Abstract
Background: Periodontitis is the second most common dental disease worldwide. TNF-α is up-regulated in periodontal disease and induces inflammation and cell apoptosis in gingival epithelial cells (GECs). miRNAs/mRNA axis play an important role in cell progression and inflammation. However, studies on the pathogenesis of periodontitisare still scarce, especially in the regulation mechanism of miRNAs. Methods: The expression and protein level of miR-16-5p, miR-145-5p, BACH2, and caspase 3 were determined by quantitative real time PCR and western blot, respectively. Cell viability was measured by MTT assay. Cell apoptosis was detected by flow cytometry. Dual-luciferase assay was applied to verify miR-16-5p and miR-145-5p target to the 3’UTR of BACH2. Results: TNF-α induced miR-16-5p, miR-145-5p and caspase 3 expression, inhibited cell viability, promoted cell apoptosis in GECs. However, down-regulated miR-16-5p and miR-145-5p can restore the effects of TNF-α on GECs. In addition, dual-luciferase assay determined that BACH2 was a common target of miR-16-5p and miR-145-5p. Knockdown of BACH2 induced GECs apoptosis. Of note, cell apoptosis induced by miR-16-5p mimic, miR-145-5p mimic, and TNF-α was significantly reversed by up-regulating BACH2. Conclusion: miR-16-5p and miR-145-5p mediate apoptosis induced by TNF-α in human gingival epithelial cells by targeting BACH2.
Keywords: miR-16-5p, miR-145-5p, BACH2, apoptosis, GECs
Introduction
Periodontal disease is usually caused by bacterial infection of the periodontal disease surrounding the teeth, leading to inflammation [1]. Tumor necrosis factor alpha (TNF-α), as one of the cytokines, is involved in systemic inflammation, which is produced by activated macrophages [2-5]. TNF-α also acts as a diagnostic marker for periodontal disease, inducing cell apoptosis and inflammation [6,7]. Fujita et al reported that TNF-α increased the permeablility of human gingival epithelial cells (GECs) and disrupted the gingival epithelial barrier [8]. However, the underlying mechanism of TNF-α has been not fully investigated.
MicroRNAs (miRNAs) are small, non-coding single-stranded RNAs of ~22 nucleotides in length that inhibit translation initiation and protein synthesis at the post-transcriptional level, or induce messenger RNA degradation [9,10]. In addition, miRNAs are associated with cell progression, inflammation, and autoimmune activation in multiple diseases, including periodontal disease [11-14]. For example, miR-146a promoted periodontal ligament cells differentiation and miR-142 triggered GECs apoptosis [14,15]. A study of Chen et al suggested that miRNAs regulate cytokine responses in gingival epithelial cells. Moreover, through sequencing normal GECs and periodontitis GECs, C. Stoecklin-Wasmer et al suggested that miRNAs and mRNAs may have influenced the occurrence of periodontitis, and be involved in homeostasis and inflammatory or immune responses [16]. However, few studies have been done on the regulatory mechanism and function of miRNAs in periodontal disease.
According to a previous study, miR-16-5p and miR-145-5p were up-regulated in inflamed gingival tissues [16]. However, the function and underlying mechanism of miR-16-5p and miR-145-5p have been barely investigated in periodontal disease. In this study, we found that miR-16-5p and miR-145-5p were induced by TNF-α in GECs, therefore, we speculated that miR-16-5p and miR-145-5p were involved in inflammatory response and development of periodontal disease.
Materials and methods
Patients
GECs collected from healthy gingival tissue were obtained from 20 chronic periodontitis patients who underwent oral surgery at University of Chinese Academy of Sciences Shenzhen Hospital. No patients underwent periodontal therapy and had other inflammatory diseases. Informed consent was obtained from each patient. This experiment was approved by institutional ethical committee and review board of University of Chinese Academy of Sciences Shenzhen Hospital.
Cell culture and TNF-α treatment
HEK293T cells were purchased from RiboBio Co. (Guangzhou, China). The GECs from gingival tissues were cultured in keratinocyte serum-free medium containing calcium chloride, bovine pituitary, epidermal growth factor, penicillin and streptomycin at 37°C with 5% of CO2. The culture medium was changed every other day. When the confluence reached up to 70-80%, cells were treated different concentration of TNF-α (PeproTech Inc., Rocky Hill, NJ, USA).
Cell transfection
NC (negative control), miR-16-5p, miR-145-5p, anti-NC, anti-miR-16-5p, anti-miR-145-5p, vector, BACH2, scramble and siBACH2 were purchased from Genecopoeia (Rockville, MD, USA). Plasmid and oligos transfection were performed using Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s protocol.
Quantitative real-time PCR
Total RNA was extracted from cells using Trizol reagent following the manufacturer’s protocol. RNA was reverse transcribed to cDNA using TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) for miRNA and using M-MLV Reverse Transcriptase (Invitrogen) for mRNA. Then, Real-time qPCR was performed using SYBR Green Real-time PCR kit (Invitrogen) according to the manufacturer’s protocol. U6 and GAPDH were employed as reference genes. Fluorescence of each sample was detected in an iQTM5 Multicolor Real-Time PCR Detection System (Bio-Rad). The primers used for qRT-PCR are: miR-16-5p F: 5’-TAGCAGCACGTAAATATTGGCG-3’, miR-16-5p R: 5’-TGCGTGTCGTGGAGTC-3’, U6 F: 5’-CTCGCTTCGGCAGCACA-3’, U6 R: 5’-AACGCTTCACGAATTTGCGT-3’, miR-145 F: 5’-GCATCTCTGGTCAGTTGGG-3’, miR-145 R: 5’-GACCTCAAGAACAGTAT-3’, BACH2 F: 5’-CAGCTTGGCAGTGTAGGC-3’, BACH2 R: 5’-CCCTGGCTGTGACCTCCTC-3’, GAPDH F: 5’-GCACCGTCAAGGCTGAGAAC-3’, GAPDH R: 5’-ATGGTGGTGAAGACGCCAGT-3’. The 2-ΔΔCt method was used to calculate the relative expression of mRNA and miRNA.
Western blot
Cells were lysed with RIPA buffer with phenylmenthylsulfonyl flourede (PMSF) and proteinase inhibitor. Nanodrop2000 (Thermo Fisher Scientific, USA) was applied to measure protein concentration. Then, 20 ug protein was loaded into the SDS-PAGE gel to run, and then transferred the gel onto the PVDF membrane. Membranes were incubated with the primary antibodies at 4°C overnight. Membranes were incubated with the HRP-conjugated secondary antibody solution for 1 h at room temperature. The blot was detected using the PierceTM ECL western blotting substrate (Thermo Fisher Scientific, 32109, USA).
Dual luciferase reporter assay
The BACH2 3’UTR and BACH2 3’UTR mutant were amplified and inserted into the pMIR-REPORT™ (Thermo Fisher Scientific, USA) to construct BACH2 wild type reporter vector (BACH2-wt) and BACH2 mutant type reporter vector (BACH2-mut). Then, BACH2-wt or BACH2-mut was co-transfected with NC or miR-16-5p and miR-145-5p into HEK293T cells using Lipofectamine 3000. After 48 h post-transfection, the luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega, USA).
Measurement of caspase 3
The activity of caspase 3 was measured using a colorimetric assay kit (BioVision Inc., Mountain View, CA, USA) according to the manufacturer’s protocol. We read the absorbance at a wavelength of 405 nm.
MTT assay
2.0×103 cells were seeded in 96-well cell culture plates (Corning Inc., Corning, NY) and incubated for 24 h. Then 20 µl MTT were added into each well and incubate for 4 h at 37°C before measuring the absorbance. 150 µL dimethyl sulfoxide (DMSO; Sigma) were added into each well and following incubation for 3 h at 37°C with 5% of CO2. Cell viability was measured using a spectrophotometric microplate reader (Beyotime Institute of Biotechnology, Haimen, China) at OD=450 nm.
Flow cytometry
Annexin V-FITC apoptosis detection kit (Beyotime, Shanghai, China) was applied to detect cell apoptosis according to the manufacturer’s protocol. Briefly, cells collected were re-suspended in binding solution (200 μl) and stained with Annexin V-FITC (10 μl) and propidium iodide (10 μl) for 15 min at 37°C. Cell apoptosis rate was monitored by flow cytometry (BD Biosciences, San Jose, CA, USA).
Statistical analysis
All data were performed as mean ± SD (standard deviation) from at least 3 times independent experiments. The Student’s t-test or one-way analysis of variance were used to analyze the differences between groups. The analysis of results was displayed using GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA). P<0.05 was regarded as significant.
Results
TNF-α induced miR-16-5p and miR-145-5p expression in GECs
To investigate the effect of TNF-α on miRNA expression, different concentrations of TNF-α was used to treat human GECs. After treatment with TNF-α at 5-20 ng/ml, the expression of miR-16-5p and miR-145-5p was significantly increased in the treatment of TNF-α compared with controls, peaking at 15 ng/ml (Figure 1A and 1C). In the following experiment, GECs were treated with 15 ng/ml TNF-α and the expression of miR-16-5p and miR-145-5p at 5 h, 10 h, 15 h, 20 h, 30 h and 45 h were detected. The results showed that miR-16-5p and miR-145-5p expression were significantly higher than that of the control group at 10 h, peaking at 20 h (Figure 1B and 1D). Therefore, miR-16-5p and miR-145-5p expression were induced by TNF-α in GECs in a dose-dependent manner.
Figure 1.
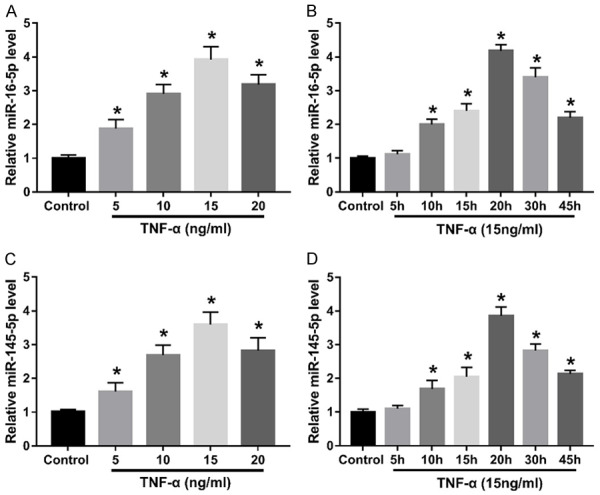
TNF-α induced miR-16-5p and miR-145-5p expression in GECs. (A, C) The expression of miR-16-5p (A) and miR-145-5p (C) was detected using qRT-PCR in GECs treated with different concentrations of TNF-α for 24 h. (B, D) The expression of miR-16-5p (B) and miR-145-5p (D) was detected using qRT-PCR in GECs at different times at 15 ng/ml TNF-α. *P<0.05.
miR-16-5p and miR-145-5p were involved in TNF-α induced apoptosis in GECs
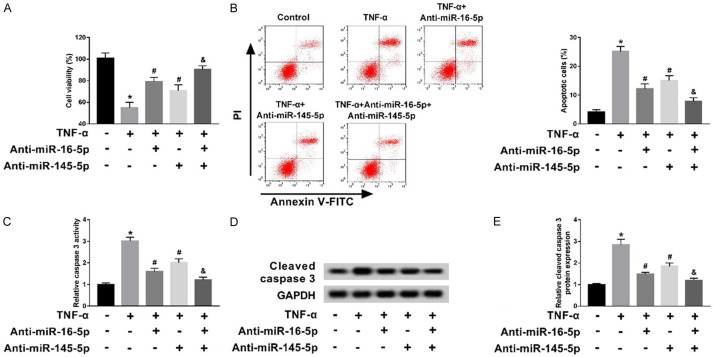
Next, we further validated the interaction of miR-16-5p and miR-145-5p with TNF-α in the biologic effects of GECs. MMT assay analysis showed that TNF-α remarkably decreased cell viability. However, transfection of anti-miR-16-5p and anti-miR-145-5p can promoted cell viability inhibited by TNF-α. Interestingly, anti-miR-16-5p and anti-miR-145-5p co-transfection significantly promoted cell viability inhibited by TNF-α compared with TNF-α + anti-miR-16-5p and TNF-α + anti-miR-145-5p groups (Figure 2A). In addition, flow cytometry demonstrated that TNF-α significantly induced cell apoptosis in comparison to the control group; nevertheless, cell apoptosis of TNF-α + anti-miR-16-5p and TNF-α + anti-miR-145-5p was lower than that in TNF-α group and was higher than that in TNF-α + anti-miR-16-5p + anti-miR-145-5p groups (Figure 2B). Moreover, caspase 3, a biomarker of apoptosis, was measured by qRT-PCR and western blot. The results showed activity of cle-caspase 3 was induced by TNF-α, but transfection of anti-miR-16-5p and anti-miR-145-5p reversed its expression in GECs respectively (Figure 2C-E). Therefore, TNF-α inhibited cell viability, induced cell apoptosis, and cle-caspase 3 activity, whereas transfection of anti-miR-16-5p and anti-miR-145-5p restored TNF-α’s effect on GECs.
Figure 2.
miR-16-5p and miR-145-5p were involved in TNF-α induced apoptosis in GECs. A. Cell viability was measured using MTT in control, TNF-α, TNF-α + anti-miR-16-5p, TNF-α + anti-miR-145-5p and TNF-α + anti-miR-16-5p + anti-miR-145-5p groups. B. Cell apoptosis was determined using flow cytometry in control, TNF-α, TNF-α + anti-miR-16-5p, TNF-α + anti-miR-145-5p and TNF-α + anti-miR-16-5p + anti-miR-145-5p groups. C. The activity of caspase 3 was measured in control, TNF-α, TNF-α + anti-miR-16-5p, TNF-α + anti-miR-145-5p and TNF-α + anti-miR-16-5p + anti-miR-145-5p groups. D and E. Western blot was applied to detect the protein level of caspase 3 in controls, TNF-α, TNF-α + anti-miR-16-5p, TNF-α + anti-miR-145-5p and TNF-α + anti-miR-16-5p + anti-miR-145-5p groups. *P<0.05, TNF-α group vs control group; #P<0.05, TNF-α + anti-miR-16-5p, TNF-α + anti-miR-145-5p groups vs TNF-α group; &P<0.05, TNF-α + anti-miR-16-5p + anti-miR-145-5p group vs TNF-α + anti-miR-16-5p, TNF-α + anti-miR-145-5p groups.
BACH2 is a target of miR-16-5p and miR-145-5p directly
To further determine the underlying regulatory mechanism of miR-16-5p and miR-145-5p, bioinformatic analysis was used to predict their downstream gene and the result showed that BACH2 was a potential target of miR-16-5p and miR-145-5p (Figure 3A and 3C). Next, luciferase reporter assay was applied to verify whether miR-16-5p and miR-145-5p binds to the 3’UTR of BACH2. The vector of BACH2 3’UTR wild type or BACH2 3’UTR mutated was co-transfected miR-16-5p or miR-145-5p into HEK293T cells respectively. The results showed that the luciferase activity of miR-16-5p binding to BACH2-wt 3’UTR was significantly decreased, but no effect with BACH2-mut 3’UTR. Similar to miR-16-5p (Figure 3B), the luciferase activity of miR-145-5p binding to BACH2-wt 3’UTR was significantly decreased, whereas there was no effect of BACH2-mut 3’UTR (Figure 3D).
Figure 3.
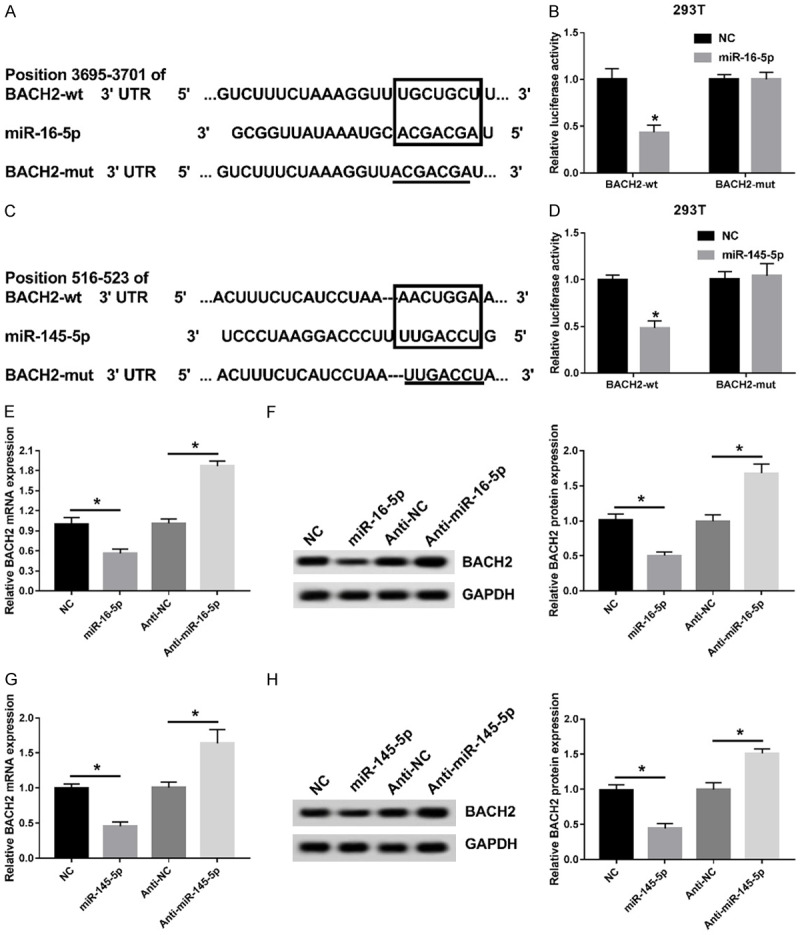
BACH2 is a target of miR-16-5p and miR-145-5p directly. (A) Prediction of a miR-16-5p binding site in the BACH2 mRNA 3’UTR. (B) The relationship between miR-145-5p and BACH2 mRNA was verified using luciferase reporter assay. (C) Prediction of a miR-145-5p binding site in the BACH2 mRNA 3’UTR. (D) The relationship between miR-145-5p and BACH2 mRNA was verified using luciferase reporter assay. (E, F) BACH2 mRNA (E) or protein level (F) was measured using qRT-PCR or western blot in NC, miR-16-5p, anti-NC and anti-miR-16-5p groups. (G, H) BACH2 mRNA (G) or protein level (H) was measured using qRT-PCR or western blot in NC, miR-145-5p, anti-NC and anti-miR-145-5p groups. *P<0.05.
Additionally, as shown in Figure 3E and 3F, up-regulated or down-regulated miR-16-5p can decrease or increase BACH2 expression. Moreover, overexpression or inhibition of miR-145-5p also can decrease or increase BACH2 expression (Figure 3G and 3H). Thus, these findings showed that BACH2 was a common target of miR-16-5p and miR-145-5p directly.
Knockdown of BACH2 promoted apoptosis in GECs
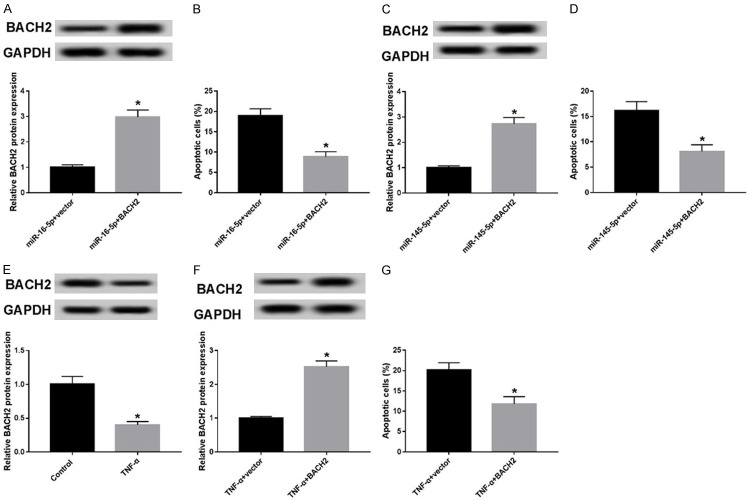
To realize the effect of BACH2 on TNF-α-induced apoptosis in GECs, we obtained the GECs lines with down-expressed BACH2. The analysis of qRT-PCR and western blot showed that siBACH2 transfection inhibited BACH2 mRNA and BACH2 protein (Figure 4A and 4B), which can be used for subsequent experiments. Then, we detected cell apoptosis in scramble and siBACH2 groups by flow cytometry. The results showed that knockdown of BACH2 remarkably raised cell apoptosis compared with the scramble group (Figure 4C). Thus, knockdown of BACH2 can promote apoptosis in GECs.
Figure 4.
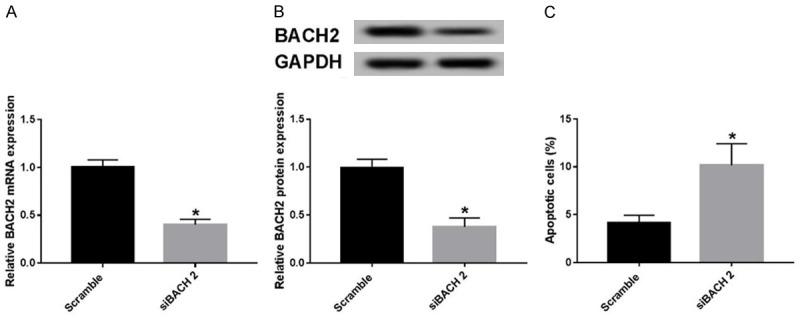
Knockdown of BACH2 promoted apoptosis in in GECs. (A, B) BACH2 mRNA (A) or protein level (B) was measured using qRT-PCR or western blot in scramble and siBACH2 groups. (C) Cell apoptosis was determined using flow cytometry in scramble and siBACH2 groups. *P<0.05.
Over-expressed BACH2 reversed apoptosis induced by miR-16-5p, miR-145-5p and TNF-α in GECs
To further explore the correlation between BACH and miR-16-5p, miR-145-5p and TNF-α, we obtained the vector overexpressed BACH2 (BACH2), and then BACH2 was co-transfected with miR-16-5p mimic or miR-145-5p mimic into GECs. The analysis of western blot displayed that the expression of BACH2 was significantly higher in miR-145-5p + BACH group or miR-16-5p + BACH group than that in miR-16-5p + vector group or miR-145-5p + vector group (Figure 5A and 5C) As shown in Figure 5B, cell apoptosis in miR-16-5p + BACH group was significantly lower than that in the miR-16-5p + vector group. Similarly, cell apoptosis in miR-145-5p + BACH group was significantly reduced compared with that in the miR-145-5p + vector group (Figure 5D). Thus, BACH2 expressed high inhibited the effect of miR-16-5p and miR-145-5p on cell apoptosis in GECs. Furthermore, Figure 5E shows that TNF-α significantly inhibited BACH expression. GECs lines overexpressing BACH2 were obtained, which were treated with TNF-α (Figure 5F). Up-regulated BACH2 could reduce cell apoptosis induced by TNF-α (Figure 5G). Taken together, overexpression of BACH2 can inhibit cell apoptosis induced by miR-16-5p, miR-145-5p, and TNF-α respectively.
Figure 5.
Over-expressed BACH2 reversed apoptosis induced by miR-16-5p, miR-145-5p or TNF-α. A. BACH2 protein level was detected using western blot in miR-16-5p + vector and miR-16-5p + BACH groups. B. Cell apoptosis was determined using flow cytometry in miR-16-5p + vector and miR-16-5p + BACH groups. C. BACH2 protein level was detected using western blot in miR-145-5p + vector and miR-145-5p + BACH groups. D. Cell apoptosis was determined using flow cytometry in miR-145-5p + vector and miR-145-5p + BACH groups. E. BACH2 protein level was measured using qRT-PCR or western blot in control and TNF-α groups. F. BACH2 protein level was measured using qRT-PCR or western blot in TNF-α + vector and TNF-α + BACH2 groups. G. Cell apoptosis was determined using flow cytometry in TNF-α + vector and TNF-α + BACH2 groups. *P<0.05.
Discussion
Periodontitis is the second most common dental disease worldwide, affecting human dental health and life quality seriously [17]. However, studies on the pathogenesis of periodontitis are still scarce, especially in the regulatory mechanism of miRNAs.
miRNAs, as an important regulatory factor in organisms, are closely related to the occurrence, development and prognosis of human diseases, including cell differentiation, cell proliferation, cell migration, apoptosis, inflammatory and immune response [12,15,18-21]. For example, miR-663 contributes to cell proliferation by targeting CDKN2A [22]. miR-21 promotes cell growth and invasion by suppression of PTEN in NSCLC [23]. However, a study showed that different expressions of miRNAs were analyzed in healthy and inflamed gingival tissues including miR-16-5p and miR-145-5p. miR-16-5p and miR-145-5p were up-regulated in inflamed gingival tissues, suggesting a possible positive correlation with inflammation. In our study, the results of the treated GECs with TNF-α showed that miR-16-5p and miR-145-5p was significantly up-regulated, suggesting TNF-α induced miR-16-5p and miR-145-5p expression.
miR-16-5p and miR-145-5p are associated with a variety of diseases. For example, miR-16-5p was down-regulated in chordoma and miR-16-5p expressed high inhibited chordoma cell proliferation, invasion and migration by targeting Smad3 [24]. Qu et al reported that up-regulation of miR-16-5p suppressed cell growth and induced apoptosis via down-regulating VEGFA in breast cancer. Otherwise, miR-16-5p/SESN1 axis regulated p53 signaling pathway, affecting myoblast proliferation, apoptosis and differentiation [25]. Similar to the function of miR-16-5p, miR-145-5p inhibited cell proliferation by inhibiting SOX2 [26]. Additionally, miR-145-5p regulated hypoxia-induced inflammatory response and apoptosis by targeting CD40 in cardiomyocytes [27]. Our functional experiments indicated that TNF-α inhibited cell viability and induced apoptosis and activated caspase 3 in GECs. However, suppression of miR-16-5p and miR-145-5p could restore the effects of TNF-α on GECs, suggesting miR-16-5p and miR-145-5p were involved in the inflammation and apoptosis of periodontitis.
To further explore the regulatory mechanism of miR-16-5p and miR-145-5p, we found that BACH2 was a target of miR-16-5p and miR-145-5p by luciferase reporter assay. Broad complex-tramtrack-bric a brac and Cap’n’collar homology 2 (BACH2), as a broad regulator of immune activation, stabilizes immunoregulatory function, and suppresses lethal inflammation to stabilize Treg-mediated immune homeostasis [28]. Otherwise, BACH2 also represses the plasma cell gene network in B cells to elevate antibody class switch and regulates the pre-B-cell receptor checkpoint gene CDKN2A and TP53 [29,30]. Moreover, BACH2 was also associated with autoimmune, inflammation, and cell progression in human diseases, such as leukemia, pancreatic carcinoma, and breast cancer [31-33]. For example, miR-148a promoted plasma cell differentiation by regulating Mitf and BACH2 [34]. miR-142 was up-regulated induced by TNF-α and promotes GECs cell apoptosis by suppression of BACH2 [14]. In our present study, we validated the function of BACH2 in GECs. The results of our experiment showed that knockdown of BACH2 could promote GECs apoptosis. In addition, to further explore the regulatory mechanism of miR-16-5p, miR-145-5p and BACH2 in GECs, BACH2 was co-transfected with miR-16-5p, miR-145-5p into GECs. The results showed that cell apoptosis induced by miR-16-5p, miR-145-5p were reversed by up-regulating BACH2. Interestingly, the effect of TNF-α on GECs apoptosis was weakened by BACH2 overexpression. These findings suggested that miR-16-5p/BACH2 and miR-145-5p/BACH2 axis are associated with GECs inflammation and cell apoptosis. Although the new regulatory mechanism of miR-16-5p, miR-145-5p and BACH2 has been verified to play an important role in GECs induced by TNF-α, it must beverified in vivo.
Conclusion
In conclusion, our findings showed that miR-16-5p and miR-145-5p was associated with GECs apoptosis induced by TNF-α via regulating BACH2, helping explain pathogenesis of periodontitis disease.
Acknowledgements
Informed consent was obtained from each patient. This experiment was approved by institutional ethical committee and review board of University of Chinese Academy of Sciences Shenzhen Hospital.
Disclosure of conflict of interest
None.
References
- 1.Nibali L, D’Aiuto F, Griffiths G, Patel K, Suvan J, Tonetti MS. Severe periodontitis is associated with systemic inflammation and a dysmetabolic status: a case-control study. J Clin Periodontol. 2007;34:931–937. doi: 10.1111/j.1600-051X.2007.01133.x. [DOI] [PubMed] [Google Scholar]
- 2.Feng L, Xie S. Puerarin prevents tumor necrosis factor-α-induced apoptosis of PC12 cells via activation of the PI3K/Akt signaling pathway. Exp Ther Med. 2017;14:813–818. doi: 10.3892/etm.2017.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murch SH, Braegger CP, Walkersmith JA, Macdonald TT. Location of tumour necrosis factor alpha by immunohistochemistry in chronic inflammatory bowel disease. Gut. 1993;34:1705–1709. doi: 10.1136/gut.34.12.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducingligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 5.Renz H, Gong JH, Schmidt A, Nain M, Gemsa D. Release of tumor necrosis factor-alpha from macrophages. Enhancement and suppression are dose-dependently regulated by prostaglandin E2 and cyclic nucleotides. J Immunol. 1988;141:2388–2393. [PubMed] [Google Scholar]
- 6.Madureira DF, Lucas De Abreu Lima I, Costa GC, Lages EMB, Martins CC, Aparecida Da Silva T. Tumor necrosis factor-alpha in gingival crevicular fluid as a diagnostic marker for periodontal diseases: a systematic review. J Evid Based Dent Pract. 2018;18:315–331. doi: 10.1016/j.jebdp.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Braegger CP, Nicholls S, Murch SH, Stephens S, Macdonald TT. Tumour necrosis factor alpha in stool as a marker of intestinal inflammation. Lancet. 1992;339:89–91. doi: 10.1016/0140-6736(92)90999-j. [DOI] [PubMed] [Google Scholar]
- 8.Fujita T, Yumoto H, Shiba H, Ouhara K, Miyagawa T, Nagahara T, Matsuda S, Kawaguchi H, Matsuo T, Murakami S. Irsogladine maleate regulates epithelial barrier function in tumor necrosis factor-α-stimulated human gingival epithelial cells. J Periodontal Res. 2012;47:55–61. doi: 10.1111/j.1600-0765.2011.01404.x. [DOI] [PubMed] [Google Scholar]
- 9.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 11.Contreras JR. Characterizing miR-146a and miR-34a in B-cell lymphomagenesis. UCLA. 2016 [Google Scholar]
- 12.Li N, Shi H, Zhang L, Li X, Gao L, Zhang G, Shi Y, Guo S. MiR-188 inhibits glioma cell proliferation and cell cycle progression through targeting β-catenin. Oncol Res. 2018;26:785–794. doi: 10.3727/096504017X15127309628257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, Liang D, He D, Wang H, Liu W. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat Med. 2012;18:1077–1086. doi: 10.1038/nm.2815. [DOI] [PubMed] [Google Scholar]
- 14.Song L, Song Z, Dong J, Rong S. microRNA-142 is upregulated by tumor necrosis factor-alpha and triggers apoptosis in human gingival epithelial cells by repressing BACH2 expression. Am J Transl Res. 2017;9:175–183. [PMC free article] [PubMed] [Google Scholar]
- 15.Hung PS, Chen FC, Kuang SH, Kao SY, Lin SC, Chang KW. miR-146a induces differentiation of periodontal ligament cells. J Dent Res. 2010;89:252–257. doi: 10.1177/0022034509357411. [DOI] [PubMed] [Google Scholar]
- 16.Stoecklin-Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN. MicroRNAs and their target genes in gingival tissues. J Dent Res. 2012;91:934–940. doi: 10.1177/0022034512456551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vos T, Flaxman AD, Naghavi M, Lozano R, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V, Abraham J, Ackerman I, Aggarwal R, Ahn SY, Ali MK, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Bahalim AN, Barker-Collo S, Barrero LH, Bartels DH, Basáñez MG, Baxter A, Bell ML, Benjamin EJ, Bennett D, Bernabé E, Bhalla K, Bhandari B, Bikbov B, Bin Abdulhak A, Birbeck G, Black JA, Blencowe H, Blore JD, Blyth F, Bolliger I, Bonaventure A, Boufous S, Bourne R, Boussinesq M, Braithwaite T, Brayne C, Bridgett L, Brooker S, Brooks P, Brugha TS, Bryan-Hancock C, Bucello C, Buchbinder R, Buckle G, Budke CM, Burch M, Burney P, Burstein R, Calabria B, Campbell B, Canter CE, Carabin H, Carapetis J, Carmona L, Cella C, Charlson F, Chen H, Cheng AT, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahiya M, Dahodwala N, Damsere-Derry J, Danaei G, Davis A, De Leo D, Degenhardt L, Dellavalle R, Delossantos A, Denenberg J, Derrett S, Des Jarlais DC, Dharmaratne SD, Dherani M, Diaz-Torne C, Dolk H, Dorsey ER, Driscoll T, Duber H, Ebel B, Edmond K, Elbaz A, Ali SE, Erskine H, Erwin PJ, Espindola P, Ewoigbokhan SE, Farzadfar F, Feigin V, Felson DT, Ferrari A, Ferri CP, Fèvre EM, Finucane MM, Flaxman S, Flood L, Foreman K, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabbe BJ, Gabriel SE, Gakidou E, Ganatra HA, Garcia B, Gaspari F, Gillum RF, Gmel G, Gosselin R, Grainger R, Groeger J, Guillemin F, Gunnell D, Gupta R, Haagsma J, Hagan H, Halasa YA, Hall W, Haring D, Haro JM, Harrison JE, Havmoeller R, Hay RJ, Higashi H, Hill C, Hoen B, Hoffman H, Hotez PJ, Hoy D, Huang JJ, Ibeanusi SE, Jacobsen KH, James SL, Jarvis D, Jasrasaria R, Jayaraman S, Johns N, Jonas JB, Karthikeyan G, Kassebaum N, Kawakami N, Keren A, Khoo JP, King CH, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lalloo R, Laslett LL, Lathlean T, Leasher JL, Lee YY, Leigh J, Lim SS, Limb E, Lin JK, Lipnick M, Lipshultz SE, Liu W, Loane M, Ohno SL, Lyons R, Ma J, Mabweijano J, MacIntyre MF, Malekzadeh R, Mallinger L, Manivannan S, Marcenes W, March L, Margolis DJ, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGill N, McGrath J, Medina-Mora ME, Meltzer M, Mensah GA, Merriman TR, Meyer AC, Miglioli V, Miller M, Miller TR, Mitchell PB, Mocumbi AO, Moffitt TE, Mokdad AA, Monasta L, Montico M, Moradi-Lakeh M, Moran A, Morawska L, Mori R, Murdoch ME, Mwaniki MK, Naidoo K, Nair MN, Naldi L, Narayan KM, Nelson PK, Nelson RG, Nevitt MC, Newton CR, Nolte S, Norman P, Norman R, O’Donnell M, O’Hanlon S, Olives C, Omer SB, Ortblad K, Osborne R, Ozgediz D, Page A, Pahari B, Pandian JD, Rivero AP, Patten SB, Pearce N, Padilla RP, Perez-Ruiz F, Perico N, Pesudovs K, Phillips D, Phillips MR, Pierce K, Pion S, Polanczyk GV, Polinder S, Pope CA 3rd, Popova S, Porrini E, Pourmalek F, Prince M, Pullan RL, Ramaiah KD, Ranganathan D, Razavi H, Regan M, Rehm JT, Rein DB, Remuzzi G, Richardson K, Rivara FP, Roberts T, Robinson C, De Leòn FR, Ronfani L, Room R, Rosenfeld LC, Rushton L, Sacco RL, Saha S, Sampson U, Sanchez-Riera L, Sanman E, Schwebel DC, Scott JG, Segui-Gomez M, Shahraz S, Shepard DS, Shin H, Shivakoti R, Singh D, Singh GM, Singh JA, Singleton J, Sleet DA, Sliwa K, Smith E, Smith JL, Stapelberg NJ, Steer A, Steiner T, Stolk WA, Stovner LJ, Sudfeld C, Syed S, Tamburlini G, Tavakkoli M, Taylor HR, Taylor JA, Taylor WJ, Thomas B, Thomson WM, Thurston GD, Tleyjeh IM, Tonelli M, Towbin JA, Truelsen T, Tsilimbaris MK, Ubeda C, Undurraga EA, van der Werf MJ, van Os J, Vavilala MS, Venketasubramanian N, Wang M, Wang W, Watt K, Weatherall DJ, Weinstock MA, Weintraub R, Weisskopf MG, Weissman MM, White RA, Whiteford H, Wiersma ST, Wilkinson JD, Williams HC, Williams SR, Witt E, Wolfe F, Woolf AD, Wulf S, Yeh PH, Zaidi AK, Zheng ZJ, Zonies D, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the global burden of disease study 2010. Lancet. 2012;380:2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Huang H, Sun L, Yang M, Pan C, Chen W, Wu D, Lin Z, Zeng C, Yao Y. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 20.Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, Croce CM. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A. 2011;108:4908–4913. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Yao K, Guo J, Shi H, Ma L, Wang Q, Liu H, Gao W, Sun A, Zou Y. miR-181a and miR-150 regulate dendritic cell immune inflammatory responses and cardiomyocyte apoptosis via targeting JAK1-STAT1/c-Fos pathway. J Cell Mol Med. 2017;21:2884–2895. doi: 10.1111/jcmm.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang S, Zhang N, Deng Y, Chen L, Zhang Y, Zheng Z, Luo W, Lv Z, Li S, Xu T. miR-663 promotes NPC cell proliferation by directly targeting CDKN2A. Mol Med Rep. 2017;16:4863–4870. doi: 10.3892/mmr.2017.7129. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Kang Y, Ren T, Yi H, Tang X, Wei G. miR-16-5p inhibits chordoma cell proliferation, invasion and metastasis by targeting Smad3. Cell Death Dis. 2018;9:680. doi: 10.1038/s41419-018-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai B, Ma M, Chen B, Li Z, Abdalla BA, Nie Q, Zhang X. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death Disease. 2018;9:367. doi: 10.1038/s41419-018-0403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozen M, Karatas OF, Gulluoglu S, Bayrak OF, Sevli S, Guzel E, Ekici ID, Caskurlu T, Solak M, Creighton CJ. Overexpression of miR-145-5p inhibits proliferation of prostate cancer cells and reduces SOX2 expression. Cancer Invest. 2015;33:251–258. doi: 10.3109/07357907.2015.1025407. [DOI] [PubMed] [Google Scholar]
- 27.Ming Y, Zhang L, Fei Y, Zhou J, Ma Y, Yang F, Ling T. MiR-145-5p regulates hypoxia-induced inflammatory response and apoptosis in cardiomyocytes by targeting CD40. Mol Cell Biochem. 2017;431:1–9. doi: 10.1007/s11010-017-2982-4. [DOI] [PubMed] [Google Scholar]
- 28.Rahul R, Kiyoshi H, Kambiz M, David C, Klebanoff CA, Michael B, Giuseppe S, Hossein Z, Golnaz V, Barbara D. Bach2 represses effector programmes to stabilize Treg-mediated immune homeostasis. Nature. 2013;498:506–510. doi: 10.1038/nature12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, Ng C, Titz B, Hurtz C, Sadiyah MF. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med. 2013;19:1014–1022. doi: 10.1038/nm.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muto A, Ochiai K, Kimura Y, Itoh-Nakadai A, Calame KL, Ikebe D, Tashiro S, Igarashi K. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2014;29:4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge Z, Zhou X, Gu Y, Han Q, Li J, Chen B, Ge Q, Dovat E, Payne JL, Sun T. Ikaros regulation of the BCL6/BACH2 axis and its clinical relevance in acute lymphoblastic leukemia. Oncotarget. 2016;8:3512–3512. doi: 10.18632/oncotarget.14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie GH, Tang ZH. Expression of a new candidate tumor suppressor gene Bach2 in pancreatic carcinoma. China Journal of Modern Medicine. 2014 [Google Scholar]
- 33.Mamasani MN, Reisi S, Peymani M. Down-regulation of BACH2 in formalin-fixed paraffin-embedded breast cancer tissue as transcriptional regulation in cancer. Iran J Obst Gynecol Infertil. 2017;20:105–113. [Google Scholar]
- 34.Porstner M, Winkelmann R, Daum P, Schmid J, Pracht K, Côrtereal J, Schreiber S, Haftmann C, Brandl A, Mashreghi MF. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur J Immunol. 2015;45:1206–1215. doi: 10.1002/eji.201444637. [DOI] [PubMed] [Google Scholar]