.080 |
| P04644 | Ribosomal protein S17 | RPS17 | 0.047 | 0.219 | −2.191 |
| P62083 | Ribosomal protein S7 | RPS7 | 0.006 | 0.000 | −12.192 |
| Q62991 | Sec 1 family domain | SCFD1 | 0.008 | 0.001 | −10.543 |
| Q09073 | Solute carrier family 25 | SLC25A5 | 0.048 | 0.621 | −0.687 |
| Q9QWN8 | Spectrin Beta, non-erythrocytic 2 | SPTBN2 | 0.018 | 2.452 | 1.294 |
| P28480 | T-complex 1 | TCP1 | 0.047 | 6.174 | 2.626 |
| Q62733 | Thymopoietin | TMPO | 0.039 | 0.223 | −2.167 |
| Q75Q39 | Translocase of outer mitochondrial membrane 70 | TOMM70 | 0.020 | 0.108 | −3.217 |
| Q66HG5 | Trans-membrane 9 superfamily member 2 | TM9SF2 | 0.044 | 0.605 | −0.726 |
| Q9Z2L0 | Voltage dependent anion channel 1 | VDAC1 | 0.013 | 0.317 | −1.656 |
Principal component analysis (PCA): To appreciate the global proteome variations in response to BK stimulation between the different groups, we conducted PCA, a multivariate data analysis approach employed to scheme and convert the global alterations in big datasets into two-dimensional plots to illustrate the similarities in the variations amid the diverse groups. PCA plots for BK treated groups 3 and 6 h, displayed a well-organized parting among the various groups, suggesting that the BK time-point stimulations produced unique proteomic profiles. In addition, we observed similarities or overlay amongst the PCA plot of the 3 h stimulation by BK and control groups (Fig. 1d).
Pathway and network analysis of proteomic profiles
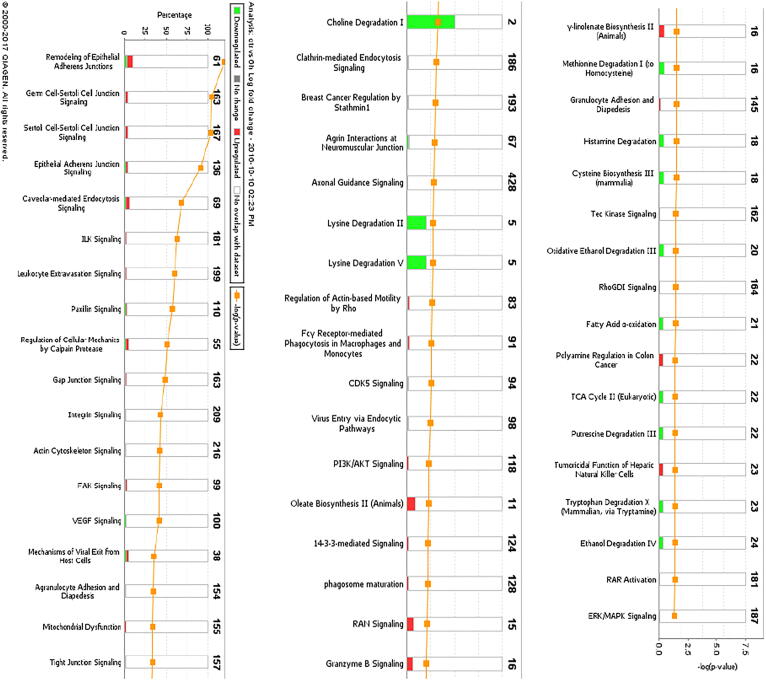
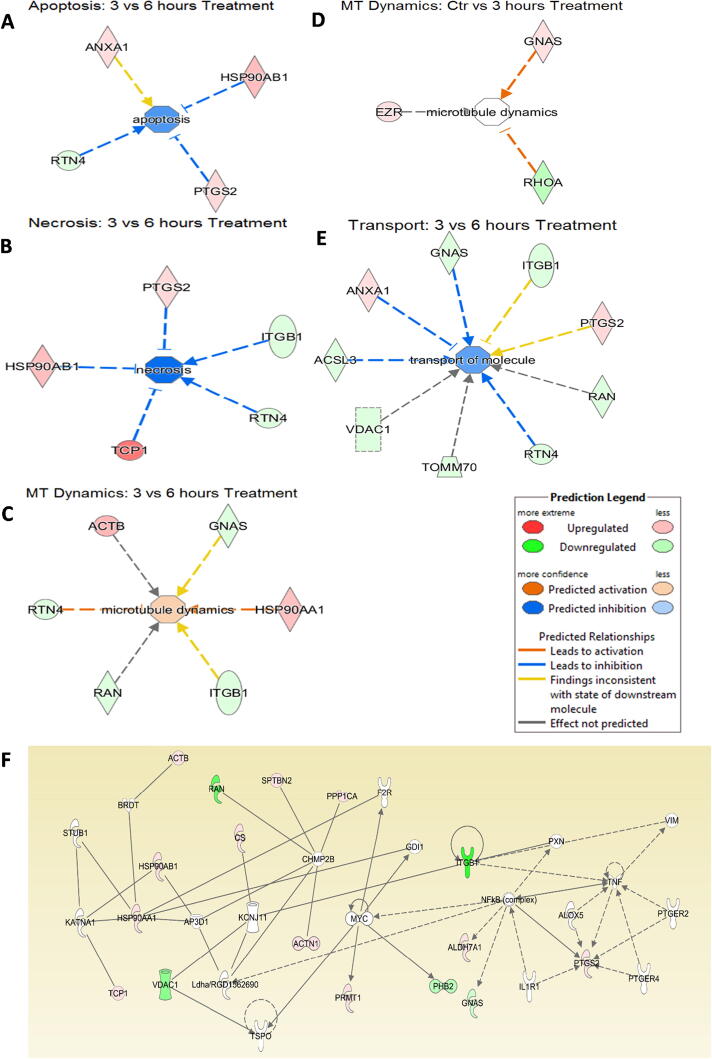
The differentially expressed proteome profiles in control and BK stimulated podocytes were analyzed by IPA. Fig. 2a–c depicts a graphical assessment of the canonical pathways of differentially (up- and down-regulated) proteins within and between each group. The canonical pathway analysis points to many intriguing processes that are suggested to be occurring with podocyte injury and apoptosis that includes upregulation of inflammatory cytokines and chemokines, Calpain signaling, Granzyme signaling and Ephrin B signaling. In addition, the PI3K pathway, PPAR signaling pathways, RhoA signaling, phospholipase C signaling, actin cytoskeleton signaling, and actin nucleation by ARP-WASP, remodeling of the epithelial adherens junctions, tight junction signaling, regulation by Rho of actin-based motility, ERK/MAPK signaling, NOS-III signaling, and hypoxia signaling in the cardiovascular system were shown to be among the top enriched pathways (Fig. 2a–c). Enrichment analysis of expressed proteins in podocytes stimulated with BK compared to control podocytes depicted that the most highly suspected diseases and biological functions to be involved fall under the umbrella of inhibition of cell death (inhibition of necrosis, cell death and apoptosis) and change in cytoskeletal elements (inhibition of transport of molecules and promotion of microtubule dynamics). Fig. 3a–d show the individual pathways from IPA’s “Diseases and Biological Functions” of the altered proteins. In addition network analysis depicted in Fig. 3f, showed that the upregulation of COX-2 is linked to the suggested expression/activity of the PGE receptors 2/4 (PTGER2 and PTGER4), the interleukin 1 receptor type 1 (IL1R1), tumor necrosis factor (TNF), NFκB, and arachidonate 5-Lipoxygenase (ALOX5).
Fig. 2.
Canonical Pathway Analysis of the comparative groups in unstimulated and BK-stimulated podocytes depicting the suggested cellular mechanisms involved. (a) Top canonical pathways related to proteins altered in the Control vs 3 h BK. (b) Top canonical pathways related to proteins altered in the Control vs 6 h BK. (c) Top canonical pathways related to proteins altered in the 3 h vs 6 h BK. Bars show the total number of proteins identified in each pathway. The green color represents the downregulated proteins, and the red color represents the upregulated proteins. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
IPA’s individual pathway analysis of different proteins from the different experimental groups, linking them to a suggested promotion or inhibition of a cellular event under the umbrella of inhibition of cell death or change in cytoskeletal activity. (a) Inhibition of Apoptosis comparing BK 6 h treated podocytes versus controls (ANXA1: Annexin A1, HSP90AB1: Hsp90 alpha family class B member 1, RTN4: Reticulon 4, PTGS-2: Prostaglandin H-Synthase 2). (b) Inhibition of Necrosis comparing BK-6h treated podocytes versus controls (ITGB1: Integrin subunit Beta 1, TCP-1: T-complex 1). (c) Activation of Microtubule Dynamics activity comparing BK- 3h treated podocytes vs BK-6h treated podocytes (ACTB: Actin Beta, GNAS: GNAS complex locus, HSP90AA1: Hsp90 alpha family class A member 1, RAN: RAN, member RAS family oncogene). (d) Suggestion of Microtubule Dynamics activity comparing BK-3h treated podocytes vs control (EZR: Ezrin, RHOA: Ras homolog family member A). (e) Inhibition of Transport of Molecules comparing BK-3h vs. Bk-6h treated podocytes (ACSL3: acyl-CoA synthetase long-chain family member 3, TOMM70: Translocase of outer mitochondrial membrane 70, VDAC1: voltage dependent anion channel 1. (f) The network titled “Cell Death and Survival, Cancer, GI Disease” of the BK -3h vs. BK-6h groups in the network analysis function of IPA showing PTGS-2/COX-2 with other proteins, expressed (colored) or suggested (uncolored). PTGS-2/COX-2 is directly in contact with the proteins ALOX5 (arachidonate-5-lipoxygenase), IL1R1 (interleukin 1 receptor type 1), PTGER2 (prostaglandin E receptor 2), PTGER4 (prostaglandin E receptor4), NFκB (complex) (nuclear factor kappa-light chain-enhancer of activated B cells), TNF (tumor necrosis factor).
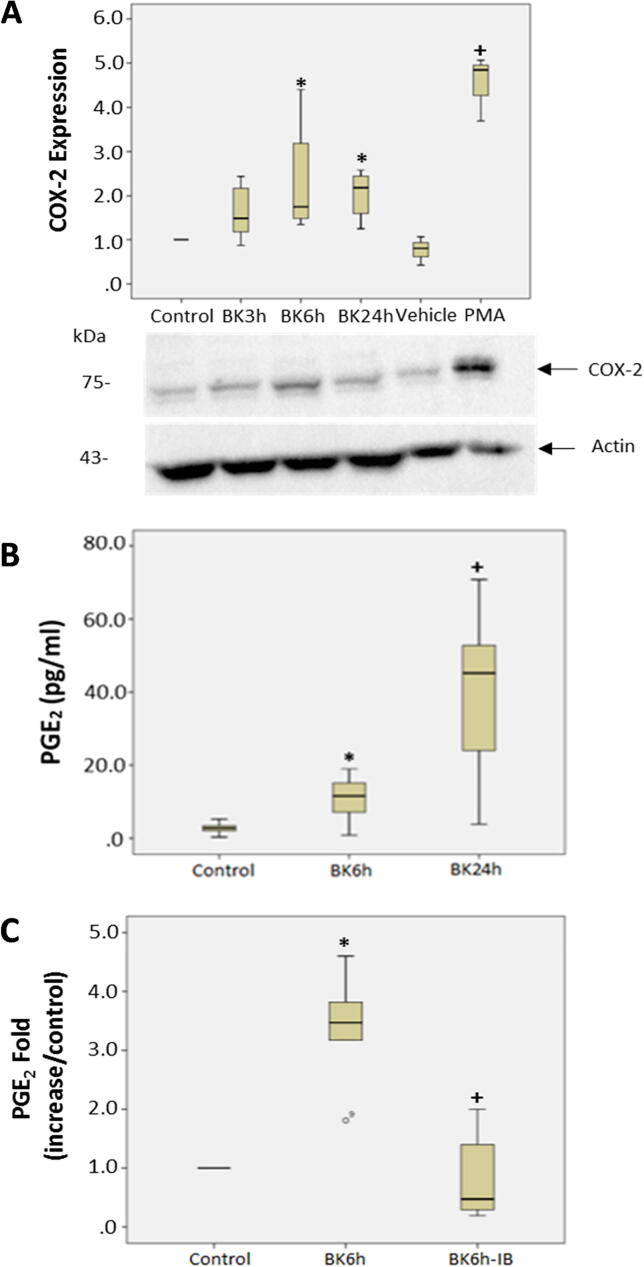
Effect of BK on COX-2 expression in podocytes
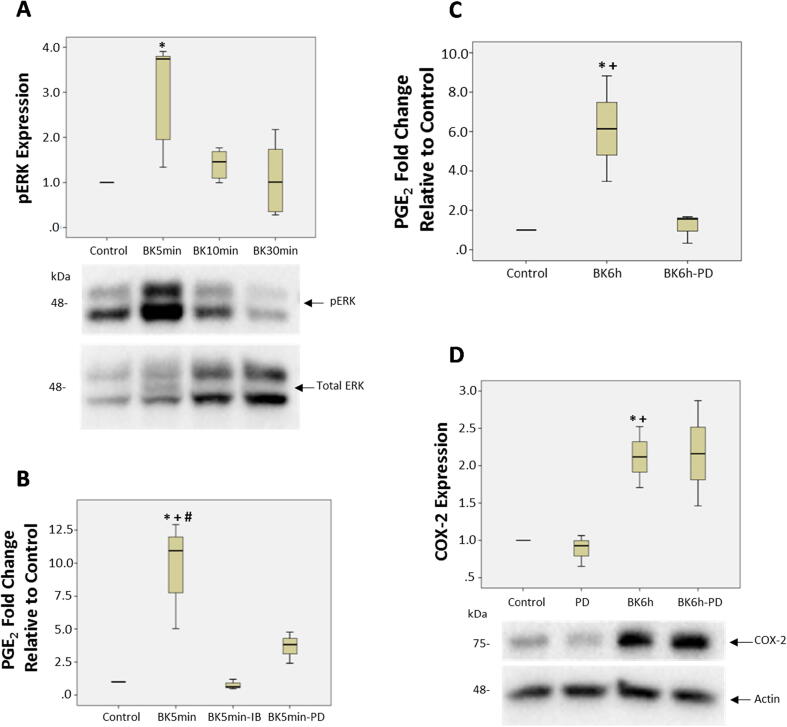
The proteomic analysis data depicting the effects of BK on COX-2 upregulation by LC-MS/MS were validated by treating quiescent podocytes with BK (10−7M) for different time points (3, 6 and 24 h). The 24 h time point was studied to assess the temporal expression of COX-2 in response to BK and to determine the time at which the expression of COX-2 peaks in response to BK. Podocytes were also stimulated with phorbol-12-myristate-13-acetate (PMA), a phorbol ester as a positive control for COX-2 expression. The results indicate that the protein levels of COX-2 are higher at 6 and 24 h post BK stimulation compared to control unstimulated cells (Fig. 4A, p < 0.008). PMA, which is a positive control, also increased the protein levels of COX-2 compared to DMSO treated podocytes (p < 0.05, Fig. 4). We next assessed if the increase in COX-2 protein level in response to BK is correlated with an increase in COX activity by measuring the production of the downstream metabolite PGE2. PGE2 levels were significantly higher in podocytes at 6 and 24 h post BK treatment compared to unstimulated cells indicating that BK increased the formation of PGE2 consequent to the induction of COX-2 expression (p < 0.0001, Fig. 4B). To further evaluate the effect of BK on COX-2, we studied the effects of a COX inhibitor on the production of PGE2 levels in response to BK. Podocytes were stimulated with BK for 6 h in the presence and absence of the COX inhibitor Ibuprofen (10−6 M). The results shown in Fig. 4C, demonstrated that the increased production in PGE2 in response to BK was significantly attenuated in the presence of Ibuprofen (p < 0.015, Fig. 4C).
Fig. 4.
Effect of BK on inflammatory signals in podocytes (a) BK stimulated COX-2 protein expression in podocytes. Quiescent podocytes were stimulated with BK (10−7M) for 3, 6, and 24 h, and with PMA (10−6M) and DMSO was used as vehicle and did not exceed 0.1% (10−6 M). Bar graph represents the fold change in COX-2 protein levels (*p-value = 0.008 vs, control; +p-value = 0.05 vs DMSO) and correspond to the average of 3 or more separate experiments. (b) BK stimulated PGE2 release in podocytes. Quiescent podocytes were stimulated with BK (10−7 M) for 3, 6, and 24 h. Bar graph represents the fold change in measured PGE2 levels (*p < 0.001 BK 6 h vs Control, +p < 0.001 BK 24 h vs Control), and is the sum of 3 or more separate experiments. (c) Ibuprofen inhibits BK-stimulated PGE2 release in podocytes. Quiescent podocytes were stimulated with BK (10−7 M) for 6 h in the presence and absence of Ibuprofen (10−6 M). PGE2 levels were measured by EIA. Bar graph represents the fold change in measured PGE2 levels (*p < 0.016 BK 6 h vs Control, +p = 0.016 BK 6 h-IB vs BK 6 h) and is the average of at least 3 separate experiments.
Role of extracellular regulated kinase ½ (ERK1/2) on COX activity in response to BK. The canonical pathway analysis generated by IPA of our proteomic data, suggested that the ERK1/2 pathway to be involved in the response of podocytes to BK treatment (Fig. 2b and 2c). To validate this finding, we first determined whether BK stimulates the activation of ERK1/2 in podocytes. Treatment of podocytes with BK (10−7M) for various times stimulated the phosphorylation of ERK1/2 compared to unstimulated cells, with a peak response at 5 min (p < 0.016, Fig. 5a). We next evaluated whether ERK1/2 plays a role in the production of PGE2 levels in response to BK treatment. PGE2 levels were measured in podocytes stimulated with BK (10−7M) for 5 min and for 6 h in the presence and absence of the MEK inhibitor PD98059 (25 μM). PD98059 is a selective and effective inhibitor of MEK 1 (MAPkinase kinase) an upstream activator of ERK1/2. PD98059 binds to MEK1 in its inactive state and hence stops its activation by c-Raf. Once inactivated, MEK1 will no longer be able to phosphorylate and active its downstream target ERK1/2. The results shown in Fig. 5B and 5C, demonstrate that the production of PGE2 levels in response to BK were significantly inhibited in the presence of the ERK1/2 inhibitor in the early time point (5 min, p < 0.024, Fig. 5B) as well as at 6 h (p < 0.05, Fig. 5C). We also evaluated the effects of Ibuprofen (10−6 M) on PGE2 production in response to BK stimulation for 5 min. The results demonstrate the PGE2 production in response to BK was significantly inhibited in the presence of the COX inhibitor (p < 0.024, Fig. 5B). Finally, we assessed whether the inhibition of the ERK1/2 pathway by PD98059 would influence the BK-induced COX-2 protein levels. The results shown in Fig. 5D, demonstrate that inhibition of ERK1/2 pathway did not significantly alter COX-2 protein levels in response to BK stimulation.
Fig. 5.
Role of extra cellular regulated kinase1/2 (ERK1/2) in BK stimulated expression of COX-2 and PGE2 production. (a) BK stimulated ERK1/2 phosphorylation (pERK 1/2) in podocytes. Quiescent podocytes were stimulated with BK (10−7 M) for 5, 10, 30 min. pERK and total ERK levels were assessed by western blot. Bar graph represents the fold change in pERK relative to total ERK protein levels and is the sum of 3 separate experiments (*p = 0.016). (b) Quiescent podocytes were stimulated with BK (10−7 M) for 5 min (5 min) in the presence and absence of PD98059 (25 μM) or Ibuprofen (10−6 M). Bar graph represents the fold change in measured PGE2 levels and is the sum of at least 3 separate experiments (*p-value = 0.004 BK 5 min vs Control; +p-value = 0.024 BK 5 min vs BK 5 min-IB; #p-value = 0.024 BK 5 min vs BK 5 min-PD). (c) Quiescent podocytes were stimulated with BK (10−7 M) for 6 h in the presence and absence of PD98059 (25 μM). Bar graph represents the fold change in measured PGE2 levels and is the sum of at least 3 separate experiments (*p < 0.03 BK 6 h vs Control, +p < 0.05 BK 6 h vs. BK 6 h-PD). (d) Quiescent podocytes were stimulated with BK (10−7 M) for 6 h in the presence and absence of PD98059 (25 μM). COX-2 and actin protein levels were measured by western blot. Bar graph represents the fold change in COX-2 proteins levels relative to β-actin protein levels and is the average of 3 separate experiments (*p-value = 0.035, BK 6 h vs. control; +p-value = 0.01 BK 6 h vs PD).
Discussion
The advent of proteomic analysis has transformed the way we understand the hidden cellular processes and networks, and by extension, the different cellular phenomena and pathologies these networks might be interrelated to. Our global proteomic analysis of podocytes using shotgun proteomics has shed light on the different cellular processes occurring in response to BK treatment that are involved in podocyte cell injury. Some of the proteins of particular interest that have been identified from canonical pathways as well as those based on the suggested interactions with other proteins and relevant cellular processes as highlighted by our systems biology techniques included; inflammatory cytokines (IL-1) and chemokines, Calpain signaling, Granzyme signaling, Ephrin B signaling, RhoA, β-catenin, integrin-β1, apoptosis-inducing factor (AIFM-1), and PTGS-2 (COX-2). Calpain signaling is linked to modulation of increased production of proinflammatory mediators and activation of podocyte apoptosis [20]. Moreover, the increase in inflammatory cytokines and chemokines in response to BK is another facet through which BK contributes to glomerular and podocyte injury [21], [22]. Enhancement of Granzyme signaling by BK, a cluster of serine proteases generated by granules in the cytoplasma with cytotoxic T cells promote podocyte cell injury and apoptosis by enhancing the activity of caspases [23]. Furthermore, BK may influence podocyte function by enhancing the Ephrin B signaling, which interfaces with nephrin to sustain the architectural integrity and role of the slit diaphragm [24]. RhoA has generally been identified as a classic player in the cytoskeletal rearrangement pathway [25], which is of importance to us considering that podocyte cell death is usually accompanied by cytoskeletal rearrangements in the form of detachment from the glomerular basement membrane [26]. This result is consistent with another study showing a downregulation of RhoA in a podocyte injury model, and where the knock-down of RhoA by siRNA has shown increased podocyte apoptosis [27]. β-Catenin is another protein implicated in cytoskeletal rearrangement activities, as it interacts with the cytoplasmic regions of cadherin, serves the crucial role of regulating the adherent junctions, as well as participating as a key player in the Wnt signaling pathway [28]. Previous research also indicated a decrease in β-catenin in a DKD model, where both the protein levels and the mRNA levels of β-catenin were decreased in hyperglycemic conditions with the presence of advanced glycation end products (AGEs) [28], consistent with the decrease observed in our proteomics data. Integrin-β1 has also been associated with podocyte biology, where a study showed that mice with specific integrin-β1 deletions cannot complete postnatal renal development, and die of proteinuria [29], suggesting a possible role of the downregulation of integrin-β1 in the progression of DKD. Of interest, AIFM-1 is a protein that is recruited to the nucleus when apoptotic signals are initiated, where it will lead to DNA fragmentation [30]. However, no studies have been conducted on the activation of AIFM-1 in podocytes, especially that podocytes undergo apoptosis in DKD [26].
Our proteomic analysis is the first to show that BK induces the expression of COX-2 in podocytes. This finding was also confirmed by assessing the protein abundance of COX-2 levels in podocytes in response to BK stimulation. Upregulation of COX-2 has been implicated in DKD, for its specific inhibition in podocytes reduced proteinuria in a glomerular injury model [31]. In addition, COX-2 upregulation was also observed in glomerular podocytes in models of DKD and the targeted overexpression of COX-2 in glomerular podocytes was associated with worsening of nephropathy in mice with type 1 diabetes [32], [33]. Treatment with COX-2 inhibitors was shown to attenuate the albuminuria observed as a result of DKD [34], [35]. Not only did BK increase the protein levels of COX-2 but PGE2 synthesis. This was demonstrated by measuring the production of PGE2 metabolite, which is one of the products of arachidonic acid metabolism generated by COX activity. The production of PGE2 in response to BK stimulation was inhibited with COX inhibitors suggesting that BK can directly or indirectly modulate COX-2 activity in podocytes.
To gain insights into the mechanisms through which BK activates COX-2, we investigated the role of ERK1/2 in this process, as suggested by the pathway enrichment analysis of our proteomic profiling analysis. Our results indicated that ERK1/2 phosphorylation was enhanced in response to BK and the increased production of PGE2 in response to BK was attenuated in the presence of ERK1/2 inhibitors. This finding implicates the ERK pathway in the mechanisms through which BK modulates the generation of PGE2 levels in podocytes and suggests that COX-2 is downstream of ERK1/2.
It is interesting to point out here that the COX metabolite PGE2 is implicated in inducing podocyte apoptosis. In this regard PGE2 has multifaceted properties depending on the context and cell types. It has been shown that PGE2 is implicated in inducing apoptosis in various cell types including T and B lymphocytes, lung fibroblasts, and mesangial cells [36], [37], [38]. In podocytes, PGE2 induced apoptosis in a Stat3-dependent pathway since inhibition of Stat3 improved the PGE2- or Adriamycin-dependent induction of podocyte injury [39], [40]. In addition, the apoptotic effects of COX-2 expression and PGE2 receptor EP1- signaling were involved in a model of calcineurin induced podocyte apoptosis and injury [41]. This is in line with the systems biology analysis that predicted physiological and pathological processes underlying the change in protein profile of podocytes in response to BK as well as the pathways linking our observed proteins with other suggested proteins. The main processes suggested to be occurring were those related to cell death (apoptosis, necrosis, and cell death) and those related to cytoskeletal remodeling (microtubule dynamics and transport of molecules). While the microtubule dynamics was suggested to be upregulated and the “Transport of Molecules” suggested to be downregulated, cytoskeletal activity plays a role in podocyte function and death, where podocyte detachment goes hand-in-hand with podocyte loss and progression of kidney disease [42]. The majority of these pathways suggest a regulation of cytoskeletal elements of the podocytes, which is of particular importance given that podocyte death is associated with detachment from the globular basement membrane.
The data described herein support the current paradigm of thinking that activation of kallikrein leads to cleavage of kininogen to release the proinflammatory peptide BK [43], [44]. BK activates its constitutively expressed B2R in the kidney in an autocrine and or paracrine fashion to generate a plethora of cellular signals that impact renal function [11]. Inactivation of B2R significantly attenuated protein excretion in diabetic mice and improved the augmented nephropathy in uninephrectomized db/db mice, lending support to the detrimental function of B2R in DKD [45]. Furthermore, diabetic B2R knockout mice exhibited decreased AER and attenuated tubular and glomerular injury, thus implicating a harmful role for B2R in DKD [10]. However, other studies have shown that BK and its B2R play a beneficial role in DKD. B2R knockout mice crossed with the insulin Akita (Ins2Akita) mice to generate the double knockout (In2Akita/B2R−/−) exhibited augmented albuminuria related to Ins2Akita mice alone [46]. Expression of B2R were shown to be attenuated in the glomeruli of STZ-diabetic rats and in podocytes cultured under hyperglycemic conditions. In addition, BK treatment attenuated the increased AER observed in diabetic rats and inhibited podocyte apoptosis [47]. The seeming changes in the function of B2R in DKD could be ascribed to variations in the DKD models studied, metabolic state and genetic background of animal models and essentials of the investigational approach and target variables evaluated.
It is important to point out here that some of the effects we observed in response to BK could conceivably be mediated via activation of B1-receptors expressed in podocytes. In this regard, it is feasible that once podocytes are exposed to BK, the peptidases that are expressed in podocytes can cleave BK to generate des-Arg9-BK, the B1-receptor ligand, which in turn will bind to its receptors in an autocrine manner to transduce its signal. This possibility will be verified in future studies.
Conclusion
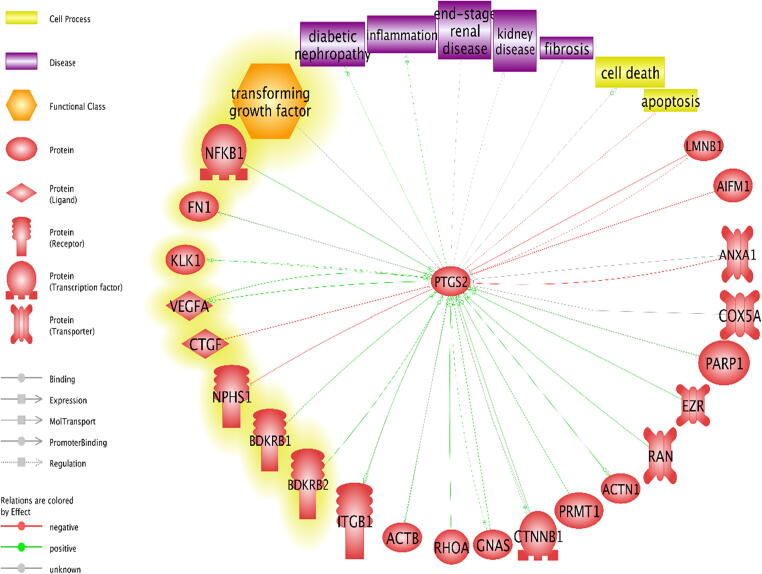
The findings of the present manuscript lends further support to our hypothesis that engagement of B2 receptors contribute to DKD development and establish that BK enhances the expression of COX-2 and stimulates that production of PGE2, factors that are associated with promotion of albuminuria and podocyte apoptosis (Fig.6). The current findings offer new understandings into mechanisms through which BK signaling processes modulate the pathogenesis of DKD and identify new targets for therapy.
Fig. 6.
Pathway Studio analysis of the relationship for PTGS-2/COX-2 to the linked proteins from the 3 experiments according to the software, and to inputted proteins highlighted in yellow, cell processes in yellow boxes, and diseases related to cell death and diabetic nephropathy in purple boxes, with the software constructing extensive links between the different elements according to its knowledgebase (NFB1: Nuclear factor Kappa B subunit 1, FN1: Fibronectin 1, KLK1: Kallikrein 1, VEGFA: Vascular endothelial growth factor A, CTGF: Connective tissue growth factor, NPHS1: Nephrin, BDKRB1: Bradykinin receptor B1, BDKRB2: Bradykinin receptor B2). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Compliance with Ethical Requirements
This article does not contain any studies with human or animal subjects
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by a grant from the Medical Practice Plan, Faculty of Medicine and from Qatar Research National Funds, NPRP 9-144-3-021. The authors thank Dr. Assaad Eid, Department of Anatomy, Cell Biology and Physiological Sciences, AUB, for providing the immortalized rat podocytes.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Bjornstad P., Cherney D., Maahs D.M. Early diabetic nephropathy in type 1 diabetes: new insights. Curr Opin Endocrinol Diabet Obes. 2014;21:279–286. doi: 10.1097/MED.0000000000000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.F.C. Sasso, P. Chiodini, O. Carbonara, L. De Nicola, G. Conte, T. Salvatore, R. Nasti, et. al. High cardiovascular risk in patients with Type 2 diabetic nephropathy: the predictive role of albuminuria and glomerular filtration rate. The NID-2 Prospective Cohort Study. Nephrol Dial Transplant, 27(2012) 2269-2274. [DOI] [PubMed]
- 3.Pagtalunan M.E., Miller P.L., Jumping-Eagle S., Nelson R.G., Myers B.D., Rennke H.G. Podocyte loss and progressive glomerular injury in type II diabetes. J Clin Invest. 1997;99:342–348. doi: 10.1172/JCI119163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stitt-Cavanagh E., MacLeod L., Kennedy C. The podocyte in diabetic kidney disease. Sci World J. 2009;9:1127–1139. doi: 10.1100/tsw.2009.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka N., Babazono T., Takagi M., Yoshida N., Toya K., Nyumura I. Albuminuria and reduced glomerular filtration rate for predicting the renal outcomes in type 2 diabetic patients. Nephrology (Carlton) 2015;20:531–538. doi: 10.1111/nep.12446. [DOI] [PubMed] [Google Scholar]
- 6.Pavenstadt H., Kriz W., Kretzler M. Cell biology of the glomerular podocyte. Physiol Rev. 2003;83:253–307. doi: 10.1152/physrev.00020.2002. [DOI] [PubMed] [Google Scholar]
- 7.Guan T.H., Chen G., Gao B., Janssen M.R., Uttarwar L., Ingram A.J. Caveolin-1 deficiency protects against mesangial matrix expansion in a mouse model of type 1 diabetic nephropathy. Diabetologia. 2013;56:2068–2077. doi: 10.1007/s00125-013-2968-z. [DOI] [PubMed] [Google Scholar]
- 8.Weil E.J., Lemley K.V., Mason C.C., Yee B., Jones L.I., Blouch K., Lovato T., Richardson M., Myers B.D., Nelson R.G. Detachment and reduced glomerular capillary endothelial fenestration promote kidney disease in type 2 diabetic nephropathy. Kidney Int. 2012;82(9):1010–1017. doi: 10.1038/ki.2012.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffa A.A., Durazo-Arvizu R., Zheng D., Lackland D.T., Srikanth S., Garvey W.T. Plasma prekallikrein: a risk marker for hypertension and nephropathy in type 1 diabetes. Diabetes. 2003;52:1215–1221. doi: 10.2337/diabetes.52.5.1215. [DOI] [PubMed] [Google Scholar]
- 10.Tan Y., Keum J.S., Wang B., McHenry M.B., Lipsitz S.R., Jaffa A.A. Targeted deletion of B2-kinin receptors protects against the development of diabetic nephropathy. Am J Physiol Renal Physiol. 2007;293:F1026–F1035. doi: 10.1152/ajprenal.00203.2007. [DOI] [PubMed] [Google Scholar]
- 11.Tan Y., Wang B., Keum J.S., Jaffa A.A. Mechanisms through which bradykinin promotes glomerular injury in diabetes. Am J Physiol Renal Physiol. 2005;288:F483–F492. doi: 10.1152/ajprenal.00165.2004. [DOI] [PubMed] [Google Scholar]
- 12.Jaffa Miran A., Kobeissy Firas, Al Hariri Moustafa, Chalhoub Hussein, Eid Assaad, Ziyadeh Fuad N., Jaffa Ayad A., Madeddu Paolo. Global Renal Gene Expression Profiling Analysis in B2-Kinin Receptor Null Mice: Impact of Diabetes. PLoS ONE. 2012;7(9):e44714. doi: 10.1371/journal.pone.0044714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Hariri M., Elmedawar M., Zhu R., Jaffa M.A., Zhao J., Mirzaei P. Proteome profiling in the aorta and kidney of type 1 diabetic rats. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0187752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hays T., Ma'ayan A., Clark N.R., Tan C.M., Teixeira A., Choi J.W. Proteomics analysis of the non-muscle myosin heavy chain IIa-enriched actin-myosin complex reveals multiple functions within the podocyte. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boerries M., Grahammer F., Eiselein S., Buck M., Meyer C., Goedel M. Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int. 2013;83:1052–1064. doi: 10.1038/ki.2012.487. [DOI] [PubMed] [Google Scholar]
- 16.Rinschen M.M., Godel M., Grahammer F., Zschiedrich S., Helmstadter M., Kretz O. A Multi-layered Quantitative In Vivo Expression Atlas of the Podocyte Unravels Kidney Disease Candidate Genes. Cell Rep. 2018;23:2495–2508. doi: 10.1016/j.celrep.2018.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pradelles P., Grassi J., Maclouf J. Enzyme immunoassays of eicosanoids using acetylcholine esterase as label: an alternative to radioimmunoassay. Anal Chem. 1985;57:1170–1173. doi: 10.1021/ac00284a003. [DOI] [PubMed] [Google Scholar]
- 18.Dagher O.K., Jaffa M.A., Habib A., Ziyadeh F.N., Jaffa A.A. Heteromerization fingerprints between bradykinin B2 and thromboxane TP receptors in native cells. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0216908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habib A., Shamseddeen I., Nasrallah M.S., Antoun T.A., Nemer G., Bertoglio J. Modulation of COX-2 expression by statins in human monocytic cells. FASEB J. 2007;21:1665–1674. doi: 10.1096/fj.06-6766com. [DOI] [PubMed] [Google Scholar]
- 20.Ji J.J., Su L., Lu Z. Critical roles of calpain in inflammation. Biomedical Rep. 2016;5:647–652. doi: 10.3892/br.2016.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders H. Of Inflammasomes and Alarmins: IL-1b and IL-1a in Kidney Disease. JASN. 2016;27:2564–2575. doi: 10.1681/ASN.2016020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber T.B., Reinhardt H.C., Exner M., Burger J.A., Kerjaschki D., Saleem M.A. Expression of Functional CCR and CXCR Chemokine Receptors in Podocytes. J Immunol. 2002;168:6244–6252. doi: 10.4049/jimmunol.168.12.6244. [DOI] [PubMed] [Google Scholar]
- 23.Bots M., Medema J.P. Granzyme at a glance. J Cell Sci. 2006;119:5011–5014. doi: 10.1242/jcs.03239. [DOI] [PubMed] [Google Scholar]
- 24.Fukusumi Y., Zhang Y., Yamagishi R., Oda K., Watanabe T., Matsui K. Nephrin-Binding Ephrin-B1 at the Slit Diaphragm Controls Podocyte Function through the JNK Pathway. JASN. 2018;29:462–1474. doi: 10.1681/ASN.2017090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jatho A., Hartmann S., Kittana N., Mugge F., Wuertz C.M., Tiburcy M. RhoA Ambivalently Controls Prominent Myofibroblast Characteritics by Involving Distinct Signaling Routes. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0137519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brosius F.C., Coward R.J. Podocytes, signaling pathways, and vascular factors in diabetic kidney disease. Adv Chronic Kidney Dis. 2014;21:304–310. doi: 10.1053/j.ackd.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Z., Zhang L., Chen Y., Zhang H., Yu C., Zhou F. RhoA deficiency disrupts podocyte cytoskeleton and induces podocyte apoptosis by inhibiting YAP/dendrin signal. BMC Nephrol. 2016;17:66. doi: 10.1186/s12882-016-0287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park S.J., Ahn E.M., Ha T.S., Shin J.I. beta-catenin and its alteration in an experimental model of diabetic nephropathy. Iran J Kidney Dis. 2014;8:299–309. [PubMed] [Google Scholar]
- 29.Kanasaki K., Kanda Y., Palmsten K., Tanjore H., Lee S.B., Lebleu V.S. Integrin beta1-mediated matrix assembly and signaling are critical for the normal development and function of the kidney glomerulus. Dev Biol. 2008;313:584–593. doi: 10.1016/j.ydbio.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kettwig M., Schubach M., Zimmermann F.A., Klinge L., Mayr J.A., Biskup S. From ventriculomegaly to severe muscular atrophy: expansion of the clinical spectrum related to mutations in AIFM1. Mitochondrion. 2015;21:12–18. doi: 10.1016/j.mito.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Cheng H., Fan X., Moeckel G.W., Harris R.C. Podocyte COX-2 exacerbates diabetic nephropathy by increasing podocyte (pro)renin receptor expression. J Am Soc Nephrol. 2011;22:1240–1251. doi: 10.1681/ASN.2010111149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komers R., Lindsley J.N., Oyama T.T., Schutzer W.E., Reed J.F., Mader S.L. Immunohistochemical and functional correlations of renal cyclooxygenase-2 in experimental diabetes. J Clin Invest. 2001;107:889–898. doi: 10.1172/JCI10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng H., Fan X., Guan Y., Moeckel G.W., Zent R., Harris R.C. Distinct roles for basal and induced COX-2 in podocyte injury. J Am Soc Nephrol. 2009;20:1953–1962. doi: 10.1681/ASN.2009010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quilley J., Santos M., Pedraza P. Renal protective effect of chronic inhibition of COX-2 with SC-58236 in streptozotocin-diabetic rats. Am J Physiol Heart Circ Physiol. 2011;300:H2316–H2322. doi: 10.1152/ajpheart.01259.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogt L., de Zeeuw D., Woittiez A.J., Navis G. Selective cyclooxygenase-2 (COX-2) inhibition reduces proteinuria in renal patients. Nephrol Dial Transplant. 2009;24:1182–1189. doi: 10.1093/ndt/gfn644. [DOI] [PubMed] [Google Scholar]
- 36.Nakanishi M., Rosenberg D.W. Multifaceted roles of PGE2 in inflammation and cancer. Semin Immunopathol. 2013;25:123–137. doi: 10.1007/s00281-012-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soleymani S.F., Jeddi M.T., Ardekani A.M. Prostaglandin E2 induces growth inhibition, apoptosis and differentiation in T and B cell-derived acute lymphoblastic leukemia cell lines (CCRF-CEM and Nalm-6) Prostaglandins Leukot Essent Fatty Acids. 2012;87:17–24. doi: 10.1016/j.plefa.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Yu X., Yang Y., Yuan H. Inhibition of COX-2/PGE2 cascade ameliorates cisplatin-induced mesangial cell apoptosis. Am J Transl Res. 2017;9:1222–1229. [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Chang J.H., Paik S.Y. Calcineurin (CN) activation promotes apoptosis of glomerular podocytes both in vitro and in vivo. Mol Endocrinol. 2011;25:1376–1386. doi: 10.1210/me.2011-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J., Wu Y., Wang L. mPGES-1-derived prostaglandin E2 stimulates Stat3 to promote podocyte apoptosis. Apoptosis. 2017;22:1431–1440. doi: 10.1007/s10495-017-1418-7. [DOI] [PubMed] [Google Scholar]
- 41.Wang L., Chang J.H., Paik S.Y., Tang Y., Eisner W., Spurney R.F. Calcineurin (CN) activation promotes apoptosis of glomerular podocytes both in vitro and in vivo. Mol Endocrinol. 2011;25:1376–1386. doi: 10.1210/me.2011-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kriz W., Lemley K.V. A potential role for mechanical forces in the detachment of podocytes and the progression of CKD. J Am Soc Nephrol. 2015;26:258–269. doi: 10.1681/ASN.2014030278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Björkqvist J., Jämsä A., Renné T. Plasma kallikrein: the bradykinin-producing enzyme. Thromb Haemost. 2013;110:399–407. doi: 10.1160/TH13-03-0258. [DOI] [PubMed] [Google Scholar]
- 44.Tang S.C.W., Yiu W.H. Innate immunity in diabetic kidney disease. Nat Rev Nephrol. 2020;16:1–17. doi: 10.1038/s41581-019-0234-4. [DOI] [PubMed] [Google Scholar]
- 45.S.C. Tang, L.Y. Chan, J.C. Leung JC, A.S. Cheng, H.Y Lan, K.N. Lai. Additive renoprotective effects of B2-kinin receptor blocker and PPAR-gamma agonist in uninephrectomized db/db mice. Laboratory investigation; a journal of technical methods and pathology 91 (2013)1351-1362 [DOI] [PubMed]
- 46.Kakoki M., Takahashi N., Jennette J.C., Smithies O. Diabetic nephropathy is markedly enhanced in mice lacking the bradykinin B2 receptor. Proc National Academy Sci USA. 2004;101(2004):13302–13305. doi: 10.1073/pnas.0405449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwak S.J., Paeng J., Kim D.H., Lee S.H., Nam B.Y., Kang H.Y. Local kallikrein–kinin system is involved in podocyte apoptosis. Apoptosis. 2011;16:478–490. doi: 10.1007/s10495-011-0585-1. [DOI] [PubMed] [Google Scholar]