Abstract
The seroprevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was examined among 105 healthcare workers (HCWs) exposed to four patients who were laboratory confirmed with coronavirus disease 2019 (COVID-19), the disease caused by SARS-CoV-2 infection. These HCWs were immediately under quarantine for 14 days as soon as they were identified as close contacts. The nasopharyngeal swab samples were collected on the first and 14th day of the quarantine, while the serum samples were obtained on the 14th day of the quarantine. With the assay of enzyme immunoassay (EIA) and microneutralization assay, 17.14% (18/105) of HCWs were seropositive, while their swab samples were found to be SARS-CoV-2 RNA negative. Risk analysis revealed that wearing face mask could reduce the infection risk (odds ratio [OR], 0.127, 95% confidence interval [CI] 0.017, 0.968), while when exposed to COVID-19 patients, doctors might have higher risk of seroconversion (OR, 346.837, 95% CI 8.924, 13479.434), compared with HCWs exposed to colleagues as well as nurses and general service assistants who exposed to patients. Our study revealed that the serological testing is useful for the identification of asymptomatic or subclinical infection of SARS-CoV-2 among close contacts with COVID-19 patients.
Keywords: COVID-19, Seroprevalence, SARS-CoV-2, Risk factors, Healthcare workers
Introduction
The ongoing pandemic of 2019 novel coronavirus, known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first reported in Wuhan, China in late Dec 2019.1 Due to its escalating spread globally, as of May 17, 2020, more than 532,224 patients were confirmed with coronavirus disease 2019 (COVID-19), the disease caused by SARS-CoV-2, across 215 countries and regions, causing a total of 24,087 deaths.
A wide spectrum of disease severity of laboratory confirmed COVID-19 has been depicted,2 , 3 including asymptomatic or minimally symptomatic cases.4 , 5 The proportion of asymptomatic SARS-CoV-2 infected patients is unknown, which remains a critical epidemiological puzzle.6 Whether one may seroconvert to SARS-CoV-2 with minimal or without symptoms still needs to be answered. Meanwhile, an efficient human-to-human transmission of SARS-CoV-2 mostly occurs among close contacts.1 Serological testing to close contacts with COVID-19 patients will help define the local transmission rate and the risk factors of infection, especially identify asymptomatic or subclinical infections.
Healthcare workers (HCWs) have been on the frontline for fighting this COVID-19 pandemic worldwide, which placed themselves at high risk of catching COVID-19. Understanding risk factors of SARS-CoV-2 infection during clinical setting is urgently needed, which not only provides the HCWs with essential guidance of self-protection, but also helps policymakers to formulate appropriate measures to control infection in hospital setting.
This present study aimed to evaluate the seroprevalence of SARS-CoV-2 in a cohort of 105 HCWs exposed to COVID-19 patients using both enzyme immunoassay (EIA) and microneutralization assay. Furthermore, risk factors of SARS-CoV-2 seroconversion among these HCWs were identified with epidemiological investigation. Our study shed lights on the subclinical infection of COVID-19 and provided information for pandemic mitigation efforts.
Materials and Methods
Subjects and samples
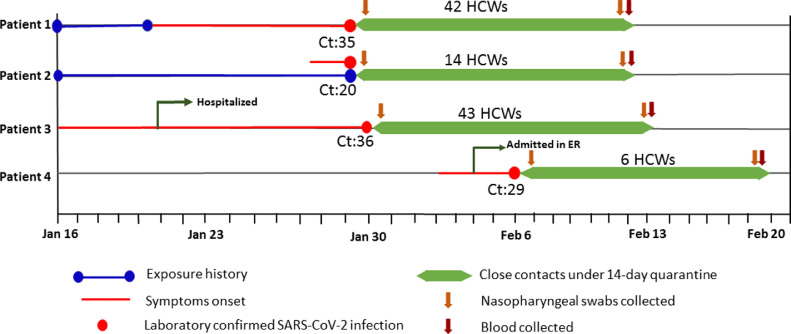
From Jan 28th to Feb 5th, 2020, four patients were diagnosed with COVID-19 in Nanjing Drum Tower Hospital, China. Patient 1 (a 30-year-old male surgeon) was occupationally exposed to SARS-CoV-2, and patient 2 (a 31-year-old female nurse) is his wife. Both of them were diagnosed with COVID-19 on Jan 29th. Patient 3 (a 54-year-old male) was diagnosed in the 10th day of his hospitalization, while patient 4 (a 35-year-old male)was in the emergency room (ER) for 2 days before diagnosis. The exposure history, the symptom onset timeline of the 4 COVID-19 patients and the cycle threshold [Ct] values of nucleic acid results were retrieved from their electronic medical records (Fig. 1 ).
Fig. 1.
Detailed timeline of exposure and illness onset of the four COVID-19 patients identified in our hospital and the quarantine timeline for the 105 healthcare workers exposed as close contacts. The cycle threshold (Ct) of values of SARS-CoV-2 RNA from nasopharyngeal swab samples at hospital admission were marked.
Because of direct contact with the four aforementioned COVID-19 patients during the past 2 weeks, the 105 HCWs were immediately quarantined for 14-day observation. At the first day of their quarantine, each HCW was asked to complete a questionnaire designed by local Center for Disease Control and Prevention (CDC) during the past two weeks. The questionnaire included clinical symptom, relationship with the patients and exposure history. The relationship between HCWs and COIVD-19 individuals was categorized as colleague, doctor, nurse, or general service assistant. The exposure history included the time of exposures, date for each exposure, activity, location, distance, duration, with or without face mask (disposable non-surgical face mask, surgical mask or N95 respiratory if wearing face mask) during the past two weeks. Disposable non-surgical face mask refers to the face mask made of non-woven textile in two or three layers, which generally lacks the capability of filtering particles, viruses and bacteria. Additionally, during their quarantine, their body temperature and clinical symptoms were also recorded twice a day. Nasopharyngeal swab specimens were collected on the first and 14th day of their quarantine by professional certified nurses who have received the training of nasopharyngeal swab collection. Blood samples were only obtained on the 14th day of the quarantine. This study was approved by ethics committee of Nanjing Drum Tower Hospital. Information consent was waived as part of a public health outbreak investigation.
Laboratory Nucleic Acid Test
Viral RNA was extracted from nasopharyngeal swab samples using the QIAamp RNA viral kit (Qiagen), and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assay was performed (BioPerfectus technologies, China). Two sets of primers and probes targeting the open reading frame1ab (ORF1ab) and nucleocapsid protein (NP) genes of SARS-CoV-2 were used as recommended by the Chinese CDC1 following WHO guidelines.7 The primers and probe set for ORF1ab are: forward primer (5′-CCCTGTGGGTTTTACACTTAA-3′); reverse primer (5′-ACGATTGTGCATCAGCTGA-3′); probe: (5ʹ-VIC-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3ʹ). The primers and probe set for NP are: forward primer (5′-GGGGAACTTCTCCTGCTAGAAT-3′); reverse primer (5′-CAGACATTTTGCTCTCAAGCTG-3′); probe: (5ʹ-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3ʹ). The thermal cycling condition was 50°C for 30 min, 95°C for 5 min, followed by 45 cycles of 95°C for 10s and 55°C for 40s. The Ct value of the amplification curve was defined as positive if less than 40 and negative if greater than 40.
Serological analysis of SARS-CoV-2
To determine the seroprevalence among close contacts with COVID-19 patients, an in-house enzyme immunoassay (EIA) was conducted as previously described.8 Two SARS-CoV-2 proteins, recombinant spike protein receptor binding domain (RBD) protein and recombinant nucleocapsid protein (NP) were used as detecting antigens, respectively. The genes encoding spike RBD (amino acid residues 319 to 541 of spike protein) and full-length NP were codon-optimized and synthesized (Genewiz, China). The gene encoding spike RBD was cloned into mammalian expression vector pcDNA3.4 in frame respective and upstream of a series of six histidine residues, and NP gene was cloned into prokaryotic expression vector pET-28(b). RBD protein was expressed in 293F cells while NP protein was expressed in Escherichia coli, followed by affinity purification. The purity of NP and RBD protein was determined with 10% sodium dodecyl sulphate (SDS) polyacrylamide gel electrophoresis. Briefly, 96-well plates were coated with 500 ng/mL of recombinant RBD or NP protein overnight, incubating with diluted serum samples at 1:20. Plates were incubated with either anti-human IgM or IgG conjugated with HRP. Optical density (OD) value (450nm-620nm) was measured. The preliminary cut-off values were calculated as the mean of the negative serum OD values plus 3 standard deviation (SD) from 90 archived healthy individuals in 2019. A close contact was considered seropositive if OD of 1:20 diluted serum was above the cut-off values for either IgM or IgG against both RBD and NP protein. Additionally, 20 serum samples from non-COVID-19 pneumonia patients were also collected and the nasopharyngeal swab samples from these patients have been repeatedly tested as negative for SARS-CoV-2 RNA at least twice at a two-day apart. Furthermore, 60 serum samples from 20 COVID-19 patients were also collected at different time points for assay validation.
Microneutralization assay
Pseudovirus expressing the SARS-CoV-2 spike protein was obtained as a general gift from the Institute of Biological Product Control from National Institute for Food and Drug Control, China. SARS-CoV-2 pseudovirus was prepared by using VSV G pseudotyped virus (G*ΔG-VSV) that packages the expression cassette for firefly luciferase instead of VSV-G in the VSV genome, and the serum neutralization capability was determined as described recently.9 Briefly, the SARS-CoV-2 pseudovirus was preincubated with serum samples at 1:20 dilution at 37°C for one hour, together with the pseudovirus control and cell control wells. Serum samples from healthy controls were served as negative control in hexaplicate. Then, the 96-well plates were seeded with 100 μg of freshly trypsinized Huh7 cells (2 × 104 cells/well). After 24 hours of incubation in a 5% CO2 environment under 37°C,the luminescence was measured using luciferase substrate (One-GloTM Luciferase assay system, Promega, E6120) and the percentage of neutralization was calculated with the following formula as: [(relative light units (RLUs) of virus control wells – RLUs in cell control wells)- (RLUs of serum incubated with virus wells- RLUs of the cell control wells)]/ (RLUs of virus control wells – RLUs of cell control wells) x100%. The percentage of neutralization over 50% was considered to have neutralization activity.
Statistical analysis
All statistical analyses were performed using SPSS 22.0. The medians (interquartile range (IQR)) were used to present the continuous variables, and the categorical variables were described as the counts and the percentages. The Man-Whitney U test (non-normal distribution) was used to compared the continuous variables between groups. Chi-square test or fisher exact test was used to compare categorical variables. Variables with p values < 0.1 in the univariate analysis were further used for a multivariate logistic regression analysis. Pearson's correlation coefficients between different assays were calculated. The threshold for statistical significance was established at a p value <0.05.
Results
Demographics characteristics and clinical symptoms of HCWs exposed to COVID-19 patients
The demographic and epidemiological characteristics of the 105 HCWs were summarized in Table 1 . The median age of these HCWs was 30 years old (IQR 26-62) and 83 (79.05%) of them were female. During the quarantine, 13 (12.38%) HCWs were reported having one or more general symptoms, including fever (2/105, 1.90%), headache (2/105, 1.90%), sore throat (3/105, 2.86%), cough (6/105, 5.71%), myalgia (1/105, 0.95%), diarrhea (3/105, 2.86%) and rhinorrhea (2/105, 1.90%). All swab specimens collected on the first and 14th day of the quarantine showed negative results for SARS-CoV-2, and none of these close contacts developed COVID-19 later.
Table 1.
The baseline characteristics and the exposure history of 105 healthcare workers as close contacts of COVID-19 patients.
| Characteristics | All close contacts (n=105) | Seropositive close contacts (n=18) | Seronegative close contacts (n=87) | p value |
|---|---|---|---|---|
| Demographic feature | ||||
| Age, years | 30.0 (26.0-39.5) | 27.0 (27.0-32.0) | 31.0 (26.0-43.0) | 0.203 |
| Sex | 0.260 | |||
| Male | 22 (20.95) | 2 (11.11) | 20 (22.99) | |
| Female | 83 (79.05) | 16 (88.89) | 67 (77.01) | |
| General Symptoms | 13 (12.38) | 4 (22.22) | 9 (10.34) | 0.164 |
| Fever | 2 (1.90) | 0 (0) | 2 (2.29) | 0.516 |
| Headache | 2 (1.90) | 0 (0) | 2 (2.29) | 0.516 |
| Sore throat | 3 (2.86) | 1 (5.56) | 2 (2.29) | 0.450 |
| Cough | 6 (5.71) | 2 (11.11) | 4 (4.60) | 0.279 |
| Myalgia | 1 (0.95) | 0 (0) | 1 (1.15) | 0.648 |
| Diarrhea | 3 (2.86) | 0 (0) | 3 (3.45) | 0.424 |
| Rhinorrhea | 2 (1.90) | 1 (5.56) | 1 (1.15) | 0.213 |
| Relationship with the COVID-19 patient | ||||
| Colleague | 52 (49.52) | 8 (44.44) | 44 (51.72) | 0.636 |
| Doctor | 17 (16.20) | 7 (38.89) | 10 (10.34) | 0.004 |
| Nurse | 25 (23.81) | 2 (11.11) | 23 (26.43) | 0.165 |
| General service assistant | 11 (10.47) | 1 (5.56) | 10 (11.49) | 0.454 |
| Exposure history | ||||
| Close contacts of Patient No. | ||||
| Patient 1 | 42 (40.01) | 5 (27.7) | 37 (43.53) | 0.245 |
| Patient 2 | 14 (13.33) | 5 (27.78) | 9 (10.34) | 0.048 |
| Patient 3 | 43 (40.95) | 7 (38.89) | 36 (41.38) | 0.845 |
| Patient 4 | 6 (5.71) | 1 (5.56) | 5 (5.75) | 0.975 |
| The extent of exposure | ||||
| ≥30 min at a distance of <1 meter | 33 (31.43) | 10 (55.56) | 23 (26.44) | 0.015 |
| Disposable non-surgical face mask wearing | 78 (74.29) | 10 (55.56) | 68 (78.16) | 0.046 |
| Swab sample collection | 5 (4.76) | 2 (11.11) | 3 (3.45) | 0.165 |
| Exposure times | ||||
| >10 | 24 (22.86) | 3 (16.67) | 21 (24.13) | 0.492 |
| 5-9 | 18 (17.14) | 3 (16.67) | 15 (17.24) | 0.953 |
| 0-4 | 63 (60.00) | 12 (66.67) | 51 (58.62) | 0.526 |
Data are median (IQR), or n/N (%). p values were calculated by Mann-Whitney U test or χ² test, as appropriate.
Serological testing using enzyme immunoassay and microneutralization assay
An EIA-based SARS-CoV-2 antibody assay was developed to detect IgM and IgG antibodies against RBD and NP protein, respectively (Fig. 2 A). Different groups of serum samples were used to determine the specificity and sensitivity of our assay. Specifically, as negative control, all 90 serum samples of healthy donors collected on October 2019 before the outbreak and all 20 serum samples from non-COVID-19 pneumonia patients were found seronegative. In addition, 56 (93.3%) out of 60 serum samples from 15 COVID-19 patients collected at various time points during hospitalization were found anti-SARS-CoV-2 positive, whereas 4 samples were negative. This was possibly because these 4 samples were collected within the first week of hospitalization and the level of anti-SARS-CoV-2 antibody was beyond detection.
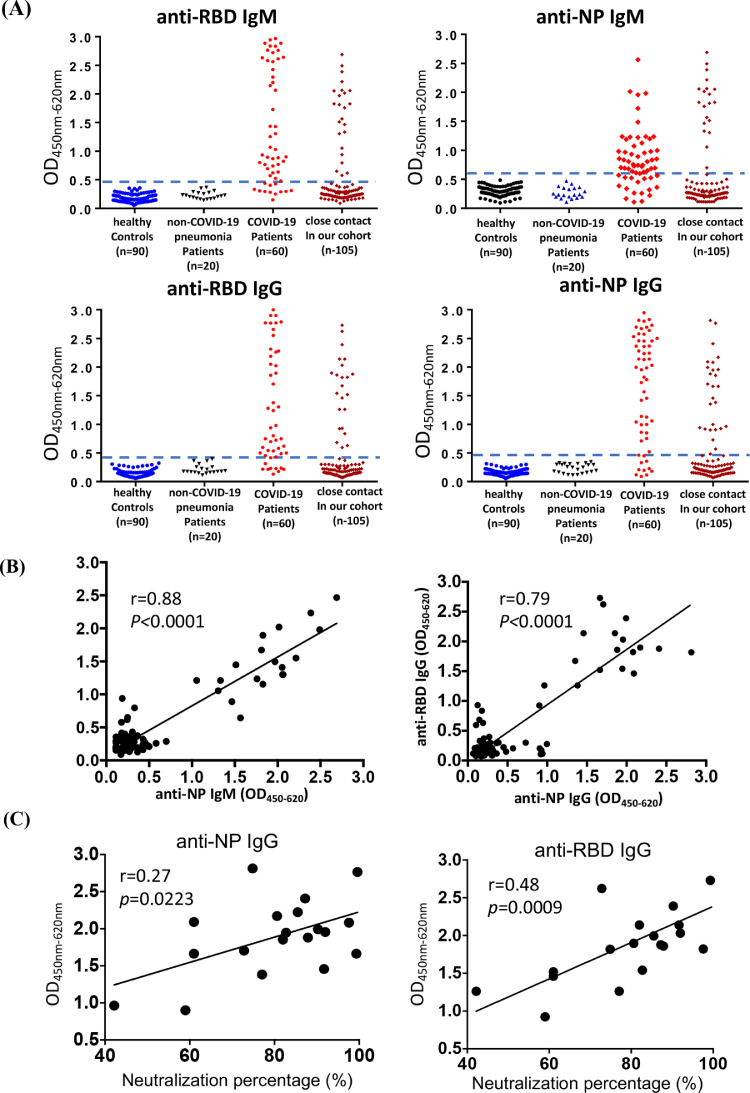
Fig. 2.
Seroprevalence analysis of SARS-CoV-2 with EIA assay and microneutralization assay. (A) The detection of IgM and IgG humoral responses against RBD and NP protein were detected by using sera samples diluted at 1:20 using EIA assay. Dashed blue line indicated cut-off value for each EIA assay, which was determined based on archived serum samples collected before COVID-19 outbreak. Sera from healthy controls (HC) collected in 2019 and the non-COVID-19 pneumonia patients were included as negative control, whereas 60 samples collected at different time points from 20 COVID-19 patients were used as positive control. Serum samples were collected on 14th day of the quarantine from 105 HCWs and they were also tested to determine IgM and IgG responses for RBD and NP protein. (B) The correlation analysis between anti-NP IgM and OD450nm-620nm value of anti-RBD IgM (left panel) and the correlation analysis between anti-NP IgG and OD450nm-620nm value of anti-RBD IgG (right panel). (C) The correlation analysis between the neutralization percentage of serum and anti-NP IgG (left panel) and the correlation analysis between the neutralization percentage of serum and OD450nm-620nm value of anti-RBD IgG (right panel). OD450nm-620nm = optical density at 450nm-620nm. NP = nucleoprotein. RBD = receptor-binding domain.
The seroprevalence rate of these 105 HCWs in our cohort was analyzed by the established EIA assays (Fig. 2B). Positive correlations between anti-RBD IgM and anti-NP IgM (r=0.88, p<0.0001) and between anti-RBD IgG and anti-NP IgM (r=0.79, p<0.0001) were identified. Beyond that, 18.10% (19/105) of contacts were showed positive IgM or IgG response for both RBD and NP protein. In order to further validate our findings with our EIA assay, a microneutralization assay was performed as recently described9 using the above 19 EIA positive samples and 20 randomly EIA selected negative samples. A total of 19 serum samples showed various degree of neutralization capability, in which from 42% to 99% of SARS-CoV-2 pseudovirus was neutralized (Fig. 2C). The percentage of neutralization was correlated with anti-NP IgG (r=0.27, p=0.02) and anti-RBD IgG (r=0.48, p=0.0009), instead of anti-NP IgM and anti-RBD IgM responses. 18 out of 19 serum samples showed neutralization percentage of over 50%, thereby being considered as seropositive. There was 1 sample that was only able to neutralize 42% of pseudovirus, concomitant with the only presence of anti-NP IgM and anti-RBD IgM response rather than anti-NP IgG or anti-RBD IgG responses. Therefore, this sample was considered as seronegative because of the low level of neutralization. In contrast, less than 20% of pseudovirus was neutralized by all 20 EIA negative sera (range, 0%-16.8%). Collectively, high rate (18/105, 17.14%) of anti-SARS-CoV-2 seroprevalence was identified among 105 HCWs in our cohort.
Risk factors for SARS-CoV-2 seroprevalence among HCWs
The exposure history of COVID-19 patients between seropositive and seronegative HCWs was summarized (Table 1). The exposure events occurred in the early period of the outbreak of COVID-19 in China, so the understanding to the SARS-CoV-2 was still limited at that time. All HCWs did not wear personal protective equipment (PPE), including N95 respirators, surgical masks, face shield or googles. First, the proportion of doctors who were exposed to patients was higher in seropositive group than in seronegative group (38.89% vs. 10.34%, p=0.004). It was identified that there was higher percentage of HCWs exposed to patient 2 in seropositive group (27.78% vs. 10.34%, p=0.048) than in seronegative group, which might be caused by the relatively high level of viral shedding from patient 2, revealed by a low CT value for her respiratory swab samples at the hospital admission. Besides, more seropositive HCWs had a previous experience of exposure for over 30 minutes within a distance of 1 meter (55.56% vs. 26.44%, p=0.015), and less seropositive HCWs contacted COVID-19 patients with disposable non-surgical face mask wearing (55.56% vs. 78.16%, p=0.046), compared to that in seronegative group. Neither swab sample collection nor multiple times of exposure with COVID-19 patients showed any differences in seroconversion.
Risk factors associated with SARS-CoV-2 seroconversion were assessed (Table 2 ). The univariate analysis showed that the exposure for more than 30 minutes at a distance of less than 1 meter (odds ratio [OR], 3.478, 95% confidence interval [CI] 1.224, 9.887), close contact with patient 2 (OR, 7.125, 95% CI 1.627, 31.210) and doctors exposed to their patient (OR, 3.850, 95% CI 1.131, 13.105) led to higher risk of seropositivity, while contact with COVID-19 patient wearing mask (OR, 0.349, 95% CI 0.121, 1.008) was associated with a reduced risk of seroconversion. In multivariate analysis, there existed higher risk of seroconversion for close contacts with patient 2 (OR, 6.605, 95% CI, 1.123, 38.830) and doctors exposed to their patient (OR, 346.837, 95% CI 8.924, 13479.434), while the lower risk of seroconversion was closely related to direct contact with COVID-19 patients wearing face mask (OR, 0.127, 95% CI 0.017, 0.968).
Table 2.
Univariate and multivariate analysis of risk factors for seroconversion of SARS-CoV-2 among HCWs.
| Variables | Univariate |
Multivariate |
||
|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | p value | |
| Age | 0.947 (0.888, 1.010) | 0.1 | ||
| Gender | ||||
| Female | Reference | |||
| Male | 0.419 (0.089, 1.978) | 0.272 | ||
| ≥30 min at a distance of <1 m | ||||
| No | Reference | |||
| Yes | 3.478 (1.224, 9.887) | 0.019 | 2.231 (0.556, 8.951) | 0.258 |
| Close contacts of Patient No. | ||||
| Patient 1 | Reference | |||
| Patient 2 | 7.125 (1.627, 31.210) | 0.009 | 6.605 (1.123, 38.830) | 0.037 |
| Patient 3 | 1.847 (0.498, 6.858) | 0.359 | 0.083 (0.004, 1.609) | 0.100 |
| Patient 4 | 1.900 (0.176, 20.559) | 0.597 | 0.722 (0.022, 24.013) | 0.856 |
| Disposable non-surgical face mask wearing | ||||
| No | Reference | |||
| Yes | 0.349 (0.121, 1.008) | 0.052 | 0.127 (0.017, 0.968) | 0.046 |
| Exposure times | ||||
| <5 | Reference | |||
| 5-9 | 0.911 (0.225, 3.680) | 0.896 | ||
| ≥10 | 0.580 (0.149, 2.259) | 0.432 | ||
| Relationship with the COVID-19 patient | ||||
| Colleague | Reference | |||
| Doctor | 3.850 (1.131, 13.105) | 0.031 | 346.837 (8.924, 13479.434) | 0.002 |
| Nurse | 0.478 (0.094, 2.440) | 0.375 | 19.523 (0.667, 571.463) | 0.085 |
| General service assistant | 0.550 (0.062, 4.911) | 0.593 | 13.294 (0.265, 666.605) | 0.195 |
Discussion
Similar to SARS-CoV, SARS-CoV-2 has been reported to be highly communicable in hospital setting.10 , 11 High attack rate of SARS-CoV-2 among healthcare workers with direct patient care has been observed worldwide, including China, Italy, and United States, etc. Our study examined the seroprevalence of the 105 HCWs exposed to COVID-19 patients without wearing PPE at early period of outbreak in a tertiary hospital of China. Of note, 56 HCWs were exposed to COVID-19 patients as colleagues, while 49 HCWs were exposed during performing direct care for patient or general service. Despite the swab samples from all HCWs collected at twice were negative for SARS-CoV-2 RNA, our serological analysis indicated 17.1% of asymptomatic or subclinical infection of SARS-CoV-2 in hospital setting.
Consistent with the efficient transmissibility of SARS-CoV-2, our serological analysis in the hospital setting highlighted a higher percentage of asymptomatic or subclinical SARS-CoV-2 infection than that of SARS-CoV12, 13, 14, 15 and MERS-CoV.16 Our data suggested that those with asymptomatic or subclinical infection of SARS-CoV-2 were able to be seroconverted. All the nasopharyngeal swab samples collected twice were found to be negative for SARS-CoV-2. This could because that the modest level of viral load from nasopharyngeal swabs was beyond detection. Also, the nasopharyngeal swab samples were not longitudinally collected and the positive samples might be missed. Therefore, serological testing is an ideal approach to assess the proportion of people who might experience the asymptomatic or subclinical infection of SARS-CoV-2, which is critical to understand the viral transmissibility and disease burden during COVID-19 pandemic.
Previous studies on human coronaviruses have shown that NP protein and RBD region are highly immunogenic, which have been successfully applied in EIA-based approach for serological analysis.14 , 17 , 18 In order to eliminate the possibility of cross-reactivity with other human coronaviruses, RBD and NP protein were used as two antigens in our assay, similar to a recent published study.19 We found that all the healthy controls in 2019 and the non-COVID-19 pneumonia patients were 100% found to be negative, while 93.3% of serum from COVID-19 patients were proved to be positive, suggesting our ELISA assay has high sensitivity and fine specificity. By using EIA assay, 19 samples were discovered positive for detectable IgM or IgG responses to both RBD and NP protein, while the antibody responses among these HCWs were relatively lower compared with COVID-19 patients. Meanwhile, 18 out of 19 sera samples displayed neutralization activities. Consistent with recent findings,19 the level of neutralization activity was associated with anti-NP IgG response and anti-RBD IgG response. Whether those seropositive HCWs acquired protective immunity from SARS- CoV-2 infection remains unknown. Active surveillance and longitudinal follow-up to close contacts should be attached more importance.
Recognizing the risk factors of SARS-CoV-2 infection has important implication to mitigate pandemic and control infection of COVID-19. Firstly, our analysis showed that the secondary attack rate to each COVID-19 patients was distinct. In our study, higher percentage of close contacts with patient 2 were seroconverted. Such efficient transmission might be caused by higher level of viral load of patient 2, which was determined by the low CT value of nucleic acid assay. It is also possible that SARS-CoV-2 was evolved in patient 2 after infection, which might further modulate the viral transmissibility and virulence.20 Besides, our data support an essential need to wear face mask, including the disposable non-surgical face mask, because it might provide effective protection against SARS-CoV-2. Of note, since these exposure events occurred at the very first of the outbreak in China, all the HCWs in our study only worn the disposable non-surgical face mask, rather than N95 respirator or surgical masks. Therefore, our data suggested that the disposable non-surgical face mask might be also beneficial to the reduction of the potential nosocomial infection. Consistently, a recent study in Hong Kong revealed that the community-wide wearing of the face mask plays a critical role in the control of COVID-19, not only by preventing the dispersal of droplets from subclinical or mild individuals with COVID-19, but also by reducing the environmental contamination of SARS-CoV-2.21 Moreover, among four relationships with the COVID-19 patients, there is higher risk for doctors who were exposed to COVID-19 patients to have seroconversion. It is possible that doctors might share more conversations with COVID-19 patients within relatively close distance, which could generate large amount of infected saliva or respiratory droplets. Our findings might shed light on the implementation of appropriate infection prevention and control measures, especially at hospital setting.
This study has several limitations. Firstly, our cohort is limited to only inclusion of 105 HCWs exposed to 4 COVID-19 patients. However, our cohort is representative since they are either colleagues or health professionals in terms of the relation with the COVID-19 patient. And different extents of exposure history in our cohort allow us to identify the potential exposure risk factors. Secondly, the serum samples were only collected on the 14th day of the quarantine, while the serum samples on the first day of the quarantine were not obtained. Therefore, the dynamic antibody responses were not determined in our study. Thirdly, the nasopharyngeal swab specimens were only collected twice on first and 14th day of the quarantine, which were not serially collected especially during the early period of the quarantine, and thus the positive nasopharyngeal swab samples might be missed.
In summary, the serological testing among 105 HCWs exposed to COVID-19 patients illustrated that 17.14% (18/105) might have experienced asymptomatic or subclinical infection of SARS-CoV-2. Our study proved that the serological testing is useful for the identification of asymptomatic or subclinical infection of SARS-CoV-2 among close contacts with COVID-19 patients. Whether these asymptomatic or subclinical infections play a role in transmission dynamics still remains to be determined. Our findings have important implications for the implementation of pandemic mitigation strategies.
Declaration of Competing Interest
The authors have declared that no conflicts of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81600201, 81672025 and 81702011), Nanjing Medical Science and Technique Development Foundation (QRX17141), Medical Science and Technology Development Foundation of Nanjing (ZDX16004), Jiangsu Provincial Medical Innovation Team (CXTDA2017005), Jiangsu Science and Technology Development Plan (BE2017605), Foundation project of Jiangsu Commission of Health (Q2017003).
Contributor Information
Han Shen, Email: shenhan10366@sina.com.
Chao Wu, Email: dr.wu@nju.edu.cn.
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X.W., Wu X.X., Jiang X.G., Xu K.J., Ying L.J., Ma C.L. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang Lin M., Wei L., Xie L., Zhu G., Dela Cruz C.S. Epidemiologic and Clinical Characteristics of Novel Coronavirus Infections Involving 13 Patients Outside Wuhan, China. JAMA. 2020;323(11):1092–1093. doi: 10.1001/jama.2020.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang R., Xia J., Chen Y., Shan C., Wu C. A family cluster of SARS-CoV-2 infection involving 11 patients in Nanjing, China. Lancet Infect Dis. 2020;20(5):534–535. doi: 10.1016/S1473-3099(20)30147-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An P., Li L., Chaolong W., Huan G., Xingjie H., Qi W. Association of Public Health Interventions With the Epidemiology of the COVID-19 Outbreak in Wuhan, China. medRxiv. 2020. JAMA. 2020;323(19):1915–1923. doi: 10.1001/jama.2020.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. Coronavirus disease (COVID-19) technical guidance: laboratory testing for 2019-nCoV in humans (https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117)
- 8.Chen Y., Vaine M., Wallace A., Han D., Wan S., Seaman M.S. A novel rabbit monoclonal antibody platform to dissect the diverse repertoire of antibody epitopes for HIV-1 Env immunogen design. J Virol. 2013;87(18):10232–10243. doi: 10.1128/JVI.00837-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams J.G., Walls R.M. Supporting the Health Care Workforce During the COVID-19 Global Epidemic. JAMA. 2020;323(15):1439–1440. doi: 10.1001/jama.2020.3972. [DOI] [PubMed] [Google Scholar]
- 11.Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA. 2020;323(16):1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow P.K., Ooi E.E., Tan H.K., Ong K.W., Sil B.K., Teo M. Healthcare worker seroconversion in SARS outbreak. Emerg Infect Dis. 2004;10(2):249–250. doi: 10.3201/eid1002.030397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan P.K., Ip M., Ng K.C., Rickjason C.W., Wu A., Lee N. Severe acute respiratory syndrome-associated coronavirus infection. Emerg Infect Dis. 2003;9(11):1453–1454. doi: 10.3201/eid0911.030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo P.C., Lau S.K., Tsoi H.W., Chan K.H., Wong B.H., Che X.Y. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363(9412):841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung G.M., Chung P.H., Tsang T., Lim W., Chan S.K., Chau P. SARS-CoV antibody prevalence in all Hong Kong patient contacts. Emerg Infect Dis. 2004;10(9):1653–1656. doi: 10.3201/eid1009.040155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C.J., Choi W.S., Jung Y., Kiem S., Seol H.Y., Woo H.J. Surveillance of the Middle East respiratory syndrome (MERS) coronavirus (CoV) infection in healthcare workers after contact with confirmed MERS patients: incidence and risk factors of MERS-CoV seropositivity. Clin Microbiol Infect. 2016;22(10):880–886. doi: 10.1016/j.cmi.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W., Wang H., Deng Y., Song T., Lan J., Wu G. Characterization of anti-MERS-CoV antibodies against various recombinant structural antigens of MERS-CoV in an imported case in China. Emerg Microbes Infect. 2016;5(11):e113. doi: 10.1038/emi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashem A.M., Al-Amri S.S., Al-Subhi T.L., Siddiq L.A., Hassan A.M., Alawi M.M. Development and validation of different indirect ELISAs for MERS-CoV serological testing. J Immunol Methods. 2019;466:41–46. doi: 10.1016/j.jim.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L. Genomic Diversity of SARS-CoV-2 in Coronavirus Disease 2019 Patients. Clin Infect Dis. 2020. Mar 4:ciaa203. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng V., Wong S., Chuang V., So S., Chen J., Sridhar S. The role of Community-Wide Wearing of Face Mask for Control of Coronavirus Disease 2019 (COVID-19) Epidemic Due to SARS-CoV-2. J Infect. 2020 Apr 23 doi: 10.1016/j.jinf.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]