Abstract
Purpose:
We evaluated whole-genome sequencing (WGS) as an alternative to multi-gene panel sequencing (PS) for genetic testing in dilated cardiomyopathy (DCM).
Methods:
Forty-two patients with familial DCM underwent PS and WGS, and detection rates of rare single nucleotide variants and small insertions/deletions in panel genes were compared. Loss-of-function variants in 406 cardiac-enriched genes were evaluated, and an assessment of structural variation was performed.
Results:
WGS provided broader and more uniform coverage than PS, with high concordance for rare variant detection in panel genes. WGS identified all PS-identified pathogenic or likely-pathogenic variants as well as two additional likely-pathogenic variants: one was missed by PS due to low coverage, the other was a known disease-causing variant in a gene not included on the panel. No loss-of-function variants in the extended gene set met clinical criteria for pathogenicity. One BAG3 structural variant was classified as pathogenic.
Conclusions –
Our data support the use of WGS for genetic testing in DCM, with high variant detection accuracy and a capacity to identify structural variants. WGS provides an opportunity to go beyond suites of established disease genes, but the incremental yield of clinically-actionable variants is limited by a paucity of genetic and functional evidence for DCM association.
Keywords: Familial dilated cardiomyopathy, panel sequencing, whole-genome sequencing, genetic testing, molecular diagnosis
INTRODUCTION
Dilated cardiomyopathy (DCM) is a common heritable heart muscle disorder that frequently has a genetic etiology.1 Although long lists of disease-associated genes have been compiled,1 genetic testing yields positive results in relatively few individuals and has not been recommended as part of routine patient care.2 Knowing genotype status has enormous potential benefit for families, permitting tailored surveillance strategies and early detection of individuals at risk.3 The lack of results for most DCM families represents an unmet clinical need and a major road-block for implementation of personalized therapy.
In recent years, next-generation sequencing has facilitated genetic testing by enabling high throughput evaluation of multiple genes, including under-investigated large genes with hundreds of coding exons. Available methods include multi-gene panel sequencing (PS), whole-exome sequencing (WES) and whole-genome sequencing (WGS). PS uses libraries enriched for protein-coding regions of disease-associated genes. There is generally high sequence coverage and comparative studies with Sanger sequencing have found excellent reproducibility for variant detection.4 PS is widely used by clinical diagnostic laboratories with cardiomyopathy panel sizes increasing over time from <20 genes to >100 genes.5,6 The finite number of genes is a major limitation, necessitating re-design of panels as new disease genes are discovered and additional costs if a second test is required in PS-negative cases. In contrast to PS, WES is not limited to specific genes and looks at all protein-coding sequences.7 Variability in breadth of coverage is a potential limiting factor for WES-based clinical testing and may necessitate extensive follow-up Sanger sequencing to fill in gaps, particularly for high-probability disease genes. In addition to protein-coding sequences, WGS uniquely provides information about the vast tracts of non-coding sequences that are increasingly implicated in human disease and enables high-resolution structural variant (SV) detection.8,9 However, since WGS typically has a lower overall sequencing depth than PS and WES, its potential sensitivity for mutation screening has been questioned.10 Golbus and colleagues11 have recently reported promising results for WGS in a pilot study of 11 DCM patients. A detailed appraisal of the role of WGS for DCM genetic testing is now timely and warranted.
Here we compare PS and WGS in a cohort of patients with familial DCM. We determined the concordance of PS and WGS for rare variant detection, evaluated loss-of-function (LOF) variants in an extended gene panel, and performed the first comprehensive evaluation of SV in DCM. Our data show that WGS is a reliable method for screening established DCM disease genes as well as providing a wealth of sequence information for ongoing data-mining in “unsolved” cases.
MATERIALS AND METHODS
Study subjects
Forty-two patients (19 [45%] males), aged 18 to 82 (mean 50) years with familial DCM were recruited from St Vincent’s Hospital and by referral from collaborating physicians. The clinical characteristics of study probands are provided in Supplementary Table S1. Familial DCM was defined by the presence of DCM and/or early (<35 years) sudden unexplained death in two or more individuals in the absence of another heritable cardiac or systemic cause. Probands and participating first-degree relatives provided informed written consent and were evaluated by history and physical examination, ECG and transthoracic echocardiography. All study subjects were of self-reported European ancestry. Protocols were approved by St Vincent’s Hospital Human Research Ethics Committee.
DNA sequencing and variant calling
See Supplementary Methods for expanded sequencing methods. Briefly, 42 patient DNA samples were newly sequenced by WGS, or previously sequenced using a custom capture panel for 67 or 69 DCM genes.12 WES data were generated in-house from the NA12878 cell line (Coriell Institute for Medical Research, Camden, NJ) using the SureSelectXT Human AllExon V5 ([SSv5], Agilent Technologies, Santa Clara, CA, n=1) and the Clinical Research Exome V2 ([CREv2], Agilent, n=13) capture kits. We also re-analyzed published data from samples that used SureSelectXT Human AllExon V6 ([SSv6], Agilent, n=6) capture kit.13 All genomic data, including previously published data, were analyzed using a GATK best practices analysis pipeline.14 Short variants were annotated, filtered and prioritized using Seave.15 Structural variants (SV) including copy number variants were identified using ClinSV (Minoche et al, manuscript in preparation) which uses a combination of discordantly mapping read pairs, split-mapping reads, and depth of coverage changes. A genomic position was defined as “covered” if the sequencing depth had ≥15 high quality reads.16 Selected variants were confirmed in probands and evaluated in family members using Sanger sequencing and/or PCR.
Variant concordance analysis
Variants that passed filters and were located within the genomic regions targeted in PS were included in this analysis. Variant concordance was assessed using bcftools (v1.2) and vcfeval from RTG-Core (Real Time Genomics, v3.4.4). In individual patients, sites at which the frequency of the two predominant alleles was < 95% (allowing for sequencing errors) were considered non bi-allelic. The concordance analysis was performed for all SNVs and indels, then separately for the subset that were annotated as high- or medium-impact.
Variant filtration and prioritization
Rare stop gain, splice donor or acceptor site loss, frame-shift indels (defined as LOF variants) and missense variants were included if the maximal minor allele frequency (MAF) in the 1000 Genomes Project, Exome Sequencing Project or Exome Aggregation Consortium (ExAC) databases was <1%. Missense variants were then excluded if they were predicted to be benign by both SIFT and PolyPhen2, or annotated as benign in ClinVar. SV were included if they had population allele frequencies <1% and overlapped with exonic regions from genes of interest (Supplementary Methods). Variants were classified into one of 5 categories: pathogenic, likely pathogenic, uncertain significance (VUS), likely benign, benign, according to recommendations for clinical reporting from the American College of Medical Genetics and Genomics (ACMG).17
Gene sets analyzed
Three sets of genes were evaluated (Supplementary Table S2). The first set included 67 “panel genes” evaluated by PS. An “extended gene set” was comprised of 406 genes, including reported DCM-associated genes that were not represented on the PS panel, genes with presumptive links to cardiac and skeletal myopathies, and cardiac-enriched genes from the human protein atlas.18 The third set included a list of 57 genes compiled by the ACMG in which incidental findings are deemed clinically-reportable.19
RESULTS
PS-identified SNV and indels
PS was performed in 42 probands with familial DCM. Seventy-eight rare (MAF <1%) LOF or potentially damaging missense variants were identified in 37 probands (Table 1). All 78 PS-identified variants were confirmed to be present in probands, and were also investigated in family members, using Sanger sequencing (Supplementary Methods). Twenty-one variants in 21 families (50%) were subsequently deemed pathogenic or likely-pathogenic based on ACMG criteria,17 including 14 LOF TTN variants that we have reported previously.12
Table 1.
Yield of rare potentially deleterious variants† in 42 probands with familial DCM.
| Proband ID | Gene | Variant | Variant Type | ACMG Classification | Detection method | |
|---|---|---|---|---|---|---|
| PS | WGS | |||||
| AA-II-3 | TTN | c.72824A>T, p.K24275I | SNV; missense | VUS | X | |
| BAG3 | Chr10:121436136–121436726 del, | SV; deletion/duplication | Pathogenic | X | ||
| Chr10:121436139–121436799 dup | ||||||
| AF-III-6 | MYH6 | c.5002G>A, p.D1668N | SNV; missense | Likely Benign | X | X |
| TTN | c.94553 T>C, p.V31518A | SNV; missense | VUS | X | ||
| SGCB | c.1_2delAT, p.M1Gfs | INDEL; frameshift deletion | Likely Pathogenic | X | ||
| AJ-II-2 | TTN | c.95722T>C, p.Y31908H | SNV; missense | VUS | X | X |
| RYR2 | c.8209_−4T>C | SNV; splice | VUS | X | ||
| AM-III-7 | DMD | c.3816G>C, p.L1272F | SNV; missense | VUS | X | X |
| TTN | c.59351delT, p.E19785fs | SNV; deletion | Pathogenic | X | X | |
| ADRA1A | c.1022_1035delGACGCCACCAGGCC, p.A469fs | INDEL; in-frame deletion | VUS | X | ||
| PLEC | c.7477C>T, p.Q2493* | SNV; stop codon | VUS | X | ||
| AP-III-4 | TTN | c.44818C>T, p.P14940S | SNV; missense | VUS | X | X |
| TTN | c.47692C>T, p.R15898* | SNV; stop codon | Likely Pathogenic | X | X | |
| AT-III-1 | DMD | c.6T>G, p.S2R | SNV; missense | VUS | X | X |
| NEB | c.8683G>A, p.D2895N | SNV; missense | VUS | X | X | |
| MYO18B | c.1692+2_1692+29delTGAGTCCCCTGTCCCGCCGTCCCCCCCAGCA | INDEL; splice deletion | VUS | X | ||
| AV-IV-2 | TTN | c.41860T>C, p.Y13954H | SNV; missense | VUS | X | X |
| TTN | c.78095_78098delGAAA, p.R26032fs | INDEL; frameshift deletion | Pathogenic | X | X | |
| TTN | c.81038G>A, p.R27013Q | SNV; missense | VUS | X | X | |
| NEB | c.16561G>A, p.D5521N | SNV; missense | VUS | X | ||
| BA-III-3 | NEBL | c.1258A>G, p.I420V | SNV; missense | VUS | X | X |
| TTN | c.13900G>T, p.E4634* | SNV; stop codon | Pathogenic | X | X | |
| TTN | c.47191C>T, p.R15731C | SNV; missense | VUS | X | X | |
| SYNE1 | c.13909G>A, p.D4637N | SNV; missense | VUS | X | X | |
| SYNE1 | c.23315G>C, p.R7772Q | SNV; missense | VUS | X | ||
| BF-III-1 | NEB | c.2533A>G, p.K845E | SNV; missense | VUS | X | X |
| BDKRB1 | c.844C>T, p.R282* | SNV; stop codon | VUS | X | ||
| BG-III-1 | NEB | c.25367C>T, p.T8456M | SNV; missense | VUS | X | X |
| MYH7 | c. 4828G>C, p.E1610Q | SNV; missense | VUS | X | X | |
| TTN | c.3002T>G, p.M1001R | SNV; missense | VUS | X | X | |
| TTN | c.30485C>T, p.T10162M | SNV; missense | VUS | X | X | |
| TRDN | Chr6:123509665–124330720dup | SV; duplication | VUS | X | ||
| BK-III-3 | ALMS1 | c.1343C>G, p.T448R | SNV; missense | Likely Benign | X | X |
| MYH7 | c.2207T>C, p.I736T | SNV; missense | Likely Pathogenic | X | X | |
| SYNM | c. 499C>T, p.R167C | SNV; missense | VUS | X | ||
| TRIM63 | c.739C>T, p.Q247* | SNV; stop codon | VUS | X | ||
| BL-III-2 | LAMP2 | c.183_+2T>C | SNV; splice | Pathogenic | X | X |
| DSC2 | Chr18:28681558–28686269del | SV; deletion | VUS | X | ||
| BM-III-25 | TTN | c.56834delC, p.G18945fs | SNV; deletion | Pathogenic | X | X |
| LAMA2 | c.1634T>A, p.L545Q | SNV; missense | VUS | X | ||
| NEB | c.5555TG, p.M1852R | SNV; missense | VUS | X | ||
| BP-III-3 | NEB | c.21685G>C, p.D7229H | SNV; missense | VUS | X | X |
| DSP | c.5874_5789delAGAAAC, p.ET1929–1930del | INDEL; in-frame deletion | Likely Benign | X | ||
| SCN4A | c.1800C>A, p.Y600* | SNV; stop codon | VUS | X | ||
| RYR2 | c.14757–7_14757–6delTCinsAT | INDEL; splice | VUS | X | ||
| BR-IV-1 | LAMA2 | c.4645A>T, p.N1549Y | SNV; missense | VUS | X | X |
| TTN | c.54710T>C, p.L18237P | SNV; missense | VUS | X | X | |
| TTN | c.78991C>T, p.R26331* | SNV; stop codon | Pathogenic | X | X | |
| SCN1B | Chr19:35531062–35531134del | SV; deletion | VUS | X | ||
| MURC | c.692_712delGAGAGAGGCTAAGGCAGTCA | INDEL; in-frame deletion | Likely Benign | X | ||
| G, p.ERLRQSG231–238del | ||||||
| TLL2 | c.1232delT, p.V411* | SNV; stop codon | VUS | X | ||
| BY-III-4 | BAG3 | c.361C>T, p.R121* | SNV; stop codon | Pathogenic | X | X |
| NEB | c.22813G>A, p.E7640K | SNV; missense | VUS | X | X | |
| SYNE1 | c.9148C>G, p.L3050V | SNV; missense | VUS | X | X | |
| DSG2 | c.473T>G, p.V158G | SNV; missense | Likely Benign | X | ||
| SYNE1 | c.1762delT, p.L588* | SNV; stop codon | VUS | X | ||
| C-II-9 | NEB | c.9352A>T, p.T3118S | SNV; missense | Likely Benign | X | X |
| SCN5A | c.5872C>T, p.R1958* | SNV; stop codon | VUS | X | X | |
| CTNNA3 | Chr10:68286145–68513397del | SV; deletion | VUS | X | ||
| CI-II-2 | TTN | c.88033A>G, p.T29345A | SNV; missense | VUS | X | X |
| CS-III-12 | CAV3 | c.216C>G, p.C72W | SNV; missense | Likely Benign | X | X |
| PSEN2 | c.211C>T, p.R104W | SNV; missense | Likely Benign | X | X | |
| TTN | c.69491–69492delTG, p.V23164fs | INDEL; frameshift deletion | Likely Pathogenic | X | X | |
| ANO5 | c.191delA, p.N64fs | SNV; deletion | VUS | X | ||
| CT-II-2 | TTN | c.54380G>C, p.G18127A | SNV; missense | VUS | X | X |
| TTN | c.76116dupT, p.N25372fs | SNV; deletion | Pathogenic | X | X | |
| CZ-III-4 | FLT1 | c.2023A>G, p.S675G | SNV; missense | VUS | X | X |
| MYH7 | c.1105G>A, p.R369Q | SNV; missense | Pathogenic | X | X | |
| TTN | c.54109C>T, p.R18037W | SNV; missense | VUS | X | X | |
| PDE4DIP | c.54G>T, p.C18* | SNV; stop codon | VUS | X | ||
| DD-III-4 | MYH7 | c.2620G>C, p.E874Q | SNV; missense | Likely Pathogenic | X | X |
| SYNE1 | c.25856T>C, p.L8619P | SNV; missense | VUS | X | X | |
| XIRP2 | Chr2:167868183–168522158del | SV; deletion | VUS | X | ||
| DF-III-1 | MYH7 | c.1578_+1G>A | SNV; splice | VUS | X | X |
| SCN5A | c.3305C>A, p.S1102Y | SNV; missense | VUS | X | X | |
| FLNC | c.7170C>A, p.C2369* | SNV; stop codon | VUS | X | ||
| DI-II-6 | TTN | c.74880_74883dupAACA, p.P24962fs | INDEL; frameshift insertion | Pathogenic | X | X |
| DO-II-4 | TTN | c.95173T>C, p.C31725R | SNV; missense | VUS | X | X |
| EMD | c.428C>T, p.S143F | SNV; missense | Likely Benign | X | ||
| NKX2–5# | c.552C>G, p.I184M | SNV; missense | Likely Pathogenic | X | ||
| SYNE2 | c.18065insT, p.A6022A* | SNV; deletion | VUS | X | ||
| EA-II-7 | NEXN | c.1955A>G, p.Y652C | SNV; missense | VUS | X | X |
| TTN | c.78991C>T, p.R26331* | SNV; stop codon | Pathogenic | X | X | |
| TTN | c.104878C>T, p.R34960C | SNV; missense | VUS | X | X | |
| ASB15 | c.1207G>T, p.G403* | SNV; stop codon | VUS | X | ||
| FJ-II-3 | ABCC9 | c.3096_+1delG | SNV; splice | Likely Pathogenic | X | X |
| DTNA | c.239G>A, p.R80H | SNV; missense | VUS | X | X | |
| FK-II-1 | ACTN2 | c.1192C>T, p.R398C | SNV; missense | VUS | X | X |
| DES | c.1193T>C, p.L398P | SNV; missense | VUS | X | X | |
| LDB3 | c.1414C>A, p.P472T | SNV; missense | VUS | X | X | |
| SYNE1 | c.14848A>G, p.N4590D | SNV; missense | VUS | X | X | |
| TTN | c.102275G>A, p.R34092H | SNV; missense | VUS | X | X | |
| FRMD3 | Chr9:85906635–85962244del | SV; deletion | VUS | X | ||
| FQ-III-11 | NEB | c.21685G>C, p.D7229H | SNV; missense | VUS | X | X |
| SCN5A | c.659C>T, p.T220I | SNV; missense | VUS | X | X | |
| TTN | c.56206delA, p.T18736fs | SNV; deletion | Likely Pathogenic | X | X | |
| LAMA2 | c.4487C>T, p.A1496V | SNV; missense | VUS | X | ||
| AK1 | Chr9:130621906–130645663dup | SV; duplication | VUS | X | ||
| GE-III-7 | DSP | c.8531G>C, p.G2844A | SNV; missense | VUS | X | X |
| TTN | c.95573A>G, p.N31858S | SNV; missense | VUS | X | X | |
| RYR1 | c.13328–13348dupGGGGGCCCCTTCCGGCCCGAA, p.GGPFRPE4443–4449dup | INDEL; in-frame deletion | Likely Benign | X | ||
| GR-II-6 | No variants met criteria | |||||
| GV-III-5 | No variants met criteria | |||||
| GW-III-1 | SYNM | c.3274C>T, p.R1092C | SNV; missense | VUS | X | X |
| LMNA | c.1567G>C, p.G523R | SNV; missense | VUS | X | ||
| HB-II-1 | HPS3 | Chr3:148874313–148882670del | SV; deletion | VUS | X | |
| HT-II-1 | No variants met criteria | |||||
| HU-II-2 | SGCA | c.929A>G, p.Y310C | SNV; missense | VUS | X | X |
| TTN | c.14486A>C, p.Q4829P | SNV; missense | VUS | X | X | |
| TTN | c.75364G>A, p.V25122M | SNV; missense | VUS | X | X | |
| ID-II-3 | SYNE1 | c.19981C>A, p.Q6661K | SNV; missense | VUS | X | X |
| KI-III-2 | TTN | c.44284C>T, p.R14762* | SNV; stop codon | Pathogenic | X | X |
| TTN | c.48953T>C, p.I16318T | SNV; missense | VUS | X | X | |
| TTN | c.97324G>A, p.A32442T | SNV; missense | VUS | X | ||
| KS-II-1 | RBM20 | c.1906C>T, p.R636C | SNV; missense | Pathogenic | X | X |
| MO-II-2 | TTN | c.49458G>A, p.W16486* | SNV; stop codon | Pathogenic | X | X |
| ACADVL | c.1077_+1G>T | SNV; splice | VUS | X | ||
| MYOM2 | c.52C>T, p.Q18* | SNV; stop codon | VUS | X | ||
| R-IV-1 | NEBL | c.2588C>G, p.S863C | SNV; missense | VUS | X | X |
| TTN | c.76116dupT, p.N25372fs | SNV; insertion | Pathogenic | X | X | |
| FLNC | c.3791_−8G>A | SNV; splice | VUS | X | ||
| S-III-1 | LAMA2 | c.7415G>T, p.G2472V | SNV; missense | VUS | X | X |
| PSEN2 | c.211C>T, p.R104W | SNV; missense | Likely Benign | X | X | |
| TTN | c.30389G>A, p.R10130H | SNV; missense | VUS | X | X | |
| TTN | c.100432T>G, p.W33478G | SNV; missense | VUS | X | X | |
| TTN | c.100447G>C, p.E33482Q | SNV; missense | VUS | X | ||
Potential disease-causing variants are shown in bold;
variant identified by candidate gene screening.
ACMG, American College of Medical Genetics; INDEL, small insertion or deletion; PS, targeted panel sequencing; SNV, single nucleotide variant; SV, structural variant; VUS, variant of unknown significance; WGS, whole-genome sequencing
WGS sequencing depth and coverage
WGS had an average depth of 34x, covering 97% of the genome, 98% of all exons, and 99% of PS gene exons (Figure 1A). In comparison, PS had a much higher average read depth (486x) but covered only 91% of its targeted regions (Figure 1A). WGS coverage was compared to a WES data-set obtained in the human NA12878 cell line using the SSv5 capture kit. In this data-set, WES had an average read depth of 150x but covered only 69% of the PS targets (Figure 1A). Similar results were found in WES data-sets for which CREv2 and SSv6 capture kits had been used, with coverage of panel targets only 64% and 56%, respectively (Figure 1A, Supplementary Table S3). The poor coverage of WES relative to PS highlights the benefit of a comprehensive disease-focussed panel, compared to a generic WES design. The remaining causes of the lower coverage of panel genes with PS and WES when compared to WGS, were incompletely- or mis-targeted exons due to biases in probe design, synthesis and hybridization (Figure 1, C and D).
Figure 1.
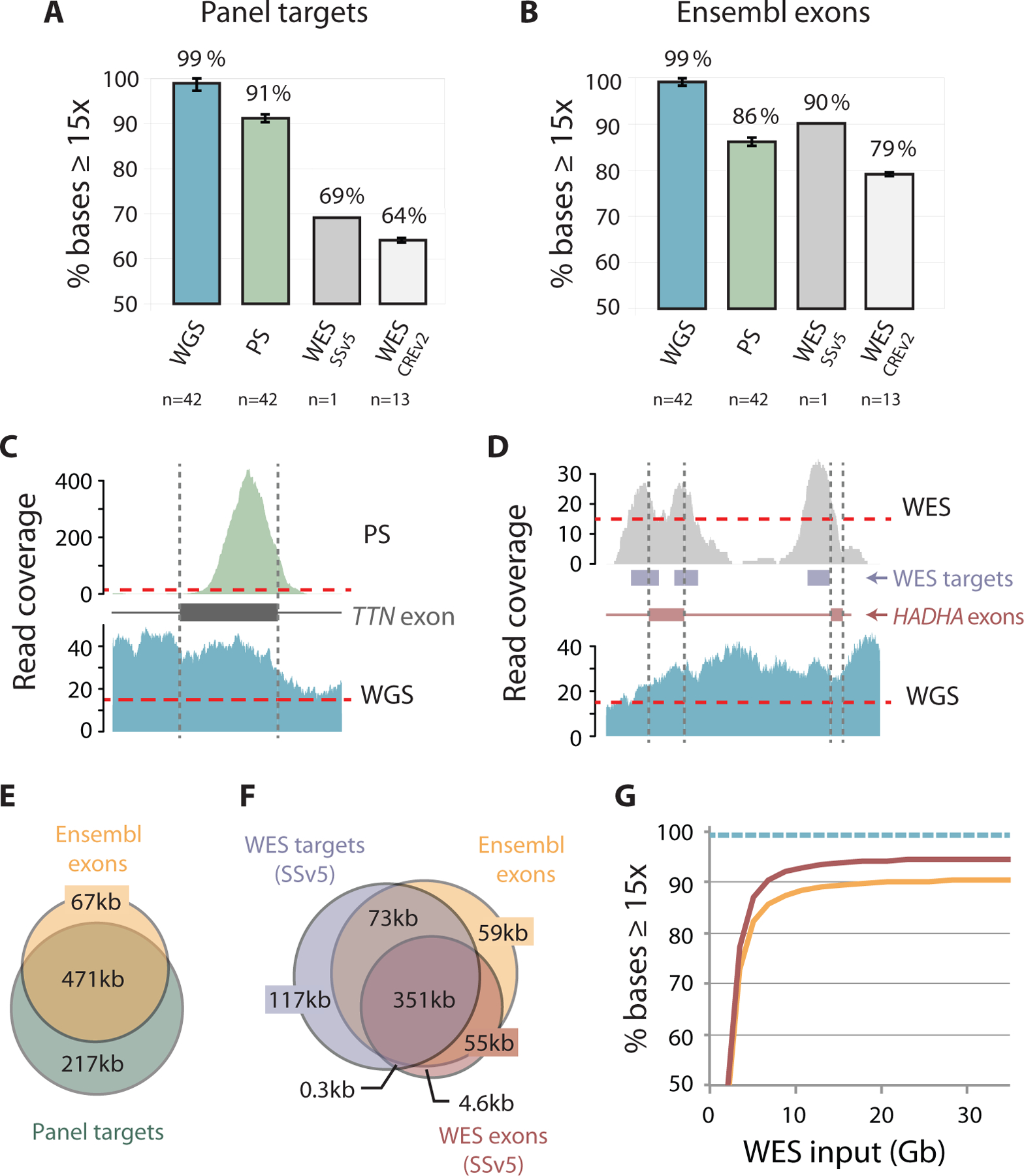
Comparative coverage across panel genes. (A) Bar graphs showing breadth of coverage, that is, the median (± 1st and 3rd quartiles) percentage of positions covered by ≥15 sequence reads with WGS, PS and WES (SSv5 and CREv2) for targeted regions on the PS panel (“panel targets”) and (B) exons in Ensembl isoforms of these genes (“Ensembl exons”). (C) Histograms comparing WGS depth of read coverage across representative gene exons with PS and (D) WES (SSv5). Note lack of uniform coverage due to capture bias in (C) and discrepancy between actual exons and WES target regions in (D). (E) Overlap of protein-coding nucleotides contained within Ensembl isoforms with regions targeted on PS panel and (F) WES (SSv5), i.e. regions targeted by WES (“WES targets”) and targeted protein-coding exons (“WES exons”). (G) Breadth of coverage across panel genes with varying WES input in Gigabases (Gb) showing WES SSv5 exons (red) and Ensembl exons (orange); WGS breadth of coverage across the same Ensembl exons is indicated by dashed line (teal), assuming the ~34x genome-wide average coverage used in thus study.
We further interrogated the reference sets of gene isoforms used in the design and analysis of target regions. WGS data were analyzed with respect to protein-coding transcripts from the comprehensive Ensembl database, reporting 1–31 isoforms per panel gene. In comparison, PS targets included exons and conserved flanking sequences of isoforms from University of California Santa Cruz (UCSC) knownGene and RefSeq, while WES SureSelect targets were based on subsets of isoforms from UCSC, RefSeq, GENCODE, and CCDS databases. Coverage of Ensembl isoforms of panel genes was high (99%) with WGS, but was 86% with PS and ranged from 90% to 79% with WES (Figure 1B, Supplementary Table S3). We found that this was mainly due to an extra 67 kb of Ensembl isoform sequences that were not included on the PS panel and 59 kb not included in WES SSv5 SureSelect targets/exons (Figure 1, E and F). Even with WES at >300x average depth of coverage, sequencing breadth plateaued at 90% for Ensembl exons and 95% for SureSelect targeted exons which was less than the 99% achieved by WGS (Figure 1G).
Concordance between WGS and PS
WGS identified on average, 3.8 × 106 SNVs and 1.2 × 106 small indels per proband genome-wide, of which 24,000 were coding and 42 were LOF and rare (MAF<1%). Extensive Sanger sequencing of a subset of 115 SNV and indels in probands and family members showed that WGS had an overall very low false-positive rate (0.81%, Supplementary Results).
For the variant concordance analysis between WGS and PS, we used the same analytical pipeline and investigated the detection rates of all SNVs and indels in PS-targeted regions irrespective of MAF and read depth. A median of 483 variants per proband were concordant between WGS and PS, with 131 variants identified exclusively by WGS and 104 variants exclusively by PS (Supplementary Table S4). 77% of WGS-specific variants were in positions that had <10 reads on PS, with the majority of these occurring at positions with no reads. In contrast, only 3% variants were missed by WGS for the same reason. Variants listed in dbSNP are more likely to be true-positives and this was the case for 87% of WGS-specific variants but only 31% of the PS-specific variants, suggesting that two-thirds of PS-specific variants were either novel or false-positives. Of these PS-specific variants, 22% were multi-allelic (vs 5% in WGS) and 53% were located at homopolymers and short tandem repeats, likely resulting from replication slippage. This can occur naturally but more often arises during PCR-mediated DNA replication.20
To further investigate discordant variants, we looked at annotated LOF and missense variants, and manually inspected the read alignments. There was an average of 82 concordant variants per proband, with 11 variants exclusive to WGS and 23 variants exclusive to PS (Supplementary Table S5). The most frequent cause of WGS-specific variants was inadequate PS read coverage (Supplementary Figure S1A), followed by missed variants occurring in Ensembl exons that were not represented on the PS panel. An additional 16% of WGS-specific variants were present in the PS reads but missed by the variant caller. In contrast, most (84%) of the PS-specific variants were sequencing artefacts due to phasing (Supplementary Figure S1B). This can occur during cycle-based sequencing when some of the DNA strands fall behind or jump ahead of the cycle.21 Other sources of discordance seen with both WGS and PS included: amplification of sequencing errors, incorrect variant calls (especially for indels), and ambiguous regions of low mapping quality (Supplementary Figure S1C). In total, there was an average of 10 likely-real LOF and missense variants per person that were missed by PS and 0.3 missed by WGS.
WGS detection of pathogenic/likely pathogenic variants in panel genes
WGS data underwent further filtering to identify rare LOF or potentially damaging missense variants in panel genes. WGS successfully detected the 78 prioritized PS variants, including all 21 of the pathogenic and likely-pathogenic variants. A frame-shift variant, SGCB p.M1Gfs, in Family AF (Table 1), identified by WGS and confirmed by Sanger sequencing was missed by PS due to lack of coverage.
LOF variants in an extended gene set
To further explore the WGS data, we compiled an extended set of 406 genes with proven and putative links to cardiomyopathy (Supplementary Table S2). WGS successfully identified a reported likely-pathogenic missense variant, p.I184M, in the NKX2–5 gene22 (Table 1). An additional 17 LOF variants in 14 families were evaluated (Table 2), all of which were validated by Sanger sequencing, but none met ACMG criteria for pathogenicity.
Table 2.
WGS-identified LOF variants in the extended gene set.
| Family ID | Gene | Variant | ExAC variant frequency; PLi score | Notes | Segregation† |
|---|---|---|---|---|---|
| Families” unsolved” after PS | |||||
| AJ | RYR2 | c.8209_−4T>C | Absent; 1.00 | Ryanodine receptor 2, SR protein involved in Ca2+ homeostasis. KO mice (−/−): embryonic lethal, (+/−): adults, normal baseline EF, ↓ contraction post-TAC; cKO mice (−/−): adults, transient ↓ contraction. CPVT families with exon 3 deletion: DCM in a subset of individuals. | 1/1 affected 0/1 unaffected. |
| AT | MYO18B | c.1692+1_1692+29del | Absent; 0 | Myosin 18B, Z-disc protein. KO mice (−/−): embryonic lethal, disordered myofibrils. | 1/2 affected 0/3 unaffected. |
| BF | BDKRB1 | p.R282* | 0.002883; 0.07 | Bradykinin receptor B1. KO mice (−/−): adults, mild ventricular dilation, normal EF. | 2/3 affected 2/10 unaffected. |
| BG | TTN# | c.31763–1G>A | 0.0002733; 0 | Titin, sarcomeric protein. Titin-truncating transgenic mice (−/−): embryonic lethal, (+/−): adults, stress-induced DCM. Zebrafish (+/−): adults, spontaneous DCM. | 3/7 affected 0/4 unaffected. |
| BP | RYR2 | c.14757–7_14757–6delTCinsAT | Absent; 1.00 | Ryanodine receptor 2 (see above). | 2/3 affected 2/5 unaffected. |
| DF | FLNC | p.C2369* | Absent; 1.00 | Filamin C, actin crosslinking protein in Z-disc and sarcolemma. KO mice (−/−): neonatal death; zebrafish MO: cardiac developmental defects. | 2/2 affected 2/3 unaffected. |
| Families with identified pathogenic/likely-pathogenic variants | |||||
| AM | ADRA1A | RRHQA341–345* | Absent; 0 | α1A adrenoceptor. KO mice (−/−): adults, normal EF. | 3/3 affected 3/12 unaffected. |
| BK | TRIM63 | p.Q247* | 0.000486; 0 | Tripartite motif containing 63, E3 ubiquitin ligase. p.Q247* variant: transfected cells, ↓auto-ubiquitination; transgenic mice, adults, ↑ LV mass, ↑ LVEDD, normal EF. |
2/2 affected 0/1 unaffected. |
| BR | TLL2 | p.V441* | Absent; 0 | Tolloid like 2, zinc-dependent metalloprotease. KO mice (−/−): adults, no heart defects reported. | 1/2 affected 1/9 unaffected. |
| CS | ANO5 | p.N64fs | 0.001027; 0 | Anoctamin 5, transmembrane protein. KO mice (−/−): adults, normal EF (baseline, isoproterenol stress). | 6/6 affected 0/6 unaffected. |
| CZ | PDE4DIP | p.C18* | 0.0000698; NA | Phosphodiesterase 4D interacting protein, A-kinase anchoring protein, contributes to phosphorylation of cMyBPC and cTNI. | 4/4 affected 0/2 unaffected. |
| DO | SYNE2 | p.A6022A* | Absent; 0 | Spectrin repeat-containing nuclear envelope protein 2, nuclear membrane protein. KO mice (−/−): adults, normal EF. | 2/3 affected 1/2 unaffected. |
| EA | ASB15 | p.G403* | 0.0000906; 0 | Ankyrin repeat and SOCS box containing 15, involved in muscle differentiation and protein turnover. | 2/3 affected 1/3 unaffected. |
| MO | |||||
| MYOM2 | p.Q18* | 0.0006355; 0 | Myomesin 2, sarcomeric protein. Neonatal rat cardiomyocytes (−/−): ↓ rate of contraction. | 1/2 affected 0/1 unaffected. |
|
| R | FLNC | c.3791_−8G>A | 0.0001607; 1.00 | Filamin C (see above). | 1/5 affected 1/2 unaffected. |
Segregation analysis did not include phenotype-negative variant carriers aged less than 40 years.
Variant absent from proband, identified by WGS of additional family members.
CPVT, catecholaminergic polymorphic ventricular tachycardia; DCM dilated cardiomyopathy; EF, ejection fraction; ExAC, Exome Aggregation Consortium; KO, knockout (cKO, cardiac-specific knockout); LV, left ventricular, LVDD, LV end-diastolic diameter; SR, sarcoplasmic reticulum; TAC, tranverse aortic constriction
Five variants, in the FLNC, ANO5, ACADVL, TRIM63, and PDE4DIP genes, were present in all (2 or more) family members with DCM in the respective kindreds (Supplementary Figure S2). In Family DF (negative after PS testing), a FLNC p.C2369* variant was identified. Variants in FLNC, that encodes the actin-binding protein filamin C, have been associated with cardiac and skeletal myopathies.23 Filamin C deficiency causes cardiac developmental defects in zebrafish and neonatal death in homozygous mice,23,24 while heterozygous mice carrying a human FLNC p.W2710X mutation show skeletal myopathy.25 To date, there is no definitive animal model evidence that heterozygous filamin C loss-of-function results in DCM. Although the nonsense FLNC variant was present in both affected individuals in Family DF (II-1, II-2), it was also present in two unaffected siblings aged 52 (II-4, II-5) and 45 years, respectively, and it was classified as a VUS. In Family CS, all affected individuals carried a likely-pathogenic LOF TTN variant and an ANO5 p.N64fs variant. ANO5 encodes anoctamin 5, a transmembrane protein with putative calcium-activated chloride channel activity. Homozygosity for LOF ANO5 variants in human subjects has been associated with limb girdle and Miyoshi muscular dystrophies.26 This phenotype is not present in Ano5 knockout mice27 and none of the CS family members had overt skeletal myopathy. Despite good family co-segregation, the ANO5 variant was called a VUS. In Family MO, individuals with a pathogenic LOF TTN variant also carried a splice acceptor site variant, ACADVL c.1077_+1G>T. The latter has been associated with a 36% reduction in very long chain acyl-CoA dehydrogenase activity but this level is predicted to be tolerated.28 Affected individuals in Family BK had a likely-pathogenic missense MYH7 variant as well as a TRIM63 p.Q274* variant, both of which have proposed associations with hypertrophic cardiomyopathy.29,30 The two variant carriers (II-3, II-4) clearly had DCM with one also showing left ventricular hypertrophy. This nonsense TRIM63 variant has been associated with reduced auto-ubiquitination in transfected cells, and increased left ventricular mass in transgenic mice.30 The four affected individuals tested in Family CZ all carried a PDE4DIP p.C18* variant in addition to a pathogenic missense MYH7 variant. PDE4DIP encodes an A-kinase anchoring protein that is involved in phosphorylation of the sarcomeric proteins, cMyBPC and cTNNI.31 This variant could plausibly modulate sarcomere kinetics but was classified as a VUS due to a lack of compelling evidence supporting PDE4DIP loss-of-function as a DCM mechanism.
Structural variants
Because WGS sequence coverage extends beyond exonic regions, comprehensive genome-wide evaluation of SV is possible. Using our in-house pipeline, ClinSV, we detected an average of 5379 SV, including 4470 copy number variants per proband, of which 232 were rare (MAF <1%), and 23 that were rare and overlapped genic regions. When the 67 panel genes were evaluated, one rare SV was identified. This was a complex BAG3 deletion/duplication that included the BAG domain and is likely to have a loss-of-function effect (Figure 2A, Table 3). This BAG3 SV was confirmed to be present in the proband using Sanger sequencing, segregated with DCM in Family AA (Supplementary Figure S2), and was deemed to be pathogenic. Numerous truncating BAG3 variants have been reported in DCM patients, with many co-segregating with disease in families.32 Eight SV were found in the extended gene set, all of which were validated by independent sequencing methods (Supplementary Methods and Results), but none met ACMG criteria for pathogenicity or were likely primary causes of DCM (Figure 2B, Table 3). For example, in the proband from Family BG we identified a rare whole gene duplication of triadin (TRDN; Figure 2B), a developmentally regulated core member of the ryanodine receptor complex, whose copy number has been conserved in mammals.33 Although present in 4 of the 6 other affected family members (Supplementary Figure S2), this variant was also seen in 4 unaffected family members, and was classified as a VUS. The remaining SV mostly showed incomplete segregation with disease, occurred in families with other identified pathogenic/likely-pathogenic variants, overlapped with SV reported in population databases, or were in genes with unknown relevance to DCM, and were all classified as VUS.
Figure 2.
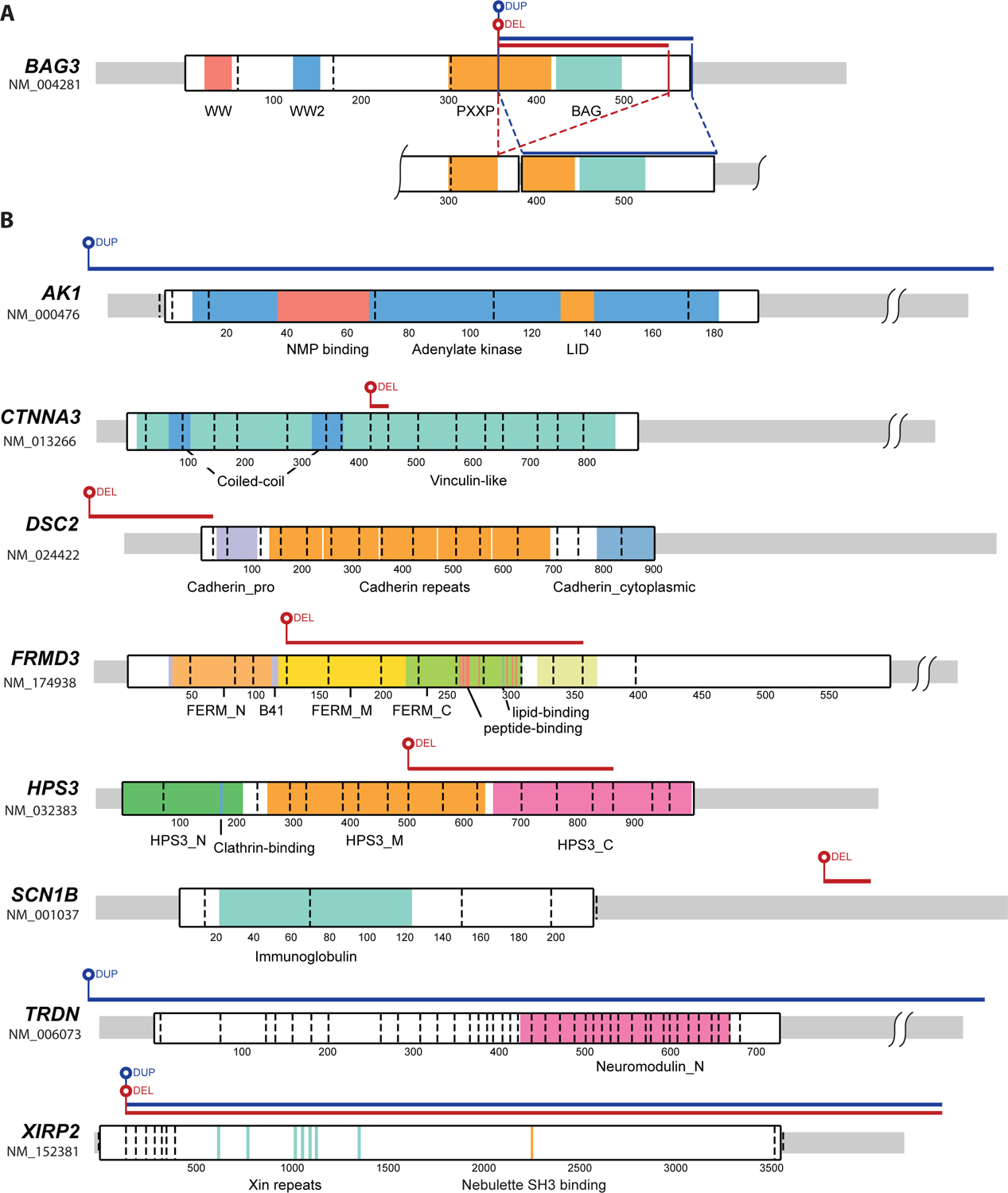
Structural variants identified in DCM probands. Schematics showing protein locations and relative size of deletions and duplications in (A) BAG3, and (B) 8 genes in the extended gene set. Protein domains are named and highlighted with colors, dashed vertical lines denote exon boundaries, and codons are numbered.
Table 3.
Structural variants identified in panel genes and in the extended gene set.
| Family ID | Gene | Variant | Database of Genomic Variants* | Notes | Segregation† |
|---|---|---|---|---|---|
| Families “unsolved” after PS | |||||
| AA | BAG3 | Chr10:121436136–121436726 del, Chr10:121436139–121436799 dup (g.30254–30844 del, 30257–30917 dup); involves exon 4 (aa.357–533) including BAG domain. | Overlap with 2 larger dup, 1 in unaffected subject. | BCL associated athanogene 3, chaperone protein. KO mice (−/−): neonatal death. Human ventricle: decreased BAG3 expression in heart failure. |
3/3 affected, 1/1 unaffected |
| BG | TRDN | Chr6:123509665–124330720 dup (821.1kb); includes TRDN (complete gene) and NKAIN2 (exon 1). | Similar dup in >10 DD patients + unaffected subjects. | Triadin, SR protein involved in Ca2+ homeostasis. Tg mice (5-fold overexpression): ↓ contraction. | 5/7 affected, 4/4 unaffected |
| C | CTNNA3 | Chr10:68286145–68513397 del (227.3kb); loss of exon 10 (aa 428–458) including vinculin-like domain. | >40 overlapping deletions, known CNV hotspot. | α3 catenin, intercalated disc protein. KO mice (−/−): adult DCM. | 4/6 affected, 4/17 unaffected |
| FK | FRMD3 | Chr9:85906635–85962244 del (55.6kb); loss of exons 5–12 (aa 125–357, includes partial FERM domain, peptide and lipid-binding regions). | Overlap with 3 del/dup. | FERM domain-containing protein 3, cytoskeletal protein. | 4/4 affected, 1/4 unaffected |
| HB | HPS3 | Chr3:148874313–148882670 del (g.31943–40298 del, 8.4 kb); loss of exon 9–14 (aa.504–863), no identified domains, | Overlap with 1 larger dup in 2 DD cases. | Hermansky-Pudlak syndrome 3 protein. | 1/2 affected 1/2 unaffected |
| Families with identified pathogenic/likely-pathogenic variants | |||||
| BL | DSC2 | Chr18:28681558–28686269 del (g.37617–42327 del, 4.7kb); loss of first exon + promoter. | Absent | Desmocollin 2, desmosomal protein. KO mice (−/−): embryonic lethal. | 2/2 affected |
| BR | SCN1B | Chr19:35531062–35531134 del (g.14470–14543 del, 73b in 3’ UTR). | Absent | β1 subunit, voltage-gated sodium channel. | 1/3 affected, 1/9 unaffected |
| DD | XIRP2 | Chr2:167868183–168522158 del/dup (654 kb); del from exon 3/dup (includes whole gene + translocated del segment) | Similar del (exon 4 on), multiple smaller CNV in same region. | Xin actin binding repeat-containing protein 2, intercalated disc protein. KO mice (−/−): postnatal death, (+/−): normal EF. Human ventricle: expression levels increased (early) or reduced (late) with myocardial stress. | 2/2 affected, 1/2 unaffected |
| FQ | AK1 | Chr9:130621906–130645663 dup (238 kb); includes complete gene. | Overlap with 3 full gene dup/multiple small del, in unaffected subjects. | Adenylate kinase 1, enzyme involved in energy metabolism. | 2/2 affected |
Database includes diverse patient and control cohorts;
Segregation analysis did not include phenotype-negative variant carriers aged less than 40 years.
CNV, copy number variant; DCM, dilated cardiomyopathy; DD, developmental delay; EF, ejection fraction; KO, knockout, PS, panel sequencing, SR, sarcoplasmic reticulum.
Value of WGS in additional family members
In two large kindreds that remained unsolved after PS and WGS testing of the proband, we undertook WGS in additional family members. In Family BP (Supplementary Figure S2), a MYBPHL p.R255* variant was identified in 3 family members but not in the proband. One of these, I-1, had been diagnosed with DCM and first degree atrioventricular block at 84 years of age. The other two variant carriers included an asymptomatic 56-year old male (II-1), and a 42-year old female with ventricular ectopy (II-6), neither of whom had DCM. MYBPHL has been recently described as a DCM disease gene, with homozygous and heterozygous knockout mice showing DCM and conduction-system abnormalities.34 The same MYBPHL p.R255* variant was seen in a family with early-onset DCM and in an unrelated individual with left ventricular dilation.34 The mutant MyBP-HL protein was function-altering, with reduced expression in human cardiomyocytes and abnormal myofilament localization in transfected neonatal mouse cardiomyocytes.34 In Family BG (Supplementary Figure S2), WGS in two of the proband’s aunts (II-2, II-4) identified a novel LOF TTN variant. Sanger sequencing of this variant in all family members confirmed its absence from the proband and presence in three affected siblings.
Overall yield of WGS and reportable incidental findings
The yield of pathogenic/likely-pathogenic variants was increased from 21 families (50%) with PS, to 24 families (57%) following WGS analysis of panel genes and preliminary mining of the extended gene panel. Interrogation of the ACMG 57-gene list17 (Supplementary Table S2) yielded three significant incidental findings. In the Family BG proband, we found a common function-altering GLA p.D313T variant associated with Fabry disease35 which did not segregate with DCM in the family (Supplementary Figure S2). Two other probands were heterozygous carriers of common missense MUTYH variants that are annotated as pathogenic in ClinVar and associated with increased cancer risk.
DISCUSSION
Our data support WGS as a viable method for genetic testing in familial DCM with high detection accuracy for rare SNVs and indels in disease-associated genes. These findings concur with recently-reported results for WGS-based testing in hypertrophic cardiomyopathy36 and extend pilot data for WGS in DCM.11 A compelling argument in favor of WGS as a first-line testing method is its potential for ongoing data mining in unsolved cases. However, in an initial analysis we found that surprisingly few variants in an extended gene set data met clinical criteria for pathogenicity.
Despite a lower overall sequencing depth than PS (and WES), WGS gave superior SNV and indel detection with less risk of variants being missed due to sequence gaps. WGS’s broad and uniform sequence coverage, even across exons targeted specifically by PS, results from sample preparation methods that avoid capture bias37 and a lack of constraint to predefined sets of transcript isoforms and target regions. Some WGS-identified variants in panel genes were missed by PS because different gene reference sets were used. This limitation of PS could potentially be reduced by optimized probe design and comprehensive representation of tissue-specific isoforms.6 The poor WES coverage observed over PS regions was recapitulated using three different exome capture kits, in data generated from two laboratories. Even at 300x depth, WES failed to achieve the same breadth of coverage as 34x WGS. Importantly we show that on target WES performance can be good, but often the target does not correspond to the nearby exon, resulting in potentially missed pathogenic variants.
Coverage uniformity gives WGS a distinctive advantage for identification of SV, that account for 0.5–1% of heritable inter-individual sequence differences (compared to 0.1% for SNVs).8 Although SV are increasingly implicated in human disorders,8 we found relatively few pathogenic SV in cardiomyopathy-associated genes. These results may be skewed by ascertainment bias, since DCM associations of panel genes have predominantly been established by profiling SNVs and indels. SVs often involve pairs of flanking low copy repeats.8 Known DCM-associated genes may lack these regions and hence be less susceptible to structural rearrangements. Extending our analysis to 406 cardiac-enriched genes resulted in the discovery of only 8 additional SV, none of which were clearly causative of DCM. One of the challenges in evaluating SV is assessment of their functional effects, especially for those variants that involve complex or partial duplications and deletions. Intuitively, changes in gene copy number would impact on gene “dose”, but this is not necessarily the case and a number of adaptive mechanisms may result in dosage compensation.8 Even full gene or balanced duplications and deletions can have unpredictable effects if there is disruption of local or long-range genomic architecture that involves gene-regulatory sequences.
To improve WGS yield, more genetic information is needed to identify patterns of differential variant prevalence in cases and controls, recurrent variant types, and mutation hotspots. Equally, more experimental data are required to identify function-altering variants and to show that these functional effects have plausible links to disease causation. In the ACMG criteria,17 segregation of variants in affected family members provides support for DCM association, but the level of evidence can increase to moderate or strong with larger family sizes and/or multiple families.
Segregation analysis in familial DCM relies on the assumption of single driver mutations but this is increasingly open to question.38 As genetic evaluation has shifted from gene candidate screening studies to genome-wide analyses, it is not uncommon to find multiple rare variants in family probands39 and, as seen with families BG and BP, the number of potential function-altering variants in any one family can increase as more individuals are studied. These combinations of variants may have additive, synergistic or neutralizing effects, and each person’s total burden of rare and common variants may determine threshold levels for myocardial dysfunction.
Should WGS be used as a first-line genetic test for DCM or reserved for PS-negative cases? The answer to this question is currently unknown and involves a dynamic interplay between cost and yield (Supplementary Figure S3). Currently, the cost of WGS-based clinical testing is more than double that of PS. With increasing customer demand and technical efficiencies, the operational costs of both WGS and PS are likely to decrease over time. For PS, these reductions will be offset by the ongoing need to design and optimize new probe sets as new disease genes are discovered. The manpower costs of expert clinical reporting are essentially equivalent for PS and WGS since the same suites of disease genes are generally evaluated, but these costs may rise as the spectrum of reportable genes and variants expands. The turn-around time for WGS is equivalent or faster than routine PS (or WES), because of the time required for targeted sequence capture, together with delays incurred by waiting for samples to be pooled. Recent data have shown that fast-tracked WES-based genetic testing in acutely ill pediatric patients is however feasible and cost-saving, due to expedited diagnosis and management.40 WGS is a storehouse of medically-relevant information that extends beyond identification of rare disease-causing variants, much of which cannot be obtained from PS or WES. For example, polygenic risk scores derived from suites of common variants (often in non-coding regions) may predict an individual’s risk of DCM complications, and pharmacogenomic associations may guide drug selection and doses.
Comprehensive economic models need to be developed that consider variables such as cost per test, number of tests ordered, yield of tests, requirement for secondary testing of probands, cascade testing of family members, and the impact of genotype information on requirements for clinical surveillance of family members. Genotype-based early interventions may also impact more broadly on long-term health outcomes and costs, potentially impacting hospitalization rates, device implantation, drug administration, workforce productivity, and use of social services. Compelling health economics data would provide a powerful argument to insurance companies or governments for the overall benefits of genetic testing and the preferred testing modality.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Kinghorn Centre for Clinical Genomics for assistance with production and processing of whole-genome sequencing data. We acknowledge Robert Graham, Matthew Grounds, John Mattick and John Schubert, as founders of the Cardiogenomics Project. This research was undertaken with the assistance of resources and services from the National Computational Infrastructure, which is supported by the Australian Government.
FUNDING SOURCES
This work was undertaken as part of the Cardiogenomics Project, and would not have been possible without the support of John Schubert, the Kinghorn Foundation, Garvan Foundation, and the Victor Chang Cardiac Research Institute. Funding support was also received from the NSW Office of Health and Medical Research Collaborative Grants Program, National Health and Medical Research Council (D.F., C.S.), Estate of the Late RT Hall (D.F.), Simon Lee Foundation (D.F.), Howard Hughes Medical Institute (C.E.S.), National Institutes of Health (J.G.S.), Leducq Foundation (J.G.S., C.E.S.), Cancer Institute NSW (M.J.C.), NSW Department of Health (M.J.C). J.I. is the recipient of a National Heart Foundation of Australia Future Leader Fellowship.
Footnotes
DISCLOSURE
M.E.D., K.W., A.L.S., and B.L. are employed by Genome.One, a clinically accredited genetic testing provider that uses whole genome sequencing.
The remaining authors declare no conflict of interest.
REFERENCES
- 1.Fatkin D, Seidman CE, Seidman JG. Genetics and disease of ventricular muscle. Cold Spring Harb Perspect Med 2014;1:a021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies. Europace 2011;13:1077–1109. [DOI] [PubMed] [Google Scholar]
- 3.Fatkin D, Johnson R, McGaughran J, Weintraub RG, Atherton JJ. Position statement on the diagnosis and management of familial dilated cardiomyopathy. Heart Lung Circ 2017;26:1127–1132. [DOI] [PubMed] [Google Scholar]
- 4.Sikkema-Raddatz B, Johansson LF, de Boer EN, et al. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat 2013;34:1035–1042. [DOI] [PubMed] [Google Scholar]
- 5.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med 2014;16:601–608. [DOI] [PubMed] [Google Scholar]
- 6.Pua CJ, Bhalshankar J, Miao K, et al. Development of a comprehensive sequencing assay for inherited cardiac condition genes. J Cardiovasc Transl Res 2016;9:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 2011;12:745–755. [DOI] [PubMed] [Google Scholar]
- 8.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet 2013;14:125–138. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Montgomery SB. Detection and impact of rare regulatory variants in human disease. Front Genet 2013;4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA 2014;311:1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golbus JR, Puckelwartz MJ, Dellefave-Castillo L, et al. Targeted analysis of whole genome sequence data to diagnose genetic cardiomyopathy. Circ Cardiovasc Genet 2014;7:751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts AM, Ware JS, Herman DS, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med 2015;7:270ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi L, Zhang Y, Feng L, et al. Multi-omics study revealing the complexity and spatial heterogeneity of tumor-infiltrating lymphocytes in primary liver carcinoma. Oncotarget 2017;8:34844–34857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gayevskiy V, Roscioli T, Dinger ME, Cowley MJ. Seave: a comprehensive web platform for storing and interrogating human genomic variation. BioRxiv doi: 10.1101/258061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehm HL, Bale SJ, Bayrak-Toydemir P, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013;15:733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlén M, Fagerberg L, Hallström BM, et al. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 19.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viguera E, Canceill D, Ehrlich SD. In vitro replication slippage by DNA polymerases from thermophilic organisms. J Mol Biol 2001;312:323–333. [DOI] [PubMed] [Google Scholar]
- 21.Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol 2009;10:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa MW, Guo G, Wolstein O, et al. Functional characterization of a novel mutation in NKX2–5 associated with congenital heart disease and dilated cardiomyopathy. Circ Cardiovasc Genet 2013;6:238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Begay RL, Tharp CA, Martin A, et al. FLNC gene splice mutations cause dilated cardiomyopathy. JACC: Basic Transl Sci. 2016;1:344–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of filamin C (FLNC) results in severe defects in myogenesis and myotube structure. Mol Cell Biol 2006;26:6522–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chevessier F, Schuld J, Orfanos Z, et al. Myofibrillar instability exacerbated by acute exercise in filaminopathy. Hum Mol Genet 2015;24:7207–7220. [DOI] [PubMed] [Google Scholar]
- 26.Bolduc V, Marlow G, Boycott KM, et al. Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am J Hum Genet 2010;86:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J, El Refaey M, Xu L, et al. Genetic disruption of Ano5 in mice does not recapitulate human Ano5-deficient muscular dystrophy. Skelet Muscle 2015;5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann L, Haussmann U, Mueller M, Spiekerkoetter U. VLCAD enzyme activity determinations in newborns identified by screening: a valuable tool for risk assessment. J Inherit Metab Dis 2012;35:269–277. [DOI] [PubMed] [Google Scholar]
- 29.Erdmann J, Daehmlow S, Wischke S, et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin Genet 2003;64:339–349. [DOI] [PubMed] [Google Scholar]
- 30.Chen SN, Czernusczewicz G, Tan Y, et al. Human molecular genetic and functional studies identify TRIM63, encoding Muscle RING Finger Protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ Res 2012;111:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uys GM, Ramburan A, Loos B, et al. Myomegalin is a novel A-kinase anchoring protein involved in the phosphorylation of cardiac myosin binding protein C. BMC Cell Biol 2011;12:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norton N, Li D, Rieder MJ, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet 2011;88:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice AM, McLysaght A. Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nat Commun 2017;8:14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barefield DY, Puckelwartz MJ, Kim EY, et al. Experimental modelling supports a role for MyBP-HL as a novel myofilament component in arrhythmia and dilated cardiomyopathy. Circulation 2017;136:1477–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froissart R, Guffon N, Vanier MT, Desnick RJ, Maire I. Fabry disease: D313Y is an α-galactosidase A sequence variant that causes pesudodeficient activity in plasma. Mol Genet Metab 2003;80:307–314. [DOI] [PubMed] [Google Scholar]
- 36.Cirino AL, Lakdawala NK, McDonough B, et al. A comparison of whole genome sequencing to multigene panel testing in hypertrophic cardiomyopathy patients. Circ Cardiovasc Genet 2017;10:e001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodi K, Perera AG, Adams PS, et al. Comparison of commercially available target enrichment methods for next-generation sequencing. J Biomol Tech 2013;24:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol 2013;10:531–547. [DOI] [PubMed] [Google Scholar]
- 39.Haas J, Frese KS, Peil B, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J 2015;36:1123–1135. [DOI] [PubMed] [Google Scholar]
- 40.Stark Z, Lunke S, Brett GR, et al. Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet Med 2018; March 15. doi: 10.1038/gim.2018.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


