Rice REPETITIVE PROLINE-RICH PROTEIN affects cytoskeletal reorganization, cell wall remodeling, and sucrose synthase activity as an adaptive response to water deficit in rice roots.
Abstract
Intrinsically disordered proteins function as flexible stress modulators in vivo through largely unknown mechanisms. Here, we elucidated the mechanistic role of an intrinsically disordered protein, REPETITIVE PRO-RICH PROTEIN (RePRP), in regulating rice (Oryza sativa) root growth under water deficit. With nearly 40% Pro, RePRP is induced by water deficit and abscisic acid (ABA) in the root elongation zone. RePRP is sufficient and necessary for repression of root development by water deficit or ABA. We showed that RePRP interacts with the highly ordered cytoskeleton components actin and tubulin both in vivo and in vitro. Binding of RePRP reduces the abundance of actin filaments, thus diminishing noncellulosic polysaccharide transport to the cell wall and increasing the enzyme activity of Suc synthase. RePRP also reorients the microtubule network, which leads to disordered cellulose microfibril organization in the cell wall. The cell wall modification suppresses root cell elongation, thereby generating short roots, whereas increased Suc synthase activity triggers starch accumulation in “heavy” roots. Intrinsically disordered proteins control cell elongation and carbon reserves via an order-by-disorder mechanism, regulating the highly ordered cytoskeleton for development of “short-but-heavy” roots as an adaptive response to water deficit in rice.
Intrinsically disordered proteins lack a defined three-dimensional structure but often contain a simple amino acid composition with repeated sequences that provide the basis for multivalent intermolecular interactions (Uversky, 2016). With their unique structural flexibility, conformational adaptability, and ability to react quickly in response to changing environments, intrinsically disordered proteins are often functional as hubs of protein-protein interaction networks in response to environmental stresses (Uversky, 2011). Recently, the crucial role of intrinsically disordered proteins in stress-triggered phase transition has been addressed (Ruff et al., 2018). As highlighted cases, tardigrade disordered proteins are essential for their high level of desiccation tolerance (Boothby et al., 2017), and disordered LATE EMBRYOGENESIS ABUNDANT (LEA) proteins often accumulate under dehydration and osmotic stresses in plants (Giarola et al., 2017).
Water deficit, caused by global climate changes and increasing world population, has been the most severe environmental stress to crop plants, affecting agricultural productivity and food security (Hu and Xiong, 2014). Water deficit triggers plants to produce abscisic acid (ABA), which orchestrates stress-specific responses and stress tolerance (Bray, 1997). The hormone ABA is also involved in seed dormancy and desiccation tolerance during seed development (Finkelstein et al., 2002). As a staple food feeding more than half of world populations, rice (Oryza sativa) requires two to three times more water than other dryland cereals do; hence, improving the adaptation of rice to water-deficit conditions is critical (Kadam et al., 2017). Because the root is one of the organs perceiving water deficit, it is essential to understand the biological mechanism regulating rice root growth under water deficit (Rellán-Álvarez et al., 2016; Buckley, 2019).
As a physical barrier surrounding every plant cell, the cell wall is inherently involved in regulating cell expansion (Bashline et al., 2014). The cell wall is composed of cellulose, noncellulosic wall polysaccharide polymers such as hemicellulose and pectin, and a small amount of protein (Bashline et al., 2014). The complexity of the cell wall is partially controlled by the dynamic intracellular cytoskeleton (Szymanski and Cosgrove, 2009; Bashline et al., 2014). Microtubules guide the orientation of cellulose synthase complexes at the plasma membrane (Paredez et al., 2006; Crowell et al., 2009). Noncellulosic polysaccharides are assembled within the Golgi, secreted and transported through Golgi-derived vesicles, and associated with newly synthesized cellulose microfibrils in the cell wall (Driouich et al., 1993). The transport of Golgi-derived vesicles containing hemicellulose and pectin is highly controlled by actin filaments (Baluska et al., 2002; Kim et al., 2005).
In animal cells, microcompartmentation of glycolytic enzymes with actin filaments generates metabolic channeling to favor the maximal reaction rates of carbohydrate metabolism (al-Habori, 1995; Real-Hohn et al., 2010). In plants, several enzymes involved in carbohydrate metabolism have been reported to interact with cytoskeleton proteins (Chuong et al., 2004; Balasubramanian et al., 2007; Garagounis et al., 2017). Among them, Suc synthase (SUS) has been shown to bind actin filaments both in vivo and in vitro (Winter et al., 1998; Duncan and Huber, 2007), but the functional significance of such binding has not been determined. SUS catalyzes the reversible conversion of Suc to/from NDP-Glc and Fru, playing an important role in conserving the energy in the nucleotide sugar products, which in turn are precursors for the synthesis of cellulose or starch (Winter and Huber, 2000; Koch, 2004). SUS exists ubiquitously in plants and is particularly active in plant sink tissues, such as roots, developing seeds, young leaves, or tubers (Winter and Huber, 2000; Koch, 2004).
Because of the versatility of intrinsically disordered proteins, they are relevant for plant adaptation and survival under changing environments (Covarrubias et al., 2017). A case in point is the LEA proteins, which are involved in water deficit responses (Candat et al., 2014). However, because of technical and experimental barriers, and lacking an in vivo mechanism and interaction evidence, research regarding intrinsically disordered proteins is limited to in vitro analyses (Covarrubias et al., 2017). Prorich repeats/proteins often provide intrinsically disordered states and serve as environmental modulators (Reiersen and Rees, 2001). One superfamily of plant cell wall proteins, the Hyp-rich glycoproteins (HRGPs), which are Prorich and contain repeated sequence motifs and extensive glycosylation, have been implicated in many biological functions in plants (Johnson et al., 2017). Both Prorich proteins and HRGPs are considered to be involved in the responses of plants to environmental factors (Sugimoto‐Shirasu et al., 2004).
We have characterized an intrinsically disordered protein, rice REPETITIVE PRO-RICH PROTEIN (OsRePRP), with extremely high Pro content (∼40%), that is distinct from the classical HRGPs (Tseng et al., 2013). OsRePRP and its homologs are only identified in monocotyledonous plants that have an adventitious root system (Tseng et al., 2013), whereas most of the other Prorich proteins with less Pro content than OsRePRP and different structural motifs are found in the dicotyledonous plants with a tap root system (Johnson et al., 2017). All four genes (OsRePRP1.1, OsRePRP1.2, OsRePRP2.1, and OsRePRP2.2) in the OsRePRP family are upregulated in roots by ABA, salinity, and water deficit (Tseng et al., 2013). OsRePRP2-overexpressing (OsRePRP2-OX) transgenic rice featured a short root and reduced cell length phenotype, resembling the repressed root growth with water deficit or ABA in wild-type rice (Tseng et al., 2013). Conversely, knockdown of both OsRePRP1.1 and OsRePRP2.1 in OsRePRP RNA-interference (OsRePRP-Ri) transgenic rice reduced the repression effect on root growth, which indicates that OsRePRPs play a necessary and sufficient role in water deficit- or ABA-regulated root development (Tseng et al., 2013).
Here we report that OsRePRPs interact with the cytoskeleton components actin and tubulin specifically, which leads to a major reorientation of cellulose microfibrils in the cell wall to inhibit the elongation of root cells but promote more starch accumulation in the “short-but-heavy” roots. Our results demonstrate a novel role of an ABA/stress-induced intrinsically disordered protein in regulating plant root growth under water deficit.
RESULTS
Recombinant OsRePRPs Bind to Actin and Tubulin In Vitro
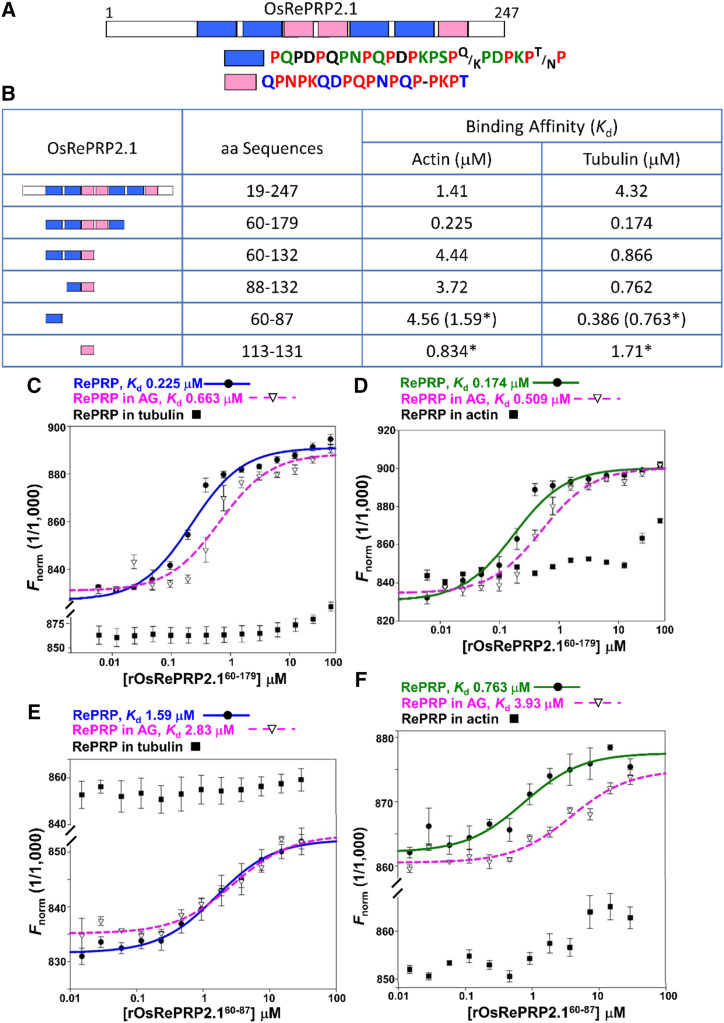
OsRePRP protein sequences are rich in the amino acid Pro, which makes up ∼40% of the total protein. When OsRePRP sequences were assessed using four different Predictor Of Natural Disordered Regions predictors, VSL2, VL3, VL-XT, and XL1-XT (Xue et al., 2010), both OsRePRP1.1 and OsRePRP2.1 showed high disorder scores (>0.5) among the major protein sequences (Supplemental Fig. S1, A and B). The internal duplication pattern was also observed in the repetitive regions of both OsRePRP1.1 and OsRePRP2.1 (Supplemental Fig. S1, C and D). Segmental repeat organization of OsRePRPs includes main repeat regions and nonrepeat regions near the termini (Supplemental Fig. S2). Using RADAR software, we dissected the repeat regions of OsRePRP1.1 and OsRePRP2.1 into several superrepeat segments (Fig. 1A; Supplemental Fig. S3A). In OsRePRP1.1, the segments are mainly composed of PEPK repeats, whereas in OsRePRP2.1, they are PQPN/PDPK repeats (Fig. 1A; Supplemental Fig. S3A). The PEPK motifs are very similar to the actin-binding PEVK repeats in animal titin, which controls the passive elasticity of the sarcomere in striated muscle cells (Gutierrez-Cruz et al., 2001). The architecture, composed of single repeats, super repeats, and nonrepeat regions, bears a remarkable resemblance to nebulin (Wang et al., 1996), a muscle actin thin filament “ruler.” Therefore, we expressed recombinant OsRePRPs (rOsRePRPs) and their serial segments in Pichia pastoris and Escherichia coli to test the binding of actin and tubulin using microscale thermophoresis (MST) technology (Wienken et al., 2010) and determine the equilibrium dissociation constant (Kd).
Figure 1.
In vitro interactions of rOsRePRP2.1 and actin/tubulin. A, Segmental repeat organization of OsRePRP2.1. The diagram shows the full-length amino acid sequences analyzed by RADAR (http://www.ebi.ac.uk/Tools/pfa/radar/). The repeat modules of OsRePRP2.1 are represented as blue and pink blocks and the nonrepeat regions as white blocks. The percentages of each amino acid in the sequences are distinguished by color and letter codes: red for 100%, green for >75%, blue for >67%, black for >50% and a dash for nonconsensus amino acids. B, Binding affinity of rOsRePRP2.1 with actin/tubulin. In vitro binding assays were conducted with MST, and the binding affinity is indicated by Kd. Asterisks indicate the binding affinity with synthetic peptides. C to F, Binding isotherms for rOsRePRP2.160–179 (C and D) and synthetic peptides OsRePRP2.160–87 (E and F) to rabbit muscle actin (C and E) and porcine brain tubulin (D and F). Rabbit muscle actin and porcine brain tubulin were fluorescently labeled with AlexaFluor 647 and kept at a constant concentration of 1.2 to 1.5 nm. The binding of OsRePRP2.1 alone to actin and tubulin is represented as blue and green curves, respectively, and the binding of OsRePRP2.1 in the presence of AG is represented as pink-dashed curves. Solid circles indicate rOsRePRP2.160–179 alone (C and D) and synthetic peptides OsRePRP2.160–87 alone (E and F). Open triangles represent rOsRePRP2.160–179 (C and D) and OsRePRP2.160–87 (E and F) in the presence of 250 μm AG. Solid squares indicate rOsRePRP2.160–179 and synthetic peptides OsRePRP2.160–87 in the presence of 25 μm porcine brain tubulin (C and E) or 57.5 μm rabbit muscle actin (D and F). Data are means ± sd of three technical repeats. G-actin and αβ-tubulin dimers were used, and at least two independent batches of recombinant proteins or synthetic peptides were subjected to the binding assays.
MST assays showed that rOsRePRP1.1 and rOsRePRP2.1 as well as their serial segmental recombinant proteins/peptides bound to rabbit muscle actin and porcine brain tubulin with various Kd (Fig. 1B; Supplemental Fig. S3B). Recombinant OsRePRP2.1 bound to actin with Kd 1.41 μm and to tubulin with Kd 4.32 μm (Fig. 1B), whereas rOsRePRP1.1 bound to actin with Kd 1.72 μm and to tubulin with Kd 6.54 μm (Supplemental Fig. S3B). The Kd of serial segmental rOsRePRP2.1 binding to actin/tubulin ranged from 0.174 to 4.56 μm and the synthetic one-superrepeat peptides of OsRePRP2.1 bound to actin and tubulin from 0.763 to 1.71 μm (Fig. 1B, asterisks). The Kd of serial segmental rOsRePRP1.1 binding to actin/tubulin ranged from 0.296 to 24.8 μm (Supplemental Fig. S3B). Consistent with our previous report (Tseng et al., 2013), rOsRePRP1.1 and rOsRePRP2.1 interacted with arabinogalactan (AG) at Kd 52.1 and 74 μm, respectively, but not with Suc (Supplemental Fig. S3, C and D). As control experiments, no binding was observed between the negative bovine serum albumin (BSA) control and rOsRePRP1.1 or rOsRePRP2.1 (Supplemental Fig. S3, E and F). We also tested two other recombinant plant proteins: recombinant RICE BIG GRAIN1 (rOsRBG1) and one intrinsically disordered LEA protein from barley, rHvHAV1. Neither showed binding to actin/tubulin (Supplemental Fig. S3, G and H). The results obtained from the control experiments suggested that the binding of OsRePRPs to actin and tubulin is not due to nonspecific interactions.
We used competition binding assays to verify segmental rOsRePRP2.160–179 binding to AG, actin, and tubulin (Fig. 1, C and D). Recombinant OsRePRP2.160–179 was first mixed with AG to measure the binding of rOsRePRP2.160–179 to actin and tubulin. In the presence of 250 μm AG, rOsRePRP2.160–179 retained the binding affinity to actin and tubulin, with Kd 0.663 and 0.509 μm, respectively (Fig. 1, C and D, pink-dashed curves). Thus, the binding of rOsRePRP2.160–179 to actin and tubulin is independent of its binding to AG in vitro. To further verify the correlation between actin and tubulin binding, rOsRePRP2.160–179 was mixed with tubulin to measure the binding of rOsRePRP2.160–179 to actin and vice versa (Fig. 1, C and D). In the presence of 25 μm tubulin, binding of rOsRePRP2.160–179 to actin was greatly reduced (Fig. 1C, solid squares), and in the presence of 57.5 μm actin, binding of rOsRePRP2.160–179 to tubulin was barely detectable (Fig. 1D, solid squares). Therefore, actin and tubulin competed with each other for binding to rOsRePRP2.160–179. Similar competition between actin and tubulin was observed in the binding of the one-superrepeat peptide OsRePRP2.160–87, and AG did not seem to affect the peptide OsRePRP2.160–87 binding to actin or tubulin much (Fig. 1, E and F).
To confirm the binding of rOsRePRP to actin filaments (F-actin) and microtubules, we used high-speed cosedimentation assays with mixtures of rOsRePRP2.160–179 and F-actin and microtubules polymerized to a steady state (Supplemental Fig. S3, I and J). High-speed centrifugation separated F-actin and microtubules in the pellet from G-actin and αβ-tubulin dimers in the supernatant. Besides binding to G-actin and αβ-tubulin dimers (Fig. 1, B–D), rOsRePRP2.160–179 also bound to F-actin and microtubules (Supplemental Fig. S3, I and J).
Colocalization of OsRePRP2 and Cytoskeleton Markers in Rice Protoplasts
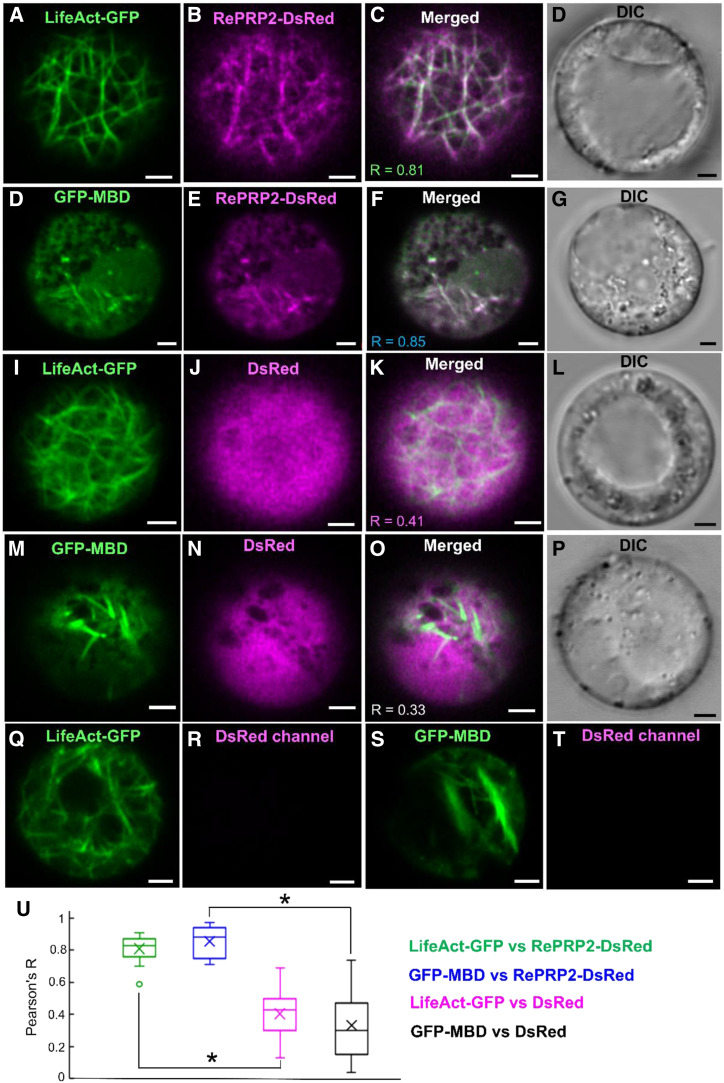
The MST binding assays suggested that OsRePRPs interact with the cytoskeleton proteins actin and tubulin in vitro (Fig. 1; Supplemental Fig. S3), so we further investigated whether OsRePRP2 interacts with the cytoskeleton proteins in vivo using the rice root protoplast transient-expression system. LifeAct has been reported to bind eukaryotic actin filaments (Riedl et al., 2008) and to be useful for studying the actin cytoskeleton in a wide range of plant lineages (Era et al., 2009). A microtubule reporter gene created by fusing the microtubule binding domain (MBD) of the mammalian microtubule-associated protein4 (MAP4) gene with the GFP gene could be used to visualize microtubule orientation in plants (Marc et al., 1998). Hence, we constructed the cytoskeleton marker LifeAct fused with C-terminal GFP (LifeAct-GFP) and MBD fused with N-terminal GFP (GFP-MBD) to verify the subcellular colocalization with OsRePRP2 fused with C-terminal DsRed (OsRePRP2-DsRed) in the rice root protoplast transient-expression system (Fig. 2). The results suggested that OsRePRP2-DsRed showed similar filamentous expression patterns and colocalized with both LifeAct-GFP and GFP-MBD in the rice root protoplasts (Fig. 2, A–H). The negative control DsRed was distributed throughout the cytosol and did not show filamentous patterns similar to those observed for LifeAct-GFP or GFP-MBD (Fig. 2, I–P). As control experiments, protoplasts expressing LifeAct-GFP and GFP-MBD did not show any fluorescence signals in the DsRed channel (Fig. 2, Q–T), excluding the possibility of channel bleeding. Pearson correlation coefficient analysis was used to quantify the colocalization (Fig. 2U): OsRePRP2-DsRed colocalized with LifeAct-GFP (R = 0.81) and with GFP-MBD (R = 0.85; Fig. 2, C, G, and U), which was distinct from the negative control DsRed correlated with LifeAct-GFP (R = 0.41) and GFP-MBD (R = 0.33; Fig. 2, K, O, and U).
Figure 2.
Interactions between OsRePRP2 and actin and tubulin in rice root protoplasts. A to P, Five-day-old wild-type rice root protoplasts coexpressing LifeAct-GFP and OsRePRP2-DsRed (A–D), GFP-MBD and OsRePRP2-DsRed (E–H), LifeAct-GFP and DsRed (I–L), and GFP-MBD and DsRed (M–P). GFP and DsRed signals are shown in green (A, E, I, and M) and magenta (B, F, J, and N), respectively; overlays of green and red channels (C, G, K, and O) and the DIC channel indicating bright-field images (D, H, L, and P) are also shown. Q to T, Five-day-old wild-type rice root protoplasts expressing LifeAct-GFP (Q and R) and GFP-MBD (S and T). GFP signals are shown in green (Q and S), whereas the DsRed signal in the respective control images is not visible (R and T). Scale bars = 2 μm. U, The Pearson's R correlation coefficients calculated by Pearson coefficient analysis are shown in a box plot. Pearson's R coefficients for LifeAct-GFP and OsRePRP2-DsRed, GFP-MBD and OsRePRP2-DsRed, LifeAct-GFP and DsRed, and GFP-MBD and DsRed are shown in green, blue, magenta, and black boxes, respectively. The line inside the box indicates the median and the ex indicates the mean. Bottom and top box edges are the 25th to 75th percentiles; whiskers indicate the range, and an outlying data point is shown as a circle. Significant differences are indicated with asterisks (P < 0.01, two-tailed Mann-Whitney U test). Data are measured from 15 protoplasts of three biological experiments. The Pearson's R correlation coefficient for each comparison is shown in C, G, K, and O.
OsRePRP Interacts with Actin and Tubulin In Vivo
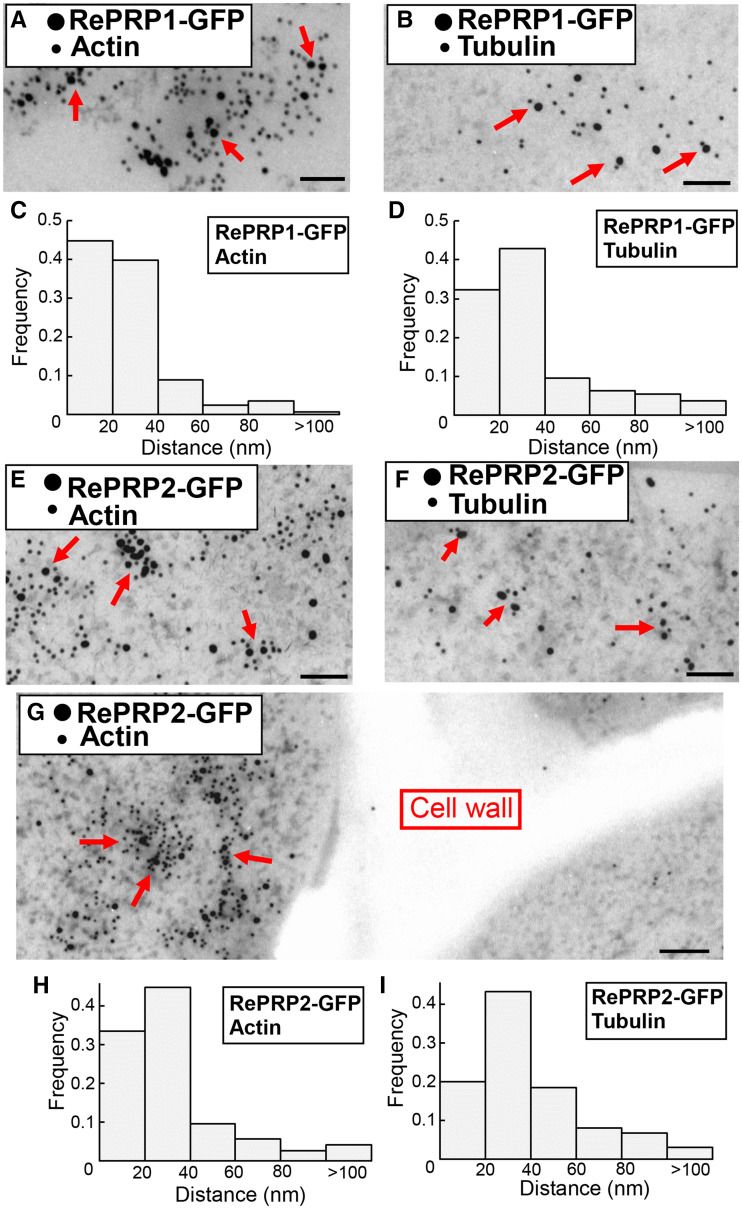
To further investigate whether OsRePRP forms complexes with actin and tubulin in planta, GFP, OsRePRP1-GFP, and OsRePRP2-GFP transgenic rice root cells were examined in coimmunoprecipitation (co-IP) assays with anti-GFP antibodies immobilized on magnetic beads. Actin and tubulin coimmunoprecipitated with anti-GFP beads in OsRePRP1-GFP and OsRePRP2-GFP but not in GFP root extracts (Supplemental Fig. S4, A and B). Detection of actin and tubulin peptides by liquid chromatography tandem mass spectrometry (LC-MS/MS) further supported that OsRePRPs may interact with actin and tubulin in planta (Supplemental Tables S1 and S2). The in vivo interaction of OsRePRP-GFP and actin and tubulin was further confirmed by immunogold-labeling transmission electron microscopy (TEM), showing OsRePRP-GFP immuno-labeled by 18-nm gold particles and actin and tubulin by 12-nm gold particles (Fig. 3). In practice, 5- to 15-nm gold particles are situated at a distance of ∼30 nm from the epitope (Bergersen et al., 2008), so a distance of <60 nm between two sizes of gold particles was assumed to be colocalization. Most of the closest distances between OsRePRP-GFP and actin/tublin were <40 nm (Fig. 3, C, D, and H–I), but most of the closest distances between GFP and actin/tubulin were >100 nm (Supplemental Fig. S4, C–F). These results also suggest that OsRePRPs interact with both types of cytoskeleton filaments in vivo.
Figure 3.
Interactions between OsRePRP and actin and tubulin in planta. A, B, and E–G, Double immunogold-labeling TEM of root elongation zone cells of 3-d-old OsRePRP1-GFP (A and B) and OsRePRP2-GFP (E–G) rice. The smaller 12-nm gold particles represent the distribution of actin (A, E, and G) and tubulin (B and F), and the larger 18-nm gold particles represent the distribution of OsRePRP1-GFP (A and B) and OsRePRP2-GFP (E–G). Arrows indicate colocalization of OsRePRP-GFP and actin (A, E, and G) or tubulin (B and F). Scale bars = 100 nm. C, D, H, and I, Distances from each OsRePRP-GFP gold particle to the closest actin or tubulin gold particle were measured as described in "Materials and Methods." The bars represent the distribution of center-to-center distances between gold particles in OsRePRP-GFP and actin or tubulin, ranging from 0 to >100 nm. Ten images were measured, and two biological replicates were performed.
Overexpression of OsRePRP2 Causes Reduced Abundance of Actin Filaments
OsRePRP2 expression was highly induced by ABA (Supplemental Fig. S5), and OsRePRP2-OX transgenic rice featured a short root and reduced cell length phenotype (Supplemental Fig. S6A; Tseng et al., 2013), resembling the repression of root growth with water deficit or ABA in wild-type rice. This phenotype was reduced in OsRePRP-Ri transgenic rice roots (Supplemental Fig. S6A), which suggests that OsRePRP is necessary and sufficient in stress/ABA repression of root elongation.
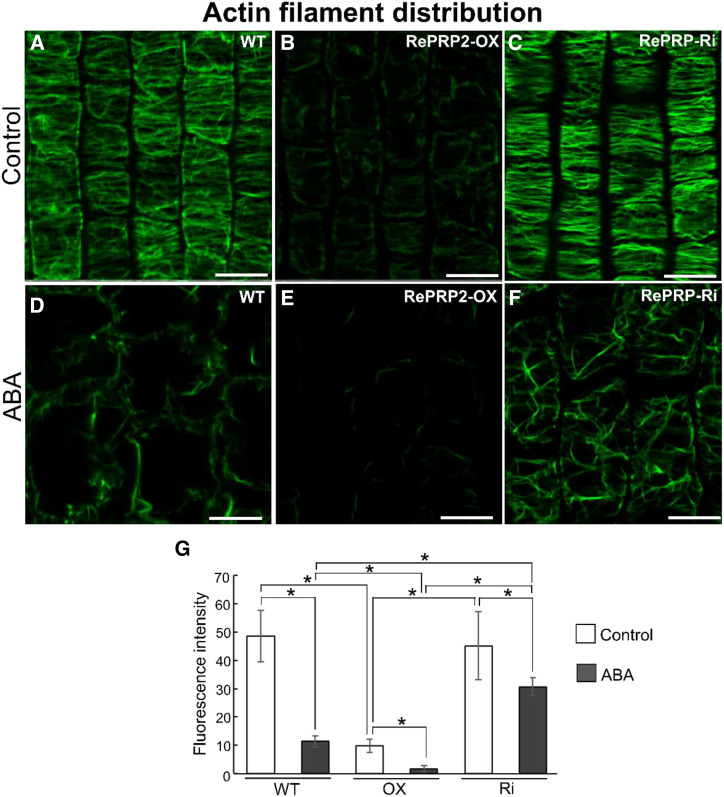
Cytoskeleton filaments play an important role in the spatial control of cell expansion (Smith and Oppenheimer, 2005). Because OsRePRPs bind to actin and tubulin in vitro (Fig. 1; Supplemental Fig. S3), we wondered whether the short root and reduced cell length phenotype of OsRePRP2-OX transgenic rice was related to cytoskeleton filaments. Hence, we examined the in vivo actin filament (F-actin) organization of wild-type, OsRePRP2-OX, and OsRePRP-Ri transgenic rice roots by Alexa Fluor 488-phalloidin staining.
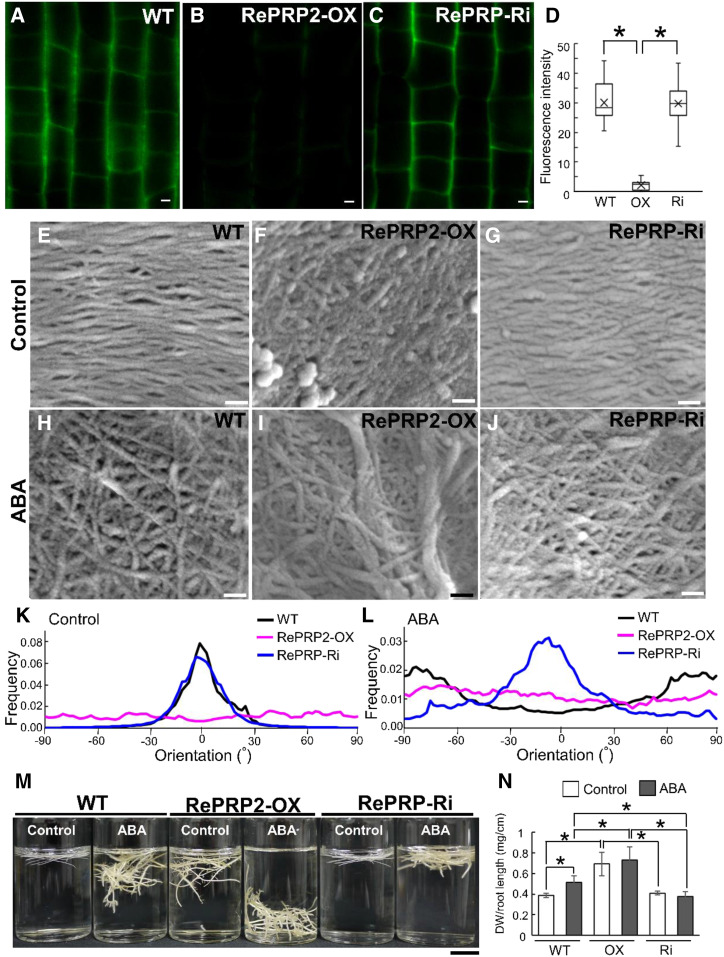
Phalloidin specifically binds to F-actin (Wulf et al., 1979), so the fluorescence intensity on staining is positively correlated with the abundance of F-actin within cells. The overall fluorescence signal of F-actin in elongation-zone cells was weaker in OsRePRP2-OX than in wild-type or OsRePRP-Ri roots (Fig. 4, A–C; Supplemental Video). The mean fluorescence intensity of F-actin was reduced by nearly 5-fold for OsRePRP2-OX, with no significant difference between wild-type and OsRePRP-Ri roots (Fig. 4G). Under ABA treatment, F-actin level was decreased in the wild type and barely visible in OsRePRP2-OX, but this phenomenon was less apparent in OsRePRP-Ri roots (Fig. 4, D–F; Supplemental Video). The mean F-actin fluorescence intensity was about 3-fold higher in OsRePRP-Ri than in wild-type roots (Fig. 4G). The same patterns were also observed in the root division and differentiation zones (Supplemental Fig. S7), and OsRePRP showed phenotypic changes with polyethylene glycol (PEG) stress treatment similar to those observed with ABA treatment (Supplemental Fig. S8). Thus, water deficit- or ABA-induced OsRePRP may be involved in regulating F-actin level primarily in the root elongation zone, but also in division and differentiation zones.
Figure 4.
OsRePRP affects F-actin organization. A to F, F-actin organization in wild-type (WT), OsRePRP2-OX, and OsRePRP-Ri root cells visualized by Alexa Fluor 488-phalloidin staining. Epidermal cells of the root elongation zones of 8-d-old seedlings (control; A–C) and 7-d-old seedlings treated with 2 μm ABA for 1 d (ABA; D–F) were observed. Scale bars = 10 μm. G, Quantitative analysis of the F-actin level in wild-type, OsRePRP2-OX (OX), and OsRePRP-Ri (Ri) 8-d-old seedlings (control) and 7-d-old seedlings under the ABA condition. Significant differences are indicated with asterisks (P < 0.01, two-tailed Mann-Whitney U test). Data for fluorescence intensity are expressed as means ± sd of 30 images. More than six biological replicates were performed and at least three independent transgenic lines for each genotype were observed (OsRePRP2-OX lines 2, 10, 19, and 24, and OsRePRP-Ri lines 5, 6, and 7).
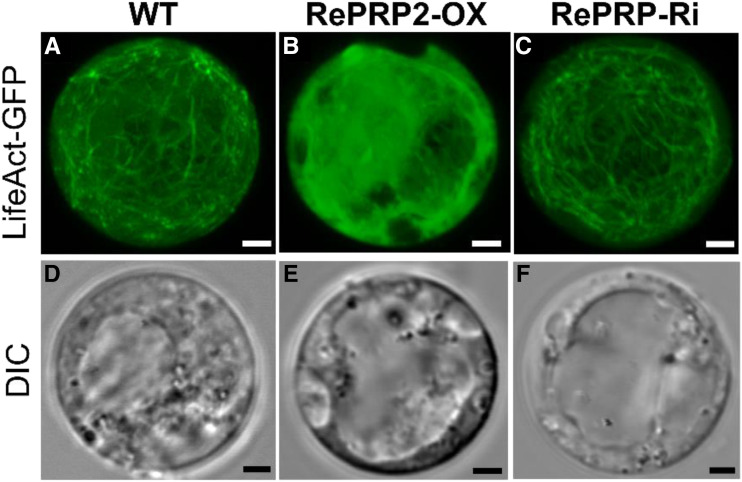
The root protoplasts of the wild type, OsRePRP2-OX, and OsRePRP-Ri were isolated and transfected with LifeAct-GFP with the DsRed transfection control (Fig. 5). In 5-d-old protoplasts, LifeAct-GFP showed filamentous patterns in both wild-type and OsRePRP-Ri protoplasts (Fig. 5, A and C), whereas LifeAct-GFP in OsRePRP2-OX protoplasts showed cytosolic patterns but few filamentous patterns (Fig. 5B). These results exclude the possibility that the reduced F-actin level in OsRePRP2-OX was an artifact caused by the differential staining of phalloidin.
Figure 5.
OsRePRP affects F-actin organization in root protoplasts. Transient expression of LifeAct-GFP in 5-d-old rice root protoplasts from the wild type (A and D), OsRePRP2-OX (B and E), and OsRePRP-Ri (C and F). GFP signals are shown in green (A–C) and the DIC channel indicates bright-field images (D–F). Images in A to C represent the maximum intensity projection of 20 optical confocal images with identical settings. Three independent transgenic lines for each genotype were observed (OsRePRP2-OX lines 2, 10, and 24 and OsRePRP-Ri lines 5–7). Scale bars = 2 μm.
Microtubule Orientation Is Altered in OsRePRP2-OX
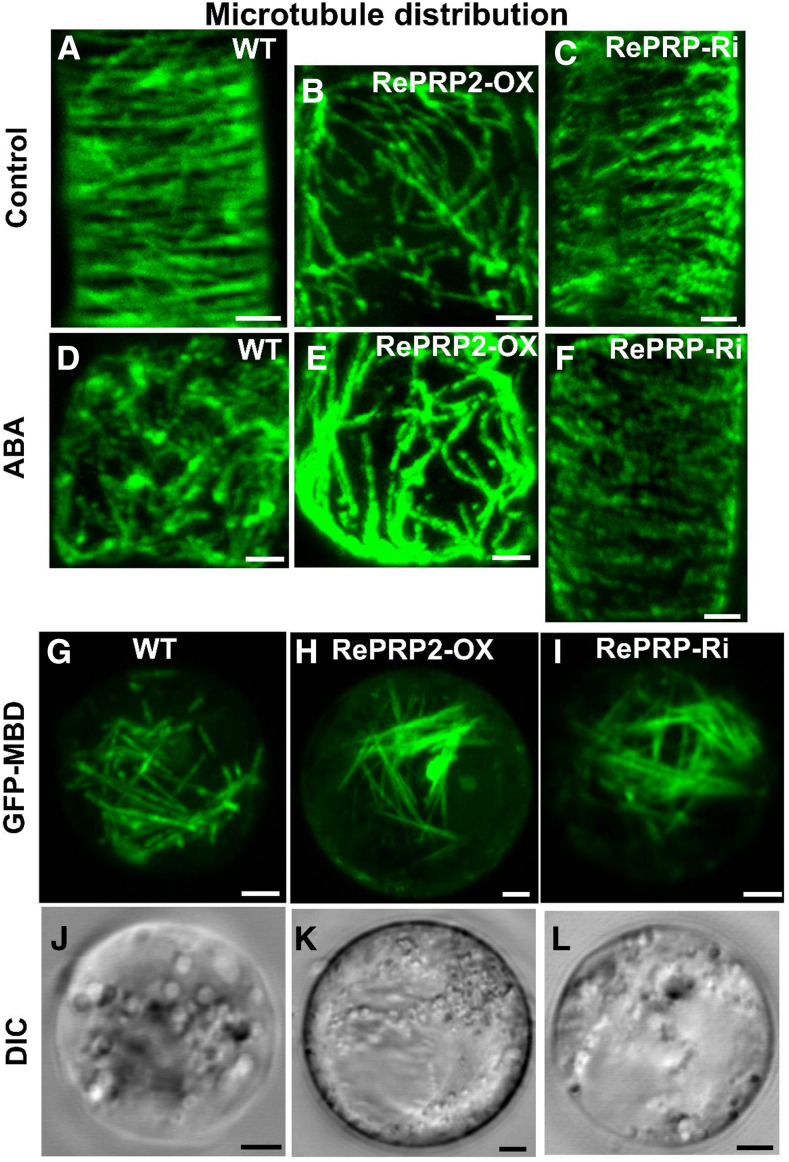
We also examined microtubule organization of wild-type, OsRePRP2-OX, and OsRePRP-Ri rice root cells under control and ABA treatment by immunofluorescence staining with anti-α-tubulin antibodies (Fig. 6, A–F). The microtubule network was less organized in OsRePRP2-OX than in wild-type or OsRePRP-Ri root cells (Fig. 6, A–C). With ABA treatment, the disordered microtubule phenotype was apparent in the wild type but less so in OsRePRP-Ri cells (Fig. 6, D and F).
Figure 6.
OsRePRP affects microtubule organization. A to C, Micrographs of microtubule staining in wild-type (WT; A), OsRePRP2-OX (B), and OsRePRP-Ri (C) root cells of elongation zones. D to F, Micrographs of microtubule staining in wild-type (D), OsRePRP2-OX (E), and OsRePRP-Ri (F) root cells after 2 μm ABA treatment for 2 d. More than three biological replicates were performed and three independent transgenic lines for each genotype were observed (OsRePRP2-OX lines 10, 19, and 24 and OsRePRP-Ri lines 5–7). G to L, Transient expression of GFP-MBD in 5-d-old rice root protoplasts from the wild type (G and J), OsRePRP2-OX (H and K), and OsRePRP-Ri (I and L). GFP signals are shown in green (G–I) and the DIC channel indicates bright-field images (J–L). Four biological replicates were performed and three independent transgenic lines for each genotype were observed (OsRePRP2-OX lines 2, 10, and 24 and OsRePRP-Ri lines 5–7). Scale bars = 2 μm.
Further study in the rice root protoplast transient-expression system indicated that GFP-MBD showed a similar filamentous expression pattern in the wild type, OsRePRP2-OX, and RePRP-Ri (Fig. 6, G–L). The microtubule organization in the rice root protoplast transient-expression system did not show the transverse arrangement seen in the whole-mount inmmunostaining of rice roots (Fig. 6A). The results suggested that without the cell wall, the microtubule orientation of OsRePRP2-OX did not differ substantially from the wild type and RePRP-Ri (Fig. 6, G–I).
OsRePRP Affects Trafficking of Noncellulosic Polysaccharides and Cell Wall Cellulose Microfibril Organization
The cytoskeleton is involved in plant cell shape determination by affecting the patterns in which cell wall materials are deposited (Smith and Oppenheimer, 2005). F-actin plays a role in vesicle trafficking of noncellulosic polysaccharides (Baluska et al., 2002; Kim et al., 2005). Microtubules maintain the cellulose synthase complexes localized at the plasma membrane to guide cellulose deposition on the cell wall (Paredez et al., 2006; Crowell et al., 2009). Because OsRePRP regulates actin filament distribution and microtubule organization in vivo, we wondered whether it affects noncellulosic polysaccharide secretion on the cell wall and cell wall cellulose microfibril arrangement. By using metabolic click-labeling with Fuc alkyne (FucAl) and Alexa Fluor 488-azide (Anderson et al., 2012), we monitored FucAl incorporation into the cell wall by confocal microscopy (Fig. 7, A–C). FucAl-associated fluorescence was observed in the wild-type and OsRePRP-Ri cell walls, but much less so in the OsRePRP2-OX cell wall (Fig. 7, A–C). The quantitative data indicate that the fluorescence signal in OsRePRP2-OX was reduced ∼15-fold compared with that in the wild type or OsRePRP-Ri (Fig. 7D). Hence, the metabolic process of FucAl incorporation in the cell wall may be blocked in OsRePRP2-OX.
Figure 7.
OsRePRP affects noncellulosic polysaccharide secretion and cell wall cellulose microfibril network. A to C, Localization of FucAl incorporation in wild-type (WT; A), OsRePRP2-OX (B), and OsRePRP-Ri (C) root elongation zone cells. Images were obtained using identical settings including laser power, pinhole, objective, zoom, and channel/filter wavelengths, as described in Materials and Methods, with no image contrast or brightness corrections. D, Quantitative analysis of FucAl-associated fluorescence signal in the wild type, OsRePRP2-OX (OX), and OsRePRP-Ri (Ri) are shown in a box plot. The line inside the box indicates the median, and the ex indicates the mean. Bottom and top box edges are the 25th to 75th percentiles; whiskers indicate the range. Significant differences are indicated with asterisks (P < 0.01, two-tailed Mann-Whitney U test). Data are mean/median (Q1–Q3) fluorescence intensity of 30 images. More than four biological replicates were performed, and at least three independent transgenic lines for each genotype were observed (OsRePRP2-OX lines 2, 10, 19, and 24 and OsRePRP-Ri lines 5–7). E to J, Field-emission scanning electron microscopy images are oriented to show cell wall cellulose microfibril alignment relative to the cell and the root long axis. Micrographs are shown of the cell wall cellulose microfibril network under the control condition (E–G) and the cell wall microfibril network after 2 μm ABA treatment for 4 d (H–J) in wild-type (E and H), OsRePRP2-OX (F and I), and OsRePRP-Ri (G and J) root elongation zone cells. Three biological replicates were performed and at least two independent transgenic lines for each genotype were observed (OsRePRP2-OX lines 10, 19, and 24 and OsRePRP-Ri lines 5 and 6). K and L, Directionality histograms of the cell wall cellulose microfibril network under the control condition (K) or after ABA treatment (L). Directionality of the wild type (black line), OsRePRP2-OX (magenta line), and OsRePRP-Ri (blue line) was analyzed using the Fiji package (Schindelin et al., 2012) as described in Materials and Methods. Data are expressed as the mean frequency of fibers in specific orientation analyzed from 15 images. M, Relative density of root segments in the wild type, OsRePRP2-OX, and OsRePRP-Ri. Two-centimeter root-tip segments from 12-d-old seedlings (Control) and 8-d-old seedlings treated with 2 μm ABA for 4 d (ABA) were cut and immersed in the distilled water. Six biological replicates were performed and three independent transgenic lines for each genotype were observed (OsRePRP2-OX lines 10, 19, and 24 and OsRePRP-Ri lines 5–7). N, Quantitative analysis of total root dry weight (DW; milligrams) divided by mean root length (centimeters) per seedling. Whole roots were harvested from wild-type, OsRePRP2-OX (OX), and OsRePRP-Ri (Ri) 14-d-old seedlings (Control) and 8-d-old seedlings treated with 2 μm ABA for 6 d (ABA). Significant differences are indicated with asterisks (P < 0.01, two-tailed Mann-Whitney U test). Fluorescence intensity of 30 images was measured in three independent transgenic lines for each genotype (OsRePRP2-OX lines 2, 10, and 24 and OsRePRP-Ri lines 5–7). Scale bars = 2 μm (A–C), 100 nm (E–J), and 1 cm (M).
Field emission scanning electron microscopy of cells in the root elongation zone revealed a transverse cell wall cellulose microfibril network in the wild type (Fig. 7E), which agrees with the previous findings of a transverse cellulose microfibril-aligned pattern perpendicular to the direction of cell elongation (Sugimoto et al., 2000). OsRePRP2-OX showed a more disorganized orientation of cellulose fibril network (Fig. 7F) compared with the transverse pattern of wild-type and OsRePRP-Ri networks (Fig. 7, E and G). Under ABA treatment, the transversely ordered cellulose fibril network of the wild type became disordered (Fig. 7H), and some thick fibrils in a disarranged cellulose network were observed in OsRePRP2-OX (Fig. 7I), whereas OsRePRP-Ri still retained some of the transversely ordered cellulose fibril network (Fig. 7J). Quantitative directionality (orientation in terms of angle) analysis has been used in a study of collagen fibril alignment (Grossman et al., 2016), and we applied it here in the quantitative estimation of the orientation of cell wall cellulose microfibrils. The analyses showed that cellulose microfibrils of both the wild type and OsRePRP-Ri were aligned in one dominant direction (nearly 0° of orientation), whereas those of OsRePRP2-OX were dispersed and oriented in a multitude of directions (Fig. 7K). After ABA treatment, the wild type and OsRePRP2-OX showed dispersed directions, but OsRePRP-Ri retained the dominant direction alignment (nearly 0° of orientation; Fig. 7L). Hence, OsRePRPs may affect root cell wall cellulose microfibril formation in vivo.
Starch Accumulation in “Heavy” Roots under PEG Stress
Besides a short root phenotype (Supplemental Fig. S6A), OsRePRP2-OX also displayed a “heavy” root phenotype (Fig. 7, M and N). In distilled water, root-tip segments of wild-type and OsRePRP-Ri plants floated on the water surface, whereas OsRePRP2-OX segments submerged beneath the surface of water, which suggests that OsRePRP2-OX segments were heavy (Fig. 7M). After ABA treatment, wild-type root-tip segments also submerged, resembling the “heavy” root phenotype of OsRePRP2-OX under the control condition (Fig. 7M). However, after ABA treatment, the “heavy” root phenotype was less apparent in OsRePRP-Ri than in the wild type (Fig. 7M). The quantification of dry weight per root length per seedling also confirmed the heavy root phenotype (Fig. 7N).
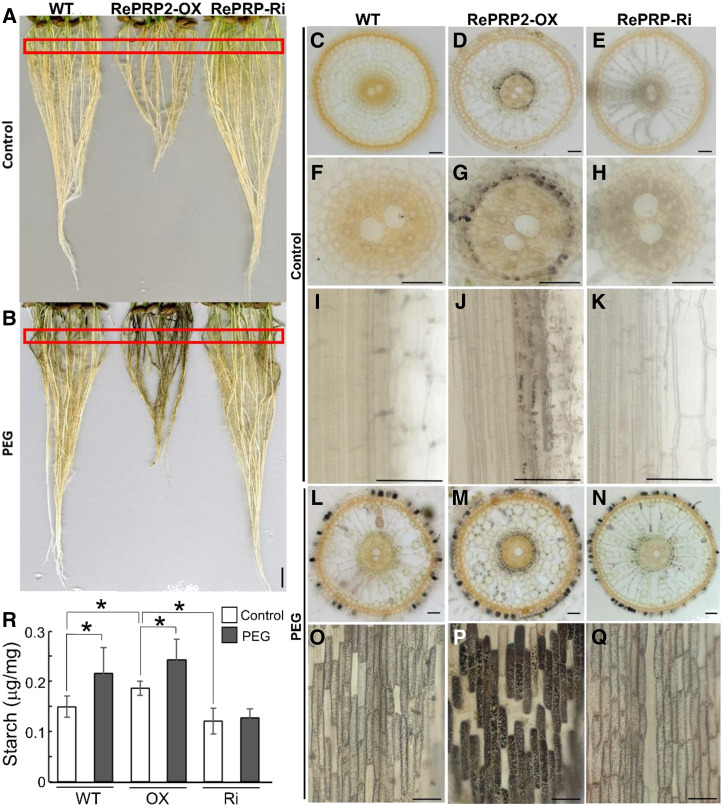
We wondered whether starch accumulation storage contributed to the biomass accumulation in OsRePRP2-OX, so we used iodine staining to address this question. Rice roots accumulated starch under PEG stress, as shown by the dark iodine-stained color (Fig. 8, A and B). OsRePRP2-OX showed more dark-stained roots than the wild type or OsRePRP-Ri after PEG treatment (Fig. 8B). Sectioning was conducted to further observe the starch accumulation phenotype (Fig. 8, C–Q; Supplemental Fig. S9). Under control conditions, some starch granules were observed in endodermal cells of OsRePRP2-OX (Fig. 8, D, G, and J; Supplemental Fig. S9B), but not in the wild type or OsRePRP-Ri (Fig. 8, C, E, F, H, I, and K; Supplemental Fig. S9, A and C). No starch granules were observed in epidermal cells in the wild type, OsRePRP2-OX, or OsRePRP-Ri (Supplemental Fig. S9, D–F). Under PEG stress, starch granules accumulated in endodermal and epidermal cells in the wild type, OsRePRP2-OX, and OsRePRP-Ri (Fig. 8, L–Q; Supplemental Fig. S9, G–I). OsRePRP2-OX showed the most severe starch accumulation phenotype (Fig. 8P; Supplemental Fig. S9H), with not much difference between the wild type and OsRePRP-Ri (Fig. 8, O and Q; Supplemental Fig. S9, G and I). In starch-accumulating cells, some very huge starch granules were observed in OsRePRP2-OX (Supplemental Fig. S9H), but not in the wild type or OsRePRP-Ri, which showed small and uniformly sized starch granules (Supplemental Fig. S9, G and I). Starch content measurements also showed high starch accumulation in OsRePRP2-OX (Fig. 8R), which suggests that OsRePRPs may play a role in storage nutrient accumulation under PEG stress.
Figure 8.
Starch accumulation in rice roots after PEG treatment. A and B, Iodine-stained rice roots from 14-d-old seedlings of the wild type (WT), OsRePRP2-OX, and OsRePRP-Ri under control (A) or PEG (B) treatments. Red rectangles indicate root areas that were sectioned for observation in C to. Four biological replicates were performed and at least two independent transgenic plants for each genotype were observed (OsRePRP2-OX lines 2 and 24 and OsRePRP-Ri lines 5–7). C to Q, Cross and longitudinal iodine-stained root sections of the wild type (C, F, I, L, and O), OsRePRP2-OX (D, G, J, M, and P), and OsRePRP-Ri (E, H, K, N, and Q) under control (C–K) or PEG (L–Q) treatments. Three biological replicates were performed, and at least two independent transgenic lines for each genotype were observed (OsRePRP2-OX lines 10, 19, and 24 and OsRePRP-Ri lines 5 and 7). R, Starch content measurements in whole roots of the wild type, OsRePRP2-OX (OX), and OsRePRP-Ri (Ri) under control or PEG treatments. Significant differences are indicated with asterisks (P < 0.05, two-tailed Mann-Whitney U test). Data are means ± sd of six technical repeats. Three biological replicates were performed and two independent transgenic lines for each genotype were measured (OsRePRP2-OX lines 10 and 24 and OsRePRP-Ri lines 5 and 6). Scale bars = 1 cm (A and B) and 50 μm (C–Q).
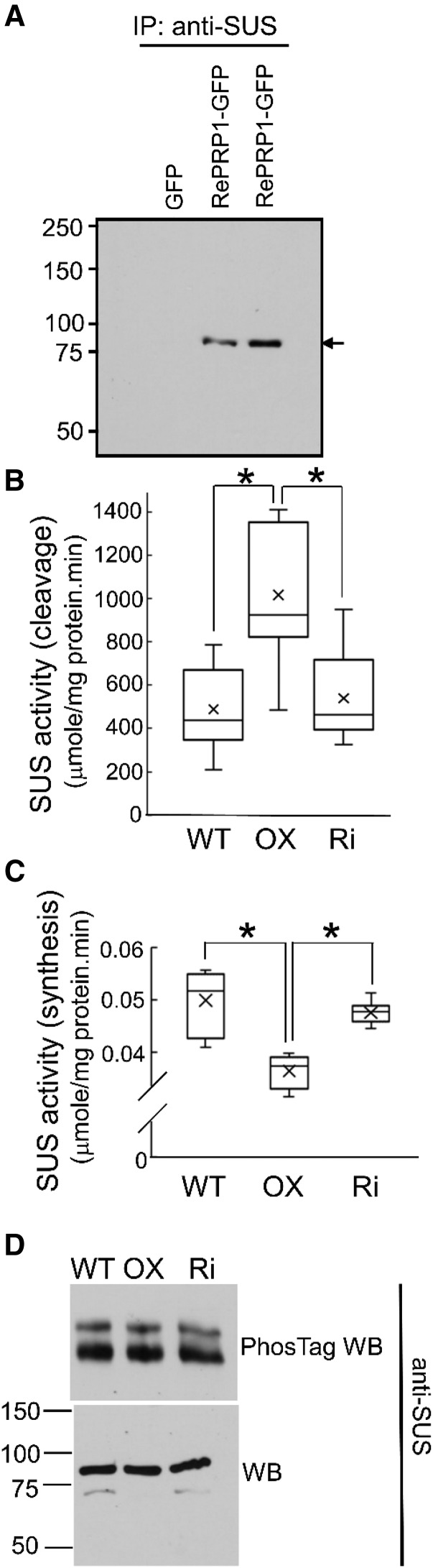
SUS Enzyme Activity Increased in OsRePRP2-OX
SUS is a key enzyme in cleaving Suc to provide NDP-Glc for starch biosynthesis in vivo (Koch, 2004). From the previous co-IP experiments using anti-GFP antibodies, peptides of SUS were detected in OsRePRP1-GFP and OsRePRP2-GFP, but not in GFP transgenic plants, by LC-MS/MS analysis (Supplemental Table S3). The co-IP of SUS in OsRePRP-GFP transgenic plants was also confirmed by western blot analysis (Fig. 9A), most likely due to SUS binding to actin filaments in vivo (Winter et al., 1998). We wondered whether OsRePRP could affect the enzyme activity of SUS by reducing the number of actin filaments. Thus, we examined the enzyme activity of SUS in both cleavage and synthetic directions of the wild type, OsRePRP2-OX, and OsRePRP-Ri (Fig. 9, B and C). The SUS activity in the cleavage direction was 2-fold higher in OsRePRP2-OX than in the wild type or OsRePRP-Ri, whereas the activity in the synthetic direction was lower in OsRePRP2-OX than in the wild type or OsRePRP-Ri (Fig. 9, B and C). Because the enzyme activity of SUS in maize (Zea mays) was associated with its phosphorylation (Huber et al., 1996), we examined the phosphorylation status of SUS in the wild type, OsRePRP2-OX, and OsRePRP-Ri using PhosTag western blot analysis (Fig. 9D). Both nonphosphorylated and phosphorylated forms of SUS were detected in the wild type, OsRePRP2-OX, and OsRePRP-Ri, with no differences in levels (Fig. 9D), which suggests that the phosphorylation status of SUS was not affected by OsRePRP. Thus, OsRePRP may affect SUS enzyme activity by interacting with actin filaments, but it does not affect SUS phosphorylation in planta.
Figure 9.
Enzyme activity of SUS was changed in OsRePRP2-OX. A, Co-IP (IP) assays of GFP, OsRePRP1-GFP, and OsRePRP2-GFP transgenic rice. Total protein was extracted from whole roots of 4-d-old seedlings treated with 2 μm ABA for 2 d. Western blot analysis involved anti-SUS antibodies. The arrow indicates 93-kD SUS. Two biological replicates were performed. B and C, Enzyme activity of SUS in the cleavage (B) and synthetic (C) directions. Total protein extract from 14-d-old whole roots of the wild type (WT), OsRePRP2-OX (OX), and OsRePRP-Ri (Ri) is shown in box plots. The line inside the box indicates the median, and the ex indicates the mean. Bottom and top box edges are the 25th to 75th percentiles, respectively, and whiskers indicate the range. Significant differences are indicated with asterisks (P < 0.01, two-tailed Mann-Whitney U test). Nine repeats from three independent biological replicates were performed. D, PhosTag western blot (WB) and WB analysis with anti-SUS antibodies. Total protein extract from 14-d-old whole roots of the wild type, OX, and Ri was assayed. Four biological replicates were performed.
DISCUSSION
Herein, we presented the unique case of an intrinsically disordered protein specifically interacting with the cytoskeleton proteins actin and tubulin to regulate development and biochemistry in rice root cells under water deficit. The studies of OsRePRP not only expand our knowledge of the roles and diversity of plant intrinsically disordered proteins but also foster communication across fields of plant and animal biology.
OsRePRPs are composed of PEPK repeats in OsRePRP1 and PQPN/PDPK repeats in OsRePRP2 (Fig. 1A; Supplemental Fig. S3A). OsRePRP1.1 and OsRePRP1.2 share 84% sequence identity and OsRePRP2.1 and OsRePRP2.2 share 94% sequence identity (Tseng et al., 2013). OsRePRPs are highly induced by ABA and water deficit, as shown by in situ hybridization studies and transcriptional analysis (Tseng et al., 2013; Supplemental Figs. S5 and S10). The induction of OsRePRP expression by ABA was highly repressed in the double-gene knockdown line OsRePRP-Ri as compared with the wild type (Supplemental Fig. S10). Thus, OsRePRP1 and OsRePRP2 may share similar functions in repression of root development by ABA/water deficit. However, we did not observe any obvious phenotypic changes of OsRePRP1-OX compared with the wild type (Supplemental Fig. S6B), and only the double-gene knockdown line OsRePRP-Ri, and not the single-gene knockdown lines OsRePRP1-Ri or OsRePRP2-Ri, showed the reduced ABA effect (Tseng et al., 2013; Supplemental Fig. S6A). Thus, we cannot rule out that OsRePRP1 may play a role similar to that of OsRePRP2, and we conclude that OsRePRPs are sufficient and necessary for ABA/water deficit repression of root development.
In the face of adversity and danger, animals can escape by using their highly modulated skeletons and muscles. Titin and nebulin rule myosin and actin contraction in muscle cells, thus controlling muscle contraction (Labeit and Kolmerer, 1995; Wang et al., 1996; Gutierrez-Cruz et al., 2001; Ma and Wang, 2002). PEVK motifs of titin are repetitive and intrinsically disordered, with highly charged residues (Labeit and Kolmerer, 1995; Gutierrez-Cruz et al., 2001). Tandem repeats of titin and nebulin provide binding sites for actin and myosin and scaffolding/crosslinking to the filamentous structures (Labeit and Kolmerer, 1995; Wang et al., 1996; Gutierrez-Cruz et al., 2001; Ma and Wang, 2002). OsRePRPs also showed repetitive (PEPK, PQPN, and PDPK), intrinsically disordered, and internal duplication patterns (Fig. 1A; Supplemental Figs. S1, S2, and S3A) similar to those of titin and nebulin. Although rooted plants are sessile and do not have muscle cells to escape from environmental stresses, disordered OsRePRPs can function to regulate highly organized cytoskeleton filaments (actin and tubulin) to adapt plant growth under stress (Figs. 4–6; Supplemental Figs. S6–S8). Similar to titin and nebulin controlling the elasticity of mammal muscle cells (Ma and Wang, 2002), OsRePRP regulates the adjustable elasticity needed for cell expansion in rice by directly binding to actin and tubulin (Fig. 1; Supplemental Fig. S3). Other than the studies on the cytoskeleton reorganization in response to abiotic and biotic stresses (Wang et al., 2011; Fujita et al., 2013; Li et al., 2015), OsRePRP is a novel case of an intrinsically disordered protein regulating the cytoskeleton in plants. Our findings suggest that these highly ordered filamentous structures are regulated by repetitive disordered proteins in both plants and animals.
Our previous study mainly emphasized the plasma membrane localization of OsRePRPs (Tseng et al., 2013). However, cytosolic localization of OsRePRPs was also evident in both cell fractionation and transient-expression experiments (Tseng et al., 2013). Since OsRePRPs contain signal peptides (Tseng et al., 2013), there are two possible explanations for their dual localization. First, signal peptides may act cotranslationally or posttranslationally (Panzner et al., 1995), leading to the presence of a protein in both the cytosol and the plasma membrane. Second, the primary amino acid sequence prediction may not precisely reflect the protein localization affected by protein-protein interactions (Koroleva et al., 2005). The immunogold-TEM observations presented here (Fig. 3; Supplemental Fig. S4G) and our previous publication (Tseng et al., 2013) both support the notion that OsRePRPs appear to be present in both the plasma membrane and the cytosol.
In the rice root protoplast transient-expression system, OsRePRP2-DsRed was primarily expressed in the cytosol with some punctuated patterns (Supplemental Fig. S11). While overexpressing the other cytoskeleton-binding proteins, LifeAct-GFP and GFP-MBD, OsRePRP2-DsRed showed more obvious filamentous patterns colocalized with LifeAct-GFP and GFP-MBD (Fig. 2). These results suggest that intrinsically disordered OsRePRP may have multifaceted functions that are affected by other cytoskeleton-binding proteins and various environmental conditions; thus, OsRePRP2-DsRed showed distinct colocalized patterns with both LifeAct-GFP and GFP-MBD. Nevertheless, our co-IP dataset did not reveal other cytoskeleton-binding proteins that may interact with OsRePRPs. The rice genome may not have been fully annotated, and more studies regarding this issue should be conducted in the future.
The coupling between the cytoskeleton and extracellular matrix in animals is well understood by the study of integrin-adhesion-receptor signaling (Schwartz and Ginsberg, 2002). However, in higher plants, critical linker molecules between the cytoskeleton and extracellular matrix (the cell wall in plants) are still missing (Baluska et al., 2003). With a reduced F-actin amount and disoriented microtubule organization (Figs. 4–6; Supplemental Fig. S7), OsRePRP2-OX transgenic rice showed impaired cell wall polysaccharide deposition and disordered cellulose microfibril organization (Fig. 7, A–L). Thus, OsRePRP may affect noncellulosic polysaccharide trafficking and cellulose patterning by altering the F-actin network and microtubule arrangement. From our results, we propose that OsRePRP is a stress-induced cytoskeleton-cell wall modulator. Unlike the transmembrane receptor integrins in animals (Schwartz and Ginsberg, 2002), OsRePRP has a distinct role in regulating the very dynamic interactions between the cytoskeleton and cell wall in plants.
Although SUS interacting with actin filaments in vivo is assumed to be a control mechanism (Koch, 2004), we lack direct evidence to support this idea. Herein, we showed increased SUS enzyme activity in OsRePRP2-OX (Fig. 9B), so reducing F-actin may affect the enzyme activity of SUS in vivo. Phosphorylation of SUS in maize has been reported to be associated with its enzyme activity (Huber et al., 1996). However, in our case, the increase in SUS enzyme activity was not associated with its phosphorylation (Fig. 9D), distinct from the regulation explored in maize SUS (Huber et al., 1996). In animals, cytoskeleton remodeling releases more free forms of aldolase, thereby increasing the glycolysis flux (Hu et al., 2016). Reducing the F-actin level in OsRePRP2-OX may increase SUS activity by releasing more free forms of SUS, similar to the case with animal aldolase (Hu et al., 2016). However, whether OsRePRP interacts with SUS directly or indirectly (through actin filaments) remains to be investigated. Our observations have to be interpreted with caution, because the cortex cells in the cross sections of the wild type, OsRePRP2-OX, and OsRePRP2-Ri look quite different in morphology (Fig. 8) probably due to the effect of ReOsPRP2 on root elongation. Although the samples were sectioned at the same distance from root tips, i.e. 0.5 cm, they may reflect different root developmental stages caused by the differential expression of OsRePRP2. Nevertheless, the differences of SUS activities can at least partially account for the different level of starch accumulation in these samples. The interaction (direct or indirect) of OsRePRPs with SUS may be related not only to starch biosynthesis but also to cell wall metabolism by providing NDP-Glc as the substrate.
Our current study explains the reduced cell length phenotype of OsRePRP2-OX found in our previous study (Tseng et al., 2013) and supports that both microtubules and actin filaments are critical for cell expansion (Smith, 2003). Based on observations of disorganization of cell wall cellulose microfibrils and cortical microtubules in OsRePRP2-OX (Figs. 6 and 7), people may expect to see a phenotype of anisotropic cell expansion rather than just reduced cell elongation. However, previous studies have shown that the degree of growth anisotropy was not correlated with the degree of alignment of either microtubules or microfibrils (Baskin et al., 1999), and cell expansion in longitudinal and radial directions can be regulated independently in roots (Liang et al., 1997; Baskin, 2005). Hence, the phenotype of OsRePRP2-OX likely supports the notion that anisotropic cell expansion and longitudinal cell elongation are regulated independently in rice roots. The ultimate goal of ABA/water deficit-induced OsRePRP regulation for cytoskeleton dynamics and cell wall organization is to aid plant survival under adversity. Because ABA plays a role in seed dormancy and desiccation tolerance during seed maturation, the function of ABA-inducible OsRePRP is analogous to the action of ABA in seeds, which halts root elongation and allows more starch accumulation (Fig. 8; Supplemental Figs.s S6A and S9), thus shifting the roots to a dormant storage organ resembling seeds.
This “short-but-heavy root” strategy is similar to the rice flooding-tolerance gene SUBMERGENCE-1, which causes growth quiescence during flooding that is associated with the capacity for regrowth on desubmergence (Fukao et al., 2006; Xu et al., 2006). Thus, the “quiescence” concept may help plants overcome water deficit and water stresses. Highly charged intrinsically disordered proteins have high hydration capacities and preference for absorption of charged solute ions such as dehydrins to protect plants against water loss (Tompa et al., 2006; Uversky, 2011). Of note, disordered OsRePRP also functions in the water deficit response.
Overall, our study highlights the importance of OsRePRP in cytoskeleton-cell wall regulation and cytoskeleton-mediated cellular processes and suggests a common “order by disorder” mechanism in both plants and animals as well as a practical basis for crop improvement designs.
MATERIALS AND METHODS
Protein Sequence Analysis
OsRePRP sequences were assessed using four different Predictor Of Natural Disordered Regions predictors, VSL2, VL3, VL-XT, and XL1-XT (Xue et al., 2010), and repeated regions were further analyzed by RADAR software to reveal superrepeat segments.
Recombinant Protein Purification
The sequences of OsRePRP1.1 from 23 to 360 amino acids and OsRePRP2.1 from 19 to 247 amino acids were amplified with the primer pairs RE-OCP-GAP-F/RE-OCP-GAP-F and OSR2.1-GAP-F/OSR2.1-GAP-R, respectively, and cloned into the expression vector pGAPZaC using 5′-EcoRI/NotI-3′ sites. Recombinant OsRePRP1.1 and OsRePRP2.1 proteins were expressed in Pichia pastoris SMD1168 according to the manufacturer’s manual (Invitrogen). The serial segments OsRePRP1.151–159, OsRePRP1.151–131, OsRePRP1.151–105, OsRePRP1.1106–131, OsRePRP2.160–179, OsRePRP2.160–132, OsRePRP2.188–132, and OsRePRP2.160–87 were amplified with the primer pairs YSC5/YSC6, YSC21/YSC22, YSC5/YSC9, YSC10/YSC11, YSC12/YSC13, YSC12/YSC15, YSC17/YSC18, and YSC12/YSC16, respectively, and cloned into the expression vector pET28a using 5′-XhoI/EcoRI-3′ sites. The segments OsRePRP1.151–131 and OsRePRP2.188–132 were cloned into the pET20b and pColdII vectors using 5′-NdeI/XhoI-3′ and 5′-XhoI/EcoRI-3′ sites, respectively. All rOsRePRP1.1 and rOsRePRP2.1 segments were expressed with His-Tag in Escherichia coli and purified using Ni Sepharose (GE Healthcare); the His-Tag was not removed. The primer sequences for constructs and induction conditions for the recombinant protein expression in E. coli are listed in Supplemental Tables S4 and S5, respectively. Full-length rOsRePRP1.1 and rOsRePRP2.1 used for AG binding assays were constructed and purified as described (Tseng et al., 2013). Purified recombinant proteins were dialyzed in 50 mm of HEPES (pH 7.5) and quantified by the standard Bradford assay (Bio-Rad).
MST Binding Assays
The binding affinity of purified rOsRePRPs to rabbit muscle actin (AKL-99, Cytoskeleton), porcine brain tubulin (T240, Cytoskeleton) and AG (10830, Sigma) was measured using MST with the Monolith NT.115 instrument (Nanotemper Technologies). For actin and tubulin binding, a serial dilution of rOsRePRPs was incubated with 1 to 2 nm of NT-647-labeled actin or tubulin for 10 min in the assay buffer (50 mm HEPES and 0.05% [v/v] Tween 20 [pH 7.5]). For AG binding, a serial dilution of AG was incubated with 3 nm NT-647-labeled rOsRePRPs for 10 min in the assay buffer (50 mm HEPES, 150 mm NaCl, 5 mm CaCl2, 1 mm MgCl2, and 0.05% [v/v] Tween 20 [pH 7.5]). The sample was loaded into the NanoTemper glass capillaries, and MST involved 20% light emission diode and 20% MST power. Kd was calculated using the mass action equation with the NanoTemper software in triplicate experiments.
Plant Materials and Growth Conditions
The rice (Oryza sativa) cultivar ‘Tainung 67’ was used throughout this study. Seeds of wild-type and transgenic rice were sterilized with 2% (v/v) sodium hypochlorite for 20 min, washed thoroughly with distilled water, soaked in distilled water at 28°C for 2 d in darkness, then germinated in petri dishes containing water with or without hygromycin B (25 μg/mL) at 28°C for 3 d. Three-day-old seedlings were transferred and cultivated in a beaker containing one-half strength Kimura B solution (Baba and Takahashi, 1956). The hydroponically cultivated seedlings were grown at 28°C and 90% relative humidity in a 14-h-light/10-h-dark condition. Transgenic rice plants of OsRePRP2-OX, OsRePRP-Ri, 35Spro::GFP, Ubipro::OsRePRP1-GFP, Ubipro::OsRePRP2-GFP, and OsRePRP2.1pro::GUS were generated as previously described (Tseng et al., 2013).
Rice Root Protoplast Transfection
The expression vectors of pSAT6-EGFP-C1/N1 and pSAT6-DsRed-C1/N1 were used in the rice root protoplast transient-expression system (Tzfira et al., 2005). Plasmids containing LifeAct-GFP with OsRePRP2-DsRed or DsRed and GFP-MBD with OsRePRP2-DsRed or DsRed were generated. The 51-bp LifeAct with the GGSG linker was produced by mixing the primer pair HAS106/HAS107 at room temperature for 30 min. LifeAct was further PCR-amplified with the primer pair HAS108/HAS109 and cloned into pSAT6-EGFP-N1 with restriction enzymes NcoI and BamHI. The 1,254-bp MBD (2048–3661 bp in mouse MAP4 complementary DNA) was amplified with the primer pair HAS140/HAS141 from the complementary DNA of C57BL/6 mice, which was a gift from Dr. Ya-Lin Lin (Institute of Biomedical Sciences, Academia Sinica), and cloned into pSAT6-EGFP-C1 with restriction enzymes HindIII and EcoRI. The 741-bp OsRePRP2 amplified by the primer pair HAS126/HAS127 was cloned into pSAT6-DsRed-N1. The 35Spro::OsRePRP2-DsRed and 35Spro::DsRed fragments amplified with the primer pair HAS142/HAS137 were further cloned into plasmids containing LifeAct-GFP and GFP-MBD using NdeI. Primer sequences are given in Supplemental Table S4. The isolation of rice root protoplasts followed the published protocol with slight modifications (Evrard et al., 2012). Whole roots harvested from 5-d-old rice seedlings were chopped in digestion buffer (400 mm mannitol, 20 mm MES hydrate, 20 mm KCl, 1.1% [v/v] cellulase R10, 0.9% cellulase RS, 0.3% [v/v] macerozyme R10, 0.12% [v/v] pectolyase Y-23, 10 mm CaCl2, 0.1% [v/v] BSA, and 2.5 mm β-mercapoethanol [pH 5.7]), vacuumed for 7 min, then incubated for 4 h with 80-rpm shaking at room temperature. The solution was filtered through a 40-μm cell strainer and protoplasts were harvested by centrifugation at 500g for 5 min. Protoplasts were washed and resuspended in WI buffer (400 mm mannitol, 20 mm MES hydrate, and 20 mm KCl [pH 5.7]). The concentration of protoplasts was adjusted to 800,000 protoplasts/mL using a hemocytometer. The transfection of rice root protoplasts involved the PEG-mediated method (Yoo et al., 2007): ∼15 μg plasmid DNA in 10 μL was mixed with 100 μL protoplasts and 110 μL PEG solution (40% [v/v] PEG 4000, 0.2 m mannitol, and 0.1 m CaCl2) and incubated at room temperature for 15 min. The transformed protoplasts were incubated in WI buffer for 16 to 24 h in the dark before observation.
Fluorescence Imaging in Rice Root Protoplasts and Analysis
The 3.5-μL protoplast suspension was placed in a well created by a 6-mm hollow circle tap pasted on slides and observed under a Zeiss LSM 880 confocal laser scanning microscope with a 40× water objective (numerical aperture [NA] 1.2). Fluorescence images were taken in the Airyscan superresolution mode and differential interference contrast (DIC) images were taken in the standard confocal mode. Samples were excited using an argon ion laser at wavelength 488 nm for GFP and a HeNe ion laser at 561 nm for DsRed. A 488/561/633 dichroic beam splitter was used to detect fluorescence; GFP fluorescence was detected with a 495- to 550-nm bandpass filter and DsRed fluorescence was detected with a 570- to 620-nm bandpass filter. The laser power for colocalization assays was 5% for GFP 488 nm and 40% for DsRed 561 nm. Image analysis was undertaken using ImageJ and the Coloc 2 plugin (Arena et al., 2017) to calculate Pearson's R correlation values. All the Pearson's R correlation values were measured over the entire protoplast and involved no region of interest selection by hand or thresholding. For observation of the LifeAct-GFP with DsRed transfection control in wild-type, OsRePRP2-OX, and OsRePRP-Ri protoplasts, images were taken under identical settings: 2% GFP 488-nm laser power, 10% DsRed 561-nm laser power, stack scanning mode, zoom 6, and image sizes 34.23 µm for x,, 34.23 µm for y, and, 5.00 µm for z, with 20 sections of 0.25-µm Zscaling. For observation of the GFP-MBD with DsRed transfection control in wild-type, OsRePRP2-OX, and OsRePRP-Ri protoplasts, the laser power was 3% for GFP 488 nm and 40% for DsRed 561 nm. Z-scaling data sets are shown in Supplemental Figures S12–S14.
Actin Filament Phalloidin Staining and F-actin Quantification
F-actin staining was performed as described (Yang et al., 2011). For F-actin observation, we used 8-d-old seedlings (control) and 7-d-old seedlings treated with 2 μm ABA for 1 d. Root tips of 1-cm segments were cut and incubated in PME buffer (100 mm PIPES, 10 mm EGTA, and 5 mm MgSO4 [pH 6.8]) containing 300 μm m-maleimidobenzoyl-N-hydroxysuccinimide ester, 1.5% (v/v) glycerol, and 0.1% (v/v) Triton X-100 with gentle shaking for 30 min. Samples were washed twice with PME buffer, then fixed in PME buffer containing 2% paraformaldehyde for 30 min. After rising thoroughly in PME buffer, samples were stained with actin-staining buffer (PME, 1.5% [v/v] glycerol, 0.1% [v/v] Triton X-100, and 66 nm Alexa Fluor 488-phalloidin; A12379, Thermo Fischer Scientific) at 4°C in the dark overnight, then washed three times in PME buffer before confocal laser microscopy observation. Images were obtained with a 63× water objective (NA 1.2) using a Zeiss LSM 880 confocal laser scanning microscope. Fluorescence was excited at 488 nm and collected with a 492- to 560-nm filter. The overall fluorescence signal of each genotype was obtained under identical staining conditions, with confocal settings 3% 488-nm laser power, 54-µm pinhole, stack scanning mode, zoom 3, and image sizes of 45 μm for x and 45 µm for y, with 10 to 40 sections of 0.42-µm Z scaling. Thirty images of at least three independent transgenic lines for each genotype were processed to determine average fluorescence intensity with ImageJ.
Immunofluorescence Staining of Microtubules
We used 6-d-old seedlings (control) and 4-d-old seedlings treated with 2 μm ABA for 2 d for microtubule assays, as described in Deng et al. (2015). Root tips of 1-cm segments were cut and fixed in 4% (v/v) paraformaldehyde in PME buffer 1 (50 mm PIPES, 2 mm MgSO4, and 2 mm EGTA [pH 6.9]) containing 0.05% (v/v) Triton X-100 for 30 min. After washing thoroughly with PME buffer 1, samples were digested with 2% (v/v) cellulase R-10 and 1% (v/v) pectolyase Y-23 (both from Yakult Pharmaceutical Industry) in PME buffer 1 at 37°C for 30 min to 1 h The softened root tips were washed gently with PME buffer 1 and frozen and thawed twice in liquid nitrogen. Samples were treated with the blocking buffer (3% [v/v] BSA in phosphate-buffered saline with Tween 20 [PBST; 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.76 mm KH2PO4, and 0.05% Triton X-100]) for 1 h at room temperature, then incubated with 1:50 diluted primary antibodies anti-α-tubulin (T9026, Sigma) in the blocking buffer at 4°C overnight. After washing with PBST five times, samples were incubated with 1:800-diluted secondary antibodies Alexa Fluor 488 goat antimouse IgG (A-11001, Thermo Fischer Scientific) in PBST at 37°C for 3 h. After washing four times with PBST and once with PBS, samples were mounted with PBS containing 50% (v/v) glycerol and 0.1% (v/v) o-phenylenediamine. Images were obtained with a 63× water objective (NA 1.2) under a Zeiss LSM 880 confocal laser scanning microscope. Fluorescence was excited at 488 nm and collected with a 492- to 560-nm filter. The overall fluorescence signal of each genotype was obtained under identical staining conditions and confocal settings 8% 488-nm laser power, 58-μm pinhole, and stack scanning mode.
Immunogold Electron Microscopy
Segments of 3-d-old root tips were cut and frozen in a high-pressure freezer (Leica EM PACT2) at 200 to 205 MPa. Freeze substitution was conducted in anhydrous acetone solution containing 0.1% uranyl acetate and 0.2% glutaraldehyde with an automatized Leica EM AFS2 system. Samples were successively kept at −85°C for 3 d, −60°C for 1 d, −20°C for 1 d, 0°C for 1 d, and at room temperature. After two acetone changes, root segments were infiltrated and embedded in LR Gold Resin. Ultrathin sections of 90 to 120 nm were cut using a Reichert Ultracut S or Leica EM UC6 microtome and collected on 100-mesh nickel grids, which were floated on PBS for 15 min, then on PBS and 1% (v/v) BSA for 15 min. The grids were incubated with the primary antibodies 1:50-diluted rabbit anti-GFP (ab6556, Abcam) and 1:20-diluted mouse anti-α-tubulin (T9026, Sigma) or mouse antiactin (A0480, Sigma) for 1 h at room temperature. After four washes with PBS, grids were incubated with 12-nm/18-nm Colloidal Gold Donkey antirabbit/antimouse IgG (Jackson ImmunoResearch) at room temperature for 1 h, then washed four times with PBS and three times with distilled water. After immunogold labeling, sections were stained with 5% (v/v) uranyl acetate for 10 min and 0.5% (v/v) lead citrate for 4 min. Sections were observed under a Tecnai G2 Spirit transmission electron microscope (FEI, Hillsboro) at 80 KV, and images were obtained with a Gatan Orius CCD camera. The distance from the 18-nm gold particle to the closest 12-nm gold particle was calculated using ImageJ, and quantification of the immunogold TEM data was performed as described (Bergersen et al., 2008).
Metabolic Click Labeling
Fuc alkyne incorporation and fluorescent labeling were performed as described, with slight modifications (Anderson et al., 2012). Seven-day-old seedlings were transferred to one-half strength Kimura B solution containing 10 μm Fuc alkyne (C10264, Thermo Fischer Scientific) and incubated for 24 h under the growth conditions described previously, then washed three times and transferred to labeling solution (one-half strength Kimura B solution containing 1 mm CuSO4, 1 mm ascorbic acid, and 0.4 μm Alexa Fluor 488-azide [A10266, Thermo Fischer Scientific]) for labeling at 28°C in the dark for 2 h. Confocal laser microscopy was performed with a 40× water objective (NA 1.2) using a Zeiss LSM 880 confocal laser scanning microscope. Fluorescence was excited at 488 nm and collected with a 492- to 560-nm filter. The overall fluorescence signal of each genotype was obtained under identical staining conditions, with confocal settings 3% 488-nm laser power, 58-µm pinhole, stack scanning mode, zoom 3, and image sizes of 45 µm for x and 45 µm for y, with 30 sections of 0.45-µm Z scaling. Thirty images of at least three independent transgenic lines for each genotype were processed to determine average fluorescence intensity with ImageJ.
Cell Wall Texture Observation
For cell wall cellulose microfibril observation, 8-d-old seedlings (control) and 4-d-old seedlings treated with 2 μm ABA for 4 d were used. Cell wall preparation was as described (Sugimoto et al., 2000), with slight modifications. The whole roots were cryoprotected in PME buffer 2 (25 mm PIPES, 0.5 mm MgSO4, and 2.5 mm EGTA [pH 7.2]) containing 25% and 50% (v/v) dimethyl sulfoxide for 10 min for each step. Root tips were excised, placed on sample carriers, and cut using a glass knife on a Leica Ultracut UCT ultramicrotome equipped with the Leica EM FCS cryo-chamber attachment at −120°C. The remaining root strips were thawed in PME buffer 2 containing 50% (v/v) dimethyl sulfoxide, then transferred to PME buffer 2. Samples were treated with acetic acid, nitric acid, and distilled water (8:1:2) for 1 h at 95°C. After a thorough washing in distilled water, samples were dehydrated with an ethanol series (30%, 50%, 70%, 95%, and 100% [v/v] three times, 30 min for each step), critical-point dried with CO2 and further mounted on carbon tape-covered stubs with the cut surface facing upward, and coated with carbon after platinum at 20 mA for 80 s. The cell wall fine structure was examined with a field-emission scanning electron microscope (JSM-7100F, JEOL) fitted with an accelerated voltage 5 kV, probe current 0.1 nA, and working distance 5 to 6 mm. The quantitative directionality analysis involved using the Fiji package (Schindelin et al., 2012). The Fourier component method was used, and orientation from −90° to 90° was analyzed using the Fiji plug-in “Directionality” according to the manufacturer’s instructions.
Starch Staining and Quantification
For starch staining, 14-d-old control seedlings and 14-d-old seedlings treated with 20% (v/v) PEG6000 for 1 h (PEG) were used. Rice roots were stained in 1:10 diluted 5% (v/v) Lugol's iodine solution for 10 min, destained with distilled water for 30 min, and scanned (Epson) for root architecture images. For sectioning, 1-cm root segments were embedded in 5% agar and cut into 100- to 120-μm sections using a DTK-100 microslicer (Dosaka EM). The sections were stained with iodine and observed under a Zeiss Axio Imager Z1 microscope. For starch quantification, 14-d-old control seedlings and 14-d-old seedlings treated with 20% (v/v) PEG6000 for 5 h were used. Whole roots of 10 to 20 seedlings were ground in liquid nitrogen with a pestle and mortar and dried at 65°C, then dry weight was measured. A starch colorimetric/fluorometric assay kit (K647, Biovision) was used to determine the level of starch.
Enzyme Activity Assays and PhosTag Western Blot Analysis
Whole roots of 10 to 20 14-d-old seedlings were ground into a fine powder in liquid nitrogen using a pestle and mortar, mixed with 1 mL ice-cold extraction buffer (100 mm HEPES-KOH [pH 7.5], 5 mm MgCl2, and 5 mm dithiothreitol), and centrifuged at 11,000g for 15 min at 4°C. The supernatant was then collected and used for the measuring SUS enzyme activity in the cleavage direction according to Sun et al. (1992). The protein extract was quantified using the Bio-Rad Protein Assay Reagent, adjusted to 1 mg mL−1, mixed with the reaction mixture (50 mm HEPES-KOH [pH 7], 2 mm MgCl2, 15 mm KCl, 50 mm Suc, 5 mm UDP, 0.5 mm NADP, 1 mm PPi, 1 U/mL phosphoglucomutase [P3397, Sigma], 2 U/mL Glc-6-P dehydrogenase [G8404, Sigma], and 0.5 U/mL UDP-Glc pyrophosphorylase [U8501, Sigma]), and incubated at 30°C for 30 min. All control reactions lacked Suc and UDP. The reaction was terminated by heating for 5 min in a boiling water bath. NADPH production was monitored at 340 nm with a SYNERY H1 microplate reader (BioTek). SUS enzyme activity in the synthetic direction was assayed using the Suc Synthase Microplate Assay Kit (MBS8243224, MyBioSource). The formation of UDP-Glc-dependent Suc was monitored at 480 nm with a SYNERY H1 microplate reader (BioTek).
For anti-SUS western blot analysis with/without PhosTag, whole roots of 14-d-old seedlings were ground, mixed with the extraction buffer (20 mm sodium phosphate [pH 7.4], 50 mm β-glycerophosphate, 100 μm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail) and centrifuged at 12,000g for 1 h at 4°C. The protein extract supernatant was quantified using Bio-Rad Protein Assay Reagent, and 2.5 μg of protein was separated in 6% (v/v) polyacrylamide gels. For PhosTag western blot analysis, the separating gel contained 50 μm Zn(NO3)2 and 50 μm PhosTag and the running buffer consisted of 100 mm Tris, 100 mm MOPS, 0.1% (v/v) SDS, and 5 mm NaHSO3 (pH 7.8). The electrophoresis was performed at constant 10 mA at 4°C for 10 h. After a washing in the transfer buffer (25 mm Tris-HCl [pH 7.4] and 192 mm Gly) containing 10 mm EDTA, the gel was transferred to the polyvinylidene difluoride membrane with the transfer buffer containing 5% (v/v) methanol and 1 mm EDTA at 24 V overnight. Immunoblot analysis was performed with 1:25,000-diluted anti-SUS antibodies (AS152830, Agrisera) and ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences). The experimental procedures of supplemental data are shown in Supplemental Methods.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/) with the following accession numbers: LOC_Os05g13900 (OsRePRP1.1), LOC_Os05g13940 (OsRePRP1.2), LOC_Os07g23660 (OsRePRP2.1), LOC_Os07g23640 (OsRePRP2.2), LOC_Os03g28330 (OsSUS1), LOC_Os06g09450 (OsSUS2), LOC_Os07g42490 (OsSUS3), LOC_Os03g22120 (OsSUS4), LOC_Os04g24430 (OsSUS5), LOC_Os02g58480 (OsSUS6), LOC_Os11g30430 (OsRBG1), LOC_Os03g08020 (OsEF1α), LOC_Os04g40950 (OsGADPH), and LOC_Os01g22490 (OsUBQ5), and in GenBank with the accession number X78205 (HvHVA1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Disorder prediction and internal duplication of OsRePRPs.
Supplemental Figure S2. Segmental repeat organization of OsRePRPs.
Supplemental Figure S3. In vitro binding assays of OsRePRPs.
Supplemental Figure S4. Co-IP assays and immunogold TEM of GFP, actin, and tubulin.
Supplemental Figure S5. Expression of OsRePRP2 in rice roots under control and ABA treatments.
Supplemental Figure S6. OsRePRP is necessary and sufficient in ABA repression of root elongation, affecting F-actin organization.
Supplemental Figure S7. OsRePRP affects F-actin organization in division and differentiation zones.
Supplemental Figure S8. OsRePRP affects F-actin organization under PEG treatment.
Supplemental Figure S9. Starch accumulation in rice roots after PEG treatment.
Supplemental Figure S10. Quantitative PCR analysis of OsRePRP expression.
Supplemental Figure S11. Subcellular localization of OsRePRP2 in rice root protoplasts.
Supplemental Figure S12. Rice root protoplasts coexpressing LifeAct-GFP and OsRePRP2-DsRed.
Supplemental Figure S13. Rice root protoplasts coexpressing GFP-MBD and OsRePRP2-DsRed.
Supplemental Figure S14. Rice root protoplasts expressing LifeAct-GFP.
Supplemental Table S1. Amino acid sequences of actin peptide hits by LC-MS/MS.
Supplemental Table S2. Amino acid sequences of tubulin peptide hits by LC-MS/MS.
Supplemental Table S3. Amino acid sequences of SUS peptide hits by LC-MS/MS.
Supplemental Table S4. Primer sequences used in plasmid construction.
Supplemental Table S5. Induction conditions of recombinant protein expression in E. coli.
Supplemental Table S6. Primer sequences used in reverse-transcription quantitative PCR analysis.
Supplemental Video. F-actin in rice root cortical cells viewed under confocal microscope from the peripheral region toward the center of the root.
Supplemental Methods. The experimental procedures of supplemental data.
Acknowledgments
The authors thank Dr. Shu-Chuan Chris Jao and Szuhuan Wang (Biophysics Core Facility, Academia Sinica), for assistance in operating the Monolith NT.115 pico instrument. Peptides were synthesized in the Institute of Biological Chemistry, Academia Sinica. Confocal images, immunogold TEM, and LC-MS/MS analysis were conducted in the Live-Cell-Imaging Core Lab, Plant Cell Biology Core Lab, and Proteomics Core Lab, respectively. Cryosection and field-emission scanning electron microscopy were done at the Imaging Core in the Institute of Cellular and Organismic Biology and the Electron Probe Micro-Analysis Lab in the Institute of Earth Sciences, respectively, Academia Sinica. They also acknowledge the In Situ Hybridization Core Facility of the Agricultural Biotechnology Research Center, Academia Sinica, for performing the in situ experiments. They thank Dr. Yoshiyuki Iizuka, Yu-Shiang Wang, Dr. Bo-Yan Lin, Yao-Kuan Huang, and Simon Wei for technical assistance and Edward Chee-Tak Yeung and Dr. Shiu-Cheung Lung for critical comments on the manuscript.
Footnotes
This research was supported by the Ministry of Science and Technology (MOST; grant nos. 106–2311–B–001–020 and 108–2311–B–005–007; postdoctoral fellowship no. 106–2811–B–001–112 to A.-S.H.) and in part by the Advanced Plant Biotechnology Center from the Featured Area Research Center Program within the framework of the Higher Education Sprout Project sponsored by the Ministry of Education (MOE) in Taiwan. A.-S.H. was also supported by a special grant contributed by T.-H.D.H.
Articles can be viewed without a subscription.
References
- al-Habori M.(1995) Microcompartmentation, metabolic channelling and carbohydrate metabolism. Int J Biochem Cell Biol 27: 123–132 [DOI] [PubMed] [Google Scholar]
- Anderson CT, Wallace IS, Somerville CR(2012) Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proc Natl Acad Sci USA 109: 1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arena ET, Rueden CT, Hiner MC, Wang S, Yuan M, Eliceiri KW(2017) Quantitating the cell: Turning images into numbers with ImageJ. Wiley Interdiscip Rev Dev Biol 6: e260. [DOI] [PubMed] [Google Scholar]
- Baba I, Takahashi Y(1956) Water and sand culture methods In Togari Y, Matsuo T, Hatamari M, Yamada N, Harada T, and Suzuki N, eds, Experimental Methods in Crop Science. Association of Agricultural Techniques, Tokyo, pp 157–185 [Google Scholar]
- Balasubramanian R, Karve A, Kandasamy M, Meagher RB, Moore B(2007) A role for F-actin in hexokinase-mediated glucose signaling. Plant Physiol 145: 1423–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F, Hlavacka A, Samaj J, Palme K, Robinson DG, Matoh T, McCurdy DW, Menzel D, Volkmann D(2002) F-actin-dependent endocytosis of cell wall pectins in meristematic root cells. Insights from brefeldin A-induced compartments. Plant Physiol 130: 422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F, Samaj J, Wojtaszek P, Volkmann D, Menzel D(2003) Cytoskeleton-plasma membrane-cell wall continuum in plants. Emerging links revisited. Plant Physiol 133: 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L, Lei L, Li S, Gu Y(2014) Cell wall, cytoskeleton, and cell expansion in higher plants. Mol Plant 7: 586–600 [DOI] [PubMed] [Google Scholar]
- Baskin TI.(2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Baskin TI, Meekes HT, Liang BM, Sharp RE(1999) Regulation of growth anisotropy in well-watered and water-stressed maize roots. II. Role of cortical microtubules and cellulose microfibrils. Plant Physiol 119: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergersen LH, Storm-Mathisen J, Gundersen V(2008) Immunogold quantification of amino acids and proteins in complex subcellular compartments. Nat Protoc 3: 144–152 [DOI] [PubMed] [Google Scholar]
- Boothby TC, Tapia H, Brozena AH, Piszkiewicz S, Smith AE, Giovannini I, Rebecchi L, Pielak GJ, Koshland D, Goldstein B(2017) Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell 65: 975–984.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA.(1997) Plant responses to water deficit. Trends Plant Sci 2: 48–54 [Google Scholar]
- Buckley TN.(2019) How do stomata respond to water status? New Phytol 224: 21–36 [DOI] [PubMed] [Google Scholar]
- Candat A, Paszkiewicz G, Neveu M, Gautier R, Logan DC, Avelange-Macherel MH, Macherel D(2014) The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 26: 3148–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong SD, Good AG, Taylor GJ, Freeman MC, Moorhead GB, Muench DG(2004) Large-scale identification of tubulin-binding proteins provides insight on subcellular trafficking, metabolic channeling, and signaling in plant cells. Mol Cell Proteomics 3: 970–983 [DOI] [PubMed] [Google Scholar]
- Covarrubias AA, Cuevas-Velazquez CL, Romero-Pérez PS, Rendón-Luna DF, Chater CCC(2017) Structural disorder in plant proteins: Where plasticity meets sessility. Cell Mol Life Sci 74: 3119–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S(2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng ZY, Liu LT, Li T, Yan S, Kuang BJ, Huang SJ, Yan CJ, Wang T(2015) OsKinesin-13A is an active microtubule depolymerase involved in glume length regulation via affecting cell elongation. Sci Rep 5: 9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich A, Faye L, Staehelin LA(1993) The plant Golgi apparatus: A factory for complex polysaccharides and glycoproteins. Trends Biochem Sci 18: 210–214 [DOI] [PubMed] [Google Scholar]
- Duncan KA, Huber SC(2007) Sucrose synthase oligomerization and F-actin association are regulated by sucrose concentration and phosphorylation. Plant Cell Physiol 48: 1612–1623 [DOI] [PubMed] [Google Scholar]
- Era A, Tominaga M, Ebine K, Awai C, Saito C, Ishizaki K, Yamato KT, Kohchi T, Nakano A, Ueda T(2009) Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol 50: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard A, Bargmann BO, Birnbaum KD, Tester M, Baumann U, Johnson AA(2012) Fluorescence-activated cell sorting for analysis of cell type-specific responses to salinity stress in Arabidopsis and rice. Methods Mol Biol 913: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD(2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl): S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S, Pytela J, Hotta T, Kato T, Hamada T, Akamatsu R, Ishida Y, Kutsuna N, Hasezawa S, Nomura Y, et al. (2013) An atypical tubulin kinase mediates stress-induced microtubule depolymerization in Arabidopsis. Curr Biol 23: 1969–1978 [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J(2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagounis C, Kostaki KI, Hawkins TJ, Cummins I, Fricker MD, Hussey PJ, Hetherington AM, Sweetlove LJ(2017) Microcompartmentation of cytosolic aldolase by interaction with the actin cytoskeleton in Arabidopsis. J Exp Bot 68: 885–898 [DOI] [PubMed] [Google Scholar]
- Giarola V, Hou Q, Bartels D(2017) Angiosperm plant desiccation tolerance: Hints from transcriptomics and genome sequencing. Trends Plant Sci 22: 705–717 [DOI] [PubMed] [Google Scholar]
- Grossman M, Ben-Chetrit N, Zhuravlev A, Afik R, Bassat E, Solomonov I, Yarden Y, Sagi I(2016) Tumor cell invasion can be blocked by modulators of collagen fibril alignment that control assembly of the extracellular matrix. Cancer Res 76: 4249–4258 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Cruz G, Van Heerden AH, Wang K(2001) Modular motif, structural folds and affinity profiles of the PEVK segment of human fetal skeletal muscle titin. J Biol Chem 276: 7442–7449 [DOI] [PubMed] [Google Scholar]
- Hu H, Juvekar A, Lyssiotis CA, Lien EC, Albeck JG, Oh D, Varma G, Hung YP, Ullas S, Lauring J, et al. (2016) Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell 164: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Xiong L(2014) Genetic engineering and breeding of drought-resistant crops. Annu Rev Plant Biol 65: 715–741 [DOI] [PubMed] [Google Scholar]
- Huber SC, Huber JL, Liao PC, Gage DA, McMichael RW Jr., Chourey PS, Hannah LC, Koch K(1996) Phosphorylation of serine-15 of maize leaf sucrose synthase. Occurrence in vivo and possible regulatory significance. Plant Physiol 112: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Cassin AM, Lonsdale A, Bacic A, Doblin MS, Schultz CJ(2017) Pipeline to identify hydroxyproline-rich glycoproteins. Plant Physiol 174: 886–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam NN, Tamilselvan A, Lawas LMF, Quinones C, Bahuguna RN, Thomson MJ, Dingkuhn M, Muthurajan R, Struik PC, Yin X, et al. (2017) Genetic control of plasticity in root morphology and anatomy of rice in response to water deficit. Plant Physiol 174: 2302–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Park M, Kim SJ, Hwang I(2005) Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell 17: 888–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K.(2004) Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Koroleva OA, Tomlinson ML, Leader D, Shaw P, Doonan JH(2005) High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J 41: 162–174 [DOI] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B(1995) Titins: Giant proteins in charge of muscle ultrastructure and elasticity. Science 270: 293–296 [DOI] [PubMed] [Google Scholar]
- Li J, Henty-Ridilla JL, Staiger BH, Day B, Staiger CJ(2015) Capping protein integrates multiple MAMP signalling pathways to modulate actin dynamics during plant innate immunity. Nat Commun 6: 7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang BM, Sharp RE, Baskin TI(1997) Regulation of growth anisotropy in well-watered and water-stressed maize roots. I. Spatial distribution of longitudinal, radial, and tangential expansion rates. Plant Physiol 115: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Wang K(2002) Interaction of nebulin SH3 domain with titin PEVK and myopalladin: Implications for the signaling and assembly role of titin and nebulin. FEBS Lett 532: 273–278 [DOI] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao T, McCubbin AG, Cyr RJ(1998) A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA(1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81: 561–570 [DOI] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW(2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Real-Hohn A, Zancan P, Da Silva D, Martins ER, Salgado LT, Mermelstein CS, Gomes AM, Sola-Penna M(2010) Filamentous actin and its associated binding proteins are the stimulatory site for 6-phosphofructo-1-kinase association within the membrane of human erythrocytes. Biochimie 92: 538–544 [DOI] [PubMed] [Google Scholar]
- Reiersen H, Rees AR(2001) The hunchback and its neighbours: Proline as an environmental modulator. Trends Biochem Sci 26: 679–684 [DOI] [PubMed] [Google Scholar]
- Rellán-Álvarez R, Lobet G, Dinneny JR(2016) Environmental control of root system biology. Annu Rev Plant Biol 67: 619–642 [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008) Lifeact: A versatile marker to visualize F-actin. Nat Methods 5: 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff KM, Roberts S, Chilkoti A, Pappu RV(2018) Advances in understanding stimulus-responsive phase behavior of intrinsically disordered protein polymers. J Mol Biol 430: 4619–4635 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MA, Ginsberg MH(2002) Networks and crosstalk: Integrin signalling spreads. Nat Cell Biol 4: E65–E68 [DOI] [PubMed] [Google Scholar]
- Smith LG.(2003) Cytoskeletal control of plant cell shape: Getting the fine points. Curr Opin Plant Biol 6: 63–73 [DOI] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG(2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO(2000) New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto‐Shirasu K, Carpita NC, McCann MC (2004) The cell wall: A sensory panel for signal transduction. In Hussey PJ, ed, The Plant Cytoskeleton in Cell Differentiation and Development, Annual Plant Reviews 10. Blackwell Publishing, Oxford, pp 176–203 [Google Scholar]
- Sun J, Loboda T, Sung SJ, Black CC(1992) Sucrose synthase in wild tomato, Lycopersicon chmielewskii, and tomato fruit sink strength. Plant Physiol 98: 1163–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB, Cosgrove DJ(2009) Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr Biol 19: R800–R811 [DOI] [PubMed] [Google Scholar]
- Tompa P, Bánki P, Bokor M, Kamasa P, Kovács D, Lasanda G, Tompa K(2006) Protein-water and protein-buffer interactions in the aqueous solution of an intrinsically unstructured plant dehydrin: NMR intensity and DSC aspects. Biophys J 91: 2243–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng IC, Hong CY, Yu SM, Ho TH(2013) Abscisic acid- and stress-induced highly proline-rich glycoproteins regulate root growth in rice. Plant Physiol 163: 118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V(2005) pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Uversky VN.(2011) Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol 43: 1090–1103 [DOI] [PubMed] [Google Scholar]
- Uversky VN.(2016) Dancing protein clouds: The strange biology and chaotic physics of intrinsically disordered proteins. J Biol Chem 291: 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Knipfer M, Huang QQ, van Heerden A, Hsu LC, Gutierrez G, Quian XL, Stedman H(1996) Human skeletal muscle nebulin sequence encodes a blueprint for thin filament architecture. Sequence motifs and affinity profiles of tandem repeats and terminal SH3. J Biol Chem 271: 4304–4314 [DOI] [PubMed] [Google Scholar]
- Wang S, Kurepa J, Hashimoto T, Smalle JA(2011) Salt stress-induced disassembly of Arabidopsis cortical microtubule arrays involves 26S proteasome-dependent degradation of SPIRAL1. Plant Cell 23: 3412–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S(2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1: 100. [DOI] [PubMed] [Google Scholar]
- Winter H, Huber JL, Huber SC(1998) Identification of sucrose synthase as an actin-binding protein. FEBS Lett 430: 205–208 [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC(2000) Regulation of sucrose metabolism in higher plants: Localization and regulation of activity of key enzymes. Crit Rev Biochem Mol Biol 35: 253–289 [DOI] [PubMed] [Google Scholar]
- Wulf E, Deboben A, Bautz FA, Faulstich H, Wieland T(1979) Fluorescent phallotoxin, a tool for the visualization of cellular actin. Proc Natl Acad Sci USA 76: 4498–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ(2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN(2010) PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 1804: 996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Ren S, Zhang X, Gao M, Ye S, Qi Y, Zheng Y, Wang J, Zeng L, Li Q, et al. (2011) BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23: 661–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J(2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]