Abstract
Echinochloa with resistance to glyphosate also contains an unreported Pro-106-Thr EPSPS target-site mutation.
Dear Editor,
Weeds can evolve resistance to glyphosate through modification of the herbicide target, 5-enolpyruvyl shikimate 3-phosphate synthase (EPSPS), primarily at the Pro-106 position or through increased production of EPSPS, both of which are referred to as target site resistance (TSR; Gaines et al., 2019). Weeds can also evolve resistance via differential interaction with glyphosate (reduced absorption or translocation, or increased metabolism of glyphosate), which is referred to as nontarget site resistance (NTSR). Recently Pan et al. (2019) reported field-evolved weed resistance to the herbicide glyphosate, putatively via an NTSR mechanism, of metabolism by aldo-keto reductase (AKR). AKR-mediated glyphosate metabolism has been previously reported, thus making AKR-based metabolism a possible route for genetic modification in glyphosate-resistant crop development (Agrawal et al., 2015; Vermanna et al., 2017). Pan et al. (2019), however, were the first to identify AKR as a route to evolution of herbicide resistance in field-treated weed species.
Preliminary work on the glyphosate-resistant Echinochloa colona population first identified by Gaines et al. (2012) on which the work of Pan et al. (2019) is based did not attribute glyphosate resistance to modification of the target site EPSPS, reduced absorption or translocation of glyphosate, or glyphosate metabolism (Goh et al., 2018). Resistant populations did not accumulate shikimate in the leaf tissue following glyphosate treatment to the level of susceptible plants, suggesting that either glyphosate did not reach the target site EPSPS or that the EPSPS target site had reduced binding affinity for glyphosate. The lack of reduced absorption or translocation, or increased metabolism in E. colona individuals, further supported a target site involvement in resistance mechanism. Thus, we hypothesized that a target site mutation had been missed by the original Goh et al. (2018) authors and was not examined using the transcriptome RNA sequencing data reported by Pan et al. (2019).
An EPSPS gene from Digitaria insularis was downloaded from NCBI (GenBank: KX108896) and used as a reference, as no Echinochloa EPSPS sequences were present in GenBank. Reads from the six different experiment accessions reported by Pan et al. (2019) within National Genomics Data Center, Beijing Institute of Genomics (http://bigd.big.ac.cn) PRJCA001826 (Table 1; CRX0654665 to CRX065470) were downloaded and mapped to the reference individually using CLC Genomics Workbench 20.0 (Qiagen). Variants were detected from the read mapping using the Basic Variant Detection tool with variants ignored with minimum nucleotide frequency of less than 35%. Nucleotide variants for EPSPS codons 101 to 110 were compiled based on read mapping and variant detection (Tables 2 and 3).
Table 1. Reads obtained from National Genomics Data Center, Beijing Institute of Genomics (https://bigd.big.ac.cn/gsa/browse/CRA002028) under accession number PRJCA001626 as reported by Pan et al. (2019).
| BioSample Accession | Experiment Accession | Run Accession | Sample Name | Archived File Name | Presumed Glyphosate Response |
|---|---|---|---|---|---|
| SAMC110955 | CRX065465 | CRR076662 | baicao-R21 | CRR076662_f1 CRR076662_r2 | Resistant |
| SAMC110956 | CRX065466 | CRR076663 | baicao-R22 | CRR076663_f1 CRR076663_r2 | Resistant |
| SAMC110957 | CRX065467 | CRR076664 | baicao-R23 | CRR076664_f1 CRR076664_r2 | Resistant |
| SAMC110958 | CRX065468 | CRR076665 | baicao-S17 | CRR076665_f1 CRR076665_r2 | Susceptible |
| SAMC110959 | CRX065469 | CRR076666 | baicao-S18 | CRR076666_f1 CRR076666_r2 | Susceptible |
| SAMC110960 | CRX065470 | CRR076667 | baicao-S19 | CRR076667_f1 CRR076667_r2 | Susceptible |
Table 2. Single nucleotide polymorphisms (SNPs) detected from read mapping of six experiment accessions of National Genome Data Center (https://bigd.big.ac.cn) Genome Sequence Archive BioProject PRJCA001826 to a Digitaria insularis EPSPS partial cds (NCBI accession KX108896) reference.
| Codon/Amino Acid Number | Reference | CRX065465 (Resistant) | CRX065466 (Resistant) | CRX065467(Resistant) | CRX065468 (Susceptible) | CRX065469 (Susceptible) | CRX065470 (Susceptible) |
|---|---|---|---|---|---|---|---|
| Codons Present | |||||||
| Translated Amino Acids | |||||||
| 101 | GGA | GGA | GGA | GGA | GGA | GGA | GGA |
| G | G | G | G | G | G | G | |
| 102 | ACG | ACT | ACT | ACT | ACT | ACT | ACT |
| T | T | T | T | T | T | T | |
| 103 | GCA | GCA | GCA | GCA | GCA | GCA | GCA |
| A | A | A | A | A | A | A | |
| 104 | ATG | ATG | ATG | ATG | ATG | ATG | ATG |
| M | M | M | M | M | M | M | |
| 105 | CGT | CGG | CGG | CGG | CGG | CGG | CGG |
| R | R | R | R | R | R | R | |
| 106 | CCA | CCA/ACA | CCA/ACA | CCA/ACA | CCA | CCA | CCA |
| P | P/T | P/T | P/T | P | P | P | |
| 107 | TTG | TTG/CCG | TTG/CCG | TTG/CCG | TTG/CCG | TTG/CCG | TTG/CCG |
| L | L | L | L | L | L | L | |
| 108 | ACA | ACA | ACA | ACA | ACA | ACA | ACA |
| T | T | T | T | T | T | T | |
| 109 | GCG | GCA | GCA | GCA | GCA | GCA | GCA |
| A | A | A | A | A | A | A | |
| 110 | GCC | GCC/GCT | GCC/GCT | GCC/GCT | GCC/GCT | GCC/GCT | GCC/GCT |
| A | A | A | A | A | A | A | |
Table 3. Reads mapped and total reads count for paired-read data sets obtained from National Genome Data Center (https://bigd.big.ac.cn) Genome Sequence Archive BioProject PRJCA001826 as reported by Pan et al. (2019).
| Experiment Accession | Presumed Glyphosate Response | Reads Mapped to Reference EPSPS Partial CDS | Total Read Count |
|---|---|---|---|
| CRX065465 | Resistant | 1,107 | 60,961,185 |
| CRX065466 | Resistant | 898 | 57,026,220 |
| CRX065467 | Resistant | 934 | 70,192,858 |
| CRX065468 | Susceptible | 276 | 62,463,728 |
| CRX065469 | Susceptible | 310 | 65,997,636 |
| CRX065470 | Susceptible | 329 | 73,336,968 |
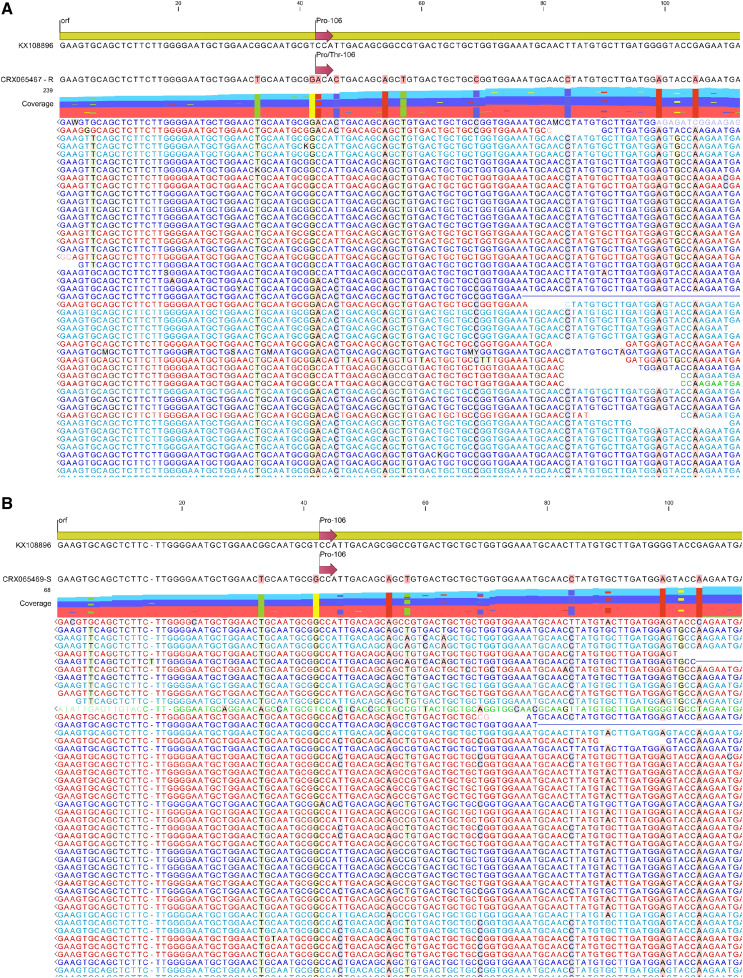
E. colona transcriptome reads (Table 1) mapped to an EPSPS reference revealed a Pro-106-Thr substitution in resistant read sets (Fig. 1; Table 2). Approximately 72% of reads that mapped in the three-glyphosate resistant read sets contained an ACA codon encoding a Thr at position 106 (data not shown). E. colona is a polyploid, which explains the ACA-Thr mutant allele on one subgenome, and the presence of the CCA-Pro allele in another subgenome. TSR-based glyphosate resistance in E. colona has been reported by Alarcon-Reverte et al. (2013). Alarcon-Reverte et al. (2013) reported 6.6 greater resistance in a Pro-106-Ser-containing E. colona biotype compared to susceptible biotype, which is comparable to the 5.6 to 8.6 more resistance compared to susceptible in the population first reported resistant by Gaines et al. (2012) and evaluated by Pan et al. (2019). Alarcon-Reverte et al. (2013) did not examine metabolism as a possible mechanism of herbicide resistance; therefore, it is possible that metabolic processes also contribute to resistance in these populations. We acknowledge that Pan et al. (2019) did observe increased metabolism in glyphosate-resistant E. colona compared to glyphosate-susceptible, which is contradictory to the initial findings of Goh et al. (2018) on this population. The ultra performace liquid chromatography tandem mass spectometry methodology used by Pan et al. (2019) likely yielded more reliable results than the C14-based thin-layer chromatography methodology used by Goh et al. (2018). Based on data provided by Pan et al. (2019), glyphosate-resistant E. colona is metabolizing glyphosate and has higher AKR transcript abundance compared to glyphosate-susceptible types. However, because the shikimate pathway remains more functional in glyphosate-resistant E. colona due to the Pro-106-Thr containing EPSPS, greater metabolism could be occurring due to a stable physiological environment that is more conducive to xenobiotic metabolism when key processes are not being fully inhibited.
Figure 1.
Graphical representation of Illumina sequencing reads for glyphosate-resistant E. colona (CRX065467; A) and glyphosate-susceptible E. colona (CRX065469; B) aligned to a partial coding sequence of Digitaria insularis as the reference EPSPS.
Secondary authors on Pan et al. (2019) have reported that EPSPS substitutions at Pro-106 are insufficient to confer resistance in E. colona specifically under some growth conditions (Han et al., 2016). Literature exists that supports the notion that EPSPS Pro-106 changes combined with secondary metabolic, translocative, or increased EPSPS expression increase the overall level of glyphosate resistance above that of target-site mutations alone (e.g. Bostamam et al., 2012; Bracamonte et al., 2016; Gherekhloo et al., 2017). It is our stance that Han et al. (2016) demonstrates that EPSPS Pro-106 amino acid changes do confer resistance and provide supporting evidence that combined target and NTSR mechanisms likely work in concert as others have reported.
We thus conclude that the glyphosate resistance in E. colona identified first by Gaines et al. (2012), evaluated by Goh et al. (2018), and on which AKR-based nontarget site glyphosate resistance evolution was asserted by Pan et al. (2019) can at least in part be explained by an undiscovered common EPSPS substitution in the resistant population. Our conclusion in no way should discount the work of previous research and the growing body of knowledge regarding AKR-based glyphosate metabolism or that AKR is playing a role in glyphosate resistance in this specific E. colona population; however, it should not be concluded that this E. colona population evolved resistance to glyphosate solely via AKR-based glyphosate metabolism.
It is our opinion that TSR can be easily overlooked, particularly in polyploid species. The current sequencing methodology to detect target site mutations is primarily PCR-based capillary sequencing of herbicide target genes. A reliance on PCR for identification of TSR may fail to adequately amplify all expressed copies of a given target site. EPSPS gene duplication has been demonstrated as a mechanism of resistance in Amaranthus spp., which further exacerbates the problem of determining if TSR is the result of gene duplication, target site mutation, or a combination of both mechanisms due to the high number of expressed gene copies that would need to be amplified (Gaines et al., 2010; Koo et al., 2018). Adequate sampling of target site loci with PCR-based methods is nontrivial, especially within weed species—the majority of which have little if any basic genomic information available, such as genome size, ploidy level, and chromosome number, let alone transcriptomic or genomic sequencing. PCR-based capillary sequencing of target site mutations obfuscates the presence of target site mutations in the presence of multiple target sites; this method as used by Goh et al. (2018) and the majority of research published to confirm TSR (even by ourselves) provides strong evidence for the presence TSR and weak evidence for its absence.
Footnotes
Articles can be viewed without a subscription.
References
- Agrawal C, Sen S, Yadav S, Rai S, Rai LC(2015) A novel aldo-keto reductase (AKR17A1) of Anabaena sp. PCC7120 degrades the rice field herbicide butachlor and confers tolerance to abiotic stresses in E. coli. PLoS One 10: e0137744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon-Reverte R, Garcia A, Urzua J, Fisher AJ(2013) Resistance to glyphosate in junglerice (Echinochloa colona) from California. Weed Sci 61: 48–54 [Google Scholar]
- Bracamonte E, Fernández-Moreno PT, Barro F, De Prado R(2016) Glyphosate-resistant Parthenium hysterophorus in the Caribbean Islands: Non target site resistance and target site resistance in relation to resistance levels. Front Plant Sci 7: 1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostamam Y, Malone JM, Dolman FC, Boutsalis P, Preston C(2012) Rigid ryegrass (Lolium rigidum) populations containing a target site mutation in EPSPS and reduced glyphosate translocation are more resistant to glyphosate. Weed Sci 60: 474–479 [Google Scholar]
- Gaines TA, Cripps A, Powles SB(2012) Evolved resistance to glyphosate in junglerice (Echinochloa colona) from the tropical ord river regions in Australia. Weed Technol 26: 480–484 [Google Scholar]
- Gaines TA, Patterson EL, Neve P(2019) Molecular mechanisms of adaptive evolution revealed by global selection for glyphosate resistance. New Phytol 223: 1770–1775 [DOI] [PubMed] [Google Scholar]
- Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, Nissen SJ, Patzoldt WL, Tranel PJ, Culpepper AS, et al. (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA 107: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherekhloo J, Fernández-Moreno PT, Alcántara-de la Cruz R, Sánchez-González E, Cruz-Hipolito HE, Domínguez-Valenzuela JA, De Prado R(2017) Pro-106-Ser mutation and EPSPS overexpression acting together simultaneously in glyphosate-resistant goosegrass (Eleusine indica). Sci Rep 7: 6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh SS, Yu Q, Han H, Vila-Aiub MM, Busi R, Powles R(2018) Non-target-site glyphosate resistance in Echinochloa colona from Western Australia. Crop Prot 112: 257–263 [Google Scholar]
- Han H, Yu Q, Widderick MJ, Powles SB(2016) Target-site EPSPS Pro-106 mutations: Sufficient to endow glyphosate resistance in polyploid Echinochloa colona? Pest Manag Sci 72: 264–271 [DOI] [PubMed] [Google Scholar]
- Koo DH, Molin WT, Saski CA, Jiang J, Putta K, Jugulam M, Friebe B, Gill BS(2018) Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. Proc Natl Acad Sci USA 115: 3332–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Yu Q, Han H, Mao L, Nyporko A, Fan L, Bai L, Powles S(2019) Aldo-keto reductase metabolizes glyphosate and confers glyphosate resistance in Echinochloa colona. Plant Physiol 181: 1519–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermanna RS, Vennapusa AR, Easwaran M, Chandrashekar BK, Rao H, Ghanti K, Sudhakar C, Mysore KS, Makarla U(2017) Aldo-keto reductase enzymes detoxify glyphosate and improve herbicide resistance in plants. Plant Biotechnol J 15: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]