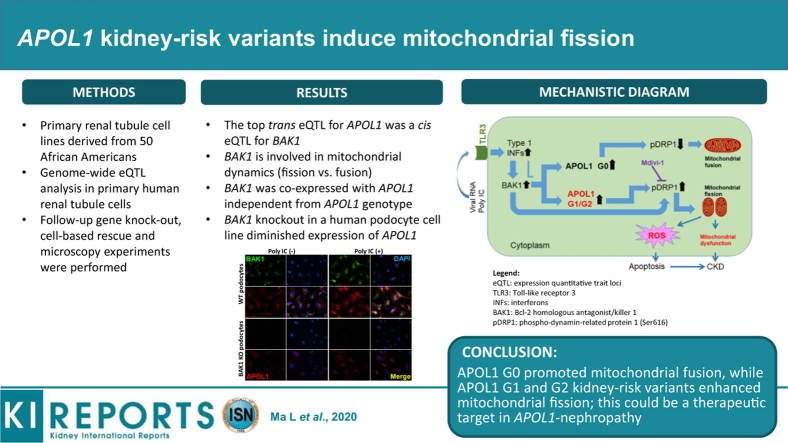
Abstract
Introduction
APOL1 G1 and G2 nephropathy-risk variants cause mitochondrial dysfunction and contribute to kidney disease. Analyses were performed to determine the genetic regulation of APOL1 and elucidate potential mechanisms in APOL1-nephropathy.
Methods
A global gene expression analysis was performed in human primary renal tubule cell lines derived from 50 African American individuals. Follow-up gene knock out, cell-based rescue, and microscopy experiments were performed.
Results
APOL1 genotypes did not alter APOL1 expression levels in the global gene expression analysis. Expression quantitative trait locus (eQTL) analysis in polyinosinic-polycytidylic acid (poly IC)–stimulated renal tubule cells revealed that single nucleotide polymorphism (SNP) rs513349 adjacent to BAK1 was a trans eQTL for APOL1 and a cis eQTL for BAK1; APOL1 and BAK1 were co-expressed in cells. BAK1 knockout in a human podocyte cell line resulted in diminished APOL1 protein, supporting a pivotal effect for BAK1 on APOL1 expression. Because BAK1 is involved in mitochondrial dynamics, mitochondrial morphology was examined in primary renal tubule cells and HEK293 Tet-on cells of various APOL1 genotypes. Mitochondria in APOL1 wild-type (G0G0) tubule cells maintained elongated morphology when stimulated by low-dose poly IC, whereas those with G1G1, G2G2, and G1G2 genotypes appeared to fragment. HEK293 Tet-on cells overexpressing APOL1 G0, G1, and G2 were created; G0 cells appeared to promote mitochondrial fusion, whereas G1 and G2 induced mitochondrial fission. The mitochondrial dynamic regulator Mdivi-1 significantly preserved cell viability and mitochondrial cristae structure and reversed mitochondrial fission induced by overexpression of G1 and G2.
Conclusion
Results suggest the mitochondrial fusion/fission pathway may be a therapeutic target in APOL1-nephropathy.
Keywords: African Americans, APOL1, chronic kidney disease, FSGS, mitochondria
Graphical abstract
Since discovery of the apolipoprotein L1 gene (APOL1) association with chronic kidney disease in populations with recent African ancestry,1,2 efforts have been undertaken to elucidate mechanisms whereby the common G1 (2 variants, S342G and I384M, in nearly perfect linkage disequilibrium) and G2 (deletion of amino acids N388 and Y389) kidney-risk variants (KRVs) cause nephropathy.3, 4, 5 Endogenous and locally acting (not circulating) APOL1 protein appears to cause nephropathy based on data from kidney transplantation, cell biology, and animal models.6, 7, 8, 9, 10, 11, 12, 13, 14, 15 Ma et al.16 and Granado et al.17 independently reported that reduced mitochondrial function caused by APOL1 G1 and G2 KRVs contribute to chronic kidney disease. Shah et al.18 reported that APOL1 KRVs induce cell death via mitochondrial translocation and opening of the inner mitochondrial membrane permeability transition pore. APOL1 is widely present in mitochondria17 and adverse effects could extend beyond permeability changes in the inner membrane. Mitochondrial dysfunction is also known to cause non–APOL1-mediated kidney diseases.19
Most individuals with 2 APOL1 KRVs do not develop nephropathy; a modifier is required.20 Proven modifiers include HIV-induced alterations in the immune response and administration of interferons.21 In these settings, APOL1 expression levels are increased via the toll-like receptor 3 (TLR3)-dependent pathway. This report assessed pathways potentially leading to APOL1-nephropathy and further implicated compromised mitochondrial function based on the results of a global gene expression analysis in primary human renal proximal tubule cells (PTCs) from African American individuals. Cells were exposed to an immune system activator (poly IC) to provide a stressor mimicking viral infection. Expression quantitative trait locus (eQTL) analysis and fluorescence microscopy were performed, with subsequent gene knockout to verify effects of an APOL1 upstream regulator identified in an eQTL analysis. To determine whether increased APOL1 G1 and G2 KRV expression directly contributed to mitochondrial effects, HEK293 Tet-on cell lines overexpressing APOL1 G0 (wild-type), G1, and G2 KRVs were used to assess the pathways affecting mitochondrial function by immunoblotting and fluorescence microscopy; these cells have minimal or no TLR3 expression. Finally, mitochondrial rescue was performed by blocking implicated pathways to confirm mechanisms underlying mitochondrial dysfunction and cell injury.
Methods
Full methods are provided in the Supplementary Materials. In brief, total RNAs from human renal PTCs obtained from 50 African American individuals with an estimated glomerular filtration rate >60 ml/min per 1.73 m2 undergoing surgical nephrectomy were isolated to perform global gene expression using Affymetrix HTA 2.0 arrays. DNA was isolated from peripheral blood, and Illumina (San Diego, CA) Multi-Ethnic Genotyping Arrays were used to genotype SNPs throughout the genome. The study was approved by the Wake Forest School of Medicine Institutional Review Board and participants provided written informed consent. Gene knockout was performed using the corresponding CRISPR/Cas9 plasmids and transfection reagents provided by Santa Cruz Biotechnology (Dallas, TX). HEK293 Tet-on APOL1 G0, G1, G2, and empty vector (EV) cells were established as previously reported.22 Reverse transcriptase-polymerase chain reaction (RT-PCR), immunoblotting, and fluorescence were performed using established protocols.10,23 Mitochondrial length was assessed using Fiji software, integrated with a plug-in macro toolset Mitochondrial Network Analysis.24 Cell viability was measured using a Cytotox 96 lactate dehydrogenase viability assay kit (Promega, Madison, WI) per manufacturer instructions.
Results
Pathway Analysis in Primary Renal PTC Lines With and Without Stimulation by Poly IC
Primary renal PTCs were treated with 2.5 μg/ml poly IC for 16 hours to stimulate the innate immune response while maintaining viability, conditions that upregulated APOL1 expression 8- to 15-fold and TLR3 expression 15- to 20-fold, with minimal changes in cell viability (data not shown). Global gene expression profiles in the 50 primary renal PTC lines from African American individuals were computed using CytoScape-based BiNGO. Among 1212 upregulated genes, 9 of the top 20 associated pathways related to immune response as anticipated with poly IC exposure. In 1060 downregulated genes, mitochondrial and related pathways were among the top 20 associated pathways (Supplementary Table S1). Index pathways were verified by Ingenuity Pathway Analysis (QIAGEN, Hilden, Germany) (Supplementary Tables S2A and S2B).
APOL1 eQTL Global Gene Expression Analyses and Genome-Wide Association Study of APOL1 mRNA Expression
To assess whether APOL1 KRVs in an additive (0 vs. 1 vs. 2) or recessive genetic model (0/1 vs. 2) affected cis and trans gene expression levels, global gene expression profiles in the African American primary renal PTC lines were estimated by Affymetrix (Santa Clara, CA) HTA 2.0 arrays. APOL1 KRVs did not significantly affect baseline or post-poly IC APOL1 expression levels, or expression of APOL2, APOL3, APOL4, APOL5, APOL6, or MYH9 (non–muscle myosin heavy chain 9 gene) (data not shown). Considering multiple testing, APOL1 KRVs did not significantly alter global trans gene expression (data not shown).
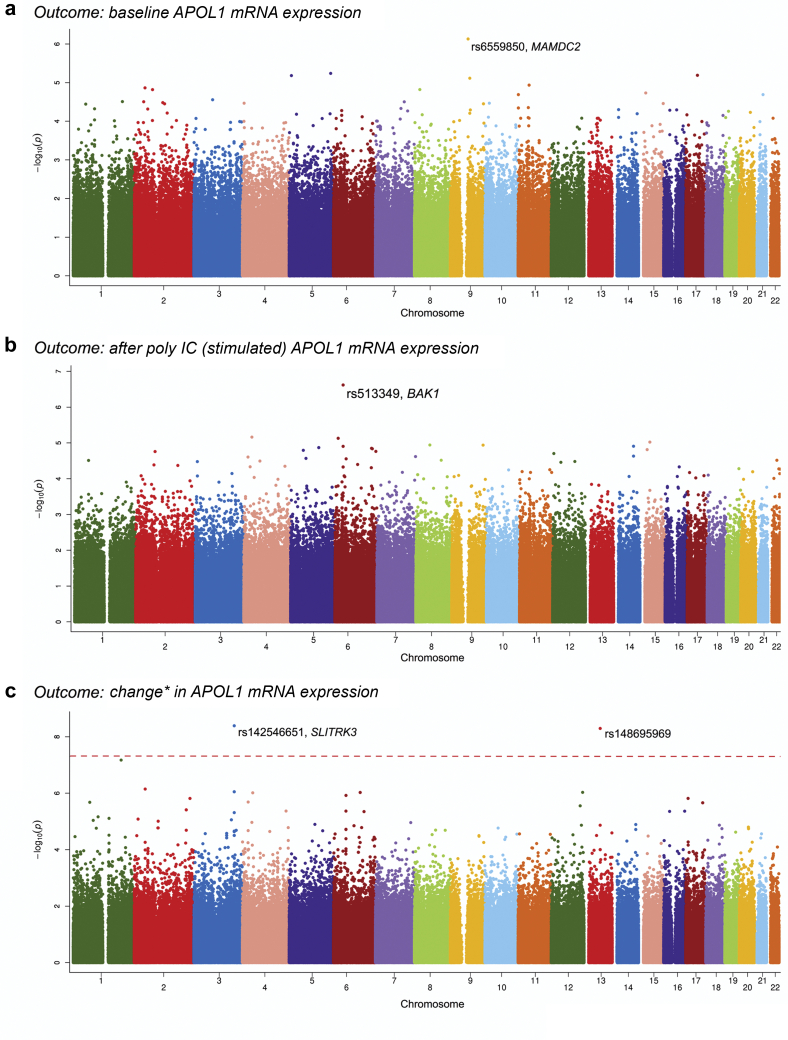
To determine whether APOL1 expression was affected by cis- or trans-regulating loci, a genome-wide association study with APOL1 expression level was performed. To maximize information from the small sample, a minor allele frequency criterion ≥0.05 was required, yielding 731,553 SNPs for analyses. Baseline, post-poly IC, and the difference between post-poly IC and baseline APOL1 mRNA levels were analyzed (Figure 1). The top associations with baseline and post-poly IC APOL1 were rs6559850 at 9q21 (P = 7.4 × 10−7) and rs513349 at 6p21 (P = 2.4 × 10−7), respectively; these did not reach the genome-wide association study significance threshold (Table 1 and Figure 1). Both rs142546651 at 3q26 and rs148695969 at 13q21 reached genome-wide association study significance for association with the difference in APOL1 expression after poly IC, compared with baseline (Figure 1c). SNP rs148695969 resides in a gene desert and follow-up for cis eQTLs was not attempted.
Figure 1.
Genome-wide association map (Manhattan Plot) showing relative APOL1 mRNA expression in 50 primary renal proximal tubule cell lines from African American individuals without nephropathy. A total of 731,553 variants with minor allele frequencies >0.05 were evaluated. X-axis refers to chromosomal single nucleotide polymorphism (SNP) location; y-axis represents negative log transformed P values for SNP association with APOL1 expression. (a) SNPs associated with baseline APOL1 expression; (b) SNPs associated with polyinosinic-polycytidylic acid (poly IC)–stimulated APOL1 expression; (c) SNPs associated with the difference in APOL1 expression (post–poly IC minus baseline). ∗Change reflects difference in APOL1 gene expression (after poly IC vs. baseline).
Table 1.
Association between top SNPs with relative APOL1 mRNA expression in primary renal PTC lines from African American individuals
| SNP | Position | Condition | Allele (M/m) | Adjacent gene ID | Model | P value | Beta (SE) | mAF | MM (n) | Mm (n) | mm (n) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs6559850 | 9:72531390 | Baseline | C/T | MAMDC2 | Dom | 7.4 × 10−7 | 1.03 (0.18) | 0.17 | 35 | 13 | 2 |
| rs513349 | 6:33541719 | Poly IC | G/A | BAK1 | Add | 2.4 × 10−7 | 0.88 (0.15) | 0.34 | 23 | 20 | 7 |
| rs142546651 | 3:164915128 | Changea | C/G | SLITRK3 | Dom | 4.1 × 10−9 | 1.37 (0.20) | 0.07 | 44 | 5 | 1 |
| rs148695969 | 13:65987966 | Changea | T/G | Dom | 5.1 × 10−9 | 1.48 (0.21) | 0.05 | 45 | 5 | 0 |
Beta, estimate was determined by the effect of minor allele; M, major allele; m, minor allele; mAF, minor allele frequency; poly IC, in polyinosinic-polycytidylic acid; PTC, proximal tubule cell; SNP, single nucleotide polymorphism.
SNP position based on human reference sequence (GRCh37). Genetic models Dom and Add refer to dominant and additive, respectively. P values adjusted for African ancestry proportion. Individuals had an estimated glomerular filtration rate >60 ml/min per 1.73 m2 (N = 50).
Change: difference between poly IC vs. baseline.
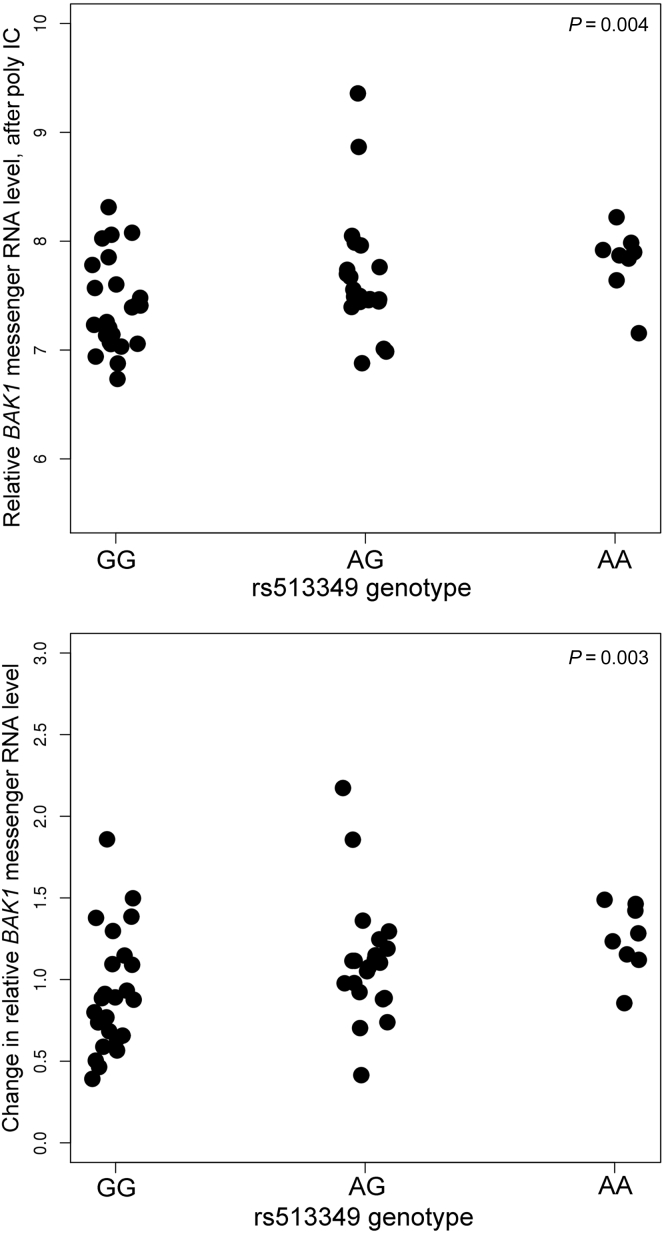
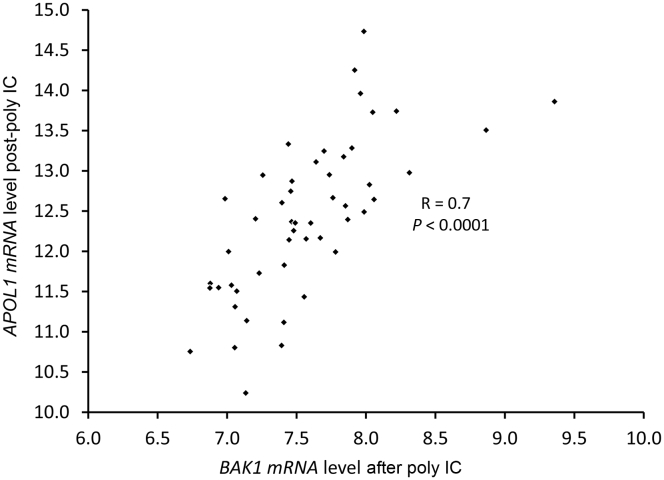
SNP rs6559850 was not a significant cis eQTL for adjacent genes (data not shown). In contrast, rs513349 was an eQTL for the adjacent BCL2 Antagonist/Killer 1 gene (BAK1) (P = 0.003–0.004) (Figure 2), and was co-expressed with APOL1 after poly IC (R = 0.7; P < 0.0001; Figure 3) independent of APOL1 genotype (t = −1.08, P = 0.29). BAK1 is a mitochondrial protein (Supplementary Figures S1 and S2) present in human renal tubule cells and podocytes (Supplementary Figure S3). At baseline (before poly IC), rs142546651 was a cis eQTL for the SLIT and NTRK-like family member 3 gene (SLITRK3). However, no association was found for SLITRK3 and APOL1 expression in primary renal PTCs at baseline, post-poly IC, or for the difference between post-poly IC and baseline. Hence, only the top trans eQTL rs513349 for APOL1 was evaluated as a cis eQTL for the adjacent BAK1 gene. This revealed that rs513349 could affect APOL1 gene expression via BAK1, particularly after poly IC exposure.
Figure 2.
Relative BAK1 expression versus single nucleotide polymorphism rs513349 in African American primary renal proximal tubule cell lines. Data expressed as mean ± SD. (a) Post– polyinosinic-polycytidylic acid (poly IC) BAK1 mRNA level versus rs513349 genotype. (b) Change in BAK1 mRNA level (difference between post– and pre–poly IC versus rs513349 genotype). P values adjusted for African ancestry proportion.
Figure 3.
Relative APOL1 versus BAK1 mRNA expression levels in African American primary renal proximal tubule cells after polyinosinic-polycytidylic acid (poly IC). Linear regression was fitted between relative APOL1 and BAK1 mRNA expression levels.
APOL1 Is Regulated by BAK1 in Human Podocytes
To verify results from the genome-wide search for eQTLs of APOL1 where BAK1 could be an upstream regulator of APOL1 expression, the BAK1 gene was knocked out in a human podocyte cell line using CRISPR/Cas9 technology. APOL1 mRNA level was less responsive to poly IC treatment in BAK1 knockout cells than in wild-type cells (Supplementary Figure S4). After BAK1 knockout, BAK1 protein was undetectable by immunoblotting and immunofluorescence (Supplementary Figures S5 and S6). APOL1 protein level was markedly reduced after BAK1 knock out (Supplementary Figures S5 and S6). Low-concentration poly IC (0.15 μg/ml) increased the expression level of BAK1 and APOL1 when BAK1 was present. When BAK1 was knocked out, the effect of poly IC on APOL1 upregulation was significantly diminished, indicating the level of APOL1 expression was largely related to presence of BAK1 (Supplementary Figures S5 and S6).
Poly IC Induces Mitochondrial Fragmentation in Primary Renal PTCs Homozygous for APOL1 G1 and G2
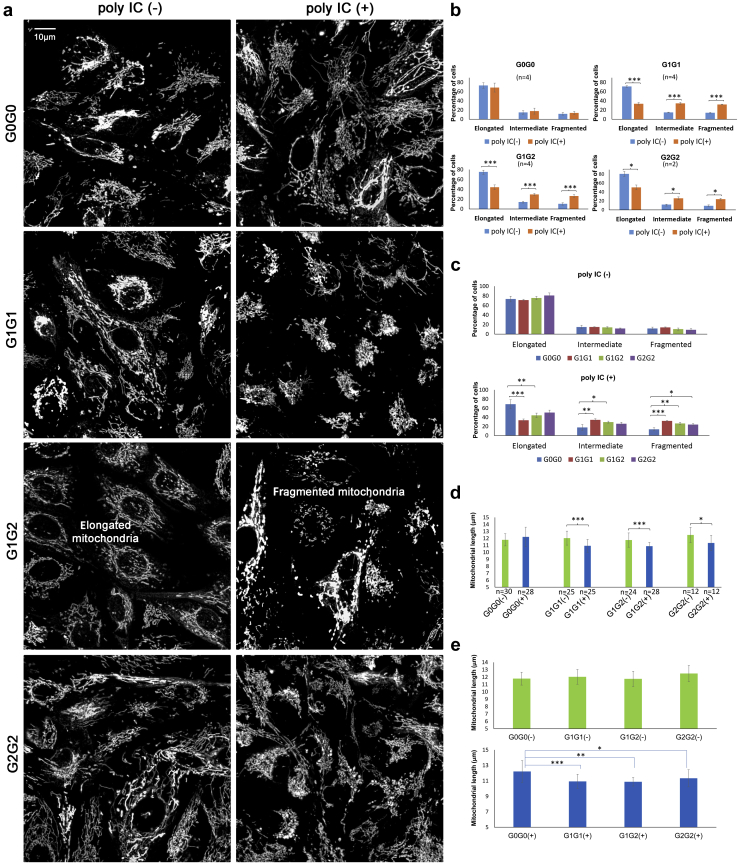
APOL1 KRVs modify mitochondrial function.16 In addition, APOL1 expression was affected by the mitochondrial protein BAK1, critical for mitochondrial dynamics.25 Therefore, mitochondrial morphology was assessed in primary renal PTCs from 50 primary PTC lines from African American individuals. Two individuals homozygous for G0, 2 homozygous for G1, 2 with the compound G1/G2 risk genotype, and the single individual homozygous for G2 were chosen to estimate mitochondrial dynamics using confocal microscopy. Among the 2 individuals with each genotype, 1 woman and 1 man were selected. Individuals with 2 APOL1 KRVs were similar in age to those without risk alleles (62.0 ± 14.7 vs. 69.0 ± 2.8 years; P = 0.56). Mitochondrial morphology was captured using MitoTracker Red (Figure 4a). Homozygous G0, G1, G2, and G1/G2 compound renal PTCs all exhibited elongated mitochondria before poly IC. However, cells with 2 copies of APOL1 KRVs exhibited mitochondrial fragmentation after exposure to low-concentration poly IC (2.5 μg/ml for 16 hours); mitochondrial morphology remained largely intact in renal PTCs from the individuals homozygous for G0 (Figure 4b–e).
Figure 4.
Mitochondrial morphology in live primary proximal tubule cell lines (PTCs) from African American individuals with different APOL1 genotypes cultured in growth media without (−) and with (+) polyinosinic-polycytidylic acid (poly IC). Primary PTC lines were from 7 African American individuals (2 homozygous for APOL1 G0, 2 homozygous for G1, 2 for compound G1/G2), and 1 homozygous for G2. Cells were grown without (−) or with (+) 2.5 μg/ml poly IC for 16 hours in full Dulbecco’s modified Eagle’s medium. Cells were subsequently incubated for 30 minutes with a final concentration of 50 nM MitoTracker Red (Invitrogen). (a) Representative images reveal that renal PTCs from the G0 homozygotes had similar mitochondrial morphology when incubated with (+) or without (−) poly IC. However, primary PTCs from G1 and G2 homozygotes and G1/G2 compound heterozygotes displayed more fragmented mitochondria when incubated with (+) poly IC than those without (−) poly IC. Each representative image was a compression of a series of z-stack scans, clearly capturing mitochondrial morphology. Binary images displayed with enhanced local contrast (CLAHE) via Fiji scoring by Mitochondrial Network Analysis. (b) Paired comparisons showing that the percentage of cells with mitochondrial elongation significantly decreased and the percentage of cells with mitochondrial fragmentation significantly increased in primary renal PTCs with (+) poly IC than without (−) poly IC in subjects homozygous for G1, homozygous for G2, and compound G1/G2 heterozygotes. The percentage of cells with mitochondrial elongation and fragmentation did not differ in primary PTCs homozygous for G0. For each primary PTC line, 2 independent experiments were performed to image mitochondrial networks with or without poly IC. Primary PTCs from 2 African American individuals (1 woman and 1 man of comparable age) were selected for homozygous G0, homozygous G1, and compound G1/G2 genotypes. Only 1 homozygous G2 (female) had available primary PTCs. (Note: n refers to number of independent cell culture experiments; ∗P < 0.05, ∗∗ P < 0.01, ∗∗∗P < 0.001 throughout Figure 4; no statistical significance was found in any other paired comparison.) (c) Paired comparisons showing that the percentage of cells with mitochondrial elongation was significantly lower, and the percentage of cells with mitochondrial fragmentation significantly higher, in primary renal PTCs from G1 homozygotes and G1/G2 compound heterozygotes versus G0 homozygotes with poly IC in growth media. The effect in G2 homozygotes may have been limited by statistical power (1 individual, 2 independent experiments). The percentage of cells with mitochondrial elongation and fragmentation did not differ in primary PTCs of different APOL1 genotypes without poly IC. (d) Paired comparisons showing that mitochondrial length significantly decreased in primary renal PTCs with (+) poly IC versus without (−) poly IC in primary PTCs homozygous for G1, homozygous for G2 and compound G1/G2 heterozygotes; however, mitochondrial length did not differ in homozygous G0 primary PTCs with (+) or without (−) poly IC. n refers to the total number of images captured for Mitochondrial Network Analysis. At least 6 z-stack images were taken for each independent cell culture experiment. For cells of each PTC line, 2 independent experiments were performed for imaging mitochondrial networks with or without poly IC. Primary PTCs from 2 different African American individuals were selected for G0 homozygotes, G1 homozygotes, and compound G1/G2 heterozygotes; no significant difference in mitochondrial morphology (mitochondrial length defined as rods/network branches in μm) was observed between the 2 individuals of the same APOL1 genotype with or without poly IC. Only 1 G2 homozygote had available primary PTCs. (e) Mitochondrial length was comparable across primary PTCs with different APOL1 genotypes without poly IC. However, when cells were incubated with low-dose poly IC (2.5 μg/ml) for 16 hours, those possessing 2 APOL1 KRVs (homozygous G1, homozygous G2, and G1/G2 compound heterozygotes) appeared to have decreased mitochondrial length (more fragmented mitochondria were present).
APOL1 G1 and G2 Variants Induce Mitochondrial Fission in HEK293 Tet-On Cell Lines
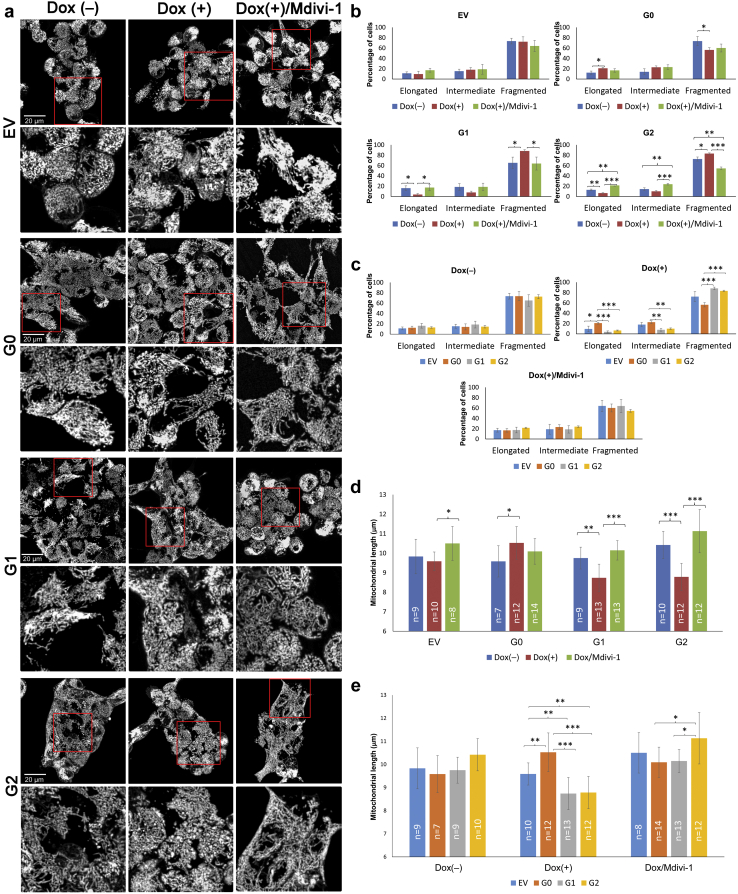
The observed mitochondrial fragmentation could relate to specific overexpression of APOL1 G1 and G2 KRVs by poly IC or as a general effect of poly IC triggering the TLR3-mediated innate immune response. Hence, we assessed HEK293 Tet-on APOL1 cell lines conditionally expressing G0, G1, and G2 variants, where the TLR3 pathway is absent.26 Cells were assessed with and without exposure to low concentrations of doxycycline (Dox) to induce moderate APOL1 expression. To clarify whether the observed fragmentation was typical G1- or G2-induced mitochondrial fission before moving into confocal imaging on HEK 293 Tet-on cells, immunoblotting was performed to determine the expression of mitochondrial fusion and fission proteins in HEK293 Tet-on APOL1 (G0, G1, and G2) cell lines with and without Dox-induced APOL1 overexpression. To mimic chronic exposure to APOL1 protein, a lower Dox concentration (∼10 ng/ml) and longer exposure time (24 hours) were used to induce APOL1 expression. Regardless of APOL1 G0, G1, or G2 genotypes, APOL1 mRNA expression levels in HEK293 Tet-on cells were ∼8-fold increased after exposure to Dox when compared with G0 cells without Dox (Supplementary Figure S7A). Comparable amounts of protein were obtained for the reference APOL1 G0 and the G1 and G2 variants (Supplementary Figure S7B). The mitochondrial fusion proteins mitochondrial dynamin like GTPase (OPA1), mitofusin-1 (MFN1), and mitofusin-2 (MFN2) and fission 1 protein (Fis1) did not display specific patterns related to APOL1 genotype or Dox-induced APOL1 overexpression. The mitochondrial fission dynamin-1-like protein (DRP1) levels were higher in Dox(+)-induced G1 and G2 cells, compared with Dox(−) G1 and G2 cells, respectively; DRP1 protein levels were lower in Dox(+)-induced G0 cells compared with Dox(−) G0 cells (Supplementary Figure S8). This suggested that G1 and G2 promoted mitochondrial fission. Because active DRP1 (phosphor-DRP1 at Ser 616, p-DRP1) is recruited to the outer mitochondrial membrane and mediates mitochondrial fission,27,28 and Mdivi-1 inhibits DRP1 by reducing p-DRP1(Ser616) on mitochondria,29 p-DRP1(Ser616) was assessed by immunofluorescence on HEK293 Tet-on APOL1 cells. G0 overexpression reduced p-DRP1(Ser616), whereas G1 and G2 overexpression increased p-DRP1(Ser616). The DRP-1 inhibitor Mdivi-1 markedly reduced the p-DRP1(Ser616) level in HEK293 Tet-on cells overexpressing G1 or G2 (Supplementary Figure S9). Hence, confocal microscopy was performed on HEK293 Tet-on cells using MitoTracker Red as a mitochondrial tracer (Figure 5a). HEK293 Tet-on EV and G0 cells exhibited elongated mitochondria before and after Dox induction; however, cells overexpressing G1 and G2 variants exhibited increased mitochondrial fragmentation after Dox induction (compared with baseline). To test the involvement of the DRP1-dependent mitochondrial fission pathway based on APOL1, the mitochondrial fission inhibitor Mdivi-1 was used to determine whether mitochondrial morphological patterns could be restored. Mdivi-1 prevented the mitochondrial fragmentation induced by KRV APOL1 overexpression in HEK293 Tet-on cells. Mdivi-1 did not alter the mitochondrial morphology in HEK293 Tet-on EV and G0 cells after treatment with Dox (Figure 5b–e).
Figure 5.
Confocal microscopy revealing altered mitochondrial morphology in HEK293 Tet-on APOL1 cell lines in response to doxycycline (Dox) and Mdivi-1. HEK293 Tet-on empty vector (EV), G0, G1, or G2 cells were grown without (−) Dox and without Mdivi-1, with Dox (+) only, and with (+) Dox/Mdivi-1 for 16 hours in full Dulbecco’s modified Eagle’s medium. The final Dox concentrations were 10, 10, 5, 10 ng/ml for EV, G0, G1, and G2 cells, respectively; these conditions maintained cell viability and produced comparable APOL1 overexpression levels in G0, G1, and G2 cells. In Dox (+) / Mdivi-1(+) cells, the final concentration for Mdivi-1 was 50 μM. Cells were subsequently incubated for 30 minutes with a final concentration of 50 nM MitoTracker Red (Invitrogen). (a) HEK293 G0 cells displayed more elongated mitochondria when exposed to Dox. In contrast, HEK293 EV cells did not change markedly in response to Dox or Mdivi-1. Dox-induced APOL1 G1 and G2 overexpression produced more fragmented mitochondria than without Dox. Addition of Mdivi-1 largely restored mitochondria to the tubular and elongated morphology in G1 and G2 cells with Dox induction. The red square was the imaged region for each cell type, detailed under the original image. Each image was representative of 18 scanned images from 3 independent experiments (binary images with enhanced local contrast [CLAHE] via Fiji scoring by Mitochondrial Network Analysis). (b) Comparison of mitochondrial morphology by APOL1 induction and additional Mdivi-1 treatment in HEK293 Tet-on cells of different APOL1 genotypes. Paired comparisons via 3 independent cell culture experiments showing that the percentage of cells with mitochondrial elongation significantly decreased and the percentage of cells with mitochondrial fragmentation significantly increased in HEK293 G1 and G2 cells with (+) Dox compared with those without (−) Dox. Mdivi-1 largely restored the mitochondrial pattern seen after Dox-induced APOL1 overexpression in G1 and G2 cells. For HEK293 G0 cells, Dox-induced overexpression of APOL1 increased the percentage of elongated cells and decreased the percentage of fragmented cells compared with those without (−) Dox, suggesting G0 may be beneficial to mitochondrial morphology; however, Mdivi-1 did not further alter the mitochondrial morphology of these cells. HEK293 EV Tet-on cells did not display changes in mitochondrial elongation or fragmentation with Dox or Mdivi-1. (Note: ∗P < 0.05, ∗∗ P < 0.01, ∗∗∗ P < 0.001 throughout Figure 5; no statistical significance found in any other paired comparison.) (c) Comparison of mitochondrial morphology by APOL1 genotype in HEK293 Tet-on cells without Dox induction, with Dox induction, and after Mdivi-1. Paired comparisons via 3 independent cell culture experiments showing that the percentage of cells with elongated and fragmented mitochondria were comparable among HEK293 cells of different APOL1 genotypes when Dox was absent. When APOL1 expression was induced by Dox, the percentage of cells with elongated mitochondria was significantly lower and the percentage of cells with fragmented mitochondria was significantly higher in HEK293 G1 and G2 cells than G0 cells. Mitochondrial morphology did not differ in Dox-induced HEK 293 EV, G0, G1, and G2 cells with addition of Mdivi-1. (d) Mitochondrial length (rods/network branches) by APOL1 genotype in HEK293 Tet-on cells without Dox induction, with Dox induction, and after Mdivi-1. Mitochondrial length, defined by rods/network branches, was estimated using Fiji integrated with the plug-in macro toolset Mitochondrial Network Analysis. Mitochondrial length did not differ in HEK293 Tet-on EV cells regardless of Dox induction or Mdivi-1 addition. Mitochondrial length increased in HEK293 Tet-on G0 cells when induced by Dox; however, it was not further affected by the addition of Mdivi-1. Mitochondrial length decreased significantly in HEK293 Tet-on G1 and G2 cells when induced by Dox, indicating enhanced mitochondrial fragmentation by overexpression of G1 and G2 variants. Reduced mitochondrial length was restored after addition of Mdivi-1. (Note: n refers to number of images.) (e) Mitochondrial branch length by Dox induction and additional Mdivi-1 treatment in HEK293 Tet-on cells of different APOL1 genotypes. Mitochondrial length was largely comparable for HEK293 Tet-on cells of different APOL1 genotypes without Dox. With Dox-induced APOL1 overexpression, G0 cells appeared to have significantly higher mitochondria length than G1 and G2, indicating APOL1 G0 promoted mitochondrial elongation, whereas G1 and G2 variants enhanced mitochondrial fragmentation Mitochondrial length was relatively comparable for Dox-induced EV, G0, G1, and G2 cells when Mdivi-1 was added, slightly longer mitochondria were seen in G2 cells.
Mdivi-1 Restores Mitochondrial Membrane Potential in HEK293 Cells Overexpressing APOL1 G1 and G2
Dox-induced APOL1 G1 and G2 overexpression in HEK 293 Tet-on APOL1 cells significantly reduced the relative mitochondrial membrane potential, defined as the fluorescence signal ratio of TMRE/MitoTracker Green. The relative mitochondrial membrane potential was partially restored in these cells after exposure to Mdivi-1 (Supplementary Figure S10). In contrast, Dox-induced overexpression in APOL1 G0 and EV cells (and subsequent exposure to Mdivi-1) did not affect the relative mitochondrial membrane potential. Mdivi-1 did not alter relative mitochondrial membrane potential in the absence of mitochondrial depolarization. Results suggest that Mdivi-1 preserves mitochondrial architecture, resulting in improved mitochondrial membrane potential.
Mdivi-1 Restores Viability of HEK293 Cells Overexpressing APOL1 G1 and G2
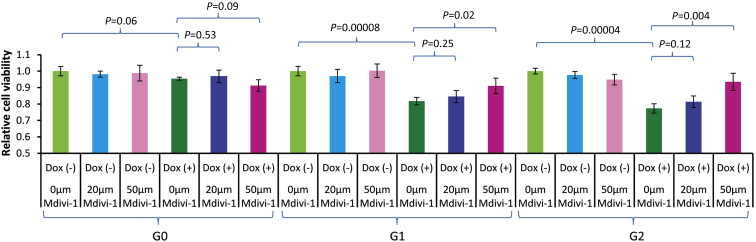
To provide proof of concept that mitochondrial fusion/fission activity can be targeted in the treatment of APOL1-nephropathy, cell viability was assessed for HEK293 Tet-on APOL1 G0, G1 and G2 cell lines at baseline, after Dox induction, and after Mdivi-1 rescue post-Dox induction. HEK293 Tet-on G0 cell (reference cells) viability was not altered after APOL1 overexpression or treatment with Mdivi-1. For HEK293 Tet-on G1 and G2 cells, Dox-induced overexpression of both KRVs significantly reduced cell viability and Mdivi-1 restored cell viability, particularly at higher concentrations (Figure 6). Dose-response curves demonstrate increased viability of G1 and G2 cells with higher Mdivi-1 concentrations (Figure 6). In contrast to Mdivi-1, the mitochondrial rescuer MitoQ did not alter the viability of cells induced by APOL1 G1 and G2 variants (Supplementary Figure S11).
Figure 6.
Cell viability in HEK293 Tet-on cells overexpressing APOL1 G0, G1, and G2 when treated with Mdivi-1. HEK293 Tet-on G0, G1, or G2 cells were grown without (−) doxycycline (Dox) and with (+) 10 ng/ml Dox for 24 hours. The lactate dehydrogenase assay was used to assess cell viability. Final concentrations of Mdivi-1 (0, 20 μm, and 50 μm) were applied to HEK293 G0, G1, and G2 cells in Dox (−) and Dox (+) conditions (n = 4 in each experiment). Dox induction did not affect cell viability in HEK293 Tet-on G0 cells; G1 and G2 cells had reduced viability after 24-hour Dox induction (P = 0.00008 and 0.00004, respectively). Mdivi-1 did not affect cell viability in HEK293 Tet-on G0, G1, and G2 cells without Dox induction. Mdivi-1 did not alter cell viability in G0 cells with or without Dox-induced APOL1 overexpression. Mdivi-1 (50 μm) restored cell viability in Dox-induced G1 and G2 cells (P = 0.02 and 0.004, respectively); a dose-dependent pattern was seen. Note: Cells with (−)/(+) refer to without or with Dox induction, respectively.
Discussion
APOL1 KRVs induce mitochondrial dysfunction16,17 and the present results support specific effects on mitochondrial dynamics that can be reversed with inhibitors of mitochondrial fission such as Mdivi-1. Global gene expression profiles were performed in primary renal PTC lines from African American individuals before and after poly IC to mimic innate immune system activation.21 APOL1 KRVs were not cis eQTLs for APOL1 or adjacent genes, nor did they contribute to differences in global transcript expression attributed to poly IC. Relationships between other SNPs and APOL1 expression were delineated using Illumina Multi-Ethnic Genotyping Arrays chip-based genome-wide genotyping. SNP rs513349 was among the top trans eQTLs for APOL1 expression. This SNP was also a cis eQTL for the adjacent BAK1 gene based on cis eQTL analyses in multiple human cells and tissues in the Genotype-Tissue Expression (GTEx) Project (in silico data retrieved December 19, 2019, from https://www.gtexportal.org/). BAK1 and APOL1 were co-expressed in primary renal PTCs after exposure to low concentrations of poly IC (R = 0.7, P < 0.0001) indicating that rs513349 acts as a trans eQTL for APOL1 via regulation of BAK1 expression on activation of the TLR3 pathway. As podocytes are critical cells in APOL1-nephropathy, BAK1 was subsequently knocked out in a human podocyte cell line.30 APOL1 protein was markedly reduced in BAK1 knockout cells, confirming the pivotal role of BAK1 in APOL1 expression. BAK1 knockout did not inhibit APOL1 messenger RNA levels when poly IC was absent; however, it did inhibit APOL1 mRNA upregulation after poly IC treatment. This supports the initial eQTL analysis results. The phenomenon appears similar to that of a BCL2 family member, BOK (BCL2-related ovarian killer), the 3ʹ untranslated region of which was modified by the presence of TRIM28. As a result, there was a lack of concordance between protein and transcript levels31 and suggests that unstable mRNA may not generate proportional protein levels. The relationship between BAK1 protein and the APOL1 regulatory region (enhancer, promoter, and 3ʹ untranslated region) requires further investigation. It would be ideal if BAK1 could be knocked out in primary tubule cells with different APOL1 genotypes. However, primary tubule cells can proliferate for only approximately 10 to 15 biological passages, typically within 3 weeks. This is approximately the time needed for cells to go through the selection stage for CRISPR knockout. Hence, we were unable to perform experiments after knockout cells were created. This is a limitation of using primary cell lines.
BAK1 is a nuclear-encoded mitochondrial outer membrane protein.32 APOL1 is also present in mitochondria and its G1 and G2 KRVs can induce mitochondrial dysfunction.16,17 Although APOL1 KRVs induce cell death by mitochondrial translocation and opening of the mitochondrial permeability transition pore on the inner membrane,18 other pathways may exist. APOL1 is co-expressed with the mitochondrial outer membrane protein BAK1, involved in mitochondrial dynamics via stabilization of phospho dynamin-related protein 1 (pDRP1) on the mitochondrial outer membrane.33 Bax/Bak-dependent release of DDP/TIMM8a protein in the mitochondrial intermembrane space promotes DRP1-mediated mitochondrial fission and mitoptosis by redistributing pDRP to the mitochondrial outer membrane and inducing mitochondrial fission.34
The link between APOL1 and BAK1 in mitochondria led us to assess whether mitochondrial dynamics could compromise mitochondrial function induced by increased expression of APOL1 KRVs. Based on known effects of BAK1, we considered whether APOL1 KRVs altered mitochondrial morphology during fusion/fission. We found that mitochondria from human primary renal PTCs with 2 APOL1 KRVs were more likely to fragment after exposure to low concentrations of poly IC than those without KRVs (homozygous G0). The poly IC concentration for induction of APOL1 (2.5 μg/ml for 16 hours) was intentionally lower than previously reported21; nonetheless, increases in APOL1 and TLR3 expression were similar to those with poly IC concentrations 5.0 or 10.0 μg/ml and cell viability was less impacted by the 2.5 μg/ml concentration (data not shown). Affymetrix HTA 2.0 array-based pathway analysis confirmed marked activation of the innate immune system with 2.5 μg/ml concentrations of poly IC (Supplementary Tables S1 and S2B). Fluorescence confocal microscopy performed on primary renal PTCs verified the mitochondrial morphology seen by transmission electron microscopy (data not shown) and confirmed increased mitochondrial fragmentation in cells with APOL1 KRVs after poly IC.
To determine whether enhanced mitochondrial fragmentation was a consequence of general innate immune system activation from poly IC or related to increased expression of APOL1 KRVs, HEK293 Tet-on APOL1 EV, G0, G1, and G2 cell lines conditionally overexpressing different APOL1 genotypes were examined. These were critical experiments because HEK293 cells lack innate immune system activation through the TLR3 pathway.26 TLR3 mRNA was measured in HEK293 Tet-on EV, G0, G1, and G2 cells with and without poly IC, and expression was virtually undetectable (data not shown). Overexpression of APOL1 G1 and G2 in HEK293 cells promoted mitochondrial fragmentation (potentially via fission) based on confocal microscopy and supported by immunoblotting and immunofluorescence. This could relate to G1- and G2-mediated increases in active DRP1 (p-DRP1 at Ser 616). In contrast, overexpression of G0 reduced p-DRP1 (Ser616) levels in mitochondria, resulting in diminished fission and elongated mitochondria. Inhibition of mitochondrial fission by the DRP1 inhibitor Mdivi-129 appeared to preserve mitochondrial morphology. Mdivi-1 led to fewer fragmented mitochondria. Restoration of cell viability in HEK293 cells overexpressing G1 and G2 cells via Mdivi-1 provides additional evidence that altered mitochondrial dynamics play a role in the compromised mitochondrial function. In addition, Mdivi-1 restored cell viability in HEK293 G1 and G2 cells in dose-dependent fashion (Figure 6). As such, the mitochondrial fusion/fission pathway may be a therapeutic target for reversing cellular dysfunction induced by APOL1 KRVs. In contrast, the mitochondrial antioxidant MitoQ, a source of coenzyme Q, did not have this effect.
The mitochondrial dynamics pathway in HEK293 Tet-on cells was assessed via immunoblotting and fluorescence microscopy after 16-hour and 24-hour Dox induction. The 16-hour Dox induction minimized the impact of cell viability (Supplementary Figure S12) when examining the mitochondrial morphology and function, whereas the 24-hour Dox induction maximized the effect of APOL1 G1 and G2 variants when most cells remained viable and cell viability could be restored. Both Dox-induction conditions produced comparable levels of APOL1 overexpression (Supplementary Figures S7 and S13).
The expected reduction in relative mitochondrial membrane potential for G1 and G2 cells was observed, reproducing the pattern in our prior report with higher concentration Dox induction and shorter exposure time.16 Mdivi-1 significantly increased the relative membrane potential in APOL1 G1 and G2 cells after low-concentration Dox induction. Mdivi-1 appeared to improve cell viability by preventing mitochondrial fragmentation and preserving membrane potentials in G1 and G2 cells, but it did not increase the membrane potential in Dox-induced G0 cells. This demonstrates specific efficacy of Mdivi-1 in mitochondrial dysfunction related to G1 and G2. Results were supported by morphological studies; however, it is unlikely that mitochondrial fission is the sole pathway compromising mitochondrial function because Mdivi-1 largely restored mitochondrial length in Dox-induced HEK293 G1 and G2 cells (Figure 5d and e) without fully restoring the mitochondrial membrane potential (Supplementary Figure S10). Results do not exclude additional effects of APOL1 on mitochondrial dynamic pathways beyond pDRP1, such as activated mitochondrial permeability transition.18 Bordt et al.35 questioned the specificity of Mdivi-1 as a mitochondrial fission inhibitor in ischemia-reperfusion injury experiments. However, Manczak et al.36 confirmed that Mdivi-1 is a DRP-1 inhibitor and directly reduces mitochondrial fission. MitoQ was developed as a mitochondrial-targeted antioxidant for diseases associated with oxidative stress. MitoQ reportedly blocks the generation of reactive oxygen species.37 Therefore, we assessed MitoQ in parallel with Mdivi-1, in rescue experiments performed in HEK293 Tet-on cells overexpressing APOL1 G0, G1, and G2. As shown in Figure 6, Mdivi-1 significantly restored cell viability, whereas MitoQ did not (Supplementary Figure S11). This indicates that mitochondrial fragmentation/fission is the likeliest pathway whereby APOL1 G1 and G2 variants induce mitochondrial dysfunction, whereas targeting reactive oxygen species may be less specific. It has been reported that mutations at Ser616 in DRP1 (phosphorylation site) or inhibiting DRP1 activity blocks mitochondrial permeability transition pore opening.38 This suggests that mitochondrial fission is an event upstream from mitochondrial permeability transition pore opening.
Removal of damaged mitochondria through autophagy or mitophagy, a process selectively removing damaged or excessive mitochondria, is critical to maintain cellular function,39 especially in podocytes40 and PTCs.41 Permanent loss of function or death of podocytes can have irreversible consequences on kidney function. Fragmented mitochondria are eliminated by autophagy when a threshold of cellular damage develops. Fission may segregate the most severely damaged mitochondria to preserve the health of the mitochondrial network.42 If G1 and G2 variant-induced mitochondrial damage/fragmentation cannot be compensated by appropriate mitophagy, possibly due to defective intracellular trafficking as proposed,5,43 cell death machinery may be activated.32
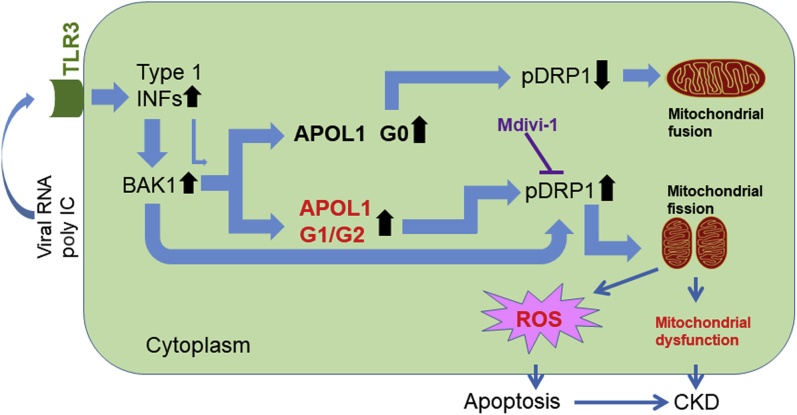
Figure 7 displays a diagram with potential mechanisms of APOL1 KRV-induced mitochondrial dysfunction. In response to environmental stressors, such as viral infection, APOL1 is upregulated through the TLR3 pathway via increased type 1 interferons.21 However, BAK1 is a more robust inducer of APOL1 than interferons; BAK1 knockout markedly reduced APOL1 expression. BAK1 promotes mitochondrial outer membrane permeabilization.44 During apoptosis, Bax/Bak-dependent release of DDP/TIMM8a protein in the mitochondrial intermembrane space promotes DRP1-mediated mitochondrial fission and mitoptosis by redistributing pDRP to the mitochondrial outer membrane and inducing mitochondrial fission.34 As a result of innate immunity, type 1 interferons increase BAK1 expression for mitoptosis and apoptosis.45 A limitation of the study is the lack of kidney sections from patients with chronic kidney disease. As such, we were unable to display BAK1 distribution in the context of kidney disease. Increased levels of APOL1 G0 protein appear to counter effects of BAK1; G0 preserves mitochondrial function by inhibiting DRP1 expression. This results in mitochondrial fusion. As APOL1 upregulation by BAK1 is independent of APOL1 genotype, increased levels of APOL1 G1 and G2 protein may reduce inhibition to DRP1 expression. These promote mitochondrial fission. Fission was reversed by the DRP1 inhibitor Mdivi-1. In addition to the deleterious effect of the KRVs on mitochondrial integrity, the “beneficial” effects of G0 upregulation on mild inhibition of p-DRP1 (Supplementary Figure S8) and decreased endoplasmic reticulum stress16 were absent for G1 or G2 KRVs. Mitochondrial membrane potentials decrease during mitochondrial fission. This effect may be independent from the mechanism proposed by Shah et al.,18 where the mitochondrial permeability transition pore opened when binding G1 or G2 variants with decreased mitochondrial membrane potential. This might explain why Mdivi-1 did not fully restore cell viability and mitochondrial membrane potential when APOL1 G1 and G2 were induced in HEK293 Tet-on cells. Although BAK1 clearly upregulates APOL1, details of this pathway require further investigation.
Figure 7.
Diagram showing potential mechanisms of APOL1 kidney-risk variant-induced mitochondrial dysfunction. CKD, chronic kidney disease; INF, interferon; pDRP1, phospho dynamin-related protein 1; poly IC, polyinosinic-polycytidylic acid; ROS, reactive oxygen species; TLR3, Toll-like receptor 3.
This series of experiments used unique human primary renal PTC lines from African American individuals to explore potential effects of APOL1 on human kidney cells, with replication in HEK293 Tet-on APOL1 cells. Results in human primary renal PTC cells with different APOL1 genotypes implicated altered mitochondrial dynamics on risk for APOL1-nephropathy. Cell viability and mitochondrial dynamics were improved in cells with APOL1 KRVs after exposure to Mdivi-1, an inhibitor of DRP1 and mitochondrial fission. Future studies involving mitochondrial dynamics and mutations could further inform the field regarding APOL1 KRV effects on nephropathy. This may lead to novel therapies for APOL1-associated disorders.
Disclosure
Wake Forest University Health Sciences and BIF have rights to an issued U.S. patent related to APOL1 genetic testing. BIF is a consultant for AstraZeneca and RenalytixAI Pharmaceuticals. All the other authors declared no competing interests.
Acknowledgments
We thank Dr. Martin R. Pollak for sharing APOL1 vectors. This work was supported by National Institutes of Health (NIH) grants R01 DK070941 (BIF), R01 DK084149 (BIF), and R01 MD009055 (JD, BIF), and P30 CA012197 (Wake Forest Comprehensive Cancer Center). The NIH GEO database access for this project is GSE119958.
Author Contributions
LM, BIF, and AJAM designed the study; LM and BIF drafted and revised the paper; JSP provided in-depth revision; LM, JAS, YAC, MSB, DC, ALC, and PJH performed the experiments; HCA, LM, JWC, CDL, and JD analyzed the data; MM and AKH recruited subjects and revised the paper; SP provided microscopy assistance and revised the paper; GAH and LDM provided global gene expression technical expertise and reviewed the manuscript; and MAS provided human immortalized podocyte and proximal tubular cell lines, helped establish cell culture conditions, and reviewed the paper. All authors approved the final version of the manuscript.
Footnotes
Supplementary Methods.
Figure S1. Presence of BAK1 protein in human renal proximal tubule cells (PTCs).
Figure S2. Presence of BAK1 protein in immortalized podocytes.
Figure S3. Presence of BAK1 on nondiseased human kidney cryosections.
Figure S4. APOL1 messenger RNA level responses to the presence of poly IC and BAK1 by RT-PCR.
Figure S5. APOL1 protein level responses to the presence of BAK1 by immunoblotting.
Figure S6. APOL1 protein level responses to the presence of BAK1 by immunofluorescence.
Figure S7. Comparison of APOL1 gene expression in HEK293 Tet-on cells.
Figure S8. Mitochondrial fission marker DRP1 increased in Dox-induced HEK293 Tet-on G1 and G2 cell lines.
Figure S9. Phospho-DRP1 at Ser 616 (pDRP1) responses to APOL1 overexpression and Mdivi-1 rescue.
Figure S10. Relative mitochondrial membrane potential was reduced in Dox-induced HEK293 Tet-on APOL1 G1 and G2 cells and rescued by Mdivi-1.
Figure S11. Cell viability in HEK293 Tet-on cells overexpressing APOL1 G0, G1, and G2 when treated with MitoQ.
Figure S12. Quantitative cell viability in HEK293 Tet-on EV, G0, G1, or G2 cells after 16-hour Dox induction.
Figure S13. Relative APOL1 expression levels were comparable in HEK293 Tet-on cells with Dox induction for 16 hours.
Table S1. Top 20 most differentially expressed pathways (1060 downregulated and 1212 upregulated) after poly IC in the 50 African American primary renal PTC lines using Cytoscape BiNGO.
Table S2A. Top canonical pathways detected based on the most downregulated genes by poly IC in 50 primary African American renal PTC lines using Ingenuity Pathway Analysis.
Table S2B. Top canonical pathways detected based on the most upregulated genes by poly IC in 50 primary African American renal PTC lines using Ingenuity Pathway Analysis.
Contributor Information
Lijun Ma, Email: lima@wakehealth.edu.
Barry I. Freedman, Email: bfreedma@wakehealth.edu.
Supplementary Material
References
- 1.Genovese G., Friedman D.J., Ross M.D. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S., Rosset S., Shemer R. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olabisi O.A., Zhang J.-Y., VerPlank L. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci U S A. 2016;113:830–837. doi: 10.1073/pnas.1522913113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y., Zhu J., Richman A. APOL1-G1 in nephrocytes induces hypertrophy and accelerates cell death. J Am Soc Nephrol. 2016;28:1106–1116. doi: 10.1681/ASN.2016050550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruzel-Davila E., Shemer R., Ofir A. APOL1–mediated cell injury involves disruption of conserved trafficking processes. J Am Soc Nephrol. 2017;28:1117–1130. doi: 10.1681/ASN.2016050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruggeman L.A., O’Toole J.F., Ross M.D. Plasma apolipoprotein L1 levels do not correlate with CKD. J Am Soc Nephrol. 2014;25:634–644. doi: 10.1681/ASN.2013070700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weckerle A., Snipes J.A., Cheng D. Characterization of circulating APOL1 protein complexes in African Americans. J Lipid Res. 2016;57:120–130. doi: 10.1194/jlr.M063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozlitina J., Zhou H., Brown P.N. Plasma levels of risk-variant APOL1 do not associate with renal disease in a population-based cohort. J Am Soc Nephrol. 2016;27:3204–3219. doi: 10.1681/ASN.2015101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madhavan S.M., O’Toole J.F., Konieczkowski M. APOL1 localization in normal kidney and nondiabetic kidney disease. J Am Soc Nephrol. 2011;22:2119–2128. doi: 10.1681/ASN.2011010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L., Shelness G.S., Snipes J.A. Localization of APOL1 protein and mRNA in the human kidney: nondiseased tissue, primary cells, and immortalized cell lines. J Am Soc Nephrol. 2015;26:339–348. doi: 10.1681/ASN.2013091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedman B.I., Pastan S.O., Israni A.K. APOL1 genotype and kidney transplantation outcomes from deceased African American donors. Transplantation. 2016;100:194–202. doi: 10.1097/TP.0000000000000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freedman B.I., Julian B.A., Pastan S.O. Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transplant. 2015;15:1615–1622. doi: 10.1111/ajt.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reeves-Daniel A.M., DePalma J.A., Bleyer A.J. The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant. 2011;11:1025–1030. doi: 10.1111/j.1600-6143.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee B.T., Kumar V., Williams T.A. The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am J Transplant. 2012;12:1924–1928. doi: 10.1111/j.1600-6143.2012.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beckerman P., Bi-Karchin J., Park A.S.D. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med. 2017;23:429–438. doi: 10.1038/nm.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma L., Chou J.W., Snipes J.A. APOL1 Renal-Risk Variants Induce Mitochondrial Dysfunction. J Am Soc Nephrol. 2017;28:1093–1105. doi: 10.1681/ASN.2016050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granado D., Müller D., Krausel V. Intracellular APOL1 risk variants cause cytotoxicity accompanied by energy depletion. J Am Soc Nephrol. 2017;28:3227–3238. doi: 10.1681/ASN.2016111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah S.S., Lannon H., Dias L. APOL1 kidney risk variants induce cell death mitochondrial translocation and opening of the mitochondrial permeability transition pore. J Am Soc Nephrol. 2019:2355–2368. doi: 10.1681/ASN.2019020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galvan D.L., Green N.H., Danesh F.R. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017;92:1051–1057. doi: 10.1016/j.kint.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freedman B.I., Skorecki K. Gene–gene and gene–environment interactions in apolipoprotein l1 gene-associated nephropathy. Clin J Am Soc Nephrol. 2014;9:2006–2013. doi: 10.2215/CJN.01330214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nichols B., Jog P., Lee J.H. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2015;87:332–342. doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng D., Weckerle A., Yu Y. Biogenesis and cytotoxicity of APOL1 renal risk variant proteins in hepatocytes and hepatoma cells. J. Lipid Res. 2015;56:1583–1593. doi: 10.1194/jlr.M059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma L., Murea M., Snipes J.A. An ACACB variant implicated in diabetic nephropathy associates with body mass index and gene expression in obese subjects. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valente A.J., Maddalena L.A., Robb E.L. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Karbowski M., Norris K.L., Cleland M.M. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 26.Sarkar S.N., Smith H.L., Rowe T.M. Double-stranded RNA signaling by Toll-like receptor 3 requires specific tyrosine residues in its cytoplasmic domain. J Biol Chem. 2002;278:4393–4396. doi: 10.1074/jbc.C200655200. [DOI] [PubMed] [Google Scholar]
- 27.Cereghetti G.M., Stangherlin A., de Brito O.M. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashatus J.A., Nascimento A., Myers L.J. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H., Lee J.Y., Park K.J. A mitochondrial division inhibitor, Mdivi-1, inhibits mitochondrial fragmentation and attenuates kainic acid-induced hippocampal cell death. BMC Neurosci. 2016;17:33. doi: 10.1186/s12868-016-0270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saleem M.A., O’Hare M.J., Reiser J. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002;13:630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Marrero Y., Bachmann D., Lauber E. Negative regulation of BOK expression by recruitment of TRIM28 to regulatory elements in its 3′ untranslated region. iScience. 2018;9:461–474. doi: 10.1016/j.isci.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boland M.L., Chourasia A.H., Macleod K.F. Mitochondrial dysfunction in cancer. Front Oncol. 2013;3:292. doi: 10.3389/fonc.2013.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wasiak S., Zunino R., McBride H.M. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnoult D., Rismanchi N., Grodet A. Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. Curr Biol. 2005;15:2112–2118. doi: 10.1016/j.cub.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 35.Bordt E.A., Clerc P., Roelofs B.A. The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev Cell. 2017;40:583–594.e6. doi: 10.1016/j.devcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manczak M., Kandimalla R., Yin X. Mitochondrial division inhibitor 1 reduces dynamin-related protein 1 and mitochondrial fission activity. Hum Mol Genet. 2019;28:177–199. doi: 10.1093/hmg/ddy335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu C., Zhang D., Whiteman M. Is antioxidant potential of the mitochondrial targeted ubiquinone derivative MitoQ conserved in cells lacking mtDNA? Antioxid Redox Signal. 2008;10:651–660. doi: 10.1089/ars.2007.1865. [DOI] [PubMed] [Google Scholar]
- 38.Xu S., Wang P., Zhang H. CaMKII induces permeability transition through Drp1 phosphorylation during chronic β-AR stimulation. Nat Commun. 2016;7:13189. doi: 10.1038/ncomms13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Um J.-H., Yun J. Emerging role of mitophagy in human diseases and physiology. BMB Rep. 2017;50:299–307. doi: 10.5483/BMBRep.2017.50.6.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartleben B., Gödel M., Meyer-Schwesinger C. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J. Clin. Invest. 2010;120:1084–1096. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang C., He L., Liu J. Mitophagy: basic mechanism and potential role in kidney diseases. Kidney Dis. 2015;1:71–79. doi: 10.1159/000381510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youle R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madhavan S.M., O’Toole J.F., Konieczkowski M. APOL1 variants change C-terminal conformational dynamics and binding to SNARE protein VAMP8. JCI Insight. 2017;2(14) doi: 10.1172/jci.insight.92581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peña-Blanco A., García-Sáez A.J. Bax, Bak and beyond—mitochondrial performance in apoptosis. FEBS J. 2018;285:416–431. doi: 10.1111/febs.14186. [DOI] [PubMed] [Google Scholar]
- 45.Fraietta J.A., Mueller Y.M., Yang G. Type I interferon upregulates Bak and contributes to T cell loss during human immunodeficiency virus (HIV) infection. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.