SUMMARY
The olfactory epithelium (OE) is a neurosensory organ required for the sense of smell. Turbinates, bony projections from the nasal cavity wall, increase the surface area within the nasal cavity lined by the OE. Here, we use engineered Fibroblast Growth Factor 20 (Fgf20) knockin alleles to identify a population of OE progenitor cells that expand horizontally during development to populate all lineages of the mature OE. We show that these Fgf20-positive, epithelium-spanning progenitor (FEP) cells are responsive to Wnt/β-Catenin signaling. Wnt signaling suppresses FEP cell differentiation into OE basal progenitors and their progeny, and positively regulates Fgf20 expression. We further show that FGF20 signals to the underlying mesenchyme to regulate the growth of turbinates. These studies thus identify a population of OE progenitor cells that function to scale OE surface area with the underlying turbinates.
Keywords: FGF20, olfactory epithelium, progenitor cell, turbinate, Wnt/β-Catenin, tissue scaling, condensation
Graphical Abstract
eTOC Blurb
Mechanisms regulating olfactory epithelium expansion and turbinate growth are poorly understood. In this issue of Developmental Cell, Yang et al. show that an Fgf20-expressing progenitor (FEP) expands the olfactory epithelium and regulates turbinate growth via FGF20. Furthermore, Wnt/β-Catenin signaling acts as a master regulator of these two processes.
INTRODUCTION
A main function of the mammalian nose is olfaction. The interior walls of the posterior nasal cavity are lined by olfactory epithelia (OE), which houses olfactory receptor neurons (ORNs) that detect odorants in inspired air. Scrolling bony plates, called turbinates, project from the walls of the nasal cavity, and greatly increase its interior surface area. In mammals, turbinate size, shape, and complexity varies dramatically among species. It has been hypothesized that the interspecies differences in turbinate complexity, and therefore surface area within the nasal cavity, correlates with interspecies differences in olfactory ability (Van Valkenburgh et al., 2014).
The mature mouse OE is a pseudostratified epithelium consisting of three main cell types, the nuclei of which are located in different layers: sustentacular cell (Sus, a supporting cell population) nuclei in the apical layer, ORN nuclei in the middle layers, and basal cell (BC, progenitors that give rise to Sus cells and ORNs) nuclei in the basal layer (Murdoch and Roskams, 2007; Schwob et al., 2017). Other OE cell types include Bowman’s duct cells and microvillar cells (Weng et al., 2016). OE development and neurogenesis occurs in two main phases: embryonic day 10 (E10) to E13, and E13 to adult (Beites et al., 2005; Ikeda et al., 2007; Smart, 1971). Neurogenesis begins at ~E10 with the invagination of the nasal pit (Treloar et al., 2010). From this stage to E13, nuclei of Sox2-expressing (Sox2+) progenitors are found throughout the thickness of the OE, particularly apically. From E13 to adult, progenitor nuclei shift to a basal position, and are referred to as BCs (Cau et al., 1997; Smart, 1971). During these stages, the OE matures into its classic pseudostratified histology. BCs continue to divide and differentiate into the overlying ORNs and Sus cells throughout life, a process termed “established neurogenesis.” It has been suggested that prior to E13, OE progenitors mainly expand, while after E13, they fill the OE with ORNs (Beites et al., 2005). Here, we use the term “immature” to refer to OE in which progenitor nuclei are located throughout the thickness of the OE, and the term “mature” to refer to OE containing progenitor (BC) nuclei in the basal layer, ORN nuclei in the middle layers, and Sus cell nuclei in the apical layer.
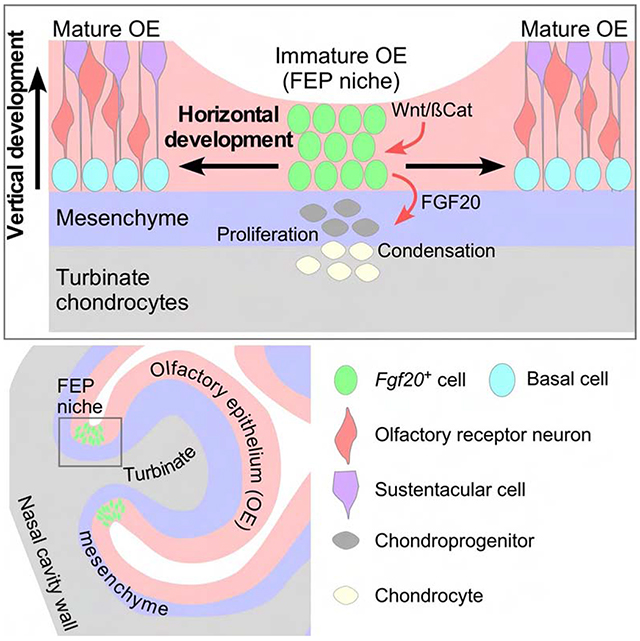
Established neurogenesis in the OE during development, adult homeostasis, and regeneration is well studied. We refer to this process as vertical development, as it involves the differentiation of BCs into cells that fill the upper OE layers. Horizontal development, or OE surface area expansion, is not well understood. Despite the importance of OE surface area to many mammals, the mechanism and a progenitor population regulating OE scaling has not been found. Moreover, besides the role of airflow in respiratory turbinate development (Coppola et al., 2014), factors that regulate turbinate development are unstudied.
A particularly interesting question is how OE expansion is scaled with turbinate growth. Crosstalk between the OE and the underlying turbinates has been predicted but not identified (Adameyko and Fried, 2016). Notably, genetic manipulations resulting in early failure of OE development also lead to disrupted formation of turbinates and other nasal structures, highlighting the importance of the OE to the growth of the nasal cavity (Duggan et al., 2008; Ikeda et al., 2007; Kawauchi et al., 2005, 2009; Kersigo et al., 2011; Laclef et al., 2003). Here, we propose that throughout embryonic and early postnatal development, there are regions of the OE that remain immature and facilitate horizontal expansion of the OE, rather than vertical development. We identify the cells residing in these regions as OE progenitors that regulate turbinate growth via Fibroblast Growth Factor 20 (FGF20). We further show that Wnt/β-Catenin (βCat, also Ctnnb1) signaling is required for the maintenance of these Fgf20-positive, epithelium-spanning progenitor (FEP) cells and their expression of Fgf20. These mechanisms regulate the overall size of the olfactory system and ensure that the OE and underlying turbinates scale proportionally.
RESULTS
In adult mice, the OE lines the nasal septum, the (olfactory) ethmo-turbinates, and the dorsal and lateral nasal cavity walls of the olfactory recess (Barrios et al., 2014). A layer of mesenchyme, called the lamina propria in the adult, separates the OE from the underlying cartilaginous or bony structures (Figure S1H). Throughout development and in the adult, the six olfactory turbinates have highly conserved branching, scrolling, and folding morphology (Figures S1I and S1I’). In order of most anterodorsal to posteroventral, they are: endoturbinate I (n1), ectoturbinate 1 (c1), endoturbinate II (n2), ectoturbinate 2 (c2), endoturbinate III (n3), and endoturbinate IV (n4). Anteriorly, n2 separates into two branches, n2’ and n2”. In this study, we focus mainly on c1. All images of turbinates presented are frontal sections through the posterior nasal cavity. We refer to “neck” (site of attachment of turbinates to the nasal cavity wall) and “tip” regions of the turbinate as shown in Figure S1H, inset. We refer to “neck” OE as the negatively-curved OE directly overlying the “neck” region of the turbinate. In the adult, “neck” regions have sometimes been referred to as the cul-de-sac of the turbinate (Greer et al., 2016).
Fgf20 is expressed in a subset of Sox2+ cells in the developing OE
Initial studies of the Fgf20βgal allele showed Fgf20 expression in progenitor-like cells in several developmental systems. To investigate the cell type marked by Fgf20 expression in the developing OE, we used the Fgf20GFP-Cre allele (Huh et al., 2015). We used an anti-GFP antibody to detect GFP-Cre expression, as native GFP fluorescence from this allele is mostly undetectable.
Fgf20GFP-Cre expression was found in the developing OE as early as E10.5, albeit very weakly, in the lateral nasal pit (Figures 1A and S1A). Throughout its expression in the OE, Fgf20 co-localized with a subset of Sox2+ cells, a marker of progenitors that fill the immature OE, and of BCs and Sus cells in the mature OE (Kawauchi et al., 2005). At E12.5, turbinates n1, n2, and n3 are composed of mesenchyme surrounded by OE and respiratory epithelia (Figure S1B, n3 not shown). At this stage, Fgf20 was mainly expressed in the lateral OE (Figure 1B). Importantly, Fgf20 was expressed in the OE overlying the site of future c1 development (Figure 1B, asterisk).
Figure 1.
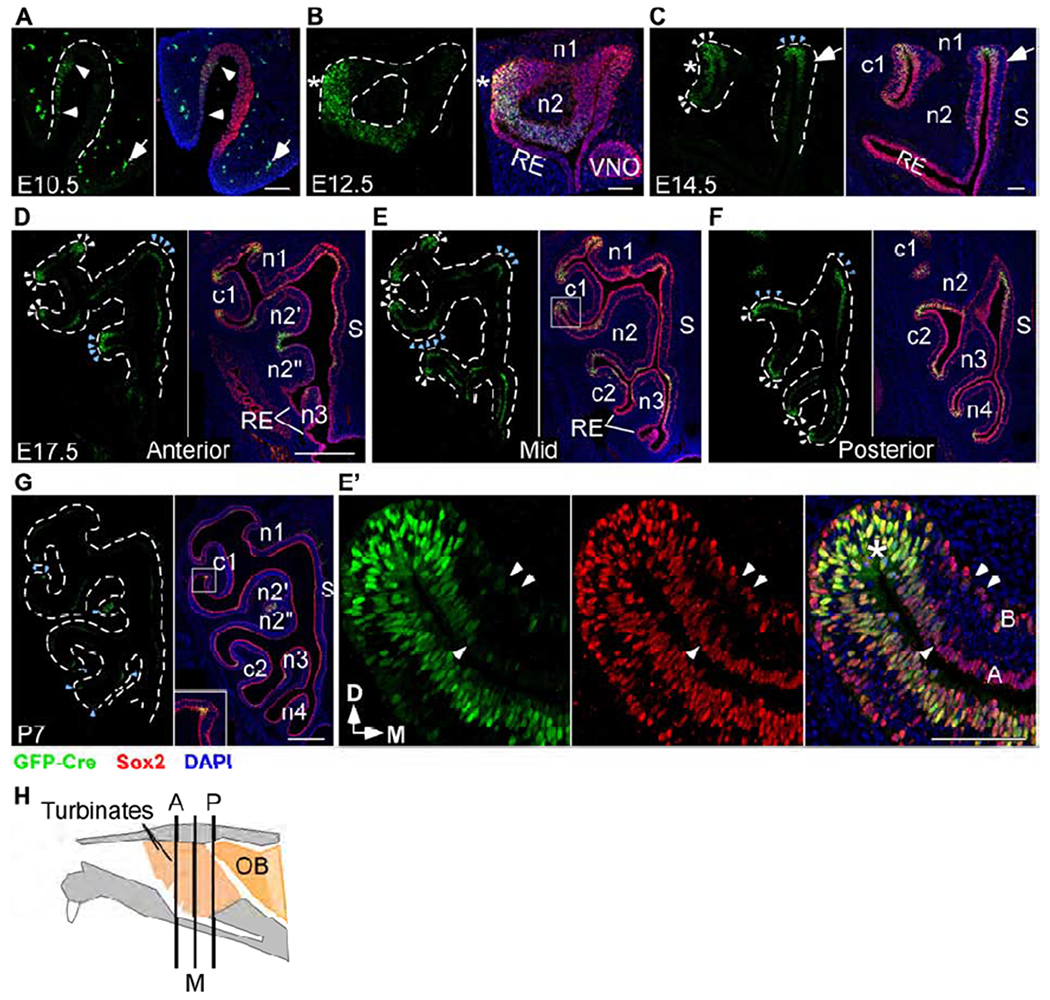
Fgf20 is expressed in a subset of Sox2+ cells in the developing OE
(A-G) Fgf20 expression detected via an anti-GFP antibody in Fgf20GFP-Cre/+ mice (green). The OE is identified via an anti-Sox2 antibody (red). Sox2 is expressed throughout the OE at E10.5 (A) and E12.5 (B), and mainly in the basal and apical layers at E14.5 (C), E17.5 (D-F), and P7 (G).
(A) Faint Fgf20GFP-Cre expression in the nasal pit at E10.5. Arrowheads indicate extent of the expression. Arrow indicates autofluorescence from red blood cells outside of the nasal pit.
(B) Fgf20GFP-Cre expression in the OE at E12.5. * indicates site of future c1 development. VNO, vomeronasal organ.
(C) Fgf20GFP-Cre expression in the OE at E14.5. White arrowheads indicate immature negatively-curved OE. Blue arrowheads indicate mature negatively-curved OE. Arrows indicate apical Fgf20 expression in OE outside of regions of negative curvature. * indicates expression in OE overlying c1.
(D-F) Fgf20GFP-Cre expression in anterior (D), mid (E), and posterior (F) sections at E17.5. White arrowheads indicate immature negatively-curved OE. Blue arrowheads indicate mature negatively-curved OE.
(E’) Magnification of the c1 neck region, boxed in (E). Arrowheads indicate fainter Fgf20GFP-Cre expression in Sox2+ basal and apical cells adjacent to negatively-curved OE (*). A, apical layer; B, basal layer indicated in the merged image.
(G) Fgf20GFP-Cre expression at P7. Arrowheads indicate apical Fgf20GFP-Cre expression at negatively-curved OE. Inset, 2.5x magnification of boxed region.
(H) Diagram of a mid-sagittal mouse nasal cavity showing locations of the three sections in (D-F). A, anterior; M, mid; P, posterior.
RE, respiratory epithelium. S, nasal septum. D, dorsal; M, medial. Dashed line, epithelial-mesenchymal boundary. DAPI, nuclei (blue). Scale bars, 100 μm (A-C, E’), 500 μm (D-G). See also Figure S1.
Turbinates develop through endochondral ossification, initiated via epithelial budding and followed by proliferation of the underlying mesenchyme (Dieulafé, 1906; Martineau-Doizé et al., 1992). At E14.5, turbinates c1, c2, and n4 appeared as mesenchymal condensations protruding from the nasal cavity wall (Figures S1C and S1C’, c2 and n4 not shown). At this stage, most of the OE was mature, with an apical and a basal layer of Sox2+ cells separated by a Sox2− middle layer of ORNs. In mature OE, Fgf20 was expressed in a subset of apical Sox2+ cells (Figure 1C, arrows). Fgf20 was also expressed in regions of negatively-curved OE formed by the developing turbinates. These regions of the OE mostly retained an immature histology, as Fgf20+/Sox2+ cell nuclei were found throughout all layers of the OE (Figure 1C, white arrowheads). Fgf20 expression was still found in the OE overlying c1 at this stage (Figure 1C, asterisk).
By E17.5, in all six turbinates, cells of the mesenchymal condensations have differentiated into chondrocytes (Figures S1D–S1F, and S1E’). At this stage, high Fgf20 expression remained localized to areas of negatively-curved OE, which surround the “neck” of each of the six turbinates (Figures 1D–1F, arrowheads). Importantly, for turbinates n1, c1, c2, n3, and n4, these areas remained immature, with Fgf20+/Sox2+ cell nuclei spanning the entire thickness of the OE. For turbinate n2, the earliest turbinate to develop, “neck” OE has become more mature by E17.5 and only retained Fgf20+/Sox2+ cell nuclei towards the apical layers of the OE (Figure 1E, blue arrowheads). The medial “neck” OE of turbinate n1, formed by the nasal septum and n1, has likewise become more mature by E17.5 (Figure 1E, blue arrowheads).
Turbinate chondrocytes undergo hypertrophy at early postnatal stages and by postnatal day 7 (P7) most of the cartilage has been replaced by ossified bone (Figures S1G and S1G’). At this stage, Fgf20 expression was still primarily found in negatively-curved “neck” OE, but at the apical surface (Figure 1G, blue arrowheads). The concentrated expression seen at earlier stages has mostly dissipated, as the “neck” OE of every turbinate has become almost completely mature by P7. After complete ossification, turbinates continue to grow, fold, and scroll until at least P30 (Figure SlI). At P30, Fgf20 expression was undetectable (data not shown).
Interestingly, at E17.5, just outside of Fgf20 expression hotspots in negatively-curved, immature OE, low GFP fluorescence was detected in the Sox2+ basal and apical cells of the mature OE (Figure 1E’, arrowheads). We suspect that this represents the capture of the transient Fgf20GFP-Cre lineage, due to GFP-Cre protein perdurance, suggesting that Fgf20+ cells give rise to adjacent Sox2+ BCs and Sus cells.
Overall in development, high Fgf20 expression was found in immature regions of the OE, in a pattern associated with the growing turbinates. Upon maturation of a region of OE, Fgf20 expression was initially shifted towards the basal and particularly apical layers before disappearing completely. Together, these observations led to the hypothesis that Fgf20+ cells are a progenitor population in the developing OE. Based on results from experiments described below, we term this population FEP (Fgf20-positive, epithelium-spanning progenitor) cells, as their nuclei are found throughout all layers of the immature OE.
Fgf20 lineage includes all major OE cell types and responds to cues for expansion
To test the hypothesis that Fgf20+ cells are OE progenitors, we first looked at proliferation and neurogenesis rates in negatively-curved “neck” OE, where FEP cells are located. At E17.5, these regions incorporated EdU at a higher rate than the OE at the turbinate “tip” (Figures S2A and S2B). This difference could be partially explained by a relative lack of post-mitotic ORNs in “neck” regions. Confirming this explanation, “neck” OE contained far fewer olfactory marker protein (OMP) positive cells (Figures S2C and S2D, arrowheads), which is a marker for mature ORNs (Hartman and Margolis, 1975). The only exception was the medial “neck” OE of n1, the most mature “neck” OE at E17.5. The distribution of Fgf20+ cells and OMP+ ORNs along the length of the c1 OE, when plotted along a straight line, showed low OMP+ ORN density in areas of high Fgf20+ cell density (Figure S2E). No cells were found to co-express Fgf20 and OMP.
In the adult, OE at “recesses of olfactory turbinates” have been shown to contain Pde2a+ ORNs, rather than OMP+ ORNs (Figure S2F; Juilfs et al., 1997). To rule out the possibility that “neck” OE contained these OMP− ORNs during embryonic stages, we examined Pde2a expression at E17.5. Pde2a+ ORNs were not found in “neck” OE or anywhere else at this stage (Figure S2G). Altogether, these results suggest that immature, “neck” OE have increased rates of proliferation and decreased rates of neurogenesis. This supports the idea that these regions host embryonic progenitors involved in horizontal, as opposed to vertical, development.
Next, we combined Fgf20GFP-Cre with Cre reporter alleles ROSAmTmG (Muzumdar et al., 2007) or ROSAtdTomato (Madisen et al., 2010) to trace the lineage of Fgf20GFP-Cre-expressing cells. The ROSAmTmG allele, in which Cre-expressing cells and their progeny express membrane-localized eGFP (mG), while all other cells express membrane-localized tdTomato (mT), showed that the Fgf20GFP-Cre lineage includes most of the postnatal OE, and the entire ventrolateral OE containing zones 2-4 (Figures 2A–2C; OE zones are described in Ressler et al., 1993; Vassar et al., 1993). Only a few spots associated with dorsomedial zone 1 were outside of the lineage (Figures 2A–2C, arrowheads). This pattern was consistent across individuals. Other olfactory structures with Fgf20GFP-Cre-lineage cells were the vomeronasal organ (VNO; Figure S2H) and septal organ (SO; Figure S2I).
Figure 2.
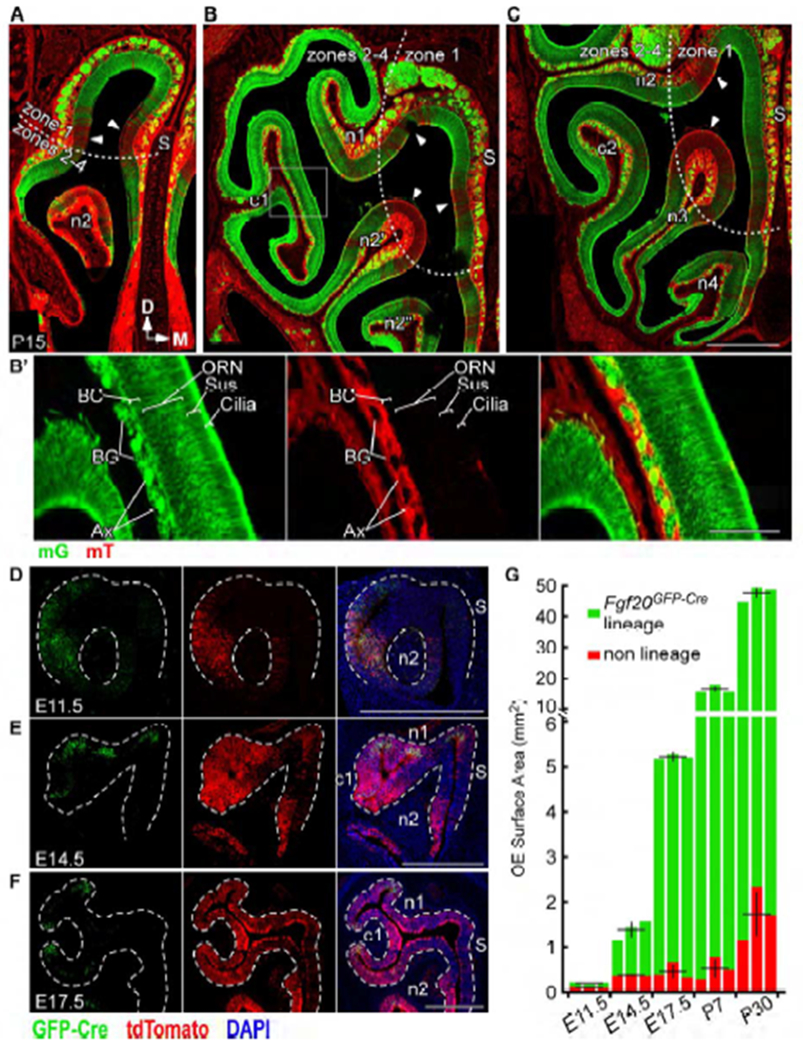
Fgf20 lineage includes all major OE cell types and responds to cues for expansion
(A-C) mG expression in the Fgf20GFP-Cre lineage and mT expression (arrowheads) in non-Fgf20GFP-Cre lineage in anterior (A), mid (B), and posterior (C) frontal sections of Fgf20GFP-Cre/+; ROSAmTmG mice at P15. S, nasal septum. Dotted line, zone 1 and zones 2-4 demarcation.
(B’) Magnification of boxed region in (B). mG expression in the basal cell (BC), olfactory receptor neuron (ORN), sustentacular cell (Sus), and cilia layers of the OE, as well as in axon bundles (Ax) and Bowman’s glands (BG) found in the lamina propria.
(D-F) Real-time Fgf20 expression (GFP-Cre) and Fgf20GFP-Cre lineage (tdTomato) in Fgf20GFP-Cre/+; ROSAtdTomato mice at E11.5 (D), E14.5 (E), and E17.5 (F). Dashed line, epithelial-mesenchymal boundary.
(G) Quantification of total Fgf20GFP-Cre lineage and non-Fgf20GFP-Cre lineage OE surface area at E11.5, E14.5, E17.5, P7, and P30. Error bars, mean ± SD; n = 3 at each stage; each bar represents one mouse
DAPI, nuclei. Scale bars, 500 μm (A-F), 100 μm (B’). See also Figure S2.
Importantly, in Fgf20GFP-Cre-lineage OE, the entire thickness of the OE (the BC, ORN, and Sus cell layers, as well as the brightly fluorescent cilia layer apical to the Sus cell layer) expressed mG, and not mT (Figure 2B’). mG expression was restricted to the OE, except for two accessory OE structures found in the lamina propria (mesenchyme): axon bundles (Ax) projected by ORNs, and Bowman’s glands (BG), which associate with Bowman’s ducts that traverse the OE. These structures can be readily identified based on morphology.
We then compared “real-time” Fgf20GFP-Cre expression, identified with an antibody to GFP-Cre, to its lineage with the ROSAtdTomato reporter at various stages of development. At E11.5, the lineage almost completely overlapped with real-time Fgf20 expression (Figure 2D). However, as the OE expands (E14.5 and E17.5), the lineage increasingly exceeded the domain of real-time expression (Figures 2E and 2F). Furthermore, quantification of surface area of OE within and outside of the Fgf20GFP-Cre lineage showed that growth of the Fgf20GFP-Cre-lineage OE far outpaced that of non-lineage OE (Figure 2G). In fact, surface area of the non-lineage OE did not significantly change from E14.5 to P7: 0.38 ± 0.02 mm2 at E14.5, 0.46 ± 0.18 mm2 at E17.5, and 0.53 ± 0.25 mm2 at P7 (n = 3, mean ± SD, p = 0.6, one-way ANOVA). The Fgf20GFP-Cre-lineage OE, on the other hand, grew significantly in surface area during this period: 1.39 ± 0.21 mm2 at E14.5, 5.21 ± 0.06 mm2 at E17.5, and 16.5 ± 1.1 mm2 at P7 (p < 0.001, one-way ANOVA). Therefore, the Fgf20GFP-Cre lineage accounted for nearly all the expanding OE from E14.5 to P7. These results support the hypothesis that FEP cells are multipotent progenitors of the OE contributing to horizontal expansion.
Fgf20-KO mice have reduced turbinate size and altered morphology
Next, we investigated the function of FGF20 signaling in olfactory system development. Because Fgf20 was expressed in a pattern associated with the developing turbinates, we compared the turbinate morphology of Fgf20-KO (Fgf20GFP-Cre/βgal) mice to those of heterozygous littermate controls (Fgf20GFP-Cre/+). Gross overview of serial frontal sections of the nasal cavity at P30 showed that most of the olfactory turbinates were smaller and/or differed in shape in Fgf20-KO mice compared to control (Figures 3A–3D). These changes were consistent across individuals. Measurement of the surface of each turbinate to estimate size (Figure 3B’, outline) showed that of the six turbinates, c1, c2, and n4 were the most reduced in size in Fgf20-KO mice; by 35%, 28%, and 34%, respectively (Figure 3E). c1 showed reduced folding and was missing its dorsal branch (Figure 3B’). n1, n2, and n3, the earliest turbinates to appear developmentally, were the least reduced in size; by 15%, 3.7%, and 7.0%, respectively (Figure 3E). In total, Fgf20-KO mice had a 17% average decrease in OE surface area within the nasal cavity, due to smaller turbinates, while septal organ surface area was not changed (Figure 3F). The septal organ is made up of olfactory epithelia, some of which is in the Fgf20GFP-Cre lineage (Figure S2I), but is only found overlying the nasal septum and not turbinates. This makes it a good internal control for turbinate surface area. The nasal cavity wall and septum did not appear to be affected by the loss of FGF20 signaling.
Figure 3.
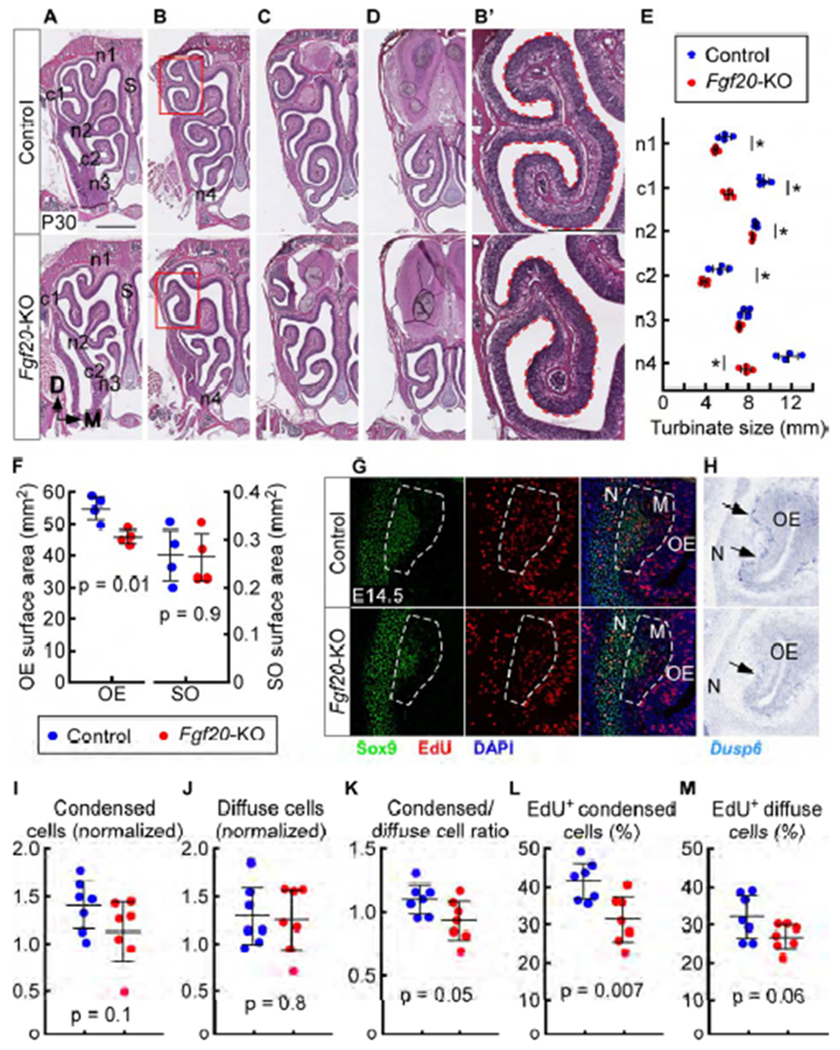
Fgf20-KO mice have reduced turbinate size and altered morphology
(A-D) H&E staining in anterior (A), mid-anterior (B), mid-posterior (C), and posterior (D) sections through the nasal cavity in control (Fgf20GFP-Cre/+) and Fgf20-KO (Fgf20GFP-Cre/βgal) mice at P30. S, nasal septum.
(B’) Magnification of boxed region in (B). Dashed outline used to estimate relative turbinate size.
(E) Quantification of turbinate size for each turbinate at P30. * indicates p < 0.05. p = 0.03 (n1), p < 0.001 (c1), p = 0.02 (n2), p = 0.02 (c2), p = 0.05 (n3), p < 0.001 (n4), n = 4, Student’s t-test.
(F) Quantification of OE surface area and septal organ (SO) surface area at P30. n = 4, Student’s t-test
(G) EdU incorporation in c1 condensed and diffuse mesenchymal cells at E14.5. Dashed outline, c1 mesenchyme (M), including both condensed (Sox9hi) and diffuse cells. N, nasal cavity wall.
(H) In situ hybridization at E14.5 showing c1 mesenchymal Dusp6 expression in control (arrows) and reduced expression in Fgf20-KO embryos (arrow). Data is representative of 7 control and 7 Fgf20-KO embryos. N, nasal cavity wall.
(I-M) Quantification of c1 normalized condensed cell number (I), normalized diffuse cell number (J), ratio of condensed cells to diffuse cells (K), and percent of EdU-incorporating condensed cells (L) and EdU-incorporating diffuse cells (M) at E14.5. n = 7, Student’s t-test.
DAPI, nuclei. Scale bars, 1 mm (A-D), 500 μm (B’), 100 μm (G, H). Error bars, mean ± SD. See also Figures S3 and S4.
We then examined Fgf20-KO mice at P0, when turbinates are still made up of chondrocytes. Many of the turbinates in Fgf20-KO mice appeared smaller than those in control (Figure S3A and S3B). Focusing on c1, there was a measurable decrease in the size of the turbinate cartilage (p = 0.009) and in the surface area overlying the turbinate (p < 0.001; Figures S3A’, S3C, and S3D). Therefore, the adult turbinate deformity can at least be partially attributed to a defect prior to osteogenesis.
Fgf20 is required for mesenchymal proliferation early in turbinate development
In the initial stages of endochondral bone development, mesenchymal cells condense to form a mesenchymal condensation, and subsequently differentiate into chondrocytes (Long and Ornitz, 2013). To determine the cause of the decrease in c1 cartilage size at P0, we examined these initial stages in Fgf20-KO embryos. Development of turbinate c1 began at around E14.5, as a mesenchymal condensation between the OE and the nasal cavity wall. The condensed mesenchymal cells had high Sox9 expression (Sox9hi; Figure 3G). Chondrocytes of the nasal cavity wall were also Sox9hi but were distinguished from the condensed mesenchymal cells by their round nuclei and large soma. Mesenchymal cells that remained diffuse (uncondensed) were found between the condensation and the OE. These cells had weak or almost undetectable Sox9 expression (Sox9low).
In Fgf20-KO embryos at E14.5, the number of condensed cells and diffuse cells were not significantly changed compared to control, although there was a trend towards fewer condensed cells (Figures 3G, 3I, and 3J). Consistent with this, the ratio of condensed cells to diffuse cells was slightly decreased compared to control (p = 0.05; Figure 3K), suggesting a defect in formation of the condensation. Analysis of EdU incorporation showed a decrease in proliferation rate in condensed cells (p = 0.007; Figures 3G and 3L) and diffuse cells (p = 0.06; Figure 3M) in Fgf20-KO embryos, although the latter was not statistically significant. To identify the cell type directly responding to FGF20, we examined Dusp6 expression by RNA in situ hybridization. Dusp6 is a downstream target of FGF signaling (Ekerot et al., 2008; Kawakami et al., 2003; Li et al., 2005, 2007; Ornitz and Itoh, 2015) and is a target of FGF20 signaling in the cochlea (unpublished data). At E14.5, Dusp6 was expressed in the c1 mesenchyme directly underneath the OE in control embryos and was markedly reduced in Fgf20-KO embryos (Figure 3H, arrows). This suggests that FGF20 is signaling to diffuse cells directly underneath the OE.
At E17.5, Sox9hi condensations were no longer observed in the turbinates. In Fgf20-KO mice, the rate of proliferation was decreased in “neck” region mesenchymal cells compared to control (Figures S4A, S4A’, and S4B), but not in “tip” region mesenchymal cells (Figure S4C). There was no change in proliferation in turbinate chondrocytes at E17.5 (Figure S4D). Together, these data suggest that Fgf20 regulates the proliferation of chondrocyte progenitors and potentially the formation of the mesenchymal condensation.
We also investigated whether Fgf20-KO mice have a defect in OE development. At P30, there was no measurable difference in OE thickness between control and Fgf20-KO mice (data not shown). Interestingly, at E14.5, there was a slight but statistically significant increase in both OE thickness (p = 0.04; Figures S3J and S3K) and the density of Sox2+ basal cells (p = 0.02; Figure S3L). The proliferation rate of Sox2+ BCs was unchanged (Figure S3M). This increase in thickness and Sox2+ BC density may be an effect of the constriction in OE surface area due to a smaller turbinate. By P0, Fgf20-KO pups had normal OE thickness (Figure S3F). Importantly, at P0, the OE of Fgf20-KO mice had a normal complement of OMP+ ORNs, Sus cells, and Sox2+ BCs (Figures S3G–S3I). In addition, neither Fgf20 expression (Figure S3G) nor FEP cell proliferation (Figures S4A’ and S4E) was affected in Fgf20-KO mice. Together, these data suggest that loss of FGF20 signaling does not affect OE vertical development.
Ectopic activation of FGF ligand expression in the OE drives mesenchymal growth
To determine whether overactivation of FGF signaling will increase proliferation in turbinate mesenchyme, we combined Fgf20GFP-Cre with the ROSArtTA(Belteki et al., 2005) and TRE-Fgf9-IRES-eGfp (White et al., 2006) alleles. We generated Fgf9-OA (Fgf20GFP-Cre/+; ROSArtTA/+; TRE-Fgf9-IRES-eGfp) mice along with littermate controls (Fgf20GFP-Cre/+; ROSArtTA/+). The Fgf9-OA mouse expresses the reverse tetracycline transactivator (rtTA) in the Fgf20GFP-Cre lineage, which drives the expression of TRE-Fgf9-IRES-eGfp upon doxycycline (Dox) induction. FGF9 is biochemically similar to FGF20 (Ornitz and Itoh, 2015), and is able to rescue and compensate for the loss of FGF20 signaling in the cochlea (Huh et al., 2012, 2015).
Pregnant dams were fed a Dox diet starting at E11.5. At E14.5, Fgf9-OA embryos showed a markedly thickened c1 mesenchymal layer (Figure S4F) and expansion and increased intensity of Dusp6 expression (Figure S4G). Compared to control, Fgf9-OA embryos had a slight but non-significant decrease in the number of condensed mesenchymal cells (p = 0.1) and a two- to three-fold increase in the number of diffuse cells (p = 0.01; Figures S4F, S4H, and S4I). Correspondingly, the ratio of condensed to diffuse cells was significantly decreased (p = 0.002; Figure S4J). However, there was no significant change in either condensed or diffuse cell proliferation (Figures S4F, S4K, and S4L).
The lack of a measurable increase in diffuse cell proliferation in Fgf9-OA embryos may be due to increased mesenchyme thickness, causing cells further away from the OE to receive less FGF signaling. Consistent with this, high levels of Dusp6 expression in Fgf9-OA embryos was only found in the mesenchyme within 58 μm of the OE (average of four samples). Therefore, we hypothesized that only diffuse cells close to the OE had an increase in proliferation in Fgf9-OA embryos. In control embryos, the diffuse mesenchymal layer averaged 69 μm thick. Quantification of proliferation of diffuse cells within 69 μm of the OE (“OE adjacent cells”) in Fgf9-OA embryos showed a significant increase compared to the total diffuse cell population in controls (Figures S4F, brackets and S4L). These data suggest that the increase in diffuse cell number in Fgf9-OA embryos likely had contributions from decreased condensation as well as increased proliferation. Unfortunately, the Fgf9-OA pups die at birth, precluding us from examining the postnatal phenotype.
Wnt activity in the developing OE coincides with Fgf20 expression
To understand the regulation of Fgf20 expression and FEP cell maintenance, we examined the canonical Wnt/βCat signaling pathway, which is a known direct transcriptional regulator of Fgf20 (Chamorro et al., 2005) and commonly involved in progenitor and stem cell maintenance (Nusse and Clevers, 2017). The Tcf/Lef:H2b-Gfp mouse, a well-described Wnt/βCat signaling reporter (Ferrer-Vaquer et al., 2010), showed an expression pattern that was similar to that of Fgf20 throughout the developing OE. H2B-GFP was detected in the lateral nasal pit at E10.5 (Figure 4A), consistent with previous reports of Wnt/βCat activity at this stage (Brugmann et al., 2007; Zhu et al., 2016). H2B-GFP was detected in the OE overlying the site of c1 development at E12.5 and E14.5 (Figures 4B, asterisk and 4C), and in negatively-curved “neck” OE at E17.5 (Figure 4D). At E17.5, H2B-GFP expression was found in FEP cell nuclei, which span all layers of the “neck” OE (Figure 4D’, arrowheads). By P7, H2B-GFP expression was less concentrated, mainly found in the apical layer at and surrounding “neck” OE (Figure 4E). Unlike Fgf20, Tcf/Lef:H2b-Gfp was also highly expressed in the nasal pit rim at E10.5 (Figure 4A, arrowhead) and in the respiratory epithelium (RE) at later stages (Figures 4B and 4C). In addition, H2B-GFP has a long half-life, likely much longer than that of GFP-Cre (Waghmare et al., 2008), resulting in a larger expression domain for Tcf/Lef:H2b-Gfp compared to Fgf20GFP-Cre. Consistent with this, the size of the Tcf/Lef:H2b-Gfp expression domain lies in between that of Fgf20GFP-Cre real-time expression and that of the Fgf20GFP-Cre-lineage. Importantly, Tcf/Lef:H2b-Gfp expression was not found in non-Fgf20GFP-Cre-lineage OE. Furthermore, products of the Tcf/Lef:H2b-Gfp and Fgf20βgal reporters appeared to have similar half-lives. Expression patterns of these two reporters more closely overlapped (Figures 4F and 4G).
Figure 4.
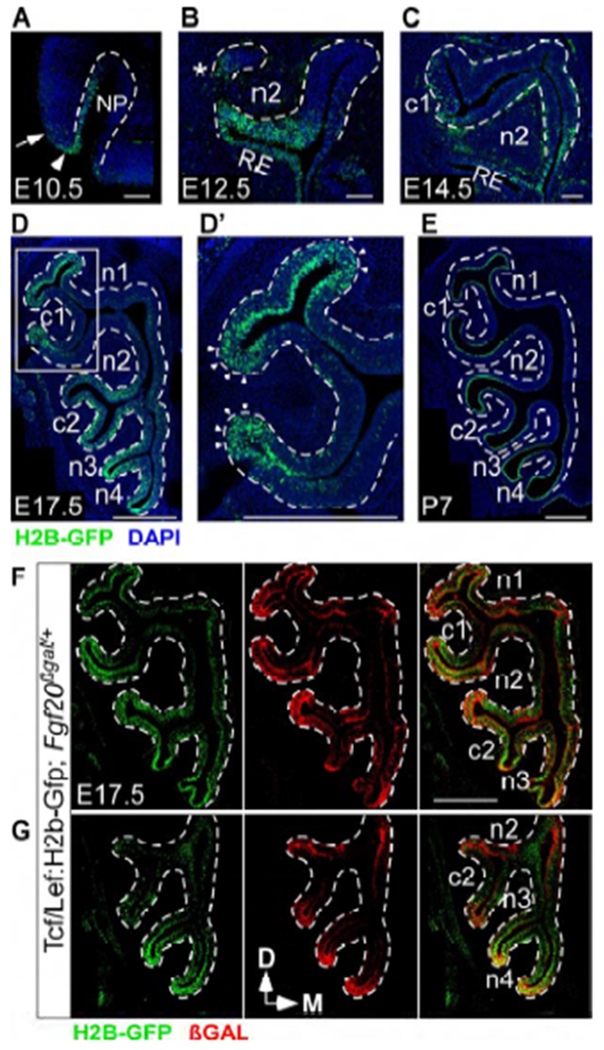
Wnt activity in the developing OE coincides with Fgf20 expression
(A-E) Tcf/Lef:H2b-Gfp expression at E10.5 (A), E12.5 (B), E14.5 (C), E17.5 (D), and P7 (E). Arrowhead indicates rim of the nasal pit (NP). Arrow indicates epithelium outside of the nasal pit. * indicates site of future c1 development. RE, respiratory epithelium.
(D’) Magnification of boxed region in (D). Arrowheads indicate immature, negatively-curved OE.
(F,G) Tcf/Lef:H2b-Gfp and Fgf20βgal expression in Tcf/Lef:H2b-Gfp; Fgf20βgal/+ mice at E17.5.
Dashed line, epithelial-mesenchymal boundary. DAPI, nuclei. Scale bar, 100 μm (A-C), 500 μm (D-G, D’).
βCat conditional deletion results in a severe deficit in turbinate development
To determine the role of Wnt/βCat signaling in the OE, we combined Fgf20GFP-Cre with the βCatfl(ex2-6) allele (Brault et al., 2001) to conditionally delete βCat. We generated βFF-CKO (Fgf20GFP-Cre/+; βCatfl(ex2-6)/fl(ex2-6)) mice along with littermate controls (Fgf20GFP-Cre/+; βCatfl(ex2-6)/+). Notably, since Fgf20 is a downstream target of Wnt/βCat signaling, Wnt/βCat signaling is predicted to activate prior to the onset of Fgf20GFP-Cre expression, leading to delayed βCat deletion. βFF-CKO pups appeared grossly normal at birth but most died by P1, with only a few surviving to adulthood. The surviving mice were smaller than littermate controls.
At P0, Fgf20 expression was nearly absent from the OE in βFF-CKO pups, as expected (Figure S5A). Therefore, a defect in turbinate development was predicted, as observed in Fgf20-KO mice. Examination of a few adult βFF-CKO mice that survived to P30 showed dramatically stunted turbinate growth of all six turbinates (Figure S5B). We focused our quantitative analysis on P0 pups, since the surviving P30 cohort was a biased sample. In addition, quantitation of turbinate size could be affected by the decreased overall size of βFF-CKO adult mice.
At P0, βFF-CKO mice had a more severe defect in turbinate development than Fgf20-KO mice (Figure 5A), with some variability across individuals. A few pups had almost no detectable structural defect compared to control, while pups with the most severe phenotype had a complete lack of c1, c2, and n4 turbinates (Figure 5A, shape 3). This variability is likely attributable to residual Wnt/βCat signaling from delayed or inefficient βCat deletion. Therefore, we believe mice with the most severe phenotypes represented the most complete βCat deletion. Focusing on c1 (Figure 5A’), we classified the varying P0 βFF-CKO phenotypes into 3 categories based on turbinate shape: shape 1 resembles control, shape 2 has a reduction in size, and shape 3 lacks c1. Shape 1 was rare and not found in every experiment.
Figure 5.
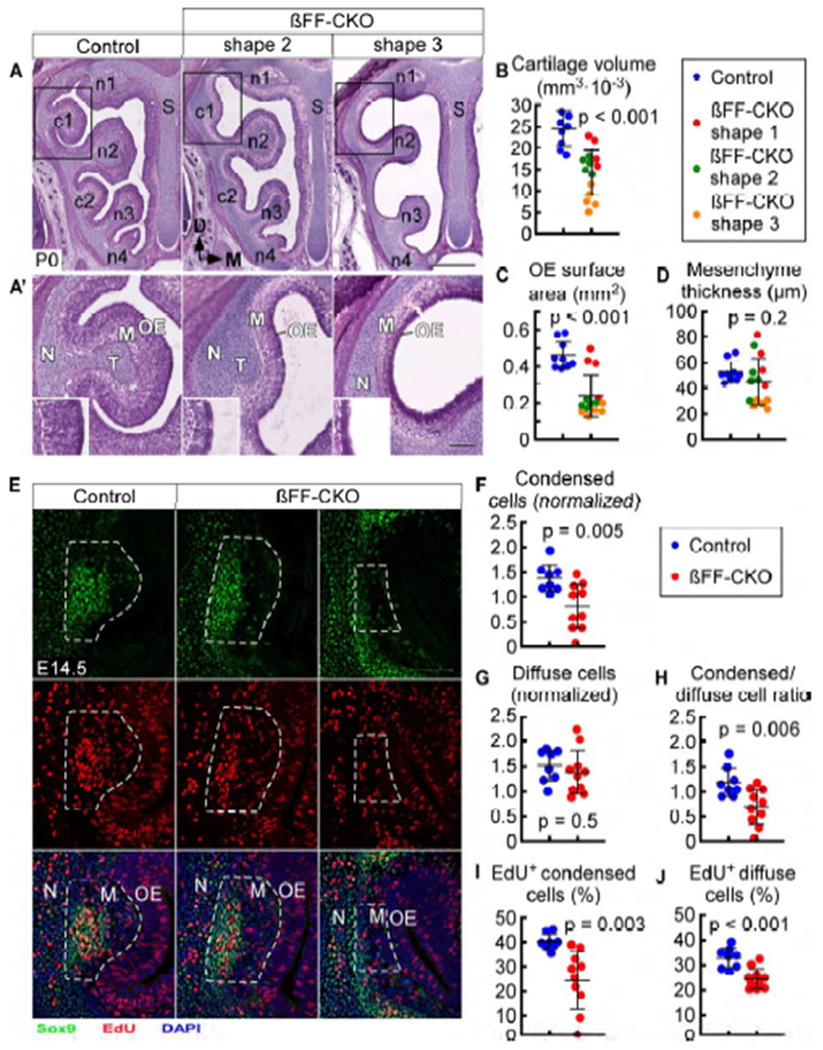
βCat conditional deletion results in a severe deficit in turbinate development
(A) H&E staining in control (Fgf20GFP-Cre/+; βCatfl(ex2-6)/+) and βFF-CKO (Fgf20GFP-Cre/+; βCatfl(ex2-6)/fl(ex2-6)) mice, with two phenotype examples (shapes 2 and 3) at P0. S, nasal septum.
(A’) Magnification of boxed region in (A). N, nasal cavity wall; T, turbinate cartilage; M, mesenchyme. Inset, 2x magnification of the OE at the turbinate tip. Dashed line, epithelial-mesenchymal boundary.
(B-D) Quantification of c1 cartilage volume (B), OE surface area (C), and mesenchyme thickness (D) at P0. βFF-CKO phenotype categorized into three shapes (1, 2, and 3). n = 9 control, 14 βFF-CKO, Welch’s t-test.
(E) EdU incorporation in c1 condensed and diffuse mesenchymal cells at E14.5, with two βFF-CKO phenotype examples. Dashed outline, c1 mesenchyme (M), including both condensed (Sox9hi) and diffuse cells. N, nasal cavity wall.
(F-J) Quantification of c1 normalized condensed cell number (F), normalized diffuse cell number (G), ratio of condensed cells to diffuse cells (H), and percent of EdU-incorporating condensed cells (I) and EdU-incorporating diffuse cells (J) in control and βFF-CKO at E14.5. n = 8 control, 10 βFF-CKO, Welch’s t-test.
DAPI, nuclei. Scale bars, 500 μm (A), 100 μm (A’, E). Error bars, mean ± SD. See also Figure S5.
Not surprisingly, c1 cartilage volume was significantly decreased in βFF-CKO mice compared to control (p < 0.001; Figure 5B), as was the overlying surface area (p < 0.001; Figure 5C). Mesenchyme thickness was noticeably decreased but only in the most severe (shape 3) βFF-CKO mice (Figure 5D). This phenotype was not observed in Fgf20-KO mice Figure S3E). OE thickness was decreased in βFF-CKO mice, also a phenotype not observed in Fgf20-KO mice (see below).
Specific disruption of βCat signaling also results in a severe deficit in turbinate development
βCat serves two major functions in epithelial cells: Wnt/βCat signaling and cell adhesion. To rule out the contribution of disrupted cell adhesion to the βFF-CKO phenotype, we used the βCatDM allele (Valenta et al., 2011) to generate a second βCat conditional deletion mouse: βDF-CKO (Fgf20GFP-Cre/+; βCatDM/fl(ex2-6)), along with littermate controls (Fgf20GFP-Cre/+; βCat+/+, Fgf20GFP-Cre/+; βCatDM/+, and Fgf20GFP-Cre/+; βCatfl(ex2-6)/+). When not specified, “control” refers to Fgf20GFP-Cre/+; βCatfl(ex2-6)/+. βDF-CKO pups appeared grossly normal at birth but almost all died by P1.
Like βFF-CKO mice, Fgf20 expression was absent from βDF-CKO pups at P0 (Figure S5A). The βDF-CKO turbinate phenotype was similar to that of βFF-CKO mice. However, the most severe βDF-CKO mice still had some observable c1, c2, and n4 development, unlike the most severe βFF-CKo mice Figures S5D and S5D’). Despite this, βDF-CKO mice (40% reduction in c1 cartilage volume, compared to control; Figure S5E) had a worse phenotype than Fgf20-KO mice (21% reduction) at P0 figure S3C). Like βFF-CKO mice, βDF-CKO mice also had decreased turbinate surface area and mesenchyme thickness, compared to littermate controls (Figures S5D’, S5F, and S5G). Also, like βFF-CKO mice, examination of a few adult βDF-CKO mice that survived to P30 showed dramatically stunted turbinate growth of all six turbinates (Figure S5B).
βCat conditional deletion causes decreased mesenchymal proliferation in developing turbinates
To explain the decreased c1 turbinate size in βFF-CKO mice, we looked at proliferation in and around the mesenchymal condensation. The earliest phenotype in βFF-CKO mice was observed at E14.5. At this stage, there was a decrease in the number of Sox9hi condensed mesenchymal cells in βFF-CKO mice compared to control (Figures 5E and 5F), but no difference in the number of diffuse cells (Figure 5G), leading to a decreased ratio of condensed cells to diffuse cells (Figure 5H). Like at P0, the phenotype at E14.5 was also variable, with the most severe having almost no condensation.
Proliferation in condensed cells was decreased in βFF-CKO mice (Figures 5E and 5I). This proliferation defect was more severe than that of Fgf20-KO mice. There was also a decrease in proliferation of diffuse cells at E14.5 (Figure 5J). The lack of a decrease in diffuse cell number at E14.5, despite the decrease in proliferation, suggests a defect in the formation of the condensation.
To directly assess the effects on FGF signaling, Dusp6 expression was examined by in situ hybridization. In the c1 mesenchymal layer in βFF-CKO mice at E14.5, Dusp6 expression was absent (Figure S5C, arrows), unlike in Fgf20-KO mice at the same stage, where some Dusp6 expression remained. Also, Dusp6 expression appeared to be slightly, yet consistently decreased in Fgf20GFP-Cre/+; βCatflox/+ controls compared to Fgf20GFP-Cre/+; βCat+/+ (compare Figures S5C and 3H). This may be attributable to heterozygosity of βCat in the OE. Overall, the βFF-CKO turbinate phenotype was similar to that of Fgf20-KO, but more severe.
βCat conditional deletion leads to premature differentiation and progenitor depletion in the OE
Interestingly, βFF-CKO mice had significantly thinner OE compared to control at P0 (p < 0.001; Figures 5A’, 6A, and 6B). In the most severe βFF-CKO mice at P0, OE at the tip of c1 was only 2-3 cell layers thick, whereas control OE was 7-9 cell layers thick in the same region. Surprisingly, βFF-CKO OE contained a normal complement of Sus cells and OMP+ ORNs (Figures 6A, 6C, and 6D). Even the most severely thinned βFF-CKO OE had a normal complement of these cells, which comprised nearly the entire OE. The presence of these differentiated OE cell types suggest that vertical development is intact in βFF-CKO mice.
Figure 6.
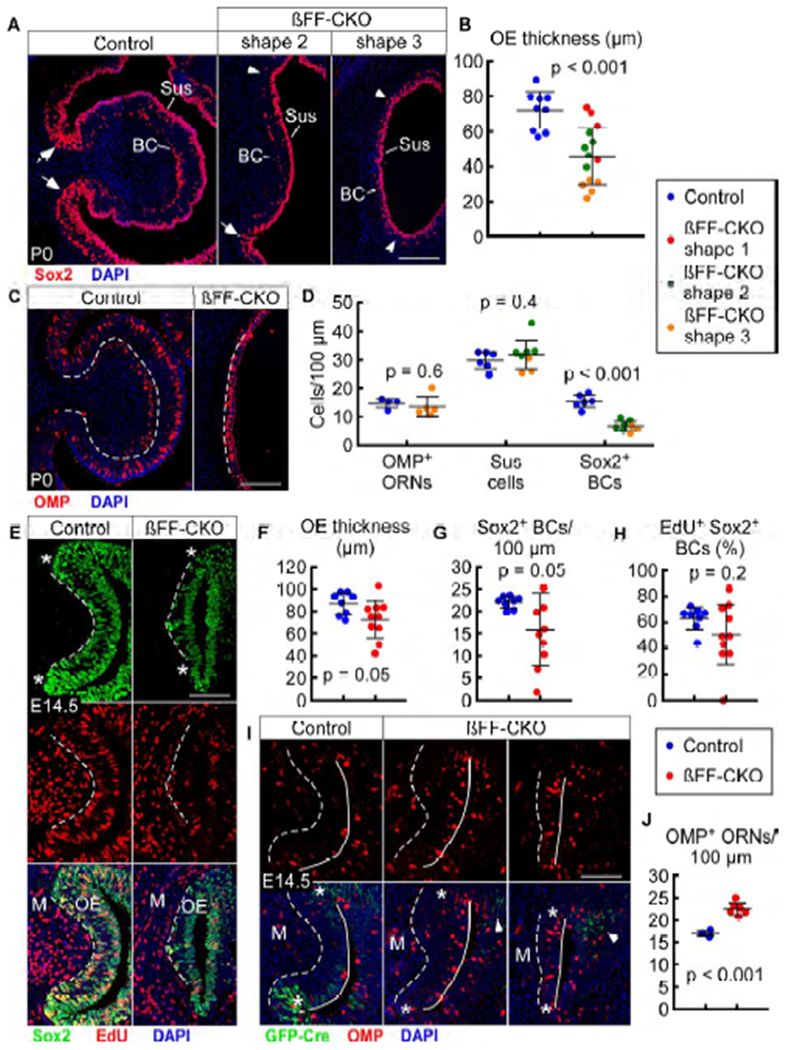
βCat conditional deletion leads to premature differentiation and progenitor depletion in the OE
(A) Sox2 expression in c1 of control (Fgf20GFP-Cre/+; βCatfl(ex2-6)/+) and βFF-CKO (Fgf20GFP-Cre/+; βCatfl(ex2-6)/fl(ex2-6)) mice at P0, with two phenotype examples (shapes 2 and 3). Arrows indicate presence of FEP cells in control and the less severe βFF-CKO phenotype. Arrowheads indicate absence of FEP cells in the severe βFF-CKO phenotype. BC, basal cells; Sus, sustentacular cells.
(B) Quantification of c1 OE thickness at P0. βFF-CKO phenotype categorized into three shapes (1, 2, and 3). n = 9 control, 14 βFF-CKO, Welch’s t-test.
(C) OMP expression in c1 at P0.
(D) Quantification of c1 OMP+ ORNs (n = 4 control, 5 βFF-CKO), Sus cells (n = 6 control, 8 βFF-CKO), and Sox2+ BCs (n = 6 control, 8 βFF-CKO) per 100 μm OE at P0. βFF-CKO phenotype categorized into two shapes (2 and 3). Welch’s t-test.
(E) EdU incorporation in c1 Sox2+ cells at E14.5. * indicates “neck” region. M, mesenchyme.
(F-H) Quantification of c1 OE thickness (H), Sox2+ BCs per 100 μm OE (F), and percent of EdU-incorporating Sox2+ BCs (G) at E14.5. n = 8 control, 10 βFF-CKO, Welch’s t-test.
(I) Fgf20GFP-Cre and OMP expression in c1 at E14.5, with two βFF-CKO phenotype examples. * indicates “neck” region. Arrowheads indicate Fgf20 expression in n1 and n2 OE. M, mesenchyme.
(J) Quantification of c1 OMP+ ORNs per 100 μm OE at E14.5. n = 3 control, 6 βFF-CKO, Welch’s t-test.
Dashed line, epithelial-mesenchymal boundary. Solid line, OE apical surface. DAPI, nuclei. Scale bars, 100 μm. Error bars, mean ± SD. See also Figure S6.
Notably, there was a lack of FEP cells in the more severe βFF-CKO mice at P0, as evidenced by an absence of Sox2+ epithelium-spanning cells and maturation of negatively-curved “neck” OE, where Sox2+ cells were found only in the apical and basal layers (Figure 6A, arrowheads). In less severe βFF-CKO mice, some Sox2+ epithelium-spanning cells were observed, resembling FEP cells in control mice (Figures S5A and 6A, arrows). However, these cells did not express Fgf20 (Figure S5A, arrows). This shows that βCat is required for the maintenance of both Fgf20 expression and FEP cells. There was also a decrease in the density of Sox2+ BCs in βFF-CKO mice (Figures 6A and 6D). Given that the severely thinned βFF-CKO OE contained a normal complement of the most differentiated cell types, this decrease in progenitor number suggests that FEP cells and Sox2+ BCs were not maintained in an undifferentiated state.
Next, we examined differentiation at an earlier stage. At E14.5, the OE was only slightly thinner in βFF-CKO embryos (p = 0.05; Figures 6E and 6F). There was also a detectable decrease in the number of Sox2+ BCs (p = 0.05; Figures 6E and 6G). Proliferation levels of Sox2+ BCs did not appear to be dramatically changed (p = 0.2; Figures 6E and 6H). There was also a noticeable decrease in the number of, or absence of, FEP cells in the c1 OE of βFF-CKO embryos, based on a lack of epithelium-spanning Sox2+ cells (Figure 6E, asterisk indicates “neck” region). At this stage, there was some residual Fgf20GFP-Cre expression in the n1 and n2 OE of βFF-CKO embryos (Figure 6I, arrowheads), likely attributable to GFP perdurance. However, there was no Fgf20 expression in the c1 OE (Figure 6I, asterisk indicates “neck” region).
Remarkably, there was an increase in the number of OMP+ ORNs in βFF-CKO mice at E14.5 compared to control (Figures 6I and 6J), despite a slight decrease in OE thickness. This supports the hypothesis that the P0 βFF-CKO phenotype was a result of premature differentiation, leading to a depletion of progenitors. At an earlier stage of development (E13.5), no obvious differences between βFF-CKO and control mice were observed (data not shown).
Examination of a few βFF-CKO mice that survived to P30 showed a lack of OE in some areas, as indicated by the absence of OMP expression. This was particularly noticeable in areas overlying and surrounding “neck” regions of turbinates (Figure S5B, arrowhead). This suggests a defect in OE expansion or maintenance in the surviving adult βFF-CKO mice.
Specific disruption of βCat signaling leads to premature OE differentiation
βDF-CKO mice also exhibited decreased OE thickness at P0 (Figures S5D’, S6A, and S6B), with a normal complement of Sus cells and OMP+ ORNs Figures S6A, S6C, and S6D). Similar to βFF-OKO mice, βDF-CKO mice also lacked FEP cells and showed early maturation of negatively-curved “neck” OE Figure S6A, arrowheads). Interestingly, there was a small, but statistically significant increase in Sox2+ BC density in βDF-CKO mice (p = 0.03; Figure S6D), opposite of the βFF-CKO phenotype.
At E17.5 in βDF-CKO mice, the OE was only slightly thinner compared to control (Figures S6E and S6F), with no change in Sox2+ BC density (Figure S6G). Sox2+ BCs were less proliferative compared to control (p = 0.01; Figure S6E and S6H), suggesting that the increase in Sox2+ BCs at P0 was not due to increased proliferation. Unlike at P0, at E17.5 some FEP cells were still found in the c1 OE of βDF-CKO mice (Figure S6E, arrow), along with Fgf20 expression (Figure S6I, arrows). The increase in Sox2+ BCs at P0 likely can be explained by premature differentiation of these FEP cells.
At E17.5, in control mice, regions of “neck” OE were filled with FEP cells, with a dearth of OMP+ ORNs. However, in βDF-CKO mice, these regions were prematurely filled with OMP+ ORNs Figure S6I, arrowheads). A distribution plot of OMP+ and Fgf20+ cells showed that in control mice, OMP+ ORN density was lowest where Fgf20+ cell density was highest (“neck” regions). In βDF-CKO mice, OMP+ ORNs were much more evenly distributed across the turbinate.
Like βFF-OKO mice, examination of a few βDF-CKO mice that survived to P30 also showed a lack of OE in areas surrounding “neck” regions of turbinates, as indicated by absence of OMP expression (Figure S5B, arrowhead).
βCat stabilization prevents differentiation of FEP cells
To determine the effects of over-activation of Wnt/βCat signaling (gain-of-function), we combined Fgf20GFP-Cre with the βCatfl(ex3) allele (Harada et al., 1999) to activate a dominant stable version of βCat. βEX3-OA (Fgf20GFP-Cre/+; βCatfl(ex3)/+) pups had pronounced growth retardation post-partum, compared to littermate controls (Fgf20GFP-Cre/+; βCat+/+) and died within a few days of birth. Some had notable abdominal distension and died at birth.
At E17.5, βEX3-OA mice had increased Fgf20 expression in the OE compared to controls. In control mice, native GFP fluorescence from the Fgf20GFP-Cre allele was barely detectable without using an antibody to GFP (Figure S7A, arrowheads). However, in βEX3-OA mice, native GFP fluorescence was broad and intense throughout the OE.
One of the most salient phenotypes in βEX3-OA mice at E17.5 was disrupted OE development Figure 7A). The c1 OE of βEX3-OA mice was very thin and contained almost exclusively Fgf20+/Sox2+ FEP cells (Figures 7A’ and 7B) and very few OMP+ ORNs or Sox2 single-positive (BC or Sus) cells (Figures 7B and 7C). This cell composition resembles that of immature “neck” OE at E17.5 wildtype mice. Interestingly, the FEP cells at E17.5 were mostly non-proliferative (Figure S7B), suggesting that the increase in the number of FEP cells was due to lack of differentiation, and not an increase in proliferation. Areas of the OE not targeted by Fgf20GFP-Cre (for example, overlying dorsomedial regions of n2) showed comparatively normal appearing OE, with Sox2+ cells in apical and basal layers, and OMP+ ORNs in between (Figures S7C and S7D).
Figure 7.
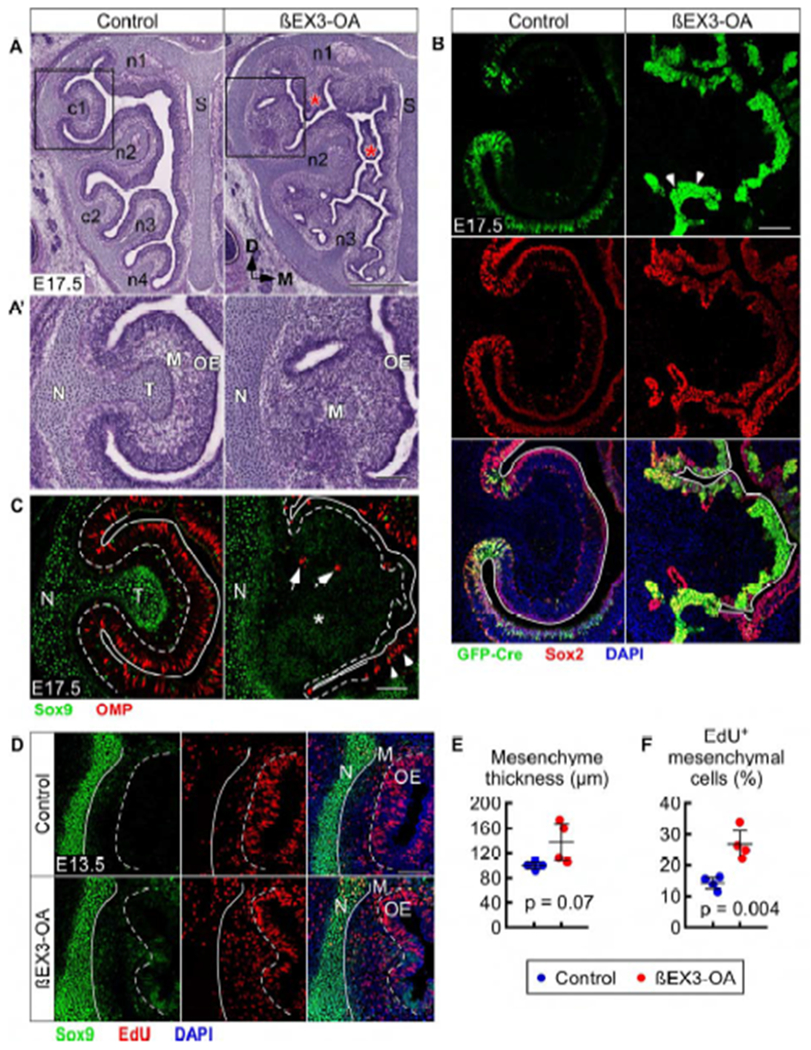
βCat stabilization prevents differentiation of FEP cells and results in mesenchyme expansion without condensation in turbinates
(A) H&E staining in control (Fgf20GFP-Cre/+; βCat+/+) and βEX3-OA (Fgf20GFP-Cre/+; βCatfl(ex3)/+) mice at E17.5. * indicate extra blebs of OE and mesenchyme. S, nasal septum.
(A’) Magnification of boxed region in (A). N, nasal cavity wall; T, turbinate cartilage; M, mesenchyme.
(B) Fgf20GFP-Cre and Sox2 expression in c1 at E17.5. Arrowheads indicate dense clumps of FEP cells. Solid line, OE apical surface.
(C) Sox9 and OMP expression in c1 at E17.5. Dashed line, epithelial-mesenchymal boundary. Solid line, OE apical surface. * indicates Sox9low mesenchyme. Arrows indicate OMP+ ORNs in the mesenchyme. Arrowheads indicate OMP+ ORNs in the n2 OE (see Figure S7D). N, nasal cavity wall; T, turbinate cartilage.
(D) EdU incorporation and Sox9 expression in c1 at E13.5. Dashed line, epithelial-mesenchymal boundary. Solid line, nasal cavity wall (N)-mesenchymal (M) boundary. Green fluorescence in the βEX3-OA OE is from Fgf20GFP-Cre.
(E,F) Quantification of c1 mesenchyme thickness (E) and percent of EdU-incorporating mesenchymal cells (F) at E13.5. n = 4, Student’s t-test.
DAPI, nuclei. Scale bars, 500 μm (A), 100 μm (A’, B-D). Error bars, mean ± SD. See also Figure S7.
In βEX3-OA mice, extra blebs of OE and mesenchyme were found throughout the nasal cavity (Figure 7A, asterisk). There were also clumps of very densely packed and highly Fgf20+ FEP cells (Figure 7B, arrowheads). These FEP cells appeared dysplastic, with irregularly shaped nuclei and a high nuclear-to-cytoplasmic ratio (Figure S7A, inset). The epithelial-mesenchymal boundary was difficult to identify in βEX3-OA mice. In fact, the OE seemed to be invading into the mesenchyme, as highlighted by the presence of OMP+ cells (Figure 7C, arrows) and duct structures (Figure S7A, arrows) in the mesenchyme.
Given the dense packing of cells, distorted cell morphology, and potentially disrupted basement membrane, it was difficult to quantify OE cells at E17.5. Therefore, we examined E13.5, the earliest stage that a phenotype was detectable. At this stage, the OE was already thinner in βEX3-OA mice (Figures S7E and S7F). There were also clumps of FEP cells (Figure S7E, arrowheads) and a measurable increase in the density of FEP cells in the OE (Figure S7G). At this stage, there was no change in FEP cell proliferation (Figures S7E and S7H), again suggesting that the increase in FEP cell number was due to inhibition of differentiation, rather than increased proliferation.
βCat stabilization results in mesenchyme expansion without condensation in turbinates
Examination of turbinate development showed a vast expansion of the mesenchymal layer in βEX3-OA mice at E17.5, at the expense of the turbinate chondrocytes (Figure 7A’). At this stage, the nasal cavity wall in βEX3-OA mice appeared relatively normal, containing Sox9hi chondrocytes (Figure 7C). However, turbinate c1 was made up entirely of Sox9low diffuse mesenchymal cells, suggesting a defect in formation of the mesenchymal condensation. Chondrocytes of n2 and n3, the least affected turbinates in Fgf20-KO and βCat loss-of-function mice, were also the least affected in βEX3-OA mice (Figure S7D). However, the shapes of these turbinates were affected along with notable expansion of the mesenchymal layer (Figure 7A).
At E13.5, control embryos showed no sign of c1 protrusion from the lateral wall (Figure 7D), which does not occur until E14.5. However, in βEX3-OA mice, a large protrusion made up of Sox9low mesenchyme could be seen in some embryos. These Sox9low cells formed the shape of a turbinate, increasing the thickness of the mesenchyme (Figures 7D and 7E). The mesenchymal cells in βEX3-OA embryos at E13.5 were more proliferative than in control embryos (Figures 7D and 7F), accounting for the mesenchymal expansion seen at E17.5. However, these mesenchymal cells remained Sox9low without showing any signs of condensation formation. Overall, the turbinate phenotype in βEX3-OA mice was similar to that of Fgf9-OA embryos, but much more severe.
DISCUSSION
FEP cells are an embryonic OE progenitor population that expands the OE
A definitive embryonic progenitor population has not been identified in the OE, although a few have been proposed. These include nestin+ (Murdoch and Roskams, 2008), Pax7+ (Murdoch et al., 2010), Meis1+ (Tucker et al., 2010), and Foxg1+ (Duggan et al., 2008; Hébert and McConnell, 2000; Kawauchi et al., 2009) cells. We propose that FEP cells are an embryonic OE progenitor population. FEP cells were identified in the early embryonic ventrolateral OE, which develops later than dorsomedial OE (Eerdunfu et al., 2017; Tucker et al., 2010). Even after the OE begins to mature and undergo established neurogenesis at E13, FEP cells continued to be localized laterally in immature and undifferentiated OE. This suggests that FEP cells define parts of the developing OE focused on progenitor expansion (horizontal development), rather than established neurogenesis (vertical development). Moreover, proliferation and neurogenesis shift from dorsomedial OE to ventrolateral OE from E12.5 to E15.5, consistent with expansion of the OE in a ventrolateral direction (Eerdunfu et al., 2017). Therefore, FEP cells occupy the correct location to account for OE expansion. Consistent with this, the Fgf20GFP-Cre lineage included the entire ventrolateral OE (zones 2-4) but spared some of the dorsomedial OE (zone 1).
The total lineage of FEP cells included all the major cell types of the OE. Transient Fgf20 lineage labeling, taking advantage of GFP-Cre perdurance, suggests that FEP cells give rise immediately to Sox2+ BCs. Sox2+ BCs, in turn, differentiate in a well-described, step-wise manner into ORNs in established neurogenesis (Calof et al., 2002). Transient Fgf20 lineage tracing also suggests that FEP cells can differentiate directly to Sus cells.
A caveat in analyzing Fgf20GFP-Cre lineage tracing data is that the lineage is cumulative. To definitively show that FEP cells give rise to the expanding OE will require an inducible system to target the FEP population at a specific time point. Nevertheless, results from the cumulative lineage strongly suggest that FEP cells contribute to OE expansion. The Fgf20GFP-Cre lineage accounted for essentially all of the OE expansion that occurred from E14.5 to P7, during which the OE surface area increased by elevenfold. This high rate of expansion coincided with the growth of pre-ossified turbinates. From P7 to P30, the OE surface area continued to expand by roughly three-fold, as ossified turbinates grow by what has been hypothesized as secondary membranous bone extensions (Martineau-Doizé et al., 1992). During these stages, Fgf20GFP-Cre-lineage OE and non-lineage OE grew proportionally, which is expected, given the absence of FEP cells at these stages. This suggests a different method of OE expansion accompanying a different method of turbinate growth.
Wnt/βCat signaling maintains FEP stemness
The “neck” OE where FEP cell were found may define a niche environment that maintains these progenitors into perinatal stages. Wnt/βCat signaling is often an important component of stem cell and progenitor niches. This signaling pathway has been studied in the OE at the nasal pit stage in the context of facial development (Brugmann et al., 2007; Mani et al., 2010; Reid et al., 2011; Zhu et al., 2016) and at the postnatal and adult stages in the context of OE stem cell maintenance (Chen et al., 2014; Wang et al., 2011). However, it has not been studied at the stages in between, except in an ectopic activation study (see below; Engel et al., 2016).
βCat loss- and gain-of-function experiments show that Wnt/βCat signaling is necessary and sufficient for the maintenance of FEP cells. Loss-of-function led to loss of FEP cells due to premature differentiation into Sox2+ BCs, and in turn, ORNs, while gain-of-function led to sustained FEP cell maintenance without differentiation into other cell types. Loss-of-function experiments also suggested that Wnt/βCat signaling is necessary for the maintenance of Sox2+ BCs, perhaps secondary to defects in the FEP cells that give rise to them.
Mechanical forces from negative curvature generation could be important for FEP cell niche maintenance. Turbinates could also be sources of Wnt ligands or other signaling factors. These possibilities suggest an interesting feedback loop in which developing turbinates help shape the niche to maintain expansive OE progenitors that, in turn, secrete signals to promote turbinate growth. It is further conceivable that the loss of FEP cells in βCat loss-of-function mice is secondary to defects in turbinate growth affecting OE negative curvature formation. However, we believe that Wnt/βCat signaling most likely acts directly to maintain FEP cells, since Wnt/βCat activity and Fgf20 expression overlap. Furthermore, overactivation of Wnt/βCat signaling was sufficient to maintain FEP cells outside of negatively-curved “neck” OE.
FEP cells have potential for dysplasia
In βCat gain-of-function mice, FEP cells became dysplastic, densely packed clumps of undifferentiated cells. A similar tumorigenic phenotype has been recently studied, in which Sox2:CreERT2 was used to activate the βCatfl(ex3) allele (Engel et al., 2016), presumably targeting FEP cells along with Sox2+ BCs and Sus cells. Notably, the strongest tumorigenic phenotype occurred when tamoxifen was given at E14.5. The phenotype was markedly and progressively less severe when tamoxifen was given at P7, P14, and P21 (no phenotype). This strongly suggests that FEP cells, which are present early in development and disappear after P7, were the cells responding to Wnt/βCat overactivation in their experiment. Therefore, our βEX3-OA experiment most likely targets the more specific cell type. Engel et al. also discuss the similarity and differences between this tumorigenic phenotype and human tumors, including olfactory neuroblastoma and sinonasal haemangiopericytoma. The results from Engel et al. indicated a turbinate phenotype, but it was not described.
Disruptions to signaling, not cell adhesion, accounts for most of the βCat loss-of-function phenotype
βFF-CKO mice have disruptions in both the signaling and cell adhesion functions of βCat. Evidences suggest that defects in cell adhesion were not responsible for the βFF-CKO phenotype. Importantly, βDF-CKO mice, in which only signaling was affected, had similar phenotypes to the complete knockout. Furthermore, a study found no defects in the number of total ORNs and Sus cells in the P0 OE in mice lacking functional αN-catenin, an important component of cell adhesion that is highly expressed in ORNs (Katsunuma et al., 2016).
The βCatDM allele is hypomorphic, and βDF-CKO mice had similar but less severe phenotypes compared to βFF-CKO mice. One difference between the two knockouts is that Sox2+ BC density was increased in βDF-CKO mice at P0 but decreased in βFF-CKO mice. This difference could be due to a later onset of premature differentiation in βDF-CKO mice. Supporting this idea, FEP cells could still be found at E17.5 in βDF-CKO mice in c1, while they were almost completely gone in the most severe βFF-CKO mice as early as E14.5.
Wnt/βCat signaling in FEP cells regulates turbinate development via epithelial-mesenchymal signaling
Mice lacking Fgf20 had decreased total OE surface area, but normal OE cellular composition. Our results suggest that the OE is not directly affected by loss of FGF20 signaling. Rather, FGF20 appears to be signaling directly to diffuse mesenchymal cells to regulate turbinate growth. The role of FGF20 in epithelial-mesenchymal signaling is consistent with findings in other developing tissues, such as the tooth, hair, and cochlea (Haara et al., 2012; Huh et al., 2013, 2015). However, we cannot rule out the possibility that FGF20 may be signaling the epithelium as well, like in the cochlea (Huh et al., 2012).
The βCat loss- and gain-of-function experiments strongly suggest that Fgf20 is a downstream target of Wnt/βCat signaling. Interestingly, in both βFF-CKO and βDF-CKO mice, turbinate phenotypes were more severe than that of Fgf20-KO mice. This indicates that other Wnt/βCat-regulated epithelial-mesenchymal signals may compensate for the loss of FGF20. We hypothesize that these signals are also expressed by FEP cells; however, we cannot rule out the possibility that they are expressed by other parts of the OE within the FEP-lineage. These signals could be other members of the FGF family, which often compensate for each other (Ornitz and Itoh, 2015). Consistent with this, some residual expression of the FGF target gene Dusp6 was found in the mesenchyme of Fgf20-KO embryos, but not of βFF-CKO embryos. We attempted to address potential redundancies of Fgf20 with Fgf9 and Fgf10. However, neither Fgf9; Fgf20 nor Fgf10; Fgf20 double-knockout mice exhibited a more severe turbinate phenotype than Fgf20-KO mice (unpublished data).
The decrease in OE surface area and turbinate size and complexity in Fgf20-KO mice makes them a potentially useful model for understanding olfaction, particularly from an evolutionary perspective. The idea that increased OE surface area results in increased olfactory ability has not been directly tested (Van Valkenburgh et al., 2014). Fgf20-KO mice are useful for testing this hypothesis. Olfactory receptors in the OE are zonally distributed (Ressler et al., 1993; Vassar et al., 1993). Fgf20-KO mice may therefore have a deficiency in specific zones of the OE, resulting in a decrease of a specific subset of olfactory receptors.
Overall, this study addresses several important gaps in knowledge in olfactory system development, including the identity of an expansive pool of OE progenitors, mechanisms regulating OE expansion, genetic regulation of turbinate development, role of Wnt/βCat signaling in olfactory development, and mechanisms linking scaling of the OE and the underlying turbinates. We do not know whether FEP cells exist in other mammalian species, but our findings have implications for mammalian evolution. Adaptive forces acting on FEP cell number and function are a potential mechanism accounting for the diversity of OE surface area and turbinate complexity seen across mammals. Furthermore, dysregulation of FEP cells or their equivalent in humans may contribute to human diseases such as anosmia and olfactory tumors. Finally, this study provides a model for tissue-scaling and progenitor niche maintenance with potential relevance to other developmental systems.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David M. Ornitz (dornitz@wustl.edu). Mouse models will require an MTA issued by Washington University in St. Louis.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were group housed with littermates, in breeding pairs, or in a breeding harem (2 females to 1 male), with food and water provided ad libitum.
For timed-pregnancy experiments, embryonic day 0.5 (E0.5) was assigned as noon of the day the vaginal plug was found. For postnatal experiments, postnatal day 0 (P0) was determined as the day of birth.
Mice were of mixed sexes and maintained on a mixed C57BL/6J x 129X1/SvJ genetic background. All mouse lines were previously reported:
Fgf20GFP-Cre: knockin allele containing a sequence encoding a GFP-Cre fusion protein replacing exon 1 of Fgf20, resulting in a null mutation (Huh et al., 2015).
Fgf20βgal: knockin allele containing a sequence encoding β-galactosidase (βgal) replacing exon 1 of Fgf20, resulting in a null mutation (Huh et al., 2012).
ROSAmTmG: knockin allele containing a sequence encoding a membrane-localized tdTomato (mT) flanked by loxP sequences, followed by a sequence encoding a membrane-localized eGFP (mG), targeted to the ubiquitously expressed ROSA26 locus. In the absence of Cre-mediated recombination, mT is expressed; upon Cre-mediated recombination, mG is alternatively expressed (Muzumdar et al., 2007).
ROSAtdTomato: Ai9 knockin allele containing a loxP-Stop-loxP sequence followed by a sequence encoding tdTomato, targeted to the ubiquitously expressed ROSA26 locus. Upon Cre-mediated recombination, tdTomato is expressed (Madisen et al., 2010).
ROSArtTA: knockin allele containing a loxP-Stop-loxP sequence followed by a sequence encoding rtTA-IRES-eGFP, targeted to the ubiquitously expressed ROSA26 locus. Upon Cre-mediated recombination, reverse tetracycline transactivator (rtTA) and eGFP are expressed (Belteki et al., 2005).
TRE-Fgf9-IRES-eGfp: transgene containing seven tetracycline-inducible regulatory elements driving the expression of FGF9-IRES-eGFP (White et al., 2006).
Tcf/Lef:H2b-Gfp: transgene containing a sequence encoding a histone 2B-eGFP (H2B-GFP) fusion protein, expressed under the control of six copies of a TCF/LEF responsive element (Ferrer-Vaquer et al., 2010). TCF/LEFs are transcription co-factors that bind β-Catenin to activate Wnt/β-Catenin regulated genes (Nusse and Clevers, 2017).
βCatfl(ex2-6): allele containing loxP sequences flanking exons 2 (containing ATG) through 6 of β-Catenin. Upon Cre-mediated recombination, produces a null mutation (Brault et al., 2001).
βCatDM: allele containing a constitutive double mutation that partially blocks the signaling capacity of β-Catenin, while preserving its function in adherens junctions (Valenta et al., 2011).
βCatfl(ex3): allele containing loxP sequences flanking exon 3 (containing all regulatory phosphorylation sites) of β-Catenin. Upon Cre-mediated recombination, produces a dominant stabilized β-Catenin to activate Wnt/β-Catenin signaling (Harada et al., 1999).
All studies performed were in accordance with the Institutional Animal Care and Use Committee at Washington University in St. Louis.
METHOD DETAILS
Doxycycline induction
For the Fgf9-OA experiment, pregnant dams were fed Dox Diet, Grain-Based Doxycycline, 200 mg/kg (Bio-Serv, S3888) ad libitum starting at noon on E11.5 until sample harvest at E14.5.
Sample preparation
Heads from mice younger than five days old were fixed in 4% PFA in PBS overnight at 4°C with gentle agitation. Samples were then washed x3 in PBS and cryoprotected in 15% sucrose in PBS overnight and then in 30% sucrose in PBS overnight. Samples were embedded in Tissue-Tek O.C.T. compound (VWR International, 4583) and frozen on dry ice. Serial frontal sections through the nasal cavity were cut at 12 μm with a cryostat, dried at room temperature, and stored at −80° until use. Mice that were five days old or older were perfused with PBS and 4% PFA in PBS prior to post-fixation in 4% PFA in PBS overnight at 4°C with gentle agitation. After P BS wash, parts of the head posterior to the maxilla and frontal bone were cut and removed, along with the mandible, hard palate, and most of the remaining exposed soft tissue, except for the olfactory bulb. Front incisors and molars, if any, were removed with rongeurs. The remaining skulls with intact nasal cavity were then decalcified by incubation in 0.5 M EDTA, pH 8 at 4°C with gentle a gitation for two days, or three days for mice two weeks old or older, with daily EDTA solution replacement. Cryoprotection, embedding, and sectioning were performed as above, except prior to embedding, samples were submerged in O.C.T. and placed in a vacuum chamber to remove air bubbles from inside the nasal cavity.
RNA in situ hybridization
Probe preparation: plasmid containing 413 bp of Dusp6 5’-UTR was a gift from Suzanne Mansour (Li et al., 2007). To make antisense probe, the plasmid was linearized with restriction enzyme Acc65I (New England Biolabs, R0599S) and transcribed with T7 RNA polymerase (New England Biolabs, M0251S) according to manufacturer’s instructions, with DIG RNA Labeling Mix (Sigma-Aldrich, 11277073910). After treatment with RNase-free DNase I (Sigma-Aldrich, 04716728001) for 15 min at 37°C, probes were hydrolyzed in hydrolysis buffer (40 mM NaHCO3, 60 mM Na2CO3) at 65°C for 30 min.
Frozen section in situ hybridization: frozen slides were warmed for 20 min at room temperature and then 5 min at 50°C on a slide warmer. Sections were fixed in 4% PFA in PBS for 20 min at room temperature, washed x2 in PBS and treated with pre-warmed 10 μg/ml Proteinase K (Sigma-Aldrich, 03115828001) in PBS for 7 min at 37°C. Sections were then fixed in 4% PFA in PBS for 15 min at room temperature, washed x2 in PBS, acetylated in 0.25% acetic anhydrate in 0.1M Triethanolamine, pH 8.0, for 10 min, and washed again in PBS. Sections were then placed in pre-warmed hybridization buffer (50% formamide, 5x SSC buffer, 5 mM EDTA, 50 μg/ml yeast tRNA) for 3 h at 60°C in humidified chamber for prehybridization. Sections were then hybridized in 10 μg/ml probe/hybridization buffer overnight (12-16 h) at 60°C. The next day, sections were washed in 1x SSC for 10 min at 60°C, followed by 1.5x SSC for 10 min at 60°C, 2x SSC for 20 min at 37°C x2, and 0.2x SSC for 30 min at 60°C x2. Sections were then washed in KTBT (0.1 M Tris, pH 7.5, 0.15 M NaCl, 5 mM KCl, 0.1% Triton X-100) at room temperature and blocked in KTBT + 20% sheep serum + 2% Blocking Reagent (Sigma-Aldrich, 11096176001) for 4 h. Blocking Reagent was dissolved in 100 mM Maleic acid, 150 mM NaCl, pH 7.5. Sections were then incubated in sheep anti-Digoxigenin-AP, Fab fragments (1:1000, Sigma-Aldrich, 11093274910) in KTBT + 20% sheep serum + 2% Blocking Reagent overnight at 4°C. Sections were then washed x3 in KTBT for 30 min at room temperature, and then washed x2 in NTMT (0.1 M Tris, pH 9.5, 0.1 M NaCl, 50 mM MgCl2, 0.1% Tween 20) for 15 min. Sections were next incubated in NTMT + 1:200 NBT/BCIP Stock Solution (Sigma-Aldrich, 11681451001) in the dark at room temperature until color appeared. Sections were then washed in PBS, post-fixed in 4% PFA in PBS for 15 min and washed x2 in PBS. Finally, sections were dehydrated in 30% and then 70% methanol, 5 min each, followed by 100% methanol for 15 min. Sections were then rehydrated in 70% and 30% methanol and then PBS, 5 min each, and mounted in 95% glycerol.
Histology and Immunofluorescence
H&E staining was done by the Washington University Developmental Biology Histology Core. Immunofluorescence: frozen slides were warmed for 30 min at room temperature and washed in PBS before incubating in PBS + 0.5% Triton X-100 (PBST) for 1 h to permeabilize the tissue. Sections were then blocked using in PBST + 5% donkey serum for 1 h and then incubated in PBST + 1% donkey serum with the primary antibody overnight at 4°C in a humidified chamber. Sections were then washed x3 in PBS and incubated in PBS + 1% Triton X-100 with the secondary antibody. After wash in PBS x3, slides were mounted in VectaShield antifade mounting medium with DAPI (Vector Labs, H-1200).
Antibodies
Antibodies for immunofluorescence were used at the following concentrations:
Rabbit anti-GFP (1:500; Life Technologies, A-11122)
Chick anti-Beta galactosidase (1:500; Abcam, ab9361)
Goat anti-Sox2 (1:200; Santa Cruz, sc-17320)
Rabbit anti-Sox9 (1:500; Millipore, AB5535)
Goat anti-OMP (1:1000; Wako Chemicals, 544-10001)
Goat anti-Pde2a (1: 100; Santa Cruz, sc-17227)
Alexa Fluor conjugated secondary antibodies (1:500; Thermo Fisher)
Sheep anti-Digoxigenin-AP, Fab fragments (1:1000, Sigma-Aldrich, 11093274910)
Cell proliferation assay
EdU (Thermo Fisher, E10187) was injected i.p. into pregnant dams at 100 μg per gram body weight. Except where noted, embryos were harvested at 1 h after injection. For 4 h EdU incorporation, 100 μg per gram body weight of EdU was injected at 4 h and 2 h before embryos were harvested. EdU was detected using the Click-iT Plus EdU Alexa Fluor 594 or 647 picolyl azide toolkit (Thermo Fisher, C10639, C10640) according to manufacturer’s instructions.
Imaging
Brightfield microscopy was done using a Hamamatsu NanoZoomer slide scanning system with a 20x objective or on a Zeiss AxioPlan 2 with a 10x or 20x objective. Images were processed with the NanoZoomer Digital Pathology (NDP.view2) software or ImageJ (imagej.nih.gov).
Fluorescent microscopy was done using a Zeiss Axio Imager Z1 with Apotome 2, with z-stack step-size determined based on objective lens type (10x or 20x), as recommended by the ZEN software (around 1 μm). Fluorescent images shown are maximum projections. Low magnification fluorescent images shown required stitching together, by hand, several images to capture the entire structure of interest. Some fluorescence microscopy was also done using a Zeiss Axio Imager Z2 with Apotome 2. In all cases, except where noted, Fgf20GFP-Cre expression was detected with an anti-GFP antibody. In all cases, mGFP, mTomato, tdTomato, and H2B-GFP expressions were detected by native fluorescence. Images were processed with ImageJ.
QUANTIFICATION AND STATISTICAL ANALYSIS
Measurements and cell quantification
Measurements and cell quantification (using the Cell Counter plugin by Kurt De Vos) were done using ImageJ.
For OE surface area measurements, length along the apical surface of the OE was measured in serial frontal sections starting just anterior to endoturbinate I and ending at the end of the nasal cavity. The measured length was then multiplied by the distance between sections to calculate surface area. OE was differentiated from respiratory epithelia based on histology (OE is pseudostratified); where ambiguous, epithelia <30 μm thick were not considered OE.
Fgf20GFP-Cre lineage quantification
OE surface area was measured as above, with the following specifics:
E11.5: measurements were done on eleven to 14 sections per sample, starting anteriorly when the VNO was reached.
E14.5: nine sections per sample, starting mid VNO, when n1 and n2 were reached.
E17.5: eleven sections per sample, starting posterior to the VNO, when n2 was reached.
P7: 14 sections per sample, starting posterior to the VNO, when n2 was reached.
P30, nine to ten sections per sample, starting posterior to the VNO, when n2 was reached.
Linear Fgf20+ and OMP+ cell distribution plot
OE of turbinate c1 was linearized in ImageJ at the level of OMP+ ORNs. The location of each Fgf20+ and OMP+ cell was marked along this line. Plots from multiple samples were scaled to their average length and combined to represent an average/combined distribution across multiple samples. Plots were made in Canvas X (ACD Systems)
Phenotype analyses at P30
Individual turbinate size: length was measured at the apical OE surface of each turbinate from two comparable sections at the anteroposterior middle of each turbinate, and then summed to estimate relative turbinate size. Outlines in Figure 3B’ indicate measurements used to estimate turbinate size.
Total surface area: OE surface area was measured as above, using 26-29 frontal sections, starting immediately posterior to the VNO for the OE, and 3-6 frontal sections spanning the entire septal organ anteroposteriorly, for the septal organ.
For the rest of this study (methods below), we focused on turbinate c1.
Phenotype analyses at E13.5 and E14.5
OE thickness: average of three measurements made on one frontal section halfway through the turbinate. Measurements were at the turbinate tip/center and 50 μm on either side. Each measurement was the shortest distance from the epithelial-mesenchymal boundary to the apical surface of the OE.
Mesenchyme thickness: as with OE thickness, average of three measurements at the turbinate tip/center and 50 μm on either side. Each measurement was the shortest distance from the epithelial-mesenchymal boundary to the nasal cavity wall.
Cell counts were made on comparable frontal sections halfway through the turbinate.
Condensed mesenchymal cells were identified by high Sox9 staining, small soma, and dense packing of cells. A few cells outside of the condensations at E14.5 (closer to the epithelium) had high Sox9 expression, but were counted as diffuse mesenchymal cells. Condensed and diffuse cell numbers were normalized to the number of Sox9+ chondrocytes in the adjacent nasal cavity wall to adjust for potential slight differences in angle of sectioning.
Quantification of “OE adjacent cell” proliferation in the Fgf9-OA experiment: average c1 diffuse mesenchyme thickness in control embryos was measured to be 69 μm. In Fgf9-OA embryos, the c1 OE was outlined at the basal side; this outline was then moved 69 μm away from the basal side of the OE. Cells between this line and the OE were considered “OE adjacent cells.” The average normalized number of “OE adjacent cells” quantified was 1.2 ± 0.1 in Fgf9-OA embryos, the same as the normalized number of diffuse cells in control (1.2 ± 0.2).
BC number was normalized to OE basal surface length. OMP+ cell number was normalized to OE apical surface length. Fgf20+ cell counts were normalized to OE area on a section.
Phenotype analyses at E17.5 and P0
Turbinate cartilage size: a curved line was drawn to the contour of the lateral nasal cavity wall dorsal and ventral to c1; the cartilage projection medial to this line was considered the turbinate. Cartilaginous area of c1 was measured in 5 to 7 serial frontal sections spanning the entire turbinate anteroposteriorly. The measured area was then multiplied by the distance between sections to calculate volume.
Turbinate OE surface area: length of the turbinate was measured along the epithelial-mesenchymal boundary in 5 to 7 serial frontal sections and multiplied by the distance between sections. Inflection points in the OE curvature at “neck” regions were used to differentiate between turbinate OE (OE overlying the turbinate) and OE overlying the nasal cavity wall. For reference, see dashed line in Figures S3H, 6C, and S6C. In cases where the turbinate was completely absent (βFF-CKO, shape 3), what was considered turbinate OE was estimated based on the height (dorsoventral length) of the turbinate in less severe βFF-CKO littermates (see Figure 6C, dashed line). Note the “neck” OE regions of c1 are further apart in βFF-CKO mice (turbinate “neck” cartilage is thicker); this was taken into consideration in the estimate.
Mesenchyme thickness: three measurements at the turbinate tip/center and 100 μm on either side, on 3 sections 96 μm apart for a total of 9 measurements (which were then averaged). Each measurement was the shortest distance from the epithelial-mesenchymal boundary to the turbinate cartilage.
OE thickness: same as mesenchyme thickness. Each measurement was the shortest distance from the epithelial-mesenchymal boundary to the apical surface of the OE.
Sox2+ and OMP+ cell count: on comparable sections at the anteroposterior middle of the turbinate. BC number was normalized to OE basal surface length. Sus cell and ORN numbers were normalized to OE apical surface length.
Mesenchyme EdU-incorporating cell count: “neck” region mesenchyme was defined as mesenchyme adjacent to FEP cell area, both dorsal and ventral to c1, combined. “Tip” region was a similarly sized area at the tip of c1.
Chondrocyte EdU-incorporating cell count: all chondrocytes of c1 (“neck” and “tip”) on a frontal section halfway through the turbinate were counted.
In situ hybridization analyses
In situ hybridization for Dusp6 was evaluated, blinded to genotype, by alkaline phosphatase color reaction intensity in the c1 mesenchyme. 2-3 serial sections per sample from Fgf20-KO (n = 7 control, 7 Fgf20-KO), Fgf9-OA (n = 4 control, 4 Fgf9-OA), and βFF-CKO (n = 4 control, 6 βFF-CKO) E14.5 embryos were examined. For Fgf20-KO samples, the genotypes of 13 out of 14 total samples were scored correctly based on Dusp6 expression intensity (one control sample was wrongly scored as Fgf20-KO). Figures 3H, S4G, and S5C show representative images of each genotype.
Statistics and plotting
All figures were made in Canvas X. Data analysis was performed using the Python programming language (python.org) in Jupyter Notebook (jupyter.org) with the following libraries: Pandas (pandas.pydata.org), NumPy (numpy.org) and SciPy (scipy.org). Plotting was done using the Matplotlib library (matplotlib.org). Statistics (t-test and one-way ANOVA) were performed using the SciPy module Stats; Tukey’s HSD was performed using the Statsmodels package (statsmodels.org).
Data from βFF-CKO mice were plotted in different colors to visualize differences in phenotype severity (shapes 1, 2, and 3). However, data from all shapes were combined for statistical analysis. Comparisons of two means in βFF-CKO experiments were performed using two-tailed, unpaired Welch’s (unequal variance) t-test, since βFF-CKO mice exhibited variability in phenotype while controls did not. All other comparisons of two means were performed using two-tailed, unpaired Student’s t-test. For comparisons of more than two means, one-way ANOVA was used; for significant results at α = 0.05, Tukey’s HSD was performed for post-hoc pair-wise analysis.
All statistical details can be found in the figures and figure legends. In all cases, sample size (n) represents the number of animals. Data are represented as mean ± standard deviation (SD).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-GFP | Thermo Fisher | Cat# A-11122; RRID:AB_221569 |
| Chick anti-Beta galactosidase | Abcam | Cat# ab9361; RRID:AB_307210 |
| Goat anti-Sox2 | Santa Cruz Biotech | Cat# sc-17320; RRID:AB_2286684 |
| Rabbit anti-Sox9 | Millipore | Cat# AB5535; RRID:AB_2239761 |
| Goat anti-OMP | Wako Chemicals | Cat# 544-10001; RRID:AB_664696 |
| Goat anti-Pde2a | Santa Cruz Biotech | Cat# sc-17227; RRID:AB_653928 |
| Donkey anti-Rabbit IgG (H+L), Alexa Fluor 488 | Thermo Fisher | Cat# A-21206; RRID:AB_2535792 |
| Donkey anti-Goat IgG (H+L); Alexa Fluor 555 | Thermo Fisher | Cat# A-21432; RRID:AB_2535853 |
| Goat anti-Rabbit IgG (H+L); Alexa Fluor 555 | Thermo Fisher | Cat# A-21428; RRID:AB_2535849 |
| Goat anti-Chicken IgY (H+L); Alexa Fluor 555 | Thermo Fisher | Cat# A-21437; RRID:AB_2535858 |
| Sheep anti-Digoxigenin-AP, Fab fragments | Sigma-Aldrich | Cat# 11093274910; RRID:AB_514497 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dox Diet, Grain-Based Doxycycline, 200 mg/kg | Bio-Serv | Cat# S3888 |
| Critical Commercial Assays | ||
| VectaShield antifade mounting medium with DAPI | Vector Labs | Cat# H-1200 |
| Click-iT Plus EdU Alexa Fluor 594 picolyl azide toolkit | Thermo Fisher | Cat# C10639 |
| Click-iT Plus EdU Alexa Fluor 647 picolyl azide toolkit | Thermo Fisher | Cat# C10640 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Fgf20GFP-Cre: B6.129-Fgf20tm2.1(cre/EGFP)Dor/J | Huh et al., 2015 | MGI:5751785 |
| Mouse: Fgf20βgal: B6.129-Fgf20tm1.1Dor/J | Huh et al., 2012 | RRID:MGI:5425887 |
| Mouse: ROSAmTmG: B6.129-Gt(ROSA)26Sortm4(ACTB-tdTomato,−EGFP)Luo/J | Muzumdar et al., 2007 | RRID:IMSR_JAX:007576 |
| Mouse: ROSAtdTomato: B6.129-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | Madisen et al., 2010 | RRID:IMSR_JAX:007905 |
| Mouse: ROSArtTA: B6.129-Gt(ROSA)26Sortm1(rtTA,EGFP)Nagy/J | Belteki et al., 2005 | RRID:IMSR_JAX:005670 |
| Mouse: TRE-Fgf9-IRES-eGfp: B6.129-Tg(tetO-Fgf9,-EGFP)#Dor/J | White et al., 2006 | MGI:5538516 |
| Mouse: Tcf/Lef:H2b-Gfp: B6.129-Tg(TCF/Lef1-HIST1H2BB/EGFP)61Hadj/J | Ferrer-Vaquer et al., 2010 | RRID:IMSR_JAX:013752 |
| Mouse: βCatfl(ex2-6): B6.129-Ctnnb1tm2Kem/J | Brault et al., 2001 | RRID:IMSR_JAX:004152 |
| Mouse: βCatDM: B6.129-Ctnnb1tm3Kba/J | Valenta et al., 2011 | MGI:5308947 |
| Mouse: βCatfl(ex3): B6.129-Ctnnb1tm1Mmt/J | Harada et al., 1999 | MGI:1858008 |
| Recombinant DNA | ||
| Plasmid: pBSKS-Dusp6 | Gift of Dr. Suzanne Mansour (Li et al., 2007) | N/A |
| Software and Algorithms | ||
| ZEN | Zeiss | https://www.zeiss.com/microscopy/us/downloads/zen.html |
| NanoZoomer Digital Pathology (NDP.view2) | Hamamatsu | https://www.hamamatsu.com/jp/en/U12388-01.html |
| Canvas X | ACD Systems | http://www.canvasgfx.com/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Cell Counter plugin for ImageJ | Kurt De Vos | https://imagej.nih.gov/ij/plugins/cell-counter.html |
| Python | Guido van Rossum and Python developers | https://www.python.org/ |
| Pandas | Pandas core team | https://pandas.pydata.org/ |
| NumPy | NumPy developers | http://www.numpy.org/ |
| SciPy | SciPy developers | https://www.scipy.org/ |
| Matplotlib | John Hunter and contributors | https://matplotlib.org/ |
| Statsmodels | Josef Perktold, Skipper Seabold, Jonathan Taylor, and statsmodels developers. | https://www.statsmodels.org/stable/index.html |
Highlights.
Fgf20-positive epithelial-spanning progenitor (FEP) cells form olfactory epithelium
FEP cells regulate turbinate growth via FGF20
Wnt/βCat signaling maintains FEP cells in an undifferentiated state
Wnt/βCat signaling regulates the expression of FGF20
ACKNOWLEDGEMENTS
We thank Y. Yin and A. Hagan for critically reading the manuscript. This work was funded by the March of Dimes Foundation, NIH grant HL111190 (D.M.O.), the Department of Developmental Biology at Washington University, DC012825 (S.H.), HOPE Center Alafi Neuroimaging Laboratory (NCRR 1S10RR027552), and Washington University Center for Cellular Imaging (Children’s Discovery Institute CDI-CORE-2015-505; NS086741).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adameyko I, and Fried K (2016). The Nervous System Orchestrates and Integrates Craniofacial Development: A Review. Front. Physiol 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios AW, Nuñez G, Sánchez Quinteiro P, and Salazar I (2014). Anatomy, histochemistry, and immunohistochemistry of the olfactory subsystems in mice. Front. Neuroanat 8, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beites CL, Kawauchi S, Crocker CE, and Calof AL (2005). Identification and molecular regulation of neural stem cells in the olfactory epithelium. Exp. Cell Res 306, 309–316. [DOI] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, and Nagy A (2005). Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 33, e51–e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, and Kemler R (2001). Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, Berge D ten, Fuerer C, Clevers H, Nusse R, and Helms JA. (2007). Wnt signaling mediates regional specification in the vertebrate face. Development 134, 3283–3295. [DOI] [PubMed] [Google Scholar]
- Calof AL, Bonnin A, Crocker C, Kawauchi S, Murray RC, Shou J, and Wu H-H (2002). Progenitor cells of the olfactory receptor neuron lineage. Microsc. Res. Tech 58, 176. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, and Guillemot F (1997). Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development 124, 1611–1621. [DOI] [PubMed] [Google Scholar]
- Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, and Varmus HE (2005). FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J. 24, 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tian S, Yang X, Lane AP, Reed RR, and Liu H (2014). Wnt-Responsive Lgr5+ Globose Basal Cells Function as Multipotent Olfactory Epithelium Progenitor Cells. J. Neurosci 34, 8268–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola DM, Craven BA, Seeger J, and Weiler E (2014). The effects of naris occlusion on mouse nasal turbinate development. J. Exp. Biol 217, 2044–2052. [DOI] [PubMed] [Google Scholar]
- Dieulafe L (1906). Morphology and Embryology of the Nasal Fossae of Vertebrates. Ann. Otol. Rhinol. Laryngol 15, 1–584. [Google Scholar]
- Duggan CD, DeMaria S, Baudhuin A, Stafford D, and Ngai J (2008). Foxg1 Is Required for Development of the Vertebrate Olfactory System. J. Neurosci 28, 5229–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eerdunfu, Ihara N, Ligao B, Ikegaya Y, and Takeuchi H (2017). Differential timing of neurogenesis underlies dorsal-ventral topographic projection of olfactory sensory neurons. Neural Develop. 12, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekerot M, Stavridis MP, Delavaine L, Mitchell MP, Staples C, Owens DM, Keenan ID, Dickinson RJ, Storey KG, and Keyse SM (2008). Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6/MKP-3 gene promoter. Biochem. J 412, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel NW, Neumann JE, Ahlfeld J, Wefers AK, Merk DJ, Ohli J, and Schuller U (2016). Canonical Wnt Signaling Drives Tumor-Like Lesions from Sox2-Positive Precursors of the Murine Olfactory Epithelium. PLoS ONE 11, e0166690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Vaquer A, Piliszek A, Tian G, Aho RJ, Dufort D, and Hadjantonakis A-K (2010). A sensitive and bright single-cell resolution live imaging reporter of Wnt/B-catenin signaling in the mouse. BMC Dev. Biol 10, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Bear DM, Lassance J-M, Bloom ML, Tsukahara T, Pashkovski SL, Masuda FK, Nowlan AC, Kirchner R, Hoekstra HE, et al. (2016). A Family of non-GPCR Chemosensors Defines an Alternative Logic for Mammalian Olfaction. Cell 165, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haara O, Harjunmaa E, Lindfors PH, Huh S-H, Fliniaux I, Aberg T, Jernvall J, Ornitz DM, Mikkola ML, and Thesleff I (2012). Ectodysplasin regulates activator-inhibitor balance in murine tooth development through Fgf20 signaling. Development 139, 3189–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, and Taketo MM (1999). Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. EMBO J. 18, 5931–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman BK, and Margolis FL (1975). Immunofluorescence localization of the olfactory marker protein. Brain Res. 96, 176–180. [DOI] [PubMed] [Google Scholar]
- Hebert JM, and McConnell SK (2000). Targeting of cre to the Foxg1 (BF-1) Locus Mediates loxP Recombination in the Telencephalon and Other Developing Head Structures. Dev. Biol 222, 296–306. [DOI] [PubMed] [Google Scholar]
- Huh S-H, Jones J, Warchol ME, and Ornitz DM (2012). Differentiation of the Lateral Compartment of the Cochlea Requires a Temporally Restricted FGF20 Signal. PLoS Biol. 10, e1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S-H, Narhi K, Lindfors PH, Haara O, Yang L, Ornitz DM, and Mikkola ML (2013). Fgf20 governs formation of primary and secondary dermal condensations in developing hair follicles. Genes Dev. 27, 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh S-H, Warchol ME, and Ornitz DM (2015). Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. ELife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ookawara S, Sato S, Ando Z, Kageyama R, and Kawakami K (2007). Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev. Biol 311, 53–68. [DOI] [PubMed] [Google Scholar]
- Juilfs DM, Fülle HJ, Zhao AZ, Houslay MD, Garbers DL, and Beavo JA (1997). A subset of olfactory neurons that selectively express cGMP-stimulated phosphodiesterase (PDE2) and guanylyl cyclase-D define a unique olfactory signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A 94, 3388–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsunuma S, Honda H, Shinoda T, Ishimoto Y, Miyata T, Kiyonari H, Abe T, Nibu K, Takai Y, and Togashi H (2016). Synergistic action of nectins and cadherins generates the mosaic cellular pattern of the olfactory epithelium. J Cell Biol 212, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Rodríguez-León J, Koth CM, Buscher D, Itoh T, Raya Á, Ng JK, Esteban CR, Takahashi S, Henrique D, et al. (2003). MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat. Cell Biol 5, 513–519. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hébert JM, McConnell SK, Mason I, and Calof AL (2005). Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development 132, 5211–5223. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Kim J, Santos R, Wu H-H, Lander AD, and Calof AL (2009). Foxg1 promotes olfactory neurogenesis by antagonizing Gdf11. Development 136, 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersigo J, D’Angelo A, Gray BD, Soukup GA, and Fritzsch B (2011). The role of sensory organs and the forebrain for the development of the craniofacial shape as revealed by Foxg1-cre-mediated microRNA loss. Genesis 49, 326–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laclef C, Souil E, Demignon J, and Maire P (2003). Thymus, kidney and craniofacial abnormalities in Six1 deficient mice. Mech. Dev 120, 669–679. [DOI] [PubMed] [Google Scholar]
- Li C, Xu X, Nelson DK, Williams T, Kuehn MR, and Deng C-X (2005). FGFR1 function at the earliest stages of mouse limb development plays an indispensable role in subsequent autopod morphogenesis. Development 132, 4755–4764. [DOI] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X, and Mansour SL (2007). Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development 134, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, and Ornitz DM (2013). Development of the endochondral skeleton. Cold Spring Harb. Perspect. Biol 5, a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani P, Jarrell A, Myers J, and Atit R (2010). Visualizing canonical Wnt signaling during mouse craniofacial development. Dev. Dyn 239, 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau-Doizé B, Caya I, and Martineau GP (1992). Osteogenesis and growth of the nasal ventral conchae of the piglet. J. Comp. Pathol 106, 323–331. [DOI] [PubMed] [Google Scholar]
- Murdoch B, and Roskams AJ (2007). Olfactory epithelium progenitors: insights from transgenic mice and in vitro biology. J. Mol. Histol 38, 581–599. [DOI] [PubMed] [Google Scholar]
- Murdoch B, and Roskams AJ (2008). A Novel Embryonic Nestin-Expressing Radial Glia-Like Progenitor Gives Rise to Zonally Restricted Olfactory and Vomeronasal Neurons. J. Neurosci 28, 4271–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch B, DelConte C, and Garcia-Castro MI (2010). Embryonic Pax7-Expressing Progenitors Contribute Multiple Cell Types to the Postnatal Olfactory Epithelium. J. Neurosci 30, 9523–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Nusse R, and Clevers H (2017). Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169, 985–999. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, and Itoh N (2015). The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol 4, 215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BS, Yang H, Melvin VS, Taketo MM, and Williams T (2011). Ectodermal Wnt/β-catenin signaling shapes the mouse face. Dev. Biol 349, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, and Buck LB (1993). A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73, 597–609. [DOI] [PubMed] [Google Scholar]
- Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, and Coleman JH (2017). The stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. J. Comp. Neurol 525, 1034–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart IH (1971). Location and orientation of mitotic figures in the developing mouse olfactory epithelium. J. Anat 109, 243–251. [PMC free article] [PubMed] [Google Scholar]
- Treloar HB, Miller AM, Ray A, and Greer CA (2010). Development of the Olfactory System In The Neurobiology of Olfaction, Menini A, ed. (Boca Raton (FL): cRc Press; ), p. Chapter 5. [PubMed] [Google Scholar]
- Tucker ES, Lehtinen MK, Maynard T, Zirlinger M, Dulac C, Rawson N, Pevny L, and LaMantia A-S (2010). Proliferative and transcriptional identity of distinct classes of neural precursors in the mammalian olfactory epithelium. Development 137, 2471–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T, Gay M, Steiner S, Draganova K, Zemke M, Hoffmans R, Cinelli P, Aguet M, Sommer L, and Basler K (2011). Probing transcription-specific outputs of β-catenin in vivo. Genes Dev. 25, 2631–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valkenburgh B, Smith TD, and Craven BA (2014). Tour of a labyrinth: exploring the vertebrate nose. Anat. Rec 297, 1975–1984. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, and Axel R (1993). Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74, 309–318. [DOI] [PubMed] [Google Scholar]
- Waghmare SK, Bansal R, Lee J, Zhang YV, McDermitt DJ, and Tumbar T (2008). Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 27, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-Z, Yamagami T, Gan Q, Wang Y, Zhao T, Hamad S, Lott P, Schnittke N, Schwob JE, and Zhou CJ (2011). Canonical Wnt signaling promotes the proliferation and neurogenesis of peripheral olfactory stem cells during postnatal development and adult regeneration. J. Cell Sci 124, 1553–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng P-L, Vinjamuri M, and Ovitt CE (2016). Ascl3 transcription factor marks a distinct progenitor lineage for non-neuronal support cells in the olfactory epithelium. Sci. Rep 6, 38199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AC, Xu J, Yin Y, Smith C, Schmid G, and Ornitz DM (2006). FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development 133, 1507–1517. [DOI] [PubMed] [Google Scholar]
- Zhu X-J, Liu Y, Yuan X, Wang M, Zhao W, Yang X, Zhang X, Hsu W, Qiu M, Zhang Z, et al. (2016). Ectodermal Wnt controls nasal pit morphogenesis through modulation of the BMP/FGF/JNK signaling axis. Dev. Dyn 245, 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


