SUMMARY
The eight metabotropic glutamate receptors (mGluRs) serve critical modulatory roles throughout the nervous system. The molecular diversity of mGluRs is thought to be further expanded by the formation of heterodimers, but the co-expression of mGluR subtypes at the cellular level and the relative propensities of heterodimer formation are not well known. Here, we analyze single-cell RNA sequencing data and find that cortical pyramidal cells express multiple mGluR subtypes with distinct profiles for different receptor combinations. We then develop quantitative, fluorescence-based assays to define the relative homo- and heterodimer propensities across group-I, -II, and -III mGluRs. We find a strong preference for heterodimerization in a number of cases, including mGluR2 with mGluR3, which we confirm in frontal cortex using in situ RNA hybridization and co-immunoprecipitation. Together, our findings support the biological relevance of mGluR heterodimerization and highlight the complex landscape of mGluR populations in the brain.
Graphical Abstract
In Brief
Lee et al. perform analysis of single-cell RNA sequencing data to reveal that metabotropic glutamate receptor (mGluR) subtypes exhibit highly overlapping expression in mouse cortical pyramidal neurons. Quantitative characterization of assembly propensities using fluorescence-based assays reveal high-efficiency heterodimerization across group-I, -II, and -III mGluRs.
INTRODUCTION
G protein-coupled receptors (GPCRs) often form families containing numerous subtypes that respond to the same ligand (Katritch et al., 2013). This apparent redundancy is explained by the high degree of specialization between different subtypes, which can include sensitivity to different ligand concentrations, signaling to different pathways, or interaction with unique regulatory partners. This phenomenon is especially critical among neuromodulatory receptors, which can respond to the same ligand in different locations and contexts throughout the nervous system. For example, within the metabotropic glutamate receptor (mGluR or mGlu receptor) family, eight different subtypes exhibit a range of glutamate sensitivities, preferred G protein effectors, ensembles of accessory regulatory proteins, and synaptic localization (Pin and Bettler, 2016; Reiner and Levitz, 2018).
Given the importance of mGluRs for modulating neuronal excitability and synaptic strength and as potential drug targets for treatment of neurological and psychiatric disorders (Niswender and Conn, 2010; Reiner and Levitz, 2018), considerable effort has been dedicated to defining the unique properties of each subtype. However, disentangling the molecular diversity of mGluRs is complicated by the fact that they form constitutive dimers (Doumazane et al., 2011; Levitz et al., 2016; Romano et al., 1996) and that dimerization is required for glutamate-driven activation (El Moustaine et al., 2012). Despite early reports that different subtypes were unable to interact (Romano et al., 1996), recent work has firmly established that mGluRs can indeed form heterodimers and that these heterodimers can have unique pharmacological and functional properties (Sevastyanova and Kammermeier, 2014; Pandya et al., 2016; Doumazane et al., 2011; Habrian et al., 2019; Levitz et al., 2016; Yin et al., 2014). For example, mGluR2/4 heterodimers form when heterologously expressed, can be co-immunoprecipitated from the brain, and can couple to G proteins primarily via the mGluR4 subunit, which leads to unique sensitivity to combinations of mGluR2 and mGluR4-targeting allosteric compounds (Liu et al., 2017; Moreno Delgado et al., 2017; Yin et al., 2014). In addition, mGluR2/3 and mGluR2/7 heterodimers show unique conformational dynamics relative to their parent dimers, thereby tuning the basal activity and glutamate-sensitivity of the heterodimer (Habrian et al., 2019; Levitz et al., 2016).
Despite the potential importance of heterodimerization to mGluR biology, the physiological relevance of such complexes remains unclear for two major reasons. (1) The overlap of receptor subtype expression at the cellular level has not been quantified, despite it being established that many brain regions express multiple subtypes (Ferraguti and Shigemoto, 2006). (2) At the molecular level, the relative propensities for homodimer versus heterodimer formation for a given subtype are not defined, leaving open the possibility that homodimers are preferentially formed with heterodimers only occurring under extreme conditions. There is general agreement that heterodimers can form between Gi/o-coupled group-II/III subtypes (mGluR2, mGluR3, mGluR4, mGluR6, mGluR7, and mGluR8) and Gq-coupled group-I subtypes (mGluR1 and mGluR5), but the relative preferences for any given pair have not been carefully characterized.
Developing methodologies for defining the co-expression and co-assembly propensities of various mGluR heterodimers is critical for identifying the biologically meaningful dimer pairs and understanding the biophysical mechanisms by which specific combinations are able to assemble. In this study, we first use analysis of single-cell RNA sequencing (scRNA-seq) data to define the expression of mGluRs in the cortex, where we find a complex pattern of highly overlapping expression between subtypes. We then develop a live-cell fluorescence assay to quantify heterodimer formation of group-II mGluRs across all mGluR subtypes, where we find a wide range of propensities including preferential heterodimerization for mGluR2 with mGluR3, which we support with single-molecule imaging experiments. Based on this, we use fluorescence in situ hybridization (FISH) to further characterize co-expression of mGluR2 and 3 and demonstrate co-immunoprecipitation of mGluR2/3 heteromers from the frontal cortex. Finally, we extend our experimental analysis across group-I, -II, and -III mGluRs, enabling quantification of relative homo-and heterodimerization propensity across all subfamilies.
RESULTS
Analysis of scRNA-Seq Data Reveals Widespread Overlap of mGluR Expression in the Cortex
A prerequisite for biologically relevant heterodimerization of mGluR subtypes is substantial co-expression in the same cells, a possibility that has not been well characterized for mGluRs. Previous studies have used in situ RNA hybridization or immunohistochemistry to reveal overlapping expression patterns across mGluR subtypes (Ferraguti and Shigemoto, 2006), demonstrating that all subtypes, with the exception of mGluR6, are found throughout the neocortex, striatum, and many other areas. However, such analysis has not typically been performed simultaneously for multiple receptor subtypes or with single-cell resolution, leaving open the possibility that receptor subtypes are segregated into different cell types, as seen with D1 and D2 dopamine receptors in layer-5 cortical pyramidal neurons (Dembrow et al., 2010; Santana et al., 2009; Vincent et al., 1993) and medium spiny neurons in the striatum (Beckstead et al., 1988; Gerfen et al., 1990; Thibault et al., 2013).
To gain further insight into the expression patterns of mGluRs in the cortex, we turned to scRNA-seq—a high throughput method that provides a quantitative snapshot of the transcriptome of individual cells (Poulin et al., 2016). The most comprehensive taxonomy of cortical neurons to date involved the sequencing of over 20,000 mouse cortical cells, which provided insight into the layer distribution and long-range projections of glutamatergic cell types (Tasic et al., 2018). We chose to use this dataset to explore the co-expression patterns of the family of eight mGluRs (gene names: grm1–grm8) across cell types in one cortical region. The database used contains 9,573 individual cells isolated from the frontal cortex (anterior lateral motor cortex, ALM), comprising a range of cell types.
Briefly, we quantified the expression level for a given transcript as copies per million (CPM) of total copies as in the original study (Tasic et al., 2018). Cells were then clustered according to the similarity of their transcriptomes and sequential splits established a hierarchical cellular categorization. The first split separated cells into three broad ‘‘classes’’: glutamatergic neurons, GABAergic neurons, and non-neuronal cells. Each broad class could be further subdivided into ‘‘subclasses’’: for glutamatergic neurons, this follows patterns of cortical layer localization, as well as long-projection targets, such as intratelencephalic (IT), corticothalamic (CT), pyramidal tract (PT), and near-projecting (NP) (Figure 1A). Likewise, further subdivisions of GABAergic neurons revealed all known inhibitory subclasses, such as somatostatin (Sst)-expressing neurons and parvalbumin (Pvalb)-expressing neurons, as well as non-neuronal cells subclasses, such as astrocytes and oligodendrocytes (Figure 1A).
Figure 1. Analysis of mGluR Co-expression across Different Cell Types in the Adult Mouse Cortex.
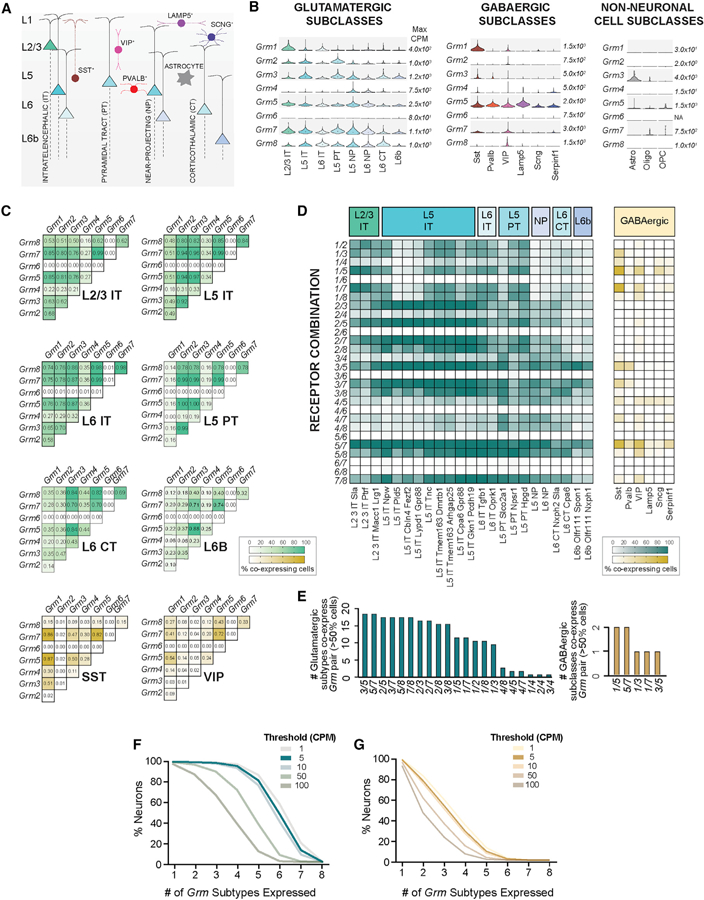
(A) Simplified schematic showing the major cortical cell subclasses. Glutamatergic neurons are named after layer (L) and main projection target whereas GABAergic neurons are named after non-overlapping molecular markers (nomenclature follows resource publication; Tasic et al., 2018).
(B) Violin plots show the distribution of the expression of each of the eight members of the Grm family across cortical subclasses, in which the different widths in each plot represent probability density (relative number of cells expressing at that range) and the black dot represents median value for each subclass. Scales represent maximum copies per million (CPM) for each gene (see also Figure S1 for relative expression of each Grm subtype). Number of cells per subclass: L2/3 IT (313), L5 IT (2387), L6 IT (387), L5 PT (367), L5 NP (294), L6 NP (241), L6 CT (343), L6b (114), Sst (1139), Pvalb (896), Vip (1224), Lamp5 (912), Sncg (148), Serpinf1 (78), astrocytes (215), oligodendrocytes (68), oligodendrocyte precursor cells (OPCs) (23).
(C) Co-expression analysis (cutoff minimum of 5 CPM) shown as heatmaps, in which color range represents proportion of cells within that subclass co-expressing each Grm pair (see also Figure S1B).
(D) As in (C), co-expression heatmaps for glutamatergic subtypes, left, and broad GABAergic subclasses, right.
(E) Bar plot showing Grm pairs (x axis) and the number of glutamatergic subtypes (green, left) or GABAergic subclasses (yellow, right), which co-express each Grm pair, assuming co-expression within a group when more than 50% of the neurons in that group expressed both Grm subtypes.
(F and G) Survival plots show, on the x-axis, the number of expressing Grms (F) (for different thresholds); and on the y-axis, the percentage of total glutamatergic neurons or (G) GABAergic neurons.
L, layer; CT, corticothalamic; IT, intratelencephalic; PT, pyramidal tract (thalamus, tectum, and pons); CPM, counts per million.
We first characterized the general expression patterns of the eight mGluRs across broad cell classes (Figure S1A). Glutamatergic neurons contain considerable mRNA levels for most subtypes, with the exception of Grm6, which is not clearly expressed (Figure S1A). In contrast, GABAergic neurons express mainly Grm5, Grm7, and Grm1, and in non-neuronal cells only Grm3 is expressed to high levels, predominantly in astrocytes (Figure S1A). As we anticipated, each Grm subtype showed a distinct pattern of expression when considering levels across the different neuronal subclasses (Figure 1B). For example, Grm1 appears enriched in layer (L) 2/3 glutamatergic neurons, while Grm4 is virtually only expressed by L5 and L6 NP neurons. Among GABAergic subclasses, Grm1 and Grm7 are nearly uniquely expressed by Sst- and VIP-expressing neurons while all subclasses express Grm5, though with variable levels. This overview of expression reveals many subclasses with potential co-expression of Grm subtypes, motivating analysis of co-expression in individual cells.
Before assessing Grm subtype co-expression based on scRNA-seq, two important technical considerations were made: (1) only 10%–20% of all mRNA molecules are thought to be recovered in this approach, and (2) only reads correctly aligned to the genome are considered for gene quantification, minimizing false positives (Islam et al., 2014). Therefore, in order to analyze the probabilities of Grm subtype co-expression we imposed a cutoff of a minimum of 5 CPM before calculating the percentages of co-expressing cells in each subclass. Co-expression analysis revealed that all glutamatergic subclasses show a high percentage of neurons co-expressing many of the 28 possible Grm pairs (Figure 1C; Figure S1B). Among GABAergic neurons, considerably smaller co-expression frequencies were observed (Figure 1C; Figure S1B), with the exception of the Sst-expressing subclass, which showed high co-expression levels for Grm1, Grm5, and Grm7.
Given the striking subtype diversity seen within neuronal subclasses (Tasic et al., 2018), which also show distinct but overlapping expression patterns across Grm subtypes (Figure S1C), we next looked at co-expression probabilities across subtypes of glutamatergic neurons. We found considerable variation among subtypes, particularly in L5 IT neurons, in which probabilities for pairs with Grm1 varied the most (Figure 1D). Yet, eight of the 28 possible Grm pairs were present in more than 50% of cells in at least 15 (of 23) glutamatergic subtypes (Figure 1E). Due to the lower Grm expression levels in GABAergic neurons and diversity of over 60 different subtypes, we did not further explore co-expression patterns in their subtypes. Still, a few pairs, including Grm1 and Grm5, were co-expressed in at least 50% of all Sst- and VIP-expressing neurons (Figure 1E).
Having seen that most neuronal subclasses express multiple Grm subtypes (Figure 1B) and co-express multiple Grm pairs (Figures 1C and 1D), we envisioned the distribution of all mGluRs within glutamatergic neurons. Therefore, we set various levels of stringencies with different cutoffs (from cutoff of minimum of one copy up to 100 CPM) and saw that even with a high threshold (minimum 50 CPM), more than 70% of all glutamatergic neurons co-express at least four members of the mGluR family (Figure 1F) in contrast to less than 20% of GABAergic neurons (Figure 1G). Together, these analyses demonstrate the substantial co-expression of the different members of the Grm family and highlight the distinct prospects for particular Grm pairs in a cell-type-specific manner, further motivating an experimental analysis of the homo- and heterodimerization propensities of mGluRs.
A Ligand Binding Domain Complementation Assay to Quantify mGluR Dimerization in Live Cells
Given the overlap in expression of mGluR subtypes at the single-cell level (Figure 1) and the lack of information available on the propensities of mGluRs to heterodimerize in different combinations, we sought to develop an assay that could allow for quantitative analysis of the relative dimerization propensities of different mGluR combinations on the surface of live cells. Interaction between extracellular ligand binding domains (LBDs) is the main driver of mGluR dimerization and the major determinant of the ability of different subtypes to heterodimerize (Levitz et al., 2016), motivating a focus on interactions at this level. Furthermore, previous studies have shown that co-expression of an isolated mGluR extracellular domain can lead to co-assembly with a full-length mGluR on the cell surface (Beqollari and Kammermeier, 2010; Robbins et al., 1999; Selkirk et al., 2002). We sought to adapt this measurement to allow for a careful control of the relative expression levels of the isolated LBD and the full-length receptor. We envisioned a three-color fluorescence assay that takes advantage of the ability to target an N-terminal benzylguanine (BG)-reactive SNAP-tag on the isolated LBD with both a membrane-impermeable fluorophore (i.e., BG-Alexa647) and a membrane-permeable fluorophore (i.e., BG-TMR) in a different color (Figure 2A).
Figure 2. A Three-Color Fluorescence-Based LBD Complementation Assay to Quantify mGluR Dimerization in Living Cells.
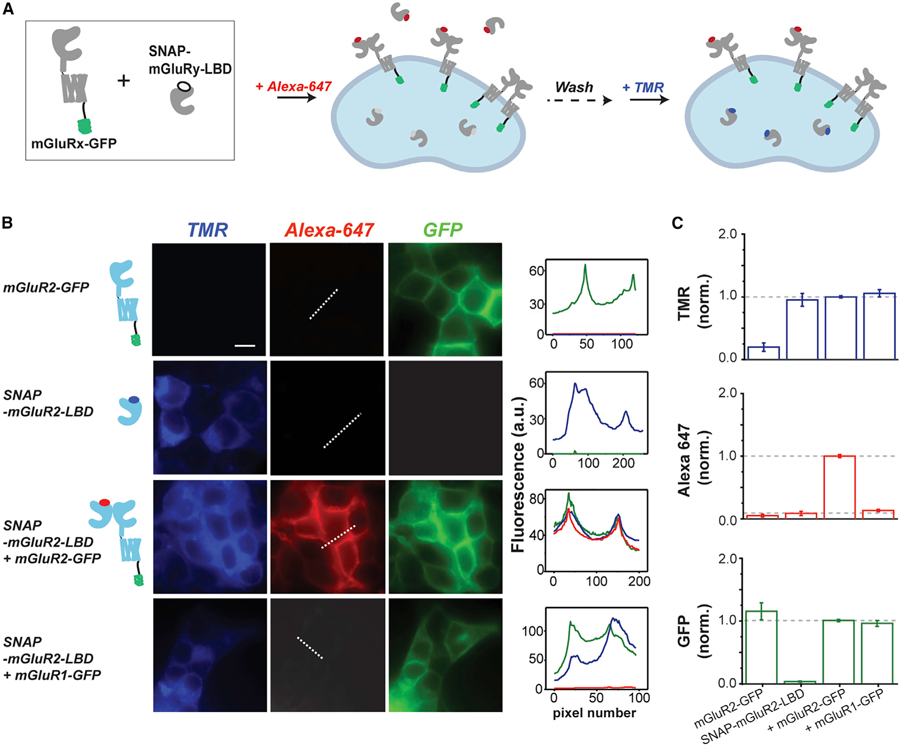
(A) Schematic showing LBD complementation assay. N-terminally SNAP-tagged mGluRy-LBD (‘‘SNAP-mGluRy-LBD’’) is co-expressed with C-terminally GFP-tagged mGluRx (‘‘mGluRx-GFP’’). Cells are first labeled with BG-Alexa647, a membrane-impermeable fluorophore that labels SNAP-mGluRy-LBD within both surface heterodimers with mGluRx-GFP and exported, soluble SNAP-mGluRy-LBD. Next, cells are labeled with BG-TMR, a membrane-permeable fluorophore, which labels intracellular SNAP-mGluRy-LBD. Following washing, soluble SNAP-mGluRy-LBD is removed and Alexa647 fluorescence intensity can be used asa measure of the capture efficiency of SNAP-mGluRy-LBD by mGluRx-GFP. TMR and GFP fluorescence is used to calibrate expression across conditions.
(B) Experimental validation of the LBD complementation assay in HEK293T cells. Only when SNAP-mGluR2-LBD is co-expressed with mGluR2-GFP is there clear Alexa647 surface fluorescence. TMR fluorescence is only seen when SNAP-mGluR2-LBD is expressed. Note that no clear Alexa647 fluorescence is observed when SNAP-mGluR2-LBD is co-expressed with mGluR1-GFP. Line scans from the dotted line are plotted in the right column. Scale bar, 10 mm. All images within the same channel are on the same intensity scale.
(C) Fluorescence intensity quantification plots for TMR, GFP, and Alexa647 channels across conditions tested in (B). Data are represented as mean ± SEM.
We focused our initial characterization on mGluR2, which serves as a standard throughout this study. Previous studies have shown that full-length SNAP-mGluR2 can be efficiently labeled with Alexa647 to allow for visualization of surface receptors (Broichhagen et al., 2015; Vafabakhsh et al., 2015). When ‘‘SNAP-mGluR2-LBD’’ was expressed alone in HEK293T cells and labeled with BG-Alexa647 (membrane-impermeable) followed by BG-TMR (membrane-permeable), fluorescence was only observed in the TMR channel (Figure 2B), suggesting that two LBD populations exist: one population that is still intracellular, likely within the secretory pathway, and another population that has been exported outside of the cell. Since the construct lacks a transmembrane domain, there is no surface population to visualize. We reasoned that co-expression with a full-length GFP-tagged receptor would capture the LBD on the surface and allow us to both quantify the dimer formation based on the Alexa647 fluorescence and normalize for expression levels using the TMR- and GFP-fluorescence levels. When we co-expressed SNAP-mGluR2-LBD with mGluR2-GFP and labeled with both fluorophores, we observed fluorescence across all three channels with clear membrane-targeting for Alexa647 (Figure 2B). As a control, when we expressed mGluR2-GFP alone and labeled it with both fluorophores, we saw no fluorescence in either the TMR- or Alexa647 channels (Figure 2B). To test the ability of our assay to probe dimer specificity, we asked if SNAP-mGluR2-LBD can co-assemble with mGluR1-GFP. Prior studies of mGluR heterodimerization have concluded that group-I and -II mGluRs are unable to co-assemble (Beqollari and Kammermeier, 2010; Doumazane et al., 2011; Levitz et al., 2016). To allow quantitative comparison between conditions, we calibrated mGluR1-GFP expression to match the green fluorescence level that was observed with mGluR2-GFP and, as expected, saw no surface fluorescence in the Alexa647 channel when SNAP-mGluR2-LBD was co-expressed (Figure 2B). In the TMR channel, a similar expression level was observed for SNAP-mGluR2-LBD compared to the homodimer experiment. Figure 2C shows the quantification of this experiment where TMR and GFP levels are similar, allowing Alexa647 to serve as a measurement of dimerization propensity for a given combination of subunits.
We further probed the sensitivity of our assay by testing two previously identified mutants that modify dimer strength. mGluR2–3xLB1 contains three alanine substitutions at conserved residues (L103A, L154A, and F158A) in the hydrophobic interface of the upper lobe of the LBD and has been shown to decrease, but not abolish, mGluR2 dimerization (Levitz et al., 2016). We introduced these mutations into the SNAP-mGluR2-LBD construct and performed our LBD assay with wild-type mGluR2-GFP. We observed a partial reduction in Alexa647 fluorescence while maintaining GFP and TMR fluorescence levels, consistent with a partial reduction in dimerization strength (Figure S2). We also tested the role of a conserved intersubunit disulfide bond in our assay. We previously showed that mGluR2-C121A has modestly decreased dimerization, but that it sensitizes the receptor to mutations elsewhere in the dimer interface (Levitz et al., 2016). Consistent with the study of Beqollari and Kammermeier (2010), we found that SNAP-mGluR2-C121A-LBD showed no surface fluorescence when co-expressed with mGluR2-GFP (Figure S2). This result suggests that the intersubunit disulfide traps the LBD dimer and that there is a sufficient on/off equilibrium between LBDs in the absence of this disulfide such that the isolated LBD can dissociate and become diluted in the extracellular media, thus preventing rebinding. Together, these data support the use of this assay for quantification of mGluR dimerization at the LBD level.
Quantifying the Homo- and Heterodimerization Propensities of Group-II mGluRs
Having established our LBD complementation assay, we next used it to ask what the relative propensity is for mGluR2 heterodimer formation with group-I, -II, and -III mGluRs (Figure 3A). For all conditions we calibrated expression levels to the point where both GFP- and TMR fluorescence intensity levels were similar (Figure S3A). We detected clear Alexa647 fluorescence for SNAP-mGluR2-LBD when co-expressed with group-II and -III mGluRs, but near-background levels with mGluR1-GFP or mGluR5-GFP (Figures 3B and 3C). Interestingly, we saw surprising differences in the level of fluorescence across conditions (Figure 3C; Table S1). For example, SNAP-mGluR2-LBD co-expression with mGluR3-GFP showed about two times higher fluorescence than the mGluR2 homodimerization condition, suggesting that the mGluR2 LBD preferentially heterodimerizes with mGluR3. Further, our data indicate that the mGluR2 LBD dimerizes with mGluR4 with a similar propensity to homodimerization, but has a clear preference for homodimerization relative to heterodimerization with mGluR7.
Figure 3. Homo- and Heterodimerization Propensities of mGluR2 with Other mGluR Subtypes.
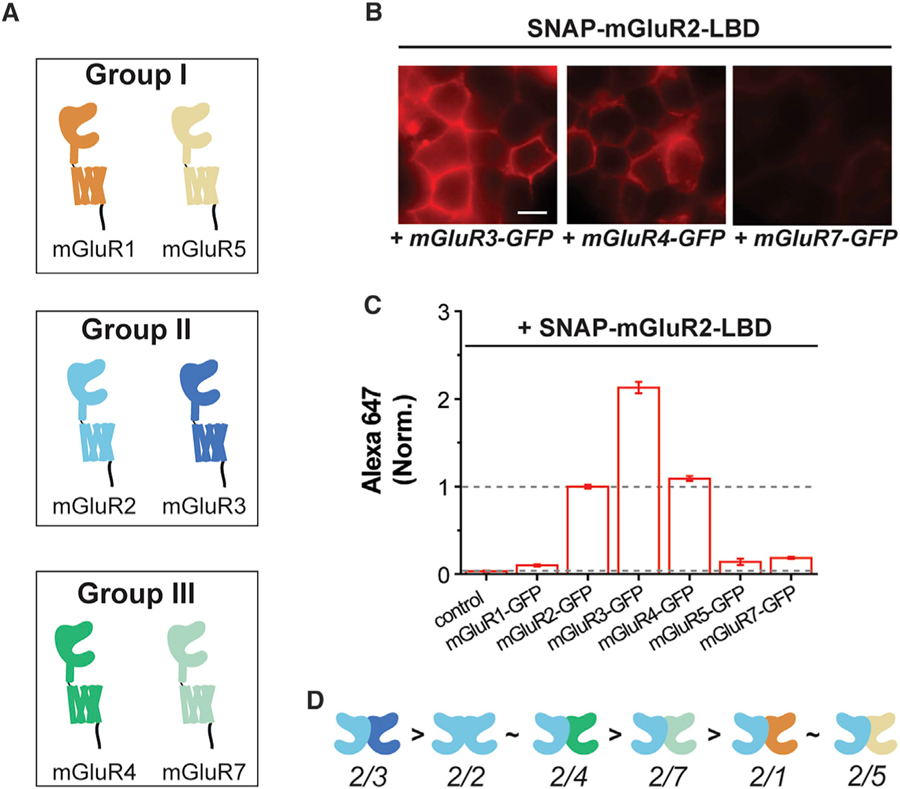
(A) Schematic showing group-I, -II, and -III mGluR subtypes tested in the LBD complementation assay.
(B) Cell images showing SNAP-mGluR2-LBD labeled with Alexa647 when co-expressed with mGluR3-GFP, mGluR4-GFP, or mGluR7-GFP (from left to right). All images are shown on the same fluorescence intensity scale. Scale bar, 10 µm.
(C) Quantification of Alexa647 fluorescence intensity when SNAP-mGluR2-LBD is co-expressed with other mGluR subtypes. Intensity values are normalized to the mGluR2/2 homodimerization condition and data are represented as mean ± SEM.
(D) Schematics showing the relative homo- and heterodimerization propensities of mGluR2.
Given the intriguing variability of dimerization strength across mGluR2 combinations, we asked if the reverse experiment would show the same trend. We produced SNAP-tagged isolated LBD constructs for mGluR1, mGluR3, mGluR4, mGluR5, and mGluR7 and co-expressed each with mGluR2-GFP (Figure S3B). Similar dimer propensities were seen for all combinations compared to the reverse experiment with SNAP-mGluR2-LBD (Figures S3B and S3C). A correlation plot comparing Alexa647 fluorescence levels normalized to the homodimer condition for both directions of the assay shows a clear linear relationship with a slope of ~1 (R2 = 0.98, Figure S3D). This supports our interpretation that mGluR2 has variable heterodimerization propensities and confirms that the direction of the LBD complementation assay does not alter the result. Additionally, we conducted the LBD complementation assay using benzylcytosine (BC)-reactive CLIP-tagged full-length mGluRs instead of the GFP-tagged constructs to allow us to limit the fluorophore labeling to only the surface full-length mGluR population. Dimerization of SNAP-mGluR2-LBD, labeled with BG-Alexa 488, shows the same relative dimerization propensities with CLIP-tagged mGluR1, mGluR2 or mGluR3, labeled with BC-Alexa647, as seen with GFP-tagged constructs (Figures S3E and S3F).
While the LBD complementation assay is quantitative and relatively high-throughput, one limitation is that it is dependent on isolation of the mGluR-LBD. To test if our results hold in full-length mGluRs, we turned to a two-color, single-molecule pull-down (SiMPull) assay. In this assay, labeled receptors from fresh HEK293T cell lysate are immobilized at a sparse level via biotinylated antibodies on the surface of a passivated coverslip and total internal reflection microscopy is used to visualize individual molecules. We previously used this method to confirm that mGluRs form dimers and to show that both inter-LBD and inter-TMD interfaces exist (Gutzeit et al., 2019; Levitz et al., 2016). Here, we co-expressed HA-SNAP-mGluR2 with CLIP-tagged mGluRs to assess relative heterodimerization propensities. Receptors were labeled with BG-LD655 for SNAP and BC-DY547 for CLIP (Figure S4A), and were immobilized via a biotinylated anti-HA antibody (Figure 4A). Expression was calibrated between conditions to maintain a similar SNAP:CLIP ratio (Figure S4B). In principle, any spots in the DY547 channel are due to co-assembly between the CLIP-tagged mGluR and HA-SNAP-mGluR2, but a small background is observed with controls with CLIP-tagged receptors only (Figure S4C). Consistent with the LBD complementation assays (Figure 3), HA-SNAP-mGluR2 pulled down more spots for CLIP-mGluR3 than CLIP-mGluR2, similar amounts for CLIP-mGluR4, less for CLIP-mGluR7, and near-background levels for CLIP-mGluR1 (Figures 4B–4D; Figure S4D). The precision of single-molecule imaging allowed us to observe co-localization of DY547 with LD655 spots and allowed us to detect photobleaching steps as a measure of the number of molecules within a diffraction-limited spot (Figure S4E; see STAR Methods). The vast majority (~90%) of co-localized spots showed photobleaching in one step per color, consistent with formation of strict heterodimers and not higher-order complexes (Figures S4E and S4F). Together, these results provide further validation for the LBD complementation technique and confirm that mGluR2 shows variable assembly propensities across group-II and -III mGluRs with a strong preference for mGluR3.
Figure 4. SiMPull Analysis of Full-Length mGluRs Confirms High mGluR2/3 Hetero-dimer Propensity.
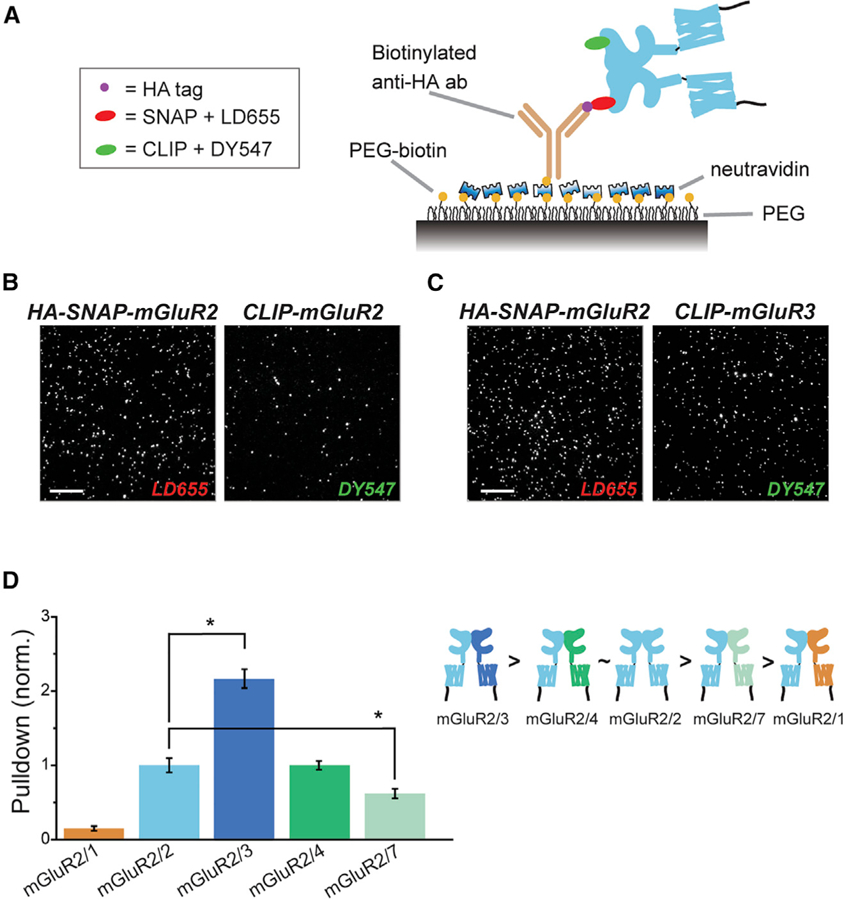
(A) Schematic of two-color SiMPull experiments. Fresh HEK293T cell lysate expressing HA-SNAP-mGluR2 with CLIP-mGluR1, 2, 3, 4, or 7 is added to the polyethylene glycol (PEG) passivated cover glass containing biotinylated anti-HA antibody. SNAP and CLIP tags are labeled with BG-LD655 and BC-DY547, respectively.
(B and C) Representative SiMPull images of HA-SNAP-mGluR2 with CLIP-mGluR2 (B) or CLIP-mGluR3 (C).
(D) Quantification of pull-down via HA-SNAP-mGluR2 normalized to the homodimer condition of HA-SNAP-mGluR2 with CLIP-mGluR2. The asterisk indicates statistical significance (unpaired t test versus mGluR2/2: p = 0.002 for mGluR2/1; p = 0.04 for mGluR2/3, p = 0.05 for mGluR2/7). Relative dimerization strength is shown in a schematic (right). Data are represented as mean ± SEM. Scale bars, 10 µm.
We next applied our assays to assess the homo- and heterodimerization propensities of the other group-II mGluR, mGluR3. Interestingly, when we screened SNAP-mGluR3-LBD across mGluR subtypes in our LBD complementation assay, we found that mGluR3 assembled with mGluR2-GFP, mGluR3-GFP, or mGluR4-GFP with similar propensities, which were higher than the assembly with mGluR7-GFP (Figures 5A and 5B; Table S2). Surprisingly, weak but substantial dimerization was observed between SNAP-mGluR3-LBD and mGluR1-GFP and, to a lesser degree, between SNAP-mGluR3-LBD and mGluR5-GFP (Figure 5B). These results suggest that the rules of assembly of mGluRs are complex and asymmetrical. For example, mGluR2 has a preference for mGluR3 but mGluR3 homodimerizes and heterodimerizes with mGluR2 with a similar efficiency. To support this interpretation, Figure S5B shows the relative fluorescence levels for all four combinations of mGluR2 and mGluR3 in this assay. To test our results in the context of full-length receptor dimers, we again turned to the SiMPull assay. We co-expressed HA-SNAP-mGluR3 with CLIP-tagged mGluR1, mGluR2, or mGluR3 and labeled with BG-LD655 and BC-DY547 (Figures S5C and S5D). HA-SNAP-mGluR3 pulled down a slightly higher, but statistically insignificant amount, of spots for CLIP-mGluR2 compared to CLIP-mGluR3, but significantly less for CLIP-mGluR1 (Figures 5C and 5D). However, the extent of pull-down of CLIP-mGluR1 was above background (Figure S5E), consistent with LBD complementation results (Figure 5B). Similar to what was observed with mGluR2, the vast majority (~90%) of co-localized spots showed photobleaching in one step per color (Figure S5F). Together, these results confirm that mGluR3 shows strong homodimer and heterodimeric assembly propensities with mGluR2.
Figure 5. Dimerization Propensity of mGluR3 via LBD Complementation and SiMPull Analysis.
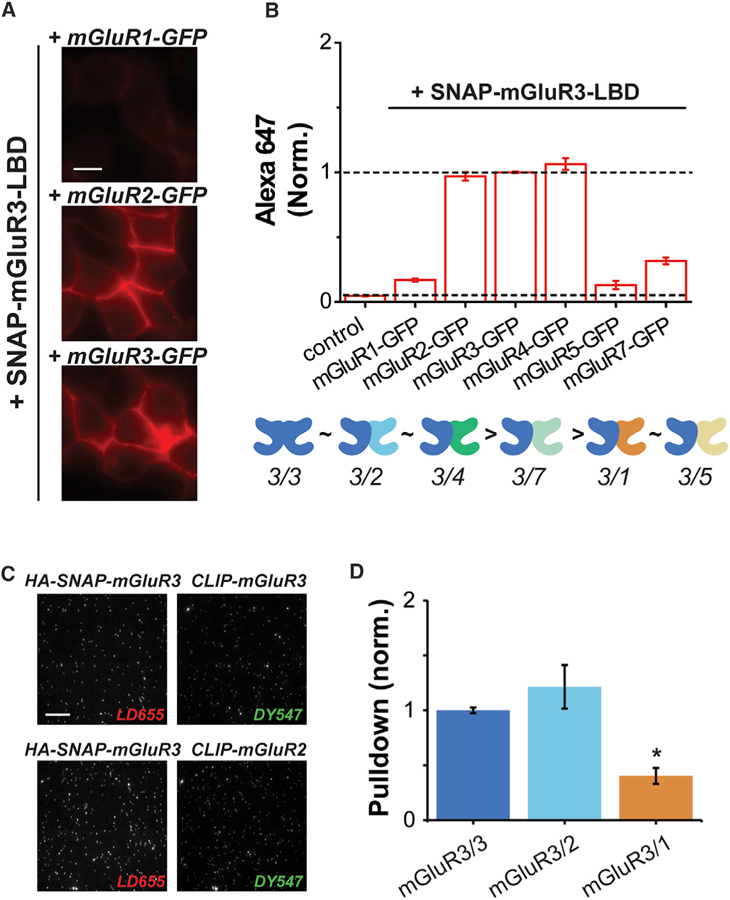
(A) Representative cell images of SNAP-mGluR3-LBD co-expressed with mGluR1-GFP, mGluR2-GFP, or mGluR3-GFP and visualized with Alexa647.
(B) Quantification of SNAP-mGluR3-LBD dimerization with other mGluR subtypes tagged with GFP. BG-Alexa647 fluorescence is normalized to the homodimer condition of SNAP-mGluR3-LBD with mGluR3-GFP and relative homo- and heterodimerization strength is shown as a cartoon below.
(C) Two-color SiMPull images of HA-SNAP-mGluR3 with CLIP-mGluR3 (top) and HA-SNAP-mGluR3 with CLIP-mGluR2 (bottom) labeled with BG-LD655 and BC-DY547, respectively.
(D) Quantification of pull-down via HA-SNAP-mGluR3 when co-expressed with CLIP-mGluR3, CLIP-mGluR2, or CLIP-mGluR1. Pull-down is normalized to the homodimer condition of mGluR3/3. The asterisk indicates statistical significance (unpaired t test versus mGluR3/3: p = 0.26 for mGluR3/2; p = 0.014 for mGluR3/1).
Data are represented as mean ± SEM. Scale bars, 10 µm.
Analysis of Native mGluR2/3 Heterodimerization
Having observed a strong propensity for mGluR2/3 heterodimerization in our assays (Figures 3, 4, and 5) and clear overlap in expression at the single-cell level in the cortex as determined by scRNA-seq (Figure 1; Figure S6A), we decided to further test for native mGluR2/3 heterodimers in the brain. To test for co-expression, we first turned to fluorescent in situ hybridization (FISH) for Grm2 and Grm3 mRNA in mouse brain slices (Figure 6A). We initially focused our analysis on the anterior secondary motor cortex and adjacent cingulate cortex (M2/Cg) subregions (+1.90 from bregma), as these areas are closely related to the area from which scRNA-seq data were collected. Importantly, control probes showed a very weak fluorescence background that did not resemble the fluorescent dots observed using experimental probes (Figure S6B). Using a minimum of five reads per cell as a cutoff, we found that ~40% of cells had co-expression of Grm2 and Grm3 and less than 10% expressed either only Grm2 or only Grm3 (Figure 6B).
Figure 6. In Situ Hybridization and Co-Immunoprecipitation (IP) Characterize Native mGluR2/3 Co-expression and Heterodimerization.
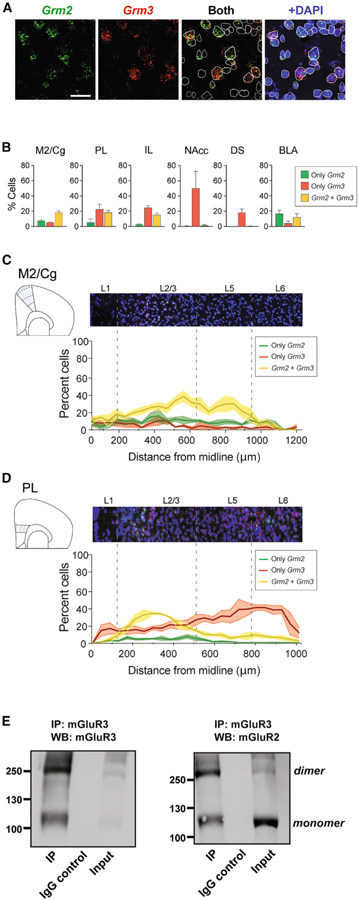
(A) Representative fluorescence in situ hybridization (FISH) for Grm2 (green) and Grm3 (red) RNA and cell delineation using DAPI (blue) in cortical cells. Scale bar, 25 µm.
(B) Bar graphs showing the percentage of cells expressing Grm2, Grm3, or both, in different brain areas.
(C and D) FISH analysis of M2/Cg (C) and PL (D) cortex with quantification of co-expression of Grm2 and Grm3 across different layers. FISH data are analyzed across three mice. Data are presented as mean ± SEM.
(E) Western blots showing that following immunoprecipitation from frontal cortex homogenate with a specific anti-mGluR3 antibody, bands can be detected for both mGluR3 (left) and mGluR2 (right). Controls using an anti-IgG antibody show no immunoprecipitation of mGluR3 or mGluR2. Representative blots are shown; the experiment was repeated three times with identical results.
M2, secondary motor cortex; Cg, cingulate cortex; PL, prelimbic cortex; IL, infralimbic cortex; NAcc, nucleus accumbens; DS, dorsal striatum; BLA, basolateral amygdala.
We next extended our FISH analysis to a set of brain regions where previous studies have identified expression of mGluR2 or mGluR3 (Shigemoto and Mizuno, 2000), although the lack of subtype-specific antibodies prevented separation of the two receptor subtypes in most studies. We first imaged in the prelimbic (PL) and infralimbic (IL) cortices, sub-regions of the medial prefrontal cortex (mPFC) where functional studies have found evidence for contribution from both mGluR2 and mGluR3 (Joffe et al., 2019a, 2019b). In both subregions, 15%–20% of the cells expressed both Grm2 and Grm3 with a similar number of cells expressing Grm3 only and very few expressing Grm2 only (Figure 6B). This suggests that the relative co-expression of mGluR2 and mGluR3 differs depending on cortical subregion. In subcortical regions, we analyzed expression in the nucleus accumbens (NAcc) and dorsal striatum (DS), where we observed a very high population of Grm3-only cells (~50%) with minimal expression of Grm2, and the basolateral amygdala (BLA), where we observed ~15% of cells with co-expression of Grm2 and Grm3 (Figure 6B; Figures S6C and S6D). Moreover, we observed that, in addition to different overall percentages of co-expressing cells in M2/Cg versus PL (Figure 6B), the location of Grm2/Grm3 co-expressing cells across cortical layers differ between M2/Cg and PL cortices (Figures 6C and 6D). Consistent with scRNA-seq data, M2/Cg cells showed high levels of Grm2/Grm3 co-expression across layers with slightly lower levels in layer 6 (Figure 6C). In contrast, in the PL, Grm2/Grm3 co-localization was concentrated to layers 2/3 with lower levels in deeper layers (Figure 6D). Scatterplots of expression level show wide variability from cell-to-cell across all brain regions, but with a higher degree of correlation between mGluR2 and mGluR3 in the cortex compared to subcortical regions (Figures S6E–S6H).
Having observed compelling evidence for co-expression of mGluR2 and mGluR3 throughout the frontal cortex, we sought to test for a direct interaction of mGluR2 and mGluR3 at the protein level using co-immunoprecipitation (coIP). We identified subtype-specific antibodies (Figure S6I) and then confirmed with heterologous expression that the anti-mGluR3 antibody could be used to co-immunoprecipitate mGluR2 from cells expressing both receptor subtypes (Figure S6J). We next performed a coIP from frontal cortex lysate and found that following IP with the anti-mGluR3 antibody, a dimeric band (250 kDa) was detected in a western blot using the anti-mGluR2 antibody, indicating that mGluR2 proteins were co-immunoprecipitated with mGluR3 (Figure 6E). Importantly, a control IP with rabbit IgG did not yield any bands when treated with either anti-mGluR2 or anti-mGluR3 antibodies (Figure 6E; Figure S6J). Together, these data show for the first time that native mGluR2 and mGluR3 heteromerize within the frontal cortex.
Global Analysis of mGluR Homo- and Heterodimerization Propensities
We decided to extend our analysis across the family of mGluRs to define the dimerization propensities of group-I and -III mGluRs using our LBD complementation assay. The LBDs of both mGluR1 and mGluR5 showed specificity for group-I mGluRs with minimal interaction with group-II/III mGluRs (Figures 7A and 7B; Table S3). However, SNAP-mGluR1-LBD showed above-background levels of interaction with mGluR3-GFP (Figure 7A), consistent with the observation of interaction between SNAP-mGluR3-LBD with mGluR1-GFP (Figure 5B). Furthermore, SNAP-mGluR1-LBD showed a preference for heterodimerization with mGluR5-GFP over homodimerization with mGluR1-GFP, providing another example of preferential heterodimerization. In contrast, SNAP-mGluR5-LBD showed no preference for assembly with mGluR1-GFP over mGluR5-GFP (Figure 7B). Group-III mGluRs also showed distinct patterns of dimerization preference. Similar to mGluR2, both SNAP-mGluR4-LBD and SNAP-mGluR7-LBD showed a strong preference for co-assembly with mGluR3-GFP, but also assembled with mGluR2-GFP and other group-III mGluRs (Figures 7C and 7D; Table S3). Additionally, two-color SiMPull of HA-SNAP-mGluR1 co-expressed with CLIP-mGluR1 or CLIP-mGluR5 confirms the preferential heterodimerization of mGluR1 with mGluR5 in full-length receptors (Figures S7E–S7I).
Figure 7. Dimerization Propensity across Group-I and Group-III mGluRs.
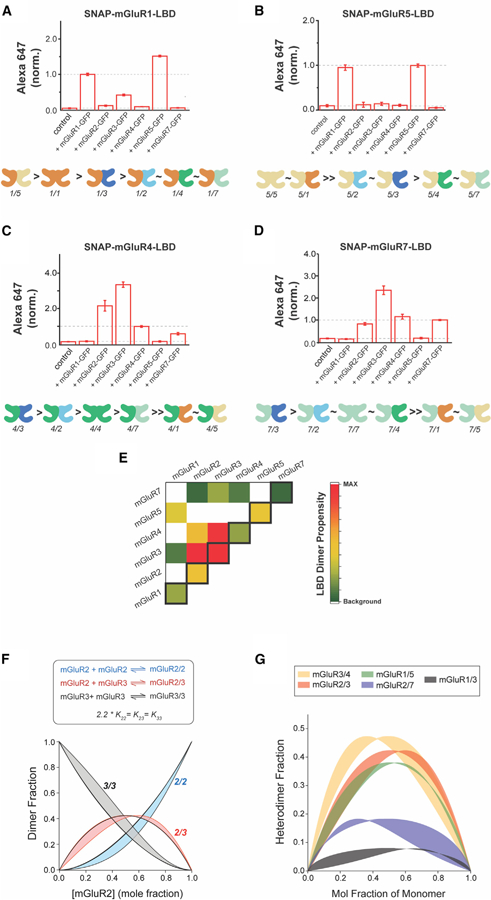
(A–D) Quantification of Alexa647 fluorescence showing relative dimerization propensities of SNAP-mGluR1-LBD (A), SNAP-mGluR5-LBD (B), SNAP-mGluR4-LBD (C), and SNAP-mGluR7-LBD (D) with other mGluR subtypes. Alexa647 fluorescence intensity is normalized to each homodimer condition and a summary of the relative homo- and heterodimerization strengths is shown as a cartoon below each bar graph. Data are represented as mean ± SEM.
(E) Heatmap providing a global summary of LBD dimerization propensities across all subtypes tested. Values come from SNAP-mGluRX-LBD dimer screens (Figures 3, 5, and 7) and are normalized to the relative homodimerization strengths obtained from side-by-side comparison in the LBD complementation assay (Figure S7J). Colored boxes show dimer combinations that show statistically significant values above background levels in both directions of the assay (i.e., SNAP-mGluRX-LBD + mGluRY-GFP and SNAP-mGluRY-LBD + mGluRX-GFP).
(F) Kinetic modeling of mGluR2 homo- and heterodimerization using a wide range of protein concentrations and absolute values for equilibrium constants while maintaining a fixed ratio among K22, K23, and K33. The black line is the simulation for the conditions outlined in Figures S7F–S7H and the envelope of distributions shows the range of results obtained when varying the total protein concentration and the raw value of equilibrium constants.
(G) Kinetic modeling of mGluR heterodimerization across five different pairs. Envelopes of distributions are shown over a wide range of conditions while maintaining the relative magnitudes of the equilibrium constants (see STAR Methods for values).
Finally, we sought to provide a global comparison of LBD dimerization propensities for all combinations tested. We first used the LBD complementation assay to compare homodimerization propensities between all subtypes when expressed to similar levels (Figure S7J; Table S3). We found a range of homodimerization propensities in this measurement, with mGluR3 showing the strongest apparent dimerization and mGluR7 showing the weakest. We then used these relative homodimer strengths to normalize the values obtained for each combination in the LBD complementation assay (Figures 3, 4, and 7) and constructed a heat-map to summarize all of the data, where we find that mGluR2/3, mGluR3/4, and mGluR1/5 are the most favorable heterodimeric combinations (Figure 7E).
Together, these data show a range of homo-and heterodimerization propensities across different dimer pairs, suggesting a complex relationship among receptor expression level, homodimer formation, and heterodimer formation that produces a different complement of receptor subtypes in any given neuron. Given the broad expression range between subtypes across different cells observed in scRNA-seq (Figure 1; Figure S1) and FISH analysis (Figure 6; Figure S6), we decided to model the relative population of homodimers and heterodimers for a given expression ratio, starting with mGluR2 and mGluR3 (see STAR Methods). mGluR2 homodimerization was analyzed over a range of protein concentration to estimate the equilibrium constant (K22) required to produce substantial (>80%) dimerization (Figure S7K). mGluR3 homodimerization and mGluR2/3 heterodimerization were then included using relative equilibrium constants based on our experimental data (Figure 7F; Figure S7L). Plots of the relative proportion of homodimers and heterodimers for a given ratio of mGluR2 subunits to mGluR3 subunits revealed a substantial population of heterodimers over a broad range of the mGluR2 mole fraction, peaking at 40% of the dimer population (Figures S7L and S7M). We then asked if the total protein concentration and absolute magnitude of the equilibrium constants are major determinants of this distribution by modeling the subunit equilibration over a wide range of each parameter while maintaining the ratio of equilibrium constants (2.2 × K22 = K23 = K33). The relative distribution of homo- and heterodimers was largely insensitive to variations of equilibrium constants or protein concentration over five orders of magnitude, indicating that the relative dimerization propensities are the major determinant of homo- and heterodimerization.
While this model does not include the formation of covalent intersubunit disulfide bonds or effects of trafficking or regulatory mechanisms at the cellular level, it provides a useful assessment of heterodimerization propensity and its sensitivity to relative abundance of the monomer subtypes with minimal assumptions. We extended this analysis to several other mGluR pairs where a range of relative heterodimer populations was observed (Figure 7G), with a maximum heterodimer population (~50%) observed for mGluR3/4 heterodimers. Together, these analyses indicate that a substantial heterodimer population can exist across a broad range of expression conditions.
DISCUSSION
This study establishes a methodological framework for quantitatively probing the co-expression and co-assembly of mGluRs into different homo- and heterodimeric pairs, and produced a number of new insights into mGluR assembly, which, together, strongly argue for the biological relevance of heterodimerization. Based on our data, we argue that mGluR2/3 heterodimers are prominent throughout the frontal cortex and form readily with similar or enhanced efficiency compared to homodimers. Together, this motivates further structural, pharmacological, and functional analysis of this complex. Previous work has shown that mGluR2/3 heterodimers show an intermediate glutamate affinity compared to parent homodimers and show a similar level of basal activity compared to mGluR3 homodimers (Levitz et al., 2016). Future work is needed to fully characterize the sensitivity of this complex to mGluR2- or mGluR3-specific drugs, identify the cellular localization of heterodimers, and probe the downstream signaling dynamics compared to parent homodimers. In addition, our data argue for prominent co-expression and co-assembly of other mGluR pairs, including mGluR2/4, mGluR1/5, mGluR3/4, and mGluR3/7, raising similar questions for each of these complexes, which have each only been partially characterized.
The ability to harness scRNA-seq data to analyze the expression patterns of cortical mGluRs provides a valuable perspective on receptor function within cortical circuits. Recent studies have begun to pinpoint the morphological and electrophysiological identities of molecularly defined neuronal subtypes within the cortex (Cadwell et al., 2016; Fuzik et al., 2016; Gouwens et al., 2019; Muñoz-Manchado et al., 2018; Naka et al., 2019), but a precise understanding of how specific signaling molecules segregate into these cellular subtypes is lacking. Here we find that all mGluR subtypes other than mGluR6 are expressed in pyramidal neurons, but a sparser expression pattern is seen in interneurons and non-neuronal cells. The most important observation enabled by scRNA-seq is the fact that many Grm subtypes are co-expressed within the same cell (Figure 1F), including the majority of pyramidal cells, which contain at least four receptor subtypes. mGluR co-expression was most prominent in L5 IT neurons, which are known to project to other cortical areas, suggesting that mGluR-mediated neuromodulation is particularly critical for shaping communication between cortical microcircuits. While expression is sparser, many interneurons also show expression of multiple Grm subtypes.
Given this extensive degree of co-expression, it is unsurprising that for any specific pair of receptors, many potential cellular sites of heterodimerization exist. For example, mGluR1 and mGluR5 are co-expressed in both glutamatergic (highest in L2/3 IT) and GABAergic (highest in Sst+) neuronal subclasses. The ability of cells to contain many G protein-coupled sensors of glutamate leads to a wide diversity of cellular responses to different patterns of excitatory neurotransmitter release that may be critical for the delicate balance of excitation and inhibition that the cortex must maintain. Interestingly, the most frequently co-expressed pair of mGluRs is mGluR5 and mGluR3, which we show do not efficiently heterodimerize (Figures 5 and 7) and are coupled to different G protein pathways, but have been suggested to functionally crosstalk in cortex (Di Menna et al., 2018). The heterodimer we focused on, mGluR2/3, showed co-expression exclusively in pyramidal neurons with the highest levels in L2/3 and L5 IT neurons and lower levels in L6 subtypes (Figure 1D), suggesting many possible locations for co-assembly of this complex.
The analysis of FISH data from many brain regions shows that group-II mGluR co-expression is prominent within the cortex and reduced in subcortical regions, with some cells (~10%) showing co-expression in the basolateral amygdala, but almost zero mGluR2 expression observed in both dorsal and ventral striatum (Figure 6). These expression patterns are generally consistent with previous studies (Ohishi et al., 1993a, 1993b), but here we provide single-cell resolution with both probes in the same preparation to allow a quantitative analysis of receptor co-expression. While all cortical regions showed prominent co-expression of mGluR2 and mGluR3, subtle differences in layer distribution between cortical areas were found, suggesting that, depending on the cortical region, the role of mGluR2/3 heterodimers in regulating both the local microcircuit and the long-distance inputs and outputs may be different. Importantly, analysis of co-expression at the RNA level still raises the questions of whether mGluR2/3 heterodimers actually form, and where, within the cell, such complexes may be localized. An electron microscopy study in the PFC of primates has shown evidence for both receptor subtypes in both pre- and post-synaptic compartments, but with a higher proportion of mGluR3 found in the post-synapse (Jin et al., 2018). Importantly, we report a coIP of mGluR2/3 from the frontal cortex at the protein level (Figure 6) to confirm that this complex indeed forms in vivo. Recent studies have shown evidence that both mGluR2 and mGluR3 provide inhibition within the PFC, with both receptors contributing to different forms of synaptic plasticity and producing anti-depressant effects in response to subtype-specific negative allosteric modulators (Joffe et al., 2019a, 2019b). Future work will be needed to disentangle the relative contributions of homo- and heterodimers in different cell types and synaptic locations to such cortical functions. It’s important to note that our expression analysis was performed in samples from mice, raising the question of how well our findings extend to humans. Unfortunately, FISH analysis of mGluRs in human samples has been limited and has lacked single-cell resolution for a thorough investigation of subtype co-expression. Studies focusing on mGluR2 and mGluR3 expression in frontal cortex (Ghose et al., 2008; Makoff et al., 1996; Ohnuma et al., 1998) show a layer distribution that broadly resembles that of our mouse cortical samples and a human in situ RNA hybridization study showed weak mGluR2 and strong mGluR3 expression in striatum (Cha et al., 1998), consistent with our data. A recent scRNA-seq study comparing human and mouse cortical expression patterns (Hodge et al., 2019) found high conservation of cell types but divergence in expression patterns for many signaling proteins, motivating future analysis of human mGluR expression patterns.
The fluorescence-based LBD complementation assay reported here provides quantitative information about the relative efficiency of dimer formation between different subtypes and should be broadly useful across the class-C GPCR family. However, it’s important to note that the dimerization signal in this assay is driven solely by interactions at the LBD level and, furthermore, is dependent on the formation of an intersubunit disulfide. This could lead to an over-reliance on the contribution of inter-LBD interactions and the formation of an intersubunit disulfide, which is not strictly necessary for dimerization in full-length mGluRs (Levitz et al., 2016). Critically, SiMPull experiments using full-length mGluRs showed the same rank order of dimer preference for mGluR2 and mGluR3 (Figures 3 and 5), confirming the general validity of the LBD complementation assay for screening. Interestingly, the mGluR2/7 interaction is somewhat stronger in the SiMPull assay suggestion (~50% of mGluR2/2 versus ~20% in the LBD assay) that the TMD may weakly enhance the relative dimer strength.
The global LBD dimerization propensities measured across all subtypes tested (Figure 7E) generally agree with previous studies that group-II/III mGluRs preferentially interact with each other over group-I mGluRs. However, we find some discrepancies compared to a previous study of mGluR heterodimerization (Doumazane et al., 2011), including more prominent heterodimerization of mGluR3. This is likely a result of the previous study depending on a FRET-based detection method that has been shown to be highly sensitive to receptor conformation (Doumazane et al., 2013). For example, high basal conformational dynamics of mGluR3-containing dimers leads to substantial population of a low FRET state (Levitz et al., 2016; Tora et al., 2018; Vafabakhsh et al., 2015), likely leading to an underestimate of dimerization in the FRET-based assay.
The kinetic modeling reported here provides further clarity about the assembly of different heterodimeric pairs with two major principles emerging. (1) It is clear that the relative values of equilibrium constants for different dimer pairs is the main determinant of the relative population of homo- and heterodimeric populations. Altering the total protein concentration or absolute value of equilibrium constants only altered the relative shape of the curves in Figure 7G, but the maximum occupancy of the heterodimer was maintained over a very wide range of values. Interestingly, our model also suggests that large changes in relative receptor expression levels are needed to substantially shift the relative population of homo- and heterodimers. For example, in the case of mGluR2/3 heterodimers, one would need to alter the heterodimer fraction from ~40% to ~25% by altering the mGluR2:mGluR3 expression ratio by a factor of 3, from 1:1 to 1:3. Such changes in expression may take place over development or in response to strong stimuli, which can then, in turn, alter the relative signaling properties of a cell by altering its distribution of receptor species. (2) In order to produce a substantial sub-population of heterodimers, it is critical that one of the subunits has a comparable, or enhanced, propensity for heterodimerization compared to homodimerization. For example, both mGluR4 and mGluR2 showed a preference for heterodimerization with mGluR3, which led to substantial population of the heterodimer (up to ~50% of the dimer population). A similar scenario is seen with group-I mGluRs, where mGluR1 shows a preference for heterodimerization with mGluR5. Interestingly, we did not identify any combinations where the heterodimer is preferred for both subunits, suggesting that mGluR homodimer populations should always be substantial. Importantly, our model fails to take into account trafficking and other regulatory mechanisms, which likely further shape the assembly of homo- and heterodimers.
Together, these results raise the question of what the biophysical mechanisms of specific and preferential assembly between subtypes are. Previous structural and mutagenesis work identified a highly conserved hydrophobic interface between the upper lobes of the LBD, which provides much of the energy for dimerization (Levitz et al., 2016). The high conservation of this interface suggests that a simple lock-and-key model of specific binding between complementary pairs is likely insufficient to explain why some combinations can or can’t form. Interestingly, mGluR3 is consistently the strongest dimerization partner within the mGluR family and is also the receptor subtype with the most basal population of the active state (Vafabakhsh et al., 2015). On the other hand, mGluR7 shows consistently weak dimerization (including homodimerization) and has been found to have the weakest population of the active state (Habrian et al., 2019). Given the more extensive dimer interface that forms in the active state (Koehl et al., 2019; Kunishima et al., 2000; Muto et al., 2007; Tsuchiya et al., 2002), it’s likely that the relative stability of this conformation contributes to dimerization. Together, these observations suggest that it is likely that there is interplay between specific interactions within the interface and some contribution from intersubunit conformational dynamics. Previous work has shown that modulating inter-LBD strength at both the covalent and non-covalent interfaces can tune receptor activation and conformational dynamics (Levitz et al., 2016), further linking receptor assembly and conformational dynamics. Ultimately, future work harnessing the power of the assays developed here, along with structural and spectroscopic studies, will be needed for deciphering the rules of engagement of mGluRs.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Joshua Levitz (jtl2003@med.cornell.edu)
Materials Availability
Requests for resources and reagents will be fulfilled by the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
This study did not generate large datasets or code. The scRNaseq database used in this study is available for download (https://portal.brain-map.org/atlases-and-data/rnaseq). All the newly generated datasets are available from the Lead Contact on request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell cultures of HEK293T
HEK293T cells were purchased from ATCC (CRL-11268), authenticated by Bio-Synthesis, Inc. and tested negative for mycoplasma using a kit (Molecular Probes). Cells were maintained in DMEM (GIBCO) supplemented with 5% fetal bovine serum and passaged by trypsin/EDTA digestion.
Mice
All animal use procedures were performed in accordance with Weill Cornell Medicine Institution Animal Care & Use Committee (IA-CUC) guidelines under approved protocol 2017–0023. Male wild-type C57BL/6J mice were purchased from Jackson Laboratory and maintained under pathogen free conditions at the Weill Cornell Medicine Animal Facility. Animals were provided food and water ad libitum and housed in a temperature and humidity controlled environment. For FISH experiments, male mice between 8–10 weeks of age were used. For co-IP experiments, male mice between 4–5 weeks of age were used.
METHOD DETAILS
Single cell RNA sequencing analysis
Details regarding preparation, processing and clustering of the cells used for scRNaseq can be found in the original resource paper (Tasic et al., 2018). All cells passed the quality control criteria and underwent subsequent hierarchical clustering according to the similarity of their individual transcriptomes. Classes, subclasses and subtypes (in the original paper named clusters) were referred to following their original nomenclature. All cells included here were dissected (and clustered) from the mouse ALM cortex: 4447 glutamatergic neurons, 4398 GABAergic interneurons and 308 non-neuronal cells. Small clusters were excluded from the entire analysis (Meis2-Adamts, 56 cells, and CR-Lhx5, 18 cells), as well as microglia and cells of the vascular system. For co-expression analysis, two subtypes with less than 15 cells were excluded (L6b Hsd17b2 and L6bP2ry12 cells). For co-expression analysis for the 28 possible Grm pairs, we took into consideration previously described aspects (see Results) of low false positives and under-sampling of a cell total mRNA with this technique, thus imposing a threshold of five copies per million (CPM). Data were analyzed using R Studio and Microsoft Excel.
Cloning
All SNAP- and CLIP-tagged mGluR clones were made by modifying previously reported constructs (Doumazane et al., 2011). The LBD constructs were made by introducing a stop codon at R521 for mGluR1 (human), A497 for mGluR2 (rat), T506 for mGluR3 (rat), R517 for mGluR4 (human), R507 for mGluR5 (rat), and S520 for mGluR7 (rat). C-terminally GFP-tagged mGluR constructs with a flexible 16 aa linker (TSGGSGGSRGSGGSGG) were made using a Gibson assembly kit (NEB). Mutations were introduced by site directed mutagenesis using Phusion High-Fidelity PCR Kit (Thermo Scientific).
Expression
HEK293T cells were seeded on 18 mm poly-L-lysine-coated coverslips in a 12-well plate and transfected using Lipofectamine 2000 (Thermo Scientific). For LBD complementation, cells were transfected with 0.6 µg/well of SNAP-tagged mGluR LBDs and 0.3–0.5 mg DNA/well of GFP-tagged mGluRs. For SiMPull experiments, cells were transfected with 0.3 µg/well of SNAP-tagged mGluRs and 0.6 µg DNA/well of CLIP-tagged mGluRs. For co-IP experiments, cells were transfected with 0.6 µg/well of wild-type, untagged mGluRs.
Live cell fluorescence imaging
After 36–48 h of expression, cells were washed with EX solution containing (in mM): 10 HEPES, 135 NaCl, 5.4 KCl, 2 CaCl2, 1 MaCl2, pH 7.4 and labeled with 1 µM BG-Alexa647 in EX for 45 min at 37°C followed by 1 µM SNAP-TMR in EX for 30 min at 37°C. An inverted microscope (IX83) was used for fluorescence imaging. 488 nm, 561 nm, and 640 nm lasers were used to excited GFP, TMR and Alexa647 and images were captured using a 60x objective (NA 1.49). Average fluorescence intensities from live cell images were measured using ImageJ by drawing a region of interest (ROI) around cell clusters for each color of fluorescence image (GFP, TMR, and Alexa647). Fluorescence intensity values from multiple images were then averaged. For each hetero-dimer condition, intensity values were normalized to the corresponding homodimer condition on the same day. Each condition was tested in at least 3 separate transfections.
Two color single molecule pulldown assay
Two color single molecule pulldown (SiMPull) was performed to visualize and quantify mGluR heterodimers. HA-tagged mGluRs were isolated on passivated glass coverslips as previously described using a biotinylated anti-HA antibody (Gutzeit et al., 2019). Prior to each experiment, flow chambers were incubated with 0.2 mg/ml NeutrAvidin for 2 min then incubated with 10 nM of antibody (abcam, ab26228) for 30 min. The flow chambers were rinsed with T50 buffer (50 mM NaCl, 10 mM Tris, pH 7.5) after each conjugation step. Cell lysate was prepared ~48 hr after transfection with HEK293T cells labeled at 37°C with 1 µM BG-LD655 followed by 1 µM CLIP-DY547 for 45 min. After washing with EX solution, cells were harvested using Ca2+ free-DPBS for 20 min at 37°C. After pelleting at 10,000 x g, 4°C for 1 min, cells were lysed using 1.2% IGEPAL detergent for 1 hour at 4°C. Next, cells were centrifuged at 16,000 x g for 20 min at 4°C and supernatant was collected. Samples were then diluted using EX buffer containing 0.1% IGEPAL and introduced to the flow chamber. After obtaining an optimal number of spots in the field of view, the chamber was washed with the dilution buffer to remove unbound proteins.
Single molecule imaging was done using a 100x objective (NA 1.49) on an inverted microscope (Olympus IX83) in total internal reflection mode at 20 Hz with 50 ms exposure times with an sCMOS camera (Hamamatsu ORCA-Flash4v3.0). Samples were excited with 561 nm and 640 nm lasers to excite BC-DY547 and BG-LD655, respectively. Briefly, we counted the total number of spots in each channel and subtracted the number of non-specific background spots determined from control pulldowns of CLIP-tagged receptor only. These values were then normalized to the corresponding homodimer condition (i.e., SNAP-mGluR22 + CLIP-mGluR2 or SNAP-mGluR3 + CLIP-mGluR3) from the same day. Values obtained from at least 3 separate experimental days were then averaged to produce bar graphs in Figures 4D and 5D. Co-localized spots were identified by overlaying movies from the same region in the DY547 and LD655 channels. Bleaching step analysis of these co-localized spots was performed as previously described (Gutzeit et al., 2019).
Fluorescence in situ hybridization
Fluorescence in situ RNA hybridization (FISH) was performed using an RNAscope Fluorescent Multiplex 2.5 labeling kit (ACD Bio). Briefly, brains were extracted and fresh frozen on dry ice before 10 µm sections were prepared using a cryostat. Probe hybridization was performed using Mm-Grm3-C2 for Grm3 (C2-Atto 550), Mm-Grm2-C3 for Grm2 (C3-Atto 647N), and DAPI counterstain was used for nuclei. Probe Mm-Grm3-C2 targets exons 1 of the Grm3 gene at base pairs 178–1183, and probe Mm-Grm2-C3 targets exons 1 and 2 of the Grm2 gene at base pairs 477–1726. RNAscope procedures were completed to manufacturers’ specifications (ACD Bio). For analysis, slides were imaged using an Olympus Confocal Laser Scanning Microscope (FV3000) with 410 nm laser for DAPI, 525 nm laser for Grm3 probe, and 625 nm laser for Grm2 probe, where all settings were maintained across different animals and brain regions. Images were taken at 10× magnification at 1048 pixels per square area. To capture entire brain regions (see Figure S6C and S6D), 6–12 images were taken per field and stitched together using the Olympus Confocal FluoViewFV3000 Software. Images from three different animals were used, and for most regions one image from each hemisphere was used from each animal for each brain region, and the results from both hemispheres were averaged. Quantification of number of reads (puncta) per cell for each probe was performed using Imaris Software (Oxford Instruments), in which settings for puncta detection varied at a minimum level across different animals. Post hoc data analysis consisted of stipulating a cutoff of minimum 5 reads throughout all brain areas, as a means to being stringent and limiting false positives. Such cutoff has been used before for highly expressed genes, such as parvalbumin (Pvalb) in inhibitory GABAergic neurons (Muñ oz-Manchado et al., 2018). For layer distribution of cells in M2/Cg and PL, cells were separated in bin according to their layer location and cell percentages were calculated out of the number of cells in each bin. Data processing and analysis were performed using Microsoft Excel, Graphpad Prism, ImageJ, and R Studio.
Co-immunoprecipitation and western blot analysis
For co-IP of heterologously-expressed receptors from HEK293T cells, lysis was performed at 4°C with lysis buffer (150mM NaCl, 10mM Tris HCl, 1mM EDTA, 1.2% IGEPAL) supplemented with protease inhibitor tablets (Pierce) and incubated for 1 hour at 4°C. The lysate was pre-cleared with protein A/G beads (Pierce) at 4°C for 1 hour. mGluR3 antibody (Abcam, ab166608) or isotype control antibodies (rabbit IgG, EPR25A) were bound to the protein A/G beads by rotating at 4°C for 1 hour. The pre-cleared lysate was then added to the antibody-bound protein A/G beads and incubated overnight, rotating at 4°C. The beads were then washed with lysis buffer and pelleted at 700 x g for 5 min. To elute the proteins, 10 µL of lysis buffer, 5 µL of NuPAGE™ LDS Sample Buffer 4x (Thermo Scientific) and 0.5 M DTT were added and the samples were heated at 95°C for 5 min. The samples were run on a gel, transferred and imaged as described below.
For co-IP of native receptors, mice were decapitated, brains were rapidly removed, and the frontal cortex was dissected out. Samples from 8–10 mice were combined and homogenized in a tissue grinder with 10 µL NP-40 lysis buffer (150mM sodium chloride, 50mM Tris hydrochloride, 1.0% NP-40, pH 8.0) per mg of cortical sample supplemented with protease inhibitor and incubated for 1 hour at 4°C. Lysate was then centrifuged at 1300 x g for 1 hour at 4°C and the supernatant was collected. The brain lysate was pre-cleared with protein A/G beads for 1 hour at 4°C. mGluR3 antibody (Abcam, ab166608) or isotype control antibodies (recombinant rabbit IgG, EPR25A) were bound to the protein A/G beads by rotating at 4°C for 1 hour. The precleared lysate was then added to the antibody-bound protein A/G beads and incubated overnight, rotating at 4°C. The beads were then washed 5 times with NP-40 lysis buffer and pelleted using 700 g spins for 5 min. Beads were washed, protein eluted and the samples were analyzed through western blotting.
Lysate protein concentration was quantified using a Bio-Rad DC Protein Assay. Lysate containing 40 µg of protein were incubated for 30 min at room temperature with NuPAGE LDS Sample Buffer 4x (NP0007) and 0.5M DL-dithiothreitol (VWR). Samples were then loaded on a Bolt 4%–12% Bis-Tris Plus gel (Thermo Fisher), the chamber was filled with 1X NuPAGE MOPS SDS running buffer (Thermo Fisher) and run at 100mV for 2.5 hours. Samples were transferred in 1X NuPAGE transfer buffer (Thermo Fisher) at 370mA for 1 hour and 50 min at 4°C onto a Bio-Rad Immun-Blot PVDF membrane (Thermo Fisher). The membrane was rinsed in TBST buffer then blocked for 30 min in TBST containing 5% Blotting-Grade Blocker (Bio-RAD) and 5% fetal bovine serum at room temperature. Anti-mGluR2 antibody (Abcam, ab15672) and anti-mGluR3 antibody (Abcam, ab166608) were diluted 1:5000 in 3% Blotting-Grade Blocker overnight at 4°C. The membrane was washed 3 times with TBST, then incubated in HRP-conjugated goat anti-mouse IgG secondary antibody (Abcam, ab6789) diluted 1:3000 in 3% Blotting-Grade Blocker or incubated in HRP-conjugated goat anti-rabbit IgG cross-adsorbed secondary antibody (Thermo Scientific) diluted 1:10,000 in 3% Blotting-Grade Blocker for 1 hour at room temperature. Membranes were then washed 3 times with 1x TBST and incubated with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific), then imaged using a Syngene G-Box Chemi XX6 imager. Images were analyzed using ImageJ. Each condition was tested in at least two separate experiments.
Kinetic modeling
Equilibrium homo- and hetero-dimer complexes were estimated using a kinetic model assuming single-step bimolecular reactions between monomers starting with a pure monomer population. For example, mGluR2 (m2) and mGluR3 (m3) dimerization reactions (homodimers R22 and R33; heterodimer R23) were defined as follows:
Distinct equilibrium constants (K22, K23, and K33) were chosen for each dimerization reaction based on the data described in Figure 7E, which determine their relative but not absolute magnitudes. Off-rates were defined based on these values: b22 = a22/K22, b23 = a23/K23, and b33 = a33/K33. The initial conditions with total monomer T and mGluR2 mole fraction x2 were defined as:
The total number of monomers is given by T = m2 + m3 + 2· (R22 + R23 + R33), and is constant throughout the simulation. This model was implemented in Igor Pro (Wavemetrics), and numerical integration (Euler method) was run until all species reached steady values using identical on-rates for all dimerizations (a22 = a23 = a33 = 1 in dimensionless units). Simulations were repeated for 100 values of x2 ranging from 0 to 1 and the equilibrium proportions of each dimer were computed. Because neither the total receptor protein concentration T nor the absolute equilibrium affinity K22 were measured in this study, the simulations were repeated across a broad range of possible T and K22 values. In dimensionless units, T ranged from 0.01 to 100 and K22 ranged from 0.5 to 5000. The heterodimer fraction (relative to the total dimer pool) was defined as R23/(R22 + R23 + R33) and plotted versus x2 for several choices of T and K22 to generate an envelope of possible values as shown in Figure 7F. Note that heterodimer propensity was largely insensitive to total protein concentration or the absolute equilibrium constant value, indicating that heterodimerization is largely determined by the relative affinities of monomers. The analysis was repeated for other dimer pairs including mGluR3/mGluR4, mGluR2/mGluR7, mGluR1/mGluR5, and mGluR1/mGluR3. The equilibrium constant ratios used in these simulations were: 2/3 (1:2.2:2.2), 3/4 (1:1:0.32), 1/5 (1:1.5:1.5), 2/7 (1:0.2:0.2), and 1/3 (1:0.33:3.67).
QUANTIFICATION AND STATISTICAL ANALYSIS
LBD complementation and SiMPull data were analyzed using ImageJ, Microsoft Excel, and Origin Pro. All conditions in both experiments were tested in at least 3 separate transfections. For fluorescence in situ hybridization, images from 3 different animals were used, and 1 image from each hemisphere was used from each animal for each brain region, and the results from both hemispheres were averaged. Analysis of number of probe hits for each cell and each probe was performed using Imaris Software (Oxford Instruments), and post hoc analysis was performed using Microsoft Excel, Graphpad Prism, ImageJ, and R Studio. For Co-IP and Western Blot Analysis, images were analyzed using ImageJ. Each condition was tested in at least two separate experiments. Kinetic modeling was performed using Igor Pro.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HA tag antibody (Biotinylated) | abcam | Cat#ab6438; RRID:AB_2115899 |
| mGluR3 antibody | abcam | Cat#ab166608; RRID:AB_2833092 |
| mGluR2 antibody | abcam | Cat#ab15672; RRID:AB_302021 |
| recombinant rabbit IgG | abcam | Cat#ab172730; RRID:AB_2687931 |
| Goat Anti-Mouse IgG H&L (HRP) | abcam | Cat#ab6789; RRID:AB_955439 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| NeutrAvidin Protein | Thermo Fisher Scientific | Cat#31000 |
| benzylguanine (BG)-LD655 | Gutzeit et al., 2019 | N/A |
| SNAP-Surface Alexa Fluor 647 | New England Biolabs | Cat#S9136S |
| CLIP-Surface 547 | New England Biolabs | Cat#S9233S |
| SNAP-Surface 488 | New England Biolabs | Cat#S9232S |
| CLIP-Surface 647 | New England Biolabs | Cat#S9234S |
| SNAP-Cell TMR star | New England Biolabs | Cat#S9105s |
| mPEG | Laysan Bio | Item# BIO- PEG-SVA-5K- And MPEG- SVA- 5K |
| biotinylated mPEG | Laysan Bio | Item# BIO- PEG-SVA-5K- 100MG and MPEG-SVA- 5K |
| IGEPAL | Sigma Aldrich | I8896–50 |
| DMEM | Thermo Fisher Scientific | Cat#11995073 |
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat#10437028 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat#11668–019 |
| Poly-L-lysine hydrobromide | Sigma Aldrich | P2636 |
| Mm-Grm2-C3 | ACD/Biotechne | Cat#317831-C3 |
| Mm-Grm3-C2 | ACD/Biotechne | Cat#317821-C2 |
| Critical Commercial Assays | ||
| RNAscope Multiplex Fluorescent Reagent Kit v2 4-plex | ACD/Biotechne | Cat#323120 |
| Experimental Models: Cell Lines | ||
| HEK293T | American Type Culture Collection (ATCC) | ATCC Cat# CRL-11268 RRID: CVCL_1926 |
| Experimental Models: Organisms/Strain | ||
| Mice C57BL/6J Strain | The Jackson Laboratory | Stock No:000664 (IMSR Cat# JAX:000664, RRID:IMSR_JAX:000664) |
| Recombinant DNA | ||
| SNAP- and CLIP-tagged mGluRs | Doumazane et al., 2011 | N/A |
| GFP-tagged mGluRs | Levitz et al., 2016 | N/A |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | RRID:SCR_003070 |
| Origin | Origin Lab Corporation | RRID:SCR_002815 |
| Microsoft Excel | Microsoft Office | RRID:SCR_016137 |
| Adobe Illustrator | Adobe | RRID:SCR_010279 |
| Olympus CellSens | Olympus | RRID:SCR_016238 |
| LabVIEW | National Instruments | RRID:SCR_014325 |
| IMARIS 9.5 | Bitplane | RRID:SCR_007370 |
| RStudio | RStudio, inc. | RRID:SCR_000432 |
| IgorPro | Wavemetrics | RRID:SCR_000325 |
Highlights.
Single-cell RNA sequencing shows overlapping mGluR expression throughout cortex
Fluorescence complementation assays enable quantification of mGluR dimerization
Many heterodimers form with higher efficiency than parent homodimers
mGluR2/3 heterodimers are found natively in mouse frontal cortex
ACKNOWLEDGMENTS
The authors thank Scott Blanchard for providing LD fluorophores and David Simon for assistance with coIP experiments. J. Levitz is supported by an R35 grant (1 R35 GM124731) from NIGMS and the Rohr Family Research Scholar Award.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107605.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Beckstead RM, Wooten GF, and Trugman JM (1988). Distribution of D1 and D2 dopamine receptors in the basal ganglia of the cat determined by quantitative autoradiography. J. Comp. Neurol 268, 131–145. [DOI] [PubMed] [Google Scholar]
- Beqollari D, and Kammermeier PJ (2010). Venus fly trap domain of mGluR1 functions as a dominant negative against group I mGluR signaling. J. Neurophysiol 104, 439–448. [DOI] [PubMed] [Google Scholar]
- Broichhagen J, Damijonaitis A, Levitz J, Sokol KR, Leippe P, Konrad D, Isacoff EY, and Trauner D (2015). Orthogonal optical control of a G protein-coupled receptor with a SNAP-tethered photochromic ligand. ACS Cent. Sci 1, 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. (2016). Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol 34, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH, Kosinski CM, Kerner JA, Alsdorf SA, Mangiarini L, Davies SW, Penney JB, Bates GP, and Young AB (1998). Altered brain neurotransmitter receptors in transgenic mice expressing a portion of an abnormal human Huntington disease gene. Proc. Natl. Acad. Sci. USA 95, 6480–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembrow NC, Chitwood RA, and Johnston D (2010). Projection-specific neuromodulation of medial prefrontal cortex neurons. J. Neurosci 30, 16922–16937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Menna L, Joffe ME, Iacovelli L, Orlando R, Lindsley CW, Mairesse J, Gresséns P, Cannella M, Caraci F, Copani A, et al. (2018). Functional partnership between mGlu3 and mGlu5 metabotropic glutamate receptors in the central nervous system. Neuropharmacology 128, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumazane E, Scholler P, Zwier JM, Trinquet E, Rondard P, and Pin J-P (2011). A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J 25, 66–77. [DOI] [PubMed] [Google Scholar]
- Doumazane E, Scholler P, Fabre L, Zwier JM, Trinquet E, Pin J-P, and Rondard P (2013). Illuminating the activation mechanisms and allosteric properties of metabotropic glutamate receptors. Proc. Natl. Acad. Sci. USA 110, E1416–E1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Moustaine D, Granier S, Doumazane E, Scholler P, Rahmeh R, Bron P, Mouillac B, Banéres J-L, Rondard P, and Pin J-P (2012). Distinct roles of metabotropic glutamate receptor dimerization in agonist activation and G-protein coupling. Proc. Natl. Acad. Sci. USA 109, 16342–16347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraguti F, and Shigemoto R (2006). Metabotropic glutamate receptors. Cell Tissue Res 326, 483–504. [DOI] [PubMed] [Google Scholar]
- Fuzik J, Zeisel A, Máté Z, Calvigioni D, Yanagawa Y, Szabó G, Linnarsson S, and Harkany T (2016). Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat. Biotechnol 34, 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ Jr., and Sibley DR (1990). D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science 250, 1429–1432. [DOI] [PubMed] [Google Scholar]
- Ghose S, Crook JM, Bartus CL, Sherman TG, Herman MM, Hyde TM, Kleinman JE, and Akil M (2008). Metabotropic glutamate receptor 2 and 3 gene expression in the human prefrontal cortex and mesencephalon in schizophrenia. Int. J. Neurosci 118, 1609–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouwens NW, Sorensen SA, Berg J, Lee C, Jarsky T, Ting J, Sunkin SM, Feng D, Anastassiou CA, Barkan E, et al. (2019). Classification of electrophysiological and morphological neuron types in the mouse visual cortex. Nat. Neurosci 22, 1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzeit VA, Thibado J, Stor DS, Zhou Z, Blanchard SC, Andersen OS, and Levitz J (2019). Conformational dynamics between transmembrane domains and allosteric modulation of a metabotropic glutamate receptor. eLife 8, e45116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habrian CH, Levitz J, Vyklicky V, Fu Z, Hoagland A, McCort-Tranchepain I, Acher F, and Isacoff EY (2019). Conformational pathway provides unique sensitivity to a synaptic mGluR. Nat. Commun 10, 5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, and Linnarsson S (2014). Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166. [DOI] [PubMed] [Google Scholar]
- Jin LE, Wang M, Galvin VC, Lightbourne TC, Conn PJ, Arnsten AFT, and Paspalas CD (2018). mGluR2 versus mGluR3 metabotropic glutamate receptors in primate dorsolateral prefrontal cortex: postsynaptic mGluR3 strengthen working memory networks. Cereb. Cortex 28, 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffe ME, Santiago CI, Oliver KH, Maksymetz J, Harris NA, Engers JL, Lindsley CW, Winder DG, and Conn PJ (2019a). mGlu2 and mGlu3 negative allosteric modulators divergently enhance thalamocortical transmission and exert rapid antidepressant-like effects. Neuron 105, 46–59.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffe ME, Santiago CI, Stansley BJ, Maksymetz J, Gogliotti RG, Engers JL, Nicoletti F, Lindsley CW, and Conn PJ (2019b). Mechanisms underlying prelimbic prefrontal cortex mGlu3/mGlu5-dependent plasticity and reversal learning deficits following acute stress. Neuropharmacology 144, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, and Stevens RC (2013). Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol 53, 531–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehl A, Hu H, Feng D, Sun B, Zhang Y, Robertson MJ, Chu M, Kobilka TS, Laeremans T, Steyaert J, et al. (2019). Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, and Morikawa K (2000). Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407, 971–977. [DOI] [PubMed] [Google Scholar]
- Levitz J, Habrian C, Bharill S, Fu Z, Vafabakhsh R, and Isacoff EY (2016). Mechanism of assembly and cooperativity of homomeric and heteromeric metabotropic glutamate receptors. Neuron 92, 143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang Z, Moreno-Delgado D, Dalton JA, Rovira X, Trapero A, Goudet C, Llebaria A, Giraldo J, Yuan Q, et al. (2017). Allosteric control of an asymmetric transduction in a G protein-coupled receptor heterodimer. eLife 6, e26985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makoff A, Volpe F, Lelchuk R, Harrington K, and Emson P (1996). Molecular characterization and localization of human metabotropic glutamate receptor type 3. Brain Res. Mol. Brain Res 40, 55–63. [DOI] [PubMed] [Google Scholar]
- Moreno Delgado D, Møller TC, Ster J, Giraldo J, Maurel D, Rovira X, Scholler P, Zwier JM, Perroy J, Durroux T, et al. (2017). Pharmacological evidence for a metabotropic glutamate receptor heterodimer in neuronal cells. eLife 6, e25233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Manchado AB, Bengtsson Gonzales C, Zeisel A, Munguba H, Bekkouche B, Skene NG, Lönnerberg P, Ryge J, Harris KD, Linnarsson S, and Hjerling-Leffler J (2018). Diversity of interneurons in the dorsal striatum revealed by single-cell RNA sequencing and PatchSeq. Cell Rep 24, 2179–2190.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto T, Tsuchiya D, Morikawa K, and Jingami H (2007). Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc. Natl. Acad. Sci. USA 104, 3759–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka A, Veit J, Shababo B, Chance RK, Risso D, Stafford D, Snyder B, Egladyous A, Chu D, Sridharan S, et al. (2019). Complementary networks of cortical somatostatin interneurons enforce layer specific control. eLife 8, e43696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, and Conn PJ (2010). Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol 50, 295–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi H, Shigemoto R, Nakanishi S, and Mizuno N (1993a). Distribution of the messenger RNA for a metabotropic glutamate receptor, mGluR2, in the central nervous system of the rat. Neuroscience 53, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Ohishi H, Shigemoto R, Nakanishi S, and Mizuno N (1993b). Distribution of the mRNA for a metabotropic glutamate receptor (mGluR3) in the rat brain: an in situ hybridization study. J. Comp. Neurol 335, 252–266. [DOI] [PubMed] [Google Scholar]
- Ohnuma T, Augood SJ, Arai H, McKenna PJ, and Emson PC (1998). Expression of the human excitatory amino acid transporter 2 and metabotropic glutamate receptors 3 and 5 in the prefrontal cortex from normal individuals and patients with schizophrenia. Brain Res. Mol. Brain Res 56, 207–217. [DOI] [PubMed] [Google Scholar]
- Pandya NJ, Klaassen RV, van der Schors RC, Slotman JA, Houtsmuller A, Smit AB, and Li KW (2016). Group 1 metabotropic glutamate receptors 1 and 5 form a protein complex in mouse hippocampus and cortex. Proteomics 16, 2698–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin J-P, and Bettler B (2016). Organization and functions of mGlu and GABAB receptor complexes. Nature 540, 60–68. [DOI] [PubMed] [Google Scholar]
- Poulin J-F, Tasic B, Hjerling-Leffler J, Trimarchi JM, and Awatramani R (2016). Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci 19, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Reiner A, and Levitz J (2018). Glutamatergic signaling in the central nervous system: ionotropic and metabotropic receptors in concert. Neuron 98, 1080–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins MJ, Ciruela F, Rhodes A, and McIlhinney RAJ (1999). Characterization of the dimerization of metabotropic glutamate receptors using an N-terminal truncation of mGluR1a. J. Neurochem 72, 2539–2547. [DOI] [PubMed] [Google Scholar]
- Romano C, Yang W-L, and O’Malley KL (1996). Metabotropic glutamate receptor 5 is a disulfide-linked dimer. J. Biol. Chem 271, 28612–28616. [DOI] [PubMed] [Google Scholar]
- Santana N, Mengod G, and Artigas F (2009). Quantitative analysis of the expression of dopamine D1 and D2 receptors in pyramidal and GABAergic neurons of the rat prefrontal cortex. Cereb. Cortex 19, 849–860. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkirk JV, Challiss RAJ, Rhodes A, and McIlhinney RAJ (2002). Characterization of an N-terminal secreted domain of the type-1 human metabotropic glutamate receptor produced by a mammalian cell line. J. Neurochem 80, 346–353. [DOI] [PubMed] [Google Scholar]
- Sevastyanova TN, and Kammermeier PJ (2014). Cooperative signaling between homodimers of metabotropic glutamate receptors 1 and 5. Mol. Pharmacol 86, 492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto R, and Mizuno N (2000). Metabotropic glutamate receptors—immunocytochemical and in situ hybridization analyses In Handbook of chemical neuroanatomy, Ottersen OP and Storm-Mathisen J, eds. (Amsterdam: Elsevier Science; ), pp. 63–98. [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault D, Loustalot F, Fortin GM, Bourque M-J, and Trudeau L-É (2013). Evaluation of D1 and D2 dopamine receptor segregation in the developing striatum using BAC transgenic mice. PLoS ONE 8, e67219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tora AS, Rovira X, Cao A-M, Cabayé A, Olofsson L, Malhaire F, Scholler P, Baik H, van Eeckhaut A, Smolders I, et al. (2018). Chloride ions stabilize the glutamate-induced active state of the metabotropic glutamate receptor 3. Neuropharmacology 140, 275–286. [DOI] [PubMed] [Google Scholar]
- Tsuchiya D, Kunishima N, Kamiya N, Jingami H, and Morikawa K (2002). Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+. Proc. Natl. Acad. Sci. USA 99, 2660–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafabakhsh R, Levitz J, and Isacoff EY (2015). Conformational dynamics of a class C G-protein-coupled receptor. Nature 524, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SL, Khan Y, and Benes FM (1993). Cellular distribution of dopamine D1 and D2 receptors in rat medial prefrontal cortex. J. Neurosci 13, 2551–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Noetzel MJ, Johnson KA, Zamorano R, Jalan-Sakrikar N, Gregory KJ, Conn PJ, and Niswender CM (2014). Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J. Neurosci. 34, 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate large datasets or code. The scRNaseq database used in this study is available for download (https://portal.brain-map.org/atlases-and-data/rnaseq). All the newly generated datasets are available from the Lead Contact on request.


