Abstract
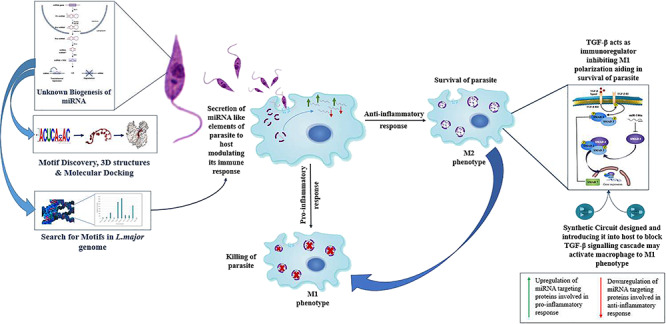
Leishmaniasis, the second most neglected tropical disease, has been reported to affect approximately 12 million people worldwide. The causative protozoan parasite Leishmania has shown drug resistance to available chemotherapies, owing to which we need to look for better approaches to deal with the clinical situations. As per recent reports, several miRNAs have been found to be differentially expressed during Leishmania major infection in host macrophages. We aim to evaluate the impact of miRNA-mediated gene regulation on the key players of inflammation and macrophage dysfunction. The origin of Leishmania miRNAs and their processing is a questionable phenomenon as of yet. Through our study, we aim to provide a framework of their characterization. We amalgamate chemical systems biology and synthetic biology approaches to identify putative miRNA targets and unravel the complexity of host–pathogen gene regulatory networks.
1. Introduction
Leishmaniasis is a zoonotic disease caused by various species of the Leishmania parasite. It affects about 12 million people and is considered a global health problem due to its diffusion in Europe, Africa, and Asia (Old World) as well as in the Americas (New World).1 It has been estimated that there are 0.7–1 million new cases of the disease every year, causing 20,000–30,000 deaths (World Health Organization, 2018). According to Leishmania species and the host response, leishmaniasis is categorized into cutaneous (CL), visceral (VL), and mucocutaneous leishmaniasis (MCL). The Leishmania parasite is an obligate intracellular parasite transmitted through female sandfly, phlebotomine-infecting mammalian host. The Leishmania parasite when residing in two different hosts shows two distinct morphological forms such as the promastigote (motile) form in the midgut of sandfly and the amastigote (amotile) form in the mammalian host. The parasite resides and proliferates inside phagocytic cells, primarily macrophages in a mammalian host.29,30
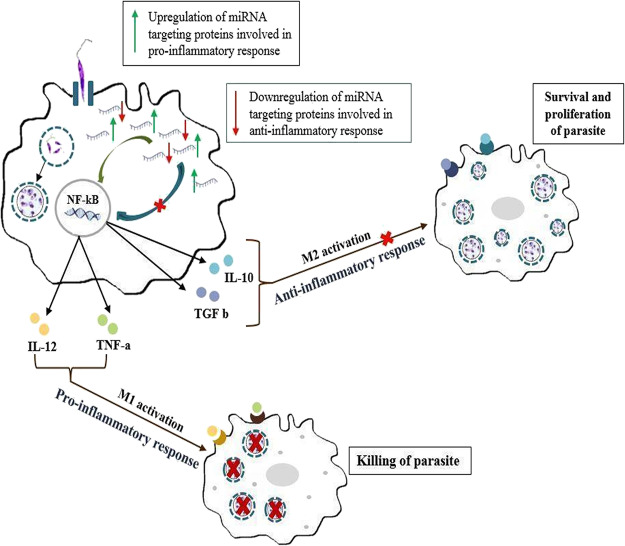
Macrophages are major effector cells in the immune system, destined for destruction of intracellular pathogens, but it is not the case for the Leishmania parasite as it infects and proliferates inside them. During the blood meal of an infected female sandfly, it injects a metacyclic promastigote form of the parasite inside the mammalian host. As an initial innate response, dendritic cells, neutrophils, and macrophages are recruited at the site of infection. Here, these promastigotes gets phagocytized inside the phagocytic cells. The parasite has developed ways to evade host immune surveillance. Several strategies have been observed for immunomodulation, adapted by the parasite for its survival and proliferation. One of them is by manipulating activation of nuclear factor kappa B (NF-κB). NF-κB is responsible for the release of pro-inflammatory cytokines as well as anti-inflammatory cytokines based on the activation of NF-κB.37 As a survival strategy, the Leishmania parasite degrades a p65 subunit (transcriptional regulator), thus inhibiting the activation of NF-κB in a classic manner.36 During Leishmania infection, specific cleavage of NF-κB p65 occurs in the cytoplasm generating a fragment of p35, which migrates into the nucleus where it binds DNA as a heterodimer with NF-κB p50, inducing chemokine gene expression. This indicates a mechanism by which a pathogen can subvert a macrophage’s regulatory pathways to alter NF-κB activity.39 In response to anti-inflammatory cytokines generated in by activation of NF-kB, macrophages get polarized into the M2 phenotype, inhibiting iNOS production with generation of polyamines, aiding in the survival and proliferation of the Leishmania parasite inside the macrophages, as shown in Figure 1. Recent reports suggest that miRNA plays a critical role in immune responses.22 During evolution, the Leishmania parasite lost its RNAi activity, but certain studies suggest the presence of miRNA-like elements.38 For the survival, some parasites export their RNA machinery such as rRNA and tRNA including miRNA in the host, interacting with host Argonaute proteins.
Figure 1.
During L. major infection in macrophages, miRNAs targeting the mRNA of proteins involved in the pro-inflammatory response gets upregulated and miRNAs targeting mRNA of proteins involved in the anti-inflammatory response gets downregulated, aiding in the survival of the parasite inside the macrophages.
MicroRNAs (miRNAs) are a class of small noncoding RNAs. These are 20–25 nucleotides, having a seed sequence that is complementary to their target mRNA. They regulate target genes post-transcriptionally. The interaction of miRNA–mRNA leads to translation inhibition and mRNA destabilization.2,11 miRNAs play an important role in the activation of macrophages and in the regulation of phagocytosis and apoptosis.3,14−16 Due to their involvement in major biological processes, they are being considered as a potential target for various diseases such as cancer, tuberculosis, osteoporosis, etc.13,17 miRNAs act as post-transcriptional regulators of gene expression and regulate many target genes, including NF-κB, IκB, IKK, and regulators in the NF-κB signaling pathway, forming positive or negative sophisticated feedback loops.40
We investigated differentially expressed miRNAs with their respective functions in the immune response and inflammation. For the said purpose, the miRNA regulatory network was generated through Cytoscape v.3.5.05 wherein key miRNAs playing a major role in inflammation and the immune response during Leishmania infection were curated. After generating the miRNA regulatory network, miR-146a-3p, miR-146a-5p, miR-155, miR-188, miR-9-3p, miR-9-5p, miR-122, and miR-147 were found to be majorly involved in the inflammatory response during Leishmania infection. As evolutionary studies have suggested, RNAi activity is lost in Leishmania major but miRNA-like elements may be present. To prove the existence of miRNA-like elements in L. major, specific patterns were searched that would act like miRNAs or provide a seed sequence of the parasite inside a human host, and 10 motifs were discovered against human miRNAs. To confirm the presence of miRNA biogenesis in L. major, patterns were searched against the L. major genome with discovered human motifs by the Knuth–Morris–Pratt (KMP) algorithm. The molecular docking studies of the modeled motifs gave us an insight into the processivity of miRNAs in the Leishmania parasite as these discovered motifs have shown an ability to bind to the human Argonaute protein with the best positioning and binding energy for their stability, and as cited before from the literature, some parasites transfer their miRNA via exosomes in the host for modulating and evading the host immune response, which helps in proving the existence of miRNAs in Leishmania.(4,12,18−28,33−35)
2. Results and Discussion
2.1. Generation of the miRNA Regulatory Network
The miRNA regulatory network was generated from Cytoscape v.3.5.0.5 The initial network produced had a huge number of nodes and edges, more precisely, 493 nodes and 1347 edges. The simulated annealing layout made the network distribution vivid for understanding, as per Table 1. Based on in and out degrees, it makes the highly connected nodes distinct. After applying in- and out-degree filters to the network, miR-146a-3p, miR-146a-5p, miR-122, miR-155-3p, miR-155-5p, miR-188, miR-9-3p, miR-9-5p, and miR-147 were found to be key miRNAs playing a role in the inflammatory response during Leishmania infection, as shown in Figure 2 a,b. Also, from a prior literature review, we had concrete evidence supporting the fact that miR-9, miR-146a, miR-155, and miR-21 have diverse roles in immune regulatory functions as well as macrophage dysfunction.10
Table 1. Statistical Analysis of miRNA Regulatory Network after Simulated Annealing Representing a Decrease in the Number of Nodes and Edges, making the Network More Robust.
| parameter | values |
|---|---|
| clustering coefficient | 0.05 |
| connected components | 20 |
| network diameter | 14 |
| network radius | 1 |
| shortest paths | 13,324 (30%) |
| characteristic path lengths | 1.864 |
| average number of neighbors | 3.131 |
| number of nodes | 557 |
| network density | 0.009 |
| isolated nodes | 0 |
| number of self-loops | 0 |
| multiedge node pairs | 15 |
| analysis time (s) | 0.099 |
Figure 2.
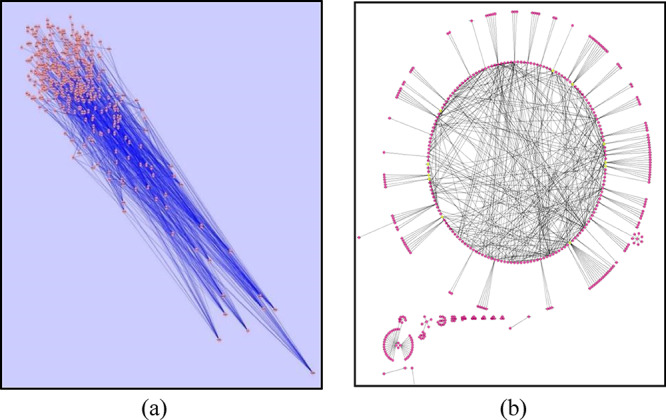
(a) Robust miRNA regulatory network generated after simulated annealing. (b) Generated network in a circular layout after applying out- and in-degree filters; miR-146a-3p, miR-146a-5p, miR-155-3p, miR-155-5p, miR-122, miR-9-3p, miR-9-5p, miR-188, and miR-147 were filtered out.
2.2. Profiling of miRNA Expression in THP-1 Cells Infected with L. major
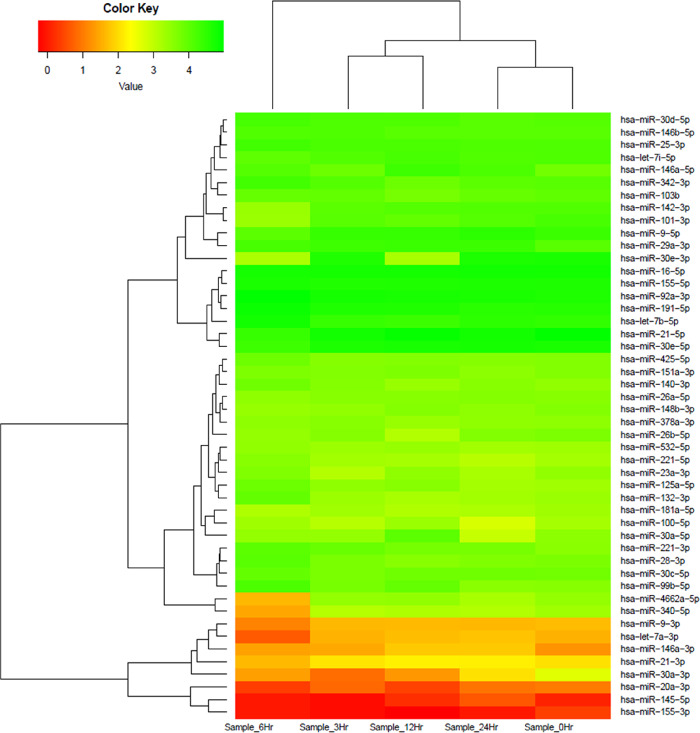
Using human miRNA arrays, a distinct miRNA expression profile was observed across different time points after infection with reference to 0 h in THP-1 cells after infection. We found several key members of miRNA families that were differentially modulated across different time points in THP-1 cells post-infection. These families included the miR-146a, let-7, miR-30, miR-9, miR-155, miR-145, and miR-21 family of miRNAs, clearly suggesting the specific role of these miRNAs during Leishmania infection and proliferation in a time-dependent manner. Figure 3 represents a heatmap of infection-specific differentially expressed miRNAs grouped in a response-specific manner.
Figure 3.
Heatmap and hierarchical clustering of infection-specific miRNAs. Heatmap and hierarchical clustering of 48 infection-specific differentially expressed key miRNAs in THP-1 cells in response to Leishmania infection at 3, 6, 12, and 24 h as compared to the infected 0 h control. The heatmap shows several members of key families of miRNAs (miR-146, let-7, miR-30, miR-155, miR-21, and miR-9) differentially modulated across different time points. The miRNA expression values are presented using a green (upregulated), yellow (median value), and red (downregulated) color scheme.
2.3. Expression of the miR-146 Family of miRNAs is Significantly Upregulated In Vitro after L. major Infection
The miR-146 subfamily of miRNAs includes several paralogs such as miR-146a-3p-5p and miR-146b-3p-5p located at different genomic positions. A considerable representation of several members of this family that were consistently upregulated across different time points was observed in our microarray-based expression analysis (P value adjusted to 1).
miR-146a-3p miRNA is two-fold upregulated in the 12 h sample as compared to the control 0 h sample.
Maximum expression of miR-146a-3p is observed at 12 and 24 h (for 12 h, ∼4.2 fold; for 24 h, ∼4 fold), and expression for miR-146a-5p is observed at 12 h only ( ∼4 fold) followed by a significant decline at 24 h (Table 2), indicating a possible regulatory role played by this noncoding RNA in human macrophages infected with L. major.
Table 2. Interpretation of Deregulated miRNAs.
| (a) | |||||
|---|---|---|---|---|---|
| miRNA | 0 h expression | 3 h expression | fold change | log 2 fold change | regulation |
| miR-146a-3p | 24.24871131 | 47.34272207 | 1.952380952 | 0.965234582 | NEUTRAL |
| miR-146b-3p | 246.8172401 | 230.9401077 | 0.935672515 | –0.09592442 | NEUTRAL |
| miR-146a-5p | 8644.665581 | 11634.76262 | 1.345889267 | 0.428559718 | NEUTRAL |
| miR-146b-5p | 15902.82449 | 23104.40307 | 1.452849026 | 0.538884792 | NEUTRAL |
| (b) | |||||
|---|---|---|---|---|---|
| miRNA | 0 h expression | 12 h expression | fold change | log 2 fold change | regulation |
| miR-146a-3p | 28 | 117 | 4.178571429 | 2.063009798 | UP |
| miR-146b-3p | 285 | 182 | 0.638596491 | –0.647023469 | NEUTRAL |
| miR-146a-5p | 9982 | 39,883 | 3.995491885 | 1.998373124 | UP |
| miR-146b-5p | 18,363 | 21,887 | 1.19190764 | 0.253272447 | NEUTRAL |
| (c) | |||||
|---|---|---|---|---|---|
| miRNA | 0 h expression | 6 h expression | fold change | log 2 fold change | regulation |
| miR-146a-3p | 16.97436036 | 18.14501362 | 1.068965972 | 0.096215929 | NEUTRAL |
| miR-146b-3p | 172.7747394 | 242.4833638 | 1.403465371 | 0.488993468 | NEUTRAL |
| miR-146a-3p | 6051.35947 | 8922.398061 | 1.474445223 | 0.560172226 | NEUTRAL |
| miR-146b-5p | 11,132.14926 | 9720.77866 | 0.873216702 | –0.195588371 | NEUTRAL |
| (d) | |||||
|---|---|---|---|---|---|
| miRNA | 0 h expression | 24 h expression | fold change | log 2 fold change | regulation |
| miR-146a-3p | 17.94244805 | 68.6639859 | 3.826901753 | 1.936176864 | UP |
| miR-146b-3p | 182.6584891 | 146.6912426 | 0.803222122 | –0.31612909 | NEUTRAL |
| miR-146a-5p | 6396.48273 | 16,864.81126 | 2.636575752 | 1.398665448 | UP |
| miR-146b-5p | 11,767.04191 | 12,670.06594 | 1.076741805 | 0.106672343 | NEUTRAL |
2.4. Pathway Enrichment Analysis
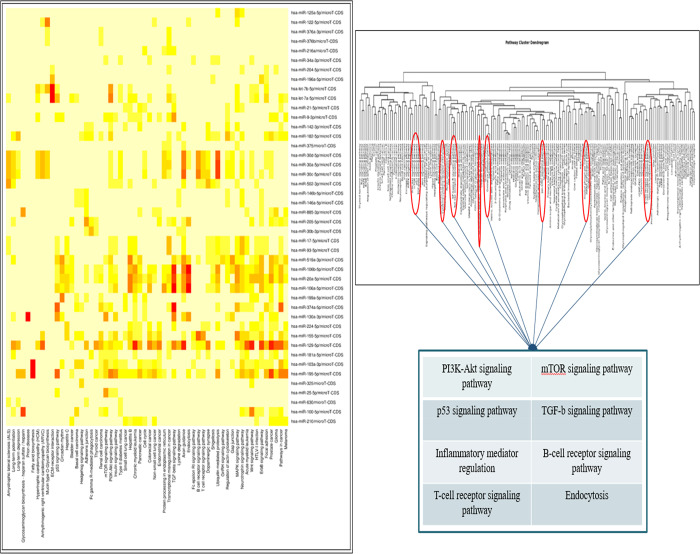
We obtained a total of 58 pathways enriched from our set of 44 miRNAs shown in Figure 4 using Diana Tools mirPath v.3 software, which uses a P value threshold of 0.05 as statistically significant. The GOSlim analysis segregated these pathways into different gene ontological processes, which fall under the three categories: biological processes, molecular function, and cellular component of gene products. The heatmap of the pathway intersection showed better enriched pathways in warmer colors. The heatmap showed involvement of key miRNAs in the immune and inflammatory response during Leishmania infection shown through Figure 4. For further analyzing miRNAs involved in enriched pathways with their interaction with a number of genes, respective P values and citations in the literature resulting from the mirPath v.3 search are tabulated and cited in Table 3. These enriched pathways were Fc-gamma R-mediated phagocytosis, mTOR signaling pathway, TGF-β signaling pathway, B-cell receptor signaling pathways, T-cell receptor signaling pathways, and MAPK signaling pathways. From a prior literature survey, miR-146a has shown its significance in modulating the immune and inflammatory responses during Leishmania infection.
Figure 4.
Heatmap generated and Pathway cluster dendrogram generated by DIANA Tools representing involvement of key miRNAs in inflammatory response such as Fc-gamma R-mediated phagocytosis, mTOR signaling pathway, TGF-β signaling pathway, B-cell receptor signaling pathways, T-cell receptor signaling pathways and MAPK signaling pathways.
Table 3. Pathway Enrichment Analysis Results (Enriched Pathways).
| target pathways | no. of miRNAs | no. of genes | P value | literature |
|---|---|---|---|---|
| Hippo signaling pathway | 40 | 101 | 1.64 × 10–08 | hsa04390 |
| TGF-β signaling pathway | 38 | 57 | 3.17 × 10–08 | PMC3697132 |
| FoxO signaling pathway | 32 | 92 | 2.72 × 10–06 | PMC5071566 |
| fatty acid biosynthesis | 14 | 8 | 1.3 × 10–07 | PMID:17363967 |
| ErbB signaling pathway | 41 | 64 | 1.49 × 10–06 | PMID:23994140 |
| apoptosis | 35 | 54 | 0.03254 | PMC1602394 |
| cAMP signaling pathway | 42 | 114 | 0.02943 | PMC3200087 |
| MAPK signaling pathway | 41 | 151 | 0.001084 | PMC4764696 |
| Ras signaling pathway | 42 | 136 | 0.0005395 | PMC2815228 |
| AMPK signaling pathway | 40 | 83 | 0.0002206 | PMC4349736 |
| PI3K-Akt signaling pathway | 43 | 201 | 0.0001588 | PMID:16889626 |
In our study, miR-146a is one of the key miRNAs from the miRNA regulatory network and is also depicting involvement in inflammatory signaling pathways, which are confirmed via heatmap generation.
Biological pathways significantly affected by miRNAs that were differentially expressed upon each intervention were identified using the hypergeometric distribution test. The P value for each pathway represented the statistical significance of the overlap between the differentially expressed mRNAs and the annotated genes in the pathway and was calculated as follows
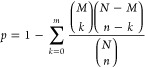 |
where N represents the total number of annotated genes in all pathways, n is the number of differentially expressed miRNAs, M is the number of annotated genes in a specific pathway, and m is the number of differentially expressed mRNAs annotated in a specific pathway. This method of hypergeometric distribution conjugates false discovery rate (FDR) correction and Bonferroni techniques. It verifies the statistical significance of P values. Our calculations showed that all the pathways had significant P values and can be taken into account for further studies.
From a prior literature survey, the TGF-β signaling pathway is majorly involved during Leishmania infection, aiding in the survival of the parasite inside macrophages. TGF-β is a known immunoregulator during Leishmania infection, inhibiting M1 activation and iNOS generation in macrophages.41 Thus, the TGF-β signaling pathway was considered to be important, although its P value during pathway enrichment analysis was not significant, considering Hippo signaling and fatty acid biosynthesis as these pathways are involved in cell proliferation and synthesis of lipids and not in the immune or inflammatory response.
2.5. Motif Identification
To have an insight into the occurrence of miRNAs in the Leishmania parasite, patterns of occurrence of specific sequences were searched against 24 human miRNAs evidently being involved in the immune and inflammatory response during Leishmania infection.7 The MEME Suite generated 10 motifs of 7 bp as output. These motifs identified may serve as a seed sequence for miRNA-like elements from the parasite or can act as a whole miRNA by binding to the host Argonaute protein. These motifs were in the 5′→3′ sense, as shown in Figure 5.
Figure 5.
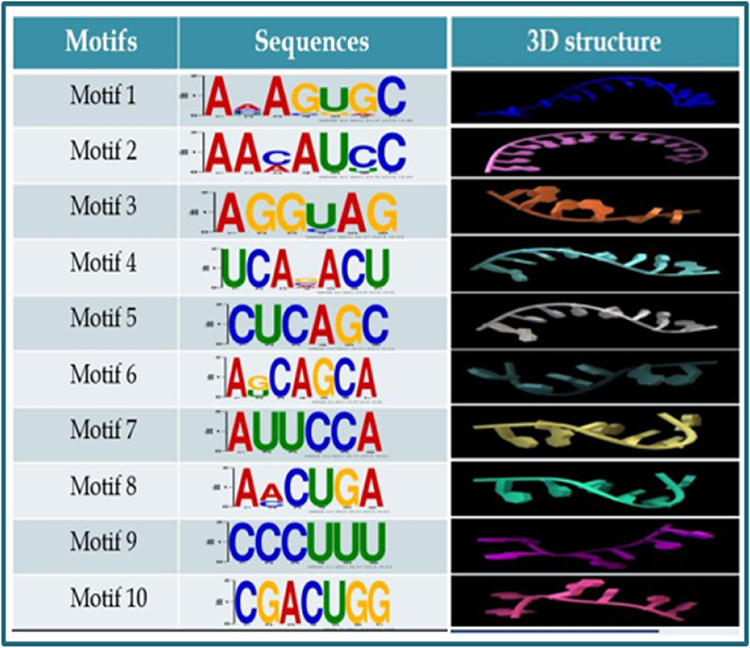
Ten motifs generated against 24 human miRNAs with their respective E values and length of the motifs and predicted 3D structures of discovered motifs generated by RNAComposer.
2.6. Structural Modeling of Motifs
For performing their activity, RNA adopts a specific structure that is important to all RNA-mediated processes. The generated 3D structure of each motif is derived from sequence and secondary structure topology. Evaluation of 3D structure motifs was done on the basis of a 3D structure energy, which was about −20 kcal/mol for each motif and its twisted helical form. The absence of palindromes and base complementarity within the sequences were a probable interpretation for this result. The predicted 3D structures of each predicted motif are represented in Figure 5.
2.7. Pattern Searching of Motif Sequences in the L. major Genome
To see the occurrence of human miRNA motifs in the Leishmania parasite, the pattern searching exercise was carried out using the KMP algorithm, which showed positive results. The human miRNA motifs were prevalent in the Leishmania major genome as well. Motif 6 (AGCAGCA), Motif 5 (CUCAGC) and Motif 9 (CCCUUU) were found to be most abundantly present as depicted in Figure 6. Motif 9 have sequence similar to a part of mir-146a sequence as per results showed by MEME output results.
Figure 6.
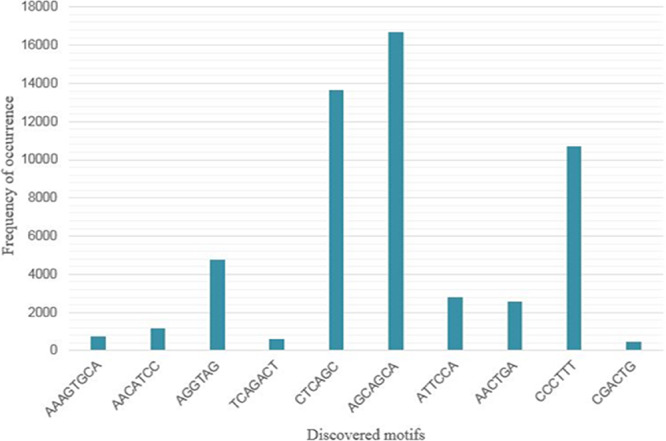
Graph depicting the frequency of occurrence of human miRNA motifs in the L. major genome. It is observed that motifs 6 (AGCAGCA), 5 (CTCAGC), and 9 (CCCTTT) are frequently occurring in the L. major genome.
2.8. Molecular Docking Analysis
Mature miRNAs in the cytoplasm bind to the Argonaute protein for stability and further recognition of their target mRNA. For representing the stability and processivity of miRNA-like elements in the Leishmania parasite, predicted motifs were blind-docked with human Argonaute 2 as a docking site for the motifs predicted is unknown. The AutoDock Vina results showed that the binding patterns of the 10 motifs to human Argonaute 2 had a stable conformity, and they were in congruence with the conformation of miRNA in the crystal structure of 4z4d, as shown in Figure 7. The configuration and the site of docking exhibited less variation. Taking into consideration their respective binding energies, motifs 5 and 7 were the most stable ligand receptors. These motifs are a representation of the miRNA-like elements likely to be processed in L. major; similar to miRNA structures, these are mainly single-stranded, facilitating a flipped or exposed base group interaction with proteins. The H-bonding and hydrophobic interactions are strong when these motifs are bound to the human Argonaute 2 protein.
Figure 7.
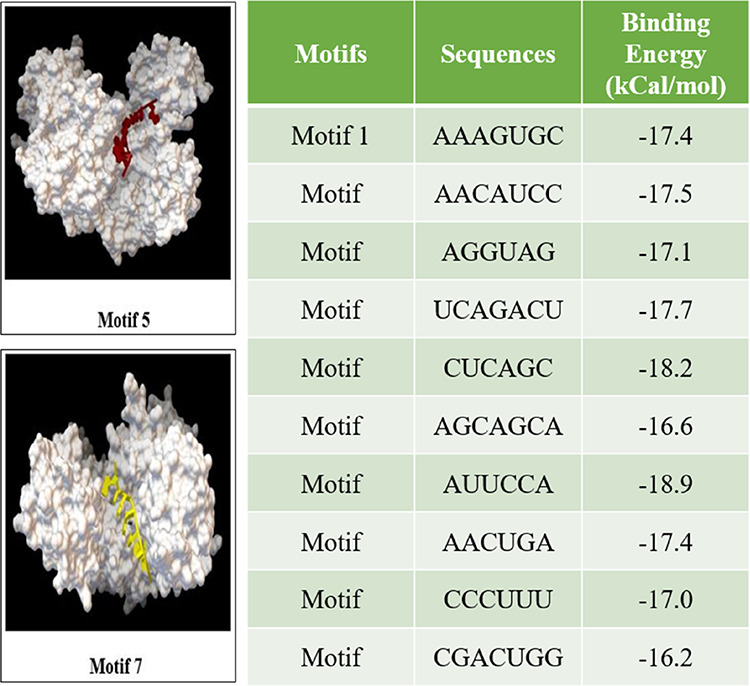
Human Argonaute 2 proteins were blind-docked with each motif from which motifs 5 and 7 had more stable ligand receptors, considering their binding energies (kCal/mol).
For modulating polarization of macrophages during Leishmania infection, i.e., conversion of M2 to M1 phenotype of macrophages, the TGF-β signaling pathway emerged as a potential target. Blocking the TGF-β signaling cascade can aid in killing of the intracellular parasite by balancing the activation of macrophages in a classical way.41−43 In relation to the miRNA of interest (i.e., miR-146a, mir-146a targets for Co-SMAD (SMAD 4), which is an essential signal transducer for receptors of the TGF-β superfamily), miR-146a will block the synthesis of SMAD 4 and due to the unavailability of SMAD 4 in the system would eventually block the TGF-β signaling cascade.
The TGF-β signaling pathway is composed of various SMAD proteins. The TGF-β ligand binding to TGFβRII phosphorylates various downstream SMAD proteins where association of SMAD 4 (Co-SMAD) and SMAD2/SMAD3 (R-SMAD) is essential for their translocation in the nucleus, binding to DNA elements for gene expression, as depicted in Figure 8. SMAD 4 is a potential target for miR-146a, and SMAD 7 is a common inhibitory SMAD protein that degrades TGFβRI and inhibits activation of R-SMAD. Targeting SMAD 4 by miR-146a and upregulating expression of SMAD 7 may help in blocking the TGF-β signaling-mediated gene expression.
Figure 8.
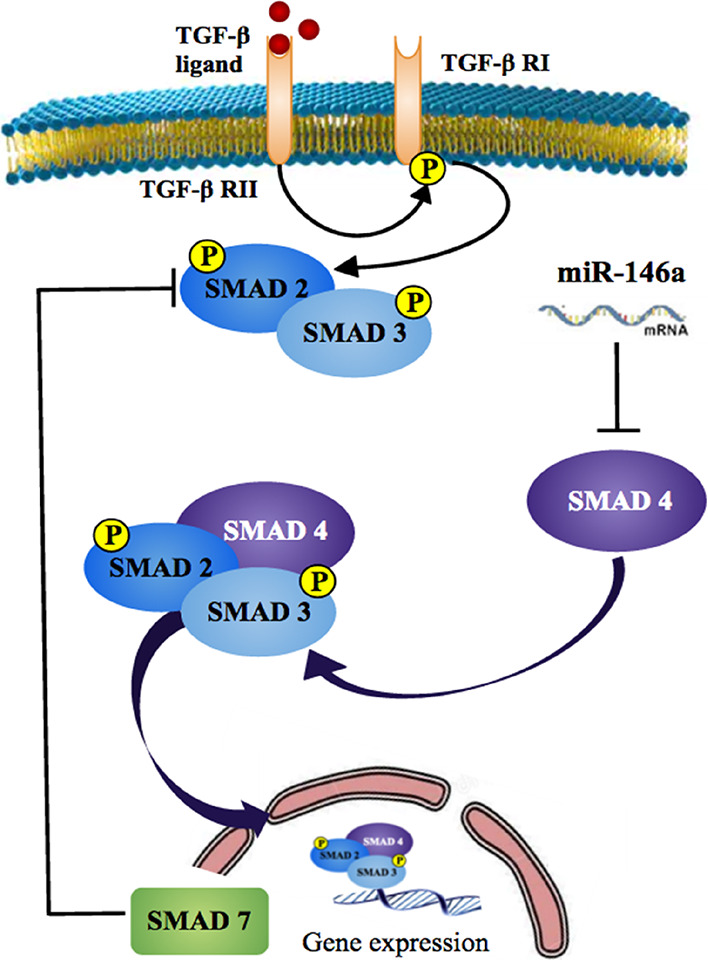
TGF-β signaling cascade in relation to the miRNA of interest (miR-146a).
To achieve this, a synthetic circuit for miR-146a and SMAD 7 was designed where miR-146a will target SMAD 4 (Co-SMAD) and SMAD 7 will target SMAD2/SMAD3 and TGFβRI, blocking the TGF-β signaling cascade in two ways.
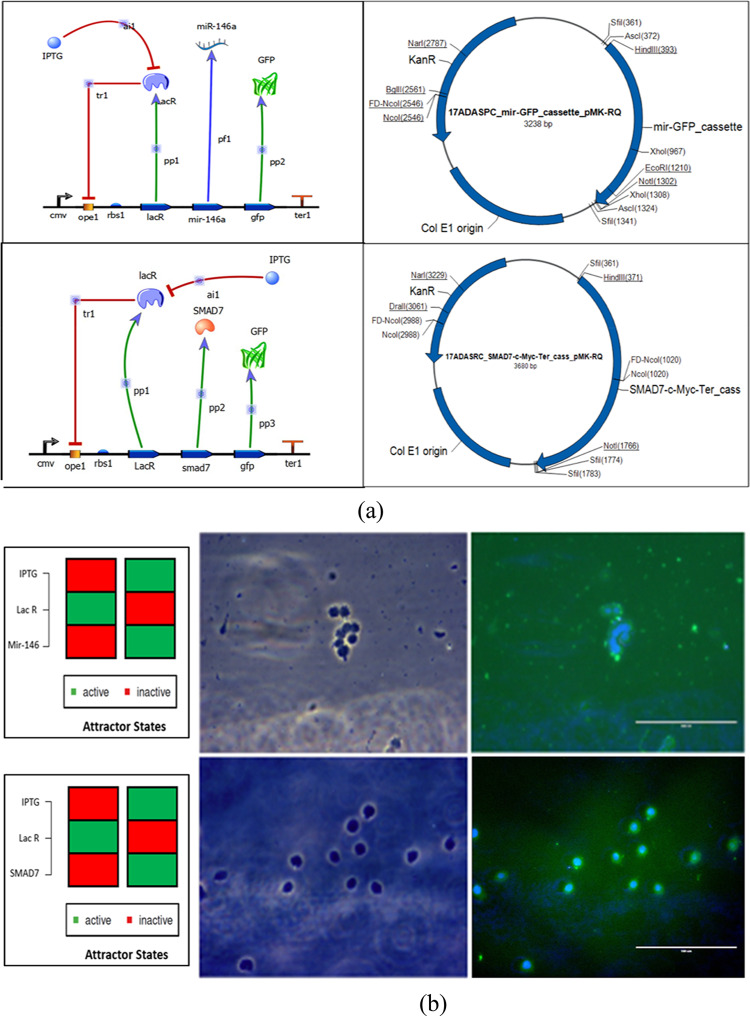
2.9. Construction of the Synthetic Circuit
Having mathematically analyzed this coherent feed forward circuit, we were interested in verifying whether the experimental data would exhibit the dynamics of the theory predicted. Hence, we went for the plasmid map construction of the biological circuit. Owing to size constraints, we decided to divide the synthetic module into two separate insets. One of the plasmids would be expressing the miRNA gene and GFP reporter protein, while the other plasmid would be expressing the target gene. Previously elaborated simulated annealing results showed that SMAD 7 attached to one of the key regulatory hubs was quite enriched in the network, so it could be one of the best possible targets to study miRNA-mediated gene suppression. Also, studies have pinpointed various functions of SMAD 7 in a large array of biological processes, including inflammation and immunity. It has both anti- and pro-inflammatory effects.42 SMAD 7 acts as an anti-apoptotic protein by targeting TGF-β signaling.43
The sequences for the CMV promoter, Lac operator, spacer, GFP fusion protein, and terminator were retrieved from the Registry of Standard Biological Parts database (http://parts.igem.org/Main_Page), SMAD 7 and c-myc tag sequences were obtained from the NCBI database, and the miR-146a sequence was retrieved from miRBase.
The designed circuits’ working mechanism is based on the Lac operon system where, in the OFF state, Lac R will remain bonded to the operator region inhibiting the expression of mir-146a and SMAD 7, while in the presence of an inducer (IPTG), Lac R will bind to the inducer, and the circuit will be in the ON state, expressing mir-146a and SMAD 7 with GFP as the reporter protein, as shown in Figure 9a. The synthetic circuits were procured in the form of plasmids from GeneArt (Invitrogen), and their representative maps are shown in Figure 9. To check the reliability and workability of the designed construct, transfection studies were conducted where THP-1 cells were transfected with each respective plasmid for 24 h using Lipofectamine 3000. Production of GFP confirmed the working condition of the designed synthetic circuit, as shown in Figure 9b.
Figure 9.
(a) Synthetic circuit design for miR-146a, SMAD 7 with their respective plasmid maps and (b) Transfection study on THP1 cells transfected with miR-146a and SMAD7 designed synthetic circuit via Lipofectamine 3000.
3. Conclusions
In the context of leishmaniasis, a robust inter-miRNA regulatory network generated along with pathway enrichment analysis, profiling of miRNA expression, differential expression of miRNAs, heatmap generation, and a pathway cluster dendrogram gave us a big picture of key miRNAs with their putative roles in the immune and inflammatory response. The resulting miR-146a was considered to have a manipulative role, targeting the TGF-β signaling cascade in a way to aid killing of the parasite inside macrophages.
To modulate polarization of macrophages from the M2 to M1 phenotype, blocking the TGF-β signaling pathway was taken under consideration as TGF-β acts as an immunoregulator, inhibiting activation of macrophages in a classic way (M1 phenotype).
Synthetic circuits were designed such as miR-146a and SMAD7 to achieve the said purpose. miR-146a has a target for SMAD 4 (Co-SMAD), and an increase in miR-146a in the system would directly lead to the unavailability of SMAD 4, resulting in the blockage of the TGF-β signaling pathway in one way. SMAD 7 is an inhibitory SMAD protein that targets SMAD2/SMAD3 (R-SMAD) and TGFβRI and thus may block TGF-β signaling in another way.
Biogenesis of miRNAs in the Leishmania parasite is not known yet, but their gene regulation and abnormal presence in a host during infection is as such aiding in its survival in macrophages, which hints for an alternative pathway with a similar function. Our in silico study prove the existence of miRNA-like elements in the Leishmania parasite, and existing literature evidence suggests that other related parasites during invasion secrete their native miRNA into the host cell, which then takes up the host machinery, i.e., the RISC complex for its functionality.45,46 Furthermore, we evoke miR-146a acting as a feedback loop with relevance of inflammation.
The current perspectives may be with respect to the multinuclease complex protein Argonaute that has not been characterized in the Leishmania proteome. Our study is in congruence with the idea that miRNA biogenesis is present in the parasite; thus, there is a possibility of Argonaute 2 (Ago2)-like proteins to be functionally active in the parasite, which is yet to be annotated and characterized. Therefore, identification and functional annotation of an Argonaute 2-like protein in Leishmania species could be a significant area of future research.
4. Experimental Section
4.1. Curation of miRNA Data
After a thorough literature survey related to protozoan diseases, miRNA intervention in such diseases, more specifically in leishmaniasis, helped in curating miRNAs with their putative roles in inflammation and immune responses in the human host.4,12,18−27,33,34
4.1.1. Construction of the Initial Network
An extensive literature survey led us to the observation that there are several miRNAs, which have been found to play regulatory roles in many immunological responses triggered during Leishmania infection. The expression profiles of miRNAs in human macrophages in response to Leishmania infection have also been reported in various instances.4 Based on their roles in inflammation and immune signaling, 48 miRNAs were selected for our study. For construction of the network, targets of shortlisted miRNAs were tabulated from mirWALK and miRBase, which are biological databases that act as an archive of microRNA sequences and annotations. With source (miRNA) and target information tabulated, construction of the network through Cytoscape v.3.5.0 was done.5 Cytoscape is an open source software project for integrating biomolecular interaction networks with high-throughput expression data and other molecular states into a unified conceptual framework. Cytoscape’s core software component provides basic functionality for integrating arbitrary data on the graph, a visual representation of the graph and integrated data, selection and filtering tools, and an interface to external methods implemented as apps.5 The inter-miRNA regulatory network thus generated was low on connectivity.
4.1.2. Simulated Annealing
Next, the tabulated source and target table were imported into Cytoscape v.2.85 to generate a robust network of miRNAs targeting proteins or transcription factors, which are upstream or downstream components of the pathways enriched and that have been hypothesized to play key regulatory roles during Leishmania infection. In the case of Cytoscape, the parameters taken into consideration are the clustering coefficient, betweenness centrality, node centrality, neighborhood connectivity, eccentricity, out degree, etc. A simulated annealing layout revealed an extensively interconnected network. In doing so, the most significant nodes that are also the connecting links between most of the cascades were segregated, making the data coherent and lucid for understanding.31
4.2. Pathway Enrichment Analysis
For this purpose, mirPATH v.3.0 under DIANA Tools6 was used. DIANA-mirPath v3.0 (http://www.microrna.gr/mirPathv3) is an online software suite dedicated to the assessment of miRNA regulatory roles and the identification of controlled pathways. The DIANA-mirPath v3.0 database and functionality has been significantly extended to support all analyses for KEGG molecular pathways as well as multiple slices of Gene Ontology (GO) in seven species (Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans, Gallus gallus, and Danio rerio). Users of DIANA-mirPath v3.0 can harness this wealth of information and substitute or combine the available in silico-predicted targets from DIANA-microT-CDS, or a unique feature of DIANA-mirPath v3.0 is its redesigned reverse search module, which enables users to identify and visualize miRNAs significantly controlling selected pathways or belonging to specific GO categories based on in silico or experimental data. Only those pathways that were relevant to the study were considered; 77 enriched pathways such as the functional and inflammatory response during Leishmania invasion were considered for further analysis. These included Hippo signaling, TGF-β signaling, MAPK pathway, ErbB signaling, FoxO signaling, mTOR signaling, Wnt signaling, PI3K-Akt signaling, Rap1 signaling, AMPK signaling, sphingolipid signaling, Ras signaling, MAPK signaling, endocytosis, cAMP signaling, and apoptosis pathways.
4.3. Statistical Significance of Pathways
Statistical significance was determined by IBM SPSS (https://www-01.ibm.com/support/docview.wss?uid=swg21476197 IBM Corp. Released 2016. IBM SPSS Statistics for Windows, Version 24.0. Armonk, NY: IBM Corp.).
4.4. Motif Identification
The mature miRNA sequences were downloaded from the miRdB database. For motif discovery, MEME Suite v.4.11.3 was used. The MEME Suite is a software toolkit with a unified web server interface that enables users to perform four types of motif analysis: motif discovery, motif–motif database searching, motif-sequence database searching, and assignment of function.7 The sequence file was uploaded into the web interface, and the parameters for the search were defined, viz., motif length,8−12 background model (zero-order model of sequences), number of occurrences of the motifs, etc. Six motifs were selected, which were six to eight nucleotides long. By exploiting the regular expression, multilevel consensus sequence, and site-specific data, we computed all the permutations of these motifs and recorded 24 such combinations.
4.5. Pattern Search against the L. major Genome
To determine the occurrence of human miRNA motifs in the Leishmania genome, the Knuth–Morris–Pratt (KMP) algorithm was used. It is a straightforward string searching algorithm, which finds all the occurrences of a pattern of length “m” within a text of length “n” in O (m + n) units of time without “backing-up” the input text. The algorithm needs only O (m) locations of internal memory if the text is read from an external file and only O (log m) units of time elapse between consecutive single-character units. The same was applied to the motif discovery problem, and we used Python for programming purposes. The output specified the position index (residue number) and the number of prevalence of a motif in a particular chromosome sequence. L. major strain Friedlin genome sequences were obtained from the NCBI database. From this data, we could derive the motifs with maximum incidence in Leishmania and the location indices might indicate regions probable of encoding for their miRNAs. The string search analysis allowed us to choose 10 motifs, which had the highest frequency of occurrence, to be considered for subsequent studies.
4.6. miRNA Modeling
For any kind of molecular- and interaction-based analysis, it is necessary to assign a well-defined structure to each of the predicted motifs.32 We used RNAfold, which is one of the core programs of the Vienna RNA package to predict the minimum free energy (MFE) secondary structure of single-motif sequences utilizing a dynamic programming algorithm.8 The dot-bracket notation output was used for 3D structure prediction. We accomplished this using RNAComposer, which is a fully automated RNA structure prediction tool. The modeled structures were downloaded as pdb files, which were subsequently used for molecular procedures.44
4.7. Molecular Docking
The mechanism of gene regulation by miRNA occurs when mature miRNAs are coupled with a multiple protein nuclease complex called the RNA-induced silencing complex (RISC). Once incorporated into an RISC, the miRNA is situated to regulate the target genes by degradation of the mRNA through direct cleavage or by inhibiting protein synthesis. The Argonaute 2 protein is a major component of the RISC complex, and its interaction with miRNA is a key ingredient of miRNA biogenesis.
For determination and validation of processivity of the modeled miRNA motifs, molecular docking techniques were used. AutoDock Vina,9 which is a program for molecular docking and virtual screening to dock the miRNA ligands to the human Argonaute 2 protein holding the PDB ID 4z4d, was used. The blind docking technique was used because all that was vividly known was the structure of the ligand and the macromolecule. The grid box was specified to cover the entire binding pocket of the protein. The docking scores were obtained, and the best docking pose for each of the ligands were considered.
4.8. Cell Culture
The human macrophage cell line, THP-1, was obtained from the NCCS Cell Repository, Pune and cultured in RPMI-1640 supplemented with 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 mg/mL) in a humidified incubator containing 5% CO2 at 37 °C.
4.9. Profiling of miRNA Expression in THP-1 Cells Infected with L. major
To investigate the modulation pattern of host miRNAs after Leishmania infection, THP-1 cells were infected with stationary-phase promastigotes of L. major for 0, 3, 6, 12, and 24 h. RNA samples were extracted from Leishmania-infected THP-1 cells post-infection. RNA derived from infected THP-1 cells (0 h) served as a control. The percentage of infected cells and parasite load at all time points was examined via microscopy, and infection was consistent across various time points.
4.10. Differential Expression (DE) Analysis
Read count across all known and novel miRNAs were generated by taking the count of reads aligning to a particular miRNA. This information is useful in understanding the expression pattern of miRNAs. Differential expression analysis was carried out using the DESeq tool. Variations in the reads are normalized by the library normalization method opted from the DESeq library. DESeq calculates the size factor, and each read count is normalized by dividing with the size factor. Mean-normalized read counts of the samples in a given condition are used for DGE calculation and the heatmap. To understand the regulation of expression between the samples, a log 2 fold of 1 was used as a cutoff. miRNAs greater than 1 were considered as “UP” regulated, miRNAs less than −1 were considered as “DOWN”, and those between 1 and −1 were flagged as “NEUTRAL”. Heatmaps were generated for each set of DGE using the top 48 miRNAs (Figure 3) (up/downregulated). An example of up- and downregulated miRNA has been shown in (Table 2). Fold changes are calculated based on “expression of the treated sample/expression of the control sample”.
4.11. Transfection Studies
miR-146a functional analyses were performed using designed synthetic circuits, i.e., miR-146a and SMAD 7. Prior to transfection, the cells were grown in the fresh culture medium at a concentration of 1 × 106 cells/mL with a PMA of 20 ng/mL. The following day, the THP-1 cells were transfected with an miR-146a- and SMAD 7-designed synthetic circuit using Lipofectamine 3000 (Invitrogen), according to the manufacturer’s instructions. Following transfection, the cells were allowed to recover for 6 h at 37 °C and a fresh RPMI-1640 medium was changed. Transfection was visualized under a fluorescence microscope after DAPI staining.
Acknowledgments
We thank intramural funding support from Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India. Prajakta Nimsarkar acknowledges her DBT-SRF (DBT/JRF/15/AL/607) and Prajakta Ingale acknowledges her ICMR SRFship (2019-3999/CMB/BMS). Authors would also like to thank NCCS Cell Repository and to the Director, NCCS for supporting the Bioinformatics and High Performance Computing Facility (BHPCF) at National Center for Cell Science (NCCS), Pune, India.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c01502.
Python codes used for the KMP algorithm for motifs pattern searching against the L. major genome (Supporting Algorithm S1), read count matrix of miRNAs differentially expressed during L. major infection on THP-1 cells at different time points (Table S1), and normalized values for miRNAs differentially expressed during L. major infection on THP-1 cells at different time points where values are average expression differential fold change (FC) versus uninfected cells (Sample_0Hr) of the same type (Table S2) (PDF)
Author Contributions
Prajakta Nimsarkar performed the in silico and in vitro experimentation and wrote the manuscript. Prajakta Ingale helped in figures and participated in comprehension of the datasets. Shailza Singh laid the design idea, conceptualization of the project, and editing of the manuscript. Prajakta Nimsarkar, Prajakta Ingale and Shailza Singh approved the current version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Akhoundi M.; Kuhls K.; Cannet A.; Votýpka J.; Marty P.; Delaunay P.; et al. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and Sandflies. PLoS Negl. Trop. Dis. 2016, 10, e0004349 10.1371/journal.pntd.0004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N. Re-thinking miRNA–mRNA interactions: intertwining issues confound target discovery. BioEssays 2015, 37, 379–388. 10.1002/bies.201400191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y.; Schober A. MicroRNA regulation of macrophages in human pathologies. Cell. Mol. Life Sci. 2016, 73, 3473–3495. 10.1007/s00018-016-2254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire J.; Mkannez G.; Guerfali F. Z.; Gustin C.; Attia H.; Sghaier R. M.; et al. MicroRNA expression profile in human macrophages in response to Leishmania major infection. PLoS Negl. Trop. Dis. 2013, 7, e2478 10.1371/journal.pntd.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos I. S.; Zagganas K.; Paraskevopoulou M. D.; Georgakilas G.; Karagkouni D.; Vergoulis T.; Dalamagas T.; Hatzigeorgiou A. G. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L.; Boden M.; Buske F. A.; Frith M.; Grant C. E.; Clementi L.; Ren J.; Li W. W.; Noble W. S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M.; Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981, 9, 133–148. 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H. F.; Zhou B.; Liu G.; Chen C.-Z. Micromanagement of the immune system by microRNAs. Nat. Rev. Immunol. 2008, 8, 120–130. 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and functionA comprehensive review that describes the genomics, biogenesis and mechanism of action of miRNAs. Cell 2004, 116, 281–297. 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel D. P.; Chen C.-Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bushati N.; Cohen S. M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Lodish H. F.; Zhou B.; Liu G.; Chen C.-Z. Erratum: Micromanagement of the immune system by microRNAs. Nat. Rev. Immunol. 2008, 8, 238. 10.1038/nri2280. [DOI] [PubMed] [Google Scholar]
- Ambros V. The evolution of our thinking about microRNAs. Nat. Med. 2008, 14, 1036–1040. 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. The perfect storm of tiny RNAs. Nat. Med. 2008, 14, 1041–1045. 10.1038/nm1008-1041. [DOI] [PubMed] [Google Scholar]
- Mendell J. T.; Olson E. N. MicroRNAs in stress signaling and human disease. Cell 2012, 148, 1172–1187. 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C.; Rajewsky K. MicroRNA control in the immune system: basic principles. Cell 2009, 136, 26–36. 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- O’Connell R. M.; Rao D. S.; Baltimore D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012, 30, 295–312. 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- Landgraf P.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkerova M.; Belickova M.; Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur. J. Haematol. 2008, 81, 304–310. 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- Lindsay M. A. microRNAs and the immune response. Trends Immunol. 2008, 29, 343–351. 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Wu H.; Neilson J. R.; Kumar P.; et al. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One 2007, 2, e1020 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 2009, 136, 215–233. 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov K. D.; Boldin M. P.; Chang K. J.; Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 12481–12486. 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham R.; Yarden Y. Regulation of signalling by microRNAs. Biochem. Soc. Trans. 2012, 26–30. 10.1042/BST20110623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X.; Xia W.; Wenzhong H. A modeled dynamic regulatory network of NF-κB and IL-6 mediated by miRNA. Biosystems. 2013, 114, 214–218. 10.1016/j.biosystems.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Agarwal G. Towards Logical Designs In Biology. Resonance 2007, 29–38. 10.1007/s12045-007-0026-1. [DOI] [Google Scholar]; Albert I et al. Boolean network simulations for life scientists (2008) Biological Medicine.
- Murray H. W.; Berman J. D.; Davies C. R.; Saravia N. G. Advances in leishmaniasis. Lancet 2005, 366, 1561–1577. 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- Olivier M.; Gregory D. J.; Forget G. Subversion mechanisms by which Leishmania parasites can escape the host immune response: a signaling point of view. Clin. Microbiol. Rev. 2005, 18, 293–305. 10.1128/CMR.18.2.293-305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S.; Heymans K.; Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Krek A.; Grün D.; Poy M. N.; Wolf R.; Rosenberg L.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Calegari-Silva T. C.; Pereira R. M. S.; De-Melo L. D. B.; Saraiva E. M.; Soares D. C.; et al. NF-κB-mediated repression of iNOS expression in Leishmania amazonensis macrophage infection. Immunol. Lett. 2009, 127, 19–26. 10.1016/j.imlet.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Bogdan C. Mechanisms and consequences of persistence of intracellular pathogens: leishmaniasis as an example. Cell. Microbiol. 2008, 10, 1221–1234. 10.1111/j.1462-5822.2008.01146.x. [DOI] [PubMed] [Google Scholar]
- Tomiotto-Pellissier F.; Bortoleti B. T. D. S.; Assolini J. P.; Gonçalves M. D.; Carloto A. C. M.; Miranda-Sapla M. M.; Conchon-Costa I.; Bordignon J.; Pavanelli W. R. Macrophage Polarization in Leishmaniasis: Broadening Horizons. Front. Immunol. 2018, 2529. 10.3389/fimmu.2018.02529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard K.; et al. The role of NF-κB activation during protection against Leishmania infection. Int. J. Med. Microbiol. 2012, 302, 230–235. 10.1016/j.ijmm.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Lawrence T. The Nuclear Factor NF-κB Pathway in Inflammation. Cold Spring Harbor Perspect. Biol. 2009, 1, a001651. 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo G. C.; Ansari M. Y.; Dikhit M. R.; Gupta N.; Rana S.; Das P. Computational Identification of microRNA-like Elements in Leishmania major. MicroRNA 2013, 225–230. 10.2174/2211536602666131203232422. [DOI] [PubMed] [Google Scholar]
- Gregory D. J.; Godbout M.; Contreras I.; Forget G.; Olivier M. A novel form of NF- κB is induced by Leishmania infection: Involvement in macrophage gene expression. Eur. J. Immunol. 2008, 38, 1071–1081. 10.1002/eji.200737586. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Wamg J. K. The functional analysis of MicroRNAs involved in NF-kB signaling. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 1764–1774. [PubMed] [Google Scholar]
- Barral A.; Teixeira M.; Reis P.; Vinhas V.; Costa J.; Less H.; Bittencourt A. L.; Reed S.; Carvalho E. M.; Barral-Netto M. Transforming growth factor-beta in human cutaneous leishmaniasis. Am. J. Pathol. 1995, 147, 947–954. [PMC free article] [PubMed] [Google Scholar]
- Zhu L.; Chen S.; Chen Y. Unraveling the biological functions of Smad7 with mouse models. Cell Biosci. 2011, 1, 44. 10.1186/2045-3701-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arase M.; Horiguchi K.; Ehata S.; et al. Transforming growth factor-β-induced lncRNA-Smad7 inhibits apoptosis of mouse breast cancer JygMC(A) cells. Cancer Sci. 2014, 105, 974–982. 10.1111/cas.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesiada M.; Purzycka K. J.; Szachniuk M.; Blazewicz J.; Adamiak R. W. Automated RNA 3D Structure Prediction with RNAComposer. Methods Mol. Biol. 2016, 1490, 199–215. 10.1007/978-1-4939-6433-8_13. [DOI] [PubMed] [Google Scholar]
- Buck A. H.; Coakley G.; Simbari F.; McSorley H. J.; Quintana J. F.; Le Bihan T.; Kumar S.; Abreu-Goodger C.; Lear M.; Harcu Y.; Ceroni A.; Babaya S. A.; Blaxter M.; Ivens A.; Maizels R. M. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 2014, 5, 5488–5488. 10.1038/ncomms6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coakley G.; Maizels R. M.; Buck A. H. Exosomes and other extracellular vesicles: the new communicators in parasite infections. Trends Parasitol. 2015, 31, 477–489. 10.1016/j.pt.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.