Abstract
Emerging evidence indicates that circular RNAs (circRNAs) are a novel class of biomarkers and therapeutic targets in malignancies. Circ_0026344 has been reported to be downregulated in colorectal cancer, and associated with prognosis. However, little is currently known regarding its expression and clinical significance in gastric cancer. In this study, we evaluated the expression level of circ_0026344 in two previous circRNAs chips (GSE78092 and GSE89143) for gastric cancer. 93 pairs of gastric cancer and adjacent non-tumour tissues were collected, and qRT-PCR was applied to determine circ_0026344 expression. The level of circ_0026344 in gastric cancer cells (MKN45 and AGS) and a normal gastric epithelial cell line (GES-1) was also analyzed. Chi-square test was used to identify the association between circ_0026344 level and clinicopathologic factors. Kaplan-Meier method with log-rank test was applied to compare survival curves. Cox regression analyses were used to assess the prognostic value of circ_0026344. In GSE24549 and GSE24550 datasets, downregulation of circ_0026344 was observed. In 93 cases of gastric cancer, circ_0026344 in cancer tissues was significantly decreased compared to adjacent non-cancerous tissues, and its level was markedly associated with tumour size, lymph node metastasis, TNM stage, and invasive depth. Furthermore, patients with lower expression of circ_0026344 showed significantly worse overall survival than those with higher expression. Additional, circ_0026344 expression was an independent predictor for overall survival. Summarily, our study highlights that downregulation of circ_0026344 is associated with tumour malignant behavior and it is a potential biomarker for gastric cancer prognosis.
Keywords: circRNAs, circ_0026344, gastric cancer, prognosis, biomarker
Introduction
Gastric cancer is one of the most common malignancies and is the second leading cause of cancer-associated mortality globally [1]. Although advancement of diagnosis and treatment of cancer improved recent decade, the prognosis of gastric cancer remains quite unsatisfactory due to recurrence, with a 5-year overall survival lower than 40% [2,3]. Additionally, many patients with gastric cancer are diagnosed at advanced stages with distant metastasis, missing the best opportunity for radical surgery [4]. Therefore, identification of novel effective biomarkers is of great significance for the improvement of diagnosis and prognosis in gastric cancer, and for the development of more efficient therapeutic strategies [5].
CircRNAs are a novel class of widespread and abundant non-coding RNAs that form a covalently closed continuous loop, which makes them much more stable than linear RNA and insusceptible to degradation by RNase R (Ribonuclease R) [6]. CircRNAs are able to modulate gene expression by sponging with microRNAs (miRNAs) or other competing endogenous RNAs [7]. With rapid advances in circRNAs chip and whole-genome sequencing technology, more and more circRNAs have been successfully identified in multiple human cancer recently [8,9]. Aberrant expression of circRNAs plays a critical role in regulation of cellular processes such as cell differentiation and growth as well as cancer progression and metastasis [10]. For instance, circ_TADA2As suppresses breast cancer progression and metastasis by targeting miR-203a-3p/SOCS3 axis [11]. CiRS-7 behavior is a promising prognostic biomarker in colorectal cancer patients and may serve as a therapeutic target for reducing EGFR-RAF1 activity [12]. Circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer [13]. Therefore, the study of circRNAs may open up a new field for molecular diagnosis and prognosis of cancer.
Circ_0026344, as a well-known circRNA, has be found to be downregulated and predicts a poor prognosis in colorectal cancer, overexpression of circ_0026344 exerted inhibitory roles by sponging miR-21 and miR-31 [14]. However, the expression and clinical significance of circ_0026344 in gastric cancer are still poorly understood. In the current study, we investigated the expression of circ_0026344 in GSE78092 and GSE89143 datasets, 93 pairs of gastric cancer and adjacent non-tumour tissues, as well as gastric cancer cells (MKN45 and AGS). We further analyzed the association between circ_0026344 expression and clinicopathologic factors and overall survival of gastric cancer patients. Our data demonstrated that circ_0026344 is a potential biomarker for gastric cancer prognosis.
Materials and methods
Patients and samples collection
A total of 93 pairs of cancer and adjacent non-tumour tissues were obtained from gastric cancer patients by gastroscopy from the Department of Digestive Medicine, Tianjin Nankai Hospital, between June 2017 and December 2019. All tissues were confirmed by two professional pathologists independently, and were immediately frozen and stored in liquid nitrogen for further use. None of the patients received radiotherapy or chemotherapy before surgery. The clinicopathologic information is shown in Table 1. Written informed consent was obtained from the patients prior to sample collection, and the study was approved by the Ethics Committee of Tianjin Nankai Hospital.
Table 1.
Relationship between circ_0026344 expression and clinicopathologic data in gastric cancer patients
| Feature | No. of patients | Circ_0026344 | P value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Gender | 0.217 | |||
| Male | 61 | 28 | 33 | |
| Female | 32 | 19 | 13 | |
| Age (years) | 0.523 | |||
| <50 | 25 | 14 | 11 | |
| ≥50 | 68 | 33 | 35 | |
| Tumor size (cm) | 0.001* | |||
| <5 | 40 | 12 | 28 | |
| ≥5 | 53 | 35 | 18 | |
| Tumor location | 0.264 | |||
| Distal/middle | 79 | 38 | 41 | |
| Proximal | 14 | 9 | 5 | |
| Differentiation | 0.536 | |||
| Well/moderate | 66 | 32 | 34 | |
| Poor | 27 | 15 | 12 | |
| Borrmann type | 0.355 | |||
| I/II | 11 | 7 | 4 | |
| III/IV | 82 | 40 | 42 | |
| CEA status | 0.962 | |||
| Negative | 16 | 8 | 8 | |
| Positive | 77 | 39 | 38 | |
| Invasive depth | 0.028* | |||
| T1/T2 | 38 | 14 | 24 | |
| T3/T4 | 55 | 33 | 22 | |
| Distant metastasis | 0.176 | |||
| Negative | 88 | 43 | 45 | |
| Positive | 5 | 4 | 1 | |
| TNM stage | 0.008* | |||
| I/II | 56 | 22 | 34 | |
| III/IV | 37 | 25 | 12 | |
| Lymph node metastasis | 0.015* | |||
| Negative | 24 | 7 | 17 | |
| Positive | 69 | 40 | 29 | |
P<0.05.
Cell lines and culture
Two human gastric cancer cell lines (MKN45 and AGS) and a normal gastric epithelial cell line (GES-1) were obtained from Shanghai Institutes for Biological Sciences, Chinese Academy of Science (Shanghai, China). All cells were cultured in DMEM with 10% FBS (Invitrogen, Grand Island, NY, USA), 100 mg/ml streptomycin and 100 U/ml penicillin at 37°C in a humidified incubator containing 5% CO2.
RNase R treatment for analysis of circ_0026344 stability
To evaluate the stability of circ_0026344 in gastric cancer cells, 4 μg RNA was treated with 5 U/μg RNase R (Takara) for 30 min at 37°C. After that, qRT-PCR was employed for analysis the expression levels of circ_0026344 and liner C-X-C motif chemokine ligand 8 (CXCL8) gene.
qRT-PCR for analysis of circ_0026344 and CXCL8 expression
Total RNA extracted by Trizol reagent (Invitrogen) was reversely transcribed into complementary DNA (cDNA) using PrimeScript™ RT Master Mix (Takara, Dalian, Japan). One Step SYBR® PrimeScript™ RT-PCR kit (Takara) and an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) was employed for PCR. The expression level of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal control for circ_0026344 and CXCL8. 2-ΔΔCT method was employed for relative gene expression determination. Oligonucleotide primers specific for GAPDH, circ_0026344 and CXCL8 were as follows: GAPDH (forward: 5’-TTGCCCTCAACGACCACTTT-3’, reverse: 5’-TGGTCCAGGGGTCTTACTCC-3’); circ_0026344 (forward: 5’-CTCAGCCTCTAGCATAAGCTC-3’, reverse: 5’-AGGCAAGAGAATGATTTGAAC-3’); CXCL8 (forward: 5’-ATTGAATGGGTTTGCTAGAATG-3’, reverse: 5’-CAAGGCACAGTGGAACAAGGAC-3’).
Statistical analysis
Data were presented as mean ± SD, and analyzed using SPSS 18.0 software (SPSS Inc., Chicago, IL). All graphs were plotted using GraphPad Prism 5.0 software (GraphPad Software, Inc., USA). The different expression of circ_0026344 between cancer and adjacent non-tumour tissues was assessed using paired Student’s t test. Chi-square test was adopted to analyze the association between circ_0026344 expression and clinicopathologic parameters. Overall survival curves were plotted using the Kaplan-Meier method and were evaluated using a log-rank test. The P<0.05 was considered significant.
Results
General characteristics of 93 patients with gastric cancer
As shown in the Table 1, most of the patients with gastric cancer were men (65.6%, 61/93), and were more than 50 years old (73.1%, 68/93). The average age was 57.6±4.2 years old. The positive rate of CEA in gastric cancer patients was 82.8% (77/93). About three-quarters of patients presented with lymph node metastasis (74.2%, 69/93). Patients without distant metastasis accounted for the vast majority (94.6%, 88/93).
Circ_0026344 expression is downregulated in gastric cancer tissues and cells
The expression level of circ_0026344 was initially examined in two previous circRNAs chips (GSE78092 and GSE89143) for gastric cancer [15,16]. As shown in Figure 1, the expression level of circ_0026344 in gastric cancer was significantly reduced (P<0.01). Subsequently, we assessed circ_0026344 expression by qRT-PCR in the 93 pairs of gastric cancer and adjacent non-tumour tissues. Results showed that the relative expression of circ_0026344 in cancer tissues was notably lower than that of non-tumour tissues (Figure 2A, P<0.01). Importantly, expression of circ_0026344 in metastatic cancer tissues was markedly lower than in non-metastatic cancer tissues (Figure 2B, P<0.01). We further examined its expression in GES-1 and two gastric cancer cell lines, and found that level of circ_0026344 in MKN45 and AGS cells was also significantly downregulated compared to GES-1 cells (Figure 2C, P<0.05 or <0.01).
Figure 1.
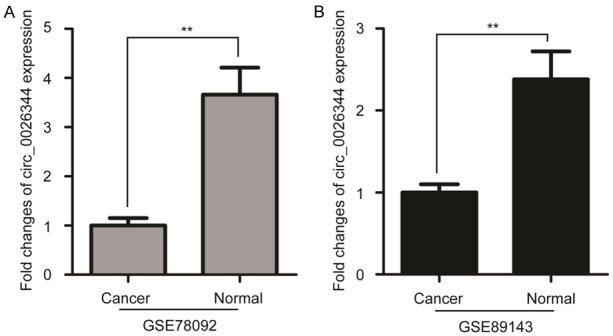
Circ_0026344 expression is reduced in circRNAs chips of gastric cancer. A. The fold changes of circ_0026344 expression in GSE78092 datasets. B. The fold changes of circ_0026344 expression in GSE89143 datasets. Error bars indicate means ± SD. **, P<0.01.
Figure 2.
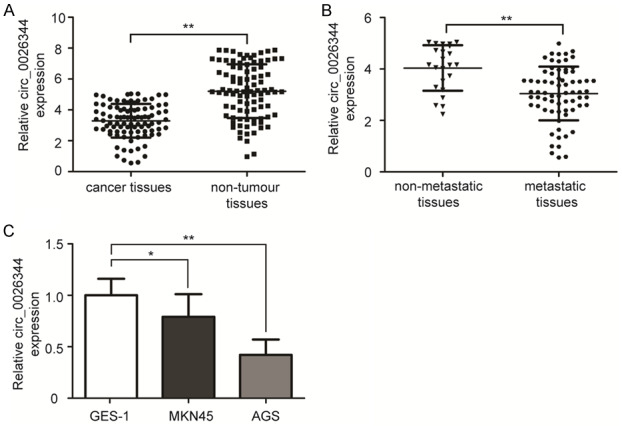
The expression of circ_0026344 level in gastric cancer tissues and cell lines. A. Relative expression of circ_0026344 in cancer tissues was significantly lower than that of non-tumour tissues. B. The relative expression of circ_0026344 in metastatic cancer tissues was notably lower than in non-metastatic tissues. C. The level of circ_0026344 in MKN45 and AGS cells was markedly downregulated compared to that in GES-1 cells. Error bars indicated means ± SD. *, P<0.05; **, P<0.01.
Low expression of circ_0026344 indicated a worse prognosis in gastric cancer patients
Using the median circ_0026344 expression level in cancer tissues as the cutoff value, the 93 patients with gastric cancer were divided into two groups, including high circ_0026344 expression group (n=46) and low circ_0026344 expression group (n=47). Kaplan-Meier method showed that the median survival time of gastric cancer patients in the high and low circ_0026344 expression group was 45.0 and 17.70 months, respectively. The 5-year overall survival rate of patients in the high and low circ_0026344 expression group was 30.43% and 12.77%, respectively (Figure 3, P<0.01), indicating that low circ_0026344 expression in gastric cancer patients was negatively associated with prognosis and displayed a poorer overall survival.
Figure 3.
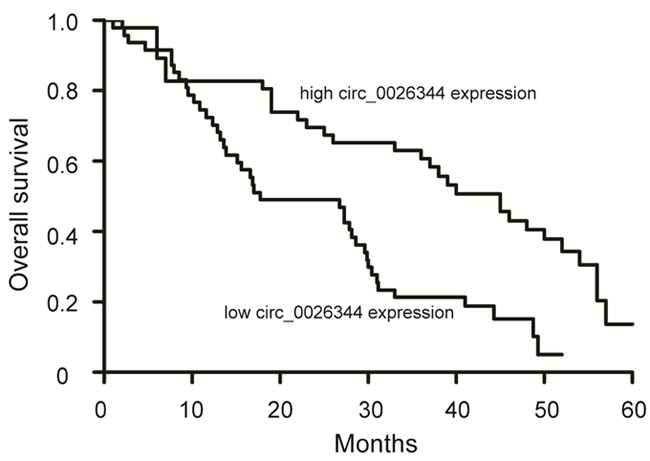
Kaplan-Meier survival analysis of association between circ_0026344 level and overall survival of gastric cancer patients. Low circ_0026344 expression correlated with a poorer overall survival.
Correlations between the circ_0026344 level and the clinicopathologic factors in gastric cancer patients
The association between the circ_0026344 expression and the clinicalpathologic data of gastric patients was analyzed by the chi-square test. As shown in Table 1, the circ_0026344 level was markedly associated with tumour size (P=0.001), lymph node metastasis (P=0.015), TNM stage (P=0.008), and invasive depth (P=0.028). However, circ_0026344 expression was not significantly related to age (P=0.523), gender (P=0.217), tumour location (P=0.264), differentiation (P=0.536), Borrmann type (P=0.355), distant metastasis (P=0.176), and CEA status (P=0.962).
Circ_0026344 is an independent prognostic indicator for overall survival of gastric cancer patients
Univariate Cox regression analyses performed to evaluate the circ_0026344 level and other clinicopathologic features on prognosis of gastric cancer patients. As shown in Table 2, it was observed that TNM stage (P<0.001), lymph node metastasis (P=0.016), distant metastasis (P<0.001), and circ_001569 level (P=0.002) were significantly correlated with overall survival of gastric cancer patients. Moreover, multivariate Cox regression analyses revealed that circ_0026344 level was an independent molecular biomarker predicting overall survival (Table 3, P=0.038).
Table 2.
Univariate Cox regression analyses of clinicopathologic factors in the prognosis of gastric cancer
| Factor | Univariate analyses | ||
|---|---|---|---|
|
| |||
| HR | 95% CI | P value | |
| Gender (Male vs. Female) | 1.14 | 0.64-1.59 | 0.771 |
| Age (<50 vs. ≥50) | 1.58 | 0.86-2.17 | 0.423 |
| Tumor size (<5 vs. ≥5) | 0.75 | 0.30-1.48 | 0.534 |
| Tumor location (Distal/middle vs. Proximal) | 1.43 | 0.81-1.90 | 0.361 |
| Differentiation (Well/moderate vs. Poor) | 2.25 | 0.97-3.74 | 0.085 |
| Borrmann type (I/II vs. III/IV) | 0.69 | 0.32-1.35 | 0.912 |
| CEA status (Negative vs. Positive) | 1.07 | 0.64-1.61 | 0.783 |
| Invasive depth (T1/T2 vs. T3/T4) | 1.34 | 0.65-1.88 | 0.302 |
| Distant metastasis (Negative vs. Positive) | 4.73 | 2.49-7.53 | <0.001* |
| TNM stage (I/II vs. III/IV) | 4.08 | 2.11-6.62 | <0.001* |
| Lymph node metastasis (Negative vs. Positive) | 2.15 | 1.23-4.09 | 0.016* |
| Circ_0026344 level (low vs. high) | 2.67 | 1.45-4.83 | 0.002* |
HR, hazard ratio; 95% CI, 95% confidence interval.
P<0.05.
Table 3.
Multivariate Cox regression analyses of clinicopathologic factors in the prognosis of gastric cancer
| Factor | Multivariate analyses | ||
|---|---|---|---|
|
| |||
| HR | 95% CI | P value | |
| Gender (Male vs. Female) | NA | ||
| Age (<50 vs. ≥50) | NA | ||
| Tumor size (<5 vs. ≥5) | NA | ||
| Tumor location (Distal/middle vs. Proximal) | NA | ||
| Differentiation (Well/moderate vs. Poor) | NA | ||
| Borrmann type (I/II vs. III/IV) | NA | ||
| CEA status (Negative vs. Positive) | NA | ||
| Invasive depth (T1/T2 vs. T3/T4) | NA | ||
| Distant metastasis (Negative vs. Positive) | 1.95 | 0.92-3.67 | 0.185 |
| TNM stage (I/II vs. III/IV) | 2.13 | 0.98-4.33 | 0.074 |
| Lymph node metastasis (Negative vs. Positive) | 1.64 | 0.77-2.46 | 0.455 |
| Circ_0026344 level (low vs. high) | 2.18 | 1.22-3.90 | 0.038* |
P<0.05.
Discussion
CircRNAs were first reported in RNA viruses as early as 1976 [17]. Although the function and molecular mechanism of most circRNAs are still not completely identified, increasing evidence has indicated that some of the circRNAs may be used as biomarkers in gastric cancers [16,18]. For example, Chen et al. [19] demonstrated that circ_0000190 is a novel non-invasive biomarker for the diagnosis of gastric cancer. Tian et al. [20] reported that circ_0003159 expression is significantly down-regulated in gastric cancer, and is negatively associated with gender, distal metastasis, and tumour-node-metastasis stage. Lu et al. [21] found that circ_0006848 acts as a novel biomarker for early gastric cancer. In addition, Hung et al. [22] illustrated that circ_0000745 plays an important role in gastric cancer, and its expression level in plasma in combination with CEA level is a promising diagnostic biomarker. In this study, we screened out the circ_0026344, which had low expression in colorectal cancer, based on our previous studies [14,23]. As a result of the study, the expression of circ_0026344 was markedly down-regulated in both gastric cancer tissues and cell lines. Furthermore, we found that low circ_0026344 expression was corrected with tumour malignant behaviors and poor prognosis of gastric cancer patients.
Circ_0026344 is a newly identified circRNA. Prior studies have shown that circ_0026344 is a tumour suppressor in the progression of colorectal cancer. Overexpression of circ_0026344 decreased cell growth and invasion while promoting apoptosis [14,23]. However, its clinical significance in gastric cancer remains unknown. Here, we found low expression of circ_0026344 in gastric cancer tissues compared to adjacent normal tissues, which is consistent with data of circ_0026344 expression in two previous circRNAs chips (GSE78092 and GSE89143) for gastric cancer [15,16]. In addition, circ_0026344 expression in metastatic cancer tissues was significantly lower than in non-metastatic tissues, providing the first evidence that downregulation of circ_0026344 is closely associated with gastric cancer.
Furthermore, we analyzed the association between circ_0026344 expression and clinicopathologic features in gastric cancer patients. The results showed that low circ_0026344 expression was markedly correlated with large tumour size, positive lymph node metastasis, advanced TNM stage, and large invasive depth. In general, patients with large tumour size and distal metastasis had a poor prognosis [24]. Interestingly, patients with low circ_0026344 expression had worse overall survival than patients with high circ_0026344 expression. Moreover, both univariate and multivariate Cox regression analyses illustrated that circ_0026344 could serve as a biomarker to predict overall survival in gastric cancer patients.
In summary, our findings revealed that circ_0026344 expression is downregulated in gastric cancer patients and cells, and low circ_0026344 expression is associated with tumour malignant behaviors. Importantly, circ_0026344 is identified as an independent diagnostic biomarker in gastric cancer patients.
Disclosure of conflict of interest
None.
References
- 1.Russo AE, Strong VE. Gastric cancer etiology and management in Asia and the West. Annu Rev Med. 2019;70:353–367. doi: 10.1146/annurev-med-081117-043436. [DOI] [PubMed] [Google Scholar]
- 2.Tan Z. Recent advances in the surgical treatment of advanced gastric cancer: a review. Med Sci Monit. 2019;25:3537–3541. doi: 10.12659/MSM.916475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biagioni A, Skalamera I, Peri S, Schiavone N, Cianchi F, Giommoni E, Magnelli L, Papucci L. Update on gastric cancer treatments and gene therapies. Cancer Metastasis Rev. 2019;38:537–548. doi: 10.1007/s10555-019-09803-7. [DOI] [PubMed] [Google Scholar]
- 4.Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, Bleotu C, Diaconu CC, Chivu-Economescu M. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25:2029–2044. doi: 10.3748/wjg.v25.i17.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang F, Hong F, Shah MW, Shen X. Circular RNAs as diagnostic biomarkers in gastric cancer: a meta-analysis review. Pathol Res Pract. 2019;215:152419. doi: 10.1016/j.prp.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, Wei F, Guo C, Wu X, Li X, Li Y, Li G, Zeng Z, Xiong W. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Liang W, Zhang P, Chen J, Qian H, Zhang X, Xu W. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36:152. doi: 10.1186/s13046-017-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu JZ, Shao CC, Wang XJ, Zhao X, Chen JQ, Ouyang YX, Feng J, Zhang F, Huang WH, Ying Q, Chen CF, Wei XL, Dong HY, Zhang GJ, Chen M. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175. doi: 10.1038/s41419-019-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang HD, Jiang LH, Hou JC, Zhong SL, Zhou SY, Zhu LP, Li J, Wang DD, Sun DW, Ji ZL, Tang JH. Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed Pharmacother. 2018;107:1342–1353. doi: 10.1016/j.biopha.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Y, Liu W, Zhang Y, Sun S. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503:870–875. doi: 10.1016/j.bbrc.2018.06.089. [DOI] [PubMed] [Google Scholar]
- 15.Huang YS, Jie N, Zou KJ, Weng Y. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16:2469–2476. doi: 10.3892/mmr.2017.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, Guo J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–1180. doi: 10.1002/cam4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547–555. doi: 10.1016/0092-8674(76)90223-3. [DOI] [PubMed] [Google Scholar]
- 18.Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17:137. doi: 10.1186/s12943-018-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32 doi: 10.1002/jcla.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J, Zhang PY, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, Li P, Zheng CH, Huang CM. Circular RNA hsa_circ_0006848 related to ribosomal protein L6 acts as a novel biomarker for early gastric cancer. Dis Markers. 2019;2019:3863458. doi: 10.1155/2019/3863458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23:6330–6338. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen T, Cheng X, Liu X, Xia C, Zhang H, Pan D, Zhang X, Li Y. Circ_0026344 restrains metastasis of human colorectal cancer cells via miR-183. Artif Cells Nanomed Biotechnol. 2019;47:4038–4045. doi: 10.1080/21691401.2019.1669620. [DOI] [PubMed] [Google Scholar]
- 24.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]


