A phytochrome B-independent pathway restricts growth at high levels of defense, and this growth inhibition is correlated with dysregulation of tryptophan biosynthesis.
Abstract
The plant hormone jasmonate (JA) promotes resistance to biotic stress by stimulating the degradation of JASMONATE ZIM-DOMAIN (JAZ) proteins, which relieves repression on MYC transcription factors that execute defense programs. JA-triggered depletion of JAZ proteins in Arabidopsis (Arabidopsis thaliana) is also associated with reduced growth and seed production, but the mechanisms underlying these pleiotropic growth effects remain unclear. Here, we investigated this question using an Arabidopsis JAZ-deficient mutant (jazD; jaz1–jaz7,jaz9, jaz10, andjaz13) that exhibits high levels of defense and strong growth inhibition. Genetic suppressor screens for mutations that uncouple growth-defense tradeoffs in the jazD mutant identified nine independent causal mutations in the red-light receptor phytochrome B (phyB). Unlike the ability of the phyB mutations to completely uncouple the mild growth-defense phenotypes in a jaz mutant (jazQ) defective in JAZ1, JAZ3, JAZ4, JAZ9, and JAZ10, phyB null alleles only weakly alleviated the growth and reproductive defects in the jazD mutant. phyB-independent growth restriction of the jazD mutant was tightly correlated with upregulation of the Trp biosynthetic pathway but not with changes in central carbon metabolism. Interestingly, jazD and jazD phyB plants were insensitive to a chemical inhibitor of Trp biosynthesis, which is a phenotype previously observed in plants expressing hyperactive MYC transcription factors that cannot bind JAZ repressors. These data provide evidence that the mechanisms underlying JA-mediated growth-defense balance depend on the level of defense, and they further establish an association between growth inhibition at high levels of defense and dysregulation of Trp biosynthesis.
Plants continuously integrate external and developmental cues to optimize their fitness in dynamic environments. Acclimation to stress is often associated with negative pleiotropic effects on plant growth and development, especially when resources are limited. Enhanced nutrient foraging and competitiveness, for example, can occur at the expense of resistance to biotic stress (Moreno et al., 2009; Ballaré, 2014). Conversely, plant resistance to herbivores and pathogens is frequently accompanied by reduced growth and reproductive output (Havko et al., 2016; Karasov et al., 2017; Züst and Agrawal, 2017). The antagonistic relationship between growth and defense has been interpreted as a symptom of metabolic competition for limited resources allocated to defense at the expense of growth, or vice versa (Herms and Mattson, 1992; Heil and Baldwin, 2002; Stamp, 2003). Recent studies, however, have challenged this simple resource-based view of tradeoffs in favor of a more complex regulatory scenario in which interactions between hormone-based signaling networks evoke transcriptional changes that downwardly adjust growth rate upon activation of defense programs (Ullmann-Zeunert et al., 2013; Huot et al., 2014; Campos et al., 2016; Kliebenstein, 2016; Karasov et al., 2017; Züst and Agrawal, 2017; Machado et al., 2017; Guo et al., 2018a; Ballaré and Austin, 2019). A better understanding of mechanisms that constrain the upper limits of growth and defense traits has potential implications for improving sustainable crop production (Ning et al., 2017; Guo et al., 2018a).
Induced resistance to biotic stress, like plant growth, is a highly complex process coordinated in large part by hormone-response pathways that integrate various developmental and environmental cues (Pieterse et al., 2009; Santner and Estelle, 2009; Bürger and Chory, 2019). Through their ability to both promote defense and inhibit growth, the lipid-derived jasmonates (JAs) exert strong control over the growth-defense balance (Baldwin, 1998; Wasternack and Hause, 2013; Guo et al., 2018a). The JA signaling pathway operates mainly in the nucleus and converges on a set of transcription factors that exert exquisite control over the amplitude of defense traits (Howe et al., 2018). In the unstressed state, JASMONATE ZIM-DOMAIN (JAZ) proteins bind to and repress the activity of cognate transcription factors such as MYC2 and its close relatives MYC3 and MYC4 (Chini et al., 2007; Dombrecht et al., 2007; Thines et al., 2007; Yan et al., 2007; Fernández-Calvo et al., 2011; Niu et al., 2011). In response to biotic challenge or developmental cues, the bioactive form of JA, jasmonoyl-l-Ile (JA-Ile), stimulates recognition of JAZ proteins by the F-box protein CORONATINE-INSENSITIVE1 (COI1), which is the specificity determinant of the Skp/Cullin/F-box (SCF)-type E3 ubiquitin ligase complex, SCFCOI1. Ubiquitylation of JAZ substrates by SCFCOI1 marks JAZs for degradation via the 26S proteasome (Thines et al., 2007; Katsir et al., 2008; Fonseca et al., 2009; Yan et al., 2009; Howe et al., 2018). Rapid, stress-induced depletion of JAZ relieves repression on MYC and other client transcription factors to execute JA-mediated defense programs and concomitant growth restriction (Yan et al., 2007; Pauwels et al., 2008; Noir et al., 2013; Attaran et al., 2014). Consistent with this model, dominant mutations that impair the ability of Arabidopsis (Arabidopsis thaliana) MYC transcription factors to bind JAZ repressors lead to activation of a subset of JA responses. For example, the atr2D allele of MYC3 causes upregulation of genes encoding enzymes in the Trp biosynthetic pathway, which gives rise to the production of defensive compounds such as indole glucosinolates and camalexin (Smolen et al., 2002; Goossens et al., 2015).
The growth-defense balance in shoot tissues is controlled in part by interactions between the JAZ-MYC pathway and various regulators of cell expansion, including light and growth hormones (Ballaré, 2014; Huot et al., 2014; Havko et al., 2016). Increasing evidence implicates MYCs as conserved regulators of JA-induced shoot growth inhibition (Zhang and Turner, 2008; Major et al., 2017; Guo et al., 2018b; Peñuelas et al., 2019). MYCs can influence leaf development by interfering with the activity of PHYTOCHROME-INTERACTING FACTORS (e.g. PIF4) and by promoting the activity of ELONGATED HYPOCOTYL5 (HY5), a central regulator of photomorphogenesis (Zhang et al., 2018; Ortigosa et al., 2019). Antagonistic signal cross talk between JA and the red-light receptor phytochrome B (phyB) is thought to allow rapid growth for improved competitiveness with neighboring plants (Moreno et al., 2009; Cerrudo et al., 2012; de Wit et al., 2013; Chico et al., 2014). The transcription factors FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED IMPAIRED RESPONSE1 (FAR1) were recently shown to connect phytochrome and JA signaling, providing a mechanism by which these pathways balance growth and defense (Liu et al., 2019). That phytochromes exert strong control over central metabolism (Yang et al., 2016; Krahmer et al., 2018) raises the additional possibility that JA and light signaling pathways interact to influence the partitioning of central metabolites during growth-to-defense transitions. Shoot growth is also modulated by antagonistic cross talk between the JA and gibberellin (GA) signaling pathways (Navarro et al., 2008; Huot et al., 2014; Machado et al., 2017). In Arabidopsis, multiple members of the JAZ family interact directly with DELLA repressors of GA signaling (Hou et al., 2010; Yang et al., 2012). JA-induced JAZ degradation can modulate the growth-defense balance by increasing the repressive activity of DELLA proteins on growth-promoting PIF transcription factors, thereby prioritizing defense over growth (Hou et al., 2010; Yang et al., 2012).
The function of JAZ proteins as negative regulators of JA responses has important implications for understanding the origins of induced resistance and its relationship to growth and reproductive success (Guo et al., 2018a; Monte et al., 2019). Consistent with the JAZ model of induced resistance (Howe and Jander, 2008; Erb and Reymond, 2019; Wang et al., 2019), the product of a single JAZ gene in the early land plant Marchantia polymorpha controls most, if not all, JA-mediated growth and defense responses (Monte et al., 2019). By comparison, the multimembered JAZ gene families of vascular plants have both overlapping and cell type-specific roles in growth- and defense-related processes (Thireault et al., 2015; Chini et al., 2016; Howe et al., 2018). In addition to hypermorphic MYC variants that are insensitive to JAZ repression (Smolen et al., 2002; Goossens et al., 2015), constitutive activation of JA responses in Arabidopsis has been achieved by combining loss-of-function mutations in multiple JAZ family members. Use of this approach to genetically “tune” JA responses in the absence of exogenous elicitors (e.g. JA treatment) provides a simple experimental system in which to understand how changes in the quantity and quality of defense impacts the growth-defense balance. For example, multimutants defective in five (jaz quintuple [jazQ]), 10 (jaz decuple [jazD]), or 11 (jaz undecuple; [jazU]) JAZ genes display increasing levels of defense concomitant with decreased growth and fecundity (Campos et al., 2016; Guo et al., 2018b). These phenotypes of JAZ deficiency provide mechanistic insight into a key prediction of the cost-benefit theory of plant defense, namely that resistance is costly in the absence of biotic stress (Simms and Rausher, 1987; Baldwin, 1998). A current gap in understanding the growth-defense balance is lack of knowledge of how JA signaling restricts biomass accretion and reproductive output, which are generally regarded as costs of defense (Heil and Baldwin, 2002; Havko et al., 2016; Züst and Agrawal, 2017). jaz mutants in which JA responses are constitutively active provide new genetic tools to address this question and, more generally, to study how variable patterns of defense influence growth and reproductive success.
Central to understanding the underlying mechanisms of the growth-defense balance is the question of how plant growth and fitness are shaped by varying levels of defense (Bergelson and Purrington, 1996; Züst and Agrawal, 2017). Using Arabidopsis as a model system, we explored this question by assessing growth and fitness parameters of jaz mutants in which the level of defense is systematically altered, and by identifying suppressor mutations that mitigate the effects of elevated defense. A genetic screen for mutations that uncouple growth-defense antagonism in the jazQ mutant showed that phyB mutation completely rescued the mild growth defect of the jazQ mutant without affecting the level of defense (Campos et al., 2016). In demonstrating that enhanced defense in the jazQ mutant is not inextricably linked to growth restriction, we initiated this study to determine whether robust growth can also be achieved at even higher levels of defense, as might be expected if JA-mediated growth inhibition is attributed solely to the phyB pathway. A genetic screen for suppressor mutations that rescue the growth deficit of the jazD mutant without compromising defense identified multiple independent mutations in PHYB, thereby validating the importance of JA-phyB cross talk in managing growth-defense phenotypes. However, unlike the jazQ phyB mutant that grows and defends well at the same time, jazD phyB plants maintained the strong defense status of the jazD mutant but displayed only weak growth recovery. Thus, growth restriction associated with high levels of defense is attributed mainly to a phyB-independent pathway. In investigating this pathway, we found that the slow growth of jazD and jazD phyB plants is correlated with upregulation of the Trp biosynthetic pathway but not with changes in central carbon metabolism. Collectively, our results indicate that the mechanisms underlying JA-mediated growth-defense tradeoffs depend on the level of defense and suggest that dysregulation of Trp biosynthesis may contribute to growth restriction at high levels of defense.
RESULTS
jaz Mutations Likely Inhibit Growth Independent of GA Signaling
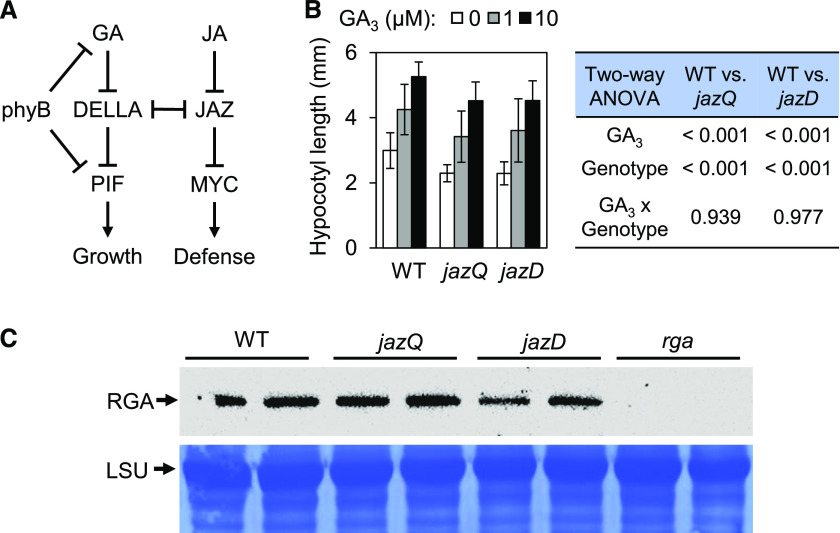
To understand the mechanism by which shoot growth is inhibited in the jazD mutant, we first tested a prevailing model (Fig. 1A) in which antagonistic cross talk between JAZ and DELLA proteins contributes to the growth-defense balance through reciprocal control of cognate MYC and PIF transcription factors (Hou et al., 2010; Yang et al., 2012). Based on this model, we hypothesized that genetic depletion of JAZs in jazQ and jazD mutants, similar to the effects of JA-induced JAZ degradation, may release DELLA proteins to increase the repression of PIFs, thereby tempering the growth of jaz mutants (Fig. 1A). To test this, we measured the sensitivity of jazQ and jazD seedlings to exogenous GA3, a bioactive GA that promotes hypocotyl elongation. In the absence of GA3, jazQ and jazD hypocotyls were both shorter than wild-type hypocotyls (Fig. 1B). However, the extent to which exogenous GA3 promoted hypocotyl elongation in jazQ and jazD seedlings was similar to that observed for wild-type seedlings (Fig. 1B). This finding suggested that moderate (jazQ) or severe (jazD) JAZ depletion does not have a major effect on GA sensitivity under these conditions. Key evidence for the JAZ-DELLA model of growth-defense tradeoffs comes from studies showing that JA-induced JAZ degradation increases the accumulation of the DELLA proteins SLENDER RICE1 (SLR1) and REPRESSOR OF GA (RGA) in rice (Oryza sativa) and Arabidopsis, respectively (Yang et al., 2012). We therefore tested whether constitutive activation of JA responses in jazQ and jazD mutants, as a consequence of JAZ depletion, is associated with elevated levels of RGA. Contrary to this expectation, immunoblot analysis showed that RGA abundance was not increased in untreated jazQ or jazD seedlings relative to the wild type (Fig. 1C). These data suggest that changes in DELLA activity do not play a major role in restricting the shoot growth of jaz mutants.
Figure 1.
Higher-order jaz mutations do not restrict growth by inhibiting gibberellic acid responses. A, Schematic diagram illustrating positive (arrows) and negative (bars) regulation of the interaction between JA and GA signaling pathways. B, Hypocotyl length of wild-type (WT), jazQ, and jazD seedlings grown on medium supplemented with various concentrations (0, 1, or 10 μm) of GA3. Data show the means ± sd (n = 10 seedlings per genotype). The effect of GA3 and genotype on hypocotyl elongation was tested by two-way ANOVA and showed that the GA3 × genotype interaction was insignificant. C, Detection of RGA protein by western blot analysis of protein extracts from wild-type, jazQ, jazD, and rga seedlings grown on agar medium. Biological replicates are shown for each genotype. Coomassie brilliant-blue staining of the PVDF membrane for the large subunit (LSU) of Rubisco is shown as a loading control.
Identification of jazD Suppressor Mutations
In the absence of evidence that the dwarf growth stature of the jazD mutant is caused by attenuation of GA-mediated growth responses, we conducted genetic suppressor screens to identify mutations that recover the growth of the jazD mutant without impeding the high level of defense. We visually screened a population of ∼20,000 ethyl methanesulfonate (EMS)-mutagenized jazD plants (M2 generation) for individuals with increased rosette size and, as a proxy for defense, persistence of elevated leaf anthocyanin content (Supplemental Fig. S1). In anticipation of a potential contribution of light signaling to the reduced growth of jazD plants (Campos et al., 2016), we screened an additional 10,000 M2 seedlings for long hypocotyls, followed by rescreening of these long-hypocotyl plants at maturity for larger rosette size and increased anthocyanin levels. These combined screens identified 13 suppressor of jazD (sjd) mutants with partially improved rosette growth relative to the jazD parental line. The partial recovery of rosette diameter, leaf area, and biomass of each of these mutant lines was heritable in subsequent generations.
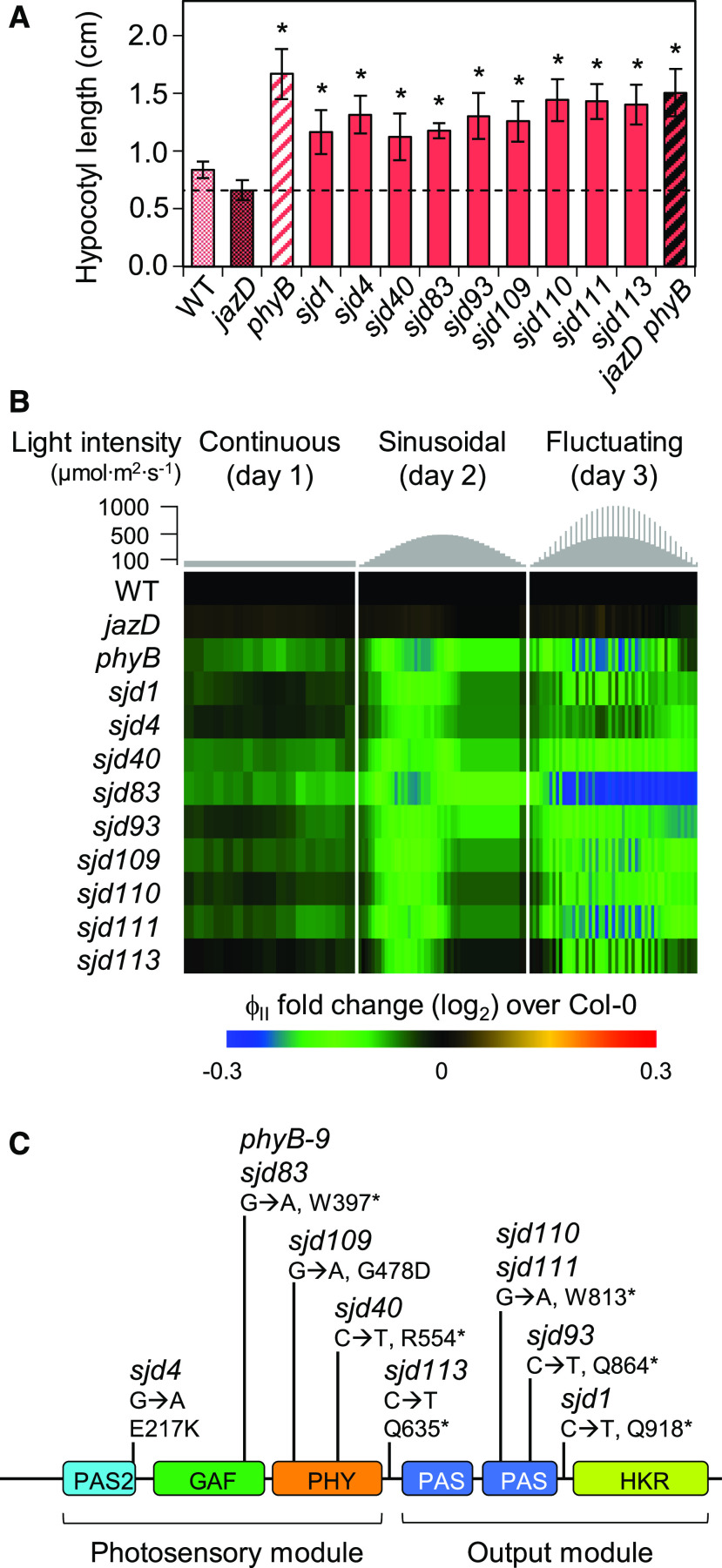
During the initial characterization of growth phenotypes, we observed that the increased leaf area and biomass in a subgroup of nine sjd mutants (sjd1, sjd4, sjd40, sjd83, sjd93, sjd109, sjd110, sjd111, and sjd113) was associated with elongated hypocotyls and petioles, the latter of which contributed to increased rosette diameter (Supplemental Fig. S2). Given these distinct morphological features and their similarity to photomorphogenic mutants, subsequent experiments were focused on this subgroup of long-hypocotyl mutants. Tests of seedling responses to monochromatic light showed that all nine sjd lines had elongated hypocotyls under continuous red light (Fig. 2A), suggesting a potential defect in phyB signaling. In support of this hypothesis, the hypocotyl growth response of all nine mutants to far red or blue light was similar to that of the jazD mutant (Supplemental Fig. S3). Because phyB mutations impair photosynthesis in mature leaves (Boccalandro et al., 2009; Campos et al., 2016), we also assessed the chlorophyll fluorescence phenotypes of sjd mutants relative to jazD plants and an authentic phyB mutant (phyB-9). In response to growth under dynamic light conditions, we found that the PSII quantum efficiency (ΦII) in all long-hypocotyl mutants was decreased in comparison to that in jazD and wild-type plants, and was in fact very similar to that of the phyB-9 mutant (Fig. 2B). Targeted DNA sequencing showed that all nine long-hypocotyl sjd mutants harbor point mutations in the PHYB gene, with most of these changes located in or near conserved domains of the protein (Fig. 2C). Allelic complementation tests further confirmed that the long-hypocotyl phenotype of sjd mutants under continuous white light was caused by the phyB mutations (Supplemental Fig. S4). To eliminate the possibility that additional EMS-induced mutations contribute to the growth phenotypes of this group of sjd mutants, we performed genetic crosses to reconstruct a jazD phyB undecuple mutant carrying the phyB-9 null allele (Fig. 2A; Supplemental Fig. S5). All subsequent experiments were performed with this genetically reconstructed jazD phyB-9 line.
Figure 2.
Long-hypocotyl sjd mutants are impaired in phytochrome B signaling. A, Hypocotyl lengths of long-hypocotyl sjd mutants under monochromatic red light. Seedlings of the indicated genotype were grown for 7 d on LS medium in continuous red light at a fluence rate of 25 μE m−2 s−1. Data points are means ± sd (n = 8–10 plants per genotype). The dashed line indicates the length of jazD hypocotyls. Asterisks denote significant differences at P < 0.05 relative to jazD by Dunnett’s Test. B, Heat map of ΦII, in which chlorophyll fluorescence values for the indicated mutants were normalized to Col-0. Photosynthetic performance was monitored over 3 d of 16 h/d light intensity regimes: constant light (day 1, left); a sinusoidal increase and decrease in light intensity (day 2, middle); and a sinusoidal light regime with higher-intensity pulses (day 3, right). C, Sequencing of the PHYB gene in red-light-insensitive sjd lines identified mutations in all nine mutants. The diagram depicts locations of mutations relative to conserved domains of phyB (colored boxes). Asterisks denote nonsense mutations. One mutant (sjd83) harbors the same G-to-A transition mutation as the phyB-9 mutant allele. The N-terminal photosensory module includes a PAS-2 (Period/Arnt/Single-minded) domain, a GAF (cGMP phosphodiesterase/adenylyl cyclase/FhlA) domain for binding the bilin chromophore, and a PHY domain that stabilizes the photoactivated Pfr state. The C-terminal output module includes two PAS domains and a regulatory His kinase-related (HKR) domain.
The phyB Mutation Does Not Compromise Defense Phenotypes in the jazD Background
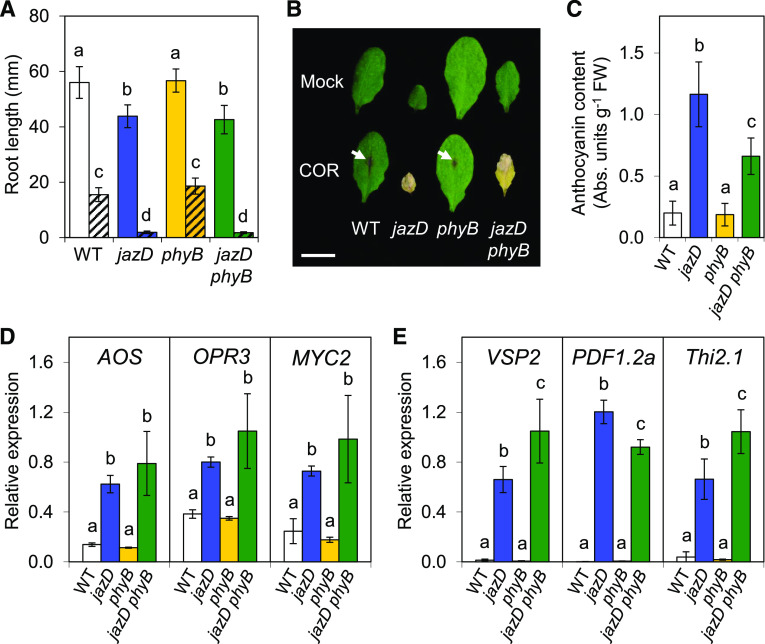
We tested whether JA signaling is altered in jazD phyB plants by examining the sensitivity of roots and shoots to exogenous JA, which elicits strong hypersensitive reactions in the jazD mutant (Guo et al., 2018b). Root growth measurements showed that phyB and wild-type roots were of similar length on JA-free medium, whereas the loss of phyB in jazD phyB plants had no effect on the constitutive short-root phenotype of the jazD mutant (Fig. 3A). On media supplemented with JA, jazD and jazD phyB roots were similarly hypersensitive to the hormone compared to wild-type or phyB plants (Fig. 3A). We also assessed leaf sensitivity to JA by treatment with coronatine, which, as an agonist of the JA-Ile receptor, elicits strong JA responses when applied to Arabidopsis shoots (Feys et al., 1994; Attaran et al., 2014). jazD and jazD phyB leaves exhibited unrestrained responses to coronatine, as seen from spreading necrosis and tissue death within 4 d of treatment. Wild-type and phyB leaves were much less sensitive to coronatine, showing only anthocyanin accumulation at the site of treatment (Fig. 3B). Together, these data indicate that the phyB mutation does not significantly alter the sensitivity of jazD roots or shoots to exogenous JA.
Figure 3.
jazD phyB plants retain extreme hypersensitivity to JA and constitutive JA responses. A, Root lengths of wild-type (WT; white bars), jazD (blue bars), phyB (yellow bars), and jazD phyB (green bars) seedlings grown on plates supplemented (hatched) or not supplemented (open) with 25 μm methyl-JA. Data points show the means ± sd (n = 16–20 seedlings per genotype). B, Leaves of wild-type, jazD, phyB, and jazD phyB plants were spotted with 5 µL of a solution containing 50 µm coronatine (COR). Four days after treatment, leaves were excised for photographing. Scale bar = 1 cm. Arrows show anthocyanin accumulation at the location of coronatine application. C, Accumulation of anthocyanins in rosette leaves of wild-type, jazD, phyB, and jazD phyB plants. Data points show the means ± sd (n = 10–13 plants per genotype). D and E, Relative gene expression level determined by RT-qPCR of JA biosynthesis and signaling genes AOS, OPR3, and MYC2 (D) and defense-associated genes VSP2, PDF1.2a, and Thi2.1 (E) in whole shoots of 28-d-old wild-type, jazD, phyB, and jazD phyB plants. Expression levels were normalized to the reference gene PP2a. Data points are means ± sd (n = 3 plants per genotype). Lowercase letters in A and C–E represent significant differences at P < 0.05 with Tukey’s honestly significant difference (HSD) mean-separation test.
We next tested whether the loss of phyB affected the high constitutive expression of various JA-responsive markers in the jazD background. Consistent with our screen for sjd mutants that retain elevated anthocyanin content, the level of anthocyanin accumulation in rosette leaves of jazD phyB plants was about 3-fold higher than in the wild type, albeit not as high as in the jazD mutant (Fig. 3C). Transcript abundance measured by reverse transcription quantitative PCR (RT-qPCR) further showed that early JA-response genes, including ALLENE OXIDE SYNTHASE (AOS), OXOPHYTODIENOATE-REDUCTASE 3 (OPR3), and MYC2, were highly expressed to similar levels in jazD and jazD phyB plants (Fig. 3D). Likewise, defense genes associated with insect and pathogen attack, including VEGETATIVE STORAGE PROTEIN2 (VSP2), PLANT DEFENSIN1.2a (PDF1.2a), and THIONIN2.1 (Thi2.1), were also highly expressed in the jazD phyB mutant (Fig. 3E). These data indicate that defense-related transcriptional programs activated in the jazD mutant remain active in the jazD phyB mutant.
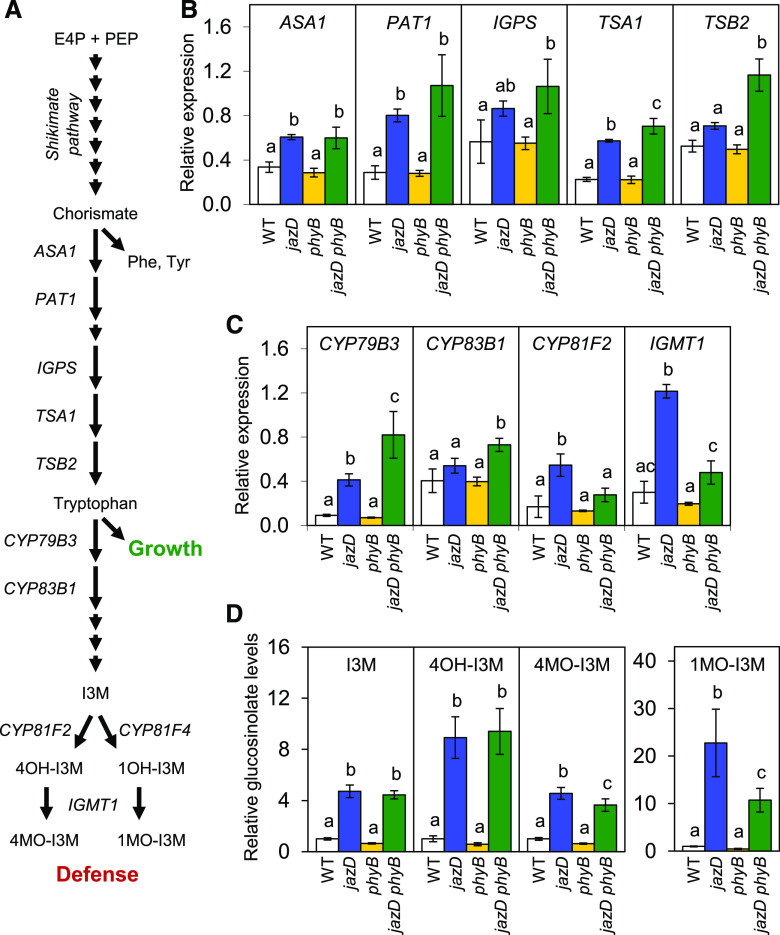
Transcriptional reprogramming in the jazD mutant involves the coordinated expression of primary and specialized metabolic genes involved in the biosynthesis of defensive compounds (Guo et al., 2018b). A prominent example is the upregulation of the Trp biosynthetic pathway, together with enhanced expression of genes encoding enzymes for the conversion of Trp to indole glucosinolates and related defense compounds (Fig. 4A). RT-qPCR analysis showed that among several Trp biosynthetic genes tested, all were upregulated in both jazD and jazD phyB plants relative to the wild-type and phyB backgrounds (Fig. 4B). Similarly, mRNAs encoding enzymes involved in the synthesis of the core glucosinolate structure (CYP79B3 and CYP83B1), as well as enzymes that modify the indole side chain (CYP81F2 and IGMT1; Fig. 4A), were generally more abundant in jazD and jazD phyB plants than in the wild type (Fig. 4C). To validate these findings, we used liquid chromatography-mass spectrometry (LC-MS) to determine the effect of the phyB mutation on the constitutive accumulation of indole glucosinolates in jazD leaves. As shown in Figure 4D (and Supplemental Fig. S6A), virtually all identifiable glucosinolates that were more abundant in the jazD mutant (relative to the wild type) were also elevated in the jazD phyB mutant, with a trend that levels in the jazD phyB mutant were slightly lower than in the jazD mutant. Principal component analysis of the complete glucosinolate profile in each genotype explained 88% of the total variance among the genotypes (Supplemental Fig. S6B). This analysis also showed that the overall glucosinolate profile of jazD phyB leaves was similar to that of jazD but distinct from that of wild-type and phyB leaves. We conclude that the constitutive production of indole glucosinolates in the jazD mutant remains largely intact in the jazD phyB mutant.
Figure 4.
Glucosinolate accumulation remains elevated in the jazD phyB mutant. A, Simplified pathway for the biosynthesis of indole glucosinolates from central carbon metabolites erythrose 4-phosphate (E4P) and phosphoenolpyruvate (PEP). As a key intermediate in this pathway, Trp is both an essential amino acid for growth and a precursor for defensive glucosinolates. B and C, RT-qPCR measurements of relative transcript levels of Trp biosynthesis genes (ASA1, PAT1, IGPS, TSA1, and TSB2; B) and indole glucosinolate biosynthetic genes (CYP79B3, CYP83B1, CYP81F2, and IGMT1; C) in rosette leaves of the indicated genotypes. Expression levels were normalized to the reference gene PP2a. Data points show the means ± sd (n = 3 plants per genotype). D, Accumulation of indole glucosinolates in rosette leaves of wild-type (WT), jazD, phyB, and jazD phyB plants. Data points are means ± sd (n = 4 plants per genotype). Lowercase letters represent a significant difference at P < 0.05, determined by Tukey’s HSD mean-separation test. ASA1, Anthranilate synthase alpha subunit 1; PAT1, anthranilate phosphoribosyltransferase1; IGPS, indole-3-glycerol-phosphate synthase; TSA1, Trp synthase α chain1; TSB2, Trp synthase β chain2; IGMT1, indole glucosinolate O-methyltransferase1; I3M, indol-3-ylmethyl (glucobrassicin); 4OH-I3M, 4-hydroxyindol-3-ylmethyl (hydroxyglucobrassicin); 4MOI3M, 4-methoxyindol-3-ylmethyl (methoxyglucobrassicin); 1OH-I3M, 1-hydroxyindol-3-ylmethyl; 1MOI3M, 1-methoxyindol-3-ylmethyl (neoglucobrassicin).
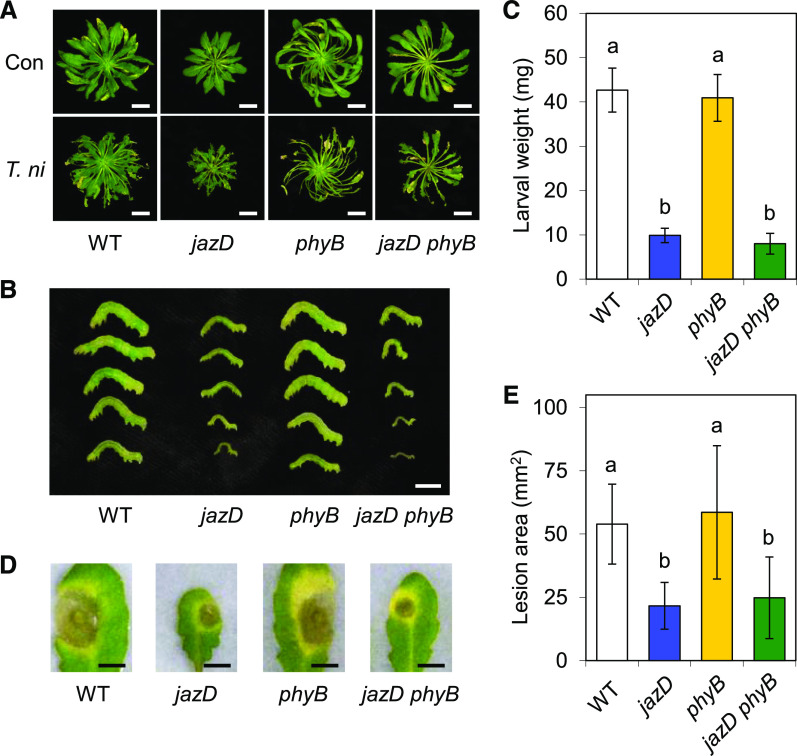
Finally, we tested whether the enhanced resistance of the jazD mutant to insect herbivory by Trichoplusia ni or infection by the necrotrophic fungal pathogen Botrytis cinerea depends on phyB signaling. jazD leaves displayed strong resistance to both T. ni and B. cinerea challenge (Fig. 5), as previously reported (Guo et al., 2018b). Whereas T. ni larvae reared on wild-type and phyB plants achieved similar weights after 10 d of feeding under these experimental conditions, insect performance on jazD phyB leaves was dramatically reduced to levels observed on jazD plants (Fig. 5, B and C). Likewise, B. cinerea infection assays showed that the size of spreading lesions on jazD and jazD phyB leaves was comparable, and much smaller than that on wild-type and phyB leaves (Fig. 5, D and E). These data indicate that the loss of phyB signaling does not significantly compromise the high level of JA-mediated resistance conferred by the jazD mutations.
Figure 5.
The enhanced resistance of the jazD mutant to biotic stress is not compromised by the loss of phyB. Plants of the indicated genotype were grown under short-day conditions and challenged with neonate T. ni larvae or B. cinerea infection. A, Photograph of control (Con) and insect-challenged (T. ni) plants at the end of the feeding trial. Scale bars = 2 cm. WT, Wild type. B and C, Photograph of representative T. ni larvae (B) and larval weights (C) measured after 10 d of feeding on the indicated genotype. Scale bar = 0.5 cm. Data points show the means ± sd (n = 14, where each sample is the mean of three larvae per plant). D and E, Photograph of representative B. cinerea lesions (D) and areas of spreading necrotic lesions (E) after 5 d of infection on detached leaves of the indicated genotype. Scale bars = 0.5 cm. Data points show the means ± sd (n = 15–18 infected leaves per genotype). Lowercase letters in C and E represent a significant difference at P < 0.05, determined by Tukey’s HSD mean-separation test.
The phyB Mutation Weakly Recovers the Growth and Reproductive Phenotypes of the jazD Mutant
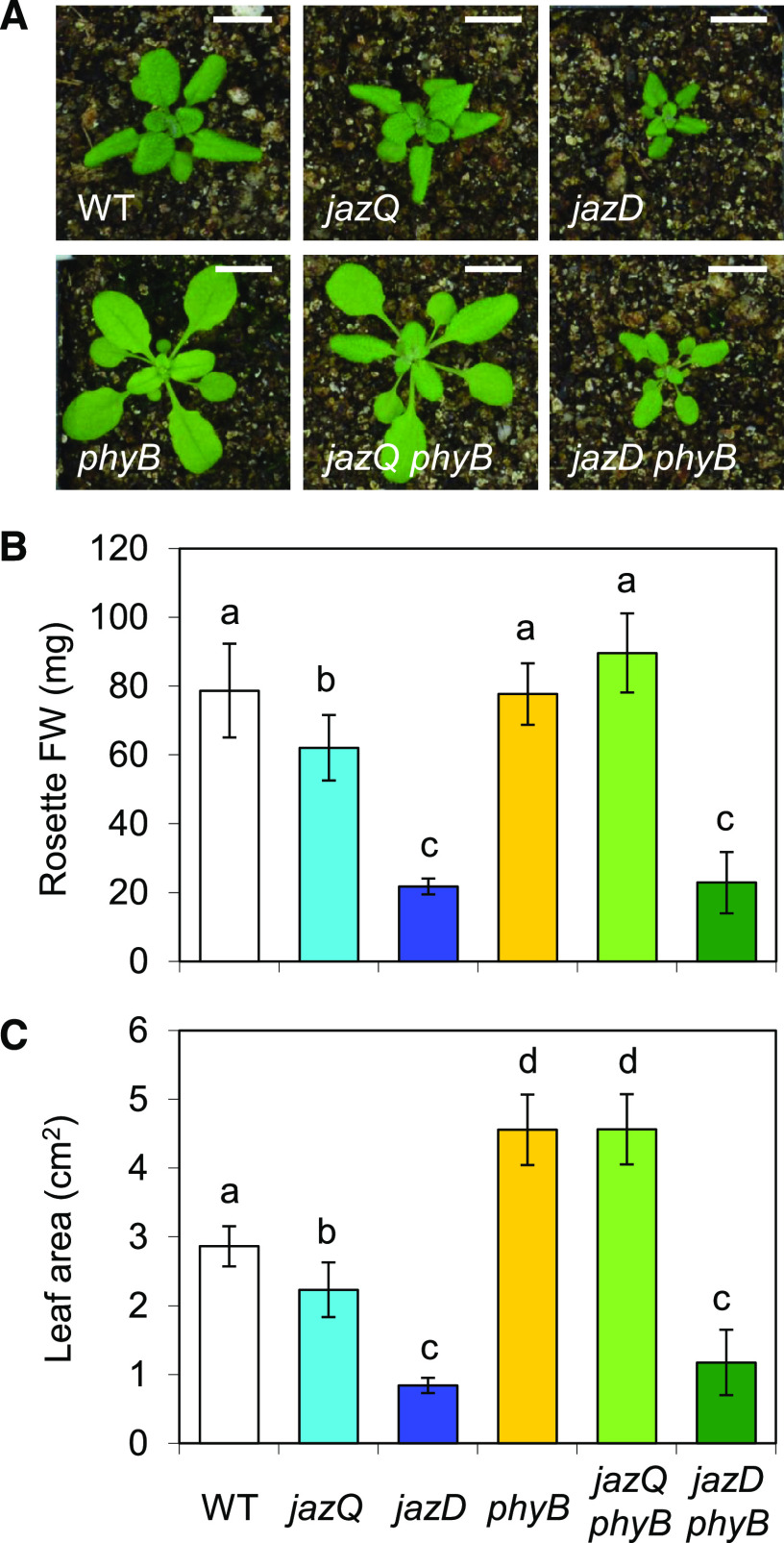
Our initial characterization of long-hypocotyl sjd mutants suggested that the phyB mutation only weakly recovered the rosette growth of the jazD mutant (Supplemental Fig. S2), in contrast to the full growth recovery reported for the jazQ phyB mutant (Campos et al., 2016). To validate this observation, we compared the growth of jazD phyB and jazQ phyB plants, together with appropriate control lines, in a set of plants grown side by side under long-day conditions. The moderate reduction in rosette biomass of jazQ plants was fully recovered by the phyB mutation, with the leaf area of the jazQ phyB mutant being even greater than that of the wild type (Fig. 6). By comparison, the rosette biomass and leaf area of the jazD phyB mutant were similar to those of the jazD mutant. Under these conditions, however, the petiole length and rosette diameter of the jazD phyB plants were greater than those of jazD plants (Supplemental Fig. S7A), consistent with our visual identification of these sjd mutants in the suppressor screen.
Figure 6.
The phyB mutation fully recovers the reduced rosette growth in the jazQ, but not the jazD, background. A, Photographs of representative 25-d-old wild-type (WT), jazQ, jazD, phyB, jazQ phyB, and jazD phyB plants. Scale bars = 1 cm. B and C, Rosette fresh weight (FW; B) and projected leaf area (C) of 25-d-old plants. Bars are means ± sd (n = 10 plants per genotype). Different letters represent a significant difference at P < 0.05 with Tukey’s HSD mean-separation test.
We noticed that jazD phyB plants grown under the short-day (8-h days) conditions used for our insect and pathogen bioassays appeared to have greater growth recovery than those grown under long-day (16-h days) conditions (Fig. 5A). Comparison of jazD and jazD phyB plants grown under long- and short-day conditions indeed showed that the growth recovery of jazD phyB plants (relative to jazD) was stronger under short days (Supplemental Fig. S7). For example, in short-day-grown jazD phyB plants, the rosette diameter and petiole length returned to wild-type levels, while the rosette fresh weight and leaf area were improved by ∼50% of that of the wild type (Supplemental Fig. S7). These data indicate that genetic interactions between jazD and phyB affect shoot growth in a photoperiod-dependent manner.
The inability of the phyB mutation to fully recover the slow shoot growth in the jazD background extended to reproductive phenotypes of the jazD mutant (Guo et al., 2018b). Specifically, we found that traits indicative of the poor reproductive performance of jazD plants, including shorter siliques and fewer seeds per silique, were only partially recovered in jazD phyB plants (Supplemental Fig. S8, A and B). We also observed that the strong delay in time to flowering in the jazD mutant was not affected by the loss of phyB in jazD phyB plants (Supplemental Fig. S8C). These results show that, similar to shoot growth phenotypes, the phyB mutation does not fully recover the reproductive phenotypes of the jazD mutant.
Reduced Growth of the jazD phyB Mutant Is Not Strongly Correlated with Changes in Central Metabolism
We next explored the hypothesis that the slow growth of jazD phyB plants is associated with changes in central metabolism resulting from strong (jazD), but not from moderate (jazQ), defense levels. We reasoned that any metabolic changes interfering with phyB-mediated growth recovery of the jazD mutant would persist in the jazD phyB plants but would be absent in jazQ and jazQ phyB mutants. Gas exchange measurements showed that the photosynthetic rate per unit leaf area relative to the wild type was not affected in the jazD mutant but was reduced in both phyB and jazD phyB plants (Supplemental Fig. S9, A and B). Analysis of the CO2 response curves further indicated that the reduced net assimilation of CO2 in jazD phyB leaves reflects limitations in the activity of Rubisco and electron transport (Supplemental Fig. S9A), which is remarkably similar to the photosynthetic phenotype of jazQ phyB plants in which growth is fully restored (Campos et al., 2016). We also employed gas exchange experiments to measure the day- and night-time respiration rates in the various genotypes. Respiration rates in the jazD mutant were slightly higher than in wild-type and phyB plants (Supplemental Fig. S9, C and D), as previously reported (Guo et al., 2018b). Respiration rates in the jazD phyB mutant were intermediate between those of the jazD and phyB mutants, but the differences were not significant. These data suggest that changes in photosynthesis and respiration do not account for the reduced growth of jazD phyB plants.
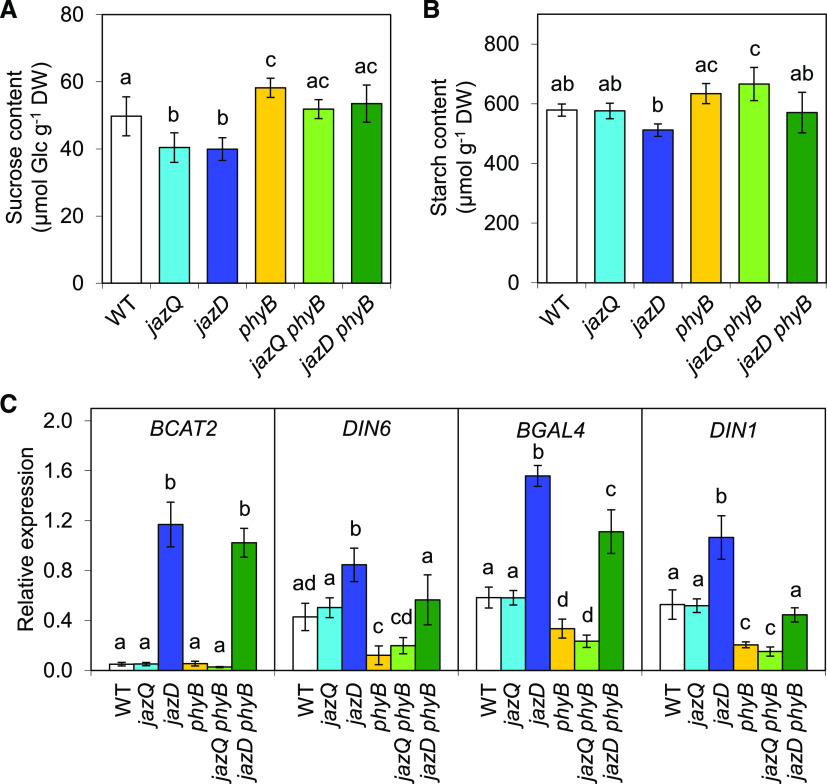
We previously observed that the heightened defense status and slow growth of the jazD mutant is associated with symptoms of carbon limitation, including increased expression of sugar starvation marker genes and modest reduction in the levels of Suc and starch (Guo et al., 2018b). In comparisons across genotypes, end-of-day Suc levels were lower in jazQ and jazD but higher in phyB plants relative to the wild type (Fig. 7A), consistent with previous studies (Guo et al., 2018b; Yang et al., 2016). Suc content was recovered in both jazQ phyB and jazD phyB plants (Fig. 7A). Thus, variations in Suc levels do not strictly correlate with growth. A similar trend was observed for starch, where the phyB mutation tended to increase starch levels in both the jazQ and jazD genetic backgrounds (Fig. 7B). We next queried the transcript abundance of four sugar starvation marker genes (BRANCHED-CHAIN AMINO ACID TRANSFERASE2 [BCAT2], DARK INDUCIBLE1 [DIN1], DARK INDUCIBLE6 [DIN6], and BETA-GALACTOSIDASE4 [BGAL4]) that are induced in response to extended darkness (Baena-González et al., 2007). In soil-grown plants maintained under our standard long-day conditions, transcripts of all four marker genes were more abundant in jazD leaves than in wild-type, phyB, and jazQ leaves (Fig. 7C). BCAT2 and BGAL4 expression remained elevated in the jazD phyB mutant, whereas that of DIN1 and DIN6 returned to wild-type levels (Fig. 7C). These findings indicate that although the expression of several marker genes for carbon limitation is elevated in jazD leaves, these markers are not invariably associated with reduced growth of the jazD phyB mutant.
Figure 7.
Symptoms of carbon limitation are partially recovered in the jazD phyB mutant. A and B, End-of-day starch (A) and Suc (B) levels in shoots of the indicated genotypes. Bars show the means ± sd (n = 5 plants per genotype). DW, Dry weight; WT, wild type. C, Expression level of sugar-starvation marker genes in shoot tissues of the indicated genotypes. mRNA levels were measured by RT-qPCR and normalized to the reference gene PP2a. Bars depict the means ± sd (n = 4 plants per genotype). Different letters represent a significant difference at P < 0.05 with Tukey’s HSD mean-separation test.
Reduced Growth of the jazD phyB Mutant Is Associated with Increased Trp Metabolism
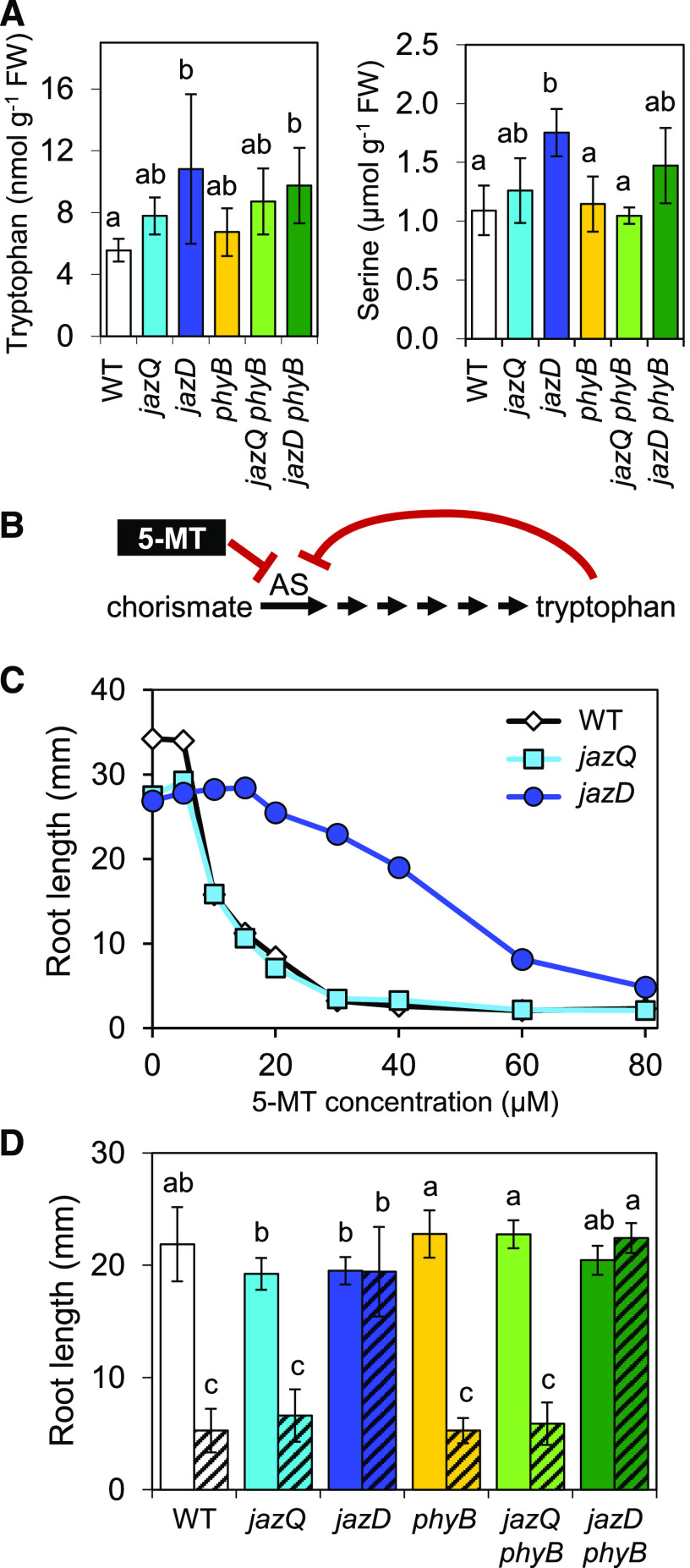
Given that the high level of defense in jazD and jazD phyB plants is associated with strong upregulation of the Trp biosynthetic pathway and accumulation of Trp-derived defense compounds (Fig. 4; Guo et al., 2018b), we considered the possibility that changes in Trp metabolism also contribute to growth inhibition of jazD and jazD phyB plants. To investigate this hypothesis, we used RT-qPCR analysis to determine the extent to which elevated expression of Trp biosynthetic genes correlates with the growth phenotype of the six relevant genotypes (wild type, jazQ, jazD, phyB, jazQ phyB, and jazD phyB). This analysis showed that the expression of genes encoding enzymes for the conversion of chorismate to Trp was elevated to similar levels in the two growth-inhibited mutants (jazD and jazD phyB) but not in the genotypes having modest (jazQ) or no (wild type, phyB, and jazQ phyB) growth reduction (Supplemental Fig. S10, A and B). Very similar results were obtained for genes in the phospho-Ser pathway, which supplies Ser for Trp biosynthesis (Supplemental Fig. S10, A and B). Consistent with these results, we also found that the levels of Trp and Ser in jazD and jazD phyB leaves were elevated relative to the other four genotypes (Fig. 8A).
Figure 8.
Elevated Trp metabolism persists in the jazD phyB mutant. A, Trp and Ser levels in rosette leaves of the indicated genotypes. Bars are means ± sd (n = 4 plants per genotype). FW, Fresh weight. B, Biosynthesis pathway for Trp from chorismate. AS is feedback inhibited by Trp and the toxic Trp analog 5-MT. C, Root lengths of wild-type (WT), jazQ, and jazD seedlings grown on agar medium containing various concentrations of 5-MT. Data points are means (n = 22–27). D, Root lengths of wild-type, jazQ, jazD, phyB, jazQ phyB, and jazD phyB seedlings grown on agar medium supplemented (hashed bars) or not (clear bars) with 15 μm 5-MT. Bars are means ± sd (n = 12–15 seedlings per genotype). Lowercase letters in A and D represent a significant difference at P < 0.05 with Tukey’s HSD mean-separation test.
The initial step in Trp biosynthesis is catalyzed by anthranilate synthase (AS), which is subject to feedback inhibition by Trp (Fig. 8B). As an independent approach to correlate changes in Trp metabolism with the strength of growth-defense tradeoffs, we tested the sensitivity of jazQ and jazD seedlings to a toxic analog of Trp, 5-methyl-Trp (5-MT). 5-MT also exerts feedback inhibition on AS, but unlike Trp, it cannot be used to support protein synthesis (Fig. 8B; Li and Last, 1996). Interestingly, we found that jazD roots were more resistant than wild-type roots to a broad range of 5-MT concentrations, whereas jazQ roots were as sensitive as wild-type roots (Fig. 8C). This finding is consistent with the fact that AS and other Trp biosynthetic enzymes are strongly upregulated in the jazD mutant, but not in the jazQ mutant (Supplemental Fig. S10; Guo et al., 2018b). Using a concentration of 5-MT (15 μm) that strongly inhibits the growth of jazQ and wild-type roots, but not jazD roots, we next compared the 5-MT sensitivity of all six genotypes. Similar to the results obtained for other Trp biosynthetic markers, jazD and jazD phyB roots were completely insensitive to 5-MT, whereas wild-type, jazQ, phyB, and jazQ phyB roots were fully sensitive to the inhibitor (Fig. 8D). These collective data show that increased Trp metabolism is associated not only with elevated production of defense compounds and biotic resistance, but also with reduced growth.
DISCUSSION
JAZ transcriptional repressors promote growth and reproductive success in plants by preventing overactivation of defense responses, a function that is conserved in ancient land plants (Guo et al., 2018a, 2018b; Monte et al., 2019). Here, we used a jaz decuple mutant (jazD) of Arabidopsis that is mostly devoid of the JAZ repressors as a model to explore the nature of growth and reproductive constraints that accompany high levels of defense. To further address how growth-defense tradeoffs are alleviated and, specifically, whether the mitigation of tradeoffs by rewiring of JA-linked transcriptional circuits is dependent on the level of defense, we employed a genetic suppressor screen of the jazD mutant. We reasoned that if genetic uncoupling of growth-defense antagonism, as observed previously in the jazQ phyB mutant (Campos et al., 2016), is independent of the level of defense, it should be possible to identify sjd mutants in which high levels of defense (i.e. jazD-like) are accompanied by robust growth (i.e. wild type-like) and reproductive performance. Although this expectation was not borne out, our identification of phyB as the causal mutation in 9 of the 13 sjd suppressors validates the role of phyB activity in JA-induced growth inhibition (Campos et al., 2016) and, given the weak growth recovery of jazD phyB mutants, also reveals a phyB-independent pathway for growth restriction. Our selection of a subset of sjd mutants for long-hypocotyl phenotypes likely biased the screen in favor of light signaling defects. Nevertheless, we note that of the four remaining (non-phyB) sjd suppressors identified, none exhibited obvious light-related phenotypes (e.g. elongated hypocotyls) or complete uncoupling of growth and defense phenotypes. Further characterization of these non-phyB suppressor mutants may provide additional insight into the underlying mechanisms of the JA-mediated growth-defense balance. Because our sjd suppressor screen was not saturated for genes other than PHYB, additional suppressor screens with the jazD mutant or other higher-order jaz mutants may be informative.
The opposing effects of the jaz and phyB mutations on leaf architecture may provide, at least in part, a physiological explanation for how loss of phyB mitigates the slow growth of constitutive JA-response mutants. The progressive negative effect of the jazQ and jazD mutations on leaf area and petiole length leads to increased leaf overlap and, as a consequence, a reduction in whole-plant leaf area available to intercept light (Guo et al., 2018b). We previously showed that phyB increases the leaf area of the jazQ mutant while also reducing leaf thickness, which reduces leaf construction costs, elevates the whole-plant photosynthetic rate, and likely contributes to growth recovery (Campos et al., 2016; Weraduwage et al., 2018). These architectural features of jazQ phyB leaves are similar to those observed here for the jazD phyB mutant, particularly the accentuated growth of jazD phyB plants relative to jazD under short-day conditions in which a longer total growth period compounds the effect of leaf area on light capture and photosynthetic performance (Weraduwage et al., 2015). Future studies are needed to better understand the mechanisms by which JAZ and phyB regulate leaf architecture, including potential impacts on cell division and cell wall remodeling (Bömer et al., 2018; Mielke and Gasperini, 2019).
The JA pathway is part of a larger, integrated signaling network that controls growth and development in response to changing environmental factors (Kazan and Manners, 2011; Ballaré, 2014; Huot et al., 2014). A key node of hormone cross talk involves the mutual antagonism between JA and GA responses, which is controlled in part by JAZ-DELLA interactions (Hou et al., 2010; Yang et al., 2012). Our results do not support the hypothesis that the growth restriction of jaz mutants results from inhibition of GA responses, consistent with other work showing that the effects of JA on growth are largely independent of GA (Zhang and Turner, 2008; Ortigosa et al., 2019). We favor an alternative hypothesis in which MYC transcription factors play a role in jaz-mediated growth inhibition, which is supported by recent studies in Arabidopsis and the liverwort Marchantia polymorpha (Major et al., 2017; Peñuelas et al., 2019). Emerging evidence further indicates that MYC transcription factors contribute to JA-phyB cross talk by influencing other photomorphogenic regulators, including PIF4, HY5, and FHY3 (Zhang et al., 2018; Chakraborty et al., 2019; Liu et al., 2019; Ortigosa et al., 2019). A better understanding of how JA and phyB signaling pathways intersect to modulate the growth-defense balance is likely to emerge from systems-level analysis of the action of cognate transcription factors, including G-box-binding MYCs and PIFs that occupy the promoter regions of many JA- and light-responsive genes (Franklin and Quail, 2010; Zander et al., 2020).
The modest recovery of rosette biomass and leaf area of the jazD phyB mutant contrasts with the restoration of wild-type-sized rosettes in jazQ phyB plants (Campos et al., 2016). The inability of the phyB mutation to fully restore growth in the jazD background indicates the existence of one or more phyB-independent pathways for growth inhibition that operate at high defense levels. A clue to the incomplete growth recovery of the jazD phyB mutant comes from the observation that this mutant maintains high levels of defense associated with major reprogramming of primary and specialized metabolism, including symptoms of carbon limitation and dysregulation of amino acid metabolism (Guo et al., 2018b). However, we found that the symptoms of carbon limitation were largely alleviated in the jazD phyB mutant, even though this mutant maintains a slow growth habit. The improved growth of jazD phyB plants under short-day photoperiod conditions is also consistent with the notion that central carbon metabolism is not limiting the growth of jazD phyB plants, because carbon limitation should be exacerbated under shorter days, thus further restricting growth (Gibon et al., 2004; Smith and Stitt, 2007).
Our finding that Trp metabolism remains elevated in jazD phyB plants but not in jazQ or jazQ phyB plants establishes a correlation between high defense levels, increased Trp metabolism, and growth constraint. Interestingly, some Trp-related phenotypes of jazD and jazD phyB plants are reminiscent of the effects of dominant mutations (e.g. atr2D) that impede the binding of MYC transcription factors to JAZ repressors, leading to hyperactive MYC activity (Smolen et al., 2002; Goossens et al., 2015). The severe growth restriction of MYC3atr2D-overexpressing plants (Smolen et al., 2002) resembles that of the jazD mutant and supports the notion of MYCs as growth inhibitory factors (Major et al., 2017). The insensitivity of jazD and MYC3atr2D mutants to 5-MT further suggests that hyperactivation of MYC3 contributes to the upregulation of Trp metabolism in jazD plants (Smolen et al., 2002; this study). It remains to be determined whether other gain-of-function MYC mutants, such as MYC2D105N (Goossens et al., 2015), are affected in 5-MT sensitivity or the production of Trp-derived defense compounds. We also note that the MYC3atr2D mutant does not recapitulate all phenotypes of the jazD mutant. For example, whereas the MYC3atr2D mutant is associated with lower levels of free Trp in leaves (Smolen et al., 2002), we found that Trp levels in jazD leaves are slightly higher than in the wild type. This observation suggests a broader regulatory function for JAZs in coordinating primary metabolism (e.g. amino acid biosynthesis) with the production of specialized defense compounds (Bolton, 2009).
Stringent control of amino acid metabolism during the growth-to-defense transition may reflect a mechanism to avoid pleiotropic negative effects on fitness while providing sufficient primary building blocks for defense. Although it is clear that Trp metabolism is required to support the production of indole glucosinolates and other defensive compounds in Arabidopsis, a direct link between altered Trp metabolism and JA-induced growth restriction remains to be established. It seems unlikely that increased levels of free Trp are responsible for the slow growth of the jazD and jazD phyB mutants, because elevated Trp levels in various Trp biosynthetic mutants (e.g. trp5) are not associated with reduced growth (Li and Last, 1996). An alternative interpretation is that the growth constraint in the jazD mutant reflects a compensatory strategy to avoid metabolic perturbations resulting from changes in primary metabolism required to supply defense pathways, analogous to amino acid biosynthetic mutants that exhibit severe growth restriction (Dong et al., 2017; de Oliveira et al., 2019). The relationship between altered Trp metabolism and growth is further complicated by potential perturbations in levels of auxin, whose predominant biosynthesis from Trp could be affected by the allocation of Trp to defense (Mashiguchi et al., 2011; Malka and Cheng, 2017) or through the formation of Trp conjugates that interfere with auxin transport (Staswick, 2009; Staswick et al., 2017). It is also possible that a metabolite derived from the Trp pathway acts via a conserved energy sensing pathway to modulate growth (Malinovsky et al., 2017).
At present, we cannot formally exclude the possibility that genetic polymorphisms introduced from non-Columbia-0 (Col-0) accessions during the construction of the jazD mutant contribute to the slow growth of this line; whereas the five jaz insertion mutations used to construct the jazQ mutant were all derived from Col-0 strains, two of the additional five jaz mutations used for construction of jazD were introgressed from other accessions (Guo et al., 2018b). Such genetic linkage effects have been implicated as a cost of resistance in breeding crop plants for R gene-mediated resistance (Karasov et al., 2017). In future studies it will be informative to determine whether the reduced growth of the jazD mutant can be recovered by loss of the COI1 receptor or the MYC transcriptional regulators, as was shown to be the case for the jazQ mutant (Major et al., 2017).
In summary, our results indicate that the mechanisms underlying JA-mediated growth-defense tradeoffs depend on the level of defense activation. As indicated by studies of the jazQ mutant (Campos et al., 2016; Major et al., 2017; Guo et al., 2018b), growth restriction at low to moderate levels of defense are not associated with major defects in reproductive output under laboratory conditions and can be suppressed by loss of phyB signaling. Moderate growth reduction at this intermediate level of defense may be an integral feature of induced resistance, which downwardly adjusts growth without necessarily penalizing fitness (Smith and Stitt, 2007; Guo et al., 2018a; Ballaré and Austin, 2019). At high levels of investment in defense, our data suggest that massive metabolic reprogramming geared toward the production of chemical defense compounds generates both growth and reproductive constraints that are independent of phyB. This form of growth-defense tradeoff may be manifested only under extreme conditions in which the high levels of defense necessitate major adjustments to primary metabolism. It is possible that strong allocation of primary metabolites to pathways for chemical defense, which in Arabidopsis are largely derived from amino acids, results in metabolic imbalances that cannot support normal growth. This hypothesis is consistent with the idea that plants have the capacity to sense changes in the availability of amino acids and other primary metabolites, and to respond by adjusting the growth rate to a level that matches the availability of resources (Smith and Stitt, 2007; Guo et al., 2018a; de Oliveira et al., 2019). A challenge for future studies will be to determine how specific changes in metabolism during the growth-to-defense transition are mechanistically linked to growth.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0), was the wild-type genetic background for all experiments. The jazD phyB mutant was constructed by combining the jazD (Guo et al., 2018b) and jazQ phyB mutants (Campos et al., 2016) as described in Supplemental Figure S5. PCR-based genotyping of the jazD phyB mutant used primer sets flanking T-DNA insertion sites, with a third primer specific for the T-DNA border (Supplemental Table S1; Campos et al., 2016; Guo et al., 2018b). The phyB-9 allele used in this study was recently shown to carry a second site phyB-nine-enhancer (bnen) mutation in the VENOSA4 gene that alters photosynthetic traits and leaf growth (Yoshida et al., 2018). We confirmed that the bnen mutation was lost during the construction of the jazQ phyB and jazD phyB mutants (Supplemental Fig. S5D). PCR reactions for genotyping were performed with GoTaq Green Master Mix (Promega) and the following programmed conditions: 5 min at 95°C, followed by 35 cycles of denaturation (30 s at 95°C), annealing (30 s at 56°C), and elongation (90 s at 72°C), with a final 10-min elongation step at 72°C. The bnen mutation was detected using a dCAPS marker. The PCR-amplified products were digested with DdeI restriction enzyme (New England Biolabs) as per the manufacturer’s instructions.
Seeds were stratified for 3 to 4 d at 4°C in the dark to improve the rate and synchrony of germination. After sowing seeds on soil, pots were covered with a transparent plastic dome for 10 d to increase humidity. Unless stated otherwise, soil-grown plants were maintained with 21°C days at a light intensity of 90–110 µmol m−2 s−1 from cool-white fluorescent lights and 20°C nights. For experiments with plate-grown seedlings, seeds were surface sterilized in a 60% (v/v) bleach solution for 10 min and washed at least six times before stratification. Seeds were sown on square culture plates (Thermo Fisher Scientific) containing one-half strength Linsmaier and Skoog (LS; Caisson Labs) salts with 0.7% (w/v) phytoblend (Caisson Labs) agar and 0.8% (w/v) Suc, unless stated otherwise. Plates were maintained at 21°C with 16 h at a light intensity of 80 µmol m−2 s−1 and 8 h dark.
jazD Suppressor Screen
Approximately 30,000 jazD seeds were mutagenized by immersion in a solution of 0.1% or 0.2% (v/v) EMS (Sigma-Aldrich) for 16 h at room temperature, with constant agitation (Campos et al., 2016). Seeds (M1 generation) were thoroughly washed with water, stratified in the dark at 4°C for 2 d and then immediately sown on soil for growth at 21°C under long-day conditions. M2 seed was collected from 24 pools of self-pollinated M1 plants (∼1,000 M1 plants/pool). Soil-grown M2 plants (∼30,000 total) were visually screened for individuals with either larger rosette size or longer hypocotyl length compared to the jazD mutant, together with persistence of anthocyanin accumulation (see Supplemental Fig. S1). Putative sjd (suppressor of jazD) mutants were rescreened in the M3 generation to confirm heritability of phenotypes.
Measurements of Shoot and Root Growth
Root growth inhibition assays were performed with seedlings grown on media oriented vertically and supplemented with the indicated concentrations of methyl-JA (Sigma-Aldrich; Shyu et al., 2012) or 5-MT dissolved in 0.5 m HCl (Sigma-Aldrich; Bender and Fink, 1998). Primary root length was determined from 9- to 11-d-old seedlings using ImageJ software (http://imagej.nih.gov/ij/). Wild-type and mutant lines were grown on the same plate to control for plate-to-plate variation. Unless stated otherwise, hypocotyl length was determined from seedlings grown on LS plates for 7 d under monochromatic light at 25 μE m−2 s−1 using ImageJ software (Warnasooriya and Montgomery, 2009; Campos et al., 2016). For rosette growth, plants were grown on soil for 4 weeks under long-day conditions (16-h day, 8-h night) or for 7 weeks under short-day conditions (8-h day, 16-h night). These plants were used for measurements of the number of rosette leaves, petiole length of the third true leaf, and fresh weight of the excised rosette. Overhead images photographed with a Nikon D80 camera were used to determine projected leaf area by GIMP (https://www.gimp.org/) and rosette diameter by ImageJ (Campos et al., 2016). For the coronatine treatment, two mature leaves from plants grown for 4 weeks on soil under long-day (16-h day, 8-h night) conditions were spotted with 5 μL of sterile water (mock) or 50 μm coronatine (C8115; Sigma-Aldrich) prepared in sterile water. Photographs were taken 4 d after treatment.
For experiments involving GA treatment, seeds were sown on one-half strength LS plates supplemented with 0.8% (w/v) Suc and various concentrations of GA3 (0, 1, and 10 μm) obtained from Caisson Labs. Plates were incubated horizontally for 7 d in growth chambers maintained at 21°C under 16 h at a light intensity of 10 μE m−2 s−1 and 8 h dark. ImageJ software was used to measure the hypocotyl length.
Plant Fecundity Measurements
Bolting time was assessed daily for plants grown on soil under long-day conditions. Seed yield was determined as described previously, with minor modifications (Guo et al., 2018b). Silique length and the number of seeds per silique were determined by sampling the ninth, 11th, and 13th siliques from the main inflorescence. Seeds collected from individual plants were dried with Drierite desiccant, and average seed mass was determined by weighing 500 seeds per plant.
Gene Expression Measurements
Rosette leaves from two 4-week-old plants grown on soil under long-day conditions were pooled and immediately frozen in liquid nitrogen and stored at −80°C until processing. Tissue was homogenized with a TissueLyser II (Qiagen) and RNA was extracted using the nucleospin plant RNA extraction kit (Macherey-Nagel) according to the manufacturer’s instructions. RNA quality was assessed by A260/A280 ratios using an ND-1000 UV Nanodrop spectrophotometer (Thermo Scientific). Complementary DNA (cDNA) was reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s protocols, and cDNA was diluted to 0.5 ng/μL with nuclease-free water. qPCR reactions consisted of 2 μL diluted cDNA template (1 ng total), 5 μL 2× Power SYBR Green Master Mix (Applied Biosystems), 1 μL of a solution containing forward and reverse primers at a concentration of 5 μm (Supplemental Table S1), and 2 μL nuclease-free water to achieve a final reaction volume of 10 μL. Reactions were run on an Applied Biosystems 7500 Fast qPCR instrument with the following conditions: 50°C for 2 min, 95°C for 10 min, then 40 cycles of denaturation (15 s at 95°C) and annealing and polymerization (60 s at 60°C). A dissociation curve was performed at the end of each reaction using default parameters (15 s at 95°C, 60 s at 60° to 95°C in 1°C increments, and 15 s at 95°C), which confirmed a single peak for each set of primers. Target gene expression was normalized to the expression of PROTEIN PHOSPHATASE 2A (PP2A), which is stable under JA-inducing conditions (Attaran et al., 2014). Primer efficiencies for each primer pair were determined with LinRegPCR v2012.0 (Ramakers et al., 2003) and were used to calculate relative expression as described previously (Vandesompele et al., 2002).
Western Blot Analysis
Total protein was extracted from shoot tissue of 16-d-old plate-grown seedlings using an extraction buffer containing 50 mm Tris (pH 8.0), 150 mm NaCl, 1% (v/v) Triton X-100, 50 μm MG-132, 1 μm phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail. The resulting protein was quantified by BCA assay (ThermoFisher). Proteins were separated on a 10% SDS-polyacrylamide gel and electro-transferred to a polyvinylidene difluoride membrane. Western blot was performed with rabbit anti-RGA primary antibody (Agrisera) and donkey antirabbit secondary antibody (IgG HRP; ThermoFisher). The blot was incubated for 5 min with SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher) and proteins were detected with a ChemiDoc MP Imaging System. Protein extracted from the transfer DNA (T-DNA) insertion mutant rga-28 (SALK_089146) was used as a control for specificity of the anti-RGA antibody.
Metabolite Measurements
Metabolites were measured from plants grown for 4 weeks on soil under long-day conditions. Rosettes were harvested, weighed, and homogenized with a TissueLyser II (Qiagen) after freezing in liquid nitrogen and were stored at −80°C until analysis. For anthocyanin measurements, tissue was extracted overnight at 4°C in MeOH containing 1% (v/v) HCl, and then clarified by centrifugation. Bulk anthocyanin content was determined spectrophotometrically by A530 – 0.25 (A657) and normalized to extracted tissue weight (Campos et al., 2016). For glucosinolate measurements, tissue was extracted in 80% (v/v) MeOH as described previously (Glauser et al., 2012). Samples were diluted 1:10 in water and analyzed in the Michigan State University Mass Spectrometry and Metabolomics Facility with a Xevo G2-XS UPLC QTOF (Waters), as described previously (Koo et al., 2009; Glauser et al., 2012). Sinigrin was used as an internal standard and glucosinolate levels were reported relative to the wild type after normalization to extracted tissue weight.
To determine levels of Trp and Ser, leaf tissue (∼10 mg) was incubated at 90°C in water containing labeled standards (13C,15N-labeled amino acids; Sigma Aldrich) for 5 min. After cooling on ice, the extract was clarified by centrifugation and filtered through 0.2-μm, low-binding hydrophilic polytetrafluoroethylene centrifugal filters (Millipore). Filtrates were diluted 2-fold in 20 mm perfluoroheptanoic acid (Sigma Aldrich) and analyzed in the Michigan State University Mass Spectrometry and Metabolomics Facility with a Quattro Micro API LC-MS/MS (Waters) equipped with an Acquity UPLC HSS T3 1.8 µm column (2.1 × 100 mm, 1.8-µm particle size; Waters) with multiple reaction monitoring, as described previously with modifications (Gu et al., 2007). The LC method was modified to a 13-min run with 10 mm perfluoroheptanoic acid solvent to better retain and separate amino acids. The MS/MS method was modified to include transitions for stable-labeled amino acid internal standards and divided into three resolved functions (0–4.5 min, 4.5–6.3 min, and 6.3–13 min) for data acquisition to allow sufficient dwell time for each analyte (Supplemental Table S2). Endogenous amino acid concentrations were determined according to external standard curves and normalized to extracted tissue weight.
For starch and Suc measurements, leaf tissue was frozen in liquid nitrogen, lyophilized, weighed, and homogenized with a TissueLyser II. Tissue was extracted in 3.5% (v/v) perchloric acid on ice for 5 min and clarified by centrifugation at 4°C. The resulting supernatant was neutralized to pH ∼7 with neutralizing buffer (2 m KOH, 150 mm HEPES, and 10 mm KCl), frozen to precipitate salts, and clarified by centrifugation. The resulting supernatant was used for Suc determination. The perchloric acid pellet was washed twice with water, twice with 80% (v/v) ethanol, resuspended in 0.2 m KOH and incubated at 95°C for 30 min. After cooling, 1 m acetic acid was added to adjust the pH to ∼5 and starch was digested to Glc by incubating with 50 U α-amylase (Megazyme) and 1 U amyloglucosidase (Megazyme) at room temperature for 2 d. Samples were clarified by centrifugation and the supernatant was used for determination of starch content. Glc content was determined spectrophotometrically with a NADP(H)-linked assay (Lowry and Passonneau, 1972). Samples were incubated until reaction completion in assay buffer (150 mm HEPES [pH 7.2], 15 mm MgCl2, and 3 mm EDTA) containing 500 nmol NADP, 500 nmol ATP, and 1 U Glc-6-phosphate dehydrogenase (Sigma). A baseline was determined at A340. Samples were then incubated until reaction completion with 1 U hexokinase (Sigma), and the absolute Glc content was determined from the increase in A340 from an extinction coefficient of 6,220 L mol−1 cm−1 for NADPH at 340 nm. To measure Suc content, samples were further incubated until reaction completion with 50 U invertase (Sigma) and the Glc content was determined from the increase in A340.
Insect and Pathogen Assays
Plants for insect feeding and pathogen infection assays were grown for 6 weeks on soil under short-day (8-h day, 16-h night) conditions. For insect feeding assays, three neonate Trichoplusia ni larvae (Benzon Research) were reared on each of at least nine plants per genotype for 10 d, after which photographs were taken of plants and larvae, and larval weights were measured (Herde et al., 2013). For pathogen infection assays, detached leaves were placed on filter paper moistened with sterile water in culture plates. Each leaf was inoculated with a 4-μL drop of Botrytis cinerea spore suspension (5,000 spores/mL in 50% [v/v] organic grape [Vitis vinifera] juice; Rowe and Kliebenstein, 2007). Leaves were photographed after 5 d and the area of spreading lesions was determined using GIMP software.
Photosynthesis and Respiration Measurements
Chlorophyll fluorescence of sjd suppressor mutants was measured in a Dynamic Environment Photosynthesis Imager (DEPI) chamber, as described previously (Cruz et al., 2016). Plants were grown for 18 d on soil under long-day conditions. Plants were exposed to three sequential days of light regimes of constant 100 µmol m−2 s−1 intensity (day 1), sinusoidal increase and decrease with maximal 500 µmol m−2 s−1 intensity at midday (day 2), and fluctuating light intensities superimposed on the day 2 sinusoidal pattern with fluctuations peaking at 1,000 µmol m−2 s−1 intensity at midday (day 3). Chlorophyll fluorescence images were processed using Visual Phenomics software (Tessmer et al., 2013). The ΦII was calculated as (FM′ − FS)/FM′, where FS is the steady‐state fluorescence and FM′ is the fluorescence maximum at steady‐state. Heat maps were generated with OLIVER (https://caapp-msu.bitbucket.io/projects/oliver/index.html).
For gas exchange measurements, plants were grown in plastic containers (“Cone-tainers”, Steuwe and Sons) with an 8-h day (120 μmol m−2 s−1, 21°C) and 16 h dark (18°C) photoperiod. Single mature rosette leaves (attached) of 8- to 10-week-old plants were assessed using a LI-6800 system (LI-COR Biosciences) outfitted with a standard leaf chamber (6-cm2 chamber area), with area corrected to the actual measured leaf area. Leaves were supplied with 400 μmol mol−1 CO2, 0.85 kPa leaf vapor pressure deficit, and light intensity of 500 μmol m−2 s−1, with leaf temperature maintained at 21°C. Leaves were acclimated under this condition for at least 20 min before the start of each experiment. Daytime respiration was determined from slope-intercept regression analysis of the intersection of five CO2 response curves (using intercellular CO2 below 10 Pa) measured at decreasing, subsaturating irradiances (Walker et al., 2016). Nighttime respiration was determined from dark-adapted plants (Weraduwage et al., 2015). Curve fitting of response curves was performed to model photosynthetic parameters (Sharkey et al., 2007).
Statistics
All experiments, except characterization of the sjd suppressors, were repeated independently at least three times and results from one representative experiment are shown. Experiments with sjd mutants were performed once to determine growth metrics (Supplemental Fig. S2), once to assess allelic complementation with the jazD phyB mutant (Supplemental Fig. S4), and twice (independent replicates) to determine hypocotyl lengths under monochromatic light (Fig. 2A; Supplemental Fig. S3) and ΦII (Fig. 2B). Unless stated otherwise, genotypes were compared using ANOVA with Tukey’s honestly significant difference (HSD) mean separation test. For principal component analysis of glucosinolate content, glucosinolate levels in mutants were normalized to that of the wild type. All analyses were performed with R software (https://www.r-project.org).
Accession Numbers
Genes described herein have the following Arabidopsis Genome Initiative (AGI) gene accession numbers: AT1G19180 (JAZ1), AT1G74950 (JAZ2), AT3G17860 (JAZ3), AT1G48500 (JAZ4), AT1G17380 (JAZ5), AT1G72450 (JAZ6), AT2G34600 (JAZ7), AT1G70700 (JAZ9), AT5G13220 (JAZ10), AT3G22275 (JAZ13), AT1G32640 (MYC2), AT5G46760 (MYC3), AT2G18790 (PHYB), AT5G42650 (AOS), AT2G06050 (OPR3), AT5G24770 (VSP2), AT5G44420 (PDF1.2a), AT1G72260 (Thi2.1), AT2G22330 (CYP79B3), AT4G31500 (CYP83B1), AT5G57220 (CYP81F2), AT1G21100 (IGMT1), AT5G05730 (ASA1), AT5G17990 (PAT1), AT2G04400 (IGPS), AT3G54640 (TSA1), AT4G27070 (TSB2), AT1G17745 (PGDH2), AT4G35630 (PSAT1), AT1G10070 (BCAT2), AT5G56870 (BGAL4), AT4G35770 (DIN1), and AT3G47340 (DIN6).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Primers used for genotyping and RT-qPCR.
Supplemental Table S2. Details of LC-MS/MS gradient and functions.
Supplemental Figure S1. Genetic screen for suppressor of jazD (sjd) mutants.
Supplemental Figure S2. Suppressor of jazD (sjd) mutants with long hypocotyls have improved growth.
Supplemental Figure S3. Hypocotyl lengths of long-hypocotyl sjd mutants under monochromatic far red and blue light.
Supplemental Figure S4. Allelic complementation of long-hypocotyl sjd mutants with jazD phyB.
Supplemental Figure S5. Genetic reconstruction of jazD phyB.
Supplemental Figure S6. jazD and jazD phyB plants have similar glucosinolate profiles.
Supplemental Figure S7. phyB partially recovers growth of jazD in a photoperiod-dependent manner.
Supplemental Figure S8. phyB mutation only weakly recovers reproductive phenotypes of jazD.
Supplemental Figure S9. jazD and phyB interact to modulate photosynthesis and respiration.
Supplemental Figure S10. Elevated Trp metabolism persists in jazD phyB.
Acknowledgments
We thank Tom Sharkey, Sean Wiese, and Alan McClain for assistance with photosynthesis and respiration measurements, Linda Savage and David Hall for help with photosynthetic phenotyping experiments and image processing, Tony Schilmiller and the Michigan State University Mass Spectrometry Facility for help with LC-MS/MS analysis of metabolites, and Sookyung Oh and Beronda Montgomery for assistance with seedling growth under monochromatic light.
Footnotes
This work was supported by the Chemical Sciences, Geosciences, and Biosciences Division, Basic Energy Sciences, Office of Science at the U.S. Department of Energy (grant no. DE–FG02–91ER20021), with additional support from the Visiting International Professional Program–Office of China (fellowship to J.Z.), the Michigan State University Plant Resilience Institute (to G.K.), and the Michigan AgBioResearch Project (grant no. MICL02278).
Articles can be viewed without a subscription.
References
- Attaran E, Major IT, Cruz JA, Rosa BA, Koo AJK, Chen J, Kramer DM, He SY, Howe GA(2014) Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol 165: 1302–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J(2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baldwin IT.(1998) Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA 95: 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL.(2014) Light regulation of plant defense. Annu Rev Plant Biol 65: 335–363 [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Austin AT(2019) Recalculating growth and defense strategies under competition: Key roles of photoreceptors and jasmonates. J Exp Bot 70: 3425–3434 [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR(1998) A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci USA 95: 5655–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Purrington CB(1996) Surveying patterns in the cost of resistance in plants. Am Nat 148: 536–558 [Google Scholar]
- Boccalandro HE, Rugnone ML, Moreno JE, Ploschuk EL, Serna L, Yanovsky MJ, Casal JJ(2009) Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis. Plant Physiol 150: 1083–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton MD.(2009) Primary metabolism and plant defense—fuel for the fire. Mol Plant Microbe Interact 22: 487–497 [DOI] [PubMed] [Google Scholar]
- Bömer M, O’Brien JA, Pérez-Salamó I, Krasauskas J, Finch P, Briones A, Daudi A, Souda P, Tsui T-L, Whitelegge JP, et al. (2018) COI1-dependent jasmonate signalling affects growth, metabolite production and cell wall protein composition in Arabidopsis. Ann Bot 122: 1117–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger M, Chory J(2019) Stressed out about hormones: How plants orchestrate immunity. Cell Host Microbe 26: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo I, Keller MM, Cargnel MD, Demkura PV, de Wit M, Patitucci MS, Pierik R, Pieterse CMJ, Ballaré CL(2012) Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol 158: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Gangappa SN, Maurya JP, Sethi V, Srivastava AK, Singh A, Dutta S, Ojha M, Gupta N, Sengupta M, et al. (2019) Functional interrelation of MYC2 and HY5 plays an important role in Arabidopsis seedling development. Plant J 99: 1080–1097 [DOI] [PubMed] [Google Scholar]
- Chico J-M, Fernández-Barbero G, Chini A, Fernández-Calvo P, Díez-Díaz M, Solano R(2014) Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26: 1967–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chini A, Gimenez-Ibanez S, Goossens A, Solano R(2016) Redundancy and specificity in jasmonate signalling. Curr Opin Plant Biol 33: 147–156 [DOI] [PubMed] [Google Scholar]
- Cruz JA, Savage LJ, Zegarac R, Hall CC, Satoh-Cruz M, Davis GA, Kovac WK, Chen J, Kramer DM(2016) Dynamic environmental photosynthetic imaging reveals emergent phenotypes. Cell Syst 2: 365–377 [DOI] [PubMed] [Google Scholar]
- de Wit M, Spoel SH, Sanchez-Perez GF, Gommers CMM, Pieterse CMJ, Voesenek LACJ, Pierik R(2013) Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J 75: 90–103 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Allboje Samami A, Wanatabe M, Sticht C, Teleman AA, et al. (2017) Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun 8: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Erb M, Reymond P(2019) Molecular interactions between plants and insect herbivores. Annu Rev Plant Biol 70: 527–557 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico J-M, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG(1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R(2009) The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr Opin Plant Biol 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH(2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios-Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Höhne M, Günther M, Stitt M(2004) Adjustment of diurnal starch turnover to short days: Depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Glauser G, Schweizer F, Turlings TCJ, Reymond P(2012) Rapid profiling of intact glucosinolates in Arabidopsis leaves by UHPLC-QTOFMS using a charged surface hybrid column. Phytochem Anal 23: 520–528 [DOI] [PubMed] [Google Scholar]
- Goossens J, Swinnen G, Vanden Bossche R, Pauwels L, Goossens A(2015) Change of a conserved amino acid in the MYC2 and MYC3 transcription factors leads to release of JAZ repression and increased activity. New Phytol 206: 1229–1237 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL(2007) LC-MS/MS assay for protein amino acids and metabolically related compounds for large-scale screening of metabolic phenotypes. Anal Chem 79: 8067–8075 [DOI] [PubMed] [Google Scholar]
- Guo Q, Major IT, Howe GA(2018a) Resolution of growth-defense conflict: Mechanistic insights from jasmonate signaling. Curr Opin Plant Biol 44: 72–81 [DOI] [PubMed] [Google Scholar]
- Guo Q, Yoshida Y, Major IT, Wang K, Sugimoto K, Kapali G, Havko NE, Benning C, Howe GA(2018b) JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc Natl Acad Sci USA 115: E10768–E10777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havko NE, Major IT, Jewell JB, Attaran E, Browse J, Howe GA(2016) Control of carbon assimilation and partitioning by jasmonate: An accounting of growth–defense tradeoffs. Plants (Basel) 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Baldwin IT(2002) Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci 7: 61–67 [DOI] [PubMed] [Google Scholar]
- Herde M, Koo AJK, Howe GA(2013) Elicitation of jasmonate-mediated defense responses by mechanical wounding and insect herbivory. Methods Mol Biol 1011: 51–61 [DOI] [PubMed] [Google Scholar]
- Herms DA, Mattson WJ(1992) The dilemma of plants: To grow or defend. Q Rev Biol 67: 283–335 [Google Scholar]
- Hou X, Lee LYC, Xia K, Yan Y, Yu H(2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G(2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Howe GA, Major IT, Koo AJ(2018) Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol 69: 387–415 [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY(2014) Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol Plant 7: 1267–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov TL, Chae E, Herman JJ, Bergelson J(2017) Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 29: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA(2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM(2011) The interplay between light and jasmonate signalling during defence and development. J Exp Bot 62: 4087–4100 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ.(2016) False idolatry of the mythical growth versus immunity tradeoff in molecular systems plant pathology. Physiol Mol Plant Pathol 95: 55–59 [Google Scholar]
- Koo AJ, Gao X, Jones AD, Howe GA(2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J 59: 974–986 [DOI] [PubMed] [Google Scholar]
- Krahmer J, Ganpudi A, Abbas A, Romanowski A, Halliday KJ(2018) Phytochrome, carbon sensing, metabolism, and plant growth plasticity. Plant Physiol 176: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Passonneau JV(1972) A Flexible System for Enzymatic Analysis. Academic Press, Orlando, FL [Google Scholar]
- Li J, Last RL(1996) The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble tryptophan. Plant Physiol 110: 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wei H, Ma M, Li Q, Kong D, Sun J, Ma X, Wang B, Chen C, Xie Y, et al. (2019) Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell 31: 2089–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado RAR, Baldwin IT, Erb M(2017) Herbivory-induced jasmonates constrain plant sugar accumulation and growth by antagonizing gibberellin signaling and not by promoting secondary metabolite production. New Phytol 215: 803–812 [DOI] [PubMed] [Google Scholar]
- Major IT, Yoshida Y, Campos ML, Kapali G, Xin X-F, Sugimoto K, de Oliveira Ferreira D, He SY, Howe GA(2017) Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol 215: 1533–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malka SK, Cheng Y(2017) Possible interactions between the biosynthetic pathways of indole glucosinolate and auxin. Front Plant Sci 8: 2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky FG, Thomsen MF, Nintemann SJ, Jagd LM, Bourgine B, Burow M, Kliebenstein DJ(2017) An evolutionarily young defense metabolite influences the root growth of plants via the ancient TOR signaling pathway. eLife 6: 29353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, McSteen P, Zhao Y, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke S, Gasperini D(2019) Interplay between plant cell walls and jasmonate production. Plant Cell Physiol 60: 2629–2637 [DOI] [PubMed] [Google Scholar]
- Monte I, Franco-Zorrilla JM, García-Casado G, Zamarreño AM, García-Mina JM, Nishihama R, Kohchi T, Solano R(2019) A single JAZ repressor controls the jasmonate pathway in Marchantia polymorpha. Mol Plant 12: 185–198 [DOI] [PubMed] [Google Scholar]
- Moreno JE, Tao Y, Chory J, Ballaré CL(2009) Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG(2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Ning Y, Liu W, Wang G-L(2017) Balancing immunity and yield in crop plants. Trends Plant Sci 22: 1069–1079 [DOI] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J(2011) Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noir S, Bömer M, Takahashi N, Ishida T, Tsui T-L, Balbi V, Shanahan H, Sugimoto K, Devoto A(2013) Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol 161: 1930–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira MVV, Jin X, Chen X, Griffith D, Batchu S, Maeda HA(2019) Imbalance of tyrosine by modulating TyrA arogenate dehydrogenases impacts growth and development of Arabidopsis thaliana. Plant J 97: 901–922 [DOI] [PubMed] [Google Scholar]
- Ortigosa A, Fonseca S, Franco-Zorrilla JM, Fernández-Calvo P, Zander M, Lewsey MG, García-Casado G, Fernández-Barbero G, Ecker JR, Solano R(2019) The JA-pathway MYC transcription factors regulate photomorphogenic responses by targeting HY5 gene expression. Plant J 102: 138–152 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A(2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñuelas M, Monte I, Schweizer F, Vallat A, Reymond P, García-Casado G, Franco-Zorrilla JM, Solano R(2019) Jasmonate-related MYC transcription factors are functionally conserved in Marchantia polymorpha. Plant Cell 31: 2491–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SC(2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM(2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rowe HC, Kliebenstein DJ(2007) Elevated genetic variation within virulence-associated Botrytis cinerea polygalacturonase loci. Mol Plant Microbe Interact 20: 1126–1137 [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M(2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Bernacchi CJ, Farquhar GD, Singsaas EL(2007) Fitting photosynthetic carbon dioxide response curves for C3 leaves. Plant Cell Environ 30: 1035–1040 [DOI] [PubMed] [Google Scholar]
- Shyu C, Figueroa P, Depew CL, Cooke TF, Sheard LB, Moreno JE, Katsir L, Zheng N, Browse J, Howe GA(2012) JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24: 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms EL, Rausher MD(1987) Costs and benefits of plant resistance to herbivory. Am Nat 130: 570–581 [Google Scholar]
- Smith AM, Stitt M(2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Smolen GA, Pawlowski L, Wilensky SE, Bender J(2002) Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161: 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamp N.(2003) Out of the quagmire of plant defense hypotheses. Q Rev Biol 78: 23–55 [DOI] [PubMed] [Google Scholar]
- Staswick PE.(2009) The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol 150: 1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P, Rowe M, Spalding EP, Splitt BL(2017) Jasmonoyl-L-tryptophan disrupts IAA activity through the AUX1 auxin permease. Front Plant Sci 8: 736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessmer OL, Jiao Y, Cruz JA, Kramer DM, Chen J(2013) Functional approach to high-throughput plant growth analysis. BMC Syst Biol 7(Suppl 6): S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J(2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thireault C, Shyu C, Yoshida Y, St Aubin B, Campos ML, Howe GA(2015) Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J 82: 669–679 [DOI] [PubMed] [Google Scholar]
- Ullmann-Zeunert L, Stanton MA, Wielsch N, Bartram S, Hummert C, Svatoš A, Baldwin IT, Groten K(2013) Quantification of growth-defense trade-offs in a common currency: Nitrogen required for phenolamide biosynthesis is not derived from ribulose-1,5-bisphosphate carboxylase/oxygenase turnover. Plant J 75: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F(2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Skabelund DC, Busch FA, Ort DR(2016) An improved approach for measuring the impact of multiple CO2 conductances on the apparent photorespiratory CO2 compensation point through slope-intercept regression. Plant Cell Environ 39: 1198–1203 [DOI] [PubMed] [Google Scholar]
- Wang J, Wu D, Wang Y, Xie D(2019) Jasmonate action in plant defense against insects. J Exp Bot 70: 3391–3400 [DOI] [PubMed] [Google Scholar]
- Warnasooriya SN, Montgomery BL(2009) Detection of spatial-specific phytochrome responses using targeted expression of biliverdin reductase in Arabidopsis. Plant Physiol 149: 424–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B(2013) Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weraduwage SM, Campos ML, Yoshida Y, Major IT, Kim Y-S, Kim S-J, Renna L, Anozie FC, Brandizzi F, Thomashow MF, et al. (2018) Molecular mechanisms affecting cell wall properties and leaf architecture In Adams WW III, and Terashima I, eds, The Leaf: A Platform for Performing Photosynthesis. Springer International Publishing, Cham, Switzerland, pp 209–253 [Google Scholar]
- Weraduwage SM, Chen J, Anozie FC, Morales A, Weise SE, Sharkey TD(2015) The relationship between leaf area growth and biomass accumulation in Arabidopsis thaliana. Front Plant Sci 6: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE(2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Seaton DD, Krahmer J, Halliday KJ(2016) Photoreceptor effects on plant biomass, resource allocation, and metabolic state. Proc Natl Acad Sci USA 113: 7667–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D-L, Yao J, Mei C-S, Tong X-H, Zeng L-J, Li Q, Xiao L-T, Sun TP, Li J, Deng X-W, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Sarmiento-Mañús R, Yamori W, Ponce MR, Micol JL, Tsukaya H(2018) The Arabidopsis phyB-9 mutant has a second-site mutation in the VENOSA4 gene that alters chloroplast size, photosynthetic traits, and leaf growth. Plant Physiol 178: 3–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander M, Lewsey MG, Clark NM, Yin L, Bartlett A, Saldierna Guzmán JP, Hann E, Langford AE, Jow B, Wise A, et al. (2020) Integrated multi-omics framework of the plant response to jasmonic acid. Nat Plants 6: 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ji Y, Xue C, Ma H, Xi Y, Huang P, Wang H, An F, Li B, Wang Y, et al. (2018) Integrated Regulation of apical hook development by transcriptional coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell 30: 1971–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Turner JG(2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst T, Agrawal AA(2017) Trade-offs between plant growth and defense against insect herbivory: An emerging mechanistic synthesis. Annu Rev Plant Biol 68: 513–534 [DOI] [PubMed] [Google Scholar]