Abstract
Up-regulated expression of INHBA has been reported in multiple malignant tumors. However, in nasopharyngeal carcinoma (NPC), the expression pattern and clinical significance of INHBA are still unclear. This study aimed to detect the expression of INHBA and its prognostic significance in NPC, then explore the tumor-associated functions of INHBA gene and the potential mechanism. The INHBA expression of mRNA levels in tumor tissues and noncancerous nasopharyngeal tissues was investigated by RT-qPCR. The protein expression in cells were detected by western blot. Cell proliferation was detected by CCK assay and cell invasion ability was evaluated by Transwell assay. The expression of INHBA in paraffin-embedded NPC tissues was detected by immunohistochemistry (IHC). Statistical analyses were further applied to assess the clinical significance of INHBA expression. The result reveals INHBA mRNA level is elevated in NPC tissues compared to those in noncancerous nasopharyngeal epithelial tissues. In paraffin-embedded NPC tissues, immunoreactivity of INHBA was primarily detected in 53.70% (58/108) of these patients. The overexpression was notably associated with the clinical stage (UICC) (P=0.048), N classification (P=0.042), carotid sheath involvement (P=0.016), and decreased disease-free survival (DFS) (P=0.004) and overall survival (OS) (P=0.010). Multivariate analysis revealed that INHBA expression was an independent prognostic factor for DFS (P=0.028). CCK assay showed SUNE1 cells’ proliferation was decreased in INHBA knockdown group than control. Transwell assay showed the invasion of SUNE1 cells was decreased in INHBA knockdown group by comparison with control. Further study showed knockdown of INHBA expression in SUNE1 cells could block the TGF-β signaling pathway. In conclusion, INHBA is up-regulated in NPC, and is significantly correlated with clinical stage (UICC), N stage, carotid sheath involvement, and survival. Knockdown INHBA in SUNE1 cells could inhibit the cells’ proliferation and invasion. The underlying mechanism may be blockade of the TGF-β signaling pathway.
Keywords: INHBA, NPC, prognosis, proliferation, invasion
Introduction
Nasopharyngeal carcinoma (NPC) is a highly invasive endemic malignancy originating from the surface epithelium of posterior nasopharynx in Southeastern Asia, North Africa, the Arctic region, and South China, especially Guangdong province [1,2]. NPC is noted for high mortality and morbidity. Most patients exhibit local or distant metastases at the time of diagnosis [2,3]. The etiology of NPC remains ambiguous. Epidemiologic studies reveal that both hereditary factors and environmental aspects play a role in its pathogenesis [4]. Racial background, infection of Epstein-Barr-Virus (EBV), and environmental carcinogens contribute to its occurrence and development [5,6]. Unlike other head-neck malignancies, NPC is highly aggressive and affected patients are predisposed to distant metastases [7]. In patients with localized disease, the survival rate is relatively satisfactory, ranging from 80% to 90%, but in those with metastatic or relapsing disease, it is usually less than 20% [8].
In accordance with the widespread use of intensity-modulated radiation therapy (IMRT) and potent chemotherapy, the local control and overall survival rate of patients with NPC have been considerably improved [9,10]. However, despite the best available treatment strategies, local recurrence and distant metastasis remain the main reasons for treatment failure, approximately 5~15% and 15~30%, respectively [11]. Therefore, a novel applicable marker of NPC is in urgent need for early diagnosis and targeted therapeutic strategies.
INHBA is located at 7p14.1, encoding Inhibin βA which is a member of the transforming growth factor β (TGF-β) superfamily [12]. It could form both activin A by homo-dimerizing and inhibin by hetero-dimerizing with inhibin βB. Activin and inhibin were observed to participate in multiple biologic activities including proliferation, differentiation, metabolism, homeostasis, apoptosis and tumor development [13] through autocrine, endocrine or paracrine mechanisms [14]. In the recent years, the up-regulated expression of INHBA has been reported in multiple malignant tumors, including lung carcinoma [15], gastric carcinoma [16], colorectal carcinoma [17], and bladder carcinoma [18]. Our previous study examined the expression level of INHBA in breast cancer, and revealed its clinical and prognostic significance [19]. However, its clinically significant and prognostic effects in NPC have not been systemically evaluated. In the current study, we attempted to determine the expression level of INHBA, evaluate its prognostic value for NPC, and explore the tumor-associated functions and the potential mechanism of the INHBA gene in an NPC cell line.
Materials and methods
Patients and specimens
A total of 108 patients with primary NPC from April 2010 to December 2017 in the First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China were enrolled in this study. In these cases male were 78 (72.2%) and female were 30 (27.8%), with median age of 48 years (range, 16~80 years). All tumors were pathologically confirmed nasopharyngeal epithelium origination. Patients were restaged basing on the 8th edition of the International Union Against Cancer (UICC) TNM staging system. There were 3, 23, 47, 25 and 4 patients respectively belonging to stage I, II, III, IV, and V category. Histological subtypes of NPC were determined according to the WHO tumor classification rules. type I: keratinizing squamous-cell carcinoma, type II: differentiated non-keratinous carcinoma, type III: undifferentiated non-keratinous carcinoma. All of these patients have received curative standardized radiotherapy, 34 of them were also treated with chemotherapy. The clinicopathologic characteristics of these patients are summarized in Table 1.
Table 1.
Correlation of INHBA expression with clinicopathologic features
| Characteristic | Total (n=108) | INHBA expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low (n=50) | High (n=58) | |||
| Age (years) | 0.805 | |||
| ≤46 | 44 (40.7%) | 21 (47.7%) | 23 (52.3%) | |
| >46 | 64 (59.3%) | 29 (45.3%) | 35 (54.7%) | |
| Gender | 0.213 | |||
| Male | 78 (72.2%) | 39 (50.0%) | 39 (50.0%) | |
| Female | 30 (27.8%) | 11 (36.7%) | 19 (63.3%) | |
| Histology | 0.994 | |||
| WHO I | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| WHO II | 41 (38.0%) | 19 (46.3%) | 22 (53.7%) | |
| WHO III | 67 (62.0%) | 31 (46.3%) | 35 (53.7%) | |
| Clinical stage (UICC) | 0.048* | |||
| I | 4 (3.7%) | 3 (75.0%) | 1 (25.0%) | |
| II | 26 (24.1%) | 17 (65.4%) | 9 (34.6%) | |
| III | 49 (45.4%) | 20 (40.8%) | 29 (59.2%) | |
| IV | 25 (23.1%) | 10 (40.0%) | 15 (60.0%) | |
| V | 4 (3.7%) | 0 (0.0%) | 4 (100.0%) | |
| T classification | 0.596 | |||
| T1 | 13 (12.0%) | 8 (61.5%) | 5 (38.5%) | |
| T2 | 42 (38.9%) | 20 (47.6%) | 22 (52.4%) | |
| T3 | 33 (30.6%) | 13 (39.4%) | 20 (60.6%) | |
| T4 | 20 (18.5%) | 9 (45.0%) | 11 (55.0%) | |
| N classification | 0.042* | |||
| N0 | 31 (28.7%) | 20 (64.5%) | 11 (35.5%) | |
| N1 | 27 (25.0%) | 13 (48.1%) | 14 (51.9%) | |
| N2 | 40 (37.0%) | 15 (37.5%) | 25 (62.5%) | |
| N3 | 10 (9.3%) | 2 (20.0%) | 8 (80.0%) | |
| Carotid sheath involvement | 0.016* | |||
| No | 18 (16.7%) | 13 (72.2%) | 5 (27.8%) | |
| Yes | 90 (83.3%) | 37 (41.1%) | 53 (58.9%) | |
| Nasal cavity involvement | 0.153 | |||
| No | 77 (71.3%) | 39 (50.6%) | 38 (49.4%) | |
| Yes | 31 (28.7%) | 11 (35.5%) | 20 (64.5%) | |
| Maxillary sinus involvement | 0.570 | |||
| No | 86 (79.6%) | 41 (47.7%) | 45 (52.3%) | |
| Yes | 22 (20.4%) | 9 (40.9%) | 13 (59.1%) | |
| Neck lymph node level involvement | 0.059 | |||
| No | 31 (28.7%) | 20 (64.5%) | 11 (35.5%) | |
| Level I-III | 67 (62.0%) | 28 (41.8%) | 39 (58.2%) | |
| Level IV-V | 10 (9.3%) | 2 (20.0%) | 8 (80.0%) | |
| Maximum neck lymph node diameter | 0.054 | |||
| <20 mm | 54 (50.0%) | 30 (55.6%) | 24 (44.4%) | |
| ≥20 mm | 54 (50.0%) | 20 (37.0%) | 34 (63.0%) | |
WHO, World Health Organization.
P≤0.05.
Follow-up time was calculated from the date of diagnosis to the date of death or relapse or the lasted census date. Overall survival (OS) was defined as the time from first diagnosis to the last follow-up or to the date of death for any reason. Disease-free survival (DFS) was defined as the time from first diagnosis to the date of death, or disease progression at locoregional and/or distant sites, or to the last follow-up. The follow-up time of the NPC cohort ranged from 3 to 89 months (median 46 months). In order to investigate the different RNA expression level of INHBA in tumor and noncancerous tissues, we also collected 40 cancerous samples and 16 noncancerous nasopharyngeal samples. These tissue samples were immediately immersed into RNAlater (Sigma-Aldrich R0901, St. Louis., MO, USA) and then stored at 4°C overnight, followed by preservation at -80°C.
Patient consents for-research use of these clinical materials were gained prior. All the protocol of this study was approved by the Institutional Research Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University.
Cell culture
SUNE1 nasopharyngeal carcinoma cells (Cell bank of the Chinese Academy of Sciences, Shanghai) were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA), penicillin (100 units/ml), and streptomycin (100 units/ml) maintained at 37°C and 5% CO2 incubator.
Reverse transcription-quantitative polymerase chain reaction (RT-PCR) analysis
Total RNA samples were extracted from cancerous tissues and noncancerous nasopharyngeal tissues using Trizol reagent (Invitrogen, CA, USA) following the manufacturer’s instructions. The RNA samples were pretreated by RNase-free DNase. 2 μg of RNA was used for cDNA synthesis. For the amplification of INHBA cDNA, an initial amplification using INHBA-specific primers was performed with a denaturation at 95°C for 10 min, followed by 40 denaturation cycles at 95°C for 20 s, then primer annealing at 58°C for 20 s, and then primer extension phase at 72°C for 20 s. The completion of these cycling steps was a final extension at 72°C for 5 min and the reaction mixture was stored at 4°C. Then RT-PCR was performed to confirm the fold increase of mRNA in each of the cancerous tissue samples and noncancerous nasopharyngeal tissue samples. The following primers were used: INHBA, 5’-CCTCGGAGATCATCACGTTT-3’ (forward) and 5’-CCCTTTAAGCCCACTTCCTC-3’ (reverse). GAPDH 5’-TGTTGCCATCAATGACCCC-3’ (forward), 5’-CTCCACGACGTACTCAGC-3’ (reverse) as an internal control. The above-mentioned primers were designed by Primer Express v 2.0 software (Applied Biosystems). In order to ensure the results were reproducible, all experiments were performed in triplicate.
Immunohistochemical (IHC) analysis
We used the methodology previously described by our breast cancer team [19]. The NPC samples were fixed with formalin and then embedded into paraffin with a tissue processor. Standard IHC analysis was performed using the primary antibody anti-INHBA rabbit polyclonal antibody (ab56057, Abcam) at a dilution of 1:100. The interpretation of immunopositivity was performed by two pathologists blinded to the clinical data. Results were assessed by initially scanning each slide under low-power field (×100) to identify regions of positive immunoreactivity, and then further evaluated at high-power (×400). The intensity of INHBA staining was classified into no staining (0,); weak staining (1, light yellow); moderate staining (2, yellow brown); and strong staining (3, brown). The percentage of immunoreactivity was classified as 0: no positive staining cells; 1, 1-25% positive staining cells; 2, 26-50% positive staining cells; 3, 51-75% positive staining cells; 4, >75% positive staining cells. The immunoreactivity score was calculated as the product of the intensity score and proportion of positive staining cells. The immunoreactivity level of INHBA was defined as: “-” (0), “+” (1-4), “++” (5-8), “+++” (9-12). The cut-off values were determined on the basis of the heterogeneity using log-rank test with respect to OS. In our study, the optimal cut-off value was determined as: a immunoreactivity index score of ≥6 indicated high expression and <6 indicated low expression. Doubtful cases were discussed by the pathologists until consensus was achieved.
Western blot analysis
Western blot was performed as previous described [20]. The blots were probed with rabbit anti-INHBA antibody (1:1000, ab56057, Abcam), anti-TGF beta 1 antibody (1:1000, ab92486, Abcam), anti p-Smad2 antibody (1:1000, ab53100, Abcam), anti p-Smad3 antibody (1:1000, ab52903, Abcam). Mouse anti-GAPDH antibody (1:5000, HC301, Transgen Biotech, Beijing, China) was used as loading control.
Short hairpin RNA (shRNA) constructs and retroviral infection
Stable knockdown of endogenous INHBA was performed using retrovirus constructs targeting INHBA with the targeting sequences: shRNA1, 5-GCTTCTGAACGCGATCAGAAA-3; shRNA2, 5-AGGCACTTTCCTACCCAATTA-3. The synthetic oligos were cloned into the pSuper-retro-puro vector after annealing. Production of retrovirus was performed according to the instructions in 293T cells. SUNE1 cells were subjected to infection of retrovirus expressing INHBA-shRNA1 and INHBA-shRNA2. An empty pSuper-retro-puro plasmid was used as a control.
Cell proliferation assay
SUNE1 INHBA-shRNA1 cells, SUNE1 INHBA-shRNA2 cells and SUNE1 control cells were planted in a 96-well plate at a density of 1000 cells/well and maintained at 37°C in 5% CO2 incubator for 1 day, 2 days, 3 days, 4 and 5 days. 10 µl of the Cell Counting Kit-8 (CCK8, Dojindo Molecular Technologies, Inc, Japan) was added and the 96-well plate was incubated for 1 h at 37°C in incubator, then the absorbance at 450 nm was measured on a microplate reader.
Boyden chamber assay
This assay measures SUNE1 cells’ invasion ability by a Matrigel matrix overlying a membrane containing 8-µm pores. 10000 cells were seeded in medium without FBS in the top chamber. Medium with FBS was added in the bottom chamber. The chambers were fixed by methyl alcohol after 24 hrs culture. 1% crystal violet was used to stain the chambers. Cell number was counted by ten random fields. Three independent experiments were performed.
Statistical analysis
Chi-square test was used to explore the difference of INHBA expression between NPC tissues and noncancerous nasopharyngeal epithelial samples. Kaplan-Meier method was used to plot survival curves, and the significance of differences were analyzed by the log-rank test [21]. Chi-square test and Fisher’s exact test was used to assess the relationship between INHBA and other clinical data. The Cox proportional hazards regression model was performed to analyze the potential prognostic factors on DFS and OS. Variables that achieved significance in univariate analysis were estimated in the following multivariable analysis. Enter method was chosen for univariate analysis and forward method was used for multivariate analysis. Statistical analysis was conducted using SPSS version 20.0 software packages (IBM Corp, Ar-monk, NY, USA). A p-value of less than 0.05 in a two-tailed test was considered significant.
Results
INHBA mRNA is overexpressed in NPC tissues
To measure whether INHBA expression are different in cancerous tissues and non-cancerous tissues, RT-PCR was performed on 40 NPC cancerous samples and 16 noncancerous tissue samples. The mean INHBA mRNA expression level in NPC cancerous tissues was higher than those in noncancerous tissues (Figure 1).
Figure 1.
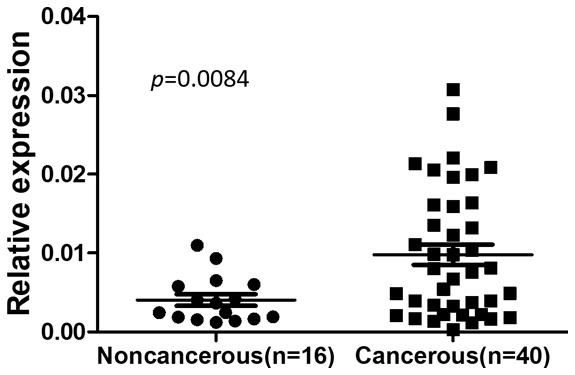
The mRNA expression of INHBA in noncancerous nasopharynx tissues and NPC tissues. Expression levels of INHBA in 40 NPC tissues and 16 noncancerous nasopharynx tissues were detected by real-time PCR.
INHBA overexpression is associated with NPC clinical features
For better understanding of the potential roles of INHBA in the development and progression of NPC, we explored the correlation of INHBA and other clinical features in 108 NPC tissue samples. In this cohort, 37 patients encountered disease failure. 30 patients died during the follow-up, all of them due to disease progression. The clinical characteristics of these patients with NPC are listed in Table 1. Among these patients, excessive expression of INHBA was observed in 58 patients (53.70%, Table 1). In the remaining 50 samples, weak or no immunoreactivity was detected (46.3%, Table 1). The primarily subcellular localization of INHBA was in cytoplasm (Figure 2). To further explore the clinical significance of INHBA in NPC, associations between INHBA expression and other clinical characteristics of patients were analyzed. INHBA was notably associated with the clinical stage (UICC) (P=0.048), N stage (P=0.042), and carotid sheath involvement (P=0.016). However, patient age, gender, histology, T classification, nasal cavity involvement, maxillary sinus involvement, neck lymph node level involvement, or maximum neck lymph node diameter showed no statistically significant correlation with INHBA as is summarized in Table 1.
Figure 2.
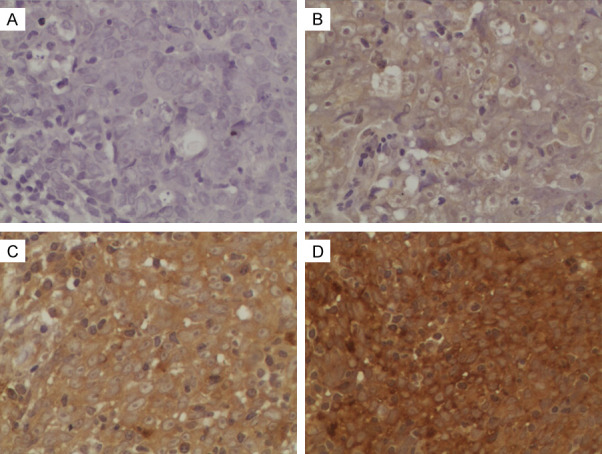
Representative IHC staining of INHBA in NPC tissue samples. INHBA expression was mainly localized in the cytoplasm. Magnification is 400×. A. Negative staining of INHBA. B. “+” (weakly positive) expression of INHBA. C. “++” (positive) expression of INHBA. D. “+++” (strongly positive) expression of INHBA.
Association between INHBA and patient prognosis
The prognostic value of INHBA was evaluated through the estimation of DFS and OS using Kaplan-Meier survival analysis and log-rank test. In survival analysis, the patients with strong INHBA expression had worse DFS (P=0.004, Figure 3A) and OS (P=0.010, Figure 3B) compared to those with moderate or negative INHBA expression. In addition, univariate Cox proportional hazard regression analysis indicated that clinical stage (UICC) (P=0.003), N classification (P=0.004), neck lymph node level involvement (P=0.010), and INHBA expression (P=0.013) were significantly associated with overall survival (Table 2). As for DFS, clinical stage (UICC) (P=0.011), N classification (P=0.005), neck lymph node level involvement (P=0.006), and INHBA (P=0.006) were significantly correlated with survival (Table 3). After multivariate adjustment for these above significant clinicopathologic indexes, only clinical stage (UICC) (P=0.025) was an independent prognostic factors for OS (Table 2), while clinical stage (UICC) (P=0.040) and INHBA (P=0.028) were independent prognostic factors for DFS (Table 3).
Figure 3.
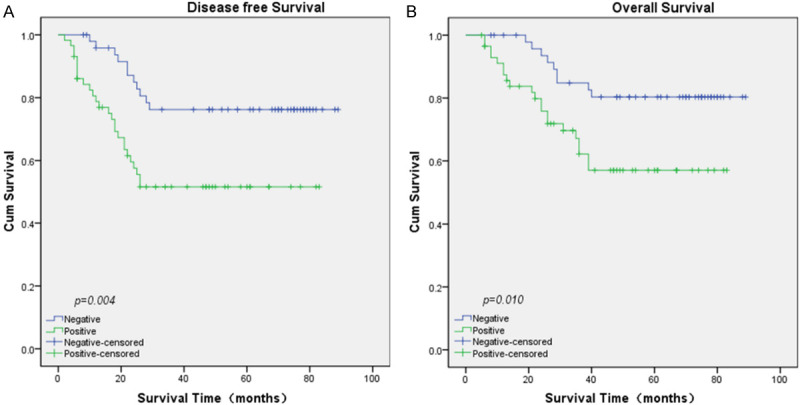
Kaplan-Meier survival curves by univariate analysis (log-rank test). A. DFS rate for NPC patients with high INHBA expression compared to those with low INHBA expression. B. OS rate for NPC patients with high INHBA expression compared to those with low INHBA expression.
Table 2.
Cox-regression analysis of factors associated with overall survival
| Factor | Univariate | Multivariate | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age | ||||
| ≤46 | Reference | |||
| >46 | 1.102 (0.535-2.268) | 0.793 | - | - |
| Gender | ||||
| Male | Reference | |||
| Female | 1.194 (0.512-2.784) | 0.682 | - | - |
| Histology | ||||
| WHO I | ||||
| WHO II | Reference | - | - | |
| WHO III | 0.700 (0.340-1.442) | 0.334 | - | - |
| Clinical stage (UICC) | ||||
| I | Reference | 0.003 | Reference | 0.025 |
| II | 0.000 (0.000-) | 0.977 | 0.000 (0.000-) | 0.981 |
| III | 0.131 (0.024-0.720) | 0.619 | 0.213 (0.027-1.674) | 0.141 |
| IV | 0.179 (0.039-0.820) | 0.027 | 0.241 (0.043-1.345) | 0.105 |
| V | 0.656 (0.149-2.896) | 0.578 | 1.046 (0.221-4.984) | 0.955 |
| T classification | ||||
| T1 | Reference | 0.124 | ||
| T2 | 0.856 (0.287-2.558) | 0.781 | - | - |
| T3 | 0.447 (0.181-1.101) | 0.080 | - | - |
| T4 | 0.340 (0.121-0.957) | 0.041 | - | - |
| N classification | ||||
| N0 | Reference | 0.004 | ||
| N1 | 0.096 (0.025-0.373) | 0.001 | - | - |
| N2 | 0.303 (0.109-0.838) | 0.021 | - | - |
| N3 | 0.286 (0.112-0.729) | 0.009 | - | - |
| Carotid sheath involvement | ||||
| No | Reference | |||
| Yes | 0.744 (0.259-2.132) | 0.582 | - | - |
| Nasal cavity involvement | ||||
| No | Reference | |||
| Yes | 0.765 (0.358-1.636) | 0.490 | - | - |
| Maxillary sinus involvement | ||||
| No | Reference | |||
| Yes | 0.595 (0.273-1.301) | 0.193 | - | - |
| Neck lymph node level involvement | ||||
| No | Reference | 0.010 | ||
| Level I | 0.096 (0.025-0.373) | 0.001 | - | - |
| Level II | 0.266 (0.033-2.168) | 0.216 | - | - |
| Level III | 0.307 (0.107-0.880) | 0.028 | - | - |
| Level IV | 0.286 (0.112-0.731) | 0.009 | - | - |
| Maximum neck lymph node diameter | ||||
| <20 mm | Reference | |||
| ≥20 mm | 0.479 (0.224-1.024) | 0.058 | - | - |
| INHBA expression | ||||
| Low | Reference | |||
| High | 2.687 (1.227-5.883) | 0.013 | - | - |
Table 3.
Cox-regression analysis of factors associated with disease-free survival
| Factor | Univariate | Multivariate | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age | ||||
| ≤46 | Reference | |||
| >46 | 1.108 (0.578-2.124) | 0.757 | - | - |
| Gender | ||||
| Male | Reference | |||
| Female | 1.152 (0.543-2.441) | 0.713 | - | - |
| Histology | ||||
| WHO I | ||||
| WHO II | Reference | - | - | |
| WHO III | 1.303 (0.676-2.513) | 0.429 | - | - |
| Clinical stage (UICC) | ||||
| I | Reference | 0.011 | Reference | 0.040 |
| II | 0.000 (0.000-) | 0.973 | 0.000 (0.000-) | 0.974 |
| III | 0.117 (0.027-0.498) | 0.004 | 0.183 (0.041-0.810) | 0.025 |
| IV | 0.203 (0.058-0.715) | 0.013 | 0.274 (0.077-0.977) | 0.046 |
| V | 0.457 (0.130-1.607) | 0.222 | 0.624 (0.174-2.232) | 0.468 |
| T classification | ||||
| T1 | Reference | 0.472 | ||
| T2 | 0.906 (0.303-2.703) | 0.589 | - | - |
| T3 | 0.511 (0.211-1.234) | 0.136 | - | - |
| T4 | 0.729 (0.307-1.731) | 0.474 | - | - |
| N classification | ||||
| N0 | Reference | 0.005 | ||
| N1 | 0.163 (0.056-0.473) | 0.001 | - | - |
| N2 | 0.293 (0.112-0.761) | 0.012 | - | - |
| N3 | 0.303 (0.126-0.727) | 0.007 | - | - |
| Carotid sheath involvement | ||||
| No | Reference | |||
| Yes | 0.564 (0.200-1.592) | 0.280 | - | - |
| Nasal cavity involvement | ||||
| No | Reference | |||
| Yes | 0.521 (0.270-1.005) | 0.052 | - | - |
| Maxillary sinus involvement | ||||
| No | Reference | |||
| Yes | 0.679 (0.328-1.403) | 0.296 | - | - |
| Neck lymph node level involvement | ||||
| No | Reference | 0.006 | ||
| Level I | 0.163 (0.056-0.471) | 0.001 | - | - |
| Level II | 0.205 (0.026-1.647) | 0.136 | - | - |
| Level III | 0.441 (0.173-1.122) | 0.086 | - | - |
| Level IV | 0.242 (0.098-0.595) | 0.002 | - | - |
| Maximum neck lymph node diameter | ||||
| <20 mm | Reference | |||
| ≥20 mm | 0.556 (0.286-1.080) | 0.083 | - | - |
| INHBA expression | ||||
| Low | Reference | Reference | ||
| High | 2.710 (1.337-5.494) | 0.006 | 0.442 (0.214-0.914) | 0.028 |
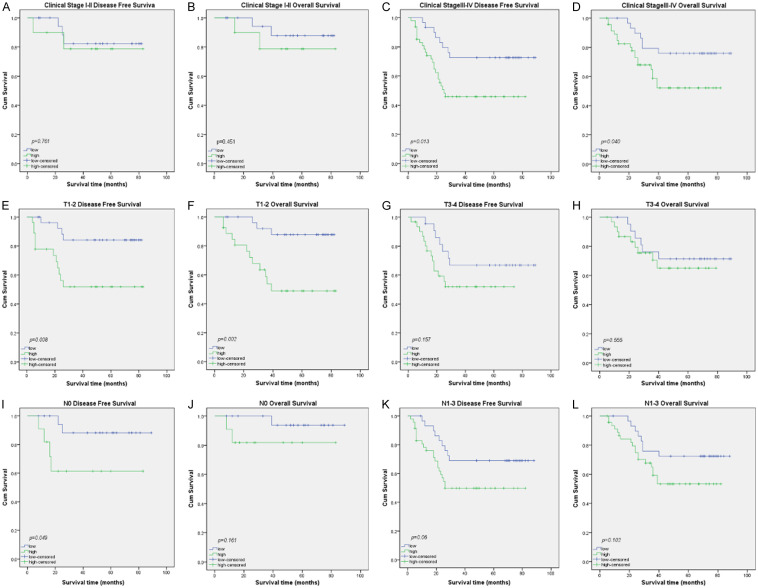
We further analyzed the prognostic significance of INHBA in specific patient subgroups stratified by clinical stage (UICC), T stage, and N stage. The expression of INHBA was closely correlated to DFS (Figure 4C, log-rank test, P=0.013) and OS (Figure 4D, log-rank test, P=0.040) in patients with advanced stage tumor, but not in patients withearly stage tumor (Figure 4A, 4B, log-rank test, P=0.451, P=0.761). INHBA expression was strongly associated with DFS (Figure 4E, log-rank test, P=0.008) and OS (Figure 4F, log-rank test, P=0.002) in patients with T12 tumor, but not in patients with T3-4 tumor (Figure 4G, 4H, log-rank test, P=0.157, P=0.555). As for N stage subgroup, the expression of INHBA was associated only with DFS (Figure 4I, log-rank test, P=0.049) but not OS (Figure 4J, log-rank test, P=0.161) in the patients with N0 tumor. For patients with N1-3 tumor, the expression of INHBA had a relationship with neither DFS (Figure 4K, log-rank test, P=0.06) or OS (Figure 4L, log-rank test, P=0.102).
Figure 4.
Kaplan-Meier survival curves by univariate analysis (log-rank test) in specific patient subgroups stratified by clinical stage (UICC), T stage, and N stage. A, B. DFS and OS rate for NPC patients of early stage tumor with high INHBA expression compared to those with low INHBA expression. C, D. DFS and OS rate for NPC patients of advanced stage tumor with high INHBA expression compared to those with low INHBA expression. E, F. DFS and OS rate for NPC patients of T1-2 tumor with high INHBA expression compared to those with low INHBA expression. G, H. DFS and OS rate for NPC patients of T3-4 tumor with high INHBA expression compared to those with low INHBA expression. I, J. DFS and OS rate for NPC patients of N0 tumor with high INHBA expression compared to those with low INHBA expression. K, L. DFS and OS rate for NPC patients of N1-3 tumor with high INHBA expression compared to those with low INHBA expression.
INHBA knockdown inhibits invasion and proliferation of SUNE1 cells in vitro
To further explore the tumor-associated functions of INHBA, we first established the stable INHBA silencing SUNE1 cell lines in view of INHBA overexpression in NPC. After the SUNE1 cells were infected with INHBA-shRNA1, INHBA-shRNA2 retrovirus, western blot analysis was performed to confirm the effect of INHBA silencing. The result showed that the INHBA expression level was decreased after INHBA silencing (Figure 6A). SUNE1 INHBA knockdown cells and control cells underwent a Transwell assay to evaluate the invasion ability. The results showed the invasive cell number was decreased in INHBA-shRNA cell group by comparison to the control group (Figure 5A, 5B), indicating that INHBA silencing could decrease invasive ability of SUNE1 cells. CCK analysis was performed to detect the cell proliferation effect of INHBA-shRNA in SUNE1 cells. The result showed a significant decrease in proliferation rate was observed in INHBA silencing cells compared to the control cells (Figure 5C), indicating that INHBA silencing could inhibit SUNE1 cell proliferation.
Figure 6.
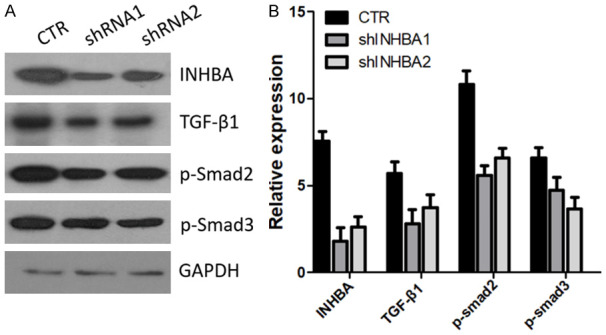
INHBA knockdown blocked the TGF-β signaling pathway in SUNE1 cells. A. Protein levels of TGF-β1, p-Smad2, p-Smad3 were assessed by western blot in SUNE1 INHBA-shRNA cells and control cells. B. Gray value of protein bands of A.
Figure 5.
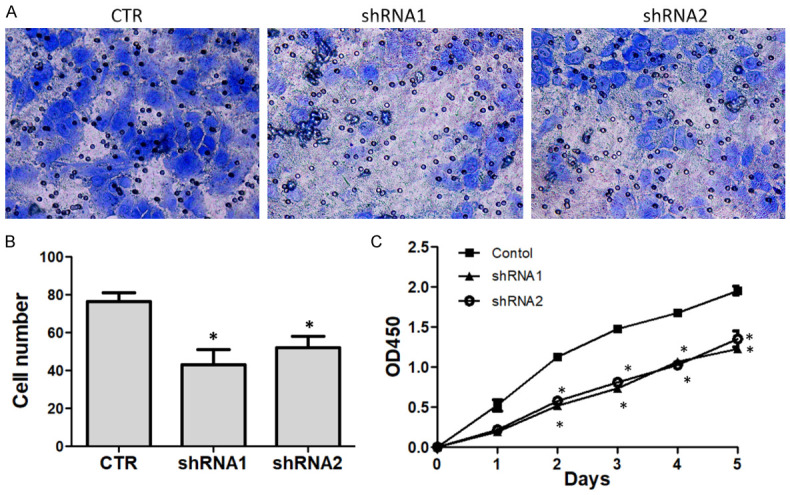
Knockdown INHBA in SUNE1 cells inhibit cell proliferation and invasion in vitro. A. Representative images of cell invasion evaluated by Transwell assay (200×). B. Quantification of the cell number invaded through the membrane. C. CCK assay was performed to determine the cell proliferation under each treatment. Decreased proliferation of SUNE1 cells was detected after INHBA knockdown. *P<0.05 versus the control group.
INHBA silencing contributes to block the TGF-β signaling pathway in SUNE1 cells
Recently a paper reported that INHBA gene could regulate the TGF-β signaling pathway [22]. We questioned whether INHBA silencing could inhibit the TGF-β signaling pathway in SUNE1 cells. Three core proteins in TGF-β signaling pathway were selected to evaluate by western blot. The result is in Figure 6. After knockdown of the expression of INHBA in SUNE1 cells, the TGF-β1, p-Smad2 and p-Smad3 expression were downregulated subsequently. Thus, INHBA may regulate the TGF-β signaling pathway in SUNE1 cells and this may be the mechanism of INHBA silencing inhibiting SUNE1 cell proliferation and invasion.
Discussion
Inhibin βA (INHBA) is a ligand belonging to the transforming growth factor beta (TGF-β) superfamily [12]. INHBA gene is located at 7p14.1 [12]. The ligand is a subunit which can form both activin A by homo-dimerizing and inhibin by hetero-dimerizing with inhibin βB. Activin and inhibin, are two tightly related glycoproteins of the TGF-β superfamily with opposite biologic functions during various different stages of cell growth, proliferation and differentiation [23-26]. Activin needs to first bind with a complex of type I and type II single transmembrane serine or threonine kinase receptors to initiate the cascade pathway. Interaction between them can trigger phosphorylation of the transmembrane receptor and initiate activation of Smad proteins. Then, the phosphorylated Smad protein complex translocates into the nucleus and binds with the promoters of multiple target genes and regulates transcription of these genes and modulates cellular functions [27]. However, inhibin exerts a completely opposing function by binding with type II transmembrane serine or threonine kinase receptors mediated by the co-receptor beta glycan. Multiple elements can effect the activin signalling pathway at different phases. The interaction of disulfide-linked homodimer of Inhibin βA constitutes activin A in the hypothalamic-pituitary-gonadal axis [28,29]. The inhibin βA may also combined with the inhibin βB and inhibin α isoforms and respectively form activin AB and inhibin A [30].
Activin A was first reported in 1978 for its function in the hypothalamic-pituitary-gonadal axis [28,29]. Since then, the involvement of activin A has been reported in a large variety of biologic activities, such as glucose metabolism [31], immune response [32], stem cell differentiation [33], and neoplastic progression [34]. Upregulated activin A in esophageal carcinoma has been reported to be associated with advanced clinical stage, N stage, and a worse OS [35]. Moreover, activin A can promote cell growth, tumorigenicity, invasion, and also induce resistance to apoptosis [36]. Furthermore, overexpression of activin A was also reported in lung [15], gastric [37], colon [38], pancreatic [39], endometrial, ovarian [40,41], cervical [42], and prostate cancers [43]. In accordance with these findings, the overexpression of INHBA was also reported in multiple types of cancers, such as esophageal adenocarcinoma [44,45], breast cancer [19,34], lung cancer [15], gastric carcinoma [16], colorectal carcinoma [17], and bladder carcinoma [18]. However, to the best of our knowledge, the expression and prognostic effects of INHBA in NPC remain unclear.
In our study of NPC in Guangdong, China, we examined the expression status of INHBA in 108 tumors and analyzed their pattern of relationship with clinicopathologic features and outcome. Based on the prevailing theory, the expression level of INHBA is likely associated with tumorigenesis and progression. The present study certified that expression level of INHBA was elevated in NPC which was in consensus with the above-mentioned reports. Our study clearly demonstrated that expression of INHBA at mRNA levels was higher in NPC lesions compared with non-cancerous nasopharyngeal epithelial samples. On the basis of these findings, we consider INHBA as a molecular biomarker of NPC that can help boost the precision of early diagnoses.
Furthermore, we analysed the intrinsic connection between INHBA and the other clinicalpathologic characteristics of NPC patients. The INHBA expression were closely related to well-established prognostic clinical characteristics such as Clinical stage (UICC) (P=0.048), N stage (P=0.042), and carotid sheath involvement (P=0.016). The INHBA expression showed no significant association with age, gender, histology, T classification, nasal cavity involvement, maxillary sinus involvement, neck lymph node level involvement, or maximum neck lymph node diameter. Interestingly, the relationship between INHBA and clinical outcome in different type of carcinomas seems to be diverse. A depressed expression of INHBA in carcinomas has been reported by some researchers. For example, Hofland et al. [46] indicated a lower expression of INHBA in adrenocortical carcinoma tissues.
In our study, as demonstrated by Kaplan-Meier analysis, patients with strong INHBA expression had worse DFS and OS compared to those with moderate or negative expression. Our data demonstrated that expression of INHBA is an indicator of poor prognosis in NPC. This is in line with our previous studies which reported the association of poor clinical outcome with strong expression of INHBA in breast cancer [19]. The exact effects of INHBA-mediation on tumor progression remain obscure. Multivariate analysis indicated that INHBA might be an independent prognostic indicator for DFS in NPC patients. This finding demonstrated the possibility of using INHBA expression as a predictor for prognosis and disease-free survival in NPC. Moreover, a sub-group analysis revealed that INHBA overexpression patients had a poor DFS and OS among the sub-group whose tumors demonstrated the features of advanced stage, and T1-2 tumors. As for N stage subgroup, the expression of INHBA was associated only with DFS duration in patients with N0 tumors.
To explore the functions and mechanisms of INHBA in NPC, a SUNE1 INHBA-shRNA cell model was constructed. We found silencing INHBA expression in SUNE1 cells could inhibit proliferation and invasion ability in vitro. We also performed colony formation and wound healing assay to determine whether INHBA could affect SUNE1 cells’ tumorigenesis and migration in vitro, but the results showed no significance (data were not show).
Chen et al. have reported that the TGF-β signaling pathway was inhibited in gastric cancer cells in response to INHBA silencing. We questioned whether there was a similar mechanism in the SUNE1 NPC cell line. Next, 3 TGF-β signaling pathway core proteins including TGF-β1, p-Smad2, and p-Smad3 were selected to detect by western blot. The result showed that in response to INHBA knockdown, the protein expression of TGF-β1, p-Smad2, and p-Smad3 were decreased, indicating that after INHBA silencing, the TGF-β signaling pathway was blocked. Thus, knockdown of INHBA expression in SUNE1 cells inhibited proliferation and invasion. The underlying mechanism may be by blocking the TGF-β signaling pathway.
In conclusion, to our knowledge, this is the first report on INHBA expression and its clinical significance in non-endemic NPC published to date. Our findings suggest that the expression of INHBA is up-regulated in NPC and is correlated with clinical stage, N classification, and carotid sheath involvement. Multivariate analysis indicated that INHBA might be an independent index for NPC prognosis. Hence, detecting the expression of INHBA may contribute to stratifying NPC patients for implementing a new therapeutic strategy. Knockdown of INHBA expression in SUNE1 cells could inhibit cell proliferation and invasion. The underlying mechanism may be blocking the TGF-β signalling pathway.
Acknowledgements
We thank the funding support of National Natural Science Foundation of China, Guangdong Province Natural Science Foundation and Guangzhou Science and Technology Project. This study was supported by grants from the National Natural Science Foundation of China (81672661), the Guangdong Province Natural Science Foundation (2017A030313858), and Guangzhou Science and Technology Project (20181A011062).
Informed consent was obtained from all individual participants included in the study. And each patient had given a written informed consent about the use of the samples for medical research.
Disclosure of conflict of interest
None.
References
- 1.Wee JT, Ha TC, Loong SL, Qian CN. Is nasopharyngeal cancer really a “Cantonese cancer”? Chin J Cancer. 2010;29:517–26. doi: 10.5732/cjc.009.10329. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Yu WM, Hussain SS. Incidence of nasopharyngeal carcinoma in Chinese immigrants, compared with Chinese in China and South East Asia: review. J Laryngol Otol. 2009;123:1067–74. doi: 10.1017/S0022215109005623. [DOI] [PubMed] [Google Scholar]
- 5.Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–1024. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 6.Huang SCM, Tsao SW, Tsang CM. Interplay of viral infection, host cell factors and tumor microenvironment in the pathogenesis of nasopharyngeal carcinoma. Cancers (Basel) 2018;10 doi: 10.3390/cancers10040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee AW, Lin JC, Ng WT. Current management of nasopharyngeal cancer. Semin Radiat Oncol. 2012;22:233–44. doi: 10.1016/j.semradonc.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Kontny U, Franzen S, Behrends U, Buhrlen M, Christiansen H, Delecluse H, Eble M, Feuchtinger T, Gademann G, Granzen B, Kratz CP, Lassay L, Leuschner I, Mottaghy FM, Schmitt C, Staatz G, Timmermann B, Vorwerk P, Wilop S, Wolff HA, Mertens R. Diagnosis and treatment of nasopharyngeal carcinoma in children and adolescents-recommendations of the GPOH-NPC study group. Klin Padiatr. 2016;228:105–12. doi: 10.1055/s-0041-111180. [DOI] [PubMed] [Google Scholar]
- 9.Lee AW, Ng WT, Chan LL, Hung WM, Chan CC, Sze HC, Chan OS, Chang AT, Yeung RM. Evolution of treatment for nasopharyngeal cancer--success and setback in the intensity-modulated radiotherapy era. Radiother Oncol. 2014;110:377–84. doi: 10.1016/j.radonc.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Zhang B, Mo Z, Du W, Wang Y, Liu L, Wei Y. Intensity-modulated radiation therapy versus 2D-RT or 3D-CRT for the treatment of nasopharyngeal carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51:1041–1046. doi: 10.1016/j.oraloncology.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Lee AW, Ma BB, Ng WT, Chan AT. Management of nasopharyngeal carcinoma: current practice and future perspective. J. Clin. Oncol. 2015;33:3356–64. doi: 10.1200/JCO.2015.60.9347. [DOI] [PubMed] [Google Scholar]
- 12.Gaddy-Kurten D, Tsuchida K, Vale W. Activins and the receptor serine kinase superfamily. Recent Prog Horm Res. 1995;50:109–29. doi: 10.1016/b978-0-12-571150-0.50010-x. [DOI] [PubMed] [Google Scholar]
- 13.Chen YG, Wang Q, Lin SL, Chang CD, Chuang J, Ying SY. Activin signaling and its role in regulation of cell proliferation, apoptosis, and carcinogenesis. Exp Biol Med (Maywood) 2006;231:534–544. doi: 10.1177/153537020623100507. [DOI] [PubMed] [Google Scholar]
- 14.Onagbesan OM, Safi M, Decuypere E, Bruggeman V. Developmental changes in inhibin alpha and inhibin/activin βA and βB mRNA levels in the gonads during post-hatch prepubertal development of male and female chickens. Mol Reprod Dev. 2004;68:319–26. doi: 10.1002/mrd.20087. [DOI] [PubMed] [Google Scholar]
- 15.Seder CW, Hartojo W, Lin L, Silvers AL, Wang Z, Thomas DG, Giordano TJ, Chen G, Chang AC, Orringer MB, Beer DG. Upregulated INHBA expression may promote cell proliferation and is associated with poor survival in lung adenocarcinoma. Neoplasia. 2009;11:388–96. doi: 10.1593/neo.81582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 17.Okano M, Yamamoto H, Ohkuma H, Kano Y, Kim H, Nishikawa S, Konno M, Kawamoto K, Haraguchi N, Takemasa I, Mizushima T, Ikeda M, Yokobori T, Mimori K, Sekimoto M, Doki Y, Mori M, Ishii H. Significance of INHBA expression in human colorectal cancer. Oncol Rep. 2013;30:2903–8. doi: 10.3892/or.2013.2761. [DOI] [PubMed] [Google Scholar]
- 18.Lee HY, Li CC, Huang CN, Li WM, Yeh HC, Ke HL, Yang KF, Liang PI, Li CF, Wu WJ. INHBA overexpression indicates poor prognosis in urothelial carcinoma of urinary bladder and upper tract. J Surg Oncol. 2015;111:414–22. doi: 10.1002/jso.23836. [DOI] [PubMed] [Google Scholar]
- 19.Wang HJ, Huang ZN, Liu RM, Huang QN, Wu JK, Liu RB. The expression status of INHBA as a prognostic marker for human breast cancer. Int J Clin Exp Pathol. 2016;9:9. [Google Scholar]
- 20.Luo XD, Yang SJ, Wang JN, Tan L, Liu D, Wang YY, Zheng RH, Wu XH, Xu LH, Tan H. Downregulation of SATB1 increases the invasiveness of jurkat cell via activation of the WNT/beta-catenin signaling pathway in vitro. Tumour Biol. 2016;37:7413–9. doi: 10.1007/s13277-015-4638-x. [DOI] [PubMed] [Google Scholar]
- 21.Pocock SJ, Clayton TC, Altman DG. Survival plots of time-to-event outcomes in clinical trials: good practice and pitfalls. Lancet. 2002;359:1686–9. doi: 10.1016/S0140-6736(02)08594-X. [DOI] [PubMed] [Google Scholar]
- 22.Chen ZL, Qin L, Peng XB, Hu Y, Liu B. INHBA gene silencing inhibits gastric cancer cell migration and invasion by impeding activation of the TGF-beta signaling pathway. J Cell Physiol. 2019;234:18065–18074. doi: 10.1002/jcp.28439. [DOI] [PubMed] [Google Scholar]
- 23.Burger HG, Igarashi M. Inhibin: definition and nomenclature, including related substances. Endocrinology. 1988;122:1701–2. doi: 10.1210/endo-122-4-1701. [DOI] [PubMed] [Google Scholar]
- 24.Murata M, Eto Y, Shibai H, Sakai M, Muramatsu M. Erythroid differentiation factor is encoded by the same mRNA as that of the inhibin β A chain. Proc Natl Acad Sci U S A. 1988;85:2434–8. doi: 10.1073/pnas.85.8.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown CW, Houston-Hawkins DE, Woodruff TK, Matzuk MM. Insertion of inhbb into the inhba locus rescues the inhba-null phenotype and reveals new activin functions. Nat Genet. 2000;25:453–7. doi: 10.1038/78161. [DOI] [PubMed] [Google Scholar]
- 26.Hofland J, van Nederveen FH, Timmerman MA, Korpershoek E, de Herder WW, Lenders JW, Verhofstad AA, de Krijger RR, de Jong FH. Expression of activin and inhibin subunits, receptors and binding proteins in human pheochromocytomas: a study based on mRNA analysis and immunohistochemistry. Clin Endocrinol (Oxf) 2007;66:335–40. doi: 10.1111/j.1365-2265.2007.02732.x. [DOI] [PubMed] [Google Scholar]
- 27.Lotinun S, Pearsall RS, Horne WC, Baron R. Activin receptor signaling: a potential therapeutic target for osteoporosis. Curr Mol Pharmacol. 2012;5:195–204. doi: 10.2174/1874467211205020195. [DOI] [PubMed] [Google Scholar]
- 28.Vale W, Rivier C, Hsueh A, Campen C, Meunier H, Bicsak T, Vaughan J, Corrigan A, Bardin W, Sawchenko P, et al. Chemical and biological characterization of the inhibin family of protein hormones. Recent Prog Horm Res. 1988;44:1–34. doi: 10.1016/b978-0-12-571144-9.50005-3. [DOI] [PubMed] [Google Scholar]
- 29.Lorenzen JR, Channing CP, Schwartz NB. Partial characterization of FSH suppressing activity (folliculostatin) in porcine follicular fluid using the metestrous rat as an in vivo bioassay model. Biol Reprod. 1978;19:635–40. doi: 10.1095/biolreprod19.3.635. [DOI] [PubMed] [Google Scholar]
- 30.Ying SY, Zhang Z, Furst B, Batres Y, Huang G, Li G. Activins and activin receptors in cell growth. Proc Soc Exp Biol Med. 1997;214:114–22. doi: 10.3181/00379727-214-44077. [DOI] [PubMed] [Google Scholar]
- 31.Totsuka Y, Tabuchi M, Kojima I, Shibai H, Ogata E. A novel action of activin A: stimulation of insulin secretion in rat pancreatic islets. Biochem Biophys Res Commun. 1988;156:335–9. doi: 10.1016/s0006-291x(88)80845-3. [DOI] [PubMed] [Google Scholar]
- 32.Robson NC, Phillips DJ, McAlpine T, Shin A, Svobodova S, Toy T, Pillay V, Kirkpatrick N, Zanker D, Wilson K, Helling I, Wei H, Chen W, Cebon J, Maraskovsky E. Activin-A: a novel dendritic cell-derived cytokine that potently attenuates CD40 ligand-specific cytokine and chemokine production. Blood. 2008;111:2733–43. doi: 10.1182/blood-2007-03-080994. [DOI] [PubMed] [Google Scholar]
- 33.Beattie GM, Lopez AD, Bucay N, Hinton A, Firpo MT, King CC, Hayek A. Activin a maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells. 2005;23:489–95. doi: 10.1634/stemcells.2004-0279. [DOI] [PubMed] [Google Scholar]
- 34.Bashir M, Damineni S, Mukherjee G, Kondaiah P. Activin-A signaling promotes epithelial-mesenchymal transition, invasion, and metastatic growth of breast cancer. NPJ Breast Cancer. 2015;1:15007. doi: 10.1038/npjbcancer.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshinaga K, Mimori K, Yamashita K, Utsunomiya T, Inoue H, Mori M. Clinical significance of the expression of activin a in esophageal carcinoma. Int J Oncol. 2003;22:75–80. [PubMed] [Google Scholar]
- 36.Yoshinaga K, Yamashita K, Mimori K, Tanaka F, Inoue H, Mori M. Activin a causes cancer cell aggressiveness in esophageal squamous cell carcinoma cells. Ann Surg Oncol. 2008;15:96–103. doi: 10.1245/s10434-007-9631-1. [DOI] [PubMed] [Google Scholar]
- 37.Wang HL, Cheng X, Ding SW, Wang DW, Chen C, Xu CL, Xie H. Molecular identification and functional characterization of the cathepsin b gene (Ab-cb-1) in the plant parasitic nematode aphelenchoides besseyi. PLoS One. 2018;13:e0199935. doi: 10.1371/journal.pone.0199935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wildi S, Kleeff J, Maruyama H, Maurer CA, Buchler MW, Korc M. Overexpression of activin a in stage IV colorectal cancer. Gut. 2001;49:409–17. doi: 10.1136/gut.49.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleeff J, Ishiwata T, Friess H, Buchler MW, Korc M. Concomitant over-expression of activin/inhibin beta subunits and their receptors in human pancreatic cancer. Int J Cancer. 1998;77:860–8. doi: 10.1002/(sici)1097-0215(19980911)77:6<860::aid-ijc11>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 40.Woodruff TK. Role of inhibins and activins in ovarian cancer. Cancer Treat Res. 2002;107:293–302. doi: 10.1007/978-1-4757-3587-1_14. [DOI] [PubMed] [Google Scholar]
- 41.Zheng W, Luo MP, Welt C, Lambert-Messerlian G, Sung CJ, Zhang Z, Ying SY, Schneyer AL, Lauchlan SC, Felix JC. Imbalanced expression of inhibin and activin subunits in primary epithelial ovarian cancer. Gynecol Oncol. 1998;69:23–31. doi: 10.1006/gyno.1998.4958. [DOI] [PubMed] [Google Scholar]
- 42.Petraglia F, Florio P, Luisi S, Gallo R, Gadducci A, Vigano P, Di Blasio AM, Genazzani AR, Vale W. Expression and secretion of inhibin and activin in normal and neoplastic uterine tissues. High levels of serum activin a in women with endometrial and cervical carcinoma. J Clin Endocrinol Metab. 1998;83:1194–200. doi: 10.1210/jcem.83.4.4689. [DOI] [PubMed] [Google Scholar]
- 43.Thomas TZ, Wang H, Niclasen P, O’Bryan MK, Evans LW, Groome NP, Pedersen J, Risbridger GP. Expression and localization of activin subunits and follistatins in tissues from men with high grade prostate cancer. J Clin Endocrinol Metab. 1997;82:3851–8. doi: 10.1210/jcem.82.11.4374. [DOI] [PubMed] [Google Scholar]
- 44.Seder CW, Hartojo W, Lin L, Silvers AL, Wang Z, Thomas DG, Giordano TJ, Chen G, Chang AC, Orringer MB, Beer DG. INHBA overexpression promotes cell proliferation and may be epigenetically regulated in esophageal adenocarcinoma. J Thorac Oncol. 2009;4:455–62. doi: 10.1097/JTO.0b013e31819c791a. [DOI] [PubMed] [Google Scholar]
- 45.Lyu S, Jiang C, Xu R, Huang Y, Yan S. INHBA upregulation correlates with poorer prognosis in patients with esophageal squamous cell carcinoma. Cancer Manag Res. 2018;10:1585–1596. doi: 10.2147/CMAR.S160186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hofland J, Timmerman MA, de Herder WW, van Schaik RH, de Krijger RR, de Jong FH. Expression of activin and inhibin subunits, receptors and binding proteins in human adrenocortical neoplasms. Clin Endocrinol (Oxf) 2006;65:792–9. doi: 10.1111/j.1365-2265.2006.02668.x. [DOI] [PubMed] [Google Scholar]