Abstract
Potassium (K) is essential for plant growth and development. Here, we show that the KUP/HAK/KT K+ transporter KUP9 controls primary root growth in Arabidopsis thaliana. Under low‐K+ conditions, kup9 mutants displayed a short‐root phenotype that resulted from reduced numbers of root cells. KUP9 was highly expressed in roots and specifically expressed in quiescent center (QC) cells in root tips. The QC acts to maintain root meristem activity, and low‐K+ conditions induced QC cell division in kup9 mutants, resulting in impaired root meristem activity. The short‐root phenotype and enhanced QC cell division in kup9 mutants could be rescued by exogenous auxin treatment or by specifically increasing auxin levels in QC cells, suggesting that KUP9 affects auxin homeostasis in QC cells. Further studies showed that KUP9 mainly localized to the endoplasmic reticulum (ER), where it mediated K+ and auxin efflux from the ER lumen to the cytoplasm in QC cells under low‐K+ conditions. These results demonstrate that KUP9 maintains Arabidopsis root meristem activity and root growth by regulating K+ and auxin homeostasis in response to low‐K+ stress.
Keywords: auxin transport, KUP9, low‐K+ stress, meristem cell, root growth
Subject Categories: Development & Differentiation, Membrane & Intracellular Transport, Plant Biology
The ER‐localized transporter KUP9 regulates K+ and auxin homeostasis in Arabidopsis quiescent center (QC) cells in response to low‐K+ stress, thus regulating meristem activity and primary root growth.

Introduction
Potassium (K+) is essential for plant growth and development, as it plays crucial roles in many physiological processes in plant cells 1. In soils, relatively little free K+ is available for plants 2, 3, and the K+ is heterogeneously distributed 4. Plants perceive external K+ levels and modulate their growth accordingly, inducing physiological and morphological alterations that help them adapt to fluctuating external K+ levels 5, including changes in root architecture 6, 7, 8.
Many of the changes in root architecture involve crosstalk between low‐K+ signaling and auxin signaling. In response to low K+, auxin signaling regulates primary root growth, lateral root formation, and root hair emergence/growth 4, 5, 6, 7, 9, 10, 11. Primary root growth relies on root meristem activity, which is mainly controlled by auxin levels 12, 13. Intracellular auxin levels are controlled by polar auxin transport, auxin compartmentation, and auxin metabolism. Polar auxin transport in roots results in the formation of an auxin gradient in the root tip, which maintains meristem activity 13, 14. The long‐distance auxin transport in roots is mediated by many different auxin carriers, such as the auxin efflux carrier PIN proteins 15, 16. Auxin compartmentalization and metabolism further fine‐tune auxin levels: When free indole‐3‐acetic acid (IAA) in the cytoplasm triggers downstream auxin signaling events 13, 17, the excess IAA can be conjugated to amino acids such as aspartate and glutamate, forming the IAA conjugates indole‐3‐acetyl‐aspartate (IA‐Asp) and indole‐3‐acetyl‐glutamate (IA‐Glu) and stored in the endoplasmic reticulum (ER) 17, 18, 19, 20, 21. The auxin efflux carriers PIN5 and PIN8 are localized in the ER, where they regulate auxin compartmentation and modulate auxin metabolism 18, 21.
The plant root tip contains mitotically inactive central cells termed the quiescent center (QC); the QC is surrounded by different types of stem cells that form a stem cell niche 22, 23, 24. The QC is essential for specification of the stem cell niche and maintenance of the undifferentiated state of stem cell initials. The auxin maximum near the QC plays a major role in the maintenance of QC and meristem activity 14, 25. The AP2‐domain transcription factors PLETHORA1 (PLT1) and PLT2 regulate QC specification and meristem activity and PLT1/2 transcript levels respond to auxin concentration 13, 14, 15, 26, 27, 28, 29, 30. The GRAS transcription factors SHORT‐ROOT (SHR) and SCARECROW (SCR) also play critical roles in QC specification and maintenance of meristem cells 25, 26, 30, 31, 32, 33, 34. However, the mechanisms by which the low‐K+ response regulates root meristem activity are still unclear.
K+ transporters are vital to regulating K+ levels and the response to low K+. The K+ in soils is absorbed by plant roots and transported into plants through different K+ channels and K+ transporters 5, 35. KUP/HAK/KT family members represent the most important K+ transporters in plants and participate in diverse processes, such as K+ uptake, K+ homeostasis, osmotic regulation, and cell growth 36, 37, 38, 39, 40. Based on the relationship between K+ uptake activity and external pH, the transporters in this family are suggested to function as H+/K+ symporters 41. Indeed, the HAK5 transporter from Venus flytrap (Dionaea muscipula) has been established as a proton‐driven, high‐affinity H+/K+ symporter, whose K+ transport activity is dependent on phosphorylation status 42.
Our previous study indicated that under low‐K+ conditions, auxin transport toward the root tip decreases, thus inhibiting primary root growth 43. The reduced auxin transport is due to the degradation of the auxin efflux carrier PIN1, and this degradation requires the K+ channel AKT1 43. In addition, the KUP/HAK/KT K+ transporter TRH1/KUP4 is involved in the regulation of root hair development and root gravitropism through auxin signaling 37, 44. Mutation of TRH1 leads to the mislocalization of PIN1 and therefore affects auxin transport 45. Although these studies have demonstrated a relationship between low‐K+ signaling and auxin signaling, how plants sense external K+ levels and regulate root growth via auxin signaling remains to be elucidated.
In the present study, we characterized the Arabidopsis KUP/HAK/KT K+ transporter KUP9, which regulates root meristem activity and modulates primary root growth in response to low‐K+ stress. Our study demonstrates that ER‐localized KUP9 directly mediates K+ and auxin efflux from the ER lumen to the cytosol to maintain K+ and auxin homeostasis in the QC cells under low‐K+ conditions, thus regulating meristem activity and primary root growth.
Results
The primary root growth of kup9 mutants is inhibited under low‐K+ conditions
Plants sense external K+ levels and modulate their growth accordingly. When plant roots encounter low‐K+ conditions, primary root growth is inhibited by reduction of root cell turgor and root tip auxin levels (43; Fig EV1). However, root growth does not cease completely; the auxin concentration in root tip can be maintained at low levels for a long time, especially in the QC cells (Fig EV1). These low auxin levels may be essential for the maintenance of meristem activity and root growth under low‐K+ conditions (Fig EV1). Therefore, plants can survive in K+‐deficient conditions.
Figure EV1. Primary root growth and auxin levels in the Arabidopsis response to LK conditions.
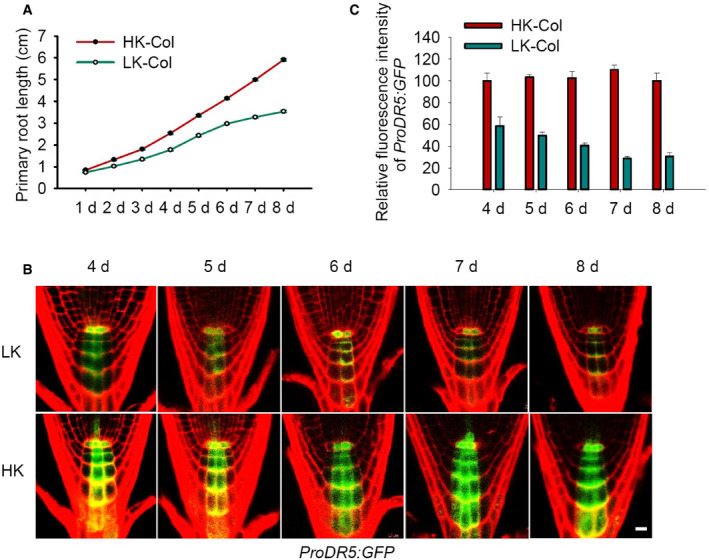
- Kinetic assay of primary root growth of wild‐type (Col) seedlings. Seeds were germinated on HK medium for 4 days, and then, the seedlings were transferred to LK or HK medium for the indicated times (n = 50, individual plants).
- Auxin levels in root tips indicated by the ProDR5:GFP marker line. Seeds were germinated on HK medium for 4 days, and then, the seedlings were transferred to LK or HK medium for the indicated times. Scale bar, 10 μm.
- Quantification of GFP fluorescence in the root tips shown in (B). Data are means ± SE (n = 20, individual plants).
Source data are available online for this figure.
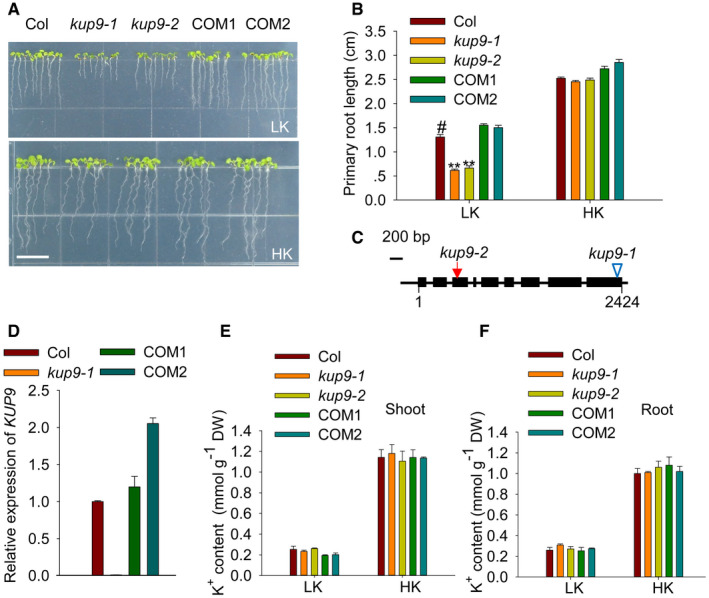
To investigate how plants maintain root growth under low‐K+ conditions, we collected Arabidopsis T‐DNA insertion mutants of nine K+ transporter genes (akt1, hak5, kup4, kup7, kup9, kup10, kup11, skor, and nrt1.5) and tested their root growth phenotypes on low‐K+ (LK, 50 μM) medium. All these genes are mainly expressed in Arabidopsis roots. We identified only the KUP/HAK/KT transporter mutant, kup9‐1, which showed a short‐root phenotype on LK medium. The primary root length of the kup9‐1 mutant was shorter than that of wild type (Col), when plants were germinated and grown on LK medium for 7 days (Fig 1A and B). The kup9‐1 allele is a T‐DNA insertion mutant; the insertion site is in the last exon of KUP9 (Fig 1C) and disrupts KUP9 transcription (Fig 1D).
Figure 1. The primary root growth of kup9 mutants is inhibited under low‐K+ conditions.
-
APhenotypes of wild type (Col), kup9 mutants (kup9‐1 and kup9‐2), and the kup9‐1/ProKUP9:KUP9 complementation lines (COM1 and COM2). Seeds were germinated and grown on low‐K+ (LK, 50 μM) or high‐K+ (HK, 5 mM) medium for 7 days. Scale bar, 1 cm.
-
BPrimary root length of the plants tested in (A). Data are means ± SE (n = 25, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
-
CDiagram of KUP9 gene structure. The T‐DNA insertion site in the kup9‐1 mutant and the mutant site in the kup9‐2 mutant are indicated by the triangle and arrow, respectively. The filled boxes and lines represent exons and introns, respectively.
-
DRT–qPCR analysis of KUP9 transcript levels in various materials. Data are means ± SE (n = 3, biological replicates; each replicate contains 120 individual plants).
-
E, FK+ content of the various plant materials tested in (A). Data are means ± SE (n = 3, biological replicates; each replicate contains 120–150 individual plants).
Source data are available online for this figure.
To verify that the mutation in KUP9 causes the short‐root phenotype, we constructed a CRISPR/Cas9 mutant, kup9‐2. We identified two single‐base insertions in the kup9‐2 mutant, which caused early termination of KUP9 translation (Fig 1C; Appendix Fig S1A). The primary root growth of kup9‐2 mutants was also inhibited on LK medium (Fig 1A and B). When grown on high‐K+ (HK, 5 mM) medium, neither of the two kup9 mutants showed a short‐root phenotype (Fig 1A and B). In addition, the short‐root phenotype of kup9 mutants could be rescued when the K+ concentration in the medium was increased (Appendix Fig S1B and C).
In addition, we constructed the complementation lines kup9‐1/ProKUP9:KUP9 (COM1 and COM2), in which the transcript levels of KUP9 were restored and even higher than that of wild‐type plants (Fig 1D). The primary root lengths of these two independent transgenic lines (COM1 and COM2) were also restored to the wild‐type level when grown on LK medium (Fig 1A and B). Therefore, the short‐primary‐root phenotype was caused by the loss of function of KUP9.
KUP9 is predicted to function as a K+ transporter; therefore, the short‐root phenotype might be due to K+ deficiency in the mutant roots. However, both the shoot and root K+ content in the two mutant lines were not significantly reduced or changed compared with wild type in plants grown on HK or LK medium (Fig 1E and F). Consistent with this finding, in a K+‐depletion assay the kup9 mutants showed similar potassium uptake rates compared with wild type (Fig EV2A).
Figure EV2. K+ uptake analyses of kup9 mutants and phenotypes of different K+ transporter mutants under LK conditions.
-
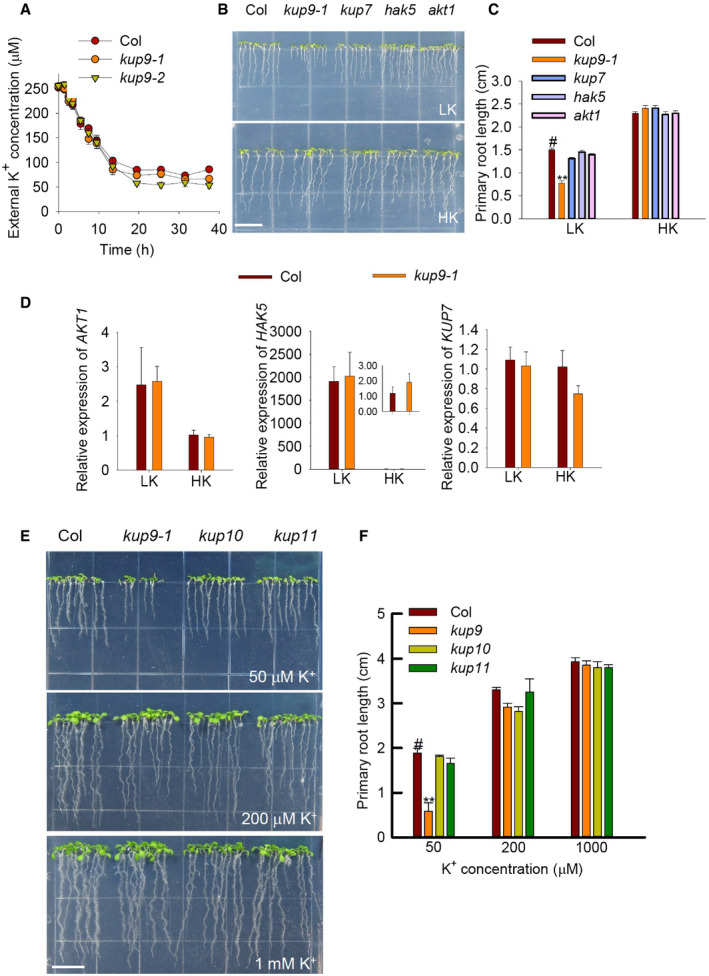
AComparison of K+ uptake capacity among various plant materials using a K+‐depletion experiment. Data are shown as means ± SE (n = 4, biological replicates; each replicate contains 1 g individual plants).
-
B, CPhenotypes and primary root lengths of akt1, hak5, and kup7 mutants. Seeds were germinated and grown on LK or HK medium for 7 days. Data are means ± SE (n = 25, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control. Scale bar, 1 cm in (B).
-
DRT–qPCR analyses of K+‐uptake related genes in wild type and kup9‐1 mutant. Data are shown as means ± SE (n = 3, biological replicates; each replicate contains 120 individual plants).
-
E, FPhenotypes and primary root lengths of kup10 and kup11 mutants. Scale bar, 1 cm in (E). Data are means ± SE (n = 15, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
Source data are available online for this figure.
In the assays, we used the akt1, hak5, and kup7 mutants as controls; these mutants lack major root K+ uptake components 39, 46, 47. None of these mutants showed a short‐root phenotype under LK conditions (Fig EV2B and C). In addition, the transcript levels of these K+‐uptake genes in the kup9‐1 mutant were not repressed upon LK treatment (Fig EV2D). KUP10 and KUP11 from the Arabidopsis KUP/HAK/KT family have high sequence similarity with KUP9, and they are located in the same clade of phylogenetic tree 35. However, neither kup10 nor kup11 mutants showed short‐root phenotype on LK medium (Fig EV2E and F). All these results indicated that KUP9 is involved in root growth regulation only under LK conditions, and this function is independent of root K+ uptake.
Root meristem activity is reduced in kup9 mutants under low‐K+ conditions
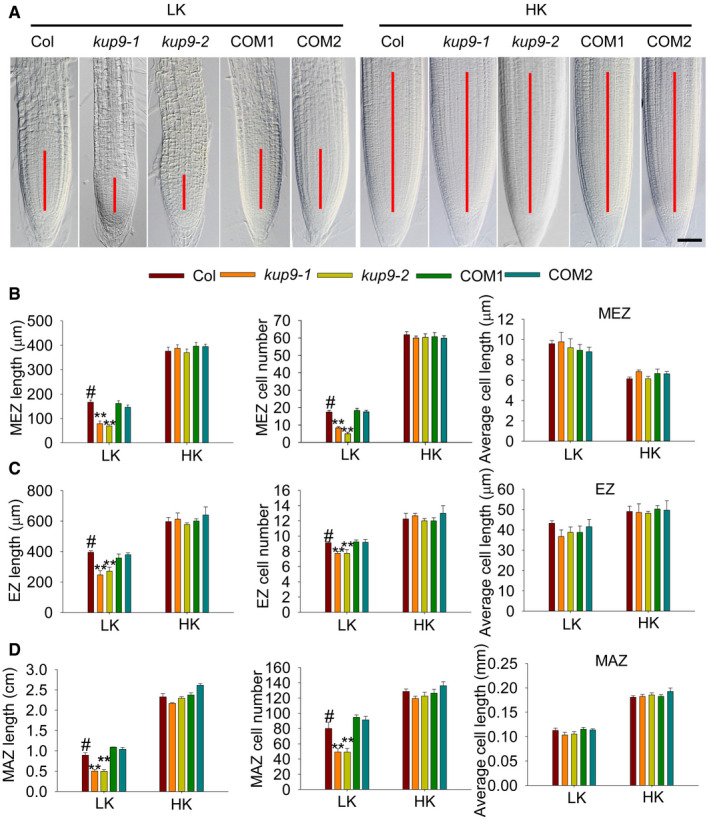
To explore why the kup9 mutants have short primary roots, we observed and measured the different zones (meristem zone, MEZ; elongation zone, EZ; maturation zone, MAZ) of primary roots. Under HK conditions, there was no difference between wild type and kup9 mutants (Fig 2). However, under LK conditions, all the tested zones were shorter in kup9 mutants than in wild type (Fig 2), due to a reduction of cell numbers rather than cell length in mutant roots (Fig 2B–D). Root growth relies on cell division and differentiation 48, 49, and the meristem is the source of all root cells 50, 51. This suggested that meristem activity may be impaired in the kup9 mutants under LK conditions (Fig 2A), which leads to the reduced root cell numbers.
Figure 2. Inhibition of meristem cell proliferation in the kup9 mutants leads to the short‐root phenotype under low‐K+ conditions.
-
ARoot meristem zones of wild type (Col), kup9 mutants (kup9‐1 and kup9‐2), and the kup9‐1/ProKUP9:KUP9 complementation lines (COM1 and COM2). Seeds were germinated and grown on LK or HK medium for 7 days. The meristem zone lengths are marked with red lines. Scale bars, 50 μm.
-
B–Drepresent different zone lengths, different zone cell numbers, and cell length of the indicated seedlings germinated and grown on LK or HK medium for 7 days. MEZ, meristem zone; EZ, elongation zone; MAZ, maturation zone. Data are means ± SE (n = 10–15, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
Source data are available online for this figure.
To test whether KUP9 was expressed in the root tip, we constructed ProKUP9:GUS plants. GUS staining showed that KUP9 was mainly expressed in Arabidopsis root at seedling stage (Fig 3A–D). Importantly, KUP9 showed specific expression in the root tip (Fig 3E and F), suggesting that KUP9 may regulate meristem activity. By contrast, both KUP10 and KUP11 were not expressed in the root tip, although they showed high expression levels in roots (Appendix Fig S2). GUS staining assays and RT–qPCR results both indicated that LK treatment did not significantly affect KUP9 transcript levels (Appendix Fig S3A and B).
Figure 3. KUP9 is mainly expressed in root cells and targets to the ER .
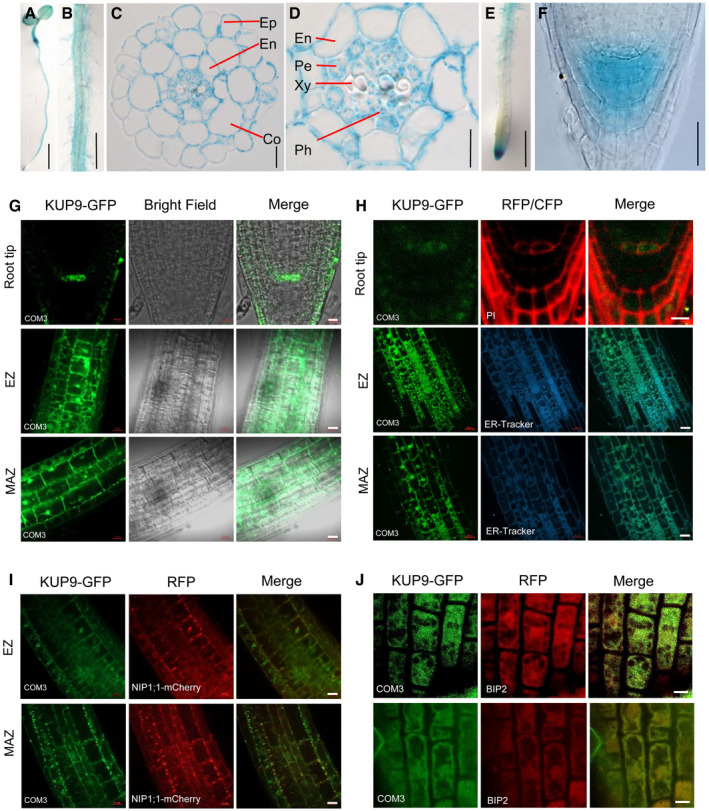
-
A–FExpression of KUP9 as determined in the ProKUP9:GUS line. GUS staining of 5‐day‐old seedling (A), root maturation zone (B), transverse section of maturation zone (C) and (D), primary root tip (E), and root apex (F). Ep, epidermis; Co, cortex; En, endodermis; Pe, pericyle; Xy, xylem; Ph, phloem. Scale bars, 5 mm in (A), 1 mm in (B), 10 μm in (C), 5 μm in (D), 1 mm in (E), and 20 μm in (F).
-
G, HExpression of KUP9 protein as determined in the kup9‐1/ProKUP9:KUP9‐GFP transgenic plants. Green fluorescence shows the subcellular localization of KUP9‐GFP. PI (propidium iodide) and ER‐Tracker were used to indicate the positions of the plasma membrane (PM) and endoplasmic reticulum (ER) and are shown as red and cyan fluorescence, respectively. Scale bars for root tip, 10 μm in (G) and 5 μm in (H). Scale bars for EZ and MAZ, 20 μm.
-
ICo‐localization analyses of KUP9‐GFP with ER marker NIP1;1‐mCherry in their crossing line. Scale bars, 20 μm.
-
JCo‐localization analyses of KUP9‐GFP with the ER marker BIP2 using immunological staining. Scale bars, 5 μm.
Source data are available online for this figure.
To further investigate the expression of KUP9 protein, we constructed the kup9‐1/ProKUP9:KUP9‐GFP complementation lines. The two independent transgenic lines (COM3 and COM4) rescued the short‐root phenotype of kup9‐1 mutant under LK conditions (Appendix Fig S4A and B), indicating that GFP fusion did not affect the function of KUP9 protein. Consistent with the GUS promoter assays, we observed KUP9‐GFP fluorescence in the root tip, elongation zone, and maturation zone (Fig 3G). Importantly, KUP9‐GFP was strictly expressed in the QC cells, but not in the surrounding stem cells (Fig 3G), suggesting that KUP9 function is tightly related to QC function.
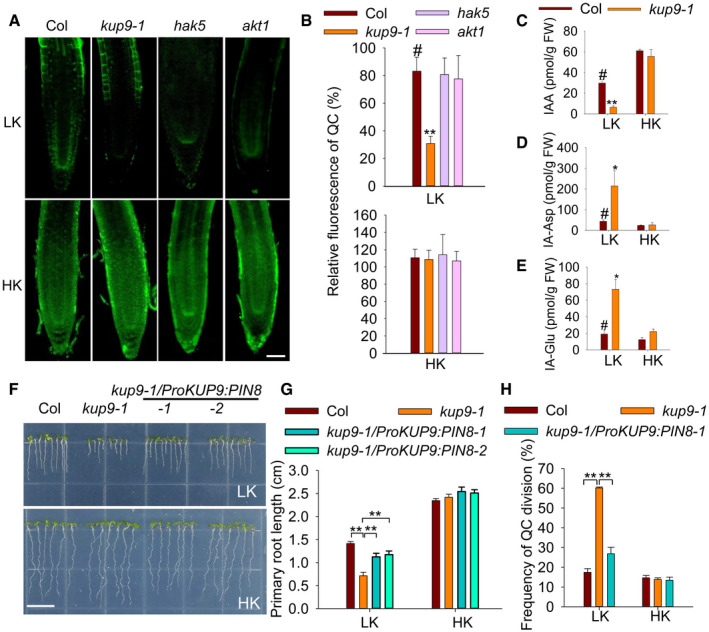
The QC is essential for the maintenance of root meristem activity 13, 14, 50, 52, and nutrient stresses affect QC size and meristem activity 53. Therefore, we examined the expression of the QC markers ProWOX5:GFP and ProQC25:GUS in kup9‐1 mutants. Under HK conditions, the QC cells in wild type and the kup9 mutants did not show any obvious difference (Fig 4A), and the division rates of the QC cells in both plants were very low (8.3–9.4%; Fig 4B). When plants were transferred to LK medium, the QC cell division rate in wild type increased slightly (to about 13.4%), but this rate was remarkably increased in the kup9‐1 and kup9‐2 mutants (to about 74.8 and 74.5%, respectively; Fig 4B). We observed more dividing QC cells in the kup9 mutants after LK treatment for 48 and 96 h (Fig 4A). These results demonstrated that KUP9 affects the maintenance of QC activity under LK stress, thus affecting meristem activity and root growth.
Figure 4. The maintenance of QC activity is defective in the kup9 mutants under LK conditions.
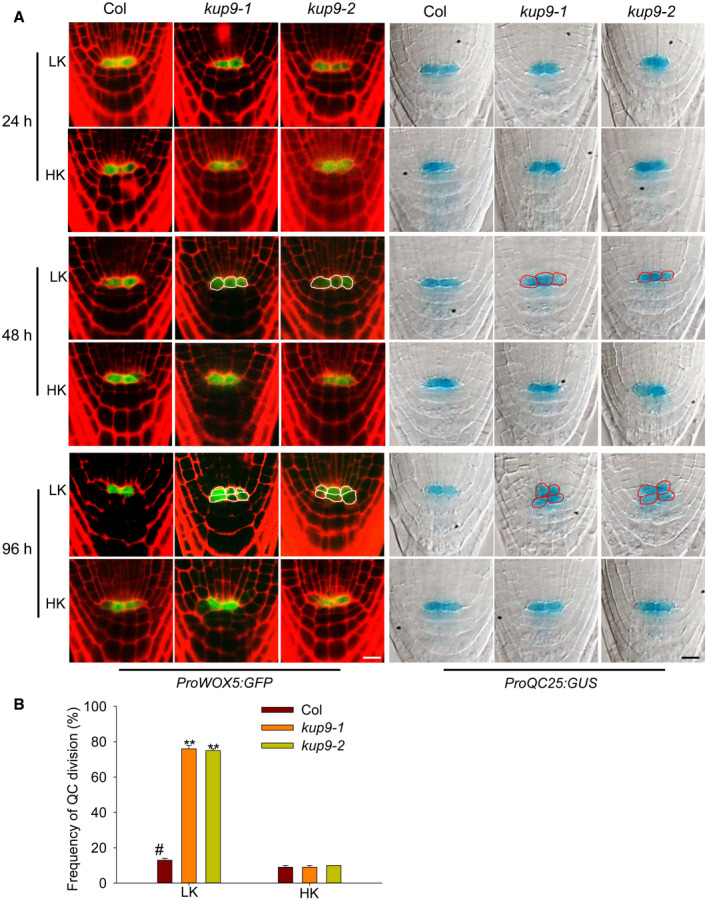
- QC cell observation in wild type and the kup9 mutants using the QC marker lines ProWOX5:GFP and ProQC25:GUS. Seeds were germinated on HK medium for 4 days, and then, the seedlings were transferred to LK or HK medium for the indicated times. Scale bars, 10 μm.
- QC division rates of wild type and the kup9 mutants after LK treatment for 96 h. The statistical method is described in Materials and Methods. Data are means ± SE (n = 150, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
Source data are available online for this figure.
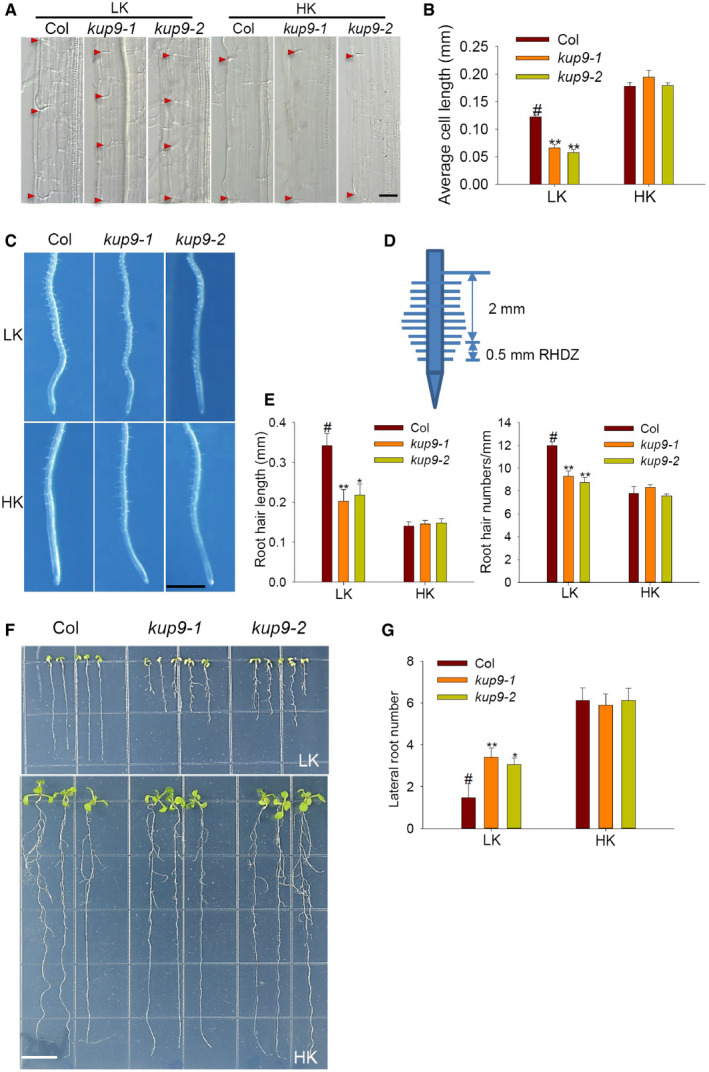
Since KUP9 is expressed in the maturation zone (Fig 3B, C, and G), we also studied KUP9 function in this region of the root. Our previous results showed that cell lengths in the maturation zone were not affected in kup9 mutants grown on LK medium for 7 days (Fig 2D). However, when LK treatment was over 10 days, the kup9 maturation zone cells were significantly shorter than those of wild type (Fig EV3A and B). In addition, the kup9 mutants had more lateral roots and shorter root hairs than wild type under LK conditions (Fig EV3C–G). These results suggested that KUP9 is also involved in the regulation of lateral root growth and cell elongation at late stages of the LK response.
Figure EV3. Phenotypes of maturation zone cells, lateral roots, and root hairs of kup9 mutants under LK conditions.
-
AMaturation zone phenotype of the cortical cells of various seedlings grown on LK or HK medium for 10 days. The red triangle showed the cell boundaries. Scale bar, 25 μm.
-
BRoot cortical cell length in the maturation zone of plants tested in (A). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control. Data are means ± SE (n = 10, individual plants).
-
C–ERoot hair phenotype of kup9 mutants under LK conditions. Data are means ± SE (n = 100, from eight individual plants for each genotype). Scale bar, 1 mm in (C). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
-
F, GLateral root phenotype and lateral root numbers of kup9 mutants under LK conditions. Scale bar, 1 cm in (F). Data are means ± SE (n = 15, individual plants). Student's t‐test (*P < 0.05, **P < 0.01) was used to analyze statistical significance, and “#” represents the control.
Source data are available online for this figure.
KUP9 localizes to the ER
Many K+ transporters in the KUP/HAK/KT family localize to the plasma membrane (PM) 35. However, observation of KUP9‐GFP suggested that KUP9 targets to the ER rather than the PM (Fig 3G). To confirm the subcellular localization, we used propidium iodide (PI) to stain the PM and BODIPY TR Glibenclamide (ER‐Tracker) to stain the ER. The KUP9‐GFP fluorescence was observed in the intracellular space of QC cells (Fig 3H). Microscopy of the elongation zone and maturation zone confirmed that the KUP9‐GFP fluorescence co‐localized with the CFP fluorescence of ER‐Tracker (Fig 3H).
To further confirm the ER localization of KUP9, the COM3 line (kup9‐1/ProKUP9:KUP9‐GFP) was crossed with the transgenic line expressing the ER marker NIP1;1‐mCherry 54. We observed the co‐localization of KUP9‐GFP and NIP1;1‐mCherry (Fig 3I). Consistent with this, immunological staining assay showed a pronounced co‐localization of KUP9‐GFP with the ER marker BIP2 (Fig 3J). All the data indicate that KUP9 localizes to the ER. In addition, we observed that both the protein level and protein localization of KUP9‐GFP were not significantly changed after LK treatment (Appendix Fig S3C and D).
KUP9 regulates primary root growth by maintaining auxin levels in the root tip under low‐K+ conditions
KUP9 is essential for the maintenance of QC and meristem activity under LK conditions, but how does it perform this function? The auxin level in root tips regulates QC and meristem activity 14, 25, 51, 52, 55, 56, 57. We speculated that KUP9 may regulate primary root growth through auxin signaling. Using DR5:GFP as an auxin response marker, we observed that, though there was no obvious difference between wild type and kup9 under HK conditions, the auxin response in the kup9 mutants was strongly repressed by LK treatment (Fig 5A). Considering the KUP9 is specifically expressed in the QC cells, the GFP fluorescence in the QC cells was quantified to indicate auxin levels. The GFP fluorescence of QC cells was slightly decreased in wild‐type plants after LK treatment, but was largely repressed in kup9 mutants (Fig 5B). These results indicated that KUP9 is involved in the maintenance of auxin level in root QC cells under LK conditions.
Figure 5. Auxin levels in the root tip of kup9 mutants are reduced under LK stress.
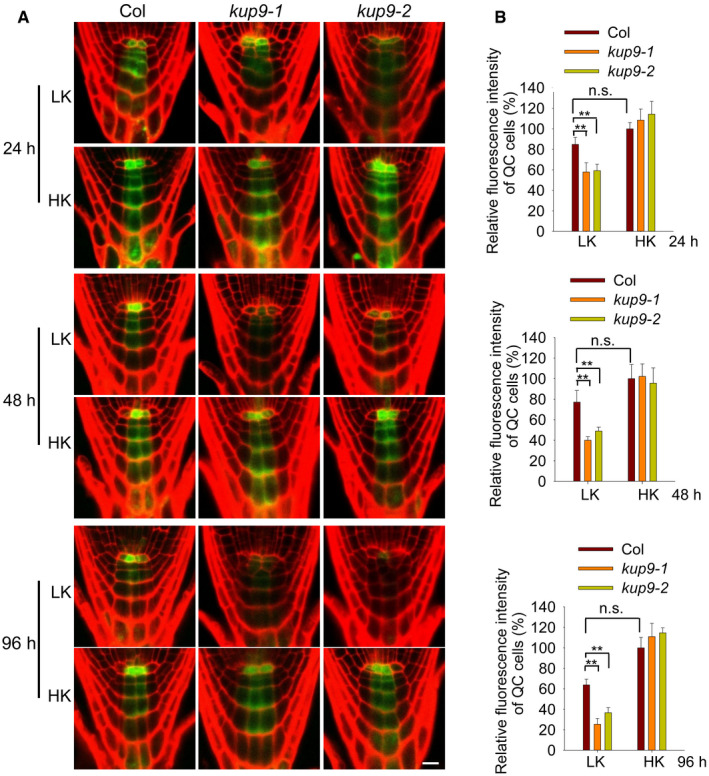
- Auxin levels in root tips, as indicated by the ProDR5:GFP reporter line. Seeds were germinated on HK medium for 4 days, and then, the seedlings were transferred to LK or HK medium for the indicated times. Scale bar, 10 μm.
- Quantification of GFP fluorescence in the QC cells shown in (A). Data are means ± SE (n = 20, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance.
Source data are available online for this figure.
To verify whether the short‐root phenotype of kup9 mutants was due to the reduction of auxin levels, we added the auxin NAA (α‐naphthalene acetic acid) to the medium. Under LK conditions, the addition of NAA repressed primary root growth of wild type and complementation lines, but promoted primary root growth of kup9 mutants (Fig 6A and B). When the NAA concentration was up to 100 nM, the test plants did not show any significant difference in primary root length. Microscopy analysis indicated that addition of NAA inhibited QC division and rescued QC activity in the kup9‐1 mutant under LK conditions (Fig 6C and D), thus restoring meristem cell numbers and meristem zone length and root length to the wild‐type level (Appendix Fig S5). These results demonstrated that KUP9 retains auxin levels in the root tip under LK conditions and maintains QC activity and primary root growth.
Figure 6. Exogenous auxin rescues the primary root growth and QC activity of kup9 mutants under LK conditions.
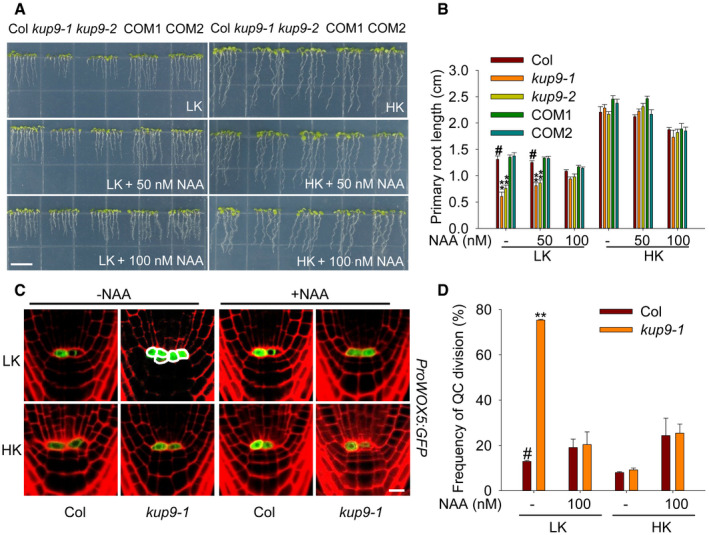
- Phenotypes of various plants. Seeds were germinated and grown on LK or HK medium with or without NAA for 7 days. Scale bar, 1 cm.
- Primary root length of the plants tested in (A). Data are means ± SE (n = 25, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
- QC cell observation using the QC marker line ProWOX5:GFP. Seeds were germinated on HK medium for 4 days, and then, the seedlings were transferred to LK or HK medium with or without NAA (100 nM) for 96 h. Scale bar, 10 μm.
- QC division rates of the plants tested in (C). The statistical method is described in Materials and Methods. Data are means ± SE (n = 50–60, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
Source data are available online for this figure.
KUP9 maintains the auxin level in QC cells independent of auxin synthesis under low‐K+ conditions
To determine whether KUP9 regulates the auxin level in QC cells, we constructed the transgenic lines kup9‐1/ProWOX5:AMI1 and kup9‐1/ProKUP9:AMI1 using QC‐specific promoter ProWOX5 and KUP9 native promoter, respectively, and compared their phenotypes. The Arabidopsis AMI (AMIDASE1) gene encodes an auxin biosynthesis enzyme, which catalyzes the conversion of IAM (indole‐3‐acetamide) to IAA 58, 59. Under LK conditions, the two independent transgenic lines of kup9‐1/ProWOX5:AMI1 showed the same short‐root phenotype as the kup9‐1 mutant in the absence of the substrate IAM (Fig 7A and B). However, addition of IAM restored the root length of transgenic plants to wild‐type level (Fig 7A and B), suggesting that newly synthesized IAA in the QC cells promotes primary root growth of the kup9‐1 mutant under LK conditions. More importantly, the kup9‐1/ProKUP9:AMI1 transgenic lines behaved similarly to the kup9‐1/ProWOX5:AMI1 plants, when IAM was added to the medium (Fig 7C and D). All these results demonstrated that KUP9 regulates primary root growth under LK conditions by controlling the auxin level specifically in the QC cells.
Figure 7. Increase of auxin level in QC cells can rescue the short‐root phenotype of kup9 mutants.
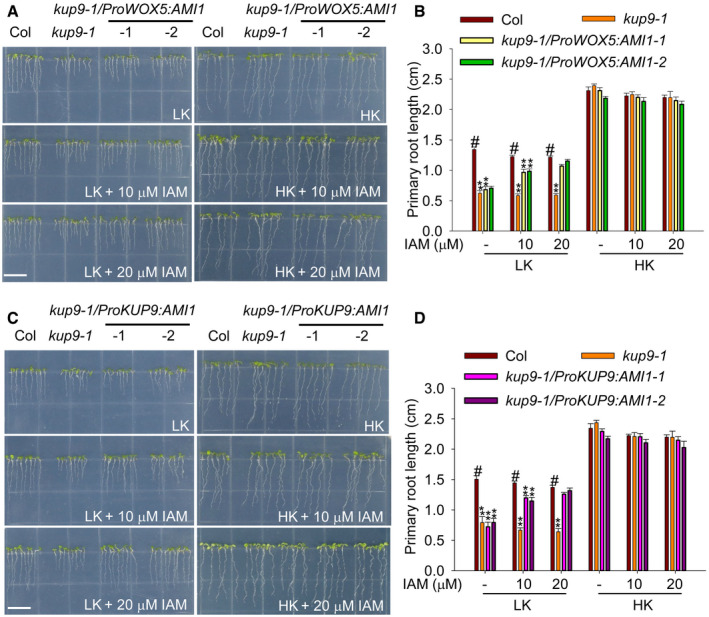
-
A, CPhenotypes of the kup9‐1/ProWOX5:AMI1 (A) and kup9‐1/ProKUP9:AMI1 (C) transgenic lines. Seeds were germinated and grown on LK or HK medium with or without IAM for 7 days. Scale bars, 1 cm.
-
B, DPrimary root length of the plants tested in (A) and (C), respectively. Data are means ± SE (n = 25, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
Source data are available online for this figure.
Auxin produced in QC cells has been suggested to be fundamental for the controlling of root meristem size 56. Here, RT–qPCR showed that the expression of auxin biosynthetic genes such as YUCs, TAA1/TAR2, ASA1, and ASB1 was not significantly affected in kup9‐1 roots under LK conditions (Appendix Fig S6). YUC1 even had an elevated transcript level in kup9‐1 mutant, which may be due to a feedback response to the reduction of auxin in mutant roots. These results suggest that KUP9 maintains the auxin level in QC cells independent of auxin synthesis.
KUP9 mediates auxin efflux in tobacco BY‐2 cells and Xenopus oocytes
Considering KUP9 is an ion transporter, we hypothesized that KUP9 may directly transport auxin. Therefore, we measured the auxin transport activity of KUP9 in tobacco (Nicotiana tabacum) BY‐2 cells, as this cell culture provides a more tractable system for measuring auxin transport than root cells. As shown in Fig 8A, when KUP9‐GFP was expressed in BY‐2 cells driven by the pSuper1300 constitutive promoter, the KUP9‐GFP protein mainly localized to the ER and also to the PM. When BY‐2 cells were cultured in medium with 3H‐IAA for 2 h, the cells expressing KUP9‐GFP protein accumulated less 3H‐IAA than the control cells expressing empty vector (Fig 8B). Here, the auxin efflux carrier PIN4 was used as a positive control 60. Similarly, the cells expressing PIN4‐GFP protein also accumulated less 3H‐IAA (Fig 8B). KUP10, the closest member of KUP9, was used as a negative control and did not affect 3H‐IAA accumulation in BY‐2 cells (Fig 8B). These results suggested that KUP9 may mediate auxin efflux from the ER into the cytoplasm and then out of the BY‐2 cells.
Figure 8. KUP9 mediates K+ and IAA efflux in BY‐2 tobacco cells and Xenopus oocytes.
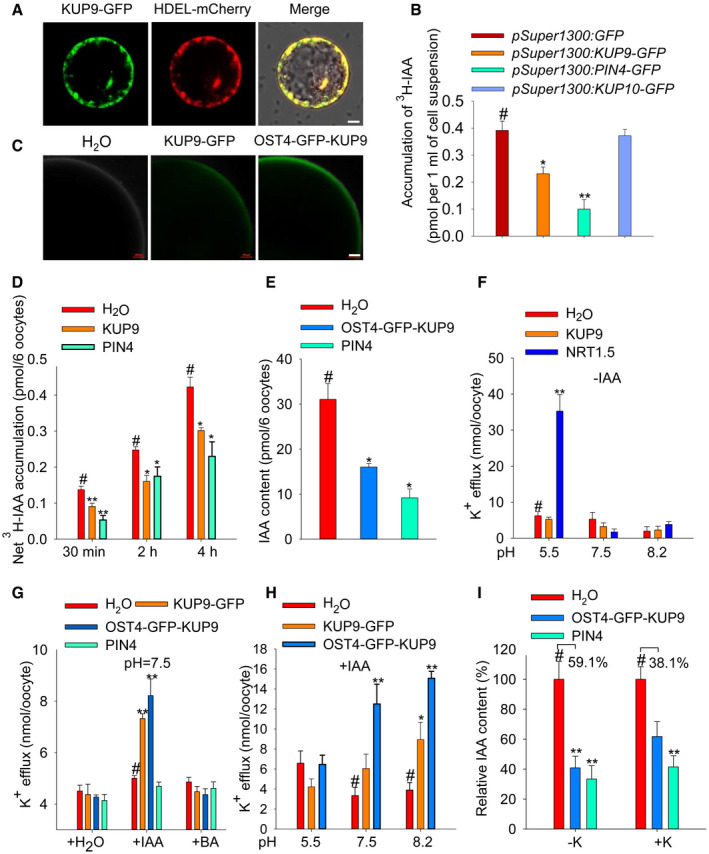
- Expression of KUP9‐GFP in BY‐2 tobacco cells. HDEL‐mCherry was used as an ER marker. Scale bar, 10 μm.
- 3H‐IAA accumulation in the BY‐2 cells expressing the indicated proteins. PIN4‐GFP and KUP10‐GFP were used as positive control and negative control, respectively. Data are means ± SE (n = 3, biological replicates; each replicate contains 3–4 ml suspension cells).
- Expression of KUP9‐GFP and OST4‐GFP‐KUP9 in Xenopus oocytes. Scale bar, 100 μm.
- 3H‐IAA accumulation in the oocytes expressing KUP9‐GFP and PIN4 at the indicated times. PIN4 was used as a positive control. The oocytes were incubated in the bath solution containing 3H‐IAA for the indicated times. Data are means ± SE (n = 6, biological replicates; each replicate contains six oocytes).
- IAA content in the oocytes expressing the indicated proteins. The oocytes were injected with the same amount of IAA and incubated in IAA‐free bath solution for 6 h. IAA content was measured using UPLC‐MS. Data are means ± SE (n = 3, biological replicates; each replicate contains six oocytes).
- K+ efflux activity of the oocytes expressing KUP9 and NRT1.5. The oocytes were not injected with IAA. The oocytes were incubated in K+‐free bath solution for 6 h. Data are means ± SE (n = 3, biological replicates; each replicate contains six oocytes).
- K+ efflux activity of the oocytes expressing the indicated proteins. The oocytes were injected with H2O, IAA, or BA. Data are means ± SE (n = 3, biological replicates; each replicate contains six oocytes).
- K+ efflux activity of the oocytes expressing the indicated proteins. IAA was injected into the oocytes. Then, the oocytes were incubated in K+‐free bath solutions at different pH for 6 h. Data are means ± SE (n = 3–6, biological replicates; each replicate contains six oocytes).
- IAA efflux activity of oocytes expressing the indicated proteins under different external K+ conditions (−K, 0 mM; +K, 50 mM). The oocytes were injected with the same amount of IAA and then incubated in IAA‐free bath solution for 6 h. IAA content was measured using UPLC‐MS. Data are means ± SE (n = 3, biological replicates; each replicate contains six oocytes).
Furthermore, we determined the auxin transport activity of KUP9 in Xenopus oocytes. KUP9‐GFP protein localized to the ER in Arabidopsis roots (Fig 3H–J); however, the KUP9‐GFP protein could also target to the PM when expressed in oocytes (Fig 8C). We observed that the oocytes expressing KUP9‐GFP or PIN4 accumulated less 3H‐IAA than the control oocytes, when the oocytes were incubated in the bath solution with 3H‐IAA for the indicated times (Fig 8D). To verify the effect of protein localization, the OST4‐GFP‐KUP9 fusion protein was constructed. OST4, a membrane anchor from yeast 61, was fused at the N‐terminus of KUP9, so that OST4‐GFP‐KUP9 protein should be recruited to the PM. Indeed, we observed the GFP fluorescence of OST4‐GFP‐KUP9 protein at the PM of oocytes and this fluorescence intensity was much stronger than that of KUP9‐GFP (Fig 8C). Then, the auxin transport activity of OST4‐GFP‐KUP9 was also tested. Oocytes were injected with IAA and incubated in the IAA‐free bath solution for 6 h; then, the IAA remained in the oocytes was measured. As shown in Fig 8E, the IAA contents in the oocytes expressing OST4‐GFP‐KUP9 and PIN4 were remarkably reduced compared with the control oocytes, suggesting OST4‐GFP‐KUP9 directly mediates auxin efflux across the PM of oocytes. As the negative control, KUP10 did not transport auxin in oocytes (Fig EV4A).
Figure EV4. IAA transport activity of PIN8 in Xenopus oocytes and phenotype of pin8 mutant under LK conditions.
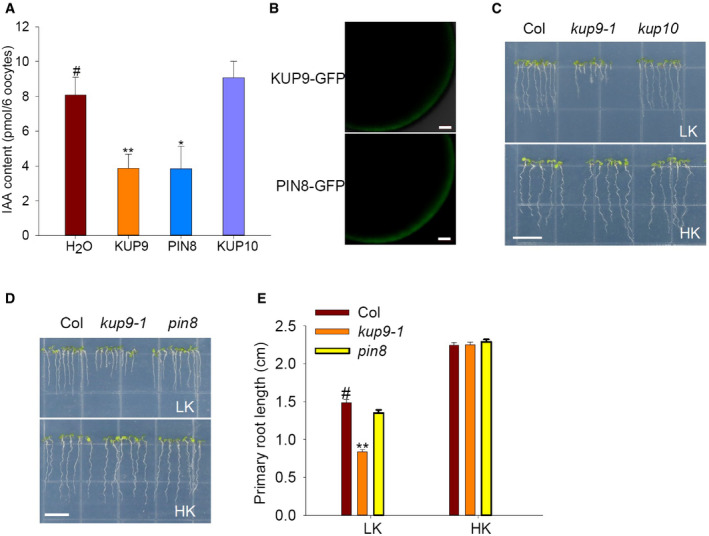
-
AIAA transport activities of KUP9, PIN8, and KUP10 in Xenopus oocytes. Data are means ± SE (n = 3, biological replicates; each replicate contains 6 oocytes). Student's t‐test (*P < 0.05, **P < 0.01) was used to analyze statistical significance, and “#” represents the control.
-
BPM localization of KUP9‐GFP and PIN8‐GFP in Xenopus oocytes. Scale bars, 100 μm.
-
C–EPrimary root phenotypes of kup10 and pin8 mutants under LK conditions. Scale bars, 1 cm in (C) and (D). Data are means ± SE (n = 15, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
Source data are available online for this figure.
KUP9 belongs to the KT/HAK/KUP K+ transporter family; therefore, we also tested the K+ transport activity of KUP9 in yeast and oocytes. As shown in Appendix Fig S7, we did not observe obvious K+ uptake activity of KUP9 in yeast, and K+ efflux activity was not detected in oocytes (Fig 8F). Considering that KUP9 could directly mediate auxin efflux, we hypothesized that the K+ transport activity of KUP9 might be linked with auxin transport. Indeed, when IAA was injected into the oocytes, the KUP9‐expressing oocytes showed K+ efflux across the PM of oocytes (Fig 8G). However, injection of BA (benzoic acid, the biologically inactive analogue of IAA) did not trigger K+ efflux in the KUP9‐expressing oocytes (Fig 8G). In addition, the IAA‐activated K+ efflux was dependent on the proton gradient across the PM of oocytes, and higher external pH enhanced K+ efflux (Fig 8H). Moreover, the K+ gradient across the PM of oocytes enhanced IAA efflux activity in the KUP9‐expressing oocytes (Fig 8I). All these results demonstrated that KUP9 may directly mediate both K+ and auxin efflux from the ER into the cytoplasm in Arabidopsis root cells.
KUP9 regulates intracellular K+ and auxin homoeostasis in the QC cells under low‐K+ stress
Since KUP9 could mediate K+ transport, we determined the intracellular K+ level of root cells in the kup9‐1 mutant using the fluorescent dye Asante Potassium Green2‐AM (APG2‐AM). Under HK conditions, the wild type and the kup9‐1 mutant showed high K+ levels in the cytoplasm of QC cells (Fig 9A and B). After LK treatment, wild type retained high K+ levels in QC cells, but the kup9‐1 mutant had lower K+ level of QC cells (Fig 9A and B). Here, the hak5 and akt1 mutants were tested as controls and these mutants showed similar K+ levels in QC cells as wild‐type plants (Fig 9A and B). The K+ level change in the kup9‐1 mutant was very similar to the auxin level change (Fig 5B), consistent with the results from oocytes that KUP9 mediates both K+ and auxin efflux (Fig 8). Therefore, KUP9 is essential for K+ and auxin homeostasis in QC cells under LK conditions.
Figure 9. KUP9 regulates intracellular K+ and auxin homoeostasis in the root tip.
-
A+ levels of various materials using the fluorescent dye Asante Potassium Green‐2 (AM). Scale bar, 50 μm.
-
BQuantification of K+ concentration under LK and HK conditions. Data are means ± SE (n = 25, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance, and “#” represents the control.
-
C–EThe contents of free IAA (C), IA‐Asp (D), and IA‐Glu (E) in the root tips of wild type and the kup9‐1 mutant. Data are means ± SE (n = 3, biological replicates; each replicate contains 1,000–1,200 individual plants). Student's t‐test (*P < 0.05, **P < 0.01) was used to analyze statistical significance, and “#” represents the control.
-
FPhenotype of the kup9‐1/ProKUP9:PIN8 transgenic lines. Seeds were germinated and grown on LK or HK medium for 7 days. Scale bar, 1 cm.
-
GPrimary root length of the plants tested in (F). Data are means ± SE (n = 25, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance.
-
HQC division rates of the indicated plants. The statistical method is described in Materials and Methods. Data are means ± SE (n = 50–60, individual plants). Student's t‐test (**P < 0.01) was used to analyze statistical significance.
Source data are available online for this figure.
Since KUP9 mediates auxin efflux in the ER, it might have a similar function to the auxin efflux carrier PIN8. The ER‐localized PIN8 mediates auxin efflux from the ER into the cytoplasm and maintains intracellular auxin homoeostasis; however, PIN8 is not expressed in Arabidopsis roots 21. To test the similarities between PIN8 and KUP9, we first compared their auxin transport activities in oocytes. PIN8‐GFP protein could also target to the PM of oocytes and mediate auxin efflux, like KUP9‐GFP (Fig EV4A and B). PIN8 is involved in controlling auxin levels and IAA metabolism 21; thus, KUP9 could have a similar function in QC cells under LK conditions. Therefore, we collected the root tips of wild type and the kup9‐1 mutant and measured the free IAA and the IAA conjugates IA‐Asp and IA‐Glu. Under HK conditions, the free IAA concentrations in wild type and the kup9‐1 mutant were not different. However, under LK conditions the free IAA concentration in the kup9‐1 mutant was much lower than that in wild type (Fig 9C). This result was consistent with the observation using the DR5:GFP plants (Fig 5A). By contrast, the IA‐Asp and IA‐Glu concentrations in the kup9‐1 mutant were much higher than that in wild type (Fig 9D and E). These results demonstrated that KUP9 regulates auxin homoeostasis and IAA metabolism in the root tip.
To further compare the functions of KUP9 and PIN8 in vivo, we expressed PIN8 under the control of the KUP9 promoter, in the kup9‐1/ProKUP9:PIN8 transgenic lines. The pin8 mutant did not show a short‐root phenotype when grown on LK medium (Fig EV4D and E). However, when PIN8 was expressed in root tip driven by the KUP9 promoter, the kup9‐1/ProKUP9:PIN8 transgenic lines could partially rescue the QC activity and short‐root phenotype on LK medium (Fig 9F–H). All these results indicated that the ER‐localized KUP9 is involved in the maintenance of K+ and auxin homoeostasis in the QC cells and thus regulates primary root growth under LK conditions.
Discussion
Multiple functions of the KUP/HAK/KT family transporters
The KUP/HAK/KT proteins are important K+ transporters in plants, and most have been reported to transport K+ and function in K+ uptake/homeostasis 35, although some are involved in osmotic regulation and cell growth/elongation 38, 40. Here, we demonstrate that Arabidopsis KUP9 is essential for the maintenance of primary root growth under LK conditions (Fig 1A), and present evidence that this function is independent of the K+ uptake or K+ content in roots (Figs 1E and F, and EV2A). Given the ER localization, KUP9 is not likely to be directly involved in the K+ uptake in Arabidopsis roots. In addition, the akt1, hak5, and kup7 mutants that are defective in root K+ uptake did not show the short‐root phenotype that the kup9 mutant did under LK conditions (Fig EV2B and C). Our data indicate that KUP9 is required for the maintenance of meristem activity and cell elongation in the maturation zone under LK conditions and thereby regulates primary root growth (Figs 2 and EV3A and B).
Previous study showed that TRH1/KUP4 regulates root hair development and root gravity response 37. Auxin transport in the trh1 mutant root is partially blocked because the polar localization of PIN1 protein is affected in the mutant root 44, 45. In addition, TRH1 can transport auxin when expressed in yeast 44. Here, we demonstrate that the K+ transport activity of KUP9 requires the presence of auxin (Fig 8G). Moreover, KUP9 directly mediates both K+ and auxin transport when expressed in tobacco cells and Xenopus oocytes (Fig 8B, D and E). By contrast, its closest family member KUP10 does not transport auxin (Figs 8B and EV4A), and the kup10 mutant does not show a short‐root phenotype on LK medium (Fig EV2E and F). We conclude that KUP9 regulates K+ and auxin homeostasis simultaneously under LK conditions and that this function is unique to KUP9 (Figs 8 and EV4). These data suggest that both TRH1/KUP4 and KUP9 are involved in Arabidopsis root growth/development and morphology by regulating auxin homeostasis; however, they display distinct functions and mechanisms.
Most of the identified KUP/HAK/KT transporters are reported to target to the PM 35; here, we determined that KUP9 is localized to the ER in Arabidopsis root cells (Figs 3H–J and EV5). In addition, KUP9 shows very specific expression in the QC cells (Fig 3G and H). Therefore, we conclude that the ER‐localized KUP9 likely mediates K+ and auxin efflux from the ER to the cytoplasm in the QC cells to maintain meristem activity in plant response to LK stress.
Figure EV5. Subcellular localization analyses of KUP9 protein.
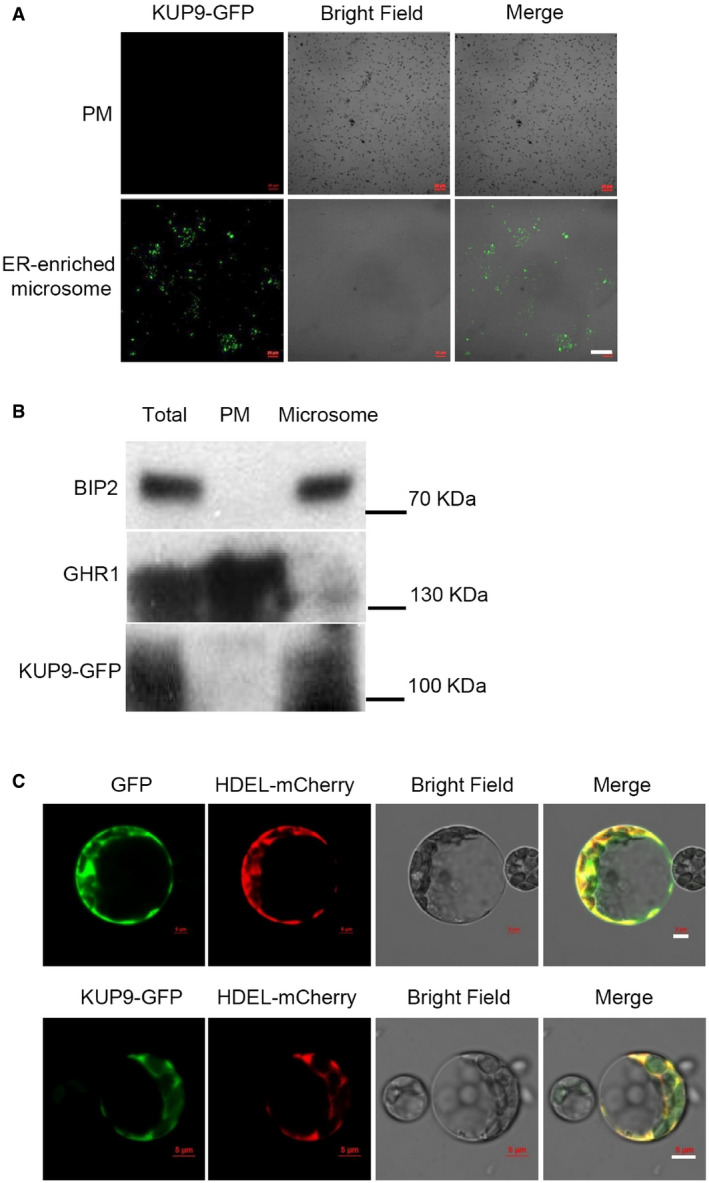
- Microscopic observation of different membrane fractions (PM and ER‐enriched microsome) isolated from the kup9‐1/ProKUP9:KUP9‐GFP transgenic plants. Different fractions were separated by density gradient centrifugation. Scale bar, 40 μm.
- Western bolt analyses of KUP9 protein in different membrane fractions. Proteins were detected by immunoblot analyses. BIP2 and GHR1 were used as ER and PM markers, respectively.
- Subcellular localization analyses of KUP9‐GFP protein in Arabidopsis mesophyll protoplasts. HDEL‐mCherry was used as an ER marker. Scale bars, 5 μm.
Since KUP9 is localized to the ER, we tried to isolate ER vesicles from Arabidopsis root in order to characterize K+ and auxin transport activities in ER vesicles. This is a challenging experiment in plants, but should be a direct evidence in vivo. Unfortunately, due to technical reasons we did not obtain appropriate ER vesicles used for transport assays.
KUP9 is involved in the responses to low‐K+ stress
In the natural environment, nutrient distribution in soils is heterogeneous 4. Plant roots must have the ability to sense external nutrient levels and regulate root growth accordingly 7, 8. How plants connect nutrient sensing with root growth regulation is a key issue in this research area. Previous studies have identified a relationship between LK perception and auxin signaling 9, 43, 44, but the molecular mechanisms remained unclear. In the present study, we demonstrated that KUP9 can respond to external LK stress and maintain root growth activity by directly regulating auxin signaling. KUP9 thus functions as a key node that directly connects K+ and auxin. However, KUP9 is not responsible for plant responses to low nitrogen and low phosphorus stresses: There was no difference in primary root lengths between wild type and kup9 mutants when grown on low nitrate ( ) or low phosphate (Pi) medium, suggesting that KUP9 is specific for the LK response (Appendix Fig S8).
When plants are subjected to LK stress, primary root growth is inhibited due to decreased auxin concentrations in the root tip (Figs 5A and 9C). We previously report that root acropetal auxin transport is inhibited under LK conditions, mainly because of degradation of PIN1 protein 43. Although the auxin polar transport is inhibited, the root tip still needs a minimum auxin level in the QC cells to maintain meristem activity. Therefore, plants undergo slow root growth under LK conditions, but rapidly resume root growth when roots enter a HK region. Accordingly, we conclude that KUP9‐mediated regulation is finely tuned to support appropriate primary root growth during plant responses to LK stress.
Based on our data, KUP9 operates under LK conditions but not HK conditions. How does KUP9 respond to LK stress? Both GUS staining and RT–qPCR results indicated that the KUP9 transcript levels were not significantly changed after LK stress (Appendix Fig S3A and B). Furthermore, neither the protein level nor protein localization of KUP9 was altered (Appendix Fig S3C and D). We therefore hypothesize that KUP9 may undergo post‐translational modification under LK conditions. Phosphorylation of the KUP/HAK/KT proteins has been widely reported; for example, HAK5, KUP6, and KUP7 are phosphorylated after stress treatment, which alters their transport activity or kinetics 40, 47, 62. LK conditions may similarly lead to the phosphorylation of KUP9 protein to activate its K+ and auxin transport activity. Alternatively, it is possible that the K+ gradient across the ER membrane may drive the auxin efflux from the ER lumen to the cytoplasm (Fig 8I). Under HK conditions, the K+ concentration in the cytoplasm of QC cells is relatively high, and the K+ gradient across the ER membrane may be insufficient to promote the K+ and auxin efflux into the cytoplasm. Under LK conditions, the low‐K+ concentration in the cytoplasm may facilitate the K+ and auxin efflux into the cytoplasm through KUP9 to maintain both K+ and auxin homeostasis in the QC cells.
ER‐localized KUP9 is involved in the local regulation of K+ and auxin homeostasis
Under LK conditions, plants enhance K+ uptake from environment and delivery of K+ ions to the cells where they are needed through intercellular K+ transport. At the same time, the K+ ions in pools such as vacuoles in root maturation cells can be also mobilized to maintain the intracellular K+ homeostasis 63, 64. The root meristem cells lack vacuoles, and we hypothesize that the ER may function as a K+ pool in the QC cells. The ER is an important auxin pool in plant root cells 20, 65. Therefore, the ER‐localized KUP9 could synchronously regulate K+ and auxin mobilization from the ER lumen and maintain their homeostasis in the cytoplasm under LK conditions.
As for auxin transport, the PM‐localized auxin carriers, such as AUX1, PIN1, 2, 3, 4, and 7, are responsible for the intercellular auxin transport 15, 16. Here, we report that none of their mutants show the short‐primary‐root phenotype on LK medium that is characteristic of the kup9‐1 mutant (Appendix Fig S9). The intracellular auxin transport related to compartmentalizing auxin is an essential component of auxin transport 20, 66, 67. Two ER‐localized auxin carriers, PIN5 and PIN8, have been identified; they show antagonistic action in the regulation of intracellular auxin homoeostasis and IAA metabolism 18, 21. They both mediate auxin transport across the ER membrane, but in opposite transport directions. Our data demonstrate that KUP9 mediates auxin transport from the ER lumen to the cytoplasm, which indicates a similar function to PIN8 (Fig EV4A and B). However, the pin8 mutant did not show a short‐root phenotype on either LK or HK medium (Fig EV4D and E), and PIN8 is not expressed in roots 21. Notably, the PIN8 gene driven by the KUP9 promoter could partially complement the short‐root phenotype of the kup9‐1 mutant (Fig 9F and G), supporting the conclusion that they do have similar molecular functions.
PIN8 is expressed in male gametophyte 21, while PIN5 is expressed in mainly the hypocotyl, leaf vasculature, and stem, but very weakly in the root 18. By contrast, KUP9 is expressed primarily in roots, specifically in the QC cells (Fig 3G and H), suggesting its specific function in the regulation of root meristem activity. We conclude that the ER‐localized PIN5, PIN8, and KUP9 all regulate intracellular auxin homeostasis, but function in different tissues and cell types. They constitute an important mechanism for the local and finely tuned regulation of intracellular auxin homoeostasis.
Taking these observations together, we propose a working model to illustrate the function of KUP9 in the regulation of primary root growth in plant response to external K+ levels (Fig 10). KUP9 is expressed in root cells and localized to the ER. When plant roots encounter a LK environment, the root acropetal auxin transport is inhibited. In addition, LK stress activates the transport activity of KUP9. Then, KUP9 activates K+ and auxin efflux from the ER lumen to the cytoplasm to regulate K+ and auxin homeostasis in the cytoplasm. K+ may help maintain the cell turgor and enzyme activities, while auxin supports the QC activity and cell elongation in maturation zone through auxin‐responsive genes. Therefore, meristem activity and cell growth are retained to continue primary root growth at a low rate. When plant roots enter a HK region, the meristem cells can be rapidly reactivated so that the primary root grows at a high rate. We conclude that KUP9 is essential for the LK response and regulates primary roots under LK conditions.
Figure 10. Working model for the KUP9‐mediated K+ and auxin homeostasis of root cells in the Arabidopsis response to low‐K+ stress.
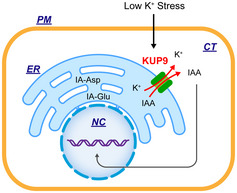
The details of this schematic model are described in the text. PM, plasma membrane; CT, cytoplasm; ER, endoplasmic reticulum; NC, nucleus.
Materials and Methods
Plant materials
The Arabidopsis Columbia ecotype (Col‐0) and T‐DNA insertion lines including kup9‐1 (SALK_108080), kup7 (CS805085), akt1 (SALK_071803), kup10 (SALK_072956), and hak5 (SALK_005604) were obtained from ABRC (Arabidopsis Biological Resource Center) or NASC (Nottingham Arabidopsis Stock Centre).
For constructing the CRISPR/Cas9 mutant kup9‐2, two targets were designed at the 866 bp and 946 bp of the genomic sequence of KUP9 on the website http://www.genome.arizona.edu/crispr/CRISPRsearch.html: one is “CCAAGAAATGGTTGGAGGGTAAA” and the other is “CCCGAATCAGCACCGTAGTGATG”. Then, the CRISPR/Cas9 construct was transformed into Col‐0 plants. The T1 generation was harvested and sequenced. The T3 generation homozygous lacking the T‐DNA was used to test phenotype. KUP9 full‐length genomic sequence was constructed into pCAMBIA1300 driven by KUP9 native promoter and transformed into kup9‐1 mutant to obtain complementation lines (COM1 and COM2). Both KUP9 coding sequence and EGFP sequence were also constructed into pCAMBIA1300 vector driven by KUP9 native promoter and transformed into kup9‐1 mutant to obtain complementation lines for observing KUP9 subcellular localization (COM3 and COM4). AMI1 and PIN8 coding sequences were constructed into pCAMBIA1300 driven by KUP9 promoter and transformed into kup9‐1 mutant to obtain homozygous lines to test phenotype. For generating kup9‐1/ProWOX5:AMI1, AMI1 coding sequence was constructed into pCAMBIA1300 driven by WOX5 promoter and transformed into kup9‐1 mutant to obtain homozygous lines to test phenotype. Arabidopsis transformation with Agrobacterium (strain GV3101) was conducted using the floral dip method 68. The T4 homozygous transgenic plants were used to test phenotype.
Phenotype analyses and plant growth conditions
For phenotype test, seeds were sterilized and treated at 4°C in darkness for 4 days. Then, the seeds were placed on HK (5 mM) or LK (50 μM) medium containing 1% (w/v) sucrose and 0.9% (w/v) agar, and grew at 22°C for 7 days with illumination at 100 μmol/m2/s for a 16‐h daily light period. The HK medium and LK medium were modified from MS (pH 5.8). NH4NO3 and KNO3 were removed, and KH2PO4 was replaced by H3PO4. The medium contained 1.5 mM MgSO4·7H2O, 85 μl H3PO4, 2.99 mM Ca(NO3)2·4H2O, 0.1 mM MnSO4·4H2O, 5 μM KI, 0.1 μM CuSO4·5H2O, 0.1 mM H3BO3, 0.1 μM CoCl2·6H2O, 0.03 mM ZnSO4·7H2O, 10 μM Na2MoO4·2H2O, 0.1 mM FeSO4·7H2O, and 0.1 mM Na2EDTA. The potassium almost came from agar in the LK medium. For preparing HK medium, KCl was supplemented until the final potassium concentration was 5 mM. NAA or IAM was added after the medium was autoclaved at 115°C for 15 min and cooled to 60°C. NAA (1‐naphthaleneacetic acid) and IAM (indole‐3‐acetamide) were ordered from Sigma.
To obtain transgenic plants and harvest seeds, Arabidopsis plants were cultured in the potting soil mixture (rich soil:vermiculite = 2:1, v/v) and kept in growth chambers at 22°C with illumination at 120 μmol/m2/s for a 16‐h daily light period. The growth chamber relative humidity remained ~ 70% (± 5%).
Primary root observation, K+ content measurement, and kinetic analysis of K+ uptake
Different materials for phenotype test were directly germinated on HK and LK medium for 7 days, and then, photographs were taken. Primary root lengths were measured using ImageJ software. At the same time, seedlings roots were harvested separately and washed thoroughly with double‐distilled water three times. For K+ content analyses, samples were dried at 80°C for 24 h to constant weight and the dry weight was measured. Then, samples were treated in a muffle furnace at 300°C for 1 h and 575°C for 5–6 h, and finally, samples were dissolved and diluted in 0.1 N HCl. The K+ concentrations were measured using 4100‐MP AES (Agilent). Three biological replicates were used in one independent experiment. About 200 individual seedlings from four plates were collected and used as one biological replicate for LK measurement; about 90–100 individual seedlings from two plates were collected and used as one biological replicate for HK measurement.
For K+‐depletion experiments, 5‐day‐old seedlings were collected (the fresh weight of each sample is 0.6 g) and pretreated in 1/4 HK solution at 22°C overnight. The seedlings were then transferred into K starvation solution (1.5 mM MgSO4·7H2O, 85 μl H3PO4, 2.99 mM Ca(NO3)2·4H2O, 5.12 mM MES, pH 5.80) for 4 days. Next, the seedlings were transferred to the K depletion solution (K starvation solution supplemented with 250 μM KCl). The experiments were conducted at 22°C in light, and all samples were shaken on a shaking table during the experiments (modified from previous method) 69. The solution samples were collected at different time points as indicated. K+ concentrations were measured using the 4100‐MP AES (Agilent).
Histochemical GUS staining and observation
The KUP9 promoter (about 2.8 kb) was cloned into pBI121 with a GUS coding region. The ProKUP9:GUS construct was then transferred into Col‐0 via Agrobacterium‐mediated transformation. About 4 independent transgenic lines expressing ProKUP9:GUS were analyzed. For GUS staining assay, seedlings were incubated in the GUS (0.5 mg/ml) staining solution (0.1 M phosphate buffer [pH 7.0], 5 mM K4Fe(CN)6·3H2O, 5 mM K3Fe(CN)6, 0.1% Triton X‐100, and 0.5 mg/ml 5‐bromo‐4‐chloro‐3‐indolyl β‐d‐glucuronic acid) from 5 to 45 min at 37°C for different zone observation of primary root and then cleared in 70% ethanol. Finally, the seedlings were photographed with Olympus SEX16 and Olympus BX53 microscope.
The transgenic plants carrying ProQC25:GUS were crossed with kup9‐1 mutants. The homozygous transgenic lines of F3 plants were selected on HK medium containing 50 mg/L kanamycin. GUS staining was carried out for 1 h at 37°C. Finally, the seedlings were photographed with Olympus BX53 microscope.
Microscopy imaging
For primary root tip observation, seedlings directly germinated on HK medium and LK medium for 7 days were imaged by Olympus SEX16 and measured the lengths using ImageJ software. The roots were placed in the solution (every 100 ml solution containing 7.5 g gum arabic, 100 g chloroacetaldehyde, 5 ml glycerol, and 60 ml ddH2O) for photographing with Olympus BX53 microscope. The meristem cell number can be measured as the number of cortex cells from the QC to the first elongated cell. The meristem cell length was measured as the distance between the QC cells and the first elongated cell 70. Elongation zone (EZ) cell number is from the first elongating cell to first cell with a root hair. Differential zone is from the first cell with a root hair to the joint of root and hypocotyl 49.
The transgenic plants carrying ProDR5:GFP and ProWOX5:GFP were crossed with kup9‐1 mutants. An Olympus SEX16 stereo fluorescence microscope was used to screen the homozygous transgenic plants. The homozygous transgenic lines of F3 plants were used for experiment. For confocal microscope images, the 4‐day‐old uniform seedlings directly germinated on HK medium were transferred to HK medium and LK medium for treating indicated time 24, 48, and 96 h. Then, the roots were incubated with 10 μM PI for 3 min before imaged by Zeiss 710 confocal microscope. PI fluorescence was used to visualize the cells in the root tip. The GFP was excited at 488 nm and detected at 505–560 nm with an argon laser (70% strength). The PI staining (excitation 536 nm, emission peak 617 nm) was detected with a ×40 (oil immersion) objective. The fluorescence intensity was measured by ImageJ program.
For quantification of the frequency of QC division, we counted the number of plants with abnormal QC division (at least one extra division) in a single medial longitudinal plane for each root. The frequency of QC division indicates the percentage of plants showing abnormal QC division in total plants. For each plant material, we analyzed at least 50 individual plants for each genotype.
Asante Potassium Green‐2 (APG‐2; TEFLabs, Austin, TX, USA) was employed to measure K+ concentrations in Arabidopsis root cells according to Wang et al 71. The indicators were dissolved in DMSO (Sigma) to a stock concentration of 1 mM. Arabidopsis seedlings directly germinated on HK medium were transferred to HK medium and LK medium for treating 24 h and then incubated in the measuring buffers (containing 1/4 HK/LK liquid medium, 5 μl 0.02% Pluronic F‐127, 2 μM APG) for 8 h in the dark at 22°C. The stained seedlings were washed in distilled water for 3 min to remove residual dyes before measuring fluorescence intensity. The GFP was excited at 488 nm and detected at 505–560 nm with an argon laser (70% strength).
For immunological staining assay, whole‐mount immunological staining on 7‐day‐old kup9‐1/ProKUP9:KUP9–GFP seedlings was done in an Intavis robot as described previously by Marvec et al 18 and Ding et al 21. Antibody was used at the following dilution: anti‐BIP2 (Hsc70), 1:200 (Stressgen Bioreagents). Anti‐rabbit antibody conjugated with fluorescein isothiocyanate (FITC; Dianova, Germany) was used at 1:600 dilutions. For ER‐Tracker Red (BODIPY TR Glibenclamide) dye labeling, kup9‐1/ProKUP9:KUP9‐GFP seedlings were mounted in water with a 1:1,000 dilution to a final ER‐Tracker dye (Invitrogen) concentration 1 μM.
Microsome and plasma membrane isolation
The method described previously 72 was used for the isolation of microsome from Arabidopsis with minor modifications. About 1 g of roots was harvested from the 10‐day‐old kup9‐1/ProKUP9:KUP9‐GFP plants. Then, the homogenates were centrifuged at 100,000 g at 4°C for 1 h. The supernatant was carefully removed, and the membrane pellets were washed with 1.5 ml wash buffer (20 mM Tris–HCl [pH 7.5], 5 mM EDTA, 5 mM EGTA, and 1 mM PMSF). Samples were centrifuged (100,000 g, 45 min), and wash buffer was discarded. Membrane pellets were dissolved in storage buffer and used for Western blot.
Plasma membrane was isolated from 10‐day‐old kup9‐1/ProKUP9:KUP9‐GFP plants using the aqueous two‐phase separation method 73, 74. About 1 g of roots were prepared. Finally, the PM was used for Western blot.
Auxin transport assays in tobacco BY‐2 cells
BY‐2 cells were treated with enzyme solution containing 1.5% (w/v) cellulase, 0.4% (w/v) macerozyme R‐10 in 0.4 M mannitol, 10 mM CaCl2, 20 mM KCl, 20 mM MES/KOH, pH 7.5, and 0.1% BSA for 4 h. The protoplast solution was filtered with nylon mesh into a round bottom tube carefully. The filtered protoplast solution was centrifuged at 100 g for 2 min, the supernatant was removed carefully, and 1 ml W5 solution (containing 154 mM NaCl, 125 mM CaCl2, 5 mM KCl and 2 mM MES/KOH, pH 7.5) was added into the tube and then gently rotated to re‐suspend the protoplasts. Twenty microliter protoplast solution was pipetted to the counting chamber to calculate protoplasts. Approximately 80,000 protoplasts were used for each transfection; then, the protoplasts were put on ice for 20 min for sedimentation. The supernatant was removed, and MMG solution (containing 400 mM mannitol, 15 mM MgCl2, 4 mM MES/KOH, pH 7.5) was added into the round bottom tube with the protoplasts. Twenty‐five microgram plasmids, 350 μl protoplast/MMG solution into the tubes, and equal column PEG solution were added into empty tubes successively, gently mixed, and reacted for 15 min at the dark. Then, 1 ml W5 solution was added into each tube to stop the reaction. After centrifugation for 2 min at 100 g, the supernatant was removed carefully and 1 ml W5 solution per transfection was added carefully. The protoplasts were incubated in the dark for 16 h.
Auxin transport assays in suspension‐cultured tobacco BY‐2 cells were carried out as described with minor modifications 60. Briefly, the accumulation was measured in 1‐ml aliquots of cell suspension. Each cell suspension was filtered, resuspended in uptake buffer (20 mM MES, 40 mM Sucrose, and 0.5 mM CaSO4, pH adjusted to 5.7 with KOH), and equilibrated for 45 min with continuous orbital shaking. Equilibrated cells were collected by filtration, resuspended in fresh uptake buffer, and incubated on the orbital shaker for 1.5 h in darkness at 25°C. 3H‐IAA was added to the cell suspension to give a final concentration of 2 nM. After a timed uptake (2 h) period, 1‐ml aliquots of suspension were withdrawn and accumulation of label was terminated by rapid filtration under reduced pressure on 22‐mm‐diameter cellulose filters. The cell cakes and filters were transferred to scintillation vials and extracted in ethanol for 30 min, and radioactivity was determined by liquid scintillation analyzer (PerkinElmer 1450 MicroBeta TriLux). Three biological replicates were used in one independent experiment. Counts were corrected for surface radioactivity by subtracting counts obtained for aliquots of cells collected immediately after the addition of 3H‐IAA.
Auxin transport assays in Xenopus oocytes
Coding sequences of KUP9, KUP10, PIN4, PIN3, D6PK, and PIN8 were cloned into the expression vector pGEMHE. OST4 (a small membrane anchor cloned from Yeast) and GFP were fused at the N‐terminus of KUP9 so that the localization of KUP9 at the PM of oocytes can be easily observed. After linearization of pGEMHE plasmids with NheI (SphI for KUP10‐pGEMHE), RNA was transcribed in vitro using an mRNA synthesis kit RiboMAX™ Large Scale RNA Production System‐T7 (Promega). cRNA concentration was adjusted to 1,000 ng/μl. Xenopus oocytes were isolated in 25 ml ND96 solution without Ca2+ containing 43 mg collagenase and 12.5 mg trypsin inhibitor at 22°C for 1.5 h and then recovered in ND96 with Ca2+ for 6 h. Stage V and VI oocytes were chosen for injection with 40 ng cRNA (for each protein) using a 10–15 μm tip diameter micropipette and a pneumatic injector and incubated at 17°C in modified Barth's solution (MBS) containing (in mM) 88 NaCl, 1.0 KCl, 2.4 NaHCO3, 0.91 CaCl2, 0.33 Ca(NO3)2, 0.82 MgSO4, 2.4 NaHCO3, and 10 HEPES‐NaOH (pH 7.5) for 48 h, supplemented with gentamycin (0.1 mg/ml) and streptomycin (0.1 mg/ml) before performing 3H‐IAA transport assays. Oocytes injected with water were used as control. The incubation solution was changed each day.
3H‐IAA transport analyses were adapted from previous publications 75, 76. Briefly, after preincubation in Ringer solution (115 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, 1 mM NaHCO3, 10 mM HEPES‐NaOH, 1 mM MgCl2) pH 6.4 for 20 min, the oocytes were transferred to Ringer solution containing 1 μM 3H‐IAA (100 nM 3H‐IAA; GE Healthcare; diluted with 900 nM cold‐IAA; Sigma) and incubated for indicated time 30 min, 2 h, and 4 h at room temperature, and the oocytes were then washed five times with Ringer solution containing 5 μM unlabeled IAA. Six oocytes incubated in each Petri dish represented one sample. Each sample was subsequently lysed in 300 μl 2% SDS for 30 min at room temperature, and the scintillation mixture was added to each sample. Incorporated radioactivity was measured by liquid scintillation analyzer (PerkinElmer 1450 MicroBeta TriLux). Three biological replicates were used in one independent experiment.
For IAA efflux assays, after incubation at 17°C for 48 h to allow for protein synthesis, oocytes were washed five times with K+‐free MBS solution. Each oocyte was injected with equal 40 pmol IAA and incubated in MBS solution without K+ or with 50 mM K+ for 6 h to allow for IAA efflux. An outside MBS buffer at pH 7.5 was chosen to prevent passive diffusion of IAA into the oocytes, which would take place at acidic pH. Six oocytes represented one sample and were harvested for IAA measurement of net accumulation. The IAA measurement detail was as below described “Quantification of IAA and IAA metabolites”. At least three biological replicates were used in one independent experiment.
K+ release assays in Xenopus oocytes
K+ release assays were carried out as described previously 77. Oocytes expressed different proteins were injected with equal 40 pmol IAA and washed five times with K+‐free MBS solution, and then, the oocytes were incubated in K+‐free MBS solution (pH 7.5) for 6 h. As described above, oocytes injected with equal volume water and BA were used as control. Six oocytes incubated in each Petri dish represented one sample for one protein. At least three biological replicates were used in one independent experiment. K+ content of the bath solution in each Petri dish was analyzed using the 4100‐MPAES system. K+ efflux activities of the oocytes were then calculated.
Quantification of IAA and IAA metabolites
Different materials were directly germinated on HK and LK medium for 7 days. Root tips were harvested using a razor blade under the microscope. For free IAA and IAA conjugate quantification, ~ 50 mg fresh weight of plant materials was taken for analysis. The materials immediately frozen in liquid nitrogen were ground with pestle and extracted for 30 min at 4°C with 500 μl cold extraction buffer (isopropanol: H2O: HCl = 2:1:0.002, v/v). Fifty microliter d2‐IAA was added as an internal standard for analysis. Each sample was added with 1 ml methenyl trichloride and shook (900 rpm) for 30 min at 4°C. Then, the samples were centrifuged at 4°C, 14,000 g for 5 min. The 1.2 ml lower liquid was transferred to 1.5‐ml Ep tubes and evaporated by nitrogen. Each sample was resuspended in precooling methanol and mixed thoroughly. After centrifugation at 4°C, 14,000 g for 5 min, each sample supernatant was filtered using a micro‐spin filter tube and quantified by ultra‐high‐performance liquid chromatography coupled to tandem mass detection (UPLC‐MS). IAA, IA‐Asp, and IA‐Glu were selected as standards. The linear range spanned at least three orders of magnitude with a correlation coefficient of 0.9997–0.9999. At least three independent biological replicates of each material were tested in this assay.
Yeast complementation assays
The coding sequences of KUP9 were constructed into p416‐GPD vector and transformed into yeast strain R5421 (trk1Δ, trk2Δ), in which two endogenous K+ transporter genes (TRK1, 2) were deleted. Yeast strain R757 was used as positive control. The yeast complementation experiment was performed as described previously 78.
RT–qPCR analysis
For RT–qPCR analyses, total RNA was extracted from roots of 7‐day‐old seedlings by using TRIzol reagent (Invitrogen) and then treated with DNase I (RNase Free, Takara) to eliminate genomic DNA contamination. The cDNA was synthesized by SuperScriptII RNase reverse transcriptase (Invitrogen). Oligo (dT) primers (Promega) were used for RT–qPCR analyses. RT–qPCR was conducted using Power SYBR Green PCR Master Mix (Applied Biosystems, USA) on a 7500 Real‐Time PCR System machine (Applied Biosystems). The amplification reactions were performed in a total volume of 20 μl, which contained 10 μl SYBR Green premix, 7 μl ddH2O, 2 μl forward and reverse primers (1 μM), and 1 μl cDNA. The PCR was programmed as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. RT–qPCR results were calculated by normalization to Actin2/8 gene. Three biological replicates were used in one independent experiment. Each replicate contained 120–150 individual seedlings. Three independent experiments were performed in one RT–qPCR analysis.
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis TAIR database (https://www.arabidopsis.org/index.jsp) under the following accession numbers: At4g19960 for KUP9, At1g31120 for KUP10, At2g35060 for KUP11, At4g13420 for HAK5, At5g09400 for KUP7, At2g26650 for AKT1, At1g73590 for PIN1, At5g57090 for PIN2, At1g70940 for PIN3, At2g01420 for PIN4, At1g23080 for PIN7, At2g38120 for AUX1, At5g15100 for PIN8, AT4G32540 for YUC1, AT4G13260 for YUC2, AT1G04610 for YUC3, AT5G11320 for YUC4, AT5G43890 for YUC5, AT5G43890 for YUC6, AT5G25620 for YUC6, AT2G33230 for YUC7, AT4G28720 for YUC8, AT1G04180 for YUC9, AT1G48910 for YUC10, AT1G70560 for TAA1, AT4G24670 for TAR2, AT1G19920 for ASA1, and AT1G25220 for ASB1.
Author contributions
MLZ, ZD, YG, WHW, and YW designed the research. M‐Z, PPH, YJ, SW, SSW, and ZL conducted the experiments. MLZ, ZD, and YW wrote and revised the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Source Data for Figure 9
Acknowledgements
We thank Dr. Qijun Chen (China Agricultural University) for providing the CRISPR/Cas9 vector. We thank Dr. Zhizhong Gong and Dr. Jing Zhang (China Agricultural University) for providing the ProDR5:GFP, ProWOX5:GFP, ProQC25:GUS plants. We also thank Dr. Keke Yi (Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences) and Dr. Eva Benková (Institute of Science and Technology Austria) for the help and discussion on this article. This work was supported by grants from the National Natural Science Foundation of China (Nos. 31761133011, 31921001, and 31622008) and Chinese Universities Scientific Fund (2019TC122 and 2019TC228).
EMBO Reports (2020) 21: e50164
References
- 1. Clarkson DT, Hanson JB (1980) The mineral nutrition of higher plants. Annu Rev Plant Physiol 31: 239–298 [Google Scholar]
- 2. Schroeder JI, Ward JM, Gassmann W (1994) Perspectives on the physiology and structure of inward‐rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu Rev Biophys Biomol Struct 23: 441–471 [DOI] [PubMed] [Google Scholar]
- 3. Maathuis FJM (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- 4. Hodge A (2004) The plastic plant: root responses to heterogeneous supplies of nutrients. New Phytol 162: 9–24 [Google Scholar]
- 5. Wang Y, Wu WH (2013) Potassium transport and signaling in higher plants. Annu Rev Plant Biol 64: 451–476 [DOI] [PubMed] [Google Scholar]
- 6. Gruber BD, Giehl RFH, Friedel S, Wirén NV (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kellermeier F, Chardon F, Amtmann A (2013) Natural variation of Arabidopsis root architecture reveals complementing adaptive strategies to potassium starvation. Plant Physiol 161: 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, Amtmann A (2014) Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao YW, Class ADM, Crawford NM (1993) Ammonium inhibition of Arabidopsis root growth can be reversed by potassium and by auxin resistance mutations aux1, axr1, and axr2 . Plant Physiol 102: 983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chérel I, Lefoulon C, Boeglin M, Sentenac H (2014) Molecular mechanisms involved in plant adaptation to low K+ availability. J Exp Bot 65: 833–848 [DOI] [PubMed] [Google Scholar]
- 12. Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J (2013) Glc‐TOR signalling leads transcriptome reprogramming and meristem activation. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang L, Yu LJ, Zhang X, Fan B, Wang FZ, Dai YS, Qi H, Zhou Y, Xie LJ, Xiao S (2019) Autophagy regulates glucose‐mediated root meristem activity by modulating ROS production in Arabidopsis . Autophagy 15: 407–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee Y, Lee WS, Kim SH (2013) Hormonal regulation of stem cell maintenance in roots. J Exp Bot 64: 1153–1165 [DOI] [PubMed] [Google Scholar]
- 15. Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- 16. Petrášek J, Friml J (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- 17. Pencík AP, Simonovik B, Petersson SV, Henyková E, Simon S, Greenham K, Zhang Y, Kowalczyk M, Estelle M, Zazímalová E et al (2013) Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole‐3‐acetic acid catabolite 2‐oxindole‐3‐acetic acid. Plant Cell 25: 3858–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mravec J, Skupa P, Bailly A, Hoyerova K, Krecek P, Bielach A, Petrasek J, Zhang J, Gaykova V, Stierhof YD et al (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER‐localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- 19. Normanly J (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wabnik K, Kleine‐Vehn J, Govaerts W, Friml J (2011) Prototype cell‐to‐cell auxin transport mechanism by intracellular auxin compartmentalization. Trends Plant Sci 16: 468–475 [DOI] [PubMed] [Google Scholar]
- 21. Ding Z, Wang BJ, Moreno L, Dupláková N, Simon S, Carraro N, Reemmer J, Pencik A, Chen X, Tejos R et al (2012) ER‐localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis . Nat Commun 3: 941 [DOI] [PubMed] [Google Scholar]
- 22. Azpeitia E, Alvarez‐Buylla ER (2012) A complex systems approach to Arabidopsis root stem‐cell niche developmental mechanisms: from molecules, to networks, to morphogenesis. Plant Mol Biol 80: 351–363 [DOI] [PubMed] [Google Scholar]
- 23. Clark NM, de Luis Balaguer MA, Sozzani R (2014) Experimental data and computational modeling link auxin gradient and development in the Arabidopsis root. Front Plant Sci 5: 328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu YT, Xu MZ, Liang NS, Zheng YH, Yu QZ, Wu S (2017) Symplastic communication spatially directs local auxin biosynthesis to maintain root stem cell niche in Arabidopsis . Proc Natl Acad Sci USA 114: 4005–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Gene Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- 27. Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose‐dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- 28. Zhou WK, Wei LR, Xu J, Zhai QZ, Jiang HL, Chen R, Chen Q, Sun JQ, Chu JF, Zhu LH et al (2010) Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mähönen AP, Tusscher K, Siligato R, Smetana O, Díaz‐Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B (2014) PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu QQ, Tian HY, Yue K, Liu JJ, Zhang B, Li XG, Ding ZJ (2016) A P‐Loop NTPase regulates quiescent center cell division and distal stem cell identity through the regulation of ROS homeostasis in Arabidopsis root. PLoS Genet 12: e1006175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- 32. Nakajima K, Sena G, Nawy T, Benfey PN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- 33. Overvoorde P, Fukaki H, Beeckman T (2010) Auxin control of root development. Cold Spring Harbor Perspect Biol 2: a001537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Véry AA, Nieves‐Cordones M, Daly M, Khan I, Fizames C, Sentenac H (2014) Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J Plant Physiol 171: 748–769 [DOI] [PubMed] [Google Scholar]
- 36. Santa‐María GE, Rubio F, Dubcovsky J, Rodríguez‐Navarro A (1997) The HAK1 gene of barley is a member of a large gene family and encodes a high‐affinity potassium transporter. Plant Cell 9: 2281–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rigas S, Debrosses G, Haralampidis K, Vicente‐Agullo F, Feldmann KA, Grabov A, Dolan A, Hatzopoulos P (2001) TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elumalai RP, Nagpal P, Reed JW (2002) A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gierth M, Mäser P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K+ deprivation‐induced high‐affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Osakabe Y, Arinaga N, Umezawa T, Katsura S, Nagamachi K, Tanaka H, Ohiraki H, Yamada K, Seo SU, Abo M et al (2013) Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis . Plant Cell 25: 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grabov A (2007) Plant KT/KUP/HAK potassium transporters: single family ‐ multiple functions. Ann Bot 99: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scherzer S, Böhm J, Krol E, Shabala L, Kreuzer I, Larisch C, Bemm F, Al‐Rasheid KAS, Shabala S, Rennenberg H et al (2015) Calcium sensor kinase activates potassium uptake systems in gland cells of Venus flytraps. Proc Natl Acad Sci USA 112: 7309–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li J, Wu WH, Wang Y (2017) Potassium channel AKT1 is involved in the auxin‐mediated root growth inhibition in Arabidopsis response to low K+ stress. J Integr Plant Biol 59: 895–909 [DOI] [PubMed] [Google Scholar]
- 44. Vicente‐Agullo F, Rigas S, Desbrosses G, Dolan L, Hatzopoulos P, Grabov A (2004) Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J 40: 523–535 [DOI] [PubMed] [Google Scholar]
- 45. Rigas S, Ditengou FA, Ljung K, Daras G, Tietz Q, Palme K, Hatzopoulos P (2013) Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytol 197: 1130–1141 [DOI] [PubMed] [Google Scholar]
- 46. Pyo YJ, Gierth M, Schroeder JI, Cho MH (2010) High‐affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low‐potassium conditions. Plant Physiol 153: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Han M, Wu W, Wu WH, Wang Y (2016) Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+‐limited conditions. Mol Plant 9: 437–446 [DOI] [PubMed] [Google Scholar]
- 48. Calderón LS, Bucio JL, López AC, Ramírez AC, Jacobo FN, Dubrovsky JG, Estrella LH (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana . Plant Cell Physiol 46: 174–184 [DOI] [PubMed] [Google Scholar]
- 49. Yang L, Zhang J, He J, Qin Y, Hua D, Duan Y, Chen Z, Gong Z (2014) ABA‐mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis . PLoS Genet 10: e1004791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B (1997) Short‐range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289 [DOI] [PubMed] [Google Scholar]
- 51. Tian H, Smet DL, Ding ZJ (2014) Shaping a root system: regulating lateral versus primary root growth. Trends Plant Sci 19: 426–431 [DOI] [PubMed] [Google Scholar]
- 52. Drisch RP, Stahl Y (2015) Function and regulation of transcription factors involved in root apical meristem and stem cell maintenance. Front Plant Sci 6: 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiang K, Feldman LJ (2005) Regulation of root apical meristem development. Annu Rev Cell Dev Biol 21: 485–509 [DOI] [PubMed] [Google Scholar]
- 54. Barozzi F, Papadia P, Stefano G, Renna L, Brandizzi F, Migoni D, Fanizzi FP, Piro G, Sansebastistiano GPD (2019) Variation in membrane trafficking linked to SNARE AtSYP51 interaction with aquaporin NIP1;1. Front Plant Sci 9: 7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ding ZJ, Friml J (2010) Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA 107: 12046–12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moubayidin L, Mambro RD, Sozzani R, Pacifici E, Salvi E, Terpstra I, Bao D, van Dijken A, Dello Ioio R, Perilli S et al (2013) Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev Cell 26: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moubayidin L, Salvi E, Giustini L, Terpstra I, Heidstra R, Costantino P, Sabatini S (2016) A SCARECROW‐based regulatory circuit controls Arabidopsis thaliana meristem size from the root endodermis. Planta 243: 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pollmann S, Neu D, Lehmann T, Berkowitz O, Schäfer T, Weiler EW (2006) Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana . Planta 224: 1241–1253 [DOI] [PubMed] [Google Scholar]
- 59. Lehmann T, Hoffmann M, Hentrich M, Pollmann S (2010) Indole‐3‐acetamide‐dependent auxin biosynthesis: a widely distributed way of indole‐3‐acetic acid production? Eur J Cell Biol 89: 895–905 [DOI] [PubMed] [Google Scholar]
- 60. Petrášek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M et al (2006) PIN proteins perform a rate‐limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- 61. Chi JH, Roos J, Dean N (1996) The OST4 gene of Saccharomyces cerevisiae encodes an unusually small protein required for normal levels of oligosaccharyltransferase activity. J Biol Chem 271: 3132–3140 [DOI] [PubMed] [Google Scholar]
- 62. Ragel P, Ródenas R, García‐Martín E, Andrés Z, Villalta I, Nieves‐Cordones M, Rivero RM, Martínez V, Pardo JM, Quintero FJ et al (2015) CIPK23 regulates HAK5‐mediated high‐affinity K+ uptake in Arabidopsis roots. Plant Physiol 169: 2863–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Leigh RA, Wyn Jones RG (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol 97: 1–13 [Google Scholar]
- 64. Walker DJ, Leigh RA, Miller AJ (1996) Potassium homeostasis in vacuolate plant cells. Proc Natl Acad Sci USA 93: 10510–10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Friml J, Jones AR (2010) Endoplasmic reticulum: the rising compartment in auxin biology. Plant Physiol 154: 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Barbez E, Laňková M, Pařezová M, Maize l, Zažímalová E, Petrášek J, Friml J (2013) Single‐cell‐based system to monitor carrier driven cellular auxin homeostasis. BMC Plant Biol 13: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ranocha P, Dima O, Nagy R, Felten J, Corratge‐Faillie C, Novak O, Morreel K, Lacombe B, Martinez Y, Pfrunder S et al (2013) Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat Commun 4: 2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- 69. Wang XP, Chen LM, Liu WX, Shen L, Wang FL, Zhou Y, Zhang Z, Wu WH, Wang Y (2016) AtKC1 and CIPK23 synergistically modulate AKT1‐mediated low potassium stress responses in Arabidopsis . Plant Physiol 170: 2264–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Perilli S, Sabatini S (2010) Analysis of root meristem size development. Methods Mol Biol 655: 177–187 [DOI] [PubMed] [Google Scholar]
- 71. Wang F, Chen ZH, Liu X, Colmer TD, Zhou M, Shabala S (2016) Tissue‐specific root ion profiling reveals essential roles of the CAX and ACA calcium transport systems in response to hypoxia in Arabidopsis . J Exp Bot 67: 3747–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Abas L, Luschnig C (2010) Maximum yields of microsomal‐type membranes from small amounts of plant material without requiring ultracentrifugation. Anal Biochem 401: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang Y, Qin Y, Xie C, Zhao F, Zhao J, Liu D, Chen S, Fuglsang AT, Palmgren MG, Schumaker KS et al (2010) The Arabidopsis chaperone J3 regulates the plasma membrane H+‐ATPase through interaction with the PKS5 kinase. Plant Cell 22: 1313–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E (2006) High‐affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127 [DOI] [PubMed] [Google Scholar]
- 76. Krouk G, Lacombe B, Bielach A, Perrine‐Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K et al (2010) Nitrate‐regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- 77. Li H, Yu M, Du XQ, Wang ZF, Wu WH, Quintero FJ, Jin XH, Li HD, Wang Y (2017) NRT1.5/NPF7.3 functions as a proton‐coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis . Plant Cell 29: 2016–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li J, Long Y, Qi GN, Li J, Xu ZJ, Wu WH, Wang Y (2014) The Os‐AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1‐CIPK23 complex. Plant Cell 26: 3387–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Source Data for Figure 9


