Sulfur deficiency in einkorn grain leads to upregulation of genes acting in sulfur transport and metabolism that may coordinate the pool of free amino acids necessary for storage protein synthesis.
Abstract
Understanding the molecular mechanisms controlling the accumulation of grain storage proteins in response to nitrogen (N) and sulfur (S) nutrition is essential to improve cereal grain nutritional and functional properties. Here, we studied the grain transcriptome and metabolome responses to postanthesis N and S supply for the diploid wheat einkorn (Triticum monococcum). During grain filling, 848 transcripts and 24 metabolites were differentially accumulated in response to N and S availability. The accumulation of total free amino acids per grain and the expression levels of 241 genes showed significant modifications during most of the grain filling period and were upregulated in response to S deficiency. Among them, 24 transcripts strongly responded to S deficiency and were identified in coexpression network analyses as potential coordinators of the grain response to N and S supply. Sulfate transporters and genes involved in sulfate and Met metabolism were upregulated, suggesting regulation of the pool of free amino acids and of the grain N-to-S ratio. Several genes highlighted in this study might limit the impact of S deficiency on the accumulation of grain storage proteins.
Improvement of the grain yield potential of wheat (Triticum monococcum), the main staple crop in many regions of the world (Shiferaw et al., 2013), is needed to ensure food security. In this context, grain end-use value must be controlled, as it is primarily determined by protein concentration, which shows a strong genetic antagonism with grain yield (Bogard et al., 2010; Aguirrezábal et al., 2015). The grain storage proteins (GSPs) account for 60% to 80% of the total protein in mature grains, and their relative amount determines the end-use value for food processing (Shewry and Halford, 2002). Therefore, understanding the molecular mechanisms that regulate GSP synthesis is essential.
The main GSP fractions in wheat grains are gliadins and glutenins, which confer the rheological properties of dough during the bread-making process (Branlard et al., 2001). GSPs are commonly categorized as sulfur (S)-rich (α/β-gliadins, γ-gliadins, and low molecular weight glutenin [LMW-GS]), and S-poor (ω1,2-gliadins, ω5-gliadins, and high molecular weight glutenins [HMW-GS]) GSPs according to Shewry et al. (2001). During S deficiency, the quantity of S-containing amino acids (Cys and Met) in mature wheat grains is decreased (Wrigley et al., 1980), resulting in a decrease in the amount of S-rich GSPs, which is compensated by an increase in the amount of S-poor GSPs (Zhao et al., 1999; Wieser et al., 2004; Zörb et al., 2010). High levels of nitrogen (N) supply increase the concentration of GSPs (Pechanek et al., 1997; Triboï et al., 2003; Chope et al., 2014). However, an excessive N supply leads to a high N-to-S ratio and results similar to those obtained in S-deficient conditions (Wieser and Seilmeier, 1998; Zörb et al., 2010). Therefore, the availability of both N and S in soils plays a major role in controlling GSP composition and grain N-to-S ratio, which generally varies from 12:1 to 25:1 (Randall et al., 1981). The effect of N and S supplies on GSP composition in mature grains can be explained by changes in their rate and duration of accumulation during grain filling (Dai et al., 2015; Bonnot et al., 2017).
In cereals, changes in GSP composition in response to nutrient availability result from alterations in transporters and metabolism. For instance, an increase in the accumulation of free Asn is observed in response to S deprivation (Granvogl et al., 2007) and is partly explained by an upregulation of Asn synthetases ASN1 and ASN2 during grain development (Gao et al., 2016; Postles et al., 2016). In Arabidopsis (Arabidopsis thaliana), several metabolic changes happen in S-deprivation conditions (Nikiforova et al., 2005) and the transcriptional regulator SULFUR LIMITATION1 (SLIM1) plays a central role in controlling both the activation of sulfate acquisition and the degradation of glucosinolates under S starvation (Maruyama-Nakashita et al., 2006, 2008). SLIM1 also regulates miR395, which is induced under low S supply (Kawashima et al., 2009), and epigenetic regulation plays a role in the response to nutrient deficiencies via the action of microRNAs (Liang et al., 2015; Paul et al., 2015; Sirohi et al., 2016). In the Brassicaceae family, the SULFUR DEFICIENCY INDUCED1 (SDI1) gene inhibits glucosinolate biosynthesis under S deficiency, leading to the prioritization of sulfate utilization for primary metabolites (Aarabi et al., 2016).
Plants have strategies to adjust their metabolism and maintain proper development in response to nutrient imbalance; however, the underlying molecular mechanisms remain poorly understood. In a previous article, we studied the responses of GSPs, nuclear proteomes, and albumin-globulin proteomes in einkorn (Triticum monococcum) grains in response to N and S supply (Bonnot et al., 2017). The objective of this work was to analyze the grain transcriptome and metabolome under the same four previously applied nutritional treatments at different time points during grain development, with the objective of identifying regulatory mechanisms that may control grain composition under S and N availability/unavailability. We hypothesized that an imbalance of the grain N-to-S ratio results in activation of mechanisms involved in responses to nutritional deficiencies. We assumed that genes acting in these processes and strongly upregulated during grain filling might be important regulators of grain composition and thus play roles in maintaining grain quality under N- and/or S-deficient conditions. Our results reveal a putative mechanism of S-deficiency response caused by an increase in the grain N-to-S ratio. This mechanism includes genes acting in sulfate transport and amino acid and S metabolism, which function to adjust the amino acid pool in grains.
RESULTS
Transcriptome and Metabolome Changes Highlight the Importance of S in the Grain Response to N Supply
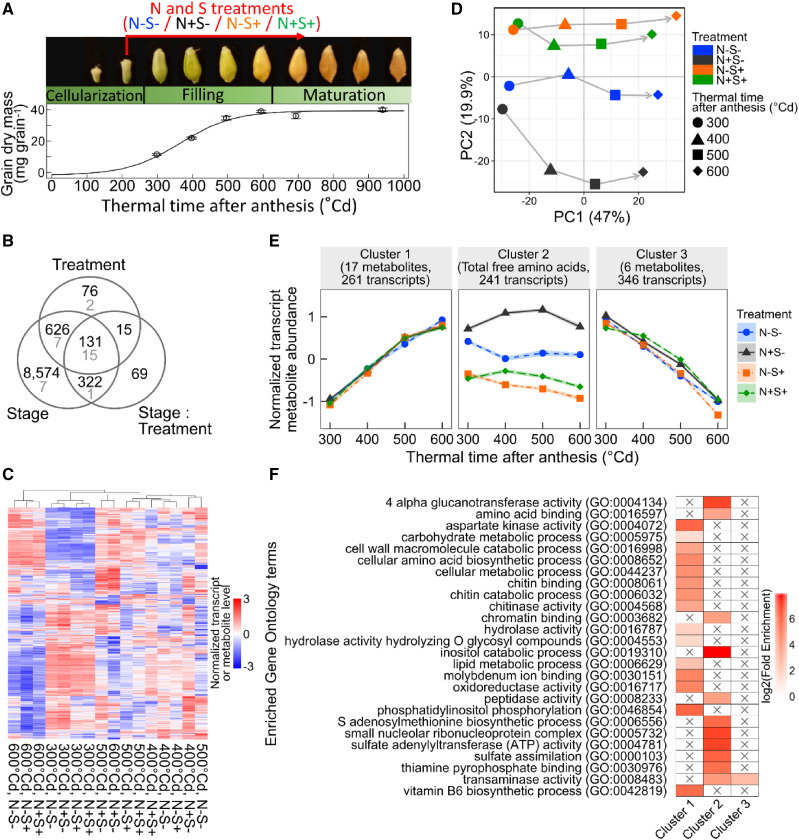
This part of the study focused on the transcriptome and metabolome responses of developing einkorn grains to N and S supply. Four treatments were applied to einkorn plants from 200°Cd after anthesis to grain maturity: N−S−, N+S−, N−S+, and N+S+, according to whether N and/or S was applied (Fig. 1A). Einkorn plants were grown in a greenhouse with a spatial arrangement similar to field conditions. To analyze the transcriptome, RNA sequencing (RNA-Seq) was performed on grain samples from 300°Cd to 600°Cd after anthesis (Supplemental Table S1). For differential expression analysis, 12,327 expressed genes were considered. Our metabolome analysis comprised the quantification of 33 metabolites and compounds during the effective grain filling and maturation periods (from 300°Cd to 950°Cd after anthesis), including 11 amino acids, five soluble sugars, five organic acids, starch, and the S-containing tripeptide glutathione (Supplemental Fig. S1; Supplemental Table S1). Statistical analyses revealed a significant effect (false discovery rate [FDR] < 0.05) of the treatments on the accumulation of 848 transcripts and 24 metabolites (Fig. 1B; Supplemental Fig. S2; Supplemental Table S2). Grain development showed significant effects in 9,653 transcripts and 30 metabolites; the accumulation of 131 transcripts and 15 metabolites was significantly affected by treatments, grain development, and interaction between the two factors (Fig. 1B). In this study, we focused on all metabolites and transcripts differentially accumulated in response to N and S treatments (24 and 848, respectively). We integrated our transcriptomic and metabolomic data obtained from 300°Cd to 600°Cd after anthesis to analyze the effects of the N and S treatments on grain metabolism during the effective grain-filling period.
Figure 1.
Differential effects of N and S treatments on the grain transcriptome and metabolome. A, Photograph of grains and accumulation of the grain dry mass during the three grain developmental phases. N and S treatments were applied from 200°Cd to 700°Cd after anthesis. B, Venn diagram depicting the overlapping transcripts (black) and metabolites (gray) with a significant (FDR < 0.05) treatment, grain development, and/or treatment × grain development interaction effect. C, Heatmap showing accumulation patterns for the 848 transcripts and 24 metabolites with a significant treatment effect in each N and S treatment and grain developmental stage. D and E, PCA (D) and hierarchical clustering on principal components (E) of the 848 transcripts and 24 metabolites with a significant treatment effect. In C and E, data are scaled (for each variable, the mean was subtracted, and the result was divided by the sd) and are means of n = 3 and 4 independent replicates for transcriptomic and metabolomic data, respectively. In each cluster, lines represent the average abundance of all cluster members in the corresponding treatment, and the shaded area represents the se. F, Enriched Gene Ontology terms in the sets of DEGs, by cluster.
Based on data from the 848 transcripts and 24 metabolites, we first assessed the overall effect of the N and S treatments on their accumulation profile (Fig. 1C). Whereas all treatments were grouped at 300°Cd after anthesis, the N+S− treatment showed a distinct effect from 400°Cd after anthesis (Fig. 1C). Consistent with this observation, principal component analysis (PCA) separated the grain developmental stages on the first principal component, whereas the second principal component clearly separated the N+S− treatment from the other three treatments (Fig. 1D). These two axes explained about 67% of the total variance. The two S+ treatments (N−S+ and N+S+) were closely projected on PCA, indicating that the second principal component is rather influenced by S availability (Fig. 1D). Transcripts and metabolites significantly affected by N and S supply were grouped into three clusters by hierarchical clustering of PCA results (Fig. 1E, Supplemental Table S3). Transcripts and metabolites in clusters 1 and 3 had almost identical accumulation profiles in the four nutritional conditions, with increased accumulation during grain filling for transcripts and metabolites in cluster 1 and the opposite pattern for those in cluster 3 (Fig. 1E). These results highlight the strong effect of grain development on the transcriptome and metabolome. Clusters 1 and 3 comprised 31% (261) and 41% (346), respectively, of the differentially expressed genes (DEGs). These two clusters also contained most of the significant metabolites (23 of 24), including eight free amino acids, glutathione, raffinose, and Suc. Interestingly, 241 transcripts and the total free amino acids in cluster 2 showed strongly increased accumulation during grain filling in the N+S− treatment (Fig. 1E). This result showed that a high N supply without S perturbed the expression of these genes and the free amino acid pool. Conversely, these transcripts and metabolites did not respond to high S supply or to high N when combined with high S supply.
To identify the biological role of genes grouped in these three clusters, a Gene Ontology (GO) enrichment was performed for each cluster (Fig. 1F; Supplemental Table S4). Cluster 1 was enriched in terms associated with metabolism (e.g. carbohydrate metabolic process, cellular amino acid biosynthetic process, and lipid metabolic process) and plant defense (e.g. chitin binding and chitinase activity; Fig. 1F), suggesting that N and S treatments disturbed grain metabolism and that grains could have perceived these treatments as stress. The molecular function “transaminase activity” was the only GO term overrepresented in cluster 3 (Fig. 1F). Interestingly, cluster 2 was enriched in the GO term “chromatin binding” and terms related to sulfate and amino acid metabolism (e.g. sulfate assimilation, sulfate adenylyltransferase (ATP) activity, amino acid binding, and transaminase activity; Fig. 1F). Therefore, transcripts and total free amino acids grouped in cluster 2 might have a role in the activation of S-related processes in response to S deficiency caused by the N+S− treatment.
Multiomics Coexpression Network Reveals the Coordination of Different Groups of Responses to N and S Supply during Grain Filling
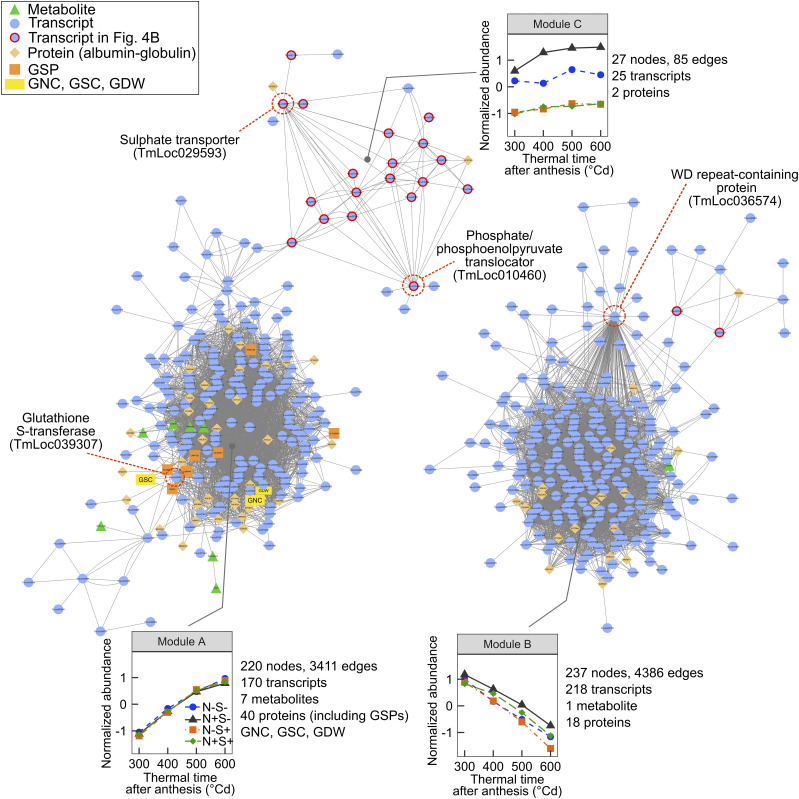
To identify genes that could coordinate the grain response to N and S supply, data for transcripts and metabolites with a significant treatment effect were combined with 132 proteomic variables (eight GSPs and 124 albumin-globulin variables) and other grain variables (N and S contents, N-to-S ratio, and dry mass per grain) previously reported in Bonnot et al. (2017). Coexpression and coaccumulation between the different variables were analyzed using a semantic rule-based network analysis (Fig. 2; Supplemental Table S5).
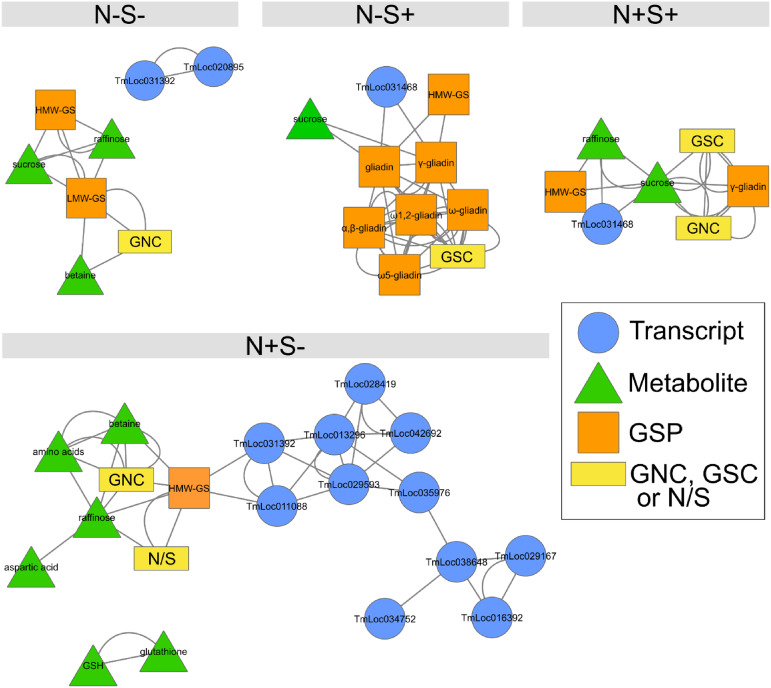
Figure 2.
Coexpression network of transcripts, metabolites, and proteins impacted by N and S supply during einkorn grain filling. Node shape and color reflect attribute category, as indicated in the key, where the yellow rectangles represent the N and S masses per grain (GNC and GSC, respectively) and dry weight (GDW). Edges between nodes represent coaccumulation links. For each module, the plot shows average abundance of the module nodes versus thermal time after anthesis for the four N and S treatments. Transcripts described in Figure 4B are encircled in red. For each module, examples of transcript nodes described in the manuscript are encircled with a dashed red line and the gene description is indicated.
Validated rules resulted in a network composed of 484 variables (nodes) and 7,882 linkages (edges), distributed into three distinct modules (Fig. 2). Module A consisted of 220 nodes highly interconnected (16 edges per node on average), including grain N and S contents; grain dry weight; GSPs; genes and proteins involved in carbohydrate metabolism, defense, and degradation inhibition processes; and metabolites including glutathione, Suc, and raffinose (Supplemental Table S5). Module B was the largest module, with 237 nodes (19 edges per node on average). Many nodes in this module were genes and proteins that play a role in DNA metabolism, RNA processing, or protein modification. This network analysis allowed the identification of similar groups compared to those revealed by clustering analysis (Figs. 1E and 2; Supplemental Table S5). Module A grouped nodes showing increased accumulation during grain filling, whereas nodes in module B had the opposite profile, as observed in clusters 1 and 3, respectively (Figs. 1E and 2). Thus, transcripts and metabolites within modules A and B strongly overlapped with cluster 1 (96%) and cluster 3 (93%), respectively (Supplemental Fig. S3). In these two modules, coexpressed transcripts, metabolites, and proteins were mostly connected because of a grain development effect. Nonetheless, several connected nodes might act together in response to N and S supply. For example, a highly connected WD-repeat-containing protein (144 connections, TmLoc036574) in module B and a glutathione S-transferase (TmLoc039307) connected to GSPs in module A could be main regulators of grain protein composition in response to nutrient availability.
In contrast to modules A and B, module C consisted of 27 nodes (three edges per node on average) with strong changes in their accumulation in response to N and S treatments (Fig. 2). These nodes were moderately accumulated in the two S+ treatments, highly accumulated in the N+S− treatment, and intermediately accumulated in the N−S− treatment, corresponding to the profile observed in cluster 2 (Figs. 1E and 2). Surprisingly, most of the cluster 2 transcripts were not grouped in module C. This result can be explained by a stronger effect of the grain development compared to the treatment effect and/or by a specific accumulation profile for many of these transcripts. Consequently, most of them were grouped in modules A and B or were absent from the network. Nonetheless, this reduced list of transcripts in module C, which were all grouped in cluster 2 (Supplemental Fig. S3), might play a significant role in the grain response to N and S availability. Particularly, a sulfate transporter (TmLoc029593) and a phosphate/phosphoenolpyruvate translocator (TmLoc010460) were the most connected within this module (18 and 17 connections, respectively; Fig. 2). Two proteins were also connected in module C, namely a NmrA domain-containing protein (M7Z7R6) and a glutathione S-transferase C-terminal domain-containing protein (M7ZKE7). Interestingly, module C is connected to module A by a gene coding for the protein SDI2 (TmLoc042692). We hypothesized that genes connected in module C could regulate grain protein accumulation under S-deficient conditions.
S Deficiency Strongly Upregulates 24 Genes during Grain Filling
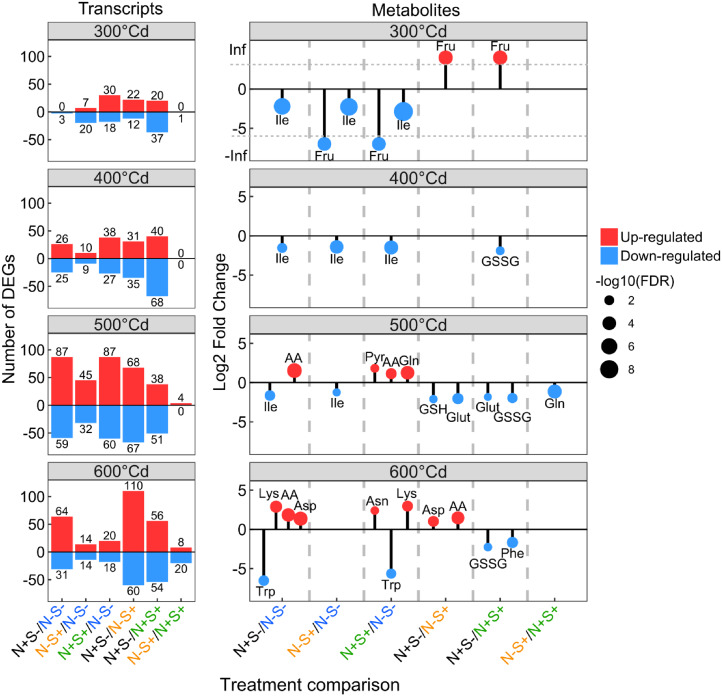
To identify transcripts and metabolites that respond strongly to N and S treatments during grain filling at multiple time points, we performed statistical analyses at each grain developmental stage from 300°Cd to 600°Cd after anthesis. We filtered the resulting datasets for transcripts and metabolites with a significant treatment effect as described above (Fig. 1) and with a log2 fold change (LFC) >|1| and FDR < 0.05 for at least one treatment comparison and at least one time point. This analysis revealed that more transcripts and metabolites were differentially accumulated at 500°Cd and 600°Cd after anthesis compared to 300°Cd and 400°Cd after anthesis (Fig. 3). At the transcriptome level, many transcripts were differentially accumulated in the N+S− treatment compared to the S+ treatments. This is particularly relevant at 600°Cd after anthesis with 170 and 110 (20% and 13%, respectively, of the 848 DEGs) significantly altered transcripts for the comparisons N+S−/N−S+ and N+S−/N+S+ (Fig. 3). At the metabolome level, treatments altered the pool of free amino acids, whereby they increased in the N+ treatments compared to the N− treatments (Fig. 3; Supplemental Figs. S1 and S2). Major effects were found for Asp, Asn, Lys, and Gln, which accumulated substantially in the N+ treatments compared to the N− treatments. Conversely, N+ treatments strongly downregulated Trp accumulation from 500°Cd after anthesis to grain maturity (Fig. 3; Supplemental Figs. S1 and S2). Overaccumulation of Ile was observed in the N−S− treatment compared to the three other treatments, especially at the beginning of grain filling (Fig. 3; Supplemental Figs. S1 and S2). The N+S− treatment is characterized by a low accumulation of oxidized and reduced forms of glutathione (Fig. 3; Supplemental Figs. S1 and S2). Interestingly, the most contrasting treatments (N−S− and N+S+) did not have the strongest effect on the grain transcriptome (Fig. 3), except at 500°Cd after anthesis. In addition, only Gln and a few transcripts (1, 4, and 28 at 300°Cd, 500°Cd, and 600°Cd after anthesis, respectively) showed differential abundances between N−S+ and N+S+ (Fig. 3). Taken together, these results showed that high N and S supply (N+S+) did not greatly perturb the grain transcriptome and metabolome compared to low-N conditions. However, high N supply without S (N+S−) had a strong effect.
Figure 3.
Effects of nitrogen and sulfur on the differentially accumulated transcripts and metabolites by grain developmental stage. On the left, histograms show the number of DEGs, and on the right, lollipop charts represent the LFC of significant metabolites, by treatment comparison and by grain developmental stage. Upregulation and downregulation of transcripts and metabolites was based on a comparison between the first and second treatments and reflect a significant higher (up-regulated) or lower (down-regulated) accumulation in the first compared to the second treatment. In the lollipop charts, dot size represents −log10 of the FDR values. AA, total free amino acids; GSH, reduced glutathione; GSSG, oxidized glutathione; Glut, glutathione; Pyr, pyrroline-5 carboxylate.
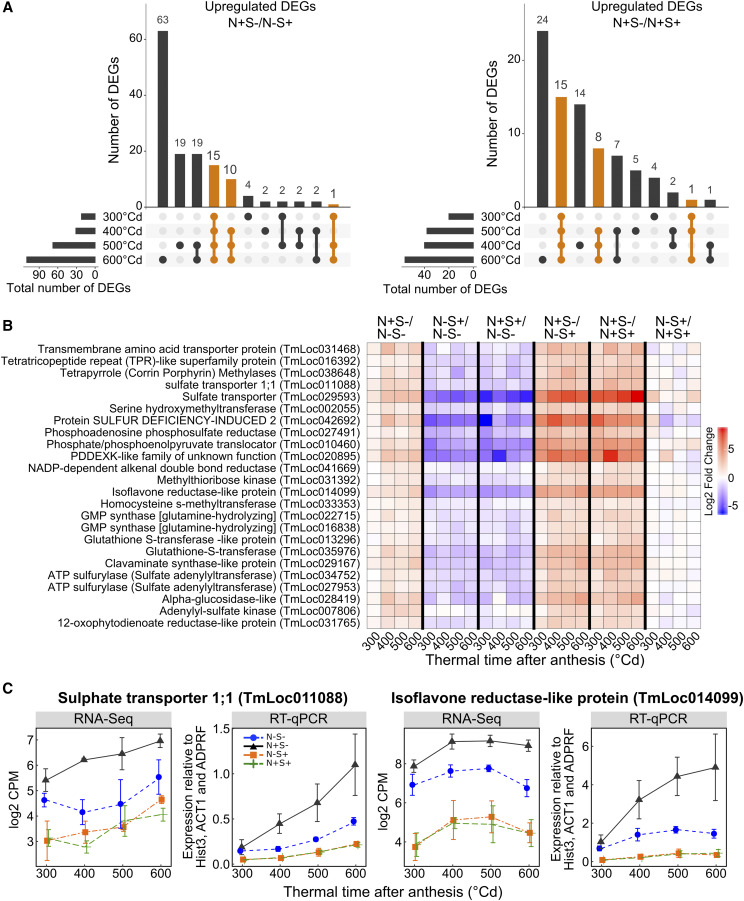
To identify genes strongly upregulated in response to N+S− treatment, which could play a role in the grain response to S deficiency, we compared the lists of upregulated transcripts obtained at the different time points for the treatment comparisons N+S−/N−S+ and N+S−/N+S+. We identified 26 and 24 transcripts upregulated at three or four grain developmental stages for the comparisons N+S−/N−S+ and N+S−/N+S+, respectively (Fig. 4A). All the 24 transcripts identified in the N+S−/N+S+ comparison overlap with the 26 transcripts in the N+S−/N−S+ comparison. We defined these 24 transcripts as strongly upregulated under S deficiency (Fig. 4B). Interestingly, these transcripts were all grouped in cluster 2 (Fig. 1E), and 19 were connected in module C in the coexpression network (Fig. 2). Three corresponded to transporters, including two sulfate transporters and an amino acid transporter (Fig. 4B). Other transcripts corresponded to genes involved in sulfate and Met metabolisms (e.g. phosphoadenosine phosphosulfate reductase, adenylyl-sulfate kinase, methythioribose kinase, and homo-Cys s-methyltransferase; Fig. 4B). Two ATP sulfurylases and two glutathione-S-transferases were also identified (Fig. 4B). Interestingly, the protein SDI2 coding gene, described above as the connector between modules A and C in the network analysis, showed high LFC values between N+S− and the S+ treatments (Fig. 4B). To validate the RNA-Seq results, transcript accumulation of two of these 24 DEGs, a sulfate transporter 1;1 (TmLoc011088) and an isoflavone reductase-like protein (TmLoc014099), were analyzed by reverse transcription quantitative PCR (RT-qPCR; Fig. 4C). Similar transcript responses were observed with both techniques (Fig. 4C). In contrast to the 24 transcripts strongly upregulated under S deficiency, 22 DEGs mainly involved in defense processes and storage accumulation, including genes encoding gliadins, showed an opposite response and were strongly downregulated by N+S− (Supplemental Fig. S4).
Figure 4.
Identification of sulfur-deficiency-induced genes. A, Unique and overlapping DEGs between grain developmental stages. Bar plots represent the number of DEGs that are unique to a developmental stage or common across different stages. DEGs upregulated in response to N+S− compared to N−S+ and N+S+ are represented on the left and the right, respectively. DEGs common across at least three grain developmental stages are represented in orange. B, Heatmap showing differential expression patterns of the 24 DEGs upregulated in at least three grain developmental stages in response to N+S− compared to both N−S+ and N+S+. The color scale depicts LFC values. C, Transcript levels quantified by RNA-Seq and RT-qPCR for two of the 24 selected DEGs identified in A. Data are means ± sd for n = 3 independent replicates.
To investigate the putative role of the 24 S deficiency-responsive genes in the regulation of grain protein composition in response to N and S supply, we analyzed the effect of the N and S treatments on the coexpression of these genes with the significant metabolites, the GSPs, and variables related to N and S masses per grain (Fig. 5). Connections in the N−S− network suggest that glutenin accumulation (LMW-GS and HMW-GS) throughout grain filling was associated in this treatment with changes in grain N mass and in three metabolites, including Suc and raffinose (Fig. 5). When S was supplied (N−S+ network), changes in gliadins and HMW-GS were highly associated with variations in grain S and Suc content (Fig. 5). In the N+S+ network, γ-gliadins and HMW-GS were connected to grain N and S contents, as well as Suc and raffinose contents (Fig. 5). In the latter two networks, an amino acid transporter was also connected (TmLoc031468), suggesting that this gene might regulate GSP accumulation under S+ conditions (Fig. 5). Remarkably, in the N+S− network, 11 of the 24 S deficiency-responsive transcripts were connected to HMW-GS. Total free amino acids, N content, and N-to-S ratio were also connected to HMW-GS. Taken together, these results suggest that under S deficiency, changes in GSP composition can be controlled by the pool of total free amino acids, the grain N-to-S ratio, and the S deficiency-responsive genes.
Figure 5.
Effects of N and S treatments on the coaccumulation of GSPs, metabolites, and sulfur-deficiency-responsive transcripts. Data for the 24 transcripts upregulated in response to S deficiency, as highlighted in Figure 4B, the 24 significant metabolites, the mass per grain of N (GNC) and S (GSC), GSPs, and the grain N-to-S ratio (N:S) were used to analyze their coaccumulation by treatment, from 300°Cd to 600°Cd after anthesis. Edges between nodes represent coaccumulation links. GSH, reduced glutathione.
DISCUSSION
Plant nutrition represents a major factor determining wheat grain yield and quality, with a well-known effect of N and S supply on GSP composition (e.g. Dupont et al., 2006; Dai et al., 2015) and consequently on flour functionality and bread-making quality (Zhao et al., 1999). To identify regulatory mechanisms that may control the grain composition under S and N availability/unavailability, we compared the impact of four postanthesis N and/or S treatments on the transcriptome and metabolome of developing grains of the diploid wheat einkorn and combined them with previously reported proteomic data (Bonnot et al., 2017). Several transcripts were strongly and durably differentially accumulated during grain filling, particularly when high N was supplied without S (that is in conditions of high N-to-S ratio and thus severe S deficiency), which could have a major role in controlling the amino acid pool in response to S deficiency.
A Putative Mechanism of Grain Response to S deficiency
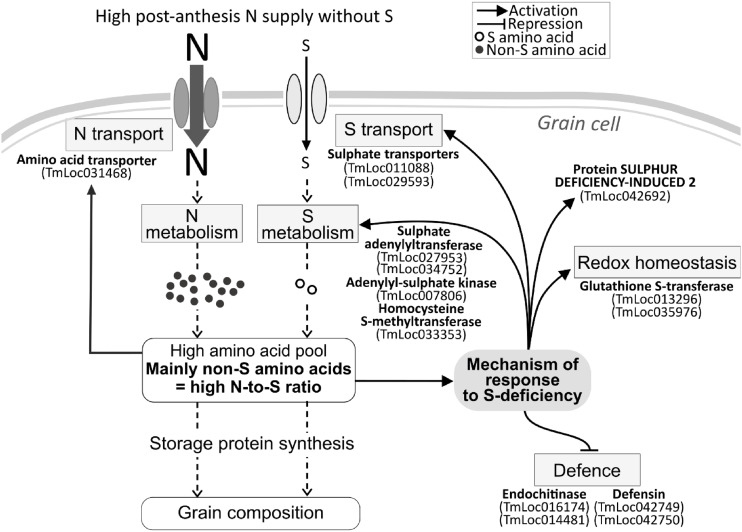
Our results highlighted an important effect of S deficiency on the grain transcriptome and metabolome. S deprivation with a high N supply resulted in a high N-to-S ratio, exceeding 25 g N g−1 S at the end of grain filling (Bonnot et al., 2017) and therefore well above the critical value for S deficiency of 17 g N g−1 S (Zhao et al., 1999). Total free amino acids and 241 transcripts were upregulated during grain filling in this nutritional condition compared with the three other treatments, including 24 transcripts defined as strongly responsive to high-N supply without S (Figs. 1E and 4B). These 24 transcripts were upregulated by high N supply but not by high N combined with high S. We hypothesized that these genes might be activated by a high N-to-S ratio. Our network analyses suggested that in the N+S− condition, the changes in grain protein composition were associated with changes in grain N-to-S ratio (Fig. 5). This is based on the principle of “guilt by association,” which can be applied to coexpression networks (Oliver, 2000; Wolfe et al., 2005). Grain N-to-S ratio was probably the main factor controlling the accumulation of GSPs in the N+S− condition. Therefore, we proposed a putative mechanism of grain response to S deficiency, governed by genes identified in the 24 selected transcripts, and that could limit the disruption of GSP accumulation (Fig. 6).
Figure 6.
Hypothetical mechanism of the grain response to S deficiency. The S-responsive mechanism was caused by an increased N-to-S ratio in the grain cell. Examples of genes involved in this mechanism are indicated in bold. They are part of the 24 genes strongly upregulated (or downregulated in the case of those involved in defense processes) in response to N+S− compared to the two S+ conditions (Fig. 4B; Supplemental Fig. S3B).
In response to a high postanthesis N supply without additional S, the amino acid pool was increased in the grain, consistent with previous reports (Howarth et al., 2008; Dai et al., 2015). This pool mainly contained non-S amino acids, which, with the decrease of glutathione (Fig. 3; Supplemental Fig. S1), caused an increase in the grain N-to-S ratio. Consequently, genes coding for S-rich GSPs like γ-gliadins were strongly repressed at the transcriptional level (Supplemental Fig. S4), leading to a drastic decrease in the rate and duration of accumulation of the corresponding proteins (Dai et al., 2015; Bonnot et al., 2017). In response to this dramatic effect on the GSP composition, sulfate transporters (TmLoc011088 and TmLoc029593) and several genes involved in sulfate and Met metabolisms were upregulated in order to activate both S transport and metabolism, therefore compensating for the S deficiency (Fig. 6). S deficiency also led to the downregulation of genes coding for S-containing proteins involved in defense processes (Fig. 6; Supplemental Fig. S4). In addition, our results highlighted a gene encoding SDI2 as one of the most upregulated DEGs in response to S deficiency (Figs. 2 and 4B). In Arabidopsis, SDI1 and SDI2 act as major repressors of glucosinolate biosynthesis under S deprivation; homologs exist in important crop species such as wheat, maize (Zea mays), and rice (Oryza sativa; Aarabi et al., 2016). Thus, a mechanism to prioritize S usage for primary metabolism probably occurred under S-deprived conditions, as previously reported (Aarabi et al., 2016).
Proteomics data, previously analyzed in Bonnot et al. (2017) and used in network analyses, revealed a NmrA domain-containing protein in module C (M7Z7R6; Fig. 2), described as a S deficiency-responsive module. In Aspergillus nidulans, transcription of genes encoding N permeases and catabolic enzymes of N metabolism requires the AreA transcription factor (TF; Marzluf, 1993). During N sufficiency, AreA is negatively regulated by NmrA (Wong et al., 2008). Thus, the NmrA domain-containing protein might repress N transport and metabolism in einkorn grain in response to a high N-to-S ratio.
Glutathione, which scavenges reactive oxygen species (Banerjee and Roychoudhury, 2019), had a lower quantity per grain in the N+S− treatment compared with the other three treatments (Supplemental Fig. S1). However, two glutathione S-transferases (TmLoc013296 and TmLoc035976) were highly upregulated in response to S deficiency, suggesting an activation of the processes of detoxification and redox buffering (Edwards et al., 2000; Nianiou-Obeidat et al., 2017). The strong S deficiency in the N+S− treatment was likely perceived as an abiotic stress and could have involved central regulators of the redox state (Gupta et al., 2016). Similar results with upregulation of genes acting in oxidation-reduction, S transport, and S metabolism processes were observed in pea (Pisum sativum) seeds under S-deficient conditions (Henriet et al., 2019).
Changes in the Quantity of Free Amino Acids per Grain Pinpoint Their Specific Sensing Role in Response to N and S Deprivation
Despite the clear role of several genes and metabolites in the grain response to S deficiency, molecular mechanisms involved in the sensing of nutrient deprivation remain unclear. A number of studies suggest that the levels of free amino acids may act as metabolic signals of grain nutritional status and directly or indirectly control the expression of several TF and kinase genes (e.g. Hernández-Sebastià et al., 2005; Ohkama-Ohtsu et al., 2008; Pandurangan et al., 2012; Cohen et al., 2016). Signals derived from overall N status play a major role in the regulation of N-responsive genes; however, some genes are differentially induced by different N sources (Kan et al., 2015), and selective sensing of amino acid precursors has been recently discovered (Dong et al., 2017). In the high-N treatments, several genes involved in amino acid transport and metabolism were upregulated and the quantity per grain of total free amino acids was increased mainly because of the higher quantity of Gln, Asn, Asp, and Lys. By contrast, the quantity of Trp was several fold higher in the low-N than in the high-N treatments. In the high-S treatments, the quantity per grain of glutathione was higher and that of Asp was lower than in the other treatments. All together, these results strongly support a role of free amino acids in the regulation of grain response to N and S availability.
In the N-to-S treatment, the quantities of branched amino acids (Ile and Val) and Thr (a precursor of Ile) per grain were much higher than in the other treatments between 300°Cd and 550°Cd after anthesis. In animals, these amino acids activate the Ser/Thr sensor-kinase TOR (Target Of Rapamycin) signaling pathway that couples amino acid availability to cell growth and autophagy (Jewell et al., 2013). In the case of Ile, it enhances Glc consumption to balance N and C metabolism (Zhang et al., 2017). TOR also controls a large diversity of physiological pathways and processes in plants (Dobrenel et al., 2016; Ryabova et al., 2019). In Arabidopsis, S availability regulates the Glc-TOR signaling pathway, whereas a decrease in amino acid production by limitation of the C/N supply is achieved by GCN2 and SnRK1 kinases (Dong et al., 2017). This mechanism allows plants to distinguish between C/N deprivation and S deprivation for amino acid biosynthesis (Dong et al., 2017). In yeast (Saccharomyces cerevisiae), GCN4 up-regulation through the activation of GCN2 protein kinase is induced by both Glc and amino acid starvation (Yang et al., 2000). Our results did not reveal a GCN4 homolog in treatment-responsive DEGs. However, a putative Ser Thr-protein kinase, GCN2 (TmLoc029081), was grouped in cluster 2 and showed higher transcript abundance in response to high N availability when S was limited. The N+S− condition also led to a lower Glc mass per grain during grain filling (Supplemental Fig. S1). Thus, in grain, the Glc-TOR signaling pathway might function in activating S-deficiency response mechanisms by sensing an excessive N-to-S ratio.
A Transient Transcriptional Reprogramming in the Early Grain Response to S deficiency?
In wheat, GSP synthesis is mainly regulated at the transcriptional level in response to N and S supply (Dai et al., 2015). Eight conserved TFs are known to be activators of GSP synthesis in cereals (Rubio-Somoza et al., 2006; Verdier and Thompson, 2008). In barley (Hordeum vulgare), the GCN4 box of C-hordein promoter, recognized by the b-ZIP TF BLZ1, is a positive or negative motif depending on N availability (Müller and Knudsen, 1993). Although these TFs are conserved in wheat, their function may be different than in barley and other cereals. In wheat, SHP, an ortholog of BLZ1 in barley and OHP in maize, has been shown to be a negative regulator of glutenin gene expression independent of N availability (Boudet et al., 2019), and results suggest that SPA, an ortholog of BLZ2 in barley and Opaque-2 in maize, is a negative regulator of gliadin gene expression (Ravel et al., 2009).
The nutritional conditions used in our study modified the level of expression of none of the known transcriptional regulators of GSP genes. However, their activity may have been regulated at the posttranslational level. For instance, phosphorylation has been shown to modulate the DNA binding capacity of Opaque2 (Ciceri et al., 1997). In addition, grains were collected from 300°Cd after anthesis, which is ∼5 d after the beginning of N and S treatment application (200°Cd) if considering a mean daily temperature of 20°C. Thus, a transcriptional regulation of these TFs may have occurred in the early grain response to N and S, specifically at the transition between the cellularization and effective grain filling stages. In this study, to identify genes and metabolites strongly influenced by nutrient deficiencies throughout grain filling, we did not focus our results on the interactions between treatment and grain development effects. Studying the temporal effect of nutrient availability would give more insights into the mechanisms regulating grain developmental processes.
Our results did not reveal TFs in DEGs strongly upregulated in response to S deprivation. However, we identified 21 (2.5% of the 848 DEGs) TFs/transcriptional regulators with a significant treatment effect (Supplemental Fig. S5). Five of these corresponded to genes encoding NAC-domain-containing proteins. This large TF family plays a role in multiple plant abiotic stress responses and includes members acting in N signaling and N-deficiency responses (He et al., 2015; Shao et al., 2015; Hsieh et al., 2018; Brooks et al., 2019). In wheat, the NAC TraesCS3A01G077900.1 has been described as a grain-specific transcript, downregulated in response to drought (Guérin et al., 2019). In our data, this transcript showed a higher accumulation in conditions lacking N and/or S (Supplemental Fig. S5). This gene showing different response profiles depending on the perceived stress might have a specific role in certain conditions including nutrient deficiencies. In addition, one NAC (TmLoc031311) was upregulated in response to N+S− compared to S+ treatments and could be a major regulator of the grain protein composition under S deficiency (Supplemental Fig. S5). Other TF families were represented in treatment-responsive TFs, including MYB, BHLH, bZIP, WRKY, and AP2/ERF, which are known to be involved in multiple abiotic stress responses (Chen et al., 2012; Roy, 2016; Das et al., 2019; Wang et al., 2019; Xie et al., 2019).
Our data also revealed five members of the RING/FYVE/PHD zinc finger protein superfamily as responsive to the experimental treatments (Supplemental Fig. S5). PHD zinc finger proteins act as epigenome readers controlling chromatin-mediated transcriptional regulation (Bienz, 2006; Sanchez and Zhou, 2011). Interestingly, two of them were upregulated in response to N+S− compared to S+ treatments (Supplemental Fig. S5). These results suggested a potential role of epigenetic mechanisms in the grain response to S deficiency. In barley, the incidence of epigenetic mechanisms in GSP synthesis and control of grain development was previously reported and suggested to play an important role in the regulation of grain composition (Sørensen et al., 1996; Radchuk et al., 2005).
CONCLUSIONS
Our results demonstrate the high impact of S deficiency on the transcriptome and metabolome of developing einkorn grains. During grain filling, high postanthesis N supply without S led to an increased pool of free amino acids, necessary for GSP synthesis. In response to the increase in the grain N-to-S ratio and the resulting grain S deficiency, sulfate transporters and genes involved in S metabolism were activated, probably to limit the impact of S deficiency on the accumulation of S-rich GSPs. Several genes identified in this study have a clear role in grain adaptation to nutritional deficiencies and could play additional key roles in controlling the grain protein composition, which is deserving of further investigation.
MATERIALS AND METHODS
Plant Material
Analyses were performed on grains collected from the same biological material used in Bonnot et al. (2017). Briefly, seeds of Triticum monococcum ssp. monococcum (accession ERGE 35821) were germinated on filter paper in petri dishes moistened with demineralized water, then seedlings were transplanted on February 4, 2014, to 50-mL PVC columns (7.5-cm i.d. × 50 cm deep; two seedlings per column) filled with a 2:1 (v/v) mixture of washed perlite:river sand. Columns were placed in a greenhouse to obtain a homogenous plant stand with a density of 512 plants m−2. The four N and S treatments were arranged in a complete randomized block design with four replicates (Supplemental Fig. S6). Air temperature, relative humidity, and photosynthetic photon flux density were measured at the height of the ears in the center of each block using a temperature and relative humidity CS215-l sensor (Campbell Scientific) and a JYP 1000 hemispherical photosynthetic photon flux density sensor (SDEC) mounted on miniature air-forced flues connected to a CR1000 data logger (Campbell Scientific). Weather variables were recorded every minute and averages over 15 min were stored. Air temperature in the greenhouse was controlled at 20°C/15°C (day/night) and air relative humidity was controlled at 55%/75% (day/night) at the top of the plant stand, with a 16-h photoperiod. During light periods plants received a mean total daily PPF of 9.68 mol m−2 d−1. Main stems were tagged when the anthers of the central florets emerged (anthesis date). Thermal time was calculated by summing average daily air temperature after anthesis (base temperature 0°C) and was expressed in degree-days (°Cd).
From seedling transplantation to anthesis, each PVC column received 167 mL d−1 of a modified Hoagland’s nutrient solution (Castle and Randall, 1987) prepared with demineralized water and containing 3 mm of N and 0.1 mm of S [1 mm KH2PO4, 1 mm KNO3, 0.5 mm Ca(NO3)2, 0.5 mm NH4NO3, 0.1 mm MgSO4, 1.9 mm MgCl2, 3.5 mm CaCl2, 4 mm KCl, 10 μm H3BO3, 0.7 μm ZnCl2, 0.4 μm CuCl2, 4.5 μm MnCl2, 0.22 μm MoO3, and 50 μm EDFS-Fe]. From anthesis to 200°Cd after anthesis, the nutrient solution was replaced by demineralized water to avoid any overaccumulation of N and S compounds in the plant or in the potting substrate. Then four treatments that contained different combinations of N and S supply were applied from 200°Cd to 700°Cd after anthesis: a treatment corresponding to a nutrient solution with no N and S (N−S−); a high-N and low-S treatment (6 mm N and no S [N+S−]); a low-N and high-S treatment (0.5 mm N and 2 mm S [N−S+]); and a high-N and high-S treatment (6 mm N and 2 mm S [N+S+]). Grains were collected every 100°Cd after anthesis from 300°Cd after anthesis to ripeness maturity (950°Cd after anthesis) for metabolite analysis, and from 100°Cd to 600°Cd after anthesis for transcriptome analysis. Grains were immediately snap-frozen in liquid N2 after harvesting, then stored at −80°C until RNA extraction, or freeze-dried and then milled to wholemeal flour in liquid N2 using a custom-made ball mill and then stored at −80°C for metabolite analyses.
RNA Extraction and RNA-Seq
RNA was extracted from developing grains according to Pont et al. (2011). Three biological replicates were used. The 54 grain samples (three replicates at 100°Cd and 200°Cd after anthesis and three replicates for each of the four treatments [N−S−, N+S−, N−S+, at N+S+] at 300°Cd, 400°Cd, 500°Cd, and 600°Cd after anthesis) were ground in liquid N2. Flour (0.5 g) was suspended in 4.5 mL of a buffer containing 10 mm Tris-HCl (pH 7.4), 1 mm EDTA, 0.1 m NaCl, 1% (w/v) SDS and 3 mL of 25:24:1 (v/v/v) phenol:chloroform:isoamyl alcohol mixture. After centrifugation at 4,420g for 20 min at 15°C, the supernatant was collected, and 3 mL of the same phenol solution was added to eliminate starch and proteins. After centrifugation in the same conditions, the aqueous phase was collected, and nucleic acids were precipitated by adding 0.1 volume of 3 m AcNA (pH 5.2) and 2 volumes of 100% (v/v) ethanol. The pellet was dried under ambient condition for 1 h and dissolved in RNase-free water. A purification was then performed using a RNase-Free DNase set (Qiagen) and then a RNeasy MinElute Cleanup kit (Qiagen). The amount and integrity of RNA were checked by agarose gel electrophoresis before library construction. The same RNA extracts were used for RNA-Seq and RT-qPCR.
RNA-Seq was done at Eurofins MWG Operon (www.eurofinsgenomics.eu). RNA strand-specific libraries were created using commercially available kits according to the manufacturer’s instructions. In brief, polyA-RNA was extracted from total RNA using an oligodT-bead based method. After mRNA fragmentation, first and deoxy-UTP-based second strand synthesis was carried out, followed by end-repair, A-tailing, ligation of the indexed Illumina Adapter, and digestion of the deoxy-UTP-strand. Size selection was done using a bead-based method targeting an average insert size of 150 to 400 bp. After PCR amplification, the resulting fragments were cleaned up, pooled, quantified and used for cluster generation. For sequencing, pooled libraries were loaded on the cBot (Illumina) and cluster generation was performed according to the manufacturer’s instructions. Paired-end sequencing using a 125-bp read length was performed on a HiSeq2500 machine (HiSeq Control Software v2.2.38) using HiSeq Flow Cell and TruSeq SBS Kit. For processing of raw data, RTA v1.18.61 and CASAVA v1.8.4 (Illumina) were used to generate FASTQ-files.
Read Alignment and Counting
Reads were mapped to the T. monococcum reference genome v1 available at the Unité de Recherche Génomique Info database (urgi.versailles.inra.fr/download/iwgsc/), using a BWA-MEM (v0.7.12-r103;, http://bio-bwa.sourceforge.net/) tool. Number of reads and percentages of mapped reads per sample are given in Supplemental Fig. S7. Only reads with a unique mapping position and a mapping quality score of at least 10 were considered for read counting. Read counting was realized using the software featureCounts from the Subread package (v1.4.6). Paired-end reads that were mapped to the same reference with the expected insert size were counted as one read. Paired-end reads that were mapped to different references or those with an unexpected insert size were counted as two reads. If only one read of a pair was mapped, it was counted as one read. Single-end reads were used straightforwardly. Only reads overlapping exon features were counted. All reads mapping to features with the same grouping tag were summed.
1H-NMR Metabolomic Profiling
Polar metabolites were extracted from 50 mg of wholemeal flour with an ethanol-water series at 80°C (adapted from Moing et al., 2004; Allwood et al., 2011). The supernatants were combined, dried under vacuum, and freeze-dried. Each freeze-dried extract was solubilized in 500 μL of 200 mm deuterated potassium phosphate buffer solution (apparent pH 6.0) containing 3 mm EDTA to chelate paramagnetic cations and improve spectrum resolution (especially in the citrate region), pH-adjusted with 1 m KOD or 0.1 m DCL solution by means of BTpH (Bruker BioSpin GmbH) to apparent pH 6.0 ± 0.02, and freeze-dried again. The freeze-dried titrated extracts were stored in darkness under vacuum at room temperature until 1H-NMR analysis was completed within 1 week.
For 1H-NMR analysis, 500 μL of D2O with sodium trimethylsilyl [2,2,3,3-d4] propionate (0.01% [w/v] final concentration for chemical shift calibration) was added to each freeze-dried titrated extract. The mixture was centrifuged at 17,700g for 5 min at room temperature. The supernatant was then transferred into a 5 mm NMR tube for acquisition. Quantitative 1H-NMR spectra were recorded at 500.162 MHz and 300 K on an Avance III spectrometer (Bruker Biospin) using a 5-mm ATMA broadband inverse probe flushed with N2 gas and an electronic reference for quantification (Digital ERETIC, Bruker TopSpin 3.0). Sixty-four scans of 32,000 data points each were acquired with a 90° pulse angle, a 6-MHz spectral width, a 2.73-s acquisition time, and a 20-s recycle delay. The assignments of metabolites in the NMR spectra were made by comparing the proton chemical shifts with literature (Likes et al., 2007) or database values from MeRy-B (a database of plant 1H-NMR metabolomic profiles), the Human Metabolome Database, the Biological Magnetic Resonance Bank, and the Madison Metabolomic Consortium Database by comparing spectra of authentic compounds and by spiking the samples. An example of an annotated NMR spectrum is given in Supplemental Figure S8. For assignment purposes, 1H-1H correlation spectra and 1H-13C heteronuclear single quantum coherence spectra were acquired for a selected sample. For absolute quantification, four calibration curves (Glc and Fru, 2.5–100 mm; Glu and Gln, 2.5–30 mm) were prepared and analyzed under the same conditions. The Glc calibration was used, for the quantification of all compounds, as a function of the number of protons of selected resonances, except for Fru, Glu, and Gln, which were quantified using their own calibration curve. The concentration of each organic or amino acid was expressed as grams of the acid form per gram of dry mass (Supplemental Table S1). The metabolite concentrations were calculated using AMIX (v3.9.14, Bruker) and Excel (Microsoft) software programs.
Targeted Analyses of Metabolites
Starch and total free amino acid assays were performed as previously described (Biais et al., 2014; Bénard et al., 2015). Four biological replicates were used. Aliquots of ∼20 mg of wholemeal flour were fractionated with ethanol/water at 95°C for 15 min, twice with 250 and 150 μL, respectively, of 80% (v/v) ethanol, and once with 250 μL of 50% (v/v) ethanol. Starch was determined on the pellet by resuspension in 100 mm NaOH and heating at 95°C for 30 min (Hendriks et al., 2003). The sum of free amino acids was determined in the ethanolic supernatant (Bantan-Polak et al., 2001; Nunes-Nesi et al., 2007). Oxidized (GSSG) and reduced (GSH) glutathione were extracted and assayed enzymatically with glutathione reductase and 5,5′-dithiobis-2-nitrobenzoic acid, as described by Griffith (1980). Extractions and assays were performed in 1.1 mL Micronic tubes (Lelystad) and 96-well polystyrene microplates (Sarstedt), respectively, using a robotized Starlet platform (Hamilton). Absorbance was read at 340 (starch) or 405 nm (glutathione assay) using a MP96 reader (SAFAS). Fluorescamine fluorescence for amino acids was determined using a Xenius reader (SAFAS) with excitation at 405 nm and detection at 485 nm.
Statistical and Data Analysis
All statistical analyses were performed using the statistical software program R v3.6.1 (R Core Team, 2018). The edgeR package v3.28 (Robinson et al., 2010) was used for differential expression analysis. The Counts Per Million method was used with a threshold of five reads per million in a third of the samples. Libraries were normalized using the trimmed mean of M-values method (Robinson and Oshlack, 2010; Dillies et al., 2013). The differential expression analysis was performed with generalized linear models. To analyze the main effects of the grain developmental stage, the treatment, and the interaction between stage and treatment, and to identify genes differentially expressed between any of the groups, an ANOVA-like test was performed by specifying multiple coefficients to the glmLRT function of the EdgeR package. Significant effects were judged at P < 0.05 after FDR correction using the Benjamini and Hochberg (1995) procedure. To identify genes differentially expressed between two different types of nutrition at each stage of development, 24 likelihood ratio tests were used (six questions per stage). The log of the average gene expression is an additive function of a replicate effect (3 modalities), a stage of development effect (4 modalities), a treatment effect (4 modalities), and an interaction between the stage and the treatment (16 modalities). The probability of significance (P-value) per contrast was adjusted using the Benjamini-Hochberg procedure. Genes with FDR < 0.05 and LFC > |1| were considered as differentially expressed in the corresponding contrast. This analysis was performed using the workspace DiCoExpress (https://www.researchsquare.com/article/84521cdd-9046-44ac-8f52-906dc21cf1a4/v1). The data, the R script, and the results are available at https://forgemia.inra.fr/GNet/einkorn_grain_transcriptome. The results of the differential analysis are in the folder /Results/Einkorn_grain/DiffAnalysis, where a subfolder is dedicated to each question. The folder /Results/Einkorn_grain/Quality_Control contains a pdf file with exploratory plots to investigate the data quality. For further explanation, we refer readers to the DiCoExpress user manual.
Differences in mass of metabolites per grain were analyzed using a two-way ANOVA with treatments and grain developmental stages (from 300°Cd to 950°Cd after anthesis) defined as factors. Effects of the grain developmental stage, the treatment, and the interaction between stage and treatment were judged to be significant at P < 0.05 after FDR correction. To identify metabolites differentially accumulated between two different types of nutrition at each stage of development, Tukey’s honestly significant difference (HSD) mean-separation tests were performed (Supplemental Table S2). For each comparison, metabolites with FDR < 0.05 and LFC > |1| were considered significant.
Transcripts and metabolites with a significant treatment effect (FDR < 0.05) were analyzed using PCA performed with the “ade4” R package (Thioulouse et al., 1997), and a hierarchical clustering on principal components was computed with the “FactomineR” package for R and the Ward’s criterion applied on the first five principal components (Husson et al., 2015; Supplemental Table S3). The number of clusters was determined with the suggested partition, that is, the one with the highest relative loss of inertia. The heatmap of significant transcripts and metabolites presented in Figure 1C was generated using the R package “pheatmap” (Kolde, 2019). Bar plots depicting overlapping transcripts between grain developmental stages were generated using the “UpSet” R package (Conway et al., 2017). All other plots were generated using the “ggplot2” R package (Wickham, 2016).
Functional Annotation and GO Enrichment Analysis
All the 12,327 transcripts considered in differential expression analyses were functionally annotated using the gene annotation pipeline TriAnnot (Leroy et al., 2012) and a custom made perl script (TriAnnotpost.pl, not published). Briefly, protein-derived gene models are reconstructed (only proteins starting with a Met and avoiding pseudogenes) based on fasta and gff files of scaffolds from the T. monococcum reference genome v 1 available at the URGI database (urgi.versailles.inra.fr/download/iwgsc/). The best alignment against a plant Poaceae protein database and the Triticum aestivum IWGSC RefSeq v1.0 annotated peptides (Appels et al., 2018) is given by TriAnnotpost.pl, as is the functional annotation including the related GO terms (molecular function, biological process, and cellular component; Supplemental Table S4). For pseudogenes, gene models with no start codon, genes corresponding to short scaffolds (<1 Kb), and genes with an unknown function, we extracted the pseudogene transcripts and performed a BLASTx against the IWGSC RefSeq v1.0 annotated peptides (Supplemental Table S4; Appels et al., 2018).
A GO enrichment analysis was performed by cluster (Fig. 1F) using all GO terms identified in the T. aestivum IWGSC RefSeq v1.0 as the reference set for comparison against GO terms associated with DEGs identified in each cluster. To identify enriched GO terms, Fisher’s exact tests were performed, and statistical differences were considered to be significant at adjusted P-value <0.05 after FDR correction (Supplemental Table S4; Benjamini and Hochberg, 1995). Enriched GO terms were visually represented as described in Bonnot et al. (2019).
Network Analysis
Coexpression/coaccumulation of genes, metabolites, and proteins that were significantly impacted by N and S treatments was investigated using the RulNet platform for network inference (http://rulnet.isima.fr; Vincent et al., 2015). Data were scaled and a semantic was written to find rules between coexpressed/coaccumulated genes, metabolites, and proteins (Supplemental Table S5; Supplemental Method S1). Three quality measures (that is support, confidence, and lift) were used to select the best rules. Threshold values for support, confidence, and lift were set at 0.3, 0.9, and 1.5, respectively, for the network presented in Figure 2. These thresholds were increased to 0.4, 1, and 2 for the networks presented in Figure 5 because of the lower number of conditions used. Validated rules were used to visualize networks using CYTOSCAPE software v3.3.0 (Smoot et al., 2011).
RT-qPCR
Samples of 5 μg of total RNA extracts were reverse transcribed with 5 μm of random hexamers (ThermoFisher Scientific) and 200 U of RevertAid Reverse Transcriptase (ThermoFisher Scientific) in a final volume of 20 μL per reaction. Transcript levels were quantified by qPCR with a Lightcycler 480 SYBR Green I Master (Roche) using 5 μL of 50× diluted complementary DNA and 0.5 μm of each primer in a final volume of 15 μL. The primer sequences used are listed in Supplemental Table S6. The mRNA expression levels relative (R) to the housekeeping genes HIST3, ACT1, and ADPRF were calculated as R = εCp−Cp*, where ε is the primer efficiency, Cp is the crossing point for the measured gene, and Cp* is the geometric mean of the crossing point for the housekeeping genes.
Accession Numbers
The RNA-Seq data analyzed in this study were deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE107807). Quantitative data of the described transcripts and metabolites are provided in Supplemental Table S1, and functional annotation of transcripts is provided in Supplemental Table S4.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Profiles of quantified metabolites in einkorn grains grown under four combinations of N and S supply.
Supplemental Figure S2. Map of the metabolites differentially accumulated during einkorn grain development in response to N and S supply.
Supplemental Figure S3. Overlap between network modules and clusters.
Supplemental Figure S4. Identification of sulfur-deficiency-repressed genes.
Supplemental Figure S5. Effects of N and S treatments on transcriptional regulator expression.
Supplemental Figure S6. Randomized block design used for T. monococcum plant cultivation.
Supplemental Figure S7. Percentage of reads sequenced and mapped in the transcriptome analysis.
Supplemental Figure S8. Representative 1D 1H 500 MHz NMR spectrum of a polar extract of T. monococcum grain at 300°Cd after anthesis and grown under N−S− treatment.
Supplemental Table S1. Quantification results.
Supplemental Table S2. Statistical analysis results.
Supplemental Table S3. Clustering analysis of the differentially accumulated transcripts and metabolites.
Supplemental Table S4. Functional annotation of the T. monococcum transcriptome.
Supplemental Table S5. RulNet rules for -omics network inference.
Supplemental Table S6. Primer sequences used in RT-qPCR.
Supplemental Method S1. Semantic used to infer regulatory networks from -omics data.
Acknowledgments
We thank Richard Blanc for his help with plant cultivation; David Alvarez and Annie Faye for organizing plant culture and planning grain sampling; Marielle Merlino, Sibille Perrochon, Julie Boudet, and Isabelle Nadaud for help with grain sampling; Mickaël Maucourt for the metabolome analysis; Florent Murat for help in supervising the read mapping step during RNA-Seq data analysis; Morgane B. Gillard for her help with statistical analyses; and Selver Babi and Romain De Oliveira for their help with data analysis.
Footnotes
This work was supported by the French Ministry for Higher Education and Research (PhD grant to T.B.), the French National Research Agency (ANR; grant nos. ANR–10–BTBR–03 BREEDWHEAT, ANR–11–INBS–0010 MetaboHUB, and ANR–10–LABX–0040–SPS), France AgriMer, the French Fund Supporting Plant Breeding (FSOV), and the European Union’s Seventh Framework Programme (FP7/2007–2013, under grant no FP7–613556).
Articles can be viewed without a subscription.
References
- Aarabi F, Kusajima M, Tohge T, Konishi T, Gigolashvili T, Takamune M, Sasazaki Y, Watanabe M, Nakashita H, Fernie AR, et al. (2016) Sulfur deficiency-induced repressor proteins optimize glucosinolate biosynthesis in plants. Sci Adv 2: e1601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirrezábal L, Martre P, Pereyra-Irujo G, Echarte MM, Izquierdo N (2015) Improving grain quality: Ecophysiological and modeling tools to develop management and breeding strategies. In Sadras VO, Calderini DF, eds, Crop Physiology: Applications for Genetic Improvement and Agronomy, Ed 2. Academic Press, Amsterdam, pp 423–465 [Google Scholar]
- Allwood JW, De Vos RCH, Moing A, Deborde C, Erban A, Kopka J, Goodacre R, Hall RD (2011) Plant metabolomics and its potential for systems biology research: Background concepts, technology, and methodology. In Jameson D, Verma M, Westerhoff HV, eds, Methods in Systems Biology, Methods in Enzymology, Vol 500. Elsevier, Amsterdam, pp 299–336 [DOI] [PubMed] [Google Scholar]
- Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A, Poland J, et al. ; International Wheat Genome Sequencing Consortium I (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Roychoudhury A(2019) Role of glutathione in plant abiotic stress tolerance In Khan NA, Singh S, and Umar S, eds, Sulfur Assimilation and Abiotic Stress in Plants. Springer, Berlin/Heidelberg, pp 207–225 [Google Scholar]
- Bantan-Polak T, Kassai M, Grant KB(2001) A comparison of fluorescamine and naphthalene-2,3-dicarboxaldehyde fluorogenic reagents for microplate-based detection of amino acids. Anal Biochem 297: 128–136 [DOI] [PubMed] [Google Scholar]
- Bénard C, Bernillon S, Biais B, Osorio S, Maucourt M, Ballias P, Deborde C, Colombié S, Cabasson C, Jacob D, et al. (2015) Metabolomic profiling in tomato reveals diel compositional changes in fruit affected by source-sink relationships. J Exp Bot 66: 3391–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y(1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Biais B, Bénard C, Beauvoit B, Colombié S, Prodhomme D, Ménard G, Bernillon S, Gehl B, Gautier H, Ballias P, et al. (2014) Remarkable reproducibility of enzyme activity profiles in tomato fruits grown under contrasting environments provides a roadmap for studies of fruit metabolism. Plant Physiol 164: 1204–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M.(2006) The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 31: 35–40 [DOI] [PubMed] [Google Scholar]
- Bogard M, Allard V, Brancourt-Hulmel M, Heumez E, Machet J-M, Jeuffroy M-H, Gate P, Martre P, Le Gouis J(2010) Deviation from the grain protein concentration-grain yield negative relationship is highly correlated to post-anthesis N uptake in winter wheat. J Exp Bot 61: 4303–4312 [DOI] [PubMed] [Google Scholar]
- Bonnot T, Bancel E, Alvarez D, Davanture M, Boudet J, Pailloux M, Zivy M, Ravel C, Martre P(2017) Grain subproteome responses to nitrogen and sulfur supply in diploid wheat Triticum monococcum ssp. monococcum. Plant J 91: 894–910 [DOI] [PubMed] [Google Scholar]
- Bonnot T, Gillard M, Nagel D(2019) A simple protocol for informative visualization of enriched Gene Ontology terms. Bio Protoc e3429 [Google Scholar]
- Boudet J, Merlino M, Plessis A, Gaudin J-C, Dardevet M, Perrochon S, Alvarez D, Risacher T, Martre P, Ravel C(2019) The bZIP transcription factor SPA Heterodimerizing Protein represses glutenin synthesis in Triticum aestivum. Plant J 97: 858–871 [DOI] [PubMed] [Google Scholar]
- Branlard G, Dardevet M, Saccomano R, Lagoutte F, Gourdon J(2001) Genetic diversity of wheat storage proteins and bread wheat quality. Euphytica 119: 59–67 [Google Scholar]
- Brooks MD, Cirrone J, Pasquino AV, Alvarez JM, Swift J, Mittal S, Juang C-L, Varala K, Gutiérrez RA, Krouk G, et al. (2019) Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nat Commun 10: 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle S, Randall P(1987) Effects of sulfur deficiency on the synthesis and accumulation of proteins in the developing wheat seed. Aust J Plant Physiol 14: 503–516 [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D(2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819: 120–128 [DOI] [PubMed] [Google Scholar]
- Chope GA, Wan Y, Penson SP, Bhandari DG, Powers SJ, Shewry PR, Hawkesford MJ(2014) Effects of genotype, season, and nitrogen nutrition on gene expression and protein accumulation in wheat grain. J Agric Food Chem 62: 4399–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciceri P, Gianazza E, Lazzari B, Lippoli G, Genga A, Hoscheck G, Schmidt RJ, Viotti A(1997) Phosphorylation of Opaque2 changes diurnally and impacts its DNA binding activity. Plant Cell 9: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H, Pajak A, Pandurangan S, Amir R, Marsolais F(2016) Higher endogenous methionine in transgenic Arabidopsis seeds affects the composition of storage proteins and lipids. Amino Acids 48: 1413–1422 [DOI] [PubMed] [Google Scholar]
- Conway JR, Lex A, Gehlenborg N(2017) UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33: 2938–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Plessis A, Vincent J, Duchateau N, Besson A, Dardevet M, Prodhomme D, Gibon Y, Hilbert G, Pailloux M, et al. (2015) Transcriptional and metabolic alternations rebalance wheat grain storage protein accumulation under variable nitrogen and sulfur supply. Plant J 83: 326–343 [DOI] [PubMed] [Google Scholar]
- Das P, Lakra N, Nutan KK, Singla-Pareek SL, Pareek A(2019) A unique bZIP transcription factor imparting multiple stress tolerance in Rice. Rice (N Y) 12: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies M-A, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, et al. ; French StatOmique Consortium (2013) A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform 14: 671–683 [DOI] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C(2016) TOR signaling and nutrient sensing. Annu Rev Plant Biol 67: 261–285 [DOI] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Allboje Samami A, Wanatabe M, Sticht C, Teleman AA, et al. (2017) Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun 8: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dupont FM, Hurkman WJ, Vensel WH, Tanaka C, Kothari KM, Chung OK, Altenbach SB(2006) Protein accumulation and composition in wheat grains: Effects of mineral nutrients and high temperature. Eur J Agron 25: 96–107 [Google Scholar]
- Edwards R, Dixon DP, Walbot V(2000) Plant glutathione S-transferases: Enzymes with multiple functions in sickness and in health. Trends Plant Sci 5: 193–198 [DOI] [PubMed] [Google Scholar]
- Gao R, Curtis TY, Powers SJ, Xu H, Huang J, Halford NG(2016) Food safety: Structure and expression of the asparagine synthetase gene family of wheat. J Cereal Sci 68: 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granvogl M, Wieser H, Koehler P, Tucher SV, Schieberle P(2007) Influence of sulfur fertilization on the amounts of free amino acids in wheat. Correlation with baking properties as well as with 3-aminopropionamide and acrylamide generation during baking. J Agric Food Chem 55: 4271–4277 [DOI] [PubMed] [Google Scholar]
- Griffith OW.(1980) Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal Biochem 106: 207–212 [DOI] [PubMed] [Google Scholar]
- Guérin C, Roche J, Allard V, Ravel C, Mouzeyar S, Bouzidi MF(2019) Genome-wide analysis, expansion and expression of the NAC family under drought and heat stresses in bread wheat (T. aestivum L.). PLoS One 14: e0213390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta DK, Palma JM, Corpas FJ(2016) Redox State as a Central Regulator of Plant-Cell Stress Responses. Springer International, Cham, Switzerland [Google Scholar]
- He X, Qu B, Li W, Zhao X, Teng W, Ma W, Ren Y, Li B, Li Z, Tong Y(2015) The nitrate inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol 169: 1991–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P(2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriet C, Aimé D, Térézol M, Kilandamoko A, Rossin N, Combes-Soia L, Labas V, Serre R-F, Prudent M, Kreplak J, et al. (2019) Water stress combined with sulfur deficiency in pea affects yield components but mitigates the effect of deficiency on seed globulin composition. J Exp Bot 70: 4287–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Sebastià C, Marsolais F, Saravitz C, Israel D, Dewey RE, Huber SC(2005) Free amino acid profiles suggest a possible role for asparagine in the control of storage-product accumulation in developing seeds of low- and high-protein soybean lines. J Exp Bot 56: 1951–1963 [DOI] [PubMed] [Google Scholar]
- Howarth JR, Parmar S, Jones J, Shepherd CE, Corol D-I, Galster AM, Hawkins ND, Miller SJ, Baker JM, Verrier PJ, et al. (2008) Co-ordinated expression of amino acid metabolism in response to N and S deficiency during wheat grain filling. J Exp Bot 59: 3675–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P-H, Kan C-C, Wu H-Y, Yang H-C, Hsieh M-H(2018) Early molecular events associated with nitrogen deficiency in rice seedling roots. Sci Rep 8: 12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson F, Josse J, Le S, Mazet J, Husson MF (2015) Package ‘FactoMineR’. https://cran.r-project.org/web/packages/FactoMineR/index.html.
- Jewell JL, Russell RC, Guan K-L(2013) Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 14: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan C-C, Chung T-Y, Juo Y-A, Hsieh M-H(2015) Glutamine rapidly induces the expression of key transcription factor genes involved in nitrogen and stress responses in rice roots. BMC Genomics 16: 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T(2009) Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J 57: 313–321 [DOI] [PubMed] [Google Scholar]
- Kolde R (2019) pheatmap: Pretty Heatmaps version 1.012 from CRAN. https://rdrr.io/cran/pheatmap.
- Leroy P, Guilhot N, Sakai H, Bernard A, Choulet F, Theil S, Reboux S, Amano N, Flutre T, Pelegrin C, et al. (2012) TriAnnot: A versatile and high performance pipeline for the automated annotation of plant genomes. Front Plant Sci 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Ai Q, Yu D(2015) Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci Rep 5: 11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likes R, Madl RL, Zeisel SH, Craig SAS(2007) The betaine and choline content of a whole wheat flour compared to other mill streams. J Cereal Sci 46: 93–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A.(2008) Transcriptional regulation of genes involved in sulfur assimilation in plants: Understanding from the analysis of high-affinity sulfate transporters. Plant Biotechnol (Tokyo) 25: 323–328 [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H(2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18: 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf GA.(1993) Regulation of sulfur and nitrogen metabolism in filamentous fungi. Annu Rev Microbiol 47: 31–55 [DOI] [PubMed] [Google Scholar]
- Moing A, Maucourt M, Renaud C, Gaudillère M, Brouquisse R, Lebouteiller B, Gousset-Dupont A, Vidal J, Granot D, Denoyes-Rothan B, et al. (2004) Quantitative metabolic profiling by 1-dimensional 1H-NMR analyses: Application to plant genetics and functional genomics. Funct Plant Biol 31: 889–902 [DOI] [PubMed] [Google Scholar]
- Müller M, Knudsen S(1993) The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. Plant J 4: 343–355 [DOI] [PubMed] [Google Scholar]
- Nianiou-Obeidat I, Madesis P, Kissoudis C, Voulgari G, Chronopoulou E, Tsaftaris A, Labrou NE(2017) Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep 36: 791–805 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R(2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138: 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR(2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Kezuka A, Onouchi H, Fujiwara T, Naito S(2008) Promoter region of the β subunit gene of β-conglycinin responds to methionine and glutathione in transient assays using Arabidopsis protoplasts. Soil Sci Plant Nutr 54: 128–132 [Google Scholar]
- Oliver S.(2000) Guilt-by-association goes global. Nature 403: 601–603 [DOI] [PubMed] [Google Scholar]
- Pandurangan S, Pajak A, Molnar SJ, Cober ER, Dhaubhadel S, Hernández-Sebastià C, Kaiser WM, Nelson RL, Huber SC, Marsolais F(2012) Relationship between asparagine metabolism and protein concentration in soybean seed. J Exp Bot 63: 3173–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Datta SK, Datta K(2015) miRNA regulation of nutrient homeostasis in plants. Front Plant Sci 6: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechanek U, Karger A, Gröger S, Charvat B, Schöggl G, Lelley T(1997) Effect of nitrogen fertilization on quantity of flour protein components, dough properties, and breadmaking quality of wheat. Cereal Chem 74: 800–805 [Google Scholar]
- Pont C, Murat F, Confolent C, Balzergue S, Salse J(2011) RNA-seq in grain unveils fate of neo- and paleopolyploidization events in bread wheat (Triticum aestivum L.). Genome Biol 12: R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postles J, Curtis TY, Powers SJ, Elmore JS, Mottram DS, Halford NG(2016) Changes in free amino acid concentration in rye grain in response to nitrogen and sulfur availability, and expression analysis of genes involved in asparagine metabolism. Front Plant Sci 7: 917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org
- Radchuk VV, Sreenivasulu N, Radchuk RI, Wobus U, Weschke W(2005) The methylation cycle and its possible functions in barley endosperm development. Plant Mol Biol 59: 289–307 [DOI] [PubMed] [Google Scholar]
- Randall P, Spencer K, Freney J(1981) Sulfur and nitrogen fertilizer effects on wheat. I. Concentrations of sulfur and nitrogen and the nitrogen to sulfur ratio in grain, in relation to the yield response. Aust J Agric Res 32: 203–212 [Google Scholar]
- Ravel C, Martre P, Romeuf I, Dardevet M, El-Malki R, Bordes J, Duchateau N, Brunel D, Balfourier F, Charmet G(2009) Nucleotide polymorphism in the wheat transcriptional activator Spa influences its pattern of expression and has pleiotropic effects on grain protein composition, dough viscoelasticity, and grain hardness. Plant Physiol 151: 2133–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK(2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A(2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.(2016) Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal Behav 11: e1117723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Martinez M, Abraham Z, Diaz I, Carbonero P(2006) Ternary complex formation between HvMYBS3 and other factors involved in transcriptional control in barley seeds. Plant J 47: 269–281 [DOI] [PubMed] [Google Scholar]
- Ryabova LA, Robaglia C, Meyer C(2019) Target of Rapamycin kinase: Central regulatory hub for plant growth and metabolism. J Exp Bot 70: 2211–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou M-M(2011) The PHD finger: A versatile epigenome reader. Trends Biochem Sci 36: 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Wang H, Tang X(2015) NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front Plant Sci 6: 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Halford NG(2002) Cereal seed storage proteins: Structures, properties and role in grain utilization. J Exp Bot 53: 947–958 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Tatham AS, Halford NG(2001) Nutritional control of storage protein synthesis in developing grain of wheat and barley. Plant Growth Regul 34: 105–111 [Google Scholar]
- Shiferaw B, Smale M, Braun H-J, Duveiller E, Reynolds M, Muricho G(2013) Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur 5: 291–317 [Google Scholar]
- Sirohi G, Pandey BK, Deveshwar P, Giri J(2016) Emerging trends in epigenetic regulation of nutrient deficiency response in plants. Mol Biotechnol 58: 159–171 [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T(2011) Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MB, Müller M, Skerritt J, Simpson D(1996) Hordein promoter methylation and transcriptional activity in wild-type and mutant barley endosperm. Mol Gen Genet 250: 750–760 [DOI] [PubMed] [Google Scholar]
- Thioulouse J, Chessel D, Doledec S, Olivier JM(1997) ADE-4: A multivariate analysis and graphical display software. Stat Comput 7: 75–83 [Google Scholar]
- Triboï E, Martre P, Triboï-Blondel A-M(2003) Environmentally-induced changes in protein composition in developing grains of wheat are related to changes in total protein content. J Exp Bot 54: 1731–1742 [DOI] [PubMed] [Google Scholar]
- Verdier J, Thompson RD(2008) Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant Cell Physiol 49: 1263–1271 [DOI] [PubMed] [Google Scholar]
- Vincent J, Martre P, Gouriou B, Ravel C, Dai Z, Petit J-M, Pailloux M(2015) RulNet: A web-oriented platform for regulatory network inference, application to wheat -omics data. PLoS One 10: e0127127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xiang L, Hong J, Xie Z, Li B(2019) Genome-wide analysis of bHLH transcription factor family reveals their involvement in biotic and abiotic stress responses in wheat (Triticum aestivum L.). 3 Biotech 9: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis, Ed 2, Springer International, Cham, Switzerland [Google Scholar]
- Wieser H, Gutser R, von Tucher S(2004) Influence of sulphur fertilisation on quantities and proportions of gluten protein types in wheat flour. J Cereal Sci 40: 239–244 [Google Scholar]
- Wieser H, Seilmeier W(1998) The influence of nitrogen fertilisation on quantities and proportions of different protein types in wheat flour. J Sci Food Agric 76: 49–55 [Google Scholar]
- Wolfe CJ, Kohane IS, Butte AJ(2005) Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Hynes MJ, Davis MA(2008) Recent advances in nitrogen regulation: A comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot Cell 7: 917–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrigley C, Cros D, Archer M, Downie P, Roxburgh C(1980) The sulfur content of wheat endosperm proteins and its relevance to grain quality. Aust J Plant Physiol 7: 755–766 [Google Scholar]
- Xie Z, Nolan TM, Jiang H, Yin Y(2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Wek SA, Wek RC(2000) Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol Cell Biol 20: 2706–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zeng X, Ren M, Mao X, Qiao S(2017) Novel metabolic and physiological functions of branched chain amino acids: A review. J Anim Sci Biotechnol 8: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Hawkesford M, McGrath S(1999) Sulphur assimilation and effects on yield and quality of wheat. J Cereal Sci 30: 1–17 [Google Scholar]
- Zörb C, Grover C, Steinfurth D, Hermann Mühling K(2010) Quantitative proteome analysis of wheat gluten as influenced by N and S nutrition. Plant Soil 327: 225–234 [Google Scholar]