Abstract
Introduction:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a recently identified coronavirus family member that triggers a respiratory disease similar to severe acute respiratory syndrome coronavirus (SARS-CoV). SARS-CoV and SARS-CoV-2 are very similar to each other in many respects, such as structure, genetics, and pathobiology. We hypothesized that coronaviruses could affect pulmonary tissues via integration with the critical immune genes after their interaction with renin–angiotensin system (RAS) elements. The aim of the present bioinformatics study was to assess expression changes of the RAS and non-RAS genes, particularly immune response genes, in the lung epithelial cells after infection with SARS-CoV.
Methods:
Linear regression, hierarchical clustering, pathway analysis, and network analysis were performed using the E-GEOD-17400 data set.
Results:
The whole-genome expression data of the lung epithelial cells infected with SARS-CoV for 12, 24, and 48 hours were analyzed, and a total of 15 RAS family and 29 immune genes were found to be highly correlated with the exposure time to the virus in the studied groups.
Conclusion:
RAS genes are important at the initiation of the infections caused by coronavirus family members and may have a strong relationship with the exchange of immune genes in due course following the infection.
Keywords: RAS, ANPEP, ACE2, EGFR, IGF2R, SARS-CoV, SARS-CoV-2, immune system genes
Introduction
Coronaviruses (CoV) are the enveloped RNA viruses that can cause enzootic fatal infections primarily in birds and mammals.1–4 Moreover, CoV has the ability to infect humankind.5–7 The presenting symptoms and signs of the infection could be chills, night sweats, persistent cough, diarrhea, and fever. However, the viral infection could cause a wide variety of clinical presentations, ranging from upper respiratory infections (URI) to lower respiratory infections (LRI), including bronchitis, pneumonia, and severe acute respiratory syndrome (SARS). CoV affects the respiratory system and specifically the lungs.5,8,9 Furthermore, the viral infection can also affect the gastrointestinal tract, as well as the neurological and hepatic systems.10 The severe acute respiratory syndrome (SARS) epidemic in 2003 and the Middle East respiratory syndrome (MERS) in 2012 revealed that CoV can be fatal when they cross the species barrier and infect humans.6 A newly identified CoV called SARS-CoV-2 (formerly entitled nCoV) appeared within the last month of 2019 in the city of Wuhan, China.11 Very early findings suggested that the virus passed from a specific animal species to humans. However, some studies then revealed that SARS-CoV-2 could also be passed from person to person, leading to serious respiratory diseases such as acute respiratory distress syndrome similar to that seen in the SARS-CoV infection.12,13 SARS-CoV-2 is an enveloped, single-stranded, positive-sense RNA (beta-) CoV. The same as SARS-CoV and MERS-CoV, the SARS-CoV-2 genome can encode structural proteins such as the spike glycoprotein, non-structural proteins, and accessory proteins. The spike glycoprotein, in which this virus has encoding functions, plays an important role in the interactions of the virus and the cell receptor during viral entry.6 More specifically, a spike protein that is associated with the envelope of SARS-CoV-2 binds first to the host receptor and then mediates the entry of the CoV into the host cells by fusing viral and host membranes.14–16 Once the fusion has taken place, the RNA virus begins to replicate its genome in the cell and makes new virions that will be secreted to infect other cells.17–19
There is a clear pathobiological association between CoV and the renin–angiotensin system (RAS). SARS-CoV (a member of the CoV family that emerged in 2002), which is very similar to SARS-CoV-2, and angiotensin converting enzyme 2 (ACE2) interactions have been extensively studied.13,16 There is a fundamental atomic interaction between the spike protein receptor binding domain of the virus and the host receptor of ACE2. SARS-CoV infections lead to ACE2 down-regulation by the binding of the SARS-CoV spike protein to ACE2.20 Thus, the SARS-CoV family, including SARS-CoV-2, utilizes ACE2 as a critical receptor to enter target host cells.13,16
The main task of ACE2 is to hydrolyze angiotensin II (Ang II) to form angiotensin (1–7) (Ang-(1–7)) that binds to the G-coupled protein receptor MAS to antagonize Ang II–mediated cellular effects. This function is very important since the increased Ang II levels are estimated to cause increased ACE2 activity. ACE2 is one of the central RAS enzymes that regulate blood pressure, fluid/electrolyte balance, and systemic vascular resistance in the tissue RAS. ACE2 is also critical for the RAS, which functions for the regulation of cell growth/proliferation, inflammation/cytokine production, blood pressure, and as a homeostatic regulator of vascular function. The RAS is highly active within the lungs where SARS-CoV-2 is estimated to be the primary target organ. Ang II can cause pulmonary vasoconstriction in response to hypoxia, which is important in patients with pneumonia or lung injury. Locally increased Ang II production increases vascular permeability, which facilitates pulmonary edema. ACE2, which plays a role in regulating the Ang II as well as Ang-(1–7) levels, may be important for those pathobiological lung events.13,21,22 ACE2 is an important negative directing factor for the severity of lung injury, and SARS-CoV spike protein–associated ACE2 inhibition has been shown to contribute to the severity of lung pathologies.20 In addition, numerous genes are involved within the interactions of ACE2, the RAS, and the pulmonary microenvironment.
The activation of the local pulmonary RAS can affect the genesis of lung damage through multiple mechanisms, such as increments in vascular permeability and alterations in alveolar epithelial cells. The pathogenesis of the CoV is a rather complicated process, which may also include local tissue RAS.13,21
The main purpose of this present in silico genomic study was to assess how the expressions of the RAS gene family changes after cellular infection with SARS-CoV in the lung epithelial cell culture. We hypothesized that CoV could affect pulmonary tissues via integration with the critical immune genes of the affected cellular microenvironment after their interaction with RAS elements. We also aimed to figure out whether these genomic alterations are related to the exposure time of the virus. In order to understand the pathobiology of SARS-CoV and SARS-CoV-2 infection better, since they share similarities within many aspects we examined the gene expression changes of other gene families as well. Elucidation of the interactions between SARS-CoV-2 and RAS genes and the description of the related genomic mechanisms are very important in the fight against this fatal viral global infection.
Methods
Obtaining and normalizing whole-genome expression data of human bronchial epithelial cells infected with SARS-CoV
Gene expression data of human bronchial epithelial cells treated with SARS-CoV for 12, 24, and 48 hours were obtained from Array Express (GSE17400).23 These data were generated by Yoshikawa et al. to characterize the dynamic, spatial, and temporal changes of the gene expression induced by SARS-CoV using microarray technology.
In order to use the obtained data in other targeted analyses, the raw data were normalized by robust multi-sequence analysis in accordance with the procedure in the Affy package in R. These data consist of 23,344 genes (54,675 probe sets). In addition, each gene has three repeated expression data values for 12, 24, and 48 hours, respectively.
Comparison of RAS gene transcripts in 12-, 24- and 48-hour SARS-CoV-infected groups
The whole normalized gene expression data of lung epithelial cells infected with SARS-CoV for 12, 24, and 48 hours were compared between different groups in order to determine significantly and differentially expressed RAS family genes. The mean value of RAS gene transcripts was determined for each group infected with SARS-CoV for 12, 24, and 48 hours. The comparison was made between the 12- and 24-hour groups as well as between the 12- and 48-hour groups, based on the RAS gene transcripts. p-Values were calculated for each gene using the t-test. The genes belonging to the RAS family with a statistically significant difference among the groups were determined (p⩽0.05).
Linear regression analysis of the genes (other than the RAS genes) with the highest variance among the groups
In order to determine the genes whose expression is highly correlated with the exposure time to the virus, we performed linear regression analysis. Genes with a standard deviation >0.9 were identified between the groups using Microsoft Excel (2016). Pearson product–moment correlation coefficient values (r) were calculated for these genes. Genes with a Pearson’s r >0.95 were selected for use in the subsequent analyses.
Hierarchical clustering, pathway analysis, and network analysis
In order to determine whether the identified genes could distinguish between the virus and infected groups, to identify the network connections between these genes and to demonstrate the pathways in which these genes were involved, we performed hierarchical clustering, pathway analysis, and network analysis.
Genes with the highest variation and Pearson’s r-value between groups were hierarchically clustered using the similarity metric parameter and the Euclidean distance Gene Cluster v3.0 program as a full link.24 Using the DAVID online tool, the pathways related to the genes were determined to elaborate on the pathway data resulting from the analysis further.25,26
In addition, the GeneMANIA application in Cytoscape was used to indicate the network and pathway relationship between the selected genes and other genes.27,28
Results
Among the three groups of human bronchial epithelial cells with different exposure times to SARS-CoV (12-, 24- and 48-hour groups), we identified the changes in expression of the RAS gene transcripts as well as the trend in expression of the most differentially expressed genes among groups other than the RAS family.
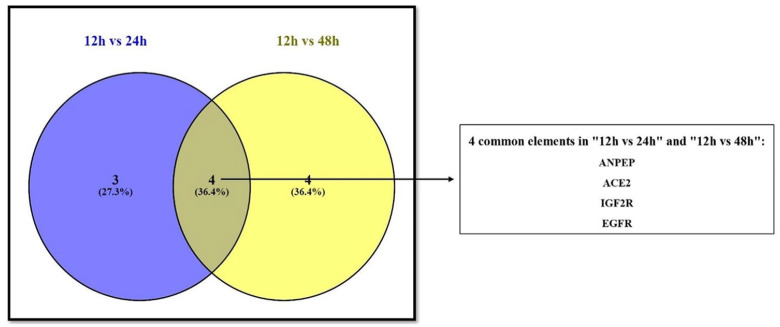
A total of seven RAS signaling pathway genes (nine probe sets; alanyl aminopeptidase (ANPEP), ACE2, angiotensin converting enzyme (ACE), insulin-like growth factor 2 receptor (IGF2R), angiotensinogen (AGT), epidermal growth factor receptor (EGFR) and membrane metalloendopeptidase (MME)) showed significantly different expression values between the 12-hour group and the 24-hour group. Comparing the 12-hour and 48-hour groups, eight genes (nine probe sets; arginyl aminopeptidase (RNPEP), epidermal growth factor receptor (EGFR), ANPEP, neurolysin (NLN), ACE2, IGF2R, leucyl and cysteinyl aminopeptidase (LNPEP) and cathepsin D (CTSD)) were statistically significantly expressed (Table 1). Four common genes (IGF2R, ANPEP, ACE2, and EGFR) in these two separate groups were identified (Figure 1).
Table 1.
The list of the RAS family genes whose expression shows a significant difference between 12-hour vs 24-hour (A) and 12-hour vs 48-hour (B) is depicted. Seven of the RAS signaling pathway genes showed different expression values in 12-hour vs 24-hour group, while 8 of the genes found significantly expressed in the 12-hour vs 48-hour group.
| A |
B |
||||
|---|---|---|---|---|---|
| Probe set ID | Gene symbol | p-Value | Probe set ID | Gene symbol | p-Value |
| 202888_s_at | ANPEP | 0.0008 | 208270_s_at | RNPEP | 0.0110 |
| 222257_s_at | ACE2 | 0.0009 | 224999_at | EGFR | 0.0112 |
| 209749_s_at | ACE | 0.0040 | 202888_s_at | ANPEP | 0.0116 |
| 219962_at | ACE2 | 0.0057 | 225943_at | NLN | 0.0283 |
| 201392_s_at | IGF2R | 0.0100 | 225944_at | NLN | 0.0296 |
| 227463_at | ACE | 0.0249 | 222257_s_at | ACE2 | 0.0371 |
| 203435_s_at | MME | 0.0309 | 201392_s_at | IGF2R | 0.0417 |
| 202834_at | AGT | 0.0481 | 207904_s_at | LNPEP | 0.0502 |
| 224999_at | EGFR | 0.0501 | 200766_at | CTSD | 0.0513 |
ANPEP: alanyl aminopeptidase; ACE2: angiotensin converting enzyme 2; ACE: angiotensin converting enzyme; IGF2R: insulin-like growth factor 2 receptor; MME: membrane metalloendopeptidase; AGT: angiotensinogen; EGFR: epidermal growth factor receptor; RNPEP: arginyl aminopeptidase; NLN: neurolysin; LNPEP: leucyl and cysteinyl aminopeptidase; CTSD: cathepsin D.
Figure 1.
Venn diagram showing common genes between the 12-hour and 24-hour groups and the 12-hour and 48-hour groups. Four genes were found to be common to both groups. As mentioned, these genes consistently showed statistically meaningful expression differences according to exposure time to the virus.
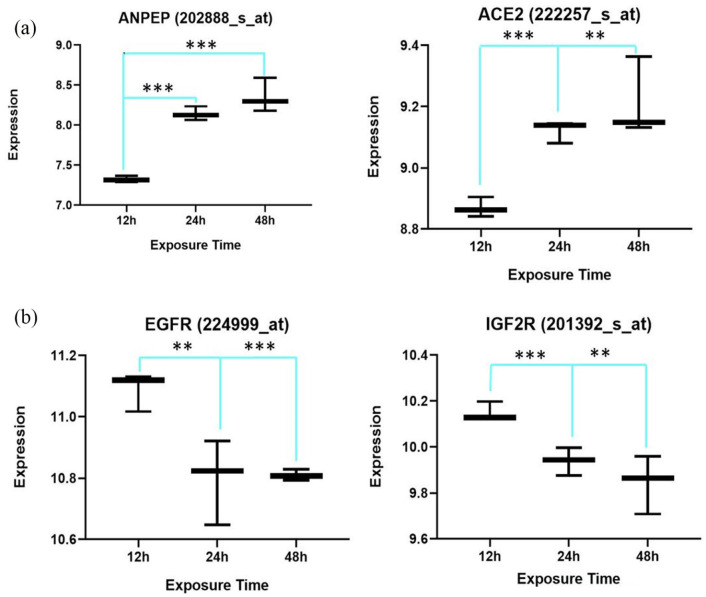
Figure 2 shows the correlation of the expression and exposure time to the virus of the statistically significant four common members of RAS genes found in the two groups (Figure 2).
Figure 2.
Two out of four common differentially expressed renin–angiotensin system (RAS) genes show a positive correlation with exposure time to the virus (a), while the other common genes, epidermal growth factor receptor (EGFR) and insulin-like growth factor 2 receptor (IGF2R), show a negative correlation (b).
**p⩽0.05; ***p⩽0.01.
In order to figure out the role of the non-RAS genes, we determined the most variant genes among these three groups. The standard deviation values of 29 genes (36 probe sets) were found to be >0.9. Linear regression analysis revealed that all these genes had a Pearson’s r >0.95 and were highly correlated with the exposure time to the virus (Table 2).
Table 2.
The most variant genes between all groups. 29 genes (36 Probe Sets) found to have >0.9 standard deviation value. All of these genes are highly correlated with exposure time to viruses an most of them are immune genes.
| Probe set ID | Gene symbol | SD | Pearson’s r |
|---|---|---|---|
| 211122_s_at | CXCL11 | 1.68295417 | 0.99402069 |
| 210163_at | CXCL11 | 1.67995636 | 0.99308306 |
| 213293_s_at | TRIM22 | 1.64689117 | 0.99812764 |
| 214079_at | DHRS2 | 1.38291569 | 0.98065023 |
| 242625_at | RSAD2 | 1.3619956 | 0.99998811 |
| 208173_at | IFNB1 | 1.32200238 | 0.96674605 |
| 213797_at | RSAD2 | 1.29016111 | 0.99987245 |
| 210029_at | IDO1 | 1.28419322 | 0.98922768 |
| 226702_at | CMPK2 | 1.23620496 | 0.99368587 |
| 204972_at | OAS2 | 1.20086416 | 0.99494266 |
| 205660_at | OASL | 1.18935939 | 0.99589149 |
| 214022_s_at | IFITM1 | 1.17991713 | 0.99401632 |
| 201601_x_at | IFITM1 | 1.17122332 | 0.99420957 |
| 206553_at | OAS2 | 1.14649914 | 0.99866159 |
| 204533_at | CXCL10 | 1.11355998 | 0.99132404 |
| 204994_at | MX2 | 1.10795097 | 0.99999559 |
| 228152_s_at | DDX60L | 1.08126568 | 0.98604577 |
| 206157_at | PTX3 | 1.06159153 | 0.97359778 |
| 228617_at | XAF1 | 1.04957699 | 0.99912443 |
| 219863_at | HERC5 | 1.0436716 | 0.99976953 |
| 204747_at | IFIT3 | 1.04191315 | 0.99999404 |
| 217502_at | IFIT2 | 1.01930351 | 0.99727623 |
| 236285_at | KLHDC7B | 1.01815991 | 0.99341293 |
| 205569_at | LAMP3 | 1.01338642 | 0.98242768 |
| 226757_at | IFIT2 | 1.0132658 | 0.99994894 |
| 229450_at | IFIT3 | 1.00446864 | 0.99482924 |
| 219211_at | USP18 | 0.99619048 | 0.9993661 |
| 218943_s_at | DDX58 | 0.94794454 | 0.99979879 |
| 218986_s_at | DDX60 | 0.94081343 | 0.99961176 |
| 210797_s_at | OASL | 0.94028093 | 0.99930186 |
| 1559883_s_at | SAMHD1 | 0.93726862 | 0.9997964 |
| 219684_at | RTP4 | 0.93351655 | 0.99821178 |
| 226603_at | SAMD9L | 0.93102808 | 0.99815238 |
| 212185_x_at | MT2A | 0.92870941 | 0.98679396 |
| 227458_at | CD274 | 0.92087529 | 0.98185747 |
| 230036_at | SAMD9L | 0.90113608 | 0.99960499 |
SD: standard deviation.
Likewise, all of these genes showed a positive correlation with exposure time to the virus. The correlation figure for each gene that was found to be highly correlated to virus infection exposure time is presented in the Supplemental Material.
In addition, as shown in Figure 3, these genes were able to be clustered for the 12-, 24- and 48-hour groups separately.
Figure 3.

Hierarchical cluster of most variant genes between three groups. As shown, determined genes were able to classify the groups clearly. In the 12-hour group, these genes show low expression, while in the 48-hour group, they all show high expression.
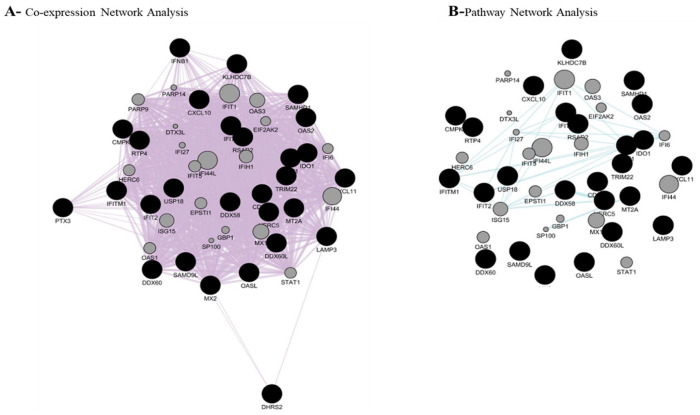
Table 3 shows the pathway analysis of all the significant genes. With the aim of discovering the relationship of these genes with each other and with other genes, a network analysis was performed (Figure 4).
Table 3.
Pathways related to non-RAS differentially expressed genes. Most of non-RAS differentially expressed genes were found to be related immune the system and pathways which could be involved during the course of the viral infectious disease.
| OAS2 2′-5′-oligoadenylate synthetase 2 (OAS2) | Related genes | Homo sapiens |
| KEGG_PATHWAY hepatitis C, measles, influenza A, herpes simplex infection | ||
| CXCL10 C-X-C motif chemokine ligand 10 (CXCL10) | Related genes | Homo sapiens |
| KEGG_PATHWAY cytokine–cytokine receptor interaction, chemokine signaling pathway, Toll-like receptor signaling pathway, RIG-I-like receptor signaling pathway, cytosolic DNA-sensing pathway, TNF signaling pathway, influenza A | ||
| CXCL11 C-X-C motif chemokine ligand 11 (CXCL11) | Related genes | Homo sapiens |
| KEGG_PATHWAY cytokine–cytokine receptor interaction, chemokine signaling pathway, Toll-like receptor signaling pathway | ||
| CD274 CD274 molecule (CD274) | Related genes | Homo sapiens |
| KEGG_PATHWAY cell adhesion molecules (CAMs) | ||
| DDX58 DExD/H-box helicase 58 (DDX58) | Related genes | Homo sapiens |
| KEGG_PATHWAY nuclear factor kappa B signaling pathway, RIG-I-like receptor signaling pathway, cytosolic DNA-sensing pathway, hepatitis C, hepatitis B, measles, influenza A, herpes simplex infection, Epstein–Barr virus infection | ||
| CMPK2 cytidine/uridine monophosphate kinase 2 (CMPK2) | Related genes | Homo sapiens |
| KEGG_PATHWAY pyrimidine metabolism, metabolic pathways | ||
| IDO1 indoleamine 2,3-dioxygenase 1 (IDO1) | Related genes | Homo sapiens |
| KEGG_PATHWAY tryptophan metabolism, metabolic pathways, African trypanosomiasis | ||
| IFNB1 interferon beta 1 (IFNB1) | Related genes | Homo sapiens |
| KEGG_PATHWAY cytokine–cytokine receptor interaction, PI3K-Akt signaling, osteoclast differentiation, Toll-like receptor signaling pathway, RIG-I-like receptor signaling pathway, cytosolic DNA-sensing pathway, Jak-STAT signaling pathway, natural killer cell-mediated cytotoxicity, Chagas disease (American trypanosomiasis), tuberculosis, hepatitis C, hepatitis B, measles, influenza A, herpes simplex infection | ||
| LAMP3 lysosomal associated membrane protein 3 (LAMP3) | Related genes | Homo sapiens |
| KEGG_PATHWAY lysosome | ||
| MT2A metallothionein 2A (MT2A) | Related genes | Homo sapiens |
| KEGG_PATHWAY mineral absorption | ||
| RSAD2 radical S-adenosyl methionine domain containing 2 (RSAD2) | Related genes | Homo sapiens |
Figure 4.
Co-expression and pathway analysis of non-RAS differentially expressed genes. The figure shows that 28/29 genes create a strong co-expression network. On the other hand, the program was able to connect some of the genes and predict some related genes in pathway network analysis.
Discussion
SARS-CoV-2 was found to be mostly similar to the SARS-CoV at the amino-acid level, despite some differences. Based on the phylogenetic analysis on all genomes of various viruses, SARS-CoV-2 is in the same beta-CoV clone as SARS-CoV, SARS-like bat CoV, and MERS-CoV. SARS-CoV-2 has the highest similarity with SARS-like bat CoV and is less associated with MERS-CoV.29
The mechanism of SARS-CoV and its association with the renin–angiotensin pathway had been defined in previous studies. The first genetic proof of the ACE2 and SARS-CoV receptor relationship was reported by Kuba et al.20 Our results showed that in lung epithelial cells, ACE2 gene expression increased between 12 and 24 hours and remained at the same level between 24 and 48 hours after infection with SARS-CoV. Complementary to our findings, Kuba et al. demonstrated a decrement in the ACE2 protein level in the lungs of SARS-CoV-infected mice on day 2. They also reported that the addition of SARS-CoV spike into mice exacerbates acute lung failure in vivo that can be reduced by inhibiting the renin–angiotensin pathway.20
Moreover, SARS-CoV-2 uses a similar mechanism as SARS-CoV. The structural analyses showed that like SARS-CoV, SARS-CoV-2 also utilizes ACE2 as the host receptor.16
Based on the evidence with regard to the similarity between SARS-CoV and SARS-CoV-2, we used data from Yoshikawa et al. In their study, all the genome expression data of the confluent 2B4 cells which were infected with SARS-CoV and grown in T-75 flasks for 12, 24 and 48 hours were used to characterize the dynamic, spatial and temporal changes of the gene expressions caused by SARS-CoV. The study was performed three times at each time point in order to meet the minimum number required for the application of statistical algorithms, and a total of nine arrays were given for SARS-CoV.23
Our current study was carried out to understand the biological mechanism of SARS-CoV-2 infection better by focusing on the similarity of SARS-CoV-2 and SARS-CoV in terms of structure, biological function, and the pathology of the infection. We proposed a hypothesis based on the genomic results obtained in the present study under three key headings (Figure 5).
Figure 5.
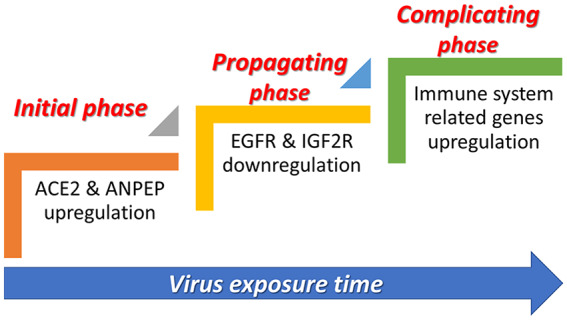
Three important phases associated with SARS-CoV infection. At the initial and propagating phases of infection, some RAS family genes were up-regulated while some others were down-regulated. At the complicating phase, there is an increase in the expression of some key immune system genes.
Initial phase
According to the results of the analysis, there was a significant up-regulation of ACE2 and ANPEP genes in human bronchial epithelial cells within 12 and 24 hours of infection. Many previous studies indicated that SARS-CoV also uses ACE2 as a receptor to enter host cells, as in SARS-CoV-2.30 The results of our present study showed the additional possible function of ANPEP (Figure 5).
The ANPEP gene acts as a receptor, especially for HCoV-229E, another member of the CoV family. This virus is predicted to induce the infection by triggering conformational changes in the spike glycoprotein, which interacts with the host ANPEP receptor and activates membrane fusion.31 Moreover, ANPEP (CD13) can act as a receptor in SARS-CoV and induce growth inhibition and apoptosis by infecting hematopoietic stem/progenitor cells.32 These findings are in concordance with our results demonstrating the up-regulation in the ACE2 and ANPEP as exposure time to the virus increased.
As suggested in many previous studies, ACE2 forms Ang-(1–7) from Ang II and binds to the MAS receptor, the specific receptor of Ang-(1–7), and then inhibits inflammatory, vascular and cellular growth mechanisms.33 In this case, it may be useful to use the agonistic peptides of Ang-(1–7) to change the functioning of ACE2 in favor of the infected host in the initial phase of infection.
Propagating phase
Based on our results, in this phase, as the exposure time to SARS-CoV increases, EGFR and IGF2R, two receptors with key roles in the RAS signaling pathway, were significantly down-regulated in the infected human bronchial epithelial cells.
Habib et al. proposed that apoptosis occurs in various cell types that require an active tyrosine kinase but do not require EGFR autophosphorylation sites by experimentally increasing the level of EGFR expression. The expression of a predominant negative RAS mutant in cells that over-express the EGFR leads to significantly enhanced EGFR-induced apoptosis.34 On the other hand, IGF2R, acting as a G-coupled protein receptor, could lead to the activation of the mitochondria-mediated apoptosis pathway, when insulin-like growth factor-II (IGF2) binds to this receptor.35 Thus, those two receptors, which are associated with cellular apoptosis, are expected to be down-regulated as the infection time of virus-infected cells progresses. This event could give the chance for the virus to have sufficient time to continue replication and increase its copy number. This phase of the viral infection might be important, particularly for hypertensive subjects.
Complicating phase
In this in silico genomic study, 29 genes (36 probe sets) were detected which showed noteworthy up-regulation in 24- versus 48-hour groups. Almost all of these are genes associated with the immune system, innate immune response, and adaptive immune response.
The relevant biological analyzers of these genes whose expressions are concordant with cell exposure time to the virus were explored via the pathway analyzers of the 29 genes. The genes involved in specific pathways related to the critical pathological events of CoV infection, including SARS-CoV and SARS-CoV-2, such as the Toll-like receptor (TLR) signaling pathway.
TLRs are essential sensor molecules of the host innate immune system. Various TLRs are involved in early interactions of invasive viruses and host cells, which affect viral pathogenesis and regulate viral replication as well as host responses. Initiation of antiviral immune responses with TLR agonists has been shown to provide protection from many different viruses, including hepatitis B virus, influenza virus, some HIV strains, and CoV. Furthermore, non-structural protein 3 of the SARS-CoV interacts with IRF3 through the papain-like protease domain. After that, the associations of the TLR and RLR pathways occur, and this binding prevents the nuclear translocation as well as the phosphorylation of IRF3.36
It has previously been observed that he TLR signal through the TRIF adapter protein protects mice from lethal SARS-CoV. Thus, a balanced immune response that works both in TRIF and MyD88-guided pathways provides the most effective host-cell intrinsic antiviral defense responses against severe SARS-CoV, and the absence of any branch of TLR signaling leads to SARS-CoV being fatal.37 Another important pathway is associated with the interferon beta 1 (IFNB1) gene identified as a result of pathway analysis is the natural killer (NK) cell-mediated cytotoxicity pathway. Interferons are considered as the screams of the affected cells during the active virus attack. IFNB1 deficiency results in partial suppression of the sterol pathway in macrophages during viral infections, thereby associating the regulation of the lipid metabolism pathway with interferon antiviral defense responses.38 Furthermore, NK cells are important in immune defense against virus infections. Enlarged virus replication and more serious diseases during encephalomyocarditis virus, Coxsackie virus, and Theiler’s murine encephalitis virus infections are linked with the reduction of NK cells or low levels of NK cell cytolytic function. NK cells are correspondingly associated with direct inhibition of virus replication and stimulation of liver damage during mouse hepatitis virus (MHV) infection. It is not known whether NK cells play a direct antiviral role in human infections with picornaviruses or CoV such as SARS.39
Lysosome, which has an important path in CoV infection, is another pathway related to the pathway analysis performed. Enveloped viruses must fuse with a host-cell membrane so as to transport their genomes to the host cell. Although some viruses fuse with the plasma membrane, many viral infections are associated with endocytosis before fusion. In the endosomal microenvironment, a particular marker induces conformational changes in viral fusion proteins, leading to viral and host membrane fusion.40 According to the De Haan et al. study, the proteins known to be important for late endosomal maturation and endosome–lysosome fusion deeply promoted infection of cells with mouse hepatitis CoV (MHV).41 In another study of SARS-CoV, SARS-CoV accessory protein open reading frames (SARS 3a) were oligomerized by dynamically inserting them into late endosomal, lysosomal and trans-Golgi network membranes.42 Furthermore, the chemotactic cytokines are engaged in a great deal of biological processes. Besides microbial infection, the infected cells have strong chemokine signals. Those signals could play significant roles in both innate and adaptive immune responses that control the growth of the invading pathogen.43 Therefore, the presence of chemokine signaling pathways parallels the other results obtained. The main limitation of this study is the lack of uninfected control data, which is subject to further experimental studies.
Conclusion
In conclusion, if the results obtained in the current study are validated by in vitro experiments and clinical samples, it can be suggested that the disruption of RAS genes may be important for the initial management of the CoV infections, particularly SARS-CoV-2. The results of the current study help us to understand better the pathobiology of SARS-CoV and other similar CoV family members such as SARS-CoV-2. Based on our results, the interactions between the SARS-CoV-2 and RAS genes, resulting in immune-related genomic disruption, lead to acquired immune deficiency states. It is hoped that critical local RAS-affecting drugs such as MAS agonists, soluble ACE2 and Ang-(1–7) will be used for the modulation of SARS-CoV-2 and RAS pathological interactions for the improvement of immune genomic states in patients infected with SARS-CoV-2 in future trials.
Supplemental Material
Supplemental material, Suppelementary_File for In vitro analysis of the renin–angiotensin system and inflammatory gene transcripts in human bronchial epithelial cells after infection with severe acute respiratory syndrome coronavirus by Can Turk, Seyhan Turk, Elif Sena Temirci, Umit Yavuz Malkan and İbrahim C. Haznedaroglu in Journal of the Renin-Angiotensin-Aldosterone System
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Supplemental material: Supplemental material for this article is available online.
ORCID iDs: Seyhan Turk  https://orcid.org/0000-0003-3843-4173
https://orcid.org/0000-0003-3843-4173
Elif Sena Temirci  https://orcid.org/0000-0001-5944-6718
https://orcid.org/0000-0001-5944-6718
Umit Yavuz Malkan  https://orcid.org/0000-0001-5444-4895
https://orcid.org/0000-0001-5444-4895
References
- 1. Dhama K, Pawaiya RVS, Chakrabort S, et al. Coronavirus infection in equines: a review. Asian J Anim Vet Adv 2014; 9: 164–176. [Google Scholar]
- 2. Fan Y, Zhao K, Shi ZL, et al. Bat coronaviruses in China. Viruses 2019; 11: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 2015; 1282: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Habibzadeh P, Stoneman EK. The novel coronavirus: a bird’s eye view. Int J Occup Environ Med 2020; 11: 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jonsdottir HR, Dijkman R. Coronaviruses and the human airway: a universal system for virus–host interaction studies. Virol J 2016; 13: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J 2019; 16: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang M, Zhou Y, Zong Z, et al. A precision medicine approach to managing Wuhan coronavirus pneumonia. Precis Clin Med 2020; 3: 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. World Health Organization. Novel coronavirus (2019-nCoV) situation report – 22. Geneva: World Health Organization, 2020. [Google Scholar]
- 12. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020; 382: 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu Y. Compensation of ACE2 function for possible clinical management of 2019-nCoV-induced acute lung injury. Virol Sin. Epub ahead of print 7 February 2020. DOI: 10.1007/s12250-020-00205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Letko M, Munster V. Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV. bioRxiv. Epub ahead of print 22 January 2020. DOI: 10.1101/2020.01.22.915660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 2016; 3: 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wan Y, Shang J, Graham R, et al. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol 2020; 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kruse RL. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res 2020; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuhn JH, Li W, Choe H, et al. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell Mol Life Sci 2004; 61: 2738–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 2005; 11: 875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tikellis C, Thomas MC. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int J Pept 2012; 2012: 256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Varagic J, Ahmad S, Nagata S, et al. ACE2: angiotensin II/angiotensin-(1–7) balance in cardiac and renal injury. Curr Hypertens Rep 2014; 16: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoshikawa T, Hill TE, Yoshikawa N, et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One 2010; 5: e8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Hoon MJ, Imoto S, Nolan J, et al. Open source clustering software. Bioinformatics 2004; 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- 25. Huang Da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 26. Huang Da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res 2009; 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucl Acids Res 2010; 38: W214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020; 27: 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffmann M, Kleine-Weber H, Krüger N, et al. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. Epub ahead of print 31 January 2020. DOI: 10.1101/2020.01.31.929042. [DOI] [Google Scholar]
- 31. Liang W, Feng Z, Rao S, et al. Diarrhea may be underestimated: a missing link in 2019 novel coronavirus. Gut. Epub ahead of print 26 February 2020. DOI: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 32. Yang M, Li C, Li K, et al. Hematological findings in SARS patients and possible mechanisms. Int J Mol Med 2004; 14: 311–315. [PubMed] [Google Scholar]
- 33. Simoes e, Silva AC, Silveira KD, Ferreira AJ, et al. ACE2, angiotensin-(1–7) and Mas receptor axis in inflammation and fibrosis. Br J Pharmacol 2013; 169: 477–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Högnason T, Chatterjee S, Vartanian T, et al. Epidermal growth factor receptor induced apoptosis: potentiation by inhibition of Ras signaling. FEBS Lett 2001; 491: 9–15. [DOI] [PubMed] [Google Scholar]
- 35. Chen WK, Kuo WW, Hsieh DJ, et al. CREB negatively regulates IGF2R gene expression and downstream pathways to inhibit hypoxia-induced H9c2 cardiomyoblast cell death. Int J Mol Sci 2015; 16: 27921–27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol 2014; 426: 1246–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Totura AL, Whitmore A, Agnihothram S, et al. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio 2015; 6: e00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dantoft W, Robertson KA, Watkins WJ, et al. Metabolic regulators Nampt and Sirt6 serially participate in the macrophage interferon antiviral cascade. Front Microbiol 2019; 10: 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waggoner SN, Reighard SD, Gyurova IE, et al. Roles of natural killer cells in antiviral immunity. Curr Opin Virol 2016; 16: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Millet JK, Whittaker GR. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology 2018; 517: 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burkard C, Verheije MH, Wicht O, et al. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog 2014; 10: e1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yue Y, Nabar NR, Shi CS, et al. SARS-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis 2018; 9: 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Skinner D, Marro BS, Lane TE. Chemokine CXCL10 and coronavirus-induced neurologic disease. Viral Immunol 2019; 32: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Suppelementary_File for In vitro analysis of the renin–angiotensin system and inflammatory gene transcripts in human bronchial epithelial cells after infection with severe acute respiratory syndrome coronavirus by Can Turk, Seyhan Turk, Elif Sena Temirci, Umit Yavuz Malkan and İbrahim C. Haznedaroglu in Journal of the Renin-Angiotensin-Aldosterone System