Abstract
Cohesin cofactors regulate the loading, maintenance, and release of cohesin complexes from chromosomes during mitosis but little is known on their role during vertebrate meiosis. One such cofactor is PDS5, which exists as two paralogs in somatic and germline cells, PDS5A and PDS5B, with unclear functions. Here, we have analyzed their distribution and functions in mouse spermatocytes. We show that simultaneous excision of Pds5A and Pds5B results in severe defects during early prophase I while their individual depletion does not, suggesting their functional redundancy. Shortened axial/lateral elements and a reduction of early recombination nodules are observed after the strong depletion of PDS5A/B proteins. Moreover, telomere integrity and their association to the nuclear envelope are severely compromised. As these defects occur without detectable reduction in chromosome‐bound cohesin, we propose that the dynamic behavior of the complex, mediated by PDS5 proteins, is key for successful completion of meiotic prophase I.
Keywords: axial elements, meiosis, mouse, PDS5, telomeres
Subject Categories: Cell Cycle; Chromatin, Epigenetics, Genomics & Functional Genomics
Analysis of spermatogenesis in mice carrying conditional knock out alleles for cohesin cofactors PDS5A and PDS5B reveals the importance of cohesin dynamics for proper assembly of the synaptonemal complex, homolog recombination and telomere integrity.

Introduction
Meiosis is a highly specialized cell division process that produces haploid gametes by performing two consecutive rounds of chromosome segregation after a single round of DNA replication. The accurate segregation of chromosomes during meiosis relies on the proper achievement of the preceding processes of homologous chromosome pairing, synapsis, and recombination 1. Meiotic recombination initiates in mammals during the leptotene stage of prophase I by the formation of programmed DNA double‐strand breaks (DSBs) by the endonuclease SPO11 2, which leads to the phosphorylation of the histone variant H2AX at serine 139 (γ‐H2AX) in the surrounding chromatin 3. After a 5′–3′ resection of DNA at DSBs, the recombinase RAD51, among other proteins, associates to the 3′ single‐stranded DNA and promotes the invasion of the double‐stranded DNA of the homolog, a process that facilitates the recognition and pairing of the homologs 4. Some of these events are finally processed as reciprocal crossovers via the participation of the DNA mismatch repair protein MLH1 5. Although meiotic recombination initiates early in meiosis, its resolution takes place at the pachytene stage after the homologs have achieved synapsis. Synapsis is mediated by the synaptonemal complex (SC), a tripartite proteinaceous structure specific of meiosis that assembles during prophase I 6. The SC assembly is initiated in leptotene, when an axial element (AE) is developed along each homolog over the trajectories of previously loaded cohesin complexes. During this stage, the telomeres attach to the nuclear envelope (NE), and by zygotene, they adopt a polarized “bouquet” configuration that promotes chromosome movements essential for accurate homolog pairing 7. The tethering of telomeres to the NE is mediated by several adaptor proteins whose deficiencies provoke alterations not only in telomere attachment but in pairing, synapsis and recombination leading in many cases to infertility 8. Subsequently, the so‐called central element (CE) is formed between the AEs of the SC. At this time, the AEs are referred as lateral elements (LEs) and the CE, composed by transverse filaments, physically connects the LEs of the two homologs. By pachytene, the homologs have achieved synapsis along their entire length and thus each bivalent presents a fully developed SC.
During the first and second meiotic divisions, recombined homologs and single chromatids migrate to opposite spindle poles, respectively, due to a sequential loss of arm and centromere sister‐chromatid cohesion 9, 10. Cohesion is mediated in somatic vertebrate cells by ring‐shaped cohesin complexes composed of two structural maintenance of chromosomes (SMC) proteins, SMC1α and SMC3, the kleisin subunit RAD21 and a STAG/SA subunit that can be SA1 or SA2 11. The dynamic loading, maintenance, and release of cohesin complexes from chromatin are mediated by cofactor proteins and some posttranslational modifications 12, 13. The functions of these cofactors have been characterized during the mitotic cycle, but much less is known of their behavior in meiosis. Once cohesin is loaded on chromatin in early G1 by the NIPBL‐MAU2 heterodimer, PDS5 and WAPL bind to the complex and promote their dissociation 14, 15. During S‐phase, as sister chromatids arise from the replication fork, cohesion is established by a fraction of cohesin complexes that are acetylated in their SMC3 subunit and bound by Sororin, which counteracts the cohesin‐releasing activity of WAPL until mitotic prophase 16, 17. Two paralogs of the PDS5 protein exist in vertebrate cells, PDS5A and PDS5B, which are required both for cohesin dissociation together with WAPL, and for cohesin stabilization together with Sororin 14, 18, 19, 20. While the two PDS5 proteins present these activities, centromeric cohesion defects are more apparent in Pds5B‐deficient cells in mitosis 19.
In addition to the canonical cohesin subunits mentioned above, several meiosis‐specific subunits have been described during mammalian meiosis. SMC1β and STAG3 are the paralogs of SMC1α and SA1/2, respectively, while REC8 and RAD21L are the meiotic counterparts of the kleisin subunit RAD21 10. The distribution and functions of the different cohesin subunits during mammalian meiosis have been extensively analyzed. All of them, both mitotic and meiosis‐specific ones, are located at AEs/LEs during mouse prophase I. Spermatocytes from mice deficient for the meiosis‐specific cohesin subunits SMC1β 21, 22, 23, 24, REC8 25, 26, RAD21L 27, and STAG3 28, 29, 30, 31 arrest at different stages of prophase I and display severe defects in the assembly and pairing of AEs/LEs, recombination and, in some cases, show altered telomere structure. However, there are few studies describing the dynamics and function of cohesin cofactors during mammalian meiosis. WAPL decorates the AEs/LEs in mouse oocytes 32 and spermatocytes, where it is involved in the removal of arm cohesion by the end of prophase I 33. Unexpectedly, Sororin has been detected at the SC central region, unlike the cohesin subunits and WAPL 34, 35.
The single orthologs of PDS5 in Sordaria macrospora (Spo76p) and budding yeast (Pds5) are located at AEs/LEs during prophase I stages 36, 37, 38. Analyses of meiosis in Pds5 mutants in Sordaria, budding and fission yeasts, and the worm Caenorhabditis elegans, indicate that this protein is involved in several processes including cohesion, formation of AEs/LEs, condensation, homolog pairing, and repair of DSBs 36, 37, 38, 39, 40, 41, 42. In contrast, simultaneous ablation of four out of the five Pds5 genes present in Arabidopsis thaliana does not have a relevant impact in meiotic chromosome structure, or the progression of meiosis 43. In mammalian meiosis, like in mitosis, both PDS5A and PDS5B are present. While nothing is known about the localization of PDS5A, PDS5B has been detected at AEs/LEs in mouse spermatocytes 44. In the present study, we report the distribution of PDS5A and PDS5B during male mouse meiosis. Moreover, taking advantage of conditional knock out (cKO) mouse models previously generated 19, we have analyzed the meiotic phenotype of spermatocytes after drastic depletion of one or both PDS5 proteins. Our results indicate that a single PDS5 protein, either PDS5A or PDS5B, is sufficient for prophase I progression. However, their simultaneous depletion results in severe meiotic defects including the formation of shortened AEs/LEs, reduced formation of early recombination nodules, and alterations in the structure of telomeres and their attachment to the NE during prophase I.
Results
Different localization and dynamics of PDS5A and PDS5B in mouse spermatocytes
We analyzed the distribution of PDS5A and PDS5B proteins on spread mouse spermatocytes by immunofluorescence using specific antibodies for these proteins (Fig EV1A–C) and for SYCP3, a structural component of the AEs/LEs, to identify the different meiotic stages. PDS5A was undetectable during leptotene when SYCP3‐labeled AEs started to organize (Fig 1A and B), but colocalized with SYCP3 along the synapsing AEs/LEs of both autosomes and sex chromosomes during zygotene (Fig 1C and D) and early pachytene (Fig 1E and F). By mid‐pachytene, however, the PDS5A labeling was dispersed throughout the chromatin and was less intense on the sex body (Fig 1G and H). In diplotene, lack of staining was additionally observed around the ends of the desynapsing autosomal LEs which corresponded to DAPI‐positive regions, namely the chromocenters (Fig 1I–L). This distribution of PDS5A persisted in diakinesis although the overall signal intensity was slightly reduced (Fig 1M and N). By metaphase I, PDS5A was found at centromeres (Fig 1O and P). In side‐viewed centromeres, PDS5A was present as a T‐shaped signal below the closely associated sister kinetochores stained with an ACA serum (Fig 1O; bottom inset), while viewed from the top PDS5A encircled sister kinetochores (Fig 1O; top inset). This localization is similar to that of SYCP3 at the inner centromere domain 45, 46, but different from that of the meiosis‐specific cohesin subunit REC8 which localized in small patches along the interchromatid domain at the arms (Fig 1Q and R). Both PDS5A and REC8 were detected at the inner centromere domain, but in side‐viewed centromeres, the PDS5A signals were larger than those of REC8 (Fig 1Q, inset). In metaphase II PDS5A was also present at the centromeres (Fig 1S and T). In side‐viewed centromeres, two PDS5A signals were detected below kinetochores (Fig 1S; bottom inset), while in top‐viewed centromeres, a single PDS5A ring encircled kinetochores (Fig 1S; top inset).
Figure EV1. Characterization of mouse models.

-
ASchematic representation of the conditional alleles (loxfrt or lox) and the null allele (−) obtained upon Cre‐mediated recombination for Pds5A (top) and Pds5B (bottom). The position of the primers used for genotyping is indicated (blue arrows).
-
BExample of genotyping PCRs for Pds5A (top) and Pds5B (bottom) alleles using DNA obtained from testis of the indicated mice as well as DNA from mouse embryo fibroblasts as control. cKO mice carry the conditional allele(s) in homozygosis and a Cre‐ERT2 transgene and have been treated with TX to promote the translocations of the Cre recombinase to the nucleus. In most cKO mice, excision of the targeted exon is not complete and a weak band corresponding to the conditional allele can be still be detected in addition to the stronger band of the null allele. The sizes of the different PCR products are as follows: Pds5A wild‐type (+) 872 bp, loxfrt 778 bp and null (−) 414 bp; for Pds5B wild‐type (+) 706 bp, lox 859 bp and null (−) 415 bp.
-
CImmunoblot analysis of total extracts prepared from testes of mice of the indicated genotypes. Decreasing amounts of extract from testes obtained from wild‐type mice (WT) were loaded for comparison. Overall, elimination of Pds5B is less efficient than elimination of Pds5A both in the single and in the double cKO mice.
-
D–MDouble immunolabeling of PDS5A (green, D–H) or PDS5B (green, I–M) and SYCP3 (red) in Pds5A cKO spread spermatocytes at the indicated prophase I stages (D–G and I–L) and in metaphase I bivalents (H and M).
-
N–WAs in (D–M), but in Pds5B cKO spread spermatocytes at the indicated prophase I stages (N–Q and S–V), and in metaphase I bivalents (R and W).
Figure 1. PDS5A distribution in mouse spermatocytes.
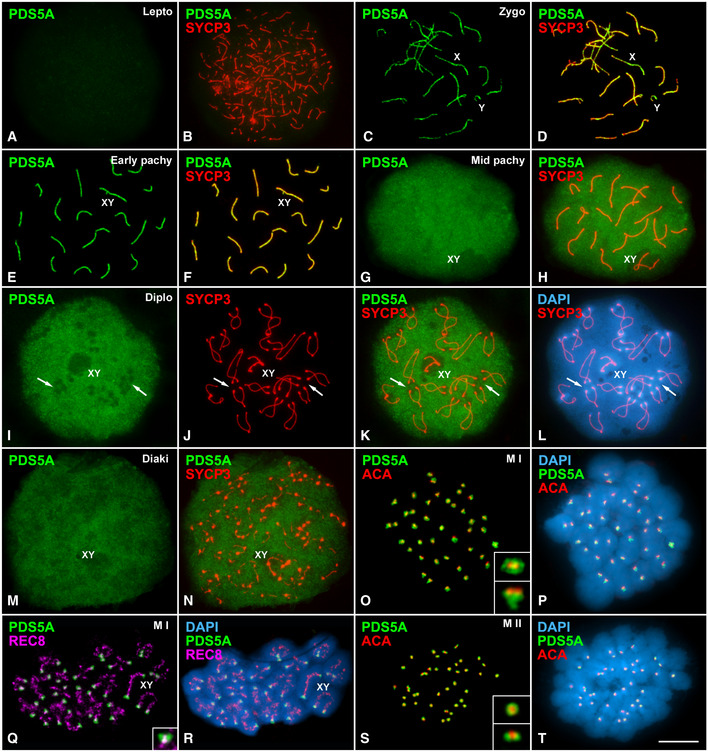
-
A–NDouble immunolabeling of PDS5A (green) and SYCP3 (red) in mouse spread spermatocytes in leptotene (A, B), zygotene (C, D), early pachytene (E, F), mid‐pachytene (G, H), diplotene (I–L), and diakinesis (M, N). Arrows in (I–L) indicate the position of some chromocenters.
-
O, PDouble immunolabeling of PDS5A (green) and kinetochores revealed with an ACA serum (red) in a metaphase I spermatocyte.
-
Q, RDouble immunolabeling of PDS5A (green) and REC8 (pseudocolored in purple) in a metaphase I spermatocyte.
-
S, TDouble immunolabeling of PDS5A (green) and kinetochores (ACA, red) in a metaphase II spermatocyte.
Unlike PDS5A, PDS5B colocalized with SYCP3 along the AEs/LEs of the autosomes and sex chromosomes from leptotene up to pachytene (Fig 2A–F). In addition, rounded PDS5B signals protruded from the AEs/LEs ends in pachytene (Fig 2E and F). REC8 colocalized with PDS5B along the AEs/LEs, but was not present at the two close PDS5B signals at SC ends (Fig 2G and H). Instead, PDS5B signals at AEs/LEs ends colocalized with TRF1, a telomere binding protein (Fig 2I and J). To validate PDS5B location at telomeres, we also analyzed its distribution in spermatocytes from mice deficient for the meiosis‐specific cohesin subunits SMC1β 23 and REC8 47, which present variable degrees of telomere abnormalities. Smc1β −/− spermatocytes present, among other phenotypes, telomere signals that are split, extended, absent, or disconnected from SC ends 23. Our results showed that in Smc1β −/− pachytene‐like spermatocytes, PDS5B localized along the shortened SCs and unsynapsed AEs/LEs (Appendix Fig S1A and B), and as altered signals at SC ends (Appendix Fig S1B and C), similar to those reported for telomeric DNA or telomere proteins in Smc1β −/− mouse spermatocytes 23. Telomere alterations are less severe in Rec8 −/− spermatocytes and only 3–4 chromosomes per spermatocyte present decreased levels of telomeric DNA and telomere proteins 47. Accordingly, in Rec8 −/− zygotene‐like spermatocytes, we found PDS5B at shortened unsynapsed AEs and at most telomeres (Appendix Fig S1D and E), and only a low number of AEs ends presented a highly reduced PDS5B labeling (Fig EV2E and F). Altogether our results indicate that PDS5B accumulates at telomeres during prophase I in mouse spermatocytes.
Figure 2. PDS5B distribution in prophase I spermatocytes.

-
A–FDouble immunolabeling of PDS5B (green) and SYCP3 (red) in spread spermatocytes at the indicated stages.
-
G–LImmunolabeling of PDS5B (green) and either REC8 (pseudocolored in blue, G, H) or SYCP3 (red) and TRF1 (pseudocolored in yellow, I, J) or ACA (pseudocolored in purple, K, L) onto a spread pachytene spermatocyte.
-
M–PDouble immunolabeling of PDS5B (green) and SYCP3 (red) on diplotene (M, N) and diakinesis (O, P) spread nuclei.
Figure EV2. Reduction of PDS5 proteins in Pds5AB cKO spermatocytes.
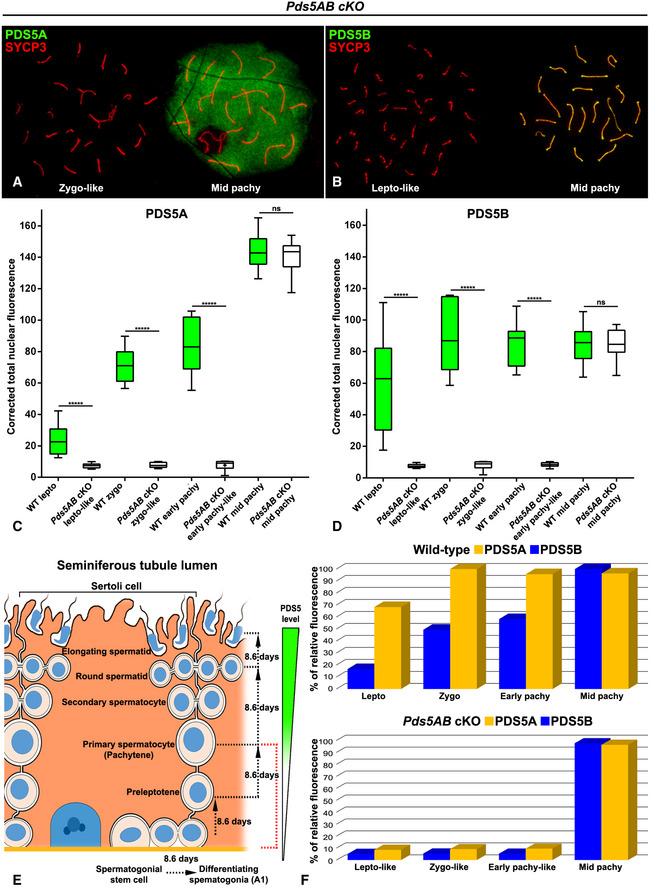
-
A, BRepresentative images of Pds5AB cKO spread spermatocytes at the indicated stages double immunolabeled for SYCP3 (red) and either PDS5A (green, A) or PDS5B (green, B).
-
C, DBox whiskers plot analyses of corrected total nuclear fluorescence of PDS5A (C) and PDS5B (D) in a given wild‐type (WT, green) or Pds5AB cKO (white) spermatocyte at the indicated stages (n = 10 for each stage). Whiskers indicate the minimum to maximum values, first and third quartiles are depicted by the box, and the horizontal lines in the boxes indicate the median value. Statistical significance was assessed using an unpaired two‐tailed t‐test function (*****P < 0.00001; ns, not significant difference).
-
ESchematic representation of cell progression and timing throughout the indicated stages of spermatogenesis in TX‐treated Pds5AB cKO testes. In mouse, the transition from a differentiating spermatogonia (A1) into a preleptotene cell lasts around 8.6, 8.6 days more to become a pachytene spermatocyte, and finally 17.2 days to progress to elongated spermatids. Note that in seminiferous tubules, this process is continuous and different cell populations coexist. Under our experimental conditions, if the targeted exons were excised in spermatogonial cells, depletion or extreme reduction of PDS5 protein levels would only be accomplished in spermatocytes up to the early pachytene stage (highlighted in red).
-
FPercentage of fluorescence intensity, relative to their respective maximum values, for PDS5A (yellow) and PDS5B (blue) in wild‐type (top) and Pds5AB cKO (bottom) spermatocytes at the indicated stages (n = 10 for each stage).
Consistent with the telocentric nature of mouse chromosomes, kinetochore signals were always internally located in relation to PDS5B telomeric ones at the centromeric end of the pachytene SCs (Fig 2K and L). By diplotene, PDS5B persisted along the desynapsing LEs and at telomeres (Fig 2M and N). In some images, we could detect two closely associated PDS5B signals at desynapsed LE ends corresponding to telomeres of sister chromatids (Fig 2M and N, insets). During diakinesis, PDS5B labeling began to disappear from the autosome LEs but not from telomeres (Fig 2O and P). In metaphase I, large PDS5B signals colocalized with SYCP3 at the inner centromere domains (Fig 3A–F). Interestingly, smaller PDS5B signals were located in pairs at the distal chromosome ends (Fig 3C–F), that are reminiscent of the location of distal telomeres in metaphase I bivalents 48. In metaphase II chromosomes, large PDS5B signals were found at each centromere between the kinetochores (Fig 3G–J). Altogether, our results indicate that PDS5A and PDS5B are present along the AEs/LEs in prophase I, similar to cohesin subunits, although the two paralogs display somehow different dynamics. Moreover, only PDS5B appears at telomeres.
Figure 3. PDS5B distribution in condensed meiotic chromosomes.

-
A–FDouble immunolabeling of PDS5B (green) and SYCP3 (red) in a metaphase I spermatocyte (A, B) and selected bivalents (C–F).The sex bivalent (XY) is indicated.
-
G–JDouble immunolabeling of PDS5B (green) and kinetochores (ACA, red) and staining of the chromatin (DAPI) in a metaphase II spermatocyte.
Conditional knock out mouse models for PDS5 protein depletion
The different localization patterns of PDS5A and PDS5B during prophase I stages suggest that they may have specific functions in meiosis. To address this possibility, we took advantage of Pds5‐deficient mice generated previously 19. Since constitutive knock out alleles for each gene are lethal in homozygosis, we used male mice carrying cKO alleles for Pds5A, Pds5B or both (Pds5AB hereafter) in combination with a Cre recombinase allele that can be induced by tamoxifen (TX) supplemented in the diet (Fig EV1A). The health condition of Pds5AB cKO mice was critically deteriorated after 2 weeks of treatment. Thus, in order to ensure the animal's welfare and to standardize the experimental conditions, mice of all genotypes were fed TX diet for 15 days, and then, testes were extracted for analyses.
Efficient, and almost complete, excision of the corresponding targeted exon(s) in testis from TX‐treated Pds5A cKO, Pds5B cKO, and Pds5AB cKO mice was confirmed by PCR. However, immunoblot analyses of testis extracts showed only a partial decrease of protein levels (Fig EV1B and C). It must be noted that testis comprises a mixture of heterogeneous and asynchronous cell populations, both at the seminiferous tubules (i.e. Sertoli cells, spermatocytes, and spermatids) and at the testis interstitium (i.e. Leydig cells and fibroblasts), which could present different turnover rates of PDS5 protein levels. To assess the depletion of PDS5 proteins in individual spermatocytes, we analyzed preparations from TX‐treated Pds5A cKO, Pds5B cKO, and Pds5AB cKO mice by immunofluorescence with antibodies against PDS5A, PDS5B, and SYCP3. In leptotene, zygotene and early pachytene spermatocytes from Pds5A cKO (Fig EV1D–F and I–K), Pds5B cKO (Fig EV1N–P and S–U), and Pds5AB cKO mice (Figs 4A–C and F–H, and EV2A and B), the levels of the PDS5 protein(s) whose gene had been presumably excised were below the detection threshold. However, PDS5A and PDS5B labeling were indistinguishable between wild‐type and Pds5 cKO spermatocytes in later meiotic stages (Figs EV1, EV2A, and B, and 4). Quantification of nuclear PDS5A and PDS5B fluorescence intensities demonstrated the strong depletion of both PDS5 proteins in individual leptotene‐like, zygotene‐like and early pachytene‐like Pds5AB cKO spermatocytes compared to wild‐type levels (Fig EV2C and D). We determined a 3.25‐, 9.14‐ and 11‐fold reduction of PDS5A fluorescence in leptotene‐like, zygotene‐like and early pachytene‐like Pds5AB cKO spermatocytes (Fig EV2C and F). Similarly, PDS5B fluorescence was 8.03‐, 11‐, and 10.08‐fold reduced in the same stages (Fig EV2D and F). These results indicate an almost complete absence of PDS5 proteins in these Pds5AB cKO early prophase I spermatocytes. On the other hand, the nuclear fluorescence levels of both PDS5 proteins in Pds5AB cKO mid‐pachytene spermatocytes were similar to those found in wild‐type ones (Fig EV2A–D and F).
Figure 4. Reduced AEs/LEs length in Pds5AB cKO spermatocytes.

-
A–JDouble immunolabeling of either PDS5A (green, A–E) or PDS5B (green, F–J) and SYCP3 (red) in Pds5AB cKO spread spermatocytes at the indicated stages.
-
K–MPreparations from the same mice were immunolabeled with the testis‐specific histone variant H1t (green) and SYCP3 (red).
-
NScatter dot‐plot graph of total SC length measured in the autosomal pairs of 33 early pachytene cells from three different individuals of each genotype. Bars indicate mean and standard error. Statistical significance was assessed using an unpaired two‐tailed t‐test function (****P < 0.0001).
We reckon that PDS5 proteins are highly stable, as previously noticed in mouse embryo fibroblasts in which protein elimination required at least 5 days following Cre activation 19. Moreover, cohesin mediating cohesion in mammalian oocytes shows very little turnover over several months 49 and the same could be true for cohesin cofactors. Thus, in A1 spermatogonia entering meiosis during the TX treatment, the corresponding targeted exon(s) would be excised and they would develop into spermatocytes in which PDS5 proteins are absent, or strongly depleted under detection limits. These spermatocytes could only progress up to early pachytene during the TX treatment, given that an A1 spermatogonia takes approximately 17 days to progress into a pachytene spermatocyte 50 (Fig EV2E). In cells already coursing meiosis at the beginning of the TX treatment, the prior synthesis of highly stable PDS5 proteins would allow them to progress normally throughout spermatogenesis (Fig EV2E). This idea, consistent with the immunofluorescence analyses, can also explain the immunoblot detection of considerable amounts of PDS5 proteins in testis extracts after the effective excision of Pds5 genes. In conclusion, two different populations of spermatocytes, clearly recognizable by immunofluorescence, coexist in Pds5A cKO, Pds5B cKO, and Pds5AB cKO testes after the 15‐day‐long TX treatment. Under our experimental design, mouse models allowed us to analyze the effects of the strong depletion of PDS5 proteins from leptotene up to early pachytene stages.
A single PDS5 paralog protein is sufficient for prophase I progression
Spermatocytes from TX‐treated Pds5A cKO and Pds5B cKO mice developed normal SYCP3‐labeled AE/LEs despite extreme reduction of PDS5A and PDS5B, respectively, observed before mid‐pachytene stages (Fig EV1D–F and N–P). In later stages of meiosis, PDS5A and PDS5B staining were indistinguishable between wild‐type and Pds5‐deficient cells (Fig EV1G, H, Q and R). Interestingly, the reduction of one PDS5 paralog did not affect the distribution of the other paralog (Fig EV1D–W). Furthermore, the analysis of seminiferous tubules from these mice treated for 15 days with TX demonstrated that spermatocytes completed all meiotic and spermiogenic stages without noticeable failures (Appendix Fig S2).
Since PDS5B has been specifically involved in centromere cohesion in MEFs 19, we next studied centromere distribution in Pds5A or Pds5B deficient spermatocytes. Our results did not evidence appreciable alterations of centromere cohesion or arrangement in those spermatocytes (Appendix Fig S3). We cannot rule out that a residual amount of PDS5 proteins could be sufficient for exerting their meiotic functions. However, since simultaneous ablation of the two Pds5 genes under identical experimental conditions does have an important impact on meiosis, as described in the next sections, we propose that the presence of normal protein levels of a single PDS5 paralog is sufficient for prophase I progression up to pachytene.
PDS5 proteins regulate AE/LE length but are not required for synapsis
We next analyzed spermatocytes from TX‐treated Pds5AB cKO animals. PDS5A protein was undetectable in leptotene‐like, zygotene‐like, and some pachytene‐like spermatocytes (Fig 4A–C). The immunofluorescence intensity of PDS5A was extremely reduced in these spermatocytes compared to wild‐type levels (Fig EV2). Thus, an almost complete depletion of PDS5A protein can be assumed in these early prophase I Pds5AB cKO spermatocytes. Interestingly, these early prophase I spermatocytes presented an obvious phenotypic alteration, namely the presence of short SYCP3‐labeled AEs/LEs and SCs (Figs 4A–C and EV3A–C), a phenotype characteristic of all meiotic cohesin subunit mutants 47, 51. In some other pachytene, spermatocytes with wild‐type levels of PDS5A SCs showed normal length (Figs 4D, and EV2A and C, and EV3D). Metaphase I bivalents also displayed wild‐type levels and distributions of PDS5A (Fig 4E), and meiotic divisions and spermiogenesis looked normal (Fig EV3E–L and N–P). Likewise, PDS5B was extremely reduced in leptotene‐like, zygotene‐like, and some pachytene‐like spermatocytes (Figs 4F–H, and EV2B and D). By contrast, pachytene spermatocytes with a normal SCs length displayed wild‐type levels of PDS5B (Figs 4I, and EV2B and D, and EV3D), which were maintained in metaphase I (Fig 4J). Thus, early prophase I Pds5AB spermatocytes with shortened AE/LEs correspond to premeiotic cells, in which the excision of the targeted genes occurred at the time of initiation of the TX treatment that have progressed through meiosis with strongly depleted PDS5 proteins (Figs 4A–C and F–H and EV2A and B). On the other hand, spermatocytes already coursing meiosis at the onset of the TX treatment, presented normal distribution of PDS5 proteins, displayed regular AE/LEs and were able to progress throughout later meiotic stages (Figs 4D, E, I and J and EV3). Staining with an antibody against the testis‐specific histone H1t, a marker of mid‐pachytene chromatin 52, showed that 95% of early pachytene‐like spermatocytes from TX‐treated Pds5AB cKO animals with short SCs (n = 200) were H1t‐negative (Fig 4K), while only the remaining 5% of cells were faintly positive for H1t labeling (Fig 4L). On the other hand, all pachytene spermatocytes with normal SC length were strongly positive for H1t (Fig 4M). These results further support the hypothesis that early pachytene‐like spermatocytes with short AEs/LEs are those generated during the TX treatment, while spermatocytes from mid‐pachytene onwards correspond to those that entered meiosis prior to the treatment. Average SC length measured in early pachytene‐like spermatocytes negative for H1t staining was significantly reduced in Pds5AB‐deficient cells compared to wild‐type early pachytene ones (80 ± 7 μm versus 154 ± 20 μm, Fig 4N), representing a reduction of around 52% of SC length. We also scored the number of spermatocytes in the different stages of prophase I (n = 1,000) and observed a > 2‐fold increase in zygotene‐like and a decrease in pachytene in Pds5AB‐deficient testes compared to wild‐type (Fig EV3M and Q). Among the Pds5AB cKO pachytene spermatocytes, only 11.6% of them displayed shortened SCs. Altogether, these results reflect a delay in synapsis and/or meiotic recombination progression.
Figure EV3. Progression of spermatogenesis in Pds5AB cKO mice.
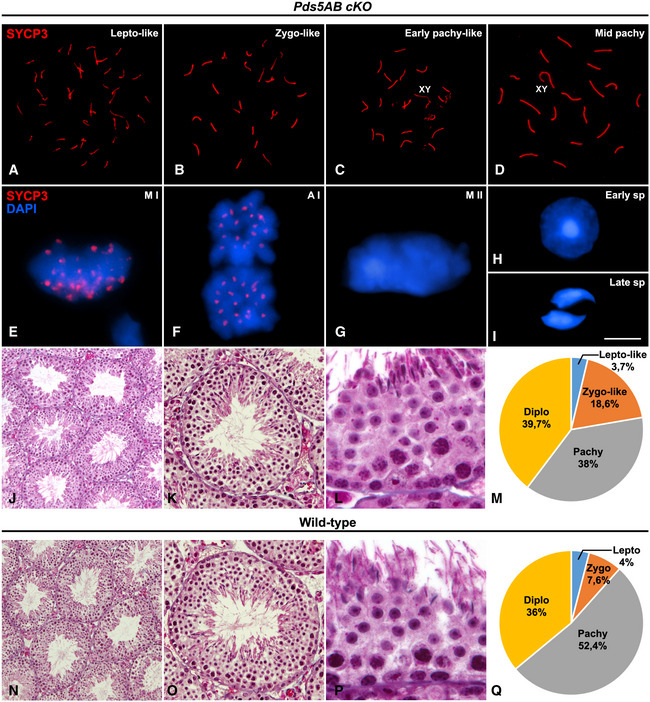
-
A–GImmunolabeling of SYCP3 (red) in Pds5AB cKO spread (A–D) and squashed (E–G) spermatocytes at the indicated stages. DAPI staining of chromatin (blue) is shown in (E–G).
-
H, IDAPI‐stained early (H) and late spermatid (I) nuclei from squashed Pds5AB cKO tubules.
-
J–LHistological sections of testes from Pds5AB cKO mice.
-
MPie graph showing the distribution of prophase I stages after analyzing 1,000 spermatocytes from three individuals of each genotype double immunolabeled for SYCP3 and SYCP1.
-
N–PHistological sections of testes from wild‐type mice.
-
QPie graph showing the distribution of prophase I stages after analyzing 1,000 spermatocytes from three individuals of each genotype double immunolabeled for SYCP3 and SYCP1.
To assess synapsis, we used double immunolabeling of SYCP3 and SYCP1. Leptotene wild‐type spermatocytes displayed long discontinuous threads of SYCP3 representing the assembly of AEs along each chromosome (Fig 5A). In early zygotene spermatocytes, the SYCP1 labeling was only located along the synapsed regions of the AEs/LEs (Fig 5B), which became longer as synapsis progressed (Fig 5C). By early pachytene, synapsis has been achieved at the entire length of the 19 autosomes and at the partially unsynapsed sex bivalent (Fig 5D). By contrast, in Pds5AB‐deficient spermatocytes 40 short individual SYCP3‐labeled AEs were found in leptotene‐like (Fig 5E). During zygotene, SYCP1 appeared along synapsed regions of the AEs/LEs, despite both the SYCP3‐labeled AEs/LEs and the SYCP1‐labeled CE were considerably shorter in the absence of both PDS5 proteins (Fig 5F and G). In early pachytene‐like spermatocytes, displaying shortened SCs, 19 fully synapsed autosomal SCs and the partially unsynapsed sex bivalent were found in Pds5AB deficient cells (Fig 5H). Double labeling of SYCP3 and HORMAD2, a protein present at unsynapsed AEs/LEs 53, further confirmed the accurate progression of synapsis in Pds5AB‐deficient spermatocytes (Fig 5I–L). We therefore conclude that PDS5 proteins are not required for synapsis of homologous chromosomes in male mouse meiosis.
Figure 5. Synapsis progression in Pds5AB cKO spermatocytes.

-
A–HDouble immunolabeling of SYCP1 (green) and SYCP3 (red) in wild‐type (A–D) and Pds5AB cKO (E–H) spread spermatocytes at the indicated stages.
-
I–LDouble immunolabeling of HORMAD2 (pseudocolored in white) and SYCP3 (red).
Normal cohesin distribution along AEs/LEs in Pds5AB cKO spermatocytes
We next analyzed whether the shortening of AEs/LEs in Pds5AB‐deficient spermatocytes was the consequence of an abnormal distribution or absence of cohesin complexes. Contrary to this possibility, the cohesin subunits SMC1α, SMC1β, SMC3, RAD21, RAD21L, REC8, and STAG3 were detected along the shortened AEs/LEs in zygotene‐like and early pachytene‐like spermatocytes (Figs 6A–I and Appendix Fig S4A–L). Cohesin cofactors Sororin and WAPL were also found along the synapsed regions (Fig 6J–L) and AEs/LEs (Fig 6M–O), respectively, as previously reported in wild‐type spermatocytes 33, 34, 35. Centromeric cohesion defects were not evident in deficient Pds5AB zygotene‐like and early pachytene‐like spermatocytes, as kinetochore signals appeared unaltered at the ends of AEs/LEs or SCs, respectively, as in Pds5AB mid‐pachytene spermatocytes (Appendix Fig S4M–O). Thus, a strong depletion of PDS5 proteins does not compromise the loading and maintenance of cohesin complexes or centromere cohesion in prophase I.
Figure 6. Distribution of cohesin subunits and regulators is not altered in Pds5AB cKO spermatocytes.

-
A–ODouble immunolabeling of SYCP3 (red) and the cohesin subunits SMC3 (green, A–C), REC8 (green, D–F), and SMC1β (green, G–I) and cohesin cofactors Sororin (green, J–L) and WAPL (green, M–O) in Pds5AB cKO spread spermatocytes at the indicated stages. Sex bivalents (XY) are indicated. Scale bar, 10 μm.
PDS5 proteins affect meiotic recombination
Since Pds5AB‐deficient spermatocytes accumulate at a zygotene‐like stage but no clear defects in synapsis could be detected, we next asked about progression of meiotic recombination. The DNA DSBs marker γ‐H2AX was found throughout the chromatin at leptotene‐like, in chromatin surrounding unsynapsed regions in zygotene‐like, and restricted to the sex body in early pachytene‐like spermatocytes with shortened SCs, as well as in mid/late pachytene spermatocytes displaying a regular SC length (Fig 7A–D). This dynamic distribution of γ‐H2AX in Pds5AB‐deficient spermatocytes is consistent with that reported for wild‐type spermatocytes 3, suggesting that DSBs were generated and repaired despite the low abundance of PDS5 proteins.
Figure 7. PDS5 proteins affect meiotic recombination.

-
A–HDouble immunolabeling of SYCP3 (red) and either γH2AX (pseudocolored in blue, A–D) or RAD51 (green, E–H) in Pds5AB cKO spread spermatocytes at the indicated stages.
-
IScatter dot‐plot graph of the quantification of RAD51 foci in 15 zygotene nuclei randomly selected from three different individuals of each genotype. Bars indicate mean and standard error, and statistical significance was assessed with an unpaired two‐tailed t‐test function (****P < 0.0001).
-
J–LDouble immunolabeling of MLH1 (pseudocolored in white) and SYCP3 (red) in Pds5AB cKO spread spermatocytes.
Next, we analyzed the distribution of RAD51, a protein that participates in homology search at the so‐called early recombination nodules along leptotene and zygotene AEs/LEs 5. RAD51 signals were indeed seen along AEs in Pds5AB cKO leptotene‐like spermatocytes and their number gradually decreased as prophase I progressed (Fig 7E–H), in a time course comparable to that found in wild‐type, Pds5A‐deficient and Pds5B‐deficient spermatocytes (Fig EV4). The average number of RAD51 foci in zygotene was 226 ± 36 in wild‐type but only 151 ± 22 in Pds5AB deficient zygotene‐like spermatocytes (Fig 7I). MLH1 foci, which mark late recombination nodules from mid‐pachytene onwards 5, could not be detected in most Pds5AB‐deficient early pachytene‐like spermatocytes with short SCs (96 out of 100; Fig 7J). In the remaining 4 spermatocytes a single focus was invariably found in autosomal bivalents and in the sex bivalent (Fig 7K), and therefore, the mean number of MLH1 foci was 20. In mid/late pachytene spermatocytes from Pds5AB cKO mice displaying normal length SCs (Fig 7L), the average number of MLH1 foci was 22 ± 1 (n = 91). Altogether, our results indicate that PDS5 proteins are not essential for initiation of meiotic recombination, but they modulate the number of early recombination nodules.
Figure EV4. A single PDS5 protein is sufficient for proper RAD51 localization in early prophase I spermatocytes.
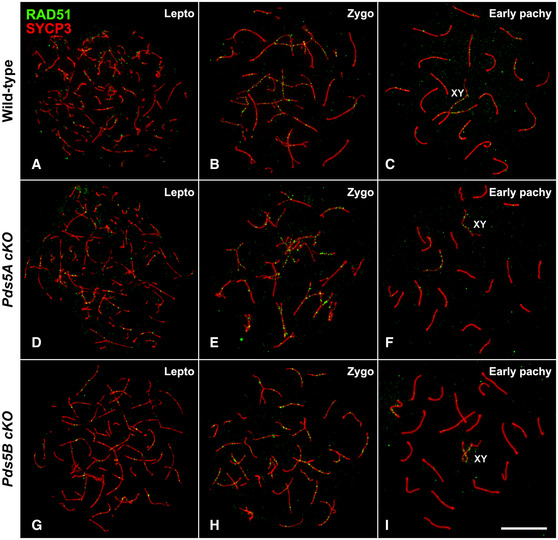
-
A–IDouble immunolabeling of RAD51 (green) and SYCP3 (red) in wild‐type (A–C), Pds5A cKO (D–F), or Pds5B cKO (G–I) spread spermatocytes at the indicated stages.
Severe telomere abnormalities in Pds5AB cKO spermatocytes
Since PDS5A and PDS5B participate in telomere cohesion in somatic cells 19, and PDS5B is present at telomeres of meiotic chromosomes (Fig 2), we then analyzed the morphology and behavior of telomeres in Pds5AB‐deficient spermatocytes. Labeling with antibodies against the telomere proteins TRF1 (Fig 8A–D) and RAP1 (Fig 8E–H) evidenced the existence of multiple morphological alterations in leptotene‐like, zygotene‐like and early pachytene‐like spermatocytes. These frequent alterations, of both TRF1 and RAP1, included stretched signals, multiple signals, a signal separated from the AEs/LEs, and AEs/LEs ends without signals (i–iv in Fig 8I and K, quantifications in Fig 8J and L, and Appendix Table S1). These alterations were not observed in mid/late pachytene Pds5AB cKO spermatocytes in which PDS5 levels are presumably normal (Fig 8D and H), or in Pds5A cKO, Pds5B cKO, or wild‐type spermatocytes (Fig EV5A–D). As a rule, more than 30% of telomeres presented an altered arrangement in zygotene‐like and early pachytene‐like Pds5AB cKO spermatocytes (Fig 8J and L, Appendix Table S1), a percentage remarkably higher than that found in mid‐pachytene spermatocytes or in wild‐type ones (Fig 8D and H, Appendix Table S1). We also studied the distribution of SUN1, an inner nuclear membrane protein that participates in the attachment of telomeres to the NE during prophase I, a crucial event for homologs pairing, recombination and synapsis in mammals 54. SUN1 signals appeared at some, but not all, AEs/LEs ends during leptotene‐like, zygotene‐like, and early pachytene‐like stages (Fig 8M–O) and displayed altered morphologies reminiscent of those observed with TRF1 and RAP1 staining (Fig 8Q and R; quantification in Fig 8S, and Appendix Table S1). Anomalous or absent SUN1 signals were observed at 36.51 and 19.27% of telomeres in Pds5AB cKO zygotene‐like and early pachytene‐like spermatocytes, respectively (Fig 8M–O, Q and R; and Appendix Table S1). By mid/late pachytene, Pds5AB cKO spermatocytes presented normal SUN1 signals at both SCs ends (Fig 8P), as in wild‐type spermatocytes (Fig EV5E–H).
Figure 8. Altered distribution of telomeric proteins in Pds5AB cKO spermatocytes.

-
A–HDouble immunolabeling of SYCP3 (red) and either TRF1 (green, A–D) or RAP1 (green, E–H) in Pds5AB cKO spread spermatocytes at the indicated stages.
-
ISelected zygotene‐like and pachytene‐like bivalents presenting altered TRF1 distribution as (from left to right) (i) telomere stretches, (ii) multiple telomere, (iii) distant telomeres, and (iv) telomere‐less. Arrowheads in (I) indicate the position of altered telomeres.
-
JQuantification of telomeres with a regular or altered disposition of TRF1 in Pds5AB cKO spermatocytes at the indicated stages (n = 1,646 telomeres in zygotene‐like spermatocytes and 738 in pachytene‐like ones).
-
K, LAs in (I, J), respectively, but for RAP1 staining (n = 1,376 telomeres in zygotene‐like spermatocytes and 328 in pachytene‐like ones). Arrowheads in (K) indicate the position of altered telomeres.
-
M–PDouble immunolabeling of SUN1 (green) and SYCP3 (red) in Pds5AB cKO spread spermatocytes at the indicated stages. 0, 1, and 2 in (N and O) indicate the number of chromosome ends labeled by SUN1 in a given bivalent.
-
QAs in (I), but for SUN1 staining. Arrowheads in (Q) indicate the position of altered telomeres.
-
RSelected zygotene‐like and early pachytene‐like bivalents from Pds5AB cKO spermatocytes immunolabeled with SYCP3 (red) and SUN1 (green), and their respective percentage of appearance.
-
SQuantification of telomeres with a regular or altered disposition of SUN1 (n = 715 telomeres in zygotene‐like spermatocytes and 410 in pachytene‐like ones).
Figure EV5. Telomere organization in wild‐type, Pds5A cKO, and Pds5B cKO spermatocytes.

-
A–HDouble immunolabeling of SYCP3 (red) and either TRF1 (green, A‐D) or SUN1 (green, E–H) in wild‐type spread spermatocytes at the indicated stages.
-
I–TImmunolabeling of SYCP3 (red) and telomere FISH (PNA‐Tel, green) in wild‐type (I–L), Pds5A cKO (M–P) or Pds5B cKO (Q–T) spread spermatocytes at the indicated stages.
Fluorescence in situ hybridization (FISH) with a telomere‐specific probe further confirmed the abnormal organization of telomeric DNA loops in Pds5AB cKO leptotene‐like, zygotene‐like, and early pachytene‐like spermatocytes (Fig 9A–T, quantification in 9E, and Appendix Table S1). By contrast, from mid/late pachytene onwards the telomere DNA showed a regular distribution at SCs ends (Fig 9U–X), as in prophase I stages from wild‐type, Pds5A‐deficient and‐Pds5B deficient spermatocytes (Fig EV5I–T). A combined analysis of telomeric DNA by FISH and immunolabeling of RAP1 and SYCP3 in Pds5AB‐deficient spermatocytes in leptotene‐like (Fig 9F–J), zygotene‐like (Fig 9K–O), and early pachytene‐like (Fig 9P–T) demonstrated that a given telomere abnormality could be sometimes equally distinguished by FISH or immunolabeling (Fig 9I and N), while in some other cases, abnormal telomere FISH signals and no RAP1 signals were observed (Fig 9J, O, S, and T). From these data, we conclude that the drastic reduction of both PDS5 proteins promotes the appearance of severe abnormalities at telomeres including aberrant telomeric DNA organization and distribution of telomere proteins.
Figure 9. Altered organization of telomeric DNA in Pds5AB cKO spermatocytes.

-
A–CImmunolabeling of SYCP3 (red) and telomere FISH (PNA‐Tel, green) in Pds5AB cKO spread spermatocytes at the indicated stages.
-
D300% magnification of selected zygotene‐like and pachytene‐like bivalents presenting regular (left) or altered (i–iv) telomere FISH signals.
-
EQuantification of telomeres with a regular or altered disposition of telomeric DNA in Pds5AB cKO spermatocytes at the indicated stages (n = 1,198 telomeres in zygotene‐like spermatocytes and 574 in pachytene‐like ones).
-
F–XDouble immunolabeling of SYCP3 (red) and RAP1 (blue) and telomere FISH (PNA‐Tel, green) in Pds5AB cKO spread spermatocytes at the indicated stages. Asterisks on (H, M, R, and W) indicate the position of the enlarged chromosomes/bivalents shown in a 300% magnification in (I, J, N, O, S, T, and X), respectively, displaying telomere stretches, multiple telomere, distant telomeres, and telomere‐less. Yellow arrowheads in (I) indicate telomeres displaying a distant telomere configuration observed both by telomere FISH and RAP1 immunolabeling. White arrowheads in (J, N, O, S, and T) indicate telomeres with an altered organization of telomeric DNA into multiple telomere signals with (N) or without RAP1 signals (J, and T), and telomere stretches (O and S) without RAP1 signals. Pink arrowheads in (T) indicate a telomere presenting a regular telomere FISH signal without a RAP1 signal.
PDS5 proteins promote proper attachment of telomeres to the NE
Given the alterations found at telomeres in Pds5AB‐deficient spermatocytes, we asked whether telomere attachment to the NE might be affected. To explore this possibility, we performed a double immunolabeling of SYCP3 and TRF1 on squashed spermatocytes, a technique that preserves the nuclear volume and chromosome positioning inside nuclei 55, 56, and collected image stacks across the entire volume of prophase I nuclei (Fig 10). TRF1 signals were found at the ends of the SYCP3‐labeled LEs and SCs in wild‐type zygotene (Fig 10A–D) and early pachytene (Fig 10I–L) spermatocytes, as well as in zygotene‐like and early pachytene‐like Pds5AB cKO spermatocytes (Fig 10E–H and M–P). Nonetheless, while TRF1 signals were invariably positioned at the nuclear periphery in wild‐type spermatocytes (Fig 10B and J), some TRF1 signals were instead found at the nuclear interior in Pds5AB cKO zygotene‐like and early pachytene‐like spermatocytes (Fig 10F and N and Movie EV1), indicating a lack of attachment of some telomeres to the NE. In 100% of Pds5AB‐deficient zygotene‐like spermatocytes analyzed (n = 25), TRF1 signals were located at the nuclear interior (from 4 to 16, mean number 12, Appendix Table S2). In 64% of Pds5AB cKO early pachytene‐like spermatocytes analyzed (n = 25), 1–4 internal TRF1 signals were observed (Appendix Table S2). Morphological alterations in telomere structure similar to those observed in spread spermatocytes could be also detected (Fig 10Q–T and Movies EV2–EV5). Interestingly, telomere‐less ends of LEs were always found at the nuclear interior and thus non‐attached to the NE (Movie EV5). Similar results were obtained when analyzing SUN1 distribution in squashed spermatocytes (Appendix Fig S5 and Movies EV6–EV8). As previously described for TRF1, SUN1 telomere‐less ends of LEs were always located at the nuclear interior (Movie EV8). Moreover, in Pds5AB‐deficient spermatocytes, 1–4 internal SUN1 signals were found in 36% and 16% of zygotene‐like and early pachytene‐like nuclei, respectively (n = 25 for each stage, Appendix Table S2), a situation not observed in wild‐type zygotene and pachytene spermatocytes 57. Consequently, our data indicate that the reduced levels of PDS5A and PDS5B cause severe abnormalities at telomeres that alter their attachment to the NE.
Figure 10. Altered attachment of telomeres to the NE in Pds5AB cKO spermatocytes.

-
A–PDouble immunolabeling of TRF1 (green) and SYCP3 (red) in squashed spermatocytes from wild‐type (A–D, I–L) or Pds5AB cKO mice (E–H, M–P). The first three columns correspond to z‐projections of 15 focal planes throughout the top, equator, and bottom regions of the nucleus. The projection of 65 focal planes across the same spermatocyte nucleus is shown at the fourth column (Z‐projection). White arrowheads in (F and N) indicate the presence of TRF1 signals non‐attached to the NE. Scale bar, 10 μm.
-
Q–TSelected bivalents from Pds5AB cKO spermatocytes displaying structural telomere abnormalities as telomere stretches (Q), multiple telomere (R), distant telomere (S), and telomere‐less (T).
Discussion
In the present study, we have examined the distribution of the cohesin cofactors PDS5A and PDS5B in mouse spermatocytes and analyzed the consequences of their depletion in order to understand their contributions to mammalian meiosis. We employed mice carrying cKO alleles for one or both genes in homozygosis and a Cre transgene induced by TX 19. Due to the deleterious effects of eliminating both PDS5 proteins in the whole organism, TX treatment was limited to 2 weeks. In this time, strong depletion of PDS5 proteins was observed in spermatocytes up to the mid‐pachytene stage, while PDS5 proteins were present in further meiotic stages. Taking into account this limitation, and considering that a residual amount of PDS5 proteins might be present and exerting some meiotic functions, we have observed that drastic reduction of either PDS5 protein does not alter progression through prophase I, while disturbance of both results in severe phenotypes on AE organization and telomere integrity. The redundancy observed here for PDS5A and PDS5B is consistent with results in A. thaliana, an organism with five different Pds5 genes, in which even quadruple Atpds5 mutants have no overt affection of meiosis 43.
PDS5A and PDS5B are located at AEs/LEs but their dynamics are different
Immunofluorescence staining with specific antibodies shows that PDS5A and PDS5B are distributed along the AEs/LEs in prophase I mouse spermatocytes, as previously described for the cohesin subunits SMC1α, SMC1β, SMC3, RAD21, RAD21L, REC8, and STAG3 10. It is likely that the presence of PDS5A and PDS5B at AEs/LEs occurs in the context of the cohesin complex, as reported for PDS5 in different organisms 36, 37, 38, 40, and consistent with the finding that PDS5B co‐immunoprecipitates with meiosis‐specific cohesin subunits REC8 and SMC1β from mouse testis extracts 44. Likewise, similar distribution patterns have been previously reported in mouse meiosis for the cohesin cofactors WAPL 32, 33, and NIPBL and MAU2 58. The exception is Sororin, which localizes at the central region of the SC 34, 35.
We have found intriguing differences between the dynamics of PDS5A and PDS5B during prophase I. While PDS5A appears at AEs/LEs by zygotene and is displaced from them by mid‐pachytene, PDS5B is visualized at AEs/LEs during all prophase I stages. Moreover, PDS5B is present at telomeres during prophase I. However, since strong depletion of either PDS5 protein does not result in defects in meiosis progression, at least up to mid‐pachytene, the functional significance of the different association/dissociation dynamics of the two PDS5 proteins is unclear. In particular, the lack of telomere defects in Pds5B‐deficient spermatocytes suggests that PDS5A can also perform the function of PDS5B required for telomere integrity even if we did not find PDS5A at telomeres either in wild‐type or in Pds5B deficient spermatocytes. We cannot disregard the possibility that the PDS5A antibody used in our study is unable to detect PDS5A at telomeres or that a residual amount of PDS5B was sufficient to ensure telomere integrity.
PDS5 proteins are required for AE organization
Pds5AB cKO spermatocytes are able to fulfill complete synapsis, but display severely shortened AEs/LEs in early prophase I stages. A similar shortening of meiotic chromosome axes in the absence of PDS5 has been described in other organisms 36, 38, 39, 41, 42. The extent of AEs/LEs shortening in Pds5AB‐deficient mouse spermatocytes is comparable, or even greater, to that found in spermatocytes from cohesin subunit knock out mouse models 47, 51. Consistent with the dependency of AE/LE assembly on previous loading of cohesin complexes along meiotic chromosomes 59, cohesin axes also appear shortened. Although it is not possible to make a quantitative assessment in our preparations, we observed that all the cohesin subunits that we tested (SMC1α, SMC1β, SMC3, REC8, RAD21, RAD21L, and STAG3), as well as the cohesin cofactors Sororin and WAPL, were present in Pds5AB‐deficient early prophase I spermatocytes.
Why do cohesin‐deficient chromosomes assemble shortened AEs/LEs? Descriptions of meiotic chromosome morphology suggest that chromatin loops emanate from a proteinaceous axial core containing cohesin and AEs/LEs proteins 60, but the actual mechanism of chromosome assembly remains unknown. A recent study reporting HiC analyses in yeast meiotic chromosomes shows that the patterns observed are reminiscent of those obtained in interphase mammalian cells after depletion of WAPL or PDS5 61, suggesting that reduced dissociation dynamics of cohesin may be an important feature of meiotic cohesin complexes 20. The HiC study further proposes that REC8‐dependent loop extrusion is responsible for these patterns and drives chromosome arm compaction while SC formation and synapsis promote additional compaction. Since SC length is restored in Smc1β −/− Sycp3 −/− double‐mutant spermatocytes, cohesin might in fact counteract chromosome axis compaction promoted by the SC 22. Whether cohesin complexes mediating sister‐chromatid cohesion and those driving loop extrusion are the same or are coordinated between them remains an open question 62, 63. In any case, PDS5 regulates both activities of cohesin, and it is therefore not surprising that its deletion results in defects similar to those reported for meiotic cohesin mutants. We envision that in both scenarios, reduced cohesin (in cohesin mutants) or reduced cohesin dynamics (after extreme reduction of PDS5), a lower number of longer chromatin loops would be formed and result in shorter AEs/LEs.
A role for PDS5 proteins in meiotic recombination?
The initiation of meiotic recombination precedes and mediates homolog synapsis in prophase I. In mouse, PDS5A and PDS5B are not required for the formation of DSBs. However, a reduced number of RAD51 foci was found along the shortened AEs/LEs of Pds5AB‐deficient spermatocytes, as well as of MLH1 foci, which might point to a decrease of recombination rates, since MLH1 is associated with late recombination nodules 5. One possibility is that the reduction in RAD51 foci might be a direct consequence of shorter AEs/LEs 64. In fact, delayed or defective DSB repair, along with AEs/LEs shortening, has been reported in meiocytes lacking the cohesin subunits REC8 25, 26, SMC1β 21, STAG3 28, 29, 30, 31, and RAD21L 27. An additional possibility is that early recombination events are impaired when the levels of PDS5 proteins are drastically reduced in mouse meiosis. A recent study in Saccharomyces cerevisiae disclosed the participation of Pds5 in both DSBs formation and in early steps of meiotic recombination 42. Although we found no effect on DSBs formation, an early recombination event such as the recruitment and/or function of RAD51 could be impaired in Pds5AB cKO spermatocytes. Moreover, in Drosophila melanogaster, Pds5 deficiency negatively affected crossover rates in oocytes 65. In this study, it was proposed that the association of Pds5 with the recombination mediator Brca2 participates in the process of homologous recombination 65. Interestingly, BRCA2 is necessary in mouse spermatocytes for the accurate loading of RAD51 and DMC1 to early recombination nodules 66. Likewise, PDS5 proteins in mouse could mediate RAD51 loading to early recombination nodules, as in vitro assays show that PDS5B interacts physically with RAD51 and stimulates RAD51‐mediated DNA strand invasion 67. In this regard, we have recently showed in human cells that PDS5A and PDS5B promote the recruitment of RAD51 and BRCA2 to stalled replication forks to protect them from excessive resection by MRE11 nuclease 68.
PDS5 proteins secure telomere integrity and attachment to the NE
In somatic cells, telomere cohesion guarantees proper replication of the telomeres. The absence of PDS5 proteins, or Sororin, or cohesin‐SA1, or treatment with low doses of the DNA replication inhibitor aphidicolin, results in altered telomere organization or telomere fragility 19, 69, 70. Cohesion is established during the premeiotic S‐phase, and it is possible that PDS5 deficiency at this time could explain the telomere defects observed in subsequent prophase I stages. However, it is important to note that telomere aberrations identical to those reported here have been observed in Smc1β −/− spermatocytes, while this meiosis‐specific cohesin subunit is not expressed in premeiotic cells 23. Moreover, an elegant study expressing SMC1α under the control of the Smc1β promoter in Smc1β −/− mice showed restoration of several meiotic phenotypes including AEs/LEs length while telomere aberrations remained uncorrected 71. It has therefore been suggested that SMC1β may facilitate the arrangement of telomere DNA into the so‐called T‐loop and/or the association of telomere factors to ensure proper telomere behavior. In this scenario, PDS5 proteins might control SMC1β function(s) at telomeres, despite our observation that they are not necessary for the loading and maintenance of SMC1β or any other cohesin subunit along chromosome axes during prophase I. Consistent with this possibility, PDS5B has been shown to co‐immunoprecipitate with SMC1β in mouse spermatocytes 44. Further studies will be necessary to elucidate whether telomere aberrations found in Pds5AB‐deficient spermatocytes are due to cohesin mis‐regulation during premeiotic DNA replication, a failure in SMC1β regulation during meiosis, or both.
In addition to alterations in telomere integrity, telomere attachment to the NE was impaired in early prophase I Pds5AB cKO spermatocytes. The frequency of telomere alterations was higher in analyses of telomeric DNA, TRF1, or RAP1 signals than in analyses of SUN1 signals, suggesting that telomeres can interact with SUN1 independently of their morphology. Similarly, in Smc1β −/−spermatocytes, SUN1 was equally detected at normal and aberrant telomeres 23. Thus, failure of telomere attachment to the NE is not solely due to alterations in telomere integrity. Higher frequencies of internal signals were detected when telomeres were labeled by TRF1 than when marked by SUN1. These variations could be due to the high percentage of SC ends without an appreciable SUN1 signal, and consequently, the real number of telomeres non‐attached to the NE would be higher. In any case, as previously suggested for Smc1β −/−spermatocytes, impaired telomere attachment to the NE could be the consequence of shortened AEs/LEs, since telomeres would have more trouble to reach the NE due to steric constrains 21. Alternatively, or in addition, PDS5 proteins could play an active role in telomere attachment to the NE, consistent with results in Drosophila oocytes describing how Pds5, in concert with BRCA2, is required to bring persistent homologous recombination sites to the NE 65. Overall, our data indicate that PDS5 proteins are crucial for the proper arrangement and functionality of telomeres in male mouse meiosis.
Materials and Methods
Animals and ethics statement
Mice carrying cKO alleles for Pds5A (Pds5Aloxfrt or Pds5A cKO) and Pds5B (Pds5Blox or Pds5B cKO) in homozygosis and a Cre‐ERT2 transgene, generated previously 19, were used for this study. After directed breeding crosses, mice carrying both cKO alleles were obtained (Pds5Aloxfrt; Pds5Blox, or Pds5AB cKO). Animals were kept at the Animal Facility of the Spanish National Cancer Research Centre (CNIO) under specific pathogen‐free conditions in accordance with the recommendation of the Federation of European Laboratory Animal Science Associations. To induce recombination, 8‐week‐old male mice were fed with TX‐containing diet for 2 weeks before being sacrificed and their testes surgically removed. All animal procedures were approved by local and regional ethics committees (Institutional Animal Care and Use Committee and Ethics Committee for Research and Animal Welfare, Instituto de Salud Carlos III) and performed according to the European Union guidelines. Genotyping was performed by PCR with the following primers, as indicated in Fig EV1A: Afw: 5′‐GGACACTTTAGCAGTTACCTCAGC‐3′, Arv1: 5′‐ACCCTAAGTCCCAATGCACC‐3′ and Arv2: 5′‐GGCGGAAAGAACCATCTAGC‐3′ for Pds5A; Bfw: 5′‐GCCCTTCTTTCATTGTTTAC‐3′, and Brv: 5′‐GGTTTGCAGAGAGTTCTAGC‐3′ for Pds5B. Protein levels were also analyzed by immunoblot of total protein extracts prepared by lysing a piece of testis tissue in RIPA buffer. Other knock out mice used in this study were as follows: Rec8 −/− (a gift from J.C. Schimenti; 25), and Smc1β −/− (a gift from R. Jessberger; 21).
Squashing and spreading of spermatocytes
For immunofluorescence analyses, testes were detunicated and seminiferous tubules processed according to previously described protocols for obtaining squashing 55, 56 or spreading 72 preparations of spermatocytes.
Immunofluorescence microscopy
After spermatocytes processing, the resulting preparations were rinsed three times for 5 min in PBS and incubated overnight at 4°C with the corresponding combinations of primary antibodies diluted in PBS. A rabbit polyclonal antibody for mouse PDS5A (C‐107) was generated by immunizing rabbits with a recombinant fragment (aa. 1,188–1,332) expressed in Escherichia coli. Two rabbit polyclonal antibodies were prepared against mouse PDS5B: for C‐100, a synthetic peptide (aa. 2–24, HSKTRTNDGKITYPPGVKEISD) was used as immunogen; for C‐102, a recombinant fragment (aa. 1,178–1,300) expressed in E. coli was used. Both antibodies against PDS5B yielded identical results in immunofluorescence staining of testes. An antibody against human PDS5B prepared by immunizing rabbits with a synthetic peptide (CEEKLGMDDLTKLVQEQKPKGSQRS, aa. 1,226–1,249) was affinity purified and used for Western blot. All primary antibodies and dilutions used are presented in Appendix Table S3. Following three washes in PBS, the slides were incubated for 45 min at room temperature with secondary antibodies at a 1:100 dilution in PBS. The appropriated combinations of the following secondary antibodies were employed for simultaneous double or triple immunolabeling: Alexa 350, Alexa 488, and Alexa 594‐conjugated donkey anti‐rabbit IgG (Molecular Probes); Alexa 350, Alexa 594, and Alexa 594‐conjugated donkey anti‐mouse IgG (Molecular Probes); Alexa 488‐conjugated donkey anti‐goat IgG (Molecular Probes); Alexa 488‐conjugated goat anti‐guinea pig IgG (Molecular Probes); Alexa 488‐conjugated donkey anti‐human IgG (Molecular Probes). Subsequently, slides were rinsed in PBS and in double‐immunolabeling experiments counterstained for 3 min with 5 μg/ml DAPI (4′, 6‐diamidino‐2‐phenylindole). After a final rinse in PBS, the slides were mounted in Vectashield (Vector Laboratories) and sealed with nail varnish.
Immunofluorescence FISH
A combination of SYCP3 immunostaining, our double immunolabeling of SYCP3 and RAP1, and telomere FISH were carried out in spread spermatocytes as previously reported 48. For this purpose, a FITC‐labeled (C3TA2)3 peptide nucleic acid (PNA) probe (Applied Biosystems) that specifically recognizes telomeric DNA was employed.
Histology
For histological sections, testes were divided into several pieces, which were fixed by immersion in Bouin's solution for 24 h. After standard washes and dehydration, Paraplast‐embedded tissue blocks were cut in 3‐μm‐thick sections in a Reichert microtome. Finally, sections were stained with conventional Mallory's trichrome stain.
Image acquisition
Observations were performed using an Olympus BX61 microscope equipped with a motorized Z‐axis and epifluorescence optics. Images were captured with an Olympus DP71 digital camera controlled by the CellP software (Olympus). For squashed spermatocytes, image stacks comprising 65 focal planes across the entire spermatocyte nuclei were captured. The complete image stacks were used to analyze the relative distribution of the proteins at a given entire nucleus and for generating animated 3D reconstructions of nuclei using the public domain software ImageJ (National Institutes of Health, USA; http://rsb.info.nih.gov/ij). For a better presentation of the results in the figures, stacks were subsequently processed for obtaining complete Z‐projections (65 focal planes) or partial Z‐projections (15 focal planes along a certain nuclear region) using the ImageJ software. Final images were processed with Adobe Photoshop 7.0 software.
Quantification and statistical analyses
Quantification of PDS5A and PDS5B immunofluorescence intensity was estimated by measuring the integrated fluorescence density in individual nuclei using ImageJ. For a given nucleus, DAPI staining was employed to create a binary mask and to calculate the corresponding nuclear area, and subsequently, the integrated density of fluorescence for PDS5A or PDS5B was obtained. Three measurements of fluorescence background were scored and the corrected total nuclear fluorescence per nucleus was calculated (corrected total nuclear fluorescence = integrated density of fluorescence − (area of nucleus × mean fluorescence of background readings)). A total amount of 10 nuclei per prophase I meiotic stage were analyzed in both wild‐type and Pds5AB‐deficient mice. For statistical analyses, we performed an unpaired two‐tailed t‐test function (confidence level: 95%) and data were presented by box whiskers plot using GraphPad Prism 6.0 software.
To measure SC length, each autosomal SC was scored in 33 early pachytene cells at random, in preparations from three different individuals of each genotype, using the “measurement” function of the ImageJ software. The SC length of the sex chromosomes was disregarded since these chromosomes do not achieve complete synapsis and might introduce errors. Since no statistical differences were found between individuals within a given genotype by a Kruskal–Wallis test, the data were grouped and analyzed by an unpaired two‐tailed t‐test function (confidence level: 95%) using GraphPad Prism 6.0 software.
In order to determine the percentage of spermatocytes in a given prophase I sub‐stage, we analyzed and classified 1,000 spermatocytes at random in three individuals for each genotype. For this purpose, spermatocytes were double immunolabeled for SYCP3 and SYCP1 to determine the progression of synapsis, and thus the meiotic staging, of each prophase I spermatocyte.
The quantification of RAD51 foci was performed in 15 zygotene nuclei, randomly selected, in preparations from three different individuals of each genotype using the “multi‐point” function of the ImageJ software. It is worth noting that only RAD51 foci located over the LEs were scored. Since no statistical differences were found between individuals within a given genotype by a Kruskal–Wallis test, the data were grouped and analyzed by an unpaired two‐tailed t‐test function (confidence level: 95%) using GraphPad Prism 6.0 software. A similar approach was employed to quantify MLH1 foci in 100 early pachytene‐like spermatocytes displaying shortened SCs and in 91 mid/late pachytene spermatocytes with regular SCs.
Telomere morphology was evaluated after the immunolabeling of the telomere associated proteins TRF1, RAP1 and SUN1, and telomeric DNA FISH. Anomalous telomere signals were classified as (i) telomere stretches: stretched signals emerging from the AE/LE ends; (ii) multiple telomere: multiple signals at a given AE/LE end; (iii) distant telomeres: a single signal separated from an AEs/LEs end; and (iv) telomere‐less: AEs/LEs ends without a detectable signal. For TRF1, we scored 1,646 telomeres in zygotene‐like spermatocytes, 738 in pachytene‐like ones, and 410 in mid‐pachytene nuclei. For RAP1, we evaluated 1,376, 328 and 410 telomeres in zygotene‐like, early pachytene‐like, and mid‐pachytene spermatocytes, respectively. A total amount of 715 zygotene and 410 early pachytene‐like and mid‐pachytene telomeres were analyzed to evaluate SUN1 morphology. Telomere FISH signals were examined in 1,198 zygotene‐like telomeres, in 574 early pachytene‐like telomeres, and in 410 mid‐pachytene ones. As controls for telomere morphology, we examined wild‐type spermatocytes, scoring 537 zygotene and 410 mid‐pachytene TRF1‐labeled telomeres, 548 zygotene and 410 mid‐pachytene SUN1‐labeled telomeres, and 568 zygotene and 410 mid‐pachytene telomeres after telomere FISH.
The quantification of telomeres detached from the NE was performed as follows. Image stacks were captured comprising 65 focal planes across entire nuclei of squashed zygotene‐like (n = 25) and early pachytene‐like (n = 25) spermatocytes immunolabeled for SYCP3 and TRF1 or SUN1. Z‐projections of 15 focal planes throughout the equator region of each nucleus were obtained using ImageJ software, and the signals located separated from the nuclear periphery were manually scored.
Author contributions
AV, AL, and JAS conceived the study; AV, IB, MR‐T, RG, AG, and JAS performed all experiments; JLB developed PDS5 antibodies C‐100, C‐102, and C‐107; AV, AL, and JAS analyzed results and wrote the paper with some contributions from the other co‐authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Review Process File
Acknowledgements
We express our sincere thanks to John Schimenti and Rolf Jessberger for providing REC8 and SMC1β knock out male mice, respectively, and to Jibak Lee, Mary Ann Handel, Attila Tóth, and Manfred Alsheimer for providing antibodies. We also thank Lorena Barreras for expert histological assistance and Miriam Rodríguez for mouse genotyping. This work was supported by the Spanish Ministry of Science and Innovation, State Research Agency (Agencia Estatal de Investigación, AEI), and European Regional Development Funds (ERDF) through grants BFU2014‐53681‐P (to JAS), BFU2016‐79841‐R (to AL), BFU2017‐89408‐R, FPI “Severo Ochoa” fellowship to MR‐T and Centro de Excelencia “Severo Ochoa” to CNIO, “Ayuda para el Fomento de la Investigación en Estudios de Máster”, and “Ayudas PostMáster del Departamento de Biología” from Universidad Autónoma de Madrid fellowships to AG. CNIO is supported by the Instituto de Salud Carlos III (ISCIII).
EMBO Reports (2020) 21: e49273
Contributor Information
Ana Losada, Email: alosada@cnio.es.
José A Suja, Email: jose.suja@uam.es.
References
- 1. Bolcun‐Filas E, Handel MA (2018) Meiosis: the chromosomal foundation of reproduction. Biol Reprod 99: 112–126 [DOI] [PubMed] [Google Scholar]
- 2. Chicheportiche A, Bernardino‐Sgherri J, de Massy B, Dutrillaux B (2007) Characterization of Spo11‐dependent and independent phospho‐H2AX foci during meiotic prophase I in the male mouse. J Cell Sci 120: 1733–1742 [DOI] [PubMed] [Google Scholar]
- 3. Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco‐Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS (2001) Recombinational DNA double‐strand breaks in mice precede synapsis. Nat Genet 27: 271–276 [DOI] [PubMed] [Google Scholar]
- 4. Grabarz A, Barascu A, Guirouilh‐Barbat J, Lopez BS (2012) Initiation of DNA double strand break repair: signaling and single‐stranded resection dictate the choice between homologous recombination, non‐homologous end‐joining and alternative end‐joining. Am J Cancer Res 2: 249–268 [PMC free article] [PubMed] [Google Scholar]
- 5. Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B (2007) Initiation and resolution of interhomolog connections: crossover and non‐crossover sites along mouse synaptonemal complexes. J Cell Sci 120: 1017–1027 [DOI] [PubMed] [Google Scholar]
- 6. Fraune J, Schramm S, Alsheimer M, Benavente R (2012) The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp Cell Res 318: 1340–1346 [DOI] [PubMed] [Google Scholar]
- 7. Shibuya H, Watanabe Y (2014) The meiosis‐specific modification of mammalian telomeres. Cell Cycle 13: 2024–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Link J, Jantsch V (2019) Meiotic chromosomes in motion: a perspective from Mus musculus and Caenorhabditis elegans . Chromosoma 128: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suja JA, Barbero JL (2009) Cohesin complexes and sister chromatid cohesion in mammalian meiosis. Genome Dyn 5: 94–116 [DOI] [PubMed] [Google Scholar]
- 10. Ishiguro KI (2019) The cohesin complex in mammalian meiosis. Genes Cells 24: 6–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morales C, Losada A (2018) Establishing and dissolving cohesion during the vertebrate cell cycle. Curr Opin Cell Biol 52: 51–57 [DOI] [PubMed] [Google Scholar]
- 12. Rankin S (2015) Complex elaboration: making sense of meiotic cohesin dynamics. FEBS J 282: 2426–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ouyang Z, Yu H (2017) Releasing the cohesin ring: a rigid scaffold model for opening the DNA exit gate by Pds5 and Wapl. BioEssays 39: 1600207 [DOI] [PubMed] [Google Scholar]
- 14. Ouyang Z, Zheng G, Tomchick DR, Luo X, Yu H (2016) Structural basis and IP6 requirement for Pds5‐dependent cohesin dynamics. Mol Cell 62: 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V et al (2013) Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501: 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shintomi K, Hirano T (2009) Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl‐Pds5 and Sgo1. Genes Dev 23: 2224–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K et al (2010) Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143: 737–749 [DOI] [PubMed] [Google Scholar]
- 18. Losada A, Yokochi T, Hirano T (2005) Functional contribution of Pds5 to cohesin‐mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci 118: 2133–2141 [DOI] [PubMed] [Google Scholar]
- 19. Carretero M, Ruiz‐Torres M, Rodriguez‐Corsino M, Barthelemy I, Losada A (2013) Pds5B is required for cohesion establishment and Aurora B accumulation at centromeres. EMBO J 32: 2938–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wutz G, Varnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ et al (2017) Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36: 3573–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R (2004) Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol 6: 555–562 [DOI] [PubMed] [Google Scholar]
- 22. Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Hoog C (2008) Cohesin Smc1β determines meiotic chromatin axis loop organization. J Cell Biol 180: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adelfalk C, Janschek J, Revenkova E, Blei C, Liebe B, Gob E, Alsheimer M, Benavente R, de Boer E, Novak I et al (2009) Cohesin SMC1β protects telomeres in meiocytes. J Cell Biol 187: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biswas U, Wetzker C, Lange J, Christodoulou EG, Seifert M, Beyer A, Jessberger R (2013) Meiotic cohesin SMC1β provides prophase I centromeric cohesion and is required for multiple synapsis‐associated functions. PLoS Genet 9: e1003985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC (2004) Positional cloning and characterization of mouse mei8, a disrupted allele of the meiotic cohesin Rec8. Genesis 40: 184–194 [DOI] [PubMed] [Google Scholar]
- 26. Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ (2005) Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell 8: 949–961 [DOI] [PubMed] [Google Scholar]
- 27. Herran Y, Gutierrez‐Caballero C, Sanchez‐Martin M, Hernandez T, Viera A, Barbero JL, de Alava E, de Rooij DG, Suja JA, Llano E et al (2011) The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J 30: 3091–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fukuda T, Fukuda N, Agostinho A, Hernandez‐Hernandez A, Kouznetsova A, Hoog C (2014) STAG3‐mediated stabilization of REC8 cohesin complexes promotes chromosome synapsis during meiosis. EMBO J 33: 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hopkins J, Hwang G, Jacob J, Sapp N, Bedigian R, Oka K, Overbeek P, Murray S, Jordan PW (2014) Meiosis‐specific cohesin component, Stag3 is essential for maintaining centromere chromatid cohesion, and required for DNA repair and synapsis between homologous chromosomes. PLoS Genet 10: e1004413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Llano E, Gomez HL, Garcia‐Tunon I, Sanchez‐Martin M, Caburet S, Barbero JL, Schimenti JC, Veitia RA, Pendas AM (2014) STAG3 is a strong candidate gene for male infertility. Hum Mol Genet 23: 3421–3431 [DOI] [PubMed] [Google Scholar]
- 31. Winters T, McNicoll F, Jessberger R (2014) Meiotic cohesin STAG3 is required for chromosome axis formation and sister chromatid cohesion. EMBO J 33: 1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang J, Hakansson H, Kuroda M, Yuan L (2008) Wapl localization on the synaptonemal complex, a meiosis‐specific proteinaceous structure that binds homologous chromosomes, in the female mouse. Reprod Domest Anim 43: 124–126 [DOI] [PubMed] [Google Scholar]
- 33. Brieno‐Enriquez MA, Moak SL, Toledo M, Filter JJ, Gray S, Barbero JL, Cohen PE, Holloway JK (2016) Cohesin removal along the chromosome arms during the first meiotic division depends on a NEK1‐PP1γ‐WAPL axis in the mouse. Cell Rep 17: 977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gomez R, Felipe‐Medina N, Ruiz‐Torres M, Berenguer I, Viera A, Perez S, Barbero JL, Llano E, Fukuda T, Alsheimer M et al (2016) Sororin loads to the synaptonemal complex central region independently of meiotic cohesin complexes. EMBO Rep 17: 695–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jordan PW, Eyster C, Chen J, Pezza RJ, Rankin S (2017) Sororin is enriched at the central region of synapsed meiotic chromosomes. Chromosome Res 25: 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Heemst D, James F, Poggeler S, Berteaux‐Lecellier V, Zickler D (1999) Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell 98: 261–271 [DOI] [PubMed] [Google Scholar]
- 37. Zhang Z, Ren Q, Yang H, Conrad MN, Guacci V, Kateneva A, Dresser ME (2005) Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion. Mol Microbiol 56: 670–680 [DOI] [PubMed] [Google Scholar]
- 38. Jin H, Guacci V, Yu HG (2009) Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol 186: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang F, Yoder J, Antoshechkin I, Han M (2003) Caenorhabditis elegans EVL‐14/PDS‐5 and SCC‐3 are essential for sister chromatid cohesion in meiosis and mitosis. Mol Cell Biol 23: 7698–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ding DQ, Sakurai N, Katou Y, Itoh T, Shirahige K, Haraguchi T, Hiraoka Y (2006) Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J Cell Biol 174: 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ding DQ, Matsuda A, Okamasa K, Nagahama Y, Haraguchi T, Hiraoka Y (2016) Meiotic cohesin‐based chromosome structure is essential for homologous chromosome pairing in Schizosaccharomyces pombe . Chromosoma 125: 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hong S, Joo JH, Yun H, Kleckner N, Kim KP (2019) Recruitment of Rec8, Pds5 and Rad61/Wapl to meiotic homolog pairing, recombination, axis formation and S‐phase. Nucleic Acids Res 47: 11691–11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pradillo M, Knoll A, Oliver C, Varas J, Corredor E, Puchta H, Santos JL (2015) Involvement of the cohesin cofactor PDS5 (SPO76) during meiosis and DNA repair in Arabidopsis thaliana . Front Plant Sci 6: 1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fukuda T, Hoog C (2010) The mouse cohesin‐associated protein PDS5B is expressed in testicular cells and is associated with the meiotic chromosome axes. Genes (Basel) 1: 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parra MT, Viera A, Gomez R, Page J, Benavente R, Santos JL, Rufas JS, Suja JA (2004) Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J Cell Sci 117: 1221–1234 [DOI] [PubMed] [Google Scholar]
- 46. Gomez R, Valdeolmillos A, Parra MT, Viera A, Carreiro C, Roncal F, Rufas JS, Barbero JL, Suja JA (2007) Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep 8: 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Biswas U, Hempel K, Llano E, Pendas A, Jessberger R (2016) Distinct roles of meiosis‐specific cohesin complexes in mammalian spermatogenesis. PLoS Genet 12: e1006389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Viera A, Parra MT, Page J, Santos JL, Rufas JS, Suja JA (2003) Dynamic relocation of telomere complexes in mouse meiotic chromosomes. Chromosome Res 11: 797–807 [DOI] [PubMed] [Google Scholar]
- 49. Burkhardt S, Borsos M, Szydlowska A, Godwin J, Williams SA, Cohen PE, Hirota T, Saitou M, Tachibana‐Konwalski K (2016) Chromosome cohesion established by Rec8‐cohesin in fetal oocytes is maintained without detectable turnover in oocytes arrested for months in mice. Curr Biol 26: 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Griswold MD (2016) Spermatogenesis: the commitment to meiosis. Physiol Rev 96: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ward A, Hopkins J, McKay M, Murray S, Jordan PW (2016) Genetic interactions between the meiosis‐specific cohesin components, STAG3, REC8, and RAD21L. G3 (Bethesda) 6: 1713–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Drabent B, Bode C, Bramlage B, Doenecke D (1996) Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem Cell Biol 106: 247–251 [DOI] [PubMed] [Google Scholar]
- 53. Wojtasz L, Daniel K, Roig I, Bolcun‐Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S et al (2009) Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA‐ATPase. PLoS Genet 5: e1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M (2007) SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell 12: 863–872 [DOI] [PubMed] [Google Scholar]
- 55. Page J, Suja JA, Santos JL, Rufas JS (1998) Squash procedure for protein immunolocalization in meiotic cells. Chromosome Res 6: 639–642 [DOI] [PubMed] [Google Scholar]
- 56. Parra MT, Page J, Yen TJ, He D, Valdeolmillos A, Rufas JS, Suja JA (2002) Expression and behaviour of CENP‐E at kinetochores during mouse spermatogenesis. Chromosoma 111: 53–61 [DOI] [PubMed] [Google Scholar]
- 57. Viera A, Alsheimer M, Gomez R, Berenguer I, Ortega S, Symonds CE, Santamaria D, Benavente R, Suja JA (2015) CDK2 regulates nuclear envelope protein dynamics and telomere attachment in mouse meiotic prophase. J Cell Sci 128: 88–99 [DOI] [PubMed] [Google Scholar]
- 58. Visnes T, Giordano F, Kuznetsova A, Suja JA, Lander AD, Calof AL, Strom L (2014) Localisation of the SMC loading complex Nipbl/Mau2 during mammalian meiotic prophase I. Chromosoma 123: 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Eijpe M, Heyting C, Gross B, Jessberger R (2000) Association of mammalian SMC1 and SMC3 proteins with meiotic chromosomes and synaptonemal complexes. J Cell Sci 113: 673–682 [DOI] [PubMed] [Google Scholar]
- 60. Zickler D, Kleckner N (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754 [DOI] [PubMed] [Google Scholar]
- 61. Schalbetter SA, Fudenberg G, Baxter J, Pollard KS, Neale MJ (2019) Principles of meiotic chromosome assembly revealed in S. cerevisiae . Nat Commun 10: 4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Muller H, Scolari VF, Agier N, Piazza A, Thierry A, Mercy G, Descorps‐Declere S, Lazar‐Stefanita L, Espeli O, Llorente B et al (2018) Characterizing meiotic chromosomes’ structure and pairing using a designer sequence optimized for Hi‐C. Mol Syst Biol 14: e8293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Holzmann J, Politi AZ, Nagasaka K, Hantsche‐Grininger M, Walther N, Koch B, Fuchs J, Durnberger G, Tang W, Ladurner R et al (2019) Absolute quantification of cohesin, CTCF and their regulators in human cells. Elife 8: e46269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kleckner N, Storlazzi A, Zickler D (2003) Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet 19: 623–628 [DOI] [PubMed] [Google Scholar]
- 65. Kusch T (2015) Brca2‐Pds5 complexes mobilize persistent meiotic recombination sites to the nuclear envelope. J Cell Sci 128: 717–727 [DOI] [PubMed] [Google Scholar]
- 66. Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S, Benedict J, Swing D, Martin BK, Tessarollo L, Evans JP et al (2004) BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131: 131–142 [DOI] [PubMed] [Google Scholar]
- 67. Couturier AM, Fleury H, Patenaude AM, Bentley VL, Rodrigue A, Coulombe Y, Niraj J, Pauty J, Berman JN, Dellaire G et al (2016) Roles for APRIN (PDS5B) in homologous recombination and in ovarian cancer prediction. Nucleic Acids Res 44: 10879–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morales C, Ruiz‐Torres M, Rodriguez‐Acebes S, Lafarga V, Rodriguez‐Corsino M, Megias D, Cisneros DA, Peters JM, Mendez J, Losada A (2020) PDS5 proteins are required for proper cohesin dynamics and participate in replication fork protection. J Biol Chem 295: 146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Remeseiro S, Cuadrado A, Carretero M, Martinez P, Drosopoulos WC, Canamero M, Schildkraut CL, Blasco MA, Losada A (2012) Cohesin‐SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J 31: 2076–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Biswas U, Stevense M, Jessberger R (2018) SMC1α substitutes for many meiotic functions of SMC1β but cannot protect telomeres from damage. Curr Biol 28: 249–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peters AH, Plug AW, van Vugt MJ, de Boer P (1997) A drying‐down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5: 66–68 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Review Process File


